Study of the Viral and Microbial Communities Associated With Crohn's Disease: A Metagenomic Approach (original) (raw)
Abstract
OBJECTIVES:
This study aimed to analyze and compare the diversity and structure of the viral and microbial communities in fecal samples from a control group of healthy volunteers and from patients affected by Crohn's disease (CD).
METHODS:
Healthy adult controls (_n=_8) and patients affected by ileocolic CD (_n=_11) were examined for the viral and microbial communities in their feces and, in one additional case, in the intestinal tissue. Using two different approaches, we compared the viral and microbial communities in several ways: by group (patients vs. controls), entity (viruses vs. bacteria), read assembly (unassembled vs. assembled reads), and methodology (our approach vs. an existing pipeline). Differences in the viral and microbial composition, and abundance between the two groups were analyzed to identify taxa that are under- or over-represented.
RESULTS:
A lower diversity but more variability between the CD samples in both virome and microbiome was found, with a clear distinction between groups based on the microbiome. Only ≈5% of the differential viral biomarkers are more represented in the CD group (Synechococcus phage S CBS1 and Retroviridae family viruses), compared with 95% in the control group. Unrelated patterns of bacteria and bacteriophages were observed.
CONCLUSIONS:
Our use of an extensive database is critical to retrieve more viral hits than in previous approaches. Unrelated patterns of bacteria and bacteriophages may be due to uneven representation of certain viruses in databases, among other factors. Further characterization of Retroviridae viruses in the CD group could be of interest, given their links with immunodeficiency and the immune responses. To conclude, some methodological considerations underlying the analysis of the viral community composition and abundance are discussed.
INTRODUCTION
The viral, bacterial, archaeal, and eukaryotic communities harbored in the human gastrointestinal tract greatly outnumber the human body cells.1 In spite of this, the known microbial biodiversity may represent only a small fraction of the actual diversity. These communities are crucial for maintaining homeostasis in such a complex ecosystem.2 In particular, although human viruses are usually pathogens associated with gastroenteritis and other acute disorders, resident intestinal bacteriophages have significant roles in host microbe mortality and genetic diversity in the gut ecosystem by predation on their bacterial hosts.3, 4 In addition, bacteriophages can hinder colonization by potential bacterial pathogens5 but can also eliminate some beneficial probiotic strains,6 or introduce new phenotypic traits, such as antibiotic resistance and the ability to produce exotoxins.7 Despite their relevance in human health, most investigations of the ecological role of viruses have focused on other environments, especially aquatic systems and sediments (see for example refs 8, 9, 10, 11, 12, 13, 14, 15, 16). In contrast, a relatively small number of studies on biological samples,17, 18, 19, 20, 21, 22 and particularly those of human origin (see for example refs 23, 24, 25), has been carried out.
The maintenance and compositional changes of the gut microbiota are known to be closely linked to human physiology, nutrition, and the prevalence of disease. Disruptions to the interactions between the microorganisms and human cells may occur due to genetic and/or environmental factors, thus disrupting homeostasis.26, 27, 28 In several complex diseases of the respiratory tract, such as asthma, or the digestive tract, such as type 1 diabetes and inflammatory bowel disease, interactions between human genotype and viral infections have been linked to autoimmune and inflammatory diseases.29 Crohn's disease (CD) is a major type of inflammatory bowel disease that affects as many as 1 in 500 individuals.30 It is a chronic disorder whose onset takes places mainly during young adulthood, with a secondary increase in older adults. Inflammation may occur in multiple discontinuous regions of the intestine, although it is more frequent in the distal ileum and colon, and may involve transmural inflammation of the intestinal wall.31 Patients typically experience episodic symptoms, including fever, abdominal pain, vomiting, diarrhea and weight loss, and may also suffer more serious gastrointestinal problems. Family and twin studies have demonstrated a strong heritable component to the disease.32, 33
Early studies based on culture methods34, 35, 36 and, more recently, molecular-based approaches37, 38 to screening for particular viruses as possible etiological agents of CD have proven negative or inconclusive. However, based on higher detection levels of bacteriophages in the mucosa of CD patients by microscopy, a role for these in CD has been postulated.39 Other studies also evidence the possible role of viruses in CD. For example, induction of intestinal pathologies in mice by the interaction between a specific virus infection and a mutation in the CD susceptibility gene Atg16L1 has been demonstrated.40 The authors provided an example of how a virus-plus-susceptibility gene interaction can, in combination with additional environmental factors and commensal bacteria, determine the phenotype of hosts carrying common risk alleles for inflammatory disease. More recently, Hubbard and Cadwell41 examined the three-way relationship between viruses, autophagy genes and CD, and discussed how host–pathogen interactions can mediate complex inflammatory disorders. They concluded that although the role of viruses in CD remains speculative, accumulating evidence indicates that this possibility requires serious consideration.
Identifying and measuring the community dynamics of viruses in the environment is complicated because less than 1% of microbial hosts have been cultivated in vitro. Furthermore, as there is no single gene common to all viral genomes, total uncultured viral diversity cannot be monitored using approaches analogous to ribosomal DNA profiling, commonly used for bacteria and archaea. Alternative approaches are therefore required for the evaluation of viral consortia in environmental samples. The development of metagenomic approaches, such as high-throughput sequencing, has allowed the exploration of viral diversity in a new way and is revolutionizing our knowledge of uncultured viral communities in a wider range of environments, including the human gastrointestinal tract. On the other hand, human gut virome studies carried out so far have been focused mainly on fecal samples from healthy adult or infant volunteers42, 43, 44, 45, 46, 47 or from children with various acute disorders.48, 49, 50
In the present study, we used 454 pyrosequencing to analyze the viral and microbial communities in fecal samples from a control group of healthy volunteers and from patients affected by CD, including an additional tissue sample from a surgical biopsy of a Crohn's patient. We have also compared the diversity and structure of some of these viral communities with that of bacterial communities from the same samples, which were determined by partially sequencing the 16S rRNA gene.
