Dissection of the Mammalian Midbody Proteome Reveals Conserved Cytokinesis Mechanisms (original) (raw)
. Author manuscript; available in PMC: 2013 Jun 12.
Published in final edited form as: Science. 2004 May 27;305(5680):61–66. doi: 10.1126/science.1097931
Abstract
Cytokinesis is the essential process that partitions cellular contents into daughter cells. To identify and characterize cytokinesis proteins rapidly, we used a functional proteomic and comparative genomic strategy. Midbodies were isolated from mammalian cells, proteins were identified by multidimensional protein identification technology (MudPIT), and protein function was assessed in Caenorhabditis elegans. Of 172 homologs disrupted by RNA interference, 58% displayed defects in cleavage furrow formation or completion, or germline cytokinesis. Functional dissection of the midbody demonstrated the importance of lipid rafts and vesicle trafficking pathways in cytokinesis, and the utilization of common membrane cytoskeletal components in diverse morphogenetic events in the cleavage furrow, the germline, and neurons.
A critical phase of cell division occurs just after segregation of the duplicated genome, when the chromosomes, cytoplasm, and organelles are partitioned to two daughter cells in a process termed cytokinesis. In animal cells, this event is driven by a cortical contraction that pinches the cell into two and requires coordination of the mitotic spindle, actin cytoskeleton, and plasma membrane. Failures in cytokinesis can cause cell death and age-related disorders or lead to a genome amplification characteristic of many cancers (1, 2). To understand the mechanisms of cytokinesis, the identity and function of proteins comprising the structures involved must be ascertained. We combined several approaches to obtain and study conserved and relevant factors, capitalizing on recent advances in proteomics and functional genomics.
To identify proteins involved in cytokinesis, we examined a transient “organelle-like” structure called the midbody, first described by Flemming in 1891 as the remnant of cell division just prior to abscission (3, 4). Morphologically, the mammalian midbody is a dense structure containing microtubules derived from the spindle midzone tightly bundled by the cytokinetic furrow (5). Although no clear function has been ascribed to the midbody, it is known to contain proteins indispensable for cytokinesis, asymmetric cell division, and chromosome segregation (6–9).
Mammalian midbody isolation
We isolated midbodies from Chinese hamster ovary (CHO) cells, adapting published procedures (5, 10) (Fig. 1). Cells synchronized in mitosis were released and monitored by phase contrast microscopy to determine when most of them had furrowed to generate midbodies (Fig. 1A). After incubation in a Taxol- and phalloidin-containing medium that stabilized microtubules and actin filaments, the cells were lysed in a buffer containing Triton X-100, solubilizing most cellular structures except midbodies, which were pelleted. Analysis of the pelleted structures by immunofluorescence microscopy revealed abundant midbodies (Fig. 1B), which contained tubulin and actin in a complex mixture of proteins (Fig. 1C). Relative to identical cytoskeletal fractions prepared from interphase cells, midbody preparations were enriched 30- to 50-fold for two known midbody proteins, tubulin and dynamin, and for two previously uncharacterized midbody resident proteins identified in this study, BiP and kinesin heavy chain (KHC) (Fig. 1D) (table S1). However, the biochemical procedure required for effective midbody isolation prevented recovery of several known midbody proteins, including INCENP (11) and PRC1 (12), which were eluted from microtubules by the Taxol treatment, and proteins such as syntaxin and RhoA, which were solubilized by the Triton X-100 treatment (13, 14).
Fig. 1.
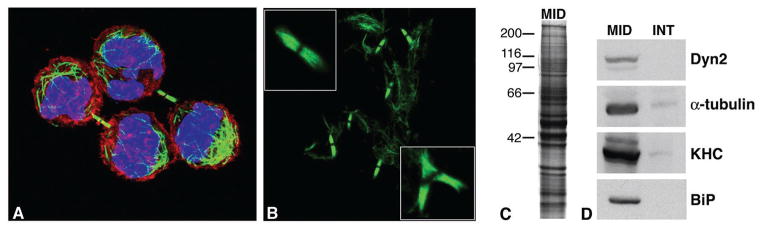
Isolation of CHO cell midbodies. (A) Synchronized CHO cells in the late stages of cytokinesis. (B) Isolated midbodies following biochemical preparation. In addition to bipolar midbodies (left, top inset), multipolar spindle midbodies (bottom inset) were also obtained, likely resulting from polyploid CHO cells. (C) Silver-stained gel showing the complex protein composition of isolated midbodies separated by SDS–polyacrylamide gel electrophoresis. (D) Western blot showing enrichment of candidate proteins in midbodies (MID) isolated from synchronized cells as compared to a fraction prepared from unsynchronized interphase cells (INT) treated in the same manner. Each lane contained 10 μg of total protein and was probed with antibodies recognizing Dyn2 (dynamin II), α-tubulin, KHC (kinesin heavy chain), or BiP.
