Transfer RNA modifications: Nature’s combinatorial chemistry playground (original) (raw)
. Author manuscript; available in PMC: 2014 Jan 1.
Published in final edited form as: Wiley Interdiscip Rev RNA. 2012 Nov 8;4(1):35–48. doi: 10.1002/wrna.1144
Abstract
Following synthesis, tRNAs are peppered by numerous chemical modifications which may differentially affect a tRNA’s structure and function. Although modifications affecting the business ends of a tRNA are predictably important for cell viability, a majority of modifications play more subtle structural roles that can affect tRNA stability and folding. The current trend is that modifications act in concert and it is in the context of the specific sequence of a given tRNA that they impart their differing effects. Recent developments in the modification field have highlighted the diversity of modifications in tRNA. From these the combinatorial nature of modifications in explaining previously described phenotypes derived from their absence has emerged as a growing theme.
Beyond the canonical nucleotides used for DNA and RNA synthesis (G, A, T, U and C), all nucleic acids in cells undergo naturally occurring post-replicative and post-transcriptional chemical modifications. As a collective, these include a myriad of chemical groups that impart distinct local effects at the site of modification but also globally affect the structure of the particular nucleic acid they target. Modifications are catalyzed by exquisitely specific enzymes with a reaction repertoire that includes deaminations, isomerizations, glycosylations, thiolation, transglcosylations, methylations, etc. Modifications sometimes even involve the attachment of complete amino acids and sugars to the bases and/or ribose. Given their extraordinary chemical diversity modifications can affect the stability, folding, transport, processing and function of nucleic acids.
In all organisms, tRNAs undergo by far the most numerous and chemically diverse post-transcriptional modifications that ensure proper structure and function. In fact, tRNA modifications are so prevalent that it has been suggested that they carry more genetic information than tRNA genes themselves. Modification content and type can vary between different tRNAs and even within similar isoacceptors. Some are common at specific positions with great degree of evolutionary conservation among tRNAs from the three domains of life, while the same modification may appear at different sites even within very similar tRNAs within a single organism. Moreover, even between different organisms within a single domain of life, modification sets can vary. A big challenge in the modification field has been the definition of the specific roles that single modifications play in tRNA function. Identifying, mapping and characterizing different modifications have proven a technical tall order. Recent developments in mass spectrometry analysis combined with the advent of genomics, have however pushed the study of post-transcriptional modifications into a blossoming field. Yet still the effect that a particular modification will have on tRNA function is hard to predict. For the most part, absence of some modifications causes such subtle phenotypic effects on organisms that it has been hard to appreciate their full impact at the cellular level. It may not be a single modification but an ensemble of modifications and how they interact in the context of a tRNA molecule and its intracellular environment that will really have the last say on a particular modification’s role on tRNA function.
In general, modifications can be divided into two major groups based on how they affect tRNA function: 1) those that affect the overall structure of the tRNA and 2) those that target the functional centers of the tRNA (anticodon sequence and/or sequences important for aminoacylation), therefore having direct effects on decoding and protein synthesis. We will highlight the diversity of modifications found in tRNA. We will focus on recent findings that illustrate themes in the tRNA modification field that may be applicable to the study or understanding the function of other modifications, even those not simply affecting tRNAs. Cases where lack of modification correlates with disease states, for example those that affect mitochondrial function; will only be covered cursorily as these have been extensively covered elsewhere in this series.
THE CHEMICAL NATURE OF MODIFIED NUCLEOTIDES
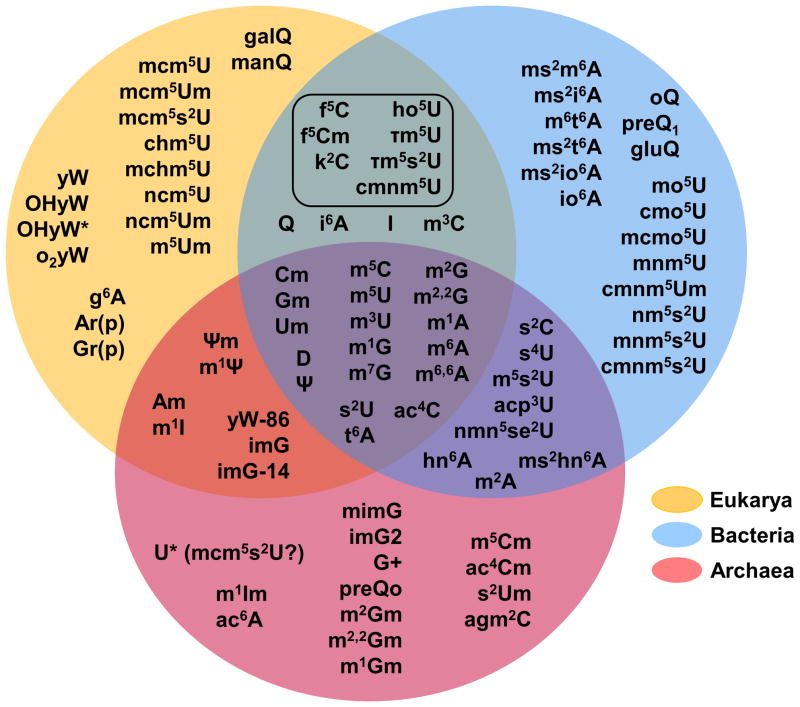
Unlike modifications found in DNA, which tend to be dominated by relatively few chemically distinct species, RNA modifications comprise a rich landscape of chemical diversity. Over 100 chemically unique modified nucleotides have been identified in RNAs from all three domains of life 1–6 (Figure 1). The prevalence of modified nucleotides in tRNAs varies between different organisms, with the density of nucleotide modifications of a given tRNA molecule generally increasing from bacterial and organellar tRNAs to eukaryotic tRNAs. There are almost 10 times more modified nucleotides in tRNA (average ~17% of the total residues are modified) than in the other abundant biological RNA, rRNA (average 1–2% of total residues are modified).
Figure 1. Extreme chemical diversity among tRNA modification in biology.
Modified nucleotides identified in at least one tRNA species are indicated in shaded spheres according to the domain(s) of life in which they are found. The modifications at the intersection of Eukarya and Bacteria enclosed by a box are modifications found in organelles, consistent with the proposed prokaryotic origins of these subcellular components. Commonly used symbols or abbreviations for the various modifications are: mnX, methylation at position n of nucleotide base X; mn, nX, dimethylation at position n of nucleotide base X; snX, replacement of oxygen with sulfur at position n of nucleotide X; Xm, 2′-O methylation of nucleotide X; D, dihydrouridine; Ψ, pseudouridine; ac4C, _N_-4 acetylcytidine; i6A, _N_-6 isopentenyladenosine; t6A, _N_-6 threonyladenosine; g6A, _N_-6 glycinylcarbamoyladenosine; io6A, _N_-6 (_cis_-hydroxyisopentenyl)adenosine; hn6A, _N_-6 hydroxynorvalylcarbamoyladenosine; ac6A, _N_-6 acetyladenosine; I, inosine (from deamination of adenosine); Xr(p), 2′-O-ribosyl phosphate derivative of nucleotide X; f5C, 5-formyl cytosine; k2C, lysidine; agm2C, agmatidine; acp3U, 3-(3-amino-3-carboxypropyl)uridine; mcm5U, _C_-5 methoxycarbonylmethyl uridine; nmn5U, C-5 carbamoylmethyl uridine; chm5U, _C_-5 carboxyhydroxymethyl uridine; ho5U, C-5 hydroxyuridine; mo5U, C-5 methoxyuridine; cmo5U, uridine 5-oxyacetic acid; mcmo5U, uridine 5-oxyacetic acid methyl ester; mnm5U,C-5 methylaminomethyluridine; cmnm5U, C-5 carboxymethylaminomethyluridine; nm5U, C-5 aminomethyluridine; Q, queosine (and related 7-deaza species oQ, preQ1, preQ0, gluQ, galQ, manQ); G+, archaeosine; yW, wybutosine (and related OHyW, OHyW*, o2yW, yW-86 species), imG, wyosine (and related imG-14, mimG and imG2 species). Various combinations of the modifications listed above are indicated with combinations of multiple symbols; i.e. nmn5s2U = 5-methylaminomethyl 2-thio uridine. This figure was adapted from figure 4 reference 2.