METHODS
Sample preparation
All procedures were reviewed and approved by the Hospital Universitari i Politècnic La Fe de Valencia (Spain) Institutional Review Board. Eight healthy adults (used as the control group) and eleven adult patients affected by CD living in Valencia, Spain, were selected for the analysis of viral and microbial diversity. Healthy participants had no known illnesses related to the gastrointestinal tract and had not undergone antibiotic treatment for at least 3 months before the sampling. Some relevant parameters (e.g., disease status, leukocyte and lymphocyte counts, therapy, etc) from the CD participants are described at the Supplementary Table S1 in the Supplementary Materials and Methods. Samples consisted of stool in all cases, except for one ileum piece, which was surgically removed from a CD patient. The fecal samples from the eight healthy control volunteers were labeled V1 to V8; those from the 10 CD affected patients were labeled C1 to C10, while the intestinal sample from a CD patient was identified by IC1. This nomenclature identified samples in both viral and bacterial analyses. The procedures used for the collection of viruses were based on previous studies17, 20 with some modifications. All samples were frozen and stored at −70 °C. An early additional step was carried out for the intestinal tissue piece, consisting of the disruption–homogenization of scalpel-excised pieces of <1 cm3, using the Tissuelyser (Qiagen Iberia SL, Madrid, Spain) at 30 Hz for 5–8 min, buffered in Hanks balanced salt solution at 4 °C. For the remaining samples, ∼10 g of frozen feces from each individual were homogenized by vigorous shaking in a final volume of 50 ml by addition of 1 × Hanks balanced salt solution (Gibco BRL, Gaithersburg, MD, USA) at 4 °C. After a single centrifugation step, at 4,000 × g, 5 min at 4 °C, 1 ml of each supernatant was transferred to 2-ml tubes and stored at −70 °C for later bacterial analysis. The remaining volume underwent three centrifugation steps at 18,000 × g, 5 min at 4 °C to pellet large particles that could clog the barrier filters. The resulting supernatants were serially filtered through 5.0-, 0.8-, 0.45-, and 0.20-μm pore size cellulose acetate filters (Sartorius Stedim Biotech, Goettingen, Germany). The viral particles contained in the resulting filtered liquid were concentrated by the serial addition of NaCl (Merk, Darmstadt, Germany) to a 1 ℳ final concentration, and then PEG-8000 (Sigma-Aldrich, St Louis, MO, USA) to a final concentration of 10% w/v. After 2-h incubation on ice, the tubes were centrifuged at 12,000 × g, 15 min at 4 °C, to pellet the virus–PEG complex. The pellets were resuspended in 500 μl of TMN buffer (10 mℳ Tris pH7.5, 10 mℳ Mg++, 10 mℳ NaCl). To remove unprotected nucleic acids of bacterial or eukaryotic origin, the resuspended pellets were treated with a cocktail of DNAses/RNAses composed of 14 U of Turbo DNAse (Ambion, Austin, TX, USA), 20 U of Benzonase (Novagen, Inc., Madison, WI, USA), and 20 U of RNAse A (Invitrogen, Carlsbad, CA, USA) in DNAse buffer (Ambion), for 120 min at 37 °C.
Viral DNA–RNA extraction and sequencing
Intact nucleic acids contained in the viral capsids were extracted using (A) a standard phenol–chloroform extraction protocol followed by an ethanol–sodium acetate precipitation for viral DNA and (B) the QiAamp viral RNA extraction kit (Qiagen), following the manufacturer's instructions for viral RNA extraction. RNAse inhibitor (Applied Biosystems, Foster City, CA, USA) was added and aliquots were stored at −70 °C for later use. Incubation at 37 °C for 30–60 min, with 2 U of Turbo DNAse (Ambion), was used to remove traces of DNA coextracted with the RNA. The synthesis of first strand of cDNA was carried out using the VersoTM cDNA Synthesis kit (Thermo Scientific, Waltham, MA, USA). Conditions were as follow: incubation at 42 °C, 60 min, and reverse transcriptase inactivation at 94 °C, 3 min. The second strand of the cDNA was then synthesized using Klenow fragment polymerase (New England Biolabs, Beverly, MA, USA). Double-stranded cDNA was incubated for 2 min at 95 °C and chilled on ice for 2 min before addition of 5 U of Klenow fragment and incubation at 37 °C for 1 h. Finally, after enzyme inactivation for 10 min at 75 °C, the reactions were purified by ethanol–sodium acetate precipitation.
To amplify the viral cDNA and genomic DNA for sequencing, a whole genome amplification strategy was carried out using the Illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Amersham, UK), incubating for 2 h at 30 °C followed by phi29 DNA polymerase inactivation for 10 min at 65 °C. The resulting DNA amplification was confirmed by fluorometric measurement using Picogreen®, and 1 μg of DNA per sample was taken. Pools of four to five samples were sequenced simultaneously using the 454 pyrosequencing Genome Sequencer FLX titanium plus on an eighth of a PicoTiterPlate device (Roche Diagnostics, Mannheim, Germany).