Identification of midbody components
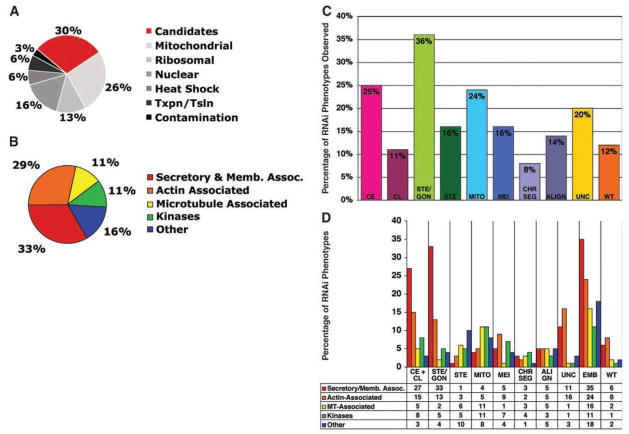
We identified the proteins present in our midbody preparations by tandem liquid chromatography and tandem mass spectrometry, taking advantage of multidimensional protein identification technology (MudPIT), a powerful method for analyzing the composition of complex protein mixtures (15). Four separate midbody preparations were analyzed to ensure comprehensive protein identification and to minimize bias against small or scarce proteins. Mud-PIT data were pooled using DTASelect and protein identification was validated, yielding 577 proteins found in all four experiments (Fig. 2A). To rapidly identify novel proteins playing specific roles in cytokinesis, we systematically eliminated several classes of midbody factors based on protein prediction analysis (417, table S2). These included ribosomal proteins (13%) and mitochondrial proteins (26%), which are important for cellular housekeeping functions, and are unlikely to play direct roles in cell division. The large number of ribosomal and mitochondrial proteins found in our midbody preparations was not surprising, because previous ultrastructural studies revealed an abundance of these organelles associated with midbodies, spindle midzones, and intercellular canals (5, 16–18). We also chose not to characterize proteins likely present due to contamination of the preparation with nuclei (1 nucleus per 60 to 70 midbodies), such as histones and chromatin-associated proteins (16%).
Fig. 2.
Distribution of functional categories of the identified proteins and RNAi phenotypes of the C. elegans homologs. (A) Pie chart displaying the categories of mammalian proteins identified by midbody mass spectrometry analysis (total number of proteins = 577). Candidate midbody proteins (160) chosen for further study are in red. Mitochondrial, ribosomal, nuclear, transcription/translation, and heat shock proteins also identified in the midbody preparations were not pursued in this study (table S2). (B) Pie chart illustrating the percentage of mammalian proteins in each functional category. (C) Bar graph displaying the percentage of C. elegans genes targeted by RNAi in each phenotype category. Some genes gave multiple phenotypes and were therefore listed in multiple categories. (D) The percentage of genes with cell division phenotypes belonging to specific functional classes.
The remaining 160 candidate midbody proteins were divided into five groups based on protein prediction analysis (Fig. 2B). The largest proportion (33%) could be categorized as secretory or membrane-trafficking proteins, and 29% were actin-associated proteins, 11% were microtubule-associated proteins, and 11% were protein kinases. A large category termed “other” contained 16% of the midbody proteins that fell into a variety of different groups difficult to classify, and included five novel proteins of unknown function. About 36% (57/160) of the candidate proteins had previously been shown to play roles in cytokinesis in one or more experimental systems, including mammalian or plant cells, Caenorhabditis elegans, Drosophila melanogaster, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Dictyostelium discoideum, and Aspergillus nidulans (table S1, Published Cytokinesis Genes). Moreover, 26% (42/160) of our identified proteins had previously been localized to cell division remnants or analogous structures (i.e., midbodies, septum, fusomes, ring canals, and phragmoplasts) in a variety of organisms, but for many of them a functional role in cytokinesis had not been described (table S1, Previously Localized.
To further validate that we had identified an enriched source of midbody proteins, we used immunofluorescence to assess the localization of 10 candidates previously uncharacterized with respect to the midbody, including a novel protein (CGI-49/AAD34044). All 10 displayed midbody localization in HeLa cells (Fig. 3), with variable staining patterns within the structure. Each protein colocalized with tubulin except for Keap1, a Kelch repeat protein, which was found in the midbody matrix, the region devoid of tubulin staining (Fig. 3), a localization pattern similar to that of CHO1/MKLP1, a kinesin-like motor (19). In summary, our midbody preparations contained known cytokinesis proteins (57/160), and 103 proteins previously uncharacterized with respect to cell division, 10 of which displayed midbody localization.
Fig. 3.
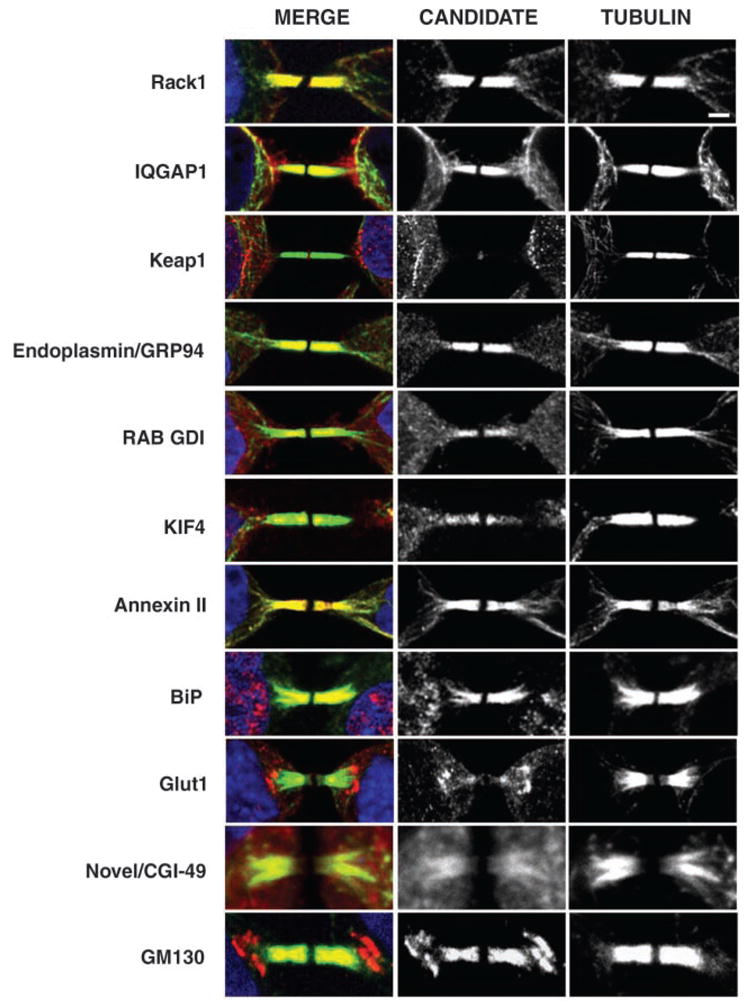
Localization of putative midbody proteins to the midbody in mammalian cells. Immunofluorescence showing 10 candidate midbody proteins previously uncharacterized with respect to the midbody. GM130 marks the localization of the Golgi in the midbody.