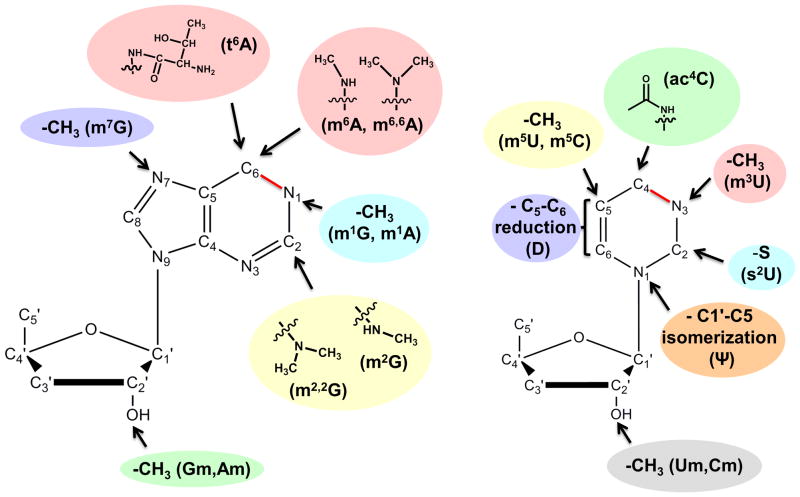
A comparison of modified tRNA species from Archaea, Bacteria and Eukarya reveals a core set of 18 “universal” modifications that occur in tRNA in all three domains of life (Figure 1). Notably, despite the chemical conservation of some modifications, for example m1G, the enzymes that synthesize them need not be evolutionary conserved and are often the result of convergent evolution, whereby enzymatic activities have been recruited for a new function as a way to cope with the selective pressure set forth by the need for a specific modification at a given nucleotide position. Regardless, this core group of modified nucleotides is generally characterized by relatively simple chemical structures, such as: the addition of one (or sometimes two) methyl groups to various positions of the nucleotide bases and or ribose sugars, replacement of oxygen with sulfur (in s2U), isomerization or reduction of the uridine base to pseudouridine or dihydrouridine, respectively, or addition of other relatively small chemical functional groups (i.e., acetylation and threonylation) (Figure 2). In comparison, the trend among modified nucleotides found in only two of the three domains (i.e., Eukarya and Archaea, but not Bacteria) or found uniquely in a single domain, is for a general increase in chemical complexity, with several examples of addition of chemical functional groups that are equivalent to or greater in size than the original purine or pyrimidine ring (Figure 3A). Many of these more complicated larger structures can be thought of as families of modifications related by a common “core” modification structure, and sharing the relevant enzymatic components of their biosynthetic pathways 7. For example, eight chemically distinct modifications of the C-5 position of wobble uridine residues (U34) are identified in eukaryotic tRNA, but 7 of these share a 5-carboxymethyl structure at their core (Figure 3B). Deletion of enzymes implicated in formation of this structure affects all eight modifications 8. It is important to note that the presence and identity of modified nucleotides in tRNA cannot be predicted from inspection of tRNA gene sequences, but only from direct investigation of the tRNA molecules themselves using biophysical, biochemical and genetic approaches. Although there are a few representative organisms for which a significant number of tRNA sequences including modifications have been determined 1, the modification status of the vast majority of tRNAs remains unknown and thus additional examples of modified nucleotides may yet to be identified.
Figure 2. Modified nucleotides found in tRNA in all three domains of life.
Chemical groups added as modifications to purine and pyrimidine rings to create the 18 “universal” tRNA modifications are shown, with arrows indicating the atom of the ring that is modified and common abbreviated name for each modified nucleotide indicated in parentheses. The C1-N6 and C4-N3 bonds of the purine and pyrimidine rings, respectively, are highlighted in red to indicate that the bonding order depends on the identity of the nucleotide base, with single bonds in the case of G and U, and double bonds in the case of A and C.
Figure 3. Additional chemical complexity of tRNA modifications is observed in selected organisms.
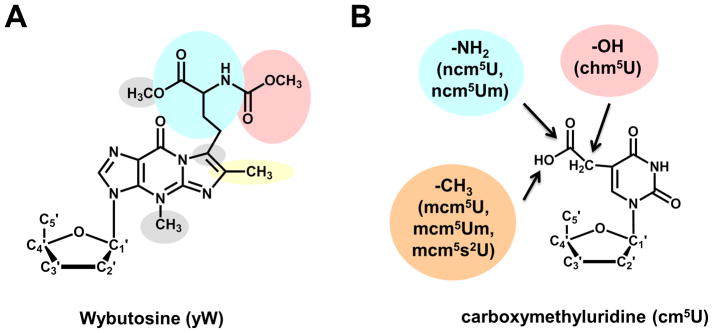
(A) The Wybutosine (yW) modification found at position 37 of eukaryotic tRNAPhe is the result of a multi-step biosynthetic process; the proposed biosynthetic pathway in yeast is indicated by the various shaded groups, each of which represents a distinct step of yW production. In higher eukaryotes and many Archaea, the yW modification is further elaborated into several derivatives, including the OHyW that is the product of TYW5 action in humans. (B) The carboxymethyluridine modification and its derivatives found at position 34 of several tRNAs in eukaryotes. Functional groups added to yield the indicated nucleotide species in parentheses are shown in colored circles. All of these species are related by the common presence of the cm5U modification, synthesized by a common set of enzymes.
BEYOND THE CLOVERLEAF: EFFECTS OF MODIFICATION CHEMISTRY ON STRUCTURE
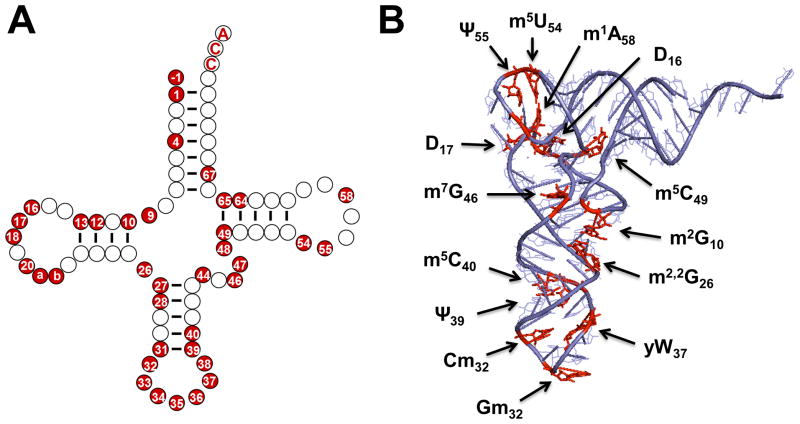
The two-dimensional “cloverleaf” structure is by now a familiar feature of tRNA, and was even proposed once the first tRNA primary sequence, of yeast tRNAAla, was deduced in 1965 9 (Figure 4A). Accumulated sequence and structural data, including the first three-dimensional tRNA structure (of yeast tRNAPhe, determined in 1974), revealed that the overall two and three-dimensional structures of tRNAs are nearly universal features 10. Deviations from the four-stem cloverleaf structure of tRNA are known, but despite even gross changes, such as loss of the entire D-stem and loop from some mitochondrial tRNAs, the overall L-shaped three dimensional tRNA structure is maintained by utilizing complex combinatorial networks of tertiary interactions. For modifications that affect the hydrogen-bonding potential of a particular nucleotide base, a role in modulating alternative secondary structures is readily imagined. For example, N-1-methylguanosine (m1G) disrupts the ability of G residues to form G-C Watson-Crick base pairs by replacing the necessary N-1 proton, preventing formation of secondary structures involving this nucleotide. Likewise, presence of m1A9 in human mitochondrial tRNALys disrupts a Watson-Crick base pair that stabilizes an alternative non-canonical elongated structure, causing the equilibrium to favor the correct cloverleaf form 11. However, despite these well-studied cases, and the possibility of similar examples of secondary structure modulation by modifications, the data suggest that most modified nucleotides do not act in this manner. Static structures of tRNA species reveal that both modified and unmodified tRNAs generally adopt similar L-shaped folds based on the cloverleaf two-dimensional structure (Figure 4B). In the most direct examination of this question to date, an X-ray structural comparison of in vitro transcribed and unmodified E. coli tRNAPhe with that of fully modified yeast tRNAPhe, revealed overall structural similarity, including a fully-formed anticodon-stem loop, albeit with some minor differences, such as a slightly increased angle between the two arms of the L-shaped structure, which appear to correlate with modification status 12.