16S rRNA gene amplification and barcoded pyrosequencing to determine bacterial diversity
Approximately 1 ml of each of the unfiltered homogenates used for viral sample preparation was set aside for analysis of bacterial diversity and stored at −70 °C. The samples were centrifuged for 5 min at 13,000 r.p.m. at 4 °C and the cellular pellets were selected for total DNA extraction using QIAamp DNA Stool Mini Kit (Qiagen) following the manufacturer's protocol. Highly variable regions, V1, V2, and V3, of the 16S rRNA gene sequences were amplified from the extracted bacterial DNA using universal bacterial forward E8F (5′-AGAGTTTGATCMTGGCTCAG-3′) and reverse B530R (5′-CCGCGGCKGCTGGCAC-3′) primers. The 5′ ends of the forward primers were tagged-barcoded with specific adapters containing 10- or 11-nucleotide Multiplex identifiers selected from those recommended by Roche to enable sample identification, followed by a unique four-nucleotide linker (TCAG). A 40-μl PCR mix was prepared using a PCR Kit (Bioline, London, UK) containing each barcode-forward-primer set. In each reaction 20 μl of Biomix®, 17 μl of H2O, 1 μl DNA, and 1 μl each of forward and reverse primers were present. PCR conditions were 95 °C for 2 min; followed by 25 cycles of 95 °C for 30 s, 55 °C for 1 min, and 72 °C for 1 min; and a final elongation step at 72 °C for 10 min. PCR products were confirmed by gel electrophoresis on a 1.4% agarose gel and purified by ultrafiltration using NucleoFast 96 PCR plates (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions. DNA concentrations were measured using Picogreen® (Invitrogen) and combined in equimolar ratios (200 ng from each sample). Finally, the pooled samples were sequenced with a 454 pyrosequencing Genome Sequencer FLX titanium on an eighth of a PicoTiterPlate device.
Bioinformatic analyses
Processing
To analyze the viral metagenomes, raw sequence reads from each sample were filtered for low-quality signal or ambiguous characters (<30 out of 40 quality units assigned by the 454) with Genome Sequencer FLX System Software Package 2.3 (Roche). Reads were assigned to their corresponding samples according to their barcode-tagged primer sequences, and primers, linkers, and adaptors were trimmed from the sequences. Exact duplicates generated by 454-based pyrosequencing were excluded using the online tool 454 replicate filter (http://microbiomes.msu.edu/replicates). Sequences shorter than 50 bp were removed using MOTHUR v.1.22.2.51 Removal of sequences of human origin was performed following a MegaBLAST search against the human genome database from the NCBI released 16/08/2011. Low-complexity filters were applied to remove highly repetitive sequences. Next, filtered reads were compared with a custom made nonredundant viral and prophage nucleotide database (Virusdb_24_04_2012), a collection composed of the complete viral genomes from the NCBI (http://www.ncbi.nlm.nih.gov/, from NCBI Refseq release 52 and Genbank release 188, March 2012), the viral genomes from the EBI (European Bioinformatics Institute, http://www.ebi.ac.uk/, release 111, March 2012), the complete and partial viral sequences from the database DDBJ (DNA Data Bank of Japan, http://www.ddbj.nig.ac.jp/, release 88.0, December 2011), and viruses and prophages from ACLAME version 0.4 (A CLAssification of Mobile genetic Elements, http://aclame.ulb.ac.be/, update 2010,52). A first MegaBLAST search was used to retrieve only those viruses that matched with expected _E_ values of <10−5. Only those sequence reads that had identities above 95% and aligned in at least the 60% of their total length were selected as viral hits at this step and retained. The remaining sequence reads were compared with a nonredundant bacterial nucleotide database composed of the bacterial reference genomes and draft genomes deposited at the NCBI site (ftp://ftp.ncbi.nih.gov/genomes/, NCBI Refseq release 51, January 2012 and Genbank release 187, December 2011). BLASTn searches with expected _E_ values of <10−5, identities of >70%, and alignment length of >60% of the query reads were assigned as candidate sequences of bacterial origin and removed from the analysis. The remaining sequences, which did not show significant matches with bacteria, were added to the viral hits previously retained (see above). The resulting bacterial-free sets of reads were further processed in parallel by two different approaches. In one of them, the reads remained unassembled for downstream analyses. In the other, they were assembled de novo into contigs using 454 Newbler Assembler53 with a criterion of at least 50-bp length overlap and minimum overlap identity of 95%.
Bacterial raw sequence reads were filtered by quality and size using MOTHUR v.1.22.2,51 discarding sequences shorter than 200 bp, and using a procedure similar to that used with the viral metagenomic sequences but with an additional step of chimera removal. Sequence reads were assigned to their original sample using the barcode-tagged primer sequences.
Viral taxonomic identification and nomenclature
Two different approaches were used with both assembled and non-assembled reads to carry out the taxonomic assignment. In the first approach, processed sequences were uploaded to the MetaVir web server, which is dedicated to the analysis of viral metagenomes (http://metavir-meb.univ-bpclermont.fr).54 This server computes the taxonomic composition via the GAAS tool55 with complete viral genomes-encoded proteins from NCBI RefSeq (release Sept. 2011) using BLASTx and four different thresholds (E value <10−3, 10−5, 10−7, and 50 on Score). In a second approach, the same set of reads was compared against Virusdb_24_04_2012 using tBLASTx with expected E values <10−3, and a set of Perl programs was used to automate the process of taxonomic binning at class, order, family, genus, and species level. Therefore, four sets of data are generated, two from MetaVir (one with unassembled and another with assembled reads) and two from our tBLASTx approach (unassembled and assembled).
The nomenclature we used for viruses and prophages was initially generated using the “Fetch taxonomic representation” tool implemented on the Galaxy platform.56 Next, we used in-house Perl scripts to convert the Galaxy output into a standardized abundance table containing four of the taxonomic levels accepted for viruses (order, family, genus, and species) by inheriting the higher or, if not possible, the lower adjacent taxonomic-level tag to fill in the missing taxonomic levels of each bin. This way a nonredundant taxonomy for all entries was generated. In addition, prophages adopted their bacterial–host taxonomy with addition of the tag “phage”.