Functional characterization of mammalian proteins in C. elegans
To rapidly and systematically screen the candidate mammalian midbody proteins for roles in cytokinesis, we identified homologous C. elegans genes and used RNA interference (RNAi) to assess their individual loss-of-function phenotypes (Fig. 2C and table S1). A total of 147 of the 160 mammalian midbody proteins (92%) had obvious C. elegans homologs (E value scores less than 10−10). Eleven additional C. elegans genes showed strong similarity (E value scores between 10−3 and 10−9) to mammalian midbody components. Only two midbody proteins identified, Annexin VI and Noggin, a vertebrate neuronal protein, failed to have a homologous or similar C. elegans counterpart. In total, we analyzed 172 C. elegans genes, which included 14 paralogs. Of the genes analyzed, 38 had previously been shown to function during mitosis or meiosis in C. elegans, 16 of these specifically in cytokinesis.
To analyze gene function, we injected double-stranded RNA corresponding to each of the C. elegans genes individually into hermaphrodite animals that expressed fluorescently tagged histone H2B and β-tubulin. The effects of RNAi were evaluated 24 to 36 hours later by time-lapse video microscopy for effects on early embryonic divisions, as well as for gonad defects of injected animals. We found that 88% of the double-stranded RNAs (141/160 mammalian genes) resulted in scoreable defects in C. elegans after RNAi. For most genes, RNAi elicited a combination of phenotypes. We used multiple descriptors for each gene (table S1), and the categories depicted in Fig. 2C therefore add up to greater than 100%.
Analysis of the distribution in phenotypes indicated that defects in cleavage furrow initiation [Cytokinesis Early (CE), 25%], cleavage furrow termination [Cytokinesis Late (CL), 11%], and germline cytokinesis [Sterile/Gonad cytokinesis defects (STE/GON), 36%] were most prevalent (combined and not overlapping = 58%) (Fig. 2C). Other phenotypes included defects in spindle alignment (14%), chromosome segregation (8%), mitosis (cell cycle progression or spindle defects, 24%) or meiosis (including polar body extrusion defects, 16%), and fertility [without gonad cytokinesis defects (STE), 16%]. In some cases referred to as EMB, we did not observe failure in cell division during the first three divisions, but embryos were inviable (Fig. 2D) (total EMB observed = 104/160, 65%; EMB with no other observable defects = 35/160, 22%). We also noticed that several genes had mutant alleles and/or RNAi phenotypes previously characterized as causing uncoordinated phenotypes possibly attributed to neuronal defects (32/160, 20%) (Fig. 2C). Depletion of many of these genes by RNAi also caused embryonic lethality and germline cytokinesis defects (28/32). Previous to our analysis, 17/160 candidate C. elegans genes had been localized to the midbody, and 16/17 had been shown to function in cytokinesis by classical genetics and genome-based RNAi screens. Thorough examination of gonads and embryos from injected animals showed that 54 genes previously reported as having wild-type RNAi phenotypes (20–23) actually caused defects in cell division, germline cytokinesis, and various other phenotypes after RNAi treatment.
Cytokinesis, gonad organization, and polar body extrusion utilize a conserved set of proteins
The large number of genes that cause defects in cytokinesis after RNAi provides a “parts list” of proteins functioning in this aspect of cell division, and suggests general roles for candidate midbody proteins in modulating membrane-cytoskeletal organization and dynamics. For example, in many RNAi-treated embryos, cleavage furrows did not form, as in arf-1/ADP ribosylation factor 1 (table S1), indicating that these components are essential for cleavage plane specification or for recruitment or assembly of contractile elements. In other cases, furrows appeared to ingress completely yet ultimately failed, as for RACK1 (Fig. 4, E to H) (Movie S2). RACK1, an anchoring receptor for activated protein kinase C (24), is suppressed in several tumor cell lines (25), as well as in the brain cortex of Down syndrome fetuses (26). Because RACK1 is thought to function as a scaffolding protein that mediates interactions among a variety of signaling molecules, the requirement for this protein suggests the necessity of regulatory cascades in furrow maintenance and/or completion.
Fig. 4.