Figure 4. Modified nucleotides in the context of two-dimensional and three-dimensional structures of tRNA.
(A) The secondary structure of a typical Type I tRNA is shown, with each ball indicating a single nucleotide. Positions that are known to be modified in S. cerevisiae cytoplasmic tRNAs are highlighted in red. Although the exact positions that are modified vary between tRNAs from different species and different domains of life, the general pattern of modification, with a high density of modifications observed in the anticodon stem loop and many fewer modified nucleotides observed in the aminoacyl-acceptor stem, is retained in most species. (B) The three dimensional structure of yeast tRNAPhe (PDB ID 4TNA), with modified nucleotides that are observed in this tRNAPhe species highlighted in red on the structure. These include modifications that are found universally in a large number of tRNAs throughout all three domains of life (such as the m5U54/Ψ55 that gives the so-called TΨ stem its name) as well as modifications such as the wybutosine base (yW37) that are more specific to tRNAPhe from Eukarya and Archaea.
Instead, tRNA modifications, particularly those found at the core of the folded RNA, are thought to predominantly affect more subtle features, including both rigidifying the overall structure as well as making tRNA more flexible 13. Increased rigidity, for example, may occur as a result of the ubiquitous pseudouridine residues found throughout tRNAs, which favor the 3′-endo sugar pucker associated with A-form RNA helices 14. Pseudouridines may also coordinate additional stabilizing water molecules using the available _N_-1 atom that no longer participates in glycosidic linkage in the isomerized form of the base. Other modifications, such as dihydrouridine, are thought to promote the opposing 2′-endo sugar pucker associated with conformational flexibility in RNA15. Maintenance of optimal tRNA structure appears to require contributions from both types of modifications, and suggests the involvement of a network of modified nucleotides in coordinately promoting alternative tRNA conformations as needed 16, 17. This idea is reinforced by studies of tRNA derived from organisms that experience extreme temperature environments. In many of these species the overall level of modification generally correlates with the expected need for tRNA stability; for example, a higher abundance of the flexibility-promoting dihydrouridine is observed in tRNA from psychrophilic archaea relative to their thermophilic archaeal counterparts 18. Moreover, modification levels have been observed to change with growth temperature in a way that also suggests the participation of a network of modifications to apparently maintain a balance between stability and flexibility of the tRNA 19, 20.
While there is a wealth of molecular detail available through X-ray crystallographic investigations of tRNA, these static structures may in fact lead to under appreciation of the dynamic features of tRNA. These are important for optimal interactions of the tRNA with various RNA and protein partners in the cell. One example of this type of structural plasticity is revealed by the interaction between tRNAVal and the archaeosine biosynthetic enzyme, ArcTGT. In this crystal structure, the tRNA adopts a highly perturbed alternative conformation (the so-called λ form) characterized by loss of base pairs in the D-stem and of the normal tertiary core interactions 21. The ability to adopt this structure is thought to expose the substrate G15 nucleotide, which would otherwise be inaccessible to the modification enzyme in the highly structured core. Similar examples of perturbations of tRNA structure, although in many cases not as extreme as in the case of tRNA-ArcTGT, have been shown to play roles in tRNA aminoacylation and interactions with the ribosome during translation. Thus, it is highly likely that the presence of both stabilizing and destabilizing forces in the form of various modified nucleotides are important for modulating structure as needed during the lifetime of the tRNA.
FUNCTION OF MODIFIED NUCLEOTIDES IN tRNA
The widespread and abundant nature of tRNA modifications, combined with significant conservation of many individual modifications across multiple species, necessarily raises questions about the biological function of tRNA modifications. The canonical four nucleotide bases generally suffice for specifying structure and function of other RNAs, including non-coding RNAs and ribozymes, so why has this additional chemical complexity evolved and been maintained at such high levels in the context of tRNA? Since for tRNA, biological function is inherently tied to its ability to adopt structures that interact in a relatively uniform way with the protein translation machinery, an obvious hypothesis is that the presence of modifications helps enhance formation of correct structures or to prevent formation of incorrect ones. As described above, this is unlikely due to gross defects in RNA structure because of lack of modifications, but to more subtle changes that affect the overall balance observed among the population of folded species.
Moreover, although effects on aminoacylation and translation due to lack of modifications have been observed (as discussed in the following sections), unmodified tRNAs lacking modifications are still generally functional in both of these critical reactions, albeit with reduced efficiency and/or fidelity in some cases. Finally, and perhaps most difficult to reconcile with the highly conserved nature of modifications, is the observation that although few of the genes that encode modification enzymes are essential for viability, in most cases deletion of single genes often causes little to no detectable growth defect. The observation of a synthetically lethal network of interactions between modification enzymes in yeast, where single deletions are viable, but deletion of combinations of tRNA modification genes are not, suggests some redundancy in the system whereby loss of single modifications can be compensated for by the presence of others 22. Taken together, these results corroborate the idea that modifications typically act in relatively subtle ways, and often in concert, to maintain functional tRNAs in the cell.
KEEPING TRANSLATION TIDY
Studies in the late sixties and early seventies revealed that a number of mutations could lead to problems with translational accuracy, where the ribosome would lose track of the proper reading frame 23. These mutations would lead to production of aberrant peptides in cases where the translational reading frame was shifted. Alternatively, mutations could lead to abnormally short versions of the intended peptide in cases where the frameshift introduces a premature stop codon. These early studies revealed aspects of the malleability of protein synthesis and brought to the forefront the question of how translational accuracy is maintained and unwanted frameshifts kept to a minimum. It quickly became clear that simple rules involving canonical Watson-Crick base pairing were not sufficient to explain translational accuracy and that to find important regions of the anticodon loop, modified positions could provide a powerful hint. Highlighting their importance in translational efficiency and accuracy, two positions in almost every tRNA are modified regardless of the organism. Position 37 and the wobble nucleotide, position 34 1 (Figure 4). In fact, these two positions, as a whole, comprise the largest chemical diversity of modified nucleotides in tRNAs.
In most tRNAs, Position 37 is an encoded purine that is almost always modified. Modifications of position 37 help maintain and open loop conformation, sterically blocking Watson-Crick pairing and thus influence frameshifting. Preventing base pairing with neighboring nucleotides on the other side of the loop (U33) also aids in the formation of the canonical U-turn structure important for anticodon-codon pairing during decoding 24, 25. Depending on the purine at position 37 cells used different modification strategies to achieve the same outcome of a canonical anticodon loop structure and may even preform the antiocodon into a “translation ready” conformation 26. In most tRNAs an encoded G37 is methylated at the base to form m1G. Lack of m1G formation leads to increased +1 frameshifting and therefore causes problems with translational accuracy 27. Since m1G formation affects many tRNAs in cells, its absence has pleiotropic effects and can lead to severe growth phenotypes 28. Because m1G is found in tRNAs in all domains of life it has been suggested that it belongs to one of the primordial modifications 27 (Figure 1). Thus m1G was apparently enlisted to prevent frameshifting early in the evolving translational machinery of the last common ancestor. In all organisms m1G37 is formed by the S-adenosyl methionine (SAM)-dependent methylation on the encoded G. However, despite its conserved chemistry, the enzymes responsible for m1G formation in Bacteria (trmD) and Eukarya (trm5) are evolutionarily unrelated 29. This again reflects the evolutionary convergence typical of many modifications and prevalent among many methyltranferases.