Viral structure and diversity estimation
MetaVir automatically generates composition and abundance tables and charts using its own taxonomic identification tools. In contrast, the resulting output files from our tBLASTx-based method used for taxonomic identification (see above) were converted into operational taxonomic units (OTUs) tables by customized Perl scripts and can thus be used for the analysis of the diversity and structure of the viral communities. The alpha (biodiversity within samples) and beta (diversity comparison between samples) diversity values were estimated using tools implemented in the QIIME pipeline version 1.3.0.57 For the alpha diversity, rarefaction curves were generated with observations from randomized OTU draws (no replacement) using 200 iterations for an increasing number of sampled sequences over 20 steps. The Observed species, Chao1 estimator and Shannon diversity index were calculated. The beta diversity was assessed by sample clustering with 10,000 iteration jackknife support. Both analyses were based on Bray–Curtis, Canberra, and Manhattan dissimilarity/distance matrices. The sample clustering was generated in R version 2.12.58
Finally, to identify a taxonomic biomarker with high stringency, we employed the linear discriminant analysis effect size (LEfSe) method,59 combining the Kruskal–Wallis and pairwise Wilcoxon tests for statistical significance with linear discriminate analysis for feature selection, to confirm the differential abundance of viral OTUs. We used default significance (alpha value=0.05) and linear discriminant analysis thresholds (2.0), at all taxonomic levels between the control group and CD patients.
Bacterial identification, taxonomic analysis and diversity estimation using 16S rRNA gene sequences
The QIIME pipeline 1.3.0 (ref. 57) was used to determine the diversity and structure of bacterial communities in the samples. Sequence clustering into OTUs was carried out with UCLUST 1.2.22q using a sequence similarity of 97% as a cut-off for phylotypes. For taxonomic classification, RDP Classifier 2.2 (ref. 60) was used with the GreenGenes database release 4_02_2011 at 97% identity. Sequences were aligned to the database entries using PyNAST 1.1, and an approximately maximum likelihood tree was constructed with FastTree 2.1.3. Next, alpha and beta diversity, sample clustering and linear discriminate analysis were estimated in the same way as for viruses (see above).
Functional analysis of the viromes
Assembled reads with homology to viral hits in the taxonomic analyses were submitted to the RAMMCAP workflow against the PFAM, TIGRFAM, and COGs databases, which is implemented in CAMERA,61 with an expected E value of <10−3 to assign a functional annotation to the reads. The lowest E value hits were selected. A hierarchical classification was used, with HMM annotation against TIGRFAM as the highest ranked supporting evidence for functional assignments, followed by HMM matches against PFAM, which was only used when no TIGRFAM match was found.
Virome and microbiome accession numbers
The viromes and bacterial 16S rDNA data sets from this survey are accessible in the EBI Short Read Archive under the study accession number ERP001706, available at the URL http://www.ebi.ac.uk/ena/data/view/ERP001706, with the following sample accession numbers, ERS161373, ERS161375, ERS161377, ERS161379, ERS161381, ERS161391, ERS161389, ERS161387, ERS161385, ERS161383, ERS161393, ERS161395, ERS161397, ERS161399, ERS161401, ERS161403, ERS161405, ERS161407, and ERS161409 for viruses and ERS161374, ERS161376, ERS161378, ERS161380, ERS161382, ERS161384, ERS161386, ERS161388, ERS161390, ERS161392, ERS161394, ERS161396, ERS161398, ERS161400, ERS161402, ERS161404, ERS161406, ERS161408, and ERS161410 for bacterial 16S rRNA genes.
RESULTS
Overview of the viral samples composition
Table 1 summarizes the sequencing depth of the total of 602,015 raw reads generated by 454 pyrosequencing before and after processing and assembling. After the preprocessing used to remove exact duplicates, short-reads, low-complexity, and human and bacterial contamination, 477,094 reads remained as putative targets to contain viral hits. Fewer than 15% of the reads showed homology to viral hits in our custom tBLASTx analyses (12.01% and 14.34% in non-assembled and assembled reads, respectively) and even lower percentages with the MetaVir approach (11.16% and 11.52%, respectively), with a majority of the remaining reads unassigned.
Table 1. Summary of the number of reads generated by 454 pyrosequencing before and after processing and assembling.
| Sample | Raw reads | Reads after removal of: exact duplicates, short-reads, low-complexity, human, and bacterial contamination | Reads after assembly (Newbler) | Viral reads tBASTX (unassembled) E value <10−3 , cut-off 0.8 | Viral reads tBASTX (assembled) E value <10−3 , cut-off 0.8 | Viral reads MetaVir (unassembled) E value <10−3 | Viral reads MetaVir (assembled) E value <10−3 |
|---|---|---|---|---|---|---|---|
| C1 | 3,196 | 901 | 605 | 272 | 129 | 195 | 98 |
| C2 | 53,420 | 40,387 | 3,088 | 3,606 | 480 | 2,641 | 374 |
| C3 | 43,543 | 33,153 | 2,060 | 3,459 | 111 | 3,411 | 78 |
| C4 | 67,822 | 50,393 | 2,758 | 4,392 | 454 | 4,087 | 355 |
| C5 | 52,747 | 44,501 | 3,241 | 5,746 | 316 | 6,444 | 277 |
| C6 | 54,838 | 49,263 | 1,813 | 5,198 | 147 | 2,769 | 101 |
| C7 | 39,990 | 23,228 | 890 | 3,343 | 111 | 1,717 | 55 |
| C8 | 62,063 | 53,874 | 2,467 | 6,214 | 323 | 6,896 | 286 |
| C9 | 26,145 | 23,325 | 870 | 1,576 | 113 | 1,899 | 116 |
| C10 | 29,035 | 19,414 | 1,540 | 2,173 | 96 | 1,266 | 46 |
| IC1 | 12,820 | 10,170 | 715 | 1,129 | 31 | 1,143 | 25 |
| V1 | 12,480 | 10,875 | 2,759 | 3,555 | 666 | 3,518 | 555 |
| V2 | 28,292 | 13,435 | 1,801 | 4,360 | 533 | 4,048 | 426 |
| V3 | 16,877 | 14,478 | 2,138 | 3,092 | 603 | 3,372 | 518 |
| V4 | 32,269 | 30,928 | 2,200 | 797 | 153 | 1,311 | 164 |
| V5 | 18,556 | 17,619 | 1,400 | 1,136 | 63 | 1,402 | 67 |
| V6 | 18,620 | 14,179 | 6,474 | 2,225 | 995 | 2,066 | 722 |
| V7 | 12,707 | 11,693 | 1,199 | 4,146 | 193 | 4,315 | 157 |
| V8 | 16,595 | 15,278 | 2,332 | 850 | 270 | 773 | 230 |
| Total | 602,015 | 477,094 | 40,350 | 57,269 | 5,787 | 53,224 | 4,648 |
The composition of viral OTUs at the species level (see Table 2) reveals that, regardless of whether the reads are assembled or not, our own approach retrieves more viral hits for the same threshold (10−3 on E value) than the MetaVir approach in all cases except for four samples (C10 unassembled, C9 assembled and V4 in both data sets). In fact, our approach retrieves, on average, 24.5 and 17.2 more viral taxa on non-assembled and assembled reads, respectively. The overall number of different viral OTUs we obtained with our approach is 958 different species, belonging to 379 genera, from 246 families, when non-assembled reads are considered. This figure is reduced to 766, 322, and 218 species, genera, and families, respectively, when assembled reads are considered instead.