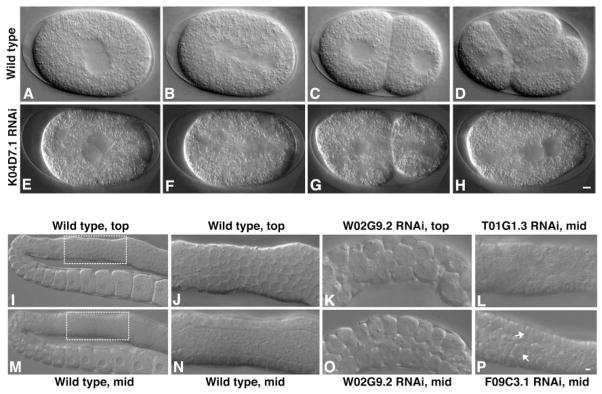
Examples of cytokinesis and germline cytokinesis defects following RNAi of midbody components. (A to D) Wild-type (WT) embryo; (E to H) K04D7.1-RACK1 RNAi. (A and E) Pronuclei meet in the center of the embryo. (B and F) Spindle sets up on the anterior-posterior axis [cytoplasmic clearing is apparent in (F)]. (C and G) Cleavage furrow ingresses. (D) WT embryo continued to divide (Movie S1). (H) K04D7.1-RACK1–inhibited embryo failed to complete cytokinesis and the furrow regressed (Movie S2). Scale bar, 5 μm. (I to P) Germline cytokinesis defects resulting from RNAi of midbody components. (I) Top focal plane, and (M) mid-focal plane of WT gonad showing organized nuclei at the periphery and a central region devoid of nuclei (the rachis) in the distal portion of the gonad. (J) Enlargement of top focal plane and (N) mid-focal plane of a WT gonad. (K) Top focal plane of W02G9.2-Keap1 gonad, showing germ cells with multiple nuclei. (O) Mid-focal plane of W02G9.2-Keap1, showing a diminished rachis and multinucleate germ cells that appear larger than normal. (L) Mid-focal plane of T01G1.3-SEC31 gonad, showing nuclei within the rachis. (P) Mid-focal plane of F09C3.1-IQGAP gonad, showing multinucleate cellularized germ cells. Scale bar, 5 μm.
A large percentage of midbody proteins functioning in cytokinesis, including RACK1, were also required for germline development. Like the blastoderm of Drosophila, the germ-line of C. elegans has a modified form of cytokinesis in which a multinucleate syncytium is formed through a series of incomplete furrows (27). Our candidate midbody list included some genes previously shown in C. elegans to cause a failure in progeny production when disrupted (21, 22, 28). In 69% of cases where RNAi treatment over long periods (≥36 hours) caused sterility, we found minor to severe defects in germline cytokinesis (57/82) (Fig. 2D and Fig. 4, I to P), which is likely responsible for their failure to produce embryos. RNAi treatment for shorter periods (~16 to 24 hours) often produced cytokinesis defects in the early embryos. In total, 67% of our genes that were scored as STE/GON (38/57) also displayed cytokinesis defects in the early embryo.
We identified 16 proteins essential for embryo and germline cytokinesis that were also required for polar body extrusion, a highly polarized and asymmetric cell division that occurs during meiosis (Fig. 5, A to C). One such protein, KEAP1, contains Kelch repeats known to bind actin and also plays a role in responding to oxidative stress (29) (W02G9.2, Fig. 5B). KEAP1 also has homology to Tea1p, a S. pombe protein that has been proposed to recruit polarity factors at the cell tip (30). IQGAP (31), a protein previously shown to function in cytokinesis in other organisms (32), is required for completion of both meiotic and mitotic divisions as well as for germline formation and maintenance (F09C3.1, Figs. 5C and 4P).
Fig. 5.
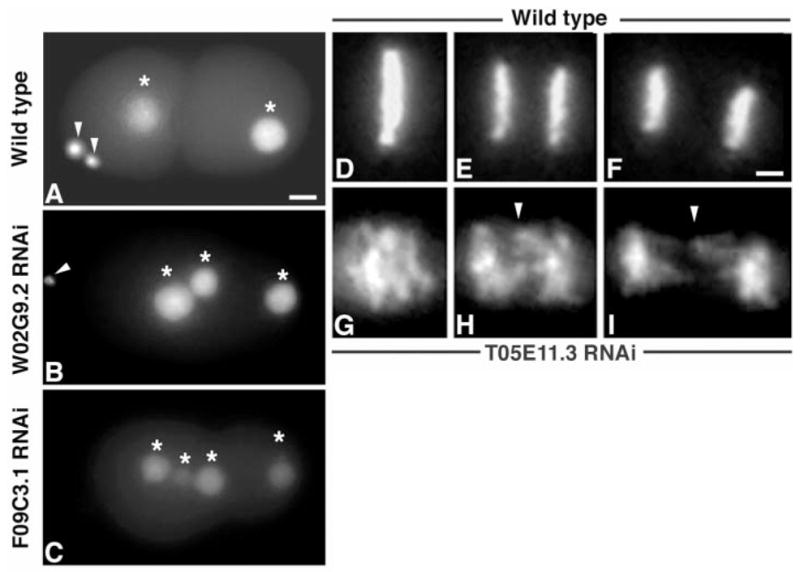
Examples of meiotic and chromosome segregation defects after RNAi treatment. (A) WT embryo with two polar bodies (arrowheads) extruded. (B) W02G9.2-Keap1–deficient embryo in which one polar body failed to be extruded, resulting in three nuclei (marked with asterisks) within the embryo. An arrowhead marks the extruded polar body. (C) F09C3.1-IQGAP–depleted embryo in which both polar bodies failed to extrude (four nuclei marked with asterisks). (D to F) Montage of metaphase through anaphase in a WT embryo expressing histone H2B::GFP (Movie S3). (D) A normal metaphase plate. (E and F) Anaphase. (G to I) Montage of T05E11.3-Endoplasmin–depleted embryo expressing histone H2B::GFP that displayed chromosome segregation defects (Movie S4). Note the lack of chromosome congression to the metaphase plate in (G) and lagging chromosomes in (H) and (I) during anaphase (marked by arrowheads). Scale bar, 5 μm.