The role of G37 in translation accuracy is not limited to methylation as the end point and in fact m1G37 is the substrate for further hypermodification in tRNAPhe of Archaea and Eukarya 1. In these organisms, m1G is the forcible intermediate in the formation of wybutosine (yW) and derivatives (wyosine and derivatives) 30, 31. Synthesis of this nucleoside(s) requires a series of enzymatic steps starting with the formation of a tricyclic ring on the methylated guanosine (Figure 3A). This reaction requires the enzyme TYW1, which as shown recently in the archaeal system, uses pyruvate as the two-carbon donor for ring closure32. Interestingly, the eukaryotic TYW1 protein contains an FMN binding domain, which is presumably important for the reduction of the essential [4Fe-4S] cluster proposed to be critical for catalysis. The archaeal counterparts, however, lack the FMN domain and it must have a yet to be defined alternative route of cluster reduction 31. Formation of the tricyclic ring is then followed by a series of SAM-dependent reactions, involving a series of methylations, hydroxylation, methoxycarbonylation, and α-amino-α-carboxy propyl group transfer from SAM 33–38. All in all, five SAM molecules are consumed in the process of wybutosine formation, all dedicated to the maturation of one position in a single tRNA in archaea and eukarya. These modifications then become part of a variably modified side chain of the purine ring. In eukarya, these differ in the extent of modification of N7, generally due to the presence or absence of TYW5 in different eukaryotes 1. In Archaea, the scenario is more complex and the reaction usually stops after the TYW3 step, but various combinations of the hypermodified nucleotide exist depending on the organisms 31, 39. All this chemical diversity leads to a single functional outcome, yW (and derivatives) help stabilize anticodon-codon interactions in the A site of the ribosome by providing a series of base stacking interactions 40. Thus playing a key function in proper frame maintenance 40. This is particular important with tRNAPhe, which decodes UUU and UUC codons, given the propensity of “slippage” when the ribosome encounters poly uridines in a message, as beautifully illustrated in the stories involving programmed frameshifting 41–44. These observations then lead to the possibility that the extent of tRNAPhe modification at position 37 may correlate with the frequency of poly uridine slippery sequences in genomes.
In most cases, when position 37 is an encoded adenosine, it is also further modified. In all domains of life this position is usually isopentenylated to form isopentenyl adenosine (i6A), while in Bacteria i6A can be further hypermodified to ms2i6A or ms2io6A depending on the tRNA and the organism 1. The i6A system of Bacteria targets tRNAs for all codons with a U at the first position while in eukarya the tRNA substrates vary but are generally restricted to a smaller set of tRNAs. In terms of substrate specificity the bacterial enzymes (encoded by the miaA gene) recognize A36A37A38 as a major determinant for i6A formation while in Eukarya what determines a good substrate depends once again on the organism revealing a remarkable plasticity in substrate recognition by this family of enzymes 45. Despite i6A constituting a bulky modification, its function differs to that of the previously described G37 modifications in that rather than playing roles in frame maintenance, A37 modifications are key determinants for codon-specific translation and therefore translation efficiency 45. In tRNAs decoding the ANN codons, A37 is also universally modified to form _N_6-threonylcarbamoyladenosine (t6A) and can be further methylated at the base (e.g. m6t6A); this modification is crucial to translational accuracy. Formation of t6A in Bacteria involves the Sua5/YrdC family of proteins while in Eukarya the Kae1/YgjD/Qri7 family 46. Significantly, mutants in these families lead to pleiotropic effects in cells including defects in transcription and genome stability 46.
At position 34, the majority of modifications ensure efficient anticodon-codon pairing and coupled with those modifications at position 37 contribute greatly to the stability of the tRNA-mRNA interaction during decoding 47. These include pseudouridylation, ribose methylation, acetylations, etc. Among all the modifications of position 34 described so far, perhaps none is better understood than the modified U34 found in various tRNAs, but highly conserved in tRNAGln, tRNAGlu and tRNALys. These U34-containing tRNAs for these 3 amino acids all undergo s2U thiolation and modification at the C5-position of the pyrimidine ring. C5 modifications may include various methylations and acetylation reactions but may also include hyper modification where entire sugars are added to the pyrimidine ring 3. These modifications in combination with s2U lead to increased anticodon rigidity and may serve as amino acylation, translation efficiency and fidelity determinants 48–52. In terms of evolution, the enzymes responsible for s2U formation evolved independently in the eukaryotic and bacterial lineage. Remarkably, both sets of enzymes exist in eukaryotic cells where the cytoplasmic pathway is uniquely eukaryotic in nature, yet the mitochondrial pathway, in line with its ancestry, is still bacterial in nature 53. Cytoplasmic thiolation requires a series of ubiquitin ligase like proteins that in an enzymatic cascade lead to formation of s2U34 53–61. In mitochondria, homologs of the bacterial system are responsible for this reaction 54–58, 60, 61. Again, regardless of which pathway is utilized the outcome is still the same and the final product of the thiolation reactions still is s2U. This raises the interesting question of why then, the two pathways differ and why s2U synthesis may have taken different evolutionary routes? These, of course are unanswered questions, but perhaps the answer lies in the nature of the surrounding intracellular environment where organisms adopted different strategies to achieve the same solution to the decoding problem.
In cases where the first position of the anticodon is a guanosine, perhaps the most prevalent and best-studied modifications are the 7-deaza guanosine derivatives. Queosine (Q) and derivatives are prevalent in bacterial and eukaryotic tRNAs, while Archaea contain archeosine (G+, G*) 62–71. In all cases these modifications again ensure translational fidelity and efficiency. Interestingly, while Bacteria and Archaea can synthesize Q and G+ de novo, eukaryotes rescue the preformed base from their nutrients 72. Enzymatically, however, in either Q or G+ synthesis involves a transglycosylation reaction performed by the enzyme tRNA-guanine transglycosylase (TGT) 64, 65, 67. This reaction involves breakage of the glycosydic bond between the base and the sugar and replacement by the modified 7-deaza derivative 62, 73. In addition, in many bacteria, Q in the anticodon of tRNAAsp can be further modified through addition of a glutamate residue by the product of yadB, a truncated glutamyl tRNA synthetase 74, 75. However, the importance of the additional glutamate beyond what Q can by itself provide is not totally clear.
There are of course many more modifications that are usually found at position 34 and 37 of tRNAs in all domains of life. We have highlighted here the few for which the biosynthesis and/or function are relatively well understood. The common underlying theme with all these is that regardless of by what evolutionary means enzymatic activities were recruited to perform a given modification, a majority of anticodon modifications, given that they affect the “business” end of the tRNA, provide anticodon loop stability and in doing so can ensure translational fidelity and efficiency during decoding.
PROGRAMMED ALTERATIONS THAT AFFECT MEANING: THE CASE OF tRNA EDITING
The combination of selective pressures set forth at all levels of genome evolution, from the necessity to make stable genomes, to coping with the inherent mutational rates of replicative polymerases, combined with ever changing environments has forced organisms to also create coding diversity as needed but without compromising translational fidelity. So far a few examples of nucleotide changes that expand the decoding capacity of tRNAs have been described. These include deaminations, aminoacylations, formylations, etc. Clearly, these changes and the specificity of the enzymes that makes them have to be carefully selected to avoid large-scale changes that could compromise translational fidelity. In this section we will highlight four examples of tRNA editing events that help expand a tRNAs decoding capacity: Lysidine/agmatidine, formylcytosine, C34/A34 deaminations and G−1 addition.
The genetic code is far from universal and a number of codons are inferred to have different meaning in different organisms. For example, in mitochondria where a number of codons have been reassigned to mean something different than in the nuclear genomes 76–79. Most conserved among these, is perhaps the use of UGA codons as tryptophan in organelles.