Table 2. Comparison of the number of different viral taxa per sample at the species level between MetaVir and our tBLASTX approach, using unassembled and assembled reads.
| Sample | Number of viral species | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MetVir | tBLASTX | |||||||||
| Unassembled | Assembled | Unassembled | Assembled | |||||||
| 10−3 | 10−5 | 10−7 | 50 on Score | 10−3 | 10−5 | 10−7 | 50 on Score | 10−3 | 10−3 | |
| C1 | 73 | 57 | 43 | 46 | 66 | 53 | 41 | 43 | 77 | 73 |
| C2 | 222 | 178 | 151 | 156 | 203 | 168 | 141 | 148 | 272 | 240 |
| C3 | 66 | 43 | 33 | 31 | 43 | 33 | 25 | 26 | 75 | 57 |
| C4 | 159 | 117 | 92 | 104 | 108 | 87 | 70 | 80 | 190 | 149 |
| C5 | 237 | 165 | 135 | 145 | 140 | 99 | 81 | 87 | 245 | 159 |
| C6 | 156 | 85 | 61 | 69 | 63 | 39 | 29 | 35 | 203 | 76 |
| C7 | 94 | 68 | 53 | 57 | 41 | 33 | 23 | 27 | 115 | 59 |
| C8 | 247 | 166 | 141 | 146 | 131 | 99 | 77 | 86 | 298 | 141 |
| C9 | 110 | 75 | 56 | 63 | 56 | 40 | 30 | 34 | 115 | 52 |
| C10 | 68 | 45 | 34 | 37 | 37 | 26 | 21 | 22 | 61 | 44 |
| IC1 | 31 | 19 | 8 | 11 | 18 | 9 | 6 | 8 | 39 | 19 |
| V1 | 313 | 236 | 195 | 213 | 248 | 188 | 153 | 168 | 363 | 284 |
| V2 | 353 | 285 | 244 | 263 | 209 | 167 | 146 | 155 | 376 | 217 |
| V3 | 315 | 255 | 222 | 237 | 233 | 186 | 163 | 176 | 371 | 270 |
| V4 | 146 | 101 | 85 | 95 | 101 | 80 | 68 | 75 | 145 | 89 |
| V5 | 34 | 19 | 17 | 18 | 18 | 14 | 13 | 14 | 34 | 20 |
| V6 | 297 | 218 | 182 | 190 | 256 | 194 | 159 | 169 | 380 | 316 |
| V7 | 61 | 41 | 31 | 34 | 45 | 38 | 30 | 32 | 70 | 59 |
| V8 | 118 | 81 | 67 | 68 | 94 | 73 | 60 | 62 | 137 | 113 |
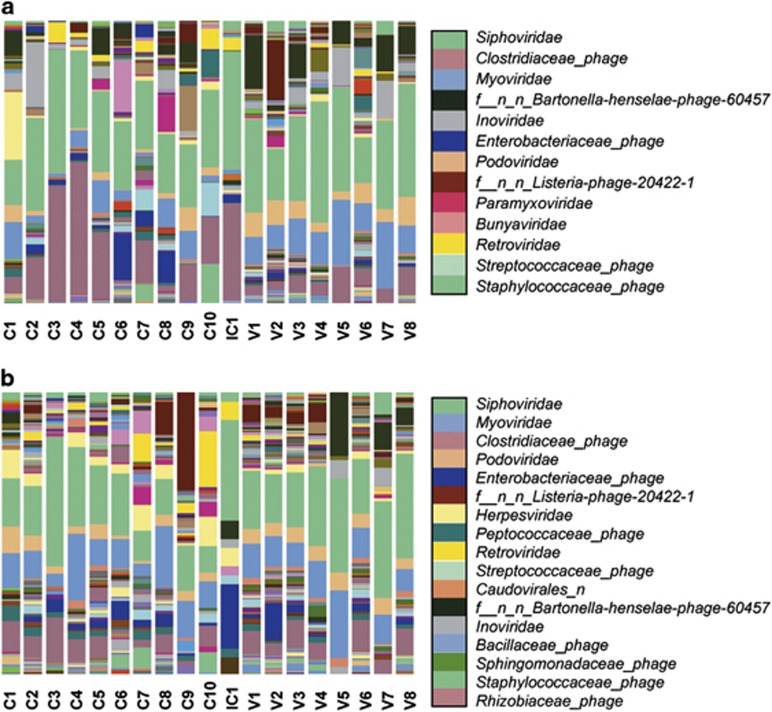
The viral taxonomic composition and abundance at the family level of the 19 samples studied are summarized in the Figure 1. Although the percentages and rank positions vary between non-assembled and assembled reads and among samples, some specific viral families remain among the most abundant. That is the case for the bacteriophages of the order Caudovirales (Siphoviridae, Myoviridae, Podoviridae, and others), as well as other bacteriophages such as the Inoviridae and certain unclassified phages, as well as certain prophages (e.g., those from Clostridiaceae or Enterobacteriaceae). Finally, viruses infecting eukaryotes had a significant presence, including Retroviridae and, more restricted to non-assembled reads, Paramyxoviridae and Bunyaviridae, or Herpesviridae in assembled contigs.