Specialized membrane domains function in cytokinesis
How membrane and cytoskeletal elements are recruited equatorially and activated at the site of cleavage furrow formation has been poorly understood. We found a large number of midbody proteins belonging to the secretory or membrane-associated (38/160) and actin-associated (20/160) classes that were essential for cytokinesis (Fig. 2D), suggesting that specialized membrane domains are involved. Plasma membranes are organized into functional domains by lipid rafts and by attachment to the underlying actin cytoskeleton (33). Lipid rafts are Triton X-100 –insoluble sphingolipid and cholesterol-rich microdomains involved in cell signaling events (34 ). Studies in S. pombe showed that sterol-rich lipid domains localized to the septum in a cell cycle–dependent manner and that disruption of these domains inhibited cytokinesis (35 ), although the protein factors involved were not known. We identified as midbody residents a large number, 25% (40/160), of membrane–cytoskeletal proteins previously known to associate with lipid rafts, 35% (14/ 40) of which gave observable cytokinesis or germline defects when inhibited. This group includes several Annexins, proteins that bind to phospholipids in a Ca2+-dependent manner and promote lipid raft assembly (36 ); dynamin, a guanosine triphosphatase (GT-Pase) that stimulates actin-based vesicle motility (37 ); and rab7, a GTPase that regulates endocytotic membrane traffic (38) (table S1). Recent studies have shown raft components associated with division membranes and midbodies in a variety of organisms (39–41). Clustering of lipid raft proteins and downstream signal transduction pathways above the spindle mid-zone could target and activate the cytoskeleton and membrane fusion machinery to induce cleavage furrow invagination. The presence of dynamin is particularly suggestive, because it could alter actin dynamics and promote cytoskeletal changes at the membrane, leading to contractile ring assembly. The localization of dynamin at cleavage membranes both in dividing embryos and the germline reinforces the concept that common mechanisms are at work (39). Several of the raft-associated factors are also required for furrow completion, suggesting that their activities might also contribute to dynamic membrane events necessary for daughter cell separation.
Similar trafficking mechanisms during animal and plant cytokinesis
Once the cleavage site has been established, new membrane must be inserted, but it is unclear how the necessary protein and membrane elements are delivered to the furrow in animal cells. Cytokinesis in both plants and animals is known to require a functional secretory pathway (42, 43), but the events are generally considered to be mechanistically distinct (44). In dividing plant cells, formation of a new cell wall occurs by the fusion of predominantly Golgi-derived vesicles to form a structure called the phragmoplast at the cell division plane (45). In animal cells, Golgi stacks normally undergo fragmentation during mitosis to promote stochastic partitioning of the organelle to each daughter cell (46). Our analysis indicates that the midbody possesses functions analogous to that of the phragmoplast, because it contains Golgiassociated components required for cytokinesis (24%, 38/160; table S1) (Fig. 3). To validate the large number of Golgi-associated proteins identified in our screen; we used the cis-Golgi matrix marker, GM130, to observe the localization of the Golgi specifically during cytokinesis. We observed a striking Golgi pattern in the midbody, as well as in patches just outside the midbody (Fig. 3), suggesting that populations of vesicles are poised for transport during daughter cell separation. In plants, transport of vesicles to the phragmoplast occurs along microtubules (47). One identified midbody component essential for cytokinesis was KIF4, a microtubule-based motor that mediates anterograde transport of vesicles in neurons (48) (table S1, Fig. 3) and could be involved in targeting vesicular transport to the ingressing furrow.
Midbody proteins function in asymmetric cell division and chromosome segregation
Spindle positioning is essential to generate cells of unequal size and to determine the geometry of the cleavage plane. In asymmetrically dividing cells like the early C. elegans embryo, the midbody is thought to provide a positional cue in the subsequent cell division by marking a site on the plasma membrane where astral microtubules will later be captured to orient the spindle (6). In addition to components of the dyneindynactin complex and G protein β (8, 49), factors known to participate in spindle alignment, we also identified membrane remodeling and vesicular trafficking proteins in the midbody that were required for proper spindle rotation in early embryonic divisions. These include Endophilin B1, Huntingtin interacting protein 1 (HIP1/DUO), Copine I, and CLIP-170 (table S1). A specific function of these proteins in either delivering cortical cues or in promoting interactions between astral microtubules and the plasma membrane can now be explored.