In Archaea and Bacteria, the challenge posed by a non-universal genetic code and years of work led to the identification of lysidine in Bacteria and more recently agmatidine in Archaea. These modifications both involve the hypermodification of C34 with the sole purpose of decoding AUA initiation codons found in some mRNAs as methionine, solving a decoding problem created by genome evolutionary dynamics. Remarkably these modifications not only convert a universal AUA codon, which should specify isoleucine, into a methionine codon, but also change the identity of the modified tRNA. Following C34 modification, what appears as an encoded isoleucine tRNA is no longer recognized by the isoleucyl tRNA synthetase and it is instead recognized by the methionyl tRNA synthetase. Because both modifications involve the addition of an amino acid to position of C34 (lysine in the case of lysidine and decarboxy arginine in the case of agmatidine) 80, 81, mechanistically these modifications must require activation of the base by an adenylation step, followed by incorporation of the amino acid. This reaction is thus somewhat analogous to that of aminoacyl tRNA synthetases and RNA ligases. Indeed this is the mechanism described for TilS the lysidine enzyme but a homolog of TilS has not been found in Archaea 82. It is possible that just like in the case of the m1G enzymes, two unrelated enzymes have been recruited to modify a potential tRNAIle into one that is recognized as methionine and still achieving the same solution to allow an encoded C34 to behave as U34 permitting efficient base pairing with an adenosine at the first position of the AUA codon.
Another interesting example is provided by the use of AUA codons as methionine in the mitochondria of a number or organisms including frogs, mammals (rat and humans), fruit flies, squid, etc. In these mitochondria, two codons are used as methionine: AUG, the standard codon and AUA, normally coding for isoleucine in the universal code. Initially it was thought that this codon reassignment could be achieved by lysidine modification of tRNAMetCAU. Since this modification had been described in Bacteria, it followed logically that mitochondrial decoding should use the same strategy. However, as mitochondrial genome sequences became available, it was apparent that no tRNAIle with anticodon UAU existed in animal mitochondria. Thus, it was proposed that the mitochondria of most organisms (with the exception of plants) might use a different strategy. It was suggested that a single tRNAMet with anticodon CAU was responsible for decoding these codons as methionine. Sequencing of the native tRNA from animal mitochondria then revealed that the first position of the anticodon of mitochondrial tRNAMet was post-transcriptionally modified to 5-formyl cytosine (f5C) 83. This modification could allow for an unusual wobble pair involving the modified C34 in the first position of the anticodon and an adenosine in the third codon position. Recent studies that measured the synthesis of polymethionine with mitochondrial ribosomes programmed with polyribonucleotides consisting of either AUG or AUA codon showed that f5C34 was absolutely required for the decoding of AUA as methionine. Significantly the presence of this modification did not interfere with AUG decoding but simply expanded the ability of the tRNA to use the additional AUA codons 84. The availability of this in vitro system should help clarify a number of observations that implicate modifications in mitochondrial defect in a more direct manner. Interestingly, the f5C system, which does not occur in Bacteria, also highlights the fact that despite their origin, years of evolution have also led mitochondria to unique solutions to the decoding problem.
By far the most widespread tRNA editing strategy is the use of deamination reactions to expand a tRNAs decoding capacity and more rare cases to change the meaning of the tRNA to recognize a new codon. Here two types of editing are at play: inosine formation (A to I editing) and cytosine deamination (C to U editing), both occurring at position 34 of some tRNAs.
Inosine, a guanosine analog that can base pair with A, C or U in the anticodon, provides one of the most extreme cases of base-pairing flexibility in protein synthesis 85. In the context of a codon-anticodon interaction, inosine at the first position of the anticodon (I34) effectively allows a single tRNA to decode three different codons for the same amino acid obviating the need for additional tRNAs to be encoded in genomes. Given the role of I34 in decoding in Bacteria and Eukarya, it is not surprising that the enzymes responsible for inosine formation are essential for viability 86–88. Currently, the best-studied example of inosine in tRNA is that occurring at the first position of the anticodon (I34) where the essential inosine has a direct bearing on decoding. In Bacteria, I34 only occurs in tRNAArgACG, which is encoded with an A at position 34 and is specified by ADATa (tadA) 86. This enzyme is dedicated to the deamination of a single adenosine in a single tRNA, but since Bacteria do not encode a G34-containing tRNA for the C-ending codon for arginine, A to I deamination is still essential. By contrast, the eukaryotic enzymes target 7 or 8 different tRNAs depending on the organism, raising questions about the evolutionary transition to this broader, multi-substrate specificity. The eukaryotic enzyme, as exemplified by the yeast and trypanosome systems, function as a heterodimer, comprised of 2 subunits, ADAT2 and ADAT3 87, 89. Finally, in Archaea inosine has not been found in the anticodon of tRNAs; instead, archaeal genomes encode G34 containing tRNAs to decode those codons that are accounted for by inosine in other systems 1. Again the reason for these two different strategies is not clear but the answer may lie once again on genome evolution, where mutational pressure at the 3rd position of codons led to the recruitment of enzymatic activities that could generate, reassign if you will, tRNAs to solve decoding problems.
Inosine formation is not relegated to the anticodon and has been described at various positions in tRNAs from a number of organisms. For example, in eukaryotic tRNAs inosine can also occur at position 37 where it may affect anticodon loop structure 90. Interestingly, the sequence of the enzyme responsible for synthesis of I37 resembles classic adenosine deaminases, while that for I34 has all the sequence signature motifs of cytidine deaminase, perhaps reflecting their different evolutionary paths. Likewise in Archaea inosine is also found in the TΨC loop of many tRNAs, but remarkably its formation requires prior methylation of the adenosine to be deaminated.
C to U editing of anticodon nucleotides is more rare and has so far being described at length in two systems: Marsupials and kinetoplastids. In the marsupial system, a single C35 to U35 editing event in the second position of the anticodon of tRNAGlyGCC converts it into a tRNA that now decodes aspartate codons (tRNAAspGUC). Significantly this single C to U editing event not only changed the meaning of the tRNA during decoding but, in a situation similar to lysidine, it also switched the recognition of the tRNA by the aminoacyl tRNA synthetase from Gly to Asp. In the kinetoplastid Leishmania tarentolae, a single tRNATrp is encoded in the nuclear genome with the anticodon CCA, which efficiently decodes the canonical UGG codons for tryptophan of nucleus-encoded mRNAs. Due to a complete lack of tRNA genes in the mitochondria genomes of these organisms, the same tRNATrp is imported into the organelle for mitochondrial protein synthesis. However, like in most eukaryotes the mitochondrial genetic code is not standard and two types of Trp codons exist in kinetoplastids: UGG and UGA. To decode the UGA codons, following import, tRNATrp then undergoes a single C34 to U34 editing event at the first position of the anticodon 91. The edited tRNA is also thiolated (s2U) at U33, thus far the only example of U33 modification in any tRNA 92. In T. brucei, expectedly from the L. tarentolae story, tRNATrp also undergo editing and thiolation 93. In these organisms there are two versions of tryptophanyl tRNA synthetase (TrpRS), one cytoplasmic and the other mitochondrial. At least in the case of T. brucei, editing and thiolation of tRNATrp serve as negative determinants for aminoacylation by the cytosolic synthetase, while the mitochondrial TrpRS does not discriminate between the edited and unedited species 93. Here again, C to U editing appears to have a say in both the ability of the tRNA to decode UGA codons and also in synthetase recognition. The full significance of this finding however is not clear given that the tRNA transits through the cytoplasm unmodified and it is only edited and thiolated inside mitochondria. Therefore the edited tRNA under normal growth conditions never comes in contact with the cytosolic enzyme. Just like inosine, C to U deamination is not limited to the anticodon and occurs at various positions in tRNAs from Archaea and Eukarya. In general the role of C to U changes that occur away from the anticodon is to regenerate stems or critical features important for proper tRNA folding. Still the enzyme(s) responsible for C to U editing of most tRNAs remain at large and it is only in Archaea where a cytosine editing deaminase specific for position 8 of tRNAs has been identified and partially characterized 94.
Perhaps the most mechanistically unusual case of editing is that involving the posttranscriptional addition of a non-encoded guanosine at the 5′-end of tRNAHis (the so-called G-1 addition) 95, 96. In a true act of enzymatic acrobatics, the enzyme responsible for this change, Thg1, does so while effectively catalyzing a polymerization reaction in the 3′ to 5′ direction 97, 98. Representing the only known case of an enzyme that can perform nucleotide polymerization in a templated manner in the opposite direction from more canonical DNA and RNA polymerases. It is worth mentioning that although G-1 addition does not alter the meaning of the tRNA in decoding, it is still a required editing step to ensure aminoacylation of tRNAHis in many organisms, including most eukaryotes. This editing event is therefore essential for viability.