Figure 1.
Taxonomic classification and relative abundance of the viral communities from the fecal CD samples (labeled as C1 to C10), fecal healthy control volunteer's samples (labeled as V1 to V8), and intestinal CD sample (labeled as IC1). The results are presented at the family level for (a) non-assembled and (b) assembled reads. Only those families with a presence ⩾1.0% in the global count are displayed in the legend, ranged by decreasing abundance.
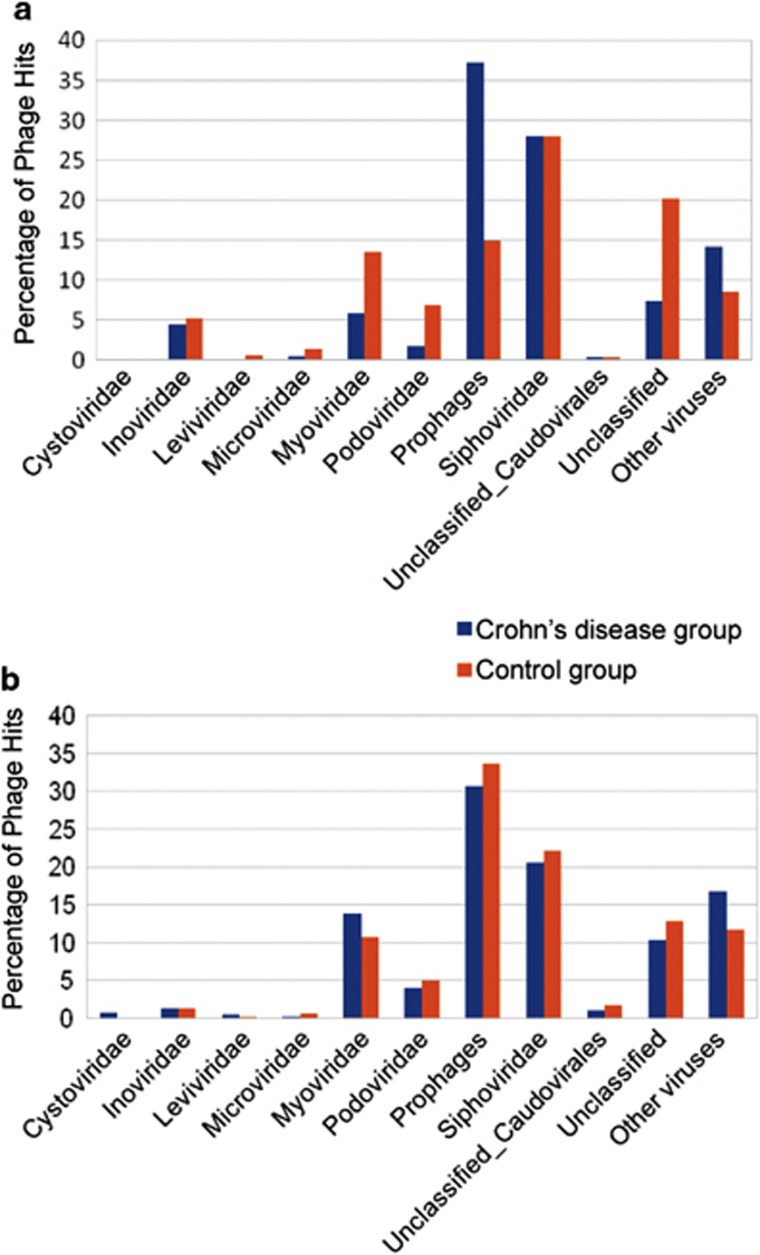
The distribution and comparison of the different bacteriophage families between the CD and control groups in both non-assembled and assembled reads are shown in Figure 2. In all cases, the families Siphoviridae, Myoviridae, and Podoviridae account for most of the viral hits. In addition there is a notable presence of bacteriophages from the family Inoviridae. Most of the observed differences between CD and control groups before read assembly, such as in prophages, unclassified phages and the families Myoviridae, Podoviridae, or Inoviridae diminish afterwards. Assembly therefore leads to reduced differences between groups, and even reverts the ratios in some cases, such as in the prophages or the Myoviridae.
Figure 2.
Comparative distribution of viruses. Comparison of the relative distribution of prophages, bacteriophage families, and other viruses between the Crohn's disease group and the control group, in non-assembled (a) and assembled reads (b).
Differences in viral taxonomic composition and abundance between CD and control groups
In non-assembled reads, the LEfSe method identifies 125 viral OTUs that show differential abundance (P<0.05) between CD and control samples at any taxonomic level (see Supplementary Table S2A in the Supplementary Materials and Methods). Of these, 120 are overrepresented in control samples whereas only 5 are overrepresented in CD samples. The latter include viruses globally classified as prophages, two unclassified prophages from Clostridiales and Alteromonadales, and Clostridium_acetobutylicum_phage, as well as one classified as virus: Synechococcus phage S CBS1.