Another spindle function of some midbody components was in chromosome segregation. One previously characterized class, called “chromosome passenger proteins,” includes a complex containing the Aurora kinase B (table S1), which initially resides at kinetochores promoting chromosome congression, but dissociates during anaphase and persists in the central spindle and midbody, where it also functions in cytokinesis (50). We have now discovered unexpected roles in chromosome segregation for several additional midbody proteins including Endoplasmin/ GRP94, the most abundant endoplasmic reticulum (ER) protein (51). Depletion of T05E11.3-Endoplasmin prevented proper chromosome alignment at the metaphase plate (Fig. 5G) and caused lagging chromosomes during anaphase (Fig. 5, H and I) (Movie S4). These phenotypes demonstrate a novel role for Endoplasmin and suggest a direct or indirect role of the ER during mitosis. In addition, we identified other factors, including F13F3.3–Flotillin 1 and M03F4.7-Calcyclin, required for proper chromosome segregation. Flotillin 1 is a lipid raft–associated integral membrane protein implicated in Alzheimer’s disease and diabetes (52, 53), and recently was localized to the midbody in hematopoietic cells (54). Calcyclin is a member of the S100 family that is elevated in a number of malignant tumors and associates with several actin-binding proteins and other lipid raft–associated Annexins in a calcium-dependent manner (55). These unpredicted results suggest functions for calcium-binding and membrane-associated factors in chromosome segregation and highlight the utility of a functional proteomic approach.
Midbody proteins function in cell cycle regulation and signaling events
The final stage of cytokinesis occurs at the midbody, where daughter cell separation occurs, but little is known about how this event is synchronized with exit from the mitotic phase of the cell cycle. As a transient structure assembled just before the terminal step in division, the midbody is a logical place to coordinate the completion of cytokinesis with regulation of the cell cycle machinery. Consistent with this hypothesis, we identified several important cell cycle regulatory proteins in the midbody, including the master mitotic kinase Cdc2, and factors required for mitotic exit, including components of the anaphasepromoting complex that targets key mitotic proteins for degradation. As expected, inhibition of these factors led to general defects in mitotic progression and embryonic lethality (table S1). Other specific activators of proteolysis, such as the ubiquitin ligase components NEDD4 and Cullin1, were also identified as essential midbody residents that could target key regulators of cytokinesis for disposal. NEDD4 has also been proposed to function in cell signaling events in association with lipid rafts (56 ). Selective degradation of cell cycle regulatory and membrane-cytoskeletal proteins may constitute an essential function at the midbody in coordinating two irreversible steps: mitotic exit and daughter cell abscission.
We were surprised to identify as midbody proteins two glucose transporters GLUT1 (Fig. 3) and GLUT4, both of which were required for the initiation of cytokinesis as well as for germline cytokinesis (table S1). During interphase these glucose transport proteins reside in vesicles that fuse with the plasma membrane in response to stimulation by insulin or other growth factors (57), and at the cell surface associate with lipid raft components implicated in signaling and membrane trafficking events (58). Although a requirement for glucose transporters in achieving cytokinesis is not easily explained, our findings raise the possibility that GLUT1 and GLUT4 are involved in the assembly or dynamics of membrane compartments necessary for furrow ingression.
Functional analysis of midbody proteins reveals conserved morphogenetic mechanisms
We used a functional proteomic strategy to identify new factors involved in cytokinesis. This approach took advantage of several tools: the ability to synchronize tissue culture cells and isolate midbodies, MudPIT technologies to characterize large protein complexes, genome databases to identify homologs, and the capacity to rapidly assess the phenotypes caused by disruption of gene function using RNAi in C. elegans. Conservation and functional relevance of the identified mammalian proteins were apparent, because 92% (147/160) had homologs in C. elegans, with 58% of these resulting in cytokinesis defects when inhibited. The diverse array of factors identified, 64% (103/ 160) of which were not previously localized to the midbody and 48% (49/103) of which are required for cytokinesis, provided an unprecedented look at the cellular machineries involved.
The predominance and nature of the membrane/vesicle and actin-binding proteins required implicate specialized membrane domains containing lipid rafts and vesicle trafficking components in establishing the cleavage furrow, mediating plasma membrane invagination and abscission. Intriguingly, in addition to cell division defects, 20% of the identified proteins also caused uncoordinated phenotypes upon depletion, indicating function during neuronal and/or muscle morphogenesis in C. elegans. The midbody may be analogous to the bundled microtubules found in neuronal extensions, because both structures possess similar proteins and signaling pathways regulating actin and microtubule dynamics, as well as vesicular trafficking events that include, for example, citron kinase, MKLP1/ CHO1, and HIP1 (59–61). Ancient and common pathways may function to establish and maintain membrane-cytoskeletal dynamics in different cell types and organisms. Fourteen percent of the mammalian proteins identified in this study are implicated in human diseases with membrane-cytoskeletal pathologies, such as Huntington’s disease, deafness, sclerosis, melanoma, and leukemia (62–66), suggesting that functional characterization of cell division proteins may help to identify mammalian disease loci and characterize pathologies. The utilization of common components in diverse dynamic membrane cytoskeletal events in the cleavage furrow, the germline, and neurons indicates ancient mechanisms mediating cell division and complex morphogenetic cellular processes critical in human development and disease.