Taken together the examples above highlight a growing theme in the tRNA editing field that may be applicable to other modifications. When put under increased selective pressure to solve a decoding problem, systems tend to recruit pre-existing activities for the purpose of survival and once recruited these become indispensable in cellular function. In the case of deaminases the activities were provided by already existing nucleotide deaminases. It presumably only took minor changes to readapt deaminases to a new function and specificity toward polymeric substrates. Even in the case of Thg1, despite its unique modus operandi, its core structure shares motifs conserved among existing polymerases.
COMBINATORIAL COMPENSATION AND ABE LINCOLN
It is exceptionally challenging in a single review to describe in great detail the function that more than 100 different modifications have in tRNAs. It has, however, become increasingly clear that with tRNA modifications the degree of chemical intricacy is so vast that indeed the roles of modifications in tRNA function are much more than currently meets the eye. There is indeed a bewildering diversity of modifications with different sets of modifications in different tRNAs of different organisms and even within single cells of single organisms. However, in general all modifications act combinatorially to achieve the common goal of permitting tRNAs with their sequence diversity to read a nearly universal genetic code.
In thinking of modifications, the field is moving away from modifications as single entities. It is in the context of the tRNA sequence, with its 4 canonical nucleotides, that the integration of positional and global structural effects of modifications impact tRNA function. Not so long ago modification aficionados were puzzled by the fact that even in the case of highly conserved modifications, their absence had negligible phenotypic effects. However, recent studies have shown that even in these cases lack of two or more modifications, which individually cause no effect in cell growth, lead to synthetic lethality. Thus modifications provide some sort of compensatory effect, which combinatorially may still maintain relatively normal tRNA function under normal conditions of growth. In addition, the redundant nature of modification enzymes themselves may help a cell survive even when single modification enzymes are missing. One must emphasize that even modifications whose absence lead to the most innocuous effects, may prove crucial to viability with changing environmental conditions such as environmental stress or nutrient limitation.
Still the challenge remains to genetically and biochemically characterize all modification enzymes and their substrates, to understand how their concerted effects are exerted on tRNA substrates. Despite overall conservation, tRNAs still provide sufficient nuances in their structures that can be exploited by modifications enzymes themselves as the source of substrate specificity. Given their diversity and their effect on both global and local tRNA structure in a context-specific manner, the lingering question remains of whether or not all modifications benefit all tRNAs 99. It is possible that a modification enzyme may become essential for one tRNA, but the same modification may be surplus on a different tRNA without affecting its function. All depends on how again the combinatorial nature of nucleotide sequences and/or additional modifications may affect substrate recognition by modification enzymes. In concluding and to answer this question, we would like to paraphrase a quote attributed to Abraham Lincoln*: Whereas some modifications may benefit all tRNAs and all modifications may benefit some tRNAs, all modifications do not benefit all tRNAs.
Acknowledgments
The authors wish to thank all members of the Jackman and Alfonzo laboratories for helpful discussions. This work was supported in part by grants R01-GM087543 to J.E.J. and R01 GM084065 to J.D.A. from the National Institutes of Health. A special thanks to H. Grosjean for help with the figures.
Footnotes
*
The original quote attributed to Abraham Lincoln stated that: “You can fool some of the people all the time, and all of the people some of the time, but you cannot fool all of the people all the time”.
References
- 1.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grosjean H. DNA and RNA modification enzymes: structure, mechanism, function and evolution. Austin, Tex: Landes Bioscience; 2009. [Google Scholar]
- 3.Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, Bujnicki JM. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rozenski J, Crain PF, McCloskey JA. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips G, de Crecy-Lagard V. Biosynthesis and function of tRNA modifications in Archaea. Curr Opin Microbiol. 2011;14:335–341. doi: 10.1016/j.mib.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011;2:611–631. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 7.Grosjean H, de Crecy-Lagard V, Marck C. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010;584:252–264. doi: 10.1016/j.febslet.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 8.Huang B, Johansson MJ, Bystrom AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holley RW, Everett GA, Madison JT, Zamir A. Nucleotide Sequences in the Yeast Alanine Transfer Ribonucleic Acid. J Biol Chem. 1965;240:2122–2128. [PubMed] [Google Scholar]
- 10.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 11.Voigts-Hoffmann F, Hengesbach M, Kobitski AY, van Aerschot A, Herdewijn P, Nienhaus GU, Helm M. A methyl group controls conformational equilibrium in human mitochondrial tRNA(Lys) J Am Chem Soc. 2007;129:13382–13383. doi: 10.1021/ja075520+. [DOI] [PubMed] [Google Scholar]
- 12.Byrne RT, Konevega AL, Rodnina MV, Antson AA. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res. 2010;38:4154–4162. doi: 10.1093/nar/gkq133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giegé R, Jühling F, Pütz J, Stadler P, Sauter C, Florentz C. Structure of transfer RNAs: similarity and variability. Wiley Interdisciplinary Reviews: RNA. 2012;3:37–61. doi: 10.1002/wrna.103. [DOI] [PubMed] [Google Scholar]
- 14.Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry. 2005;44:8078–8089. doi: 10.1021/bi050343f. [DOI] [PubMed] [Google Scholar]
- 15.Dalluge JJ, Hashizume T, McCloskey JA. Quantitative measurement of dihydrouridine in RNA using isotope dilution liquid chromatography-mass spectrometry (LC/MS) Nucleic Acids Res. 1996;24:3242–3245. doi: 10.1093/nar/24.16.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zagryadskaya EI, Kotlova N, Steinberg SV. Key elements in maintenance of the tRNA L-shape. J Mol Biol. 2004;340:435–444. doi: 10.1016/j.jmb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Steinberg S, Cedergren R. A correlation between N2-dimethylguanosine presence and alternate tRNA conformers. Rna. 1995;1:886–891. [PMC free article] [PubMed] [Google Scholar]
- 18.Edmonds CG, Crain PF, Gupta R, Hashizume T, Hocart CH, Kowalak JA, Pomerantz SC, Stetter KO, McCloskey JA. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria) J Bacteriol. 1991;173:3138–3148. doi: 10.1128/jb.173.10.3138-3148.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishida K, Kunibayashi T, Tomikawa C, Ochi A, Kanai T, Hirata A, Iwashita C, Hori H. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 2011;39:2304–2318. doi: 10.1093/nar/gkq1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agris PF, Koh H, Soll D. The effect of growth temperatures on the in vivo ribose methylation of Bacillus stearothermophilus transfer RNA. Arch Biochem Biophys. 1973;154:277–282. doi: 10.1016/0003-9861(73)90058-1. [DOI] [PubMed] [Google Scholar]
- 21.Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell. 2003;113:383–394. doi: 10.1016/s0092-8674(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 22.Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 23.Yourno J, Tanemura S. Restoration of in-phase translation by an unlinked suppressor of a frameshift mutation in Salmonella typhimurium. Nature. 1970;225:422–426. doi: 10.1038/225422a0. [DOI] [PubMed] [Google Scholar]