In the assembled reads, the LEfSe method shows 57 differentially abundant viral OTUs (P<0.05) between CD and control samples at any taxonomic level (see Supplementary Table S2B in the Supplementary Materials and Methods). Of these, 54 are overrepresented in control samples whereas only 3 are overrepresented in CD samples. The latter consist of two taxa: the family (and its unnamed order) Retroviridae and the species Synechococcus phage S CBS1. In contrast, the 54 OTUs whose relative abundance is significantly higher in the control samples include 43 different viruses and 11 prophage-like viruses.
Overview of the bacterial samples composition
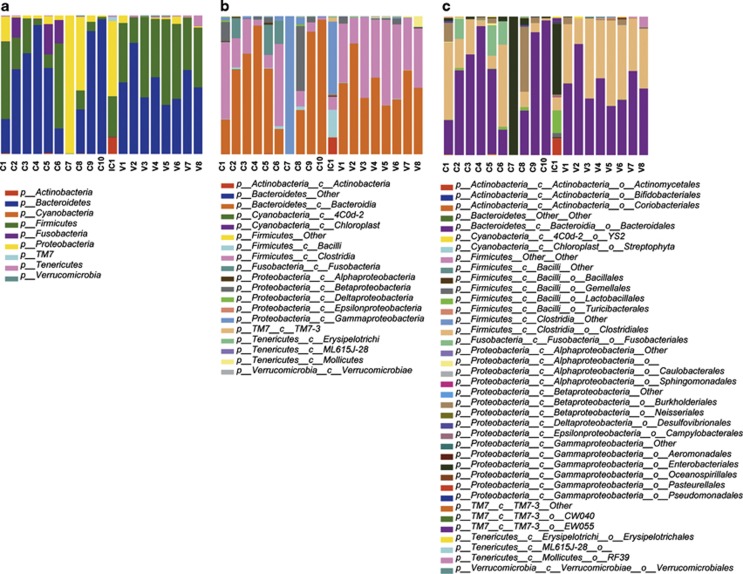
16S rDNA sampling depth is summarized in Table 3. The global number of distinct bacterial OTUs generated in our analyses totals 252, with the composition and abundance shown in Figure 3. The taxonomic assignment reveals the presence of 252 species, 147 genera, 72 families, 38 orders, 19 classes, and 9 phyla. The Bacteroidetes and Firmicutes account for 54.1%and 32.5% of the OTUs, respectively, followed by the Proteobacteria (9.3%), Fusobacteria (2.6%), lower numbers of Tenericutes (0.8%), Actinobacteria (0.4%), Verrucomicrobia,TM7 (0.1%), and Cyanobacteria (<0.1%).
Table 3. Sampling depth found by amplicon 454 pyrosequencing V1V2 and part of V3 regions.
| Sample | Raw reads | Reads>200 bp | Chimera-free reads | Different OTUs |
|---|---|---|---|---|
| C1 | 9,085 | 5,644 | 5,587 | 4,896 |
| C2 | 7,803 | 6,158 | 5,431 | 4,530 |
| C3 | 8,048 | 6,693 | 6,609 | 5,664 |
| C4 | 8,241 | 6,921 | 6,338 | 5,057 |
| C5 | 5,616 | 4,559 | 4,378 | 3,696 |
| C6 | 9,970 | 8,174 | 8,091 | 6,679 |
| C7 | 5,232 | 3,726 | 3,611 | 2,870 |
| C8 | 6,968 | 5,826 | 5,728 | 4,873 |
| C9 | 10,863 | 8,889 | 8,267 | 6,867 |
| C10 | 9,278 | 7,503 | 7,352 | 6,045 |
| IC1 | 13,427 | 5,737 | 5,680 | 2,276 |
| V1 | 10,117 | 8,449 | 7,416 | 6,497 |
| V2 | 8,832 | 7,331 | 7,056 | 6,125 |
| V3 | 5,502 | 4,038 | 3,839 | 3,339 |
| V4 | 10,125 | 8,414 | 7,892 | 6,889 |
| V5 | 8,205 | 6,910 | 5,804 | 5,174 |
| V6 | 12,110 | 9,958 | 9,387 | 8,108 |
| V7 | 9,031 | 7,496 | 7,017 | 6,097 |
| V8 | 9,935 | 8,150 | 7,070 | 6,130 |
Figure 3.
Taxonomic classification and relative abundance of the microbial communities from the fecal CD samples (labeled as C1 to C10), fecal healthy control volunteer's samples (labeled as V1 to V8), and intestinal CD sample (labeled as IC1). Three taxonomic levels are shown: (a) phylum, (b) class, and (c) order.