Supplementary Material
Original Supplement
Revised Supplemental Data File
Acknowledgments
We thank J. White, M. Mullins, R. Kuriyama, M. Welch, B. Mohler, K. O’Connell, E. Ralston, D. Chu, A. Severson, R. Chan, J. Banks, A. Van Hooser, K. Hagstrom, D. Reiner, and T. Wu for support during midbody isolation, RNAi analysis, and/or for reading the manuscript. We thank M. McNiven for antibodies to Dynamin II and to KHC, T. Hasson for antibody to KEAP1, and A. Caceres for antibody to KIF4. A.R.S. is supported by NIH grant F32 GM64159-01. R.H. is supported by the NIH, The Pew Charitable Trust, and The Cancer Research Coordinating Committee. B.J.M. is an investigator for the Howard Hughes Medical Institute. J.Y. and H.L. are supported by the Yeast Resource Center RR1823 and Office of Naval Research grant N00014-00-0421. This paper is dedicated to K. Van Doren.
Footnotes
References and Notes
- 1.Bhalla KN. Oncogene. 2003;22:9075. doi: 10.1038/sj.onc.1207233. [DOI] [PubMed] [Google Scholar]
- 2.Carmena M, Earnshaw WC. Nat Rev Mol Cell Biol. 2003;4:842. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 3.Flemming W. Arch Mikrosk Anat. 1891;37:685. [Google Scholar]
- 4.Paweletz N. Nat Rev Mol Cell Biol. 2001;2:72. doi: 10.1038/35048077. [DOI] [PubMed] [Google Scholar]
- 5.Mullins JM, McIntosh JR. J Cell Biol. 1982;94:654. doi: 10.1083/jcb.94.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hyman AA. J Cell Biol. 1989;109:1185. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dan K, Tanaka Y. Ann N Y Acad Sci. 1990;582:108. doi: 10.1111/j.1749-6632.1990.tb21672.x. [DOI] [PubMed] [Google Scholar]
- 8.Skop AR, White JG. Curr Biol. 1998;8:1110. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller RK, Cheng SC, Rose MD. Mol Biol Cell. 2000;11:2949. doi: 10.1091/mbc.11.9.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sellitto C, Kuriyama R. J Cell Biol. 1988;106:431. doi: 10.1083/jcb.106.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheatley SP, Kandels-Lewis SE, Adams RR, Ainsztein AM, Earnshaw WC. Exp Cell Res. 2001;262:122. doi: 10.1006/excr.2000.5088. [DOI] [PubMed] [Google Scholar]
- 12.Mollinari C, et al. J Cell Biol. 2002;157:1175. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lang T, et al. EMBO J. 2001;20:2202. doi: 10.1093/emboj/20.9.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming IN, Elliott CM, Exton JH. J Biol Chem. 1996;271:33067. doi: 10.1074/jbc.271.51.33067. [DOI] [PubMed] [Google Scholar]
- 15.Link AJ, et al. Nat Biotechnol. 1999;17:676. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 16.Pickett-Heaps J, Kowalski SE. Eur J Cell Biol. 1981;25:150. [PubMed] [Google Scholar]
- 17.Brawley SH, Quatrano RS, Wetherbee R. J Cell Sci. 1977;24:275. doi: 10.1242/jcs.24.1.275. [DOI] [PubMed] [Google Scholar]
- 18.Bieliavsky N, Geuskens M. J Submicrosc Cytol Pathol. 1990;22:445. [PubMed] [Google Scholar]
- 19.Matuliene J, Kuriyama R. Mol Biol Cell. 2002;13:1832. doi: 10.1091/mbc.01-10-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamath RS, et al. Nature. 2003;421:231. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 21.Fraser AG, et al. Nature. 2000;408:325. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 22.Gonczy P, et al. Nature. 2000;408:331. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 23.Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Curr Biol. 2001;11:171. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- 24.Won M, et al. Biochem Biophys Res Commun. 2001;282:10. doi: 10.1006/bbrc.2001.4535. [DOI] [PubMed] [Google Scholar]
- 25.Seidl P, Huettinger R, Knuechel R, Kunz-Schughart LA. Int J Cancer. 2002;102:129. doi: 10.1002/ijc.10675. [DOI] [PubMed] [Google Scholar]
- 26.Peyril A, Weitzdoerfer R, Gulesserian T, Fountoulakis M, Lubec G. Electrophoresis. 2002;23:152. doi: 10.1002/1522-2683(200201)23:1<152::AID-ELPS152>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 27.Sisson JC, Rothwell WF, Sullivan W. Cell Biol Int. 1999;23:871. doi: 10.1006/cbir.1999.0484. [DOI] [PubMed] [Google Scholar]
- 28.Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ. Curr Biol. 2000;10:1619. doi: 10.1016/s0960-9822(00)00869-1. [DOI] [PubMed] [Google Scholar]
- 29.Velichkova M, et al. Cell Motil Cytoskelet. 2002;51:147. doi: 10.1002/cm.10025. [DOI] [PubMed] [Google Scholar]
- 30.Chang F. Cell Struct Funct. 2001;26:539. doi: 10.1247/csf.26.539. [DOI] [PubMed] [Google Scholar]
- 31.Machesky LM. Curr Biol. 1998;8:R202. doi: 10.1016/s0960-9822(98)70125-3. [DOI] [PubMed] [Google Scholar]