- 24.Bilbille Y, Vendeix FA, Guenther R, Malkiewicz A, Ariza X, Vilarrasa J, Agris PF. The structure of the human tRNALys3 anticodon bound to the HIV genome is stabilized by modified nucleosides and adjacent mismatch base pairs. Nucleic Acids Res. 2009;37:3342–3353. doi: 10.1093/nar/gkp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabello-Villegas J, Winkler ME, Nikonowicz EP. Solution conformations of unmodified and A(37)N(6)-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNA(Phe) J Mol Biol. 2002;319:1015–1034. doi: 10.1016/S0022-2836(02)00382-0. [DOI] [PubMed] [Google Scholar]
- 26.Vendeix FA, Murphy FVt, Cantara WA, Leszczynska G, Gustilo EM, Sproat B, Malkiewicz A, Agris PF. Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism. J Mol Biol. 2012;412:467–85. doi: 10.1016/j.jmb.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. A primordial tRNA modification required for the evolution of life? Embo J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. Embo J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christian T, Hou YM. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J Mol Biol. 2007;373:623–632. doi: 10.1016/j.jmb.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noma A, Suzuki T. Ribonucleome analysis identified enzyme genes responsible for wybutosine synthesis. Nucleic Acids Symp Ser (Oxf) 2006:65–66. doi: 10.1093/nass/nrl032. [DOI] [PubMed] [Google Scholar]
- 31.de Crecy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, et al. Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in Archaea. Mol Biol Evol. 2010;27:2062–2077. doi: 10.1093/molbev/msq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young AP, Bandarian V. Pyruvate is the source of the two carbons that are required for formation of the imidazoline ring of 4-demethylwyosine. Biochemistry. 2011;50:10573–10575. doi: 10.1021/bi2015053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kan J, Suzuki MT, Wang K, Evans SE, Chen F. High temporal but low spatial heterogeneity of bacterioplankton in the Chesapeake Bay. Appl Environ Microbiol. 2007;73:6776–6789. doi: 10.1128/AEM.00541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umitsu M, Nishimasu H, Noma A, Suzuki T, Ishitani R, Nureki O. Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2. Proc Natl Acad Sci U S A. 2009;106:15616–15621. doi: 10.1073/pnas.0905270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki Y, Noma A, Suzuki T, Senda M, Senda T, Ishitani R, Nureki O. Crystal structure of the radical SAM enzyme catalyzing tricyclic modified base formation in tRNA. J Mol Biol. 2007;372:1204–1214. doi: 10.1016/j.jmb.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Goto-Ito S, Ishii R, Ito T, Shibata R, Fusatomi E, Sekine SI, Bessho Y, Yokoyama S. Structure of an archaeal TYW1, the enzyme catalyzing the second step of wye-base biosynthesis. Acta Crystallogr D Biol Crystallogr. 2007;63:1059–1068. doi: 10.1107/S0907444907040668. [DOI] [PubMed] [Google Scholar]
- 37.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. Embo J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki Y, Noma A, Suzuki T, Ishitani R, Nureki O. Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4. Nucleic Acids Res. 2009;37:2910–2925. doi: 10.1093/nar/gkp158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCloskey JA, Graham DE, Zhou S, Crain PF, Ibba M, Konisky J, Soll D, Olsen GJ. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 2001;29:4699–4706. doi: 10.1093/nar/29.22.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urbonavicius J, Stahl G, Durand JM, Ben Salem SN, Qian Q, Farabaugh PJ, Bjork GR. Transfer RNA modifications that alter +1 frameshifting in general fail to affect -1 frameshifting. Rna. 2003;9:760–768. doi: 10.1261/rna.5210803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atkins JF, Bjork GR. A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 2009;73:178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farabaugh PJ. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog Nucleic Acid Res Mol Biol. 2000;64:131–170. doi: 10.1016/s0079-6603(00)64004-7. [DOI] [PubMed] [Google Scholar]
- 43.Farabaugh PJ. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dinman JD. Control of gene expression by translational recoding. Adv Protein Chem Struct Biol. 2012;86:129–149. doi: 10.1016/B978-0-12-386497-0.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamichhane TN, Blewett NH, Maraia RJ. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. Rna. 2011;17:1846–1857. doi: 10.1261/rna.2628611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D, Murzin AG, de Crecy-Lagard V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. Embo J. 2011;30:882–893. doi: 10.1038/emboj.2010.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 48.Tisne C, Rigourd M, Marquet R, Ehresmann C, Dardel F. NMR and biochemical characterization of recombinant human tRNA(Lys)3 expressed in Escherichia coli: identification of posttranscriptional nucleotide modifications required for efficient initiation of HIV-1 reverse transcription. Rna. 2000;6:1403–1412. doi: 10.1017/s1355838200000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madore E, Florentz C, Giege R, Sekine S, Yokoyama S, Lapointe J. Effect of modified nucleotides on Escherichia coli tRNAGlu structure and on its aminoacylation by glutamyl-tRNA synthetase. Predominant and distinct roles of the mnm5 and s2 modifications of U34 Eur J Biochem. 1999;266:1128–1135. doi: 10.1046/j.1432-1327.1999.00965.x. [DOI] [PubMed] [Google Scholar]
- 50.Ashraf SS, Sochacka E, Cain R, Guenther R, Malkiewicz A, Agris PF. Single atom modification (O-->S) of tRNA confers ribosome binding. Rna. 1999;5:188–194. doi: 10.1017/s1355838299981529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bjork GR, Huang B, Persson OP, Bystrom AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. Rna. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 54.Sasarman F, Antonicka H, Horvath R, Shoubridge EA. The 2-thiouridylase function of the human MTU1 (TRMU) enzyme is dispensable for mitochondrial translation. Hum Mol Genet. 2011;20:4634–4643. doi: 10.1093/hmg/ddr397. [DOI] [PubMed] [Google Scholar]
- 55.Paris Z, Changmai P, Rubio MA, Zikova A, Stuart KD, Alfonzo JD, Lukes J. The Fe/S cluster assembly protein Isd11 is essential for tRNA thiolation in Trypanosoma brucei. J Biol Chem. 2011;285:22394–22402. doi: 10.1074/jbc.M109.083774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bruske EI, Sendfeld F, Schneider A. Thiolated tRNAs of Trypanosoma brucei are imported into mitochondria and dethiolated after import. J Biol Chem. 2009;284:36491–36499. doi: 10.1074/jbc.M109.064527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wohlgamuth-Benedum JM, Rubio MA, Paris Z, Long S, Poliak P, Lukes J, Alfonzo JD. Thiolation controls cytoplasmic tRNA stability and acts as a negative determinant for tRNA editing in mitochondria. J Biol Chem. 2009;284:23947–23953. doi: 10.1074/jbc.M109.029421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pedrioli PG, Leidel S, Hofmann K. Urm1 at the crossroad of modifications. ‘Protein Modifications: Beyond the Usual Suspects’ Review Series EMBO Rep. 2008;9:1196–1202. doi: 10.1038/embor.2008.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shigi N, Sakaguchi Y, Asai S, Suzuki T, Watanabe K. Common thiolation mechanism in the biosynthesis of tRNA thiouridine and sulphur-containing cofactors. Embo J. 2008;27:3267–3278. doi: 10.1038/emboj.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dewez M, Bauer F, Dieu M, Raes M, Vandenhaute J, Hermand D. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc Natl Acad Sci U S A. 2008;105:5459–5464. doi: 10.1073/pnas.0709404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwata-Reuyl D. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorg Chem. 2003;31:24–43. doi: 10.1016/s0045-2068(02)00513-8. [DOI] [PubMed] [Google Scholar]
- 63.Watanabe M, Matsuo M, Tanaka S, Akimoto H, Asahi S, Nishimura S, Katze JR, Hashizume T, Crain PF, McCloskey JA, et al. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, proceeds via a pathway involving base replacement on the tRNA polynucleotide chain. J Biol Chem. 1997;272:20146–20151. doi: 10.1074/jbc.272.32.20146. [DOI] [PubMed] [Google Scholar]