- 32.Osman MA, Cerione RA. J Cell Biol. 1998;142:443. doi: 10.1083/jcb.142.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simons K, Ikonen E. Nature. 1997;387:569. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 34.Ikonen E. Curr Opin Cell Biol. 2001;13:470. doi: 10.1016/s0955-0674(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 35.Wachtler V, Rajagopalan S, Balasubramanian MK. J Cell Sci. 2003;116:867. doi: 10.1242/jcs.00299. [DOI] [PubMed] [Google Scholar]
- 36.Babiychuk EB, Draeger A. J Cell Biol. 2000;150:1113. doi: 10.1083/jcb.150.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merrifield CJ, Feldman ME, Wan L, Almers W. Nat Cell Biol. 2002;4:691. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 38.Li N, et al. Proteomics. 2003;3:536. doi: 10.1002/pmic.200390067. [DOI] [PubMed] [Google Scholar]
- 39.Thompson HM, Skop AR, Euteneuer U, Meyer BJ, McNiven MA. Curr Biol. 2002;12:2111. doi: 10.1016/s0960-9822(02)01390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng B, Schwarz H, Jesuthasan S. Exp Cell Res. 2002;279:14. doi: 10.1006/excr.2002.5579. [DOI] [PubMed] [Google Scholar]
- 41.Kuwabara PE, Lee MH, Schedl T, Jefferis GS. Genes Dev. 2000;14:1933. [PMC free article] [PubMed] [Google Scholar]
- 42.Skop AR, Bergmann D, Mohler WA, White JG. Curr Biol. 2001;11:735. doi: 10.1016/s0960-9822(01)00231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staehelin LA, Hepler PK. Cell. 1996;84:821. doi: 10.1016/s0092-8674(00)81060-0. [DOI] [PubMed] [Google Scholar]
- 44.Glotzer M. Annu Rev Cell Dev Biol. 2001;17:351. doi: 10.1146/annurev.cellbio.17.1.351. [DOI] [PubMed] [Google Scholar]
- 45.Nebenfuhr A, Frohlick JA, Staehelin LA. Plant Physiol. 2000;124:135. doi: 10.1104/pp.124.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucocq JM, Berger EG, Warren G. J Cell Biol. 1989;109:463. doi: 10.1083/jcb.109.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bednarek SY, Falbel TG. Traffic. 2002;3:621. doi: 10.1034/j.1600-0854.2002.30904.x. [DOI] [PubMed] [Google Scholar]
- 48.Peretti D, Peris L, Rosso S, Quiroga S, Caceres A. J Cell Biol. 2000;149:141. doi: 10.1083/jcb.149.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gotta M, Ahringer J. Nat Cell Biol. 2001;3:297. doi: 10.1038/35060092. [DOI] [PubMed] [Google Scholar]
- 50.Earnshaw WC, Cooke CA. J Cell Sci. 1991;98:443. doi: 10.1242/jcs.98.4.443. [DOI] [PubMed] [Google Scholar]
- 51.Koch GL. J Cell Sci. 1987;87:491. doi: 10.1242/jcs.87.4.491. [DOI] [PubMed] [Google Scholar]
- 52.Girardot N, et al. Neuropathol Appl Neurobiol. 2003;29:451. doi: 10.1046/j.1365-2990.2003.00479.x. [DOI] [PubMed] [Google Scholar]
- 53.Jiang M, et al. Biochem Biophys Res Commun. 2003;309:196. doi: 10.1016/s0006-291x(03)01559-6. [DOI] [PubMed] [Google Scholar]
- 54.Rajendran L, et al. Proc Natl Acad Sci USA. 2003;100:8241. doi: 10.1073/pnas.1331629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Breen EC, Tang K. J Cell Biochem. 2003;88:848. doi: 10.1002/jcb.10398. [DOI] [PubMed] [Google Scholar]
- 56.Lafont F, Simons K. Proc Natl Acad Sci USA. 2001;98:3180. doi: 10.1073/pnas.051003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thurmond DC, Pessin JE. Mol Membr Biol. 2001;18:237. doi: 10.1080/09687680110082400. [DOI] [PubMed] [Google Scholar]
- 58.Chamberlain LH, Gould GW. J Biol Chem. 2002;277:49750. doi: 10.1074/jbc.M206936200. [DOI] [PubMed] [Google Scholar]
- 59.Aumais JP, et al. J Cell Sci. 2003;116:1991. doi: 10.1242/jcs.00412. [DOI] [PubMed] [Google Scholar]
- 60.LoTurco JJ, Sarkisian MR, Cosker L, Bai J. Cereb Cortex. 2003;13:588. doi: 10.1093/cercor/13.6.588. [DOI] [PubMed] [Google Scholar]
- 61.Sharp DJ, Kuriyama R, Essner R, Baas PW. J Cell Sci. 1997;110:2373. doi: 10.1242/jcs.110.19.2373. [DOI] [PubMed] [Google Scholar]
- 62.Walling HW, Baldassare JJ, Westfall TC. J Neurosci Res. 1998;54:301. doi: 10.1002/(SICI)1097-4547(19981101)54:3<301::AID-JNR1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 63.Melchionda S, et al. Am J Hum Genet. 2001;69:635. doi: 10.1086/323156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brandt R. Cell Tissue Res. 2001;305:255. doi: 10.1007/s004410000334. [DOI] [PubMed] [Google Scholar]
- 65.Legg JW, Lewis CA, Parsons M, Ng T, Isacke CM. Nat Cell Biol. 2002;4:399. doi: 10.1038/ncb797. [DOI] [PubMed] [Google Scholar]
- 66.Basu J, Kundu M, Rakshit MM, Chakrabarti P. Biochim Biophys Acta. 1988;945:121. doi: 10.1016/0005-2736(88)90474-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Original Supplement
Revised Supplemental Data File