- 64.Boland C, Hayes P, Santa-Maria I, Nishimura S, Kelly VP. Queuosine formation in eukaryotic tRNA occurs via a mitochondria-localized heteromeric transglycosylase. J Biol Chem. 2009;284:18218–18227. doi: 10.1074/jbc.M109.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iwata-Reuyl D. An embarrassment of riches: the enzymology of RNA modification. Curr Opin Chem Biol. 2008;12:126–133. doi: 10.1016/j.cbpa.2008.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee BW, Van Lanen SG, Iwata-Reuyl D. Mechanistic studies of Bacillus subtilis QueF, the nitrile oxidoreductase involved in queuosine biosynthesis. Biochemistry. 2007;46:12844–12854. doi: 10.1021/bi701265r. [DOI] [PubMed] [Google Scholar]
- 67.Garcia GA, Kittendorf JD. Transglycosylation: a mechanism for RNA modification (and editing? ) Bioorg Chem. 2005;33:229–251. doi: 10.1016/j.bioorg.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vinayak M, Pathak C. Queuosine modification of tRNA: its divergent role in cellular machinery. Biosci Rep. 2009;30:135–148. doi: 10.1042/BSR20090057. [DOI] [PubMed] [Google Scholar]
- 69.Gregson JM, Crain PF, Edmonds CG, Gupta R, Hashizume T, Phillipson DW, McCloskey JA. Structure of the archaeal transfer RNA nucleoside G*-15 (2-amino-4,7-dihydro- 4-oxo-7-beta-D-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximi dam ide (archaeosine)) J Biol Chem. 1993;268:10076–10086. [PubMed] [Google Scholar]
- 70.El Yacoubi B, Phillips G, Blaby IK, Haas CE, Cruz Y, Greenberg J, de Crecy-Lagard V. A Gateway platform for functional genomics in Haloferax volcanii: deletion of three tRNA modification genes. Archaea. 2009;2:211–219. doi: 10.1155/2009/428489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Phillips G, El Yacoubi B, Lyons B, Alvarez S, Iwata-Reuyl D, de Crecy-Lagard V. Biosynthesis of 7-deazaguanosine-modified tRNA nucleosides: a new role for GTP cyclohydrolase I. J Bacteriol. 2008;190:7876–7884. doi: 10.1128/JB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Katze JR, Gunduz U, Smith DL, Cheng CS, McCloskey JA. Evidence that the nucleic acid base queuine is incorporated intact into tRNA by animal cells. Biochemistry. 1984;23:1171–1176. doi: 10.1021/bi00301a022. [DOI] [PubMed] [Google Scholar]
- 73.Van Lanen SG, Iwata-Reuyl D. Kinetic mechanism of the tRNA-modifying enzyme S-adenosylmethionine:tRNA ribosyltransferase-isomerase (QueA) Biochemistry. 2003;42:5312–5320. doi: 10.1021/bi034197u. [DOI] [PubMed] [Google Scholar]
- 74.Salazar JC, Ambrogelly A, Crain PF, McCloskey JA, Soll D. A truncated aminoacyl-tRNA synthetase modifies RNA. Proc Natl Acad Sci U S A. 2004;101:7536–7541. doi: 10.1073/pnas.0401982101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blaise M, Becker HD, Keith G, Cambillau C, Lapointe J, Giege R, Kern D. A minimalist glutamyl-tRNA synthetase dedicated to aminoacylation of the tRNAAsp QUC anticodon. Nucleic Acids Res. 2004;32:2768–2775. doi: 10.1093/nar/gkh608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jukes TH, Osawa S. Evolutionary changes in the genetic code. Comp Biochem Physiol B. 1993;106:489–494. doi: 10.1016/0305-0491(93)90122-l. [DOI] [PubMed] [Google Scholar]
- 77.Jukes TH, Osawa S. The genetic code in mitochondria and chloroplasts. Experientia. 1990;46:1117–1126. doi: 10.1007/BF01936921. [DOI] [PubMed] [Google Scholar]
- 78.Yokobori S, Suzuki T, Watanabe K. Genetic code variations in mitochondria: tRNA as a major determinant of genetic code plasticity. J Mol Evol. 2001;53:314–326. doi: 10.1007/s002390010221. [DOI] [PubMed] [Google Scholar]
- 79.Jukes TH, Osawa S. CUG codons in Candida spp. J Mol Evol. 1996;42:321–322. doi: 10.1007/BF02198859. [DOI] [PubMed] [Google Scholar]
- 80.Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, Ote T, Kato J, Watanabe K, Sekine Y, Suzuki T. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol Cell. 2003;12:689–698. doi: 10.1016/s1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- 81.Mandal D, Kohrer C, Su D, Russell SP, Krivos K, Castleberry CM, Blum P, Limbach PA, Soll D, RajBhandary UL. Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc Natl Acad Sci U S A. 2010;107:2872–2877. doi: 10.1073/pnas.0914869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ikeuchi Y, Soma A, Ote T, Kato J, Sekine Y, Suzuki T. molecular mechanism of lysidine synthesis that determines tRNA identity and codon recognition. Mol Cell. 2005;19:235–246. doi: 10.1016/j.molcel.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 83.Moriya J, Yokogawa T, Wakita K, Ueda T, Nishikawa K, Crain PF, Hashizume T, Pomerantz SC, McCloskey JA, Kawai G, et al. A novel modified nucleoside found at the first position of the anticodon of methionine tRNA from bovine liver mitochondria. Biochemistry. 1994;33:2234–2239. doi: 10.1021/bi00174a033. [DOI] [PubMed] [Google Scholar]
- 84.Takemoto C, Spremulli LL, Benkowski LA, Ueda T, Yokogawa T, Watanabe K. Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 2009;37:1616–1627. doi: 10.1093/nar/gkp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murphy FVt, Ramakrishnan V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat Struct Mol Biol. 2004;11:1251–1252. doi: 10.1038/nsmb866. [DOI] [PubMed] [Google Scholar]
- 86.Wolf J, Gerber AP, Keller W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. Embo J. 2002;21:3841–3851. doi: 10.1093/emboj/cdf362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gerber AP, Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999;286:1146–1149. doi: 10.1126/science.286.5442.1146. [DOI] [PubMed] [Google Scholar]
- 88.Rubio MA, Pastar I, Gaston KW, Ragone FL, Janzen CJ, Cross GA, Papavasiliou FN, Alfonzo JD. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc Natl Acad Sci U S A. 2007;104:7821–7826. doi: 10.1073/pnas.0702394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Auxilien S, Crain PF, Trewyn RW, Grosjean H. Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J Mol Biol. 1996;262:437–458. doi: 10.1006/jmbi.1996.0527. [DOI] [PubMed] [Google Scholar]
- 90.Gerber A, Grosjean H, Melcher T, Keller W. Tad1p, a yeast tRNA-specific adenosine deaminase, is related to the mammalian pre-mRNA editing enzymes ADAR1 and ADAR2. Embo J. 1998;17:4780–4789. doi: 10.1093/emboj/17.16.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alfonzo JD, Blanc V, Estevez AM, Rubio MA, Simpson L. C to U editing of the anticodon of imported mitochondrial tRNA(Trp) allows decoding of the UGA stop codon in Leishmania tarentolae. Embo J. 1999;18:7056–7062. doi: 10.1093/emboj/18.24.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crain PF, Alfonzo JD, Rozenski J, Kapushoc ST, McCloskey JA, Simpson L. Modification of the universally unmodified uridine-33 in a mitochondria-imported edited tRNA and the role of the anticodon arm structure on editing efficiency. Rna. 2002;8:752–761. doi: 10.1017/s1355838202022045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Charriere F, Helgadottir S, Horn EK, Soll D, Schneider A. Dual targeting of a single tRNA(Trp) requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2006;103:6847–6852. doi: 10.1073/pnas.0602362103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Randau L, Stanley BJ, Kohlway A, Mechta S, Xiong Y, Soll D. A cytidine deaminase edits C to U in transfer RNAs in Archaea. Science. 2009;324:657–659. doi: 10.1126/science.1170123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Williams JB, Cooley L, Soll D. Enzymatic addition of guanylate to histidine transfer RNA. Methods Enzymol. 1990;181:451–462. doi: 10.1016/0076-6879(90)81143-i. [DOI] [PubMed] [Google Scholar]
- 96.Pande S, Jahn D, Soll D. Histidine tRNA guanylyltransferase from Saccharomyces cerevisiae. I. Purification and physical properties. J Biol Chem. 1991;266:22826–22831. [PubMed] [Google Scholar]
- 97.Jackman JE, Phizicky EM. tRNAHis guanylyltransferase catalyzes a 3′-5′ polymerization reaction that is distinct from G-1 addition. Proc Natl Acad Sci U S A. 2006;103:8640–8645. doi: 10.1073/pnas.0603068103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM. tRNAHis maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev. 2003;17:2889–2901. doi: 10.1101/gad.1148603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Phizicky EM, Alfonzo JD. Do all modifications benefit all tRNAs? FEBS Lett. 2009;584:265–271. doi: 10.1016/j.febslet.2009.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]