Differences in bacterial taxonomic composition and abundance between CD and control groups, and between fecal and tissue samples
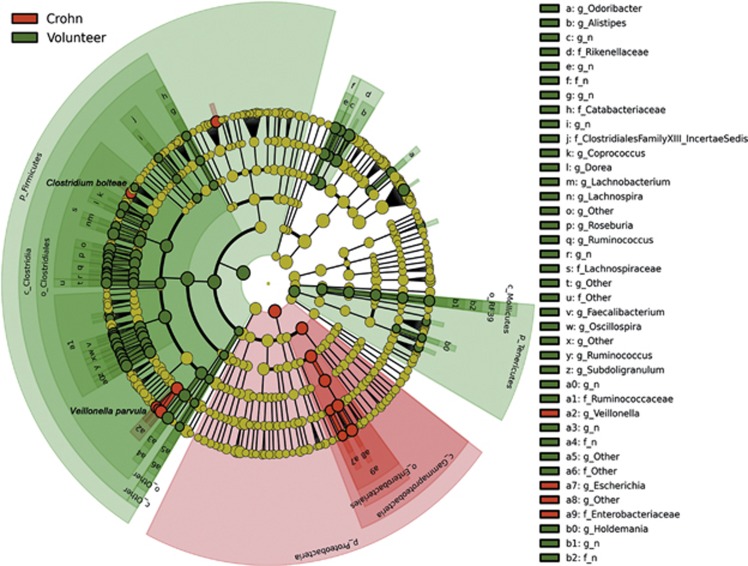
The LEfSe method identifies 97 bacterial OTUs that show differential abundance between CD and control samples at any taxonomic level. Of these, 84 are overrepresented in control samples whereas only 13 are overrepresented in CD samples. The cladogram (see Figure 4) shows a higher abundance of some members of the phylum Proteobacteria in the CD group (eight OTUs), particularly within the family Enterobacteriaceae, such as Escherichia coli, as well as two groups of Clostridiales: the genus Veillonella (four OTUs) and Clostridium bolteae. This result contrasts with the relative impoverishment in CD samples of the phylum Firmicutes (61 OTUs), particularly of the Clostridia class (59 OTUs), as well as the phylum Tenericutes (10 OTUs), the order Bacteroidales (12 OTUs) and one species within Actinobacteria (Collinsella aerofaciens). The tissue sample IC1 has Veillonella parvula followed by the family Enterobacteriaceae as the most significant taxa. In the case of V. parvula, the highest relative abundance among all samples is detected in the intestinal tissue, whereas the genus Escherichia and other Enterobacteriaceae are only exceeded by one other CD sample (C7), which after two independent amplifications exhibits an extreme bias toward the family Enterobacteriaceae (with a 99.6% of the reads), thus differing from the rest of the samples in our study. Owing to the discrepancy shown by sample C7 in terms of phylogenetic composition, which could be the result of an unexpected analytical problem such as prolonged storage in room temperature and aerobic conditions or a bacterial contamination, this extreme outlier was discarded for subsequent analyses.
Figure 4.
Cladogram representing the features that are discriminative with respect to the classes “Crohn” and “Volunteer” using the linear discriminant analysis model results on the bacterial hierarchy.
When looking at higher taxonomic levels (e.g., Order) in the classification of the metagenomic sequences of bacteria (Figure 3), other OTUs specifically abundant in IC1 are the Actinomycetales, with 11.4% of the reads, compared with 0.0–0.1% in the remaining samples; the Lactobacillales, Bacillales, and Gamellales, which represent almost 20.0% compared with 0.0–2.4% of the other samples; the Enterobacteriales with 51.1% compared with 0.0–4.9% (with the previously mentioned exception of sample C7), or two orders within the TM7-3 class: EW055 with 2.6% compared with 0.0–0.1%, and CW040 with 0.4% compared with <0.1% in the remaining samples.
Viral and microbial diversity within and between samples
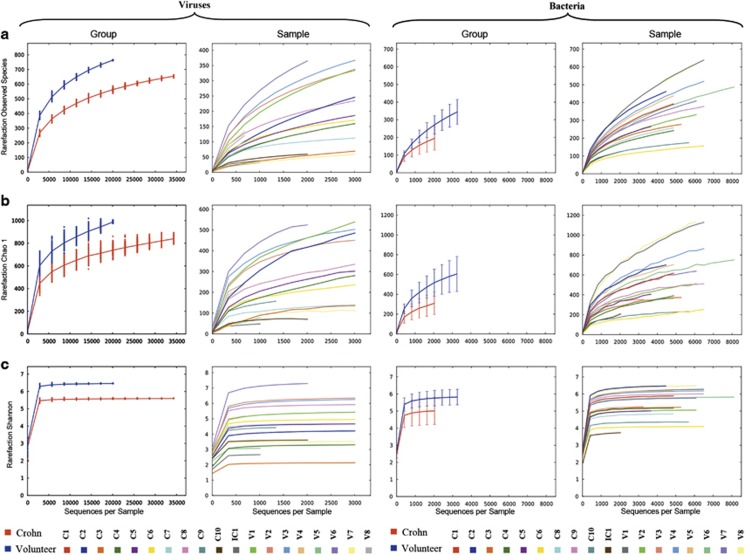
The viral diversity within samples was measured with a variety of alpha diversity metrics: the observed species, the Chao1 estimator, and Shannon's diversity index (Figure 5). Owing to the heterogeneous number of reads among samples, direct comparisons are more difficult and thus the evidence is weaker. In spite of this, generally speaking higher values are obtained for control samples (Shannon index >5.0), with the exceptions of V5 and V7 (around 3–3.5), than in CD samples with only one sample (C8) above this Shannon's index value, and several of them do not even reach the threshold of 4.0. The intestinal sample (IC1) exhibits one of the lowest diversity indices (with Shannon index <2.7).
Figure 5.
Diversity within samples based on the abundance of viral (left) and microbial (right) operational taxonomic units within the community, plotted by rarefaction curves. Three different diversity metrics are used: (a) Observed number of species, (b) Chao1 estimator, and (c) Shannon diversity index. Viral CD groups and individual samples are plotted on the first and second columns, respectively. Microbial CD and control groups and individual samples are plotted on the third and fourth columns, respectively.
To assess the microbial diversity within each of the samples, the alpha-diversity metrics observed species, Chao1 estimator, Shannon's diversity index, and Simpson's diversity index were calculated. The first three are displayed in Figure 5. The CD samples are less diverse than the control samples when considered as a group, but they also show a much higher heterogeneity when compared with the more homogeneous control samples, as they encompass some of the samples with the highest diversity indices (such as C1 or C2) but also the ones with the lowest values (C6, C8, and C10). Low diversity is also identified for the tissue sample (IC1), which exhibits the lowest values for Shannon and Simpson diversity indices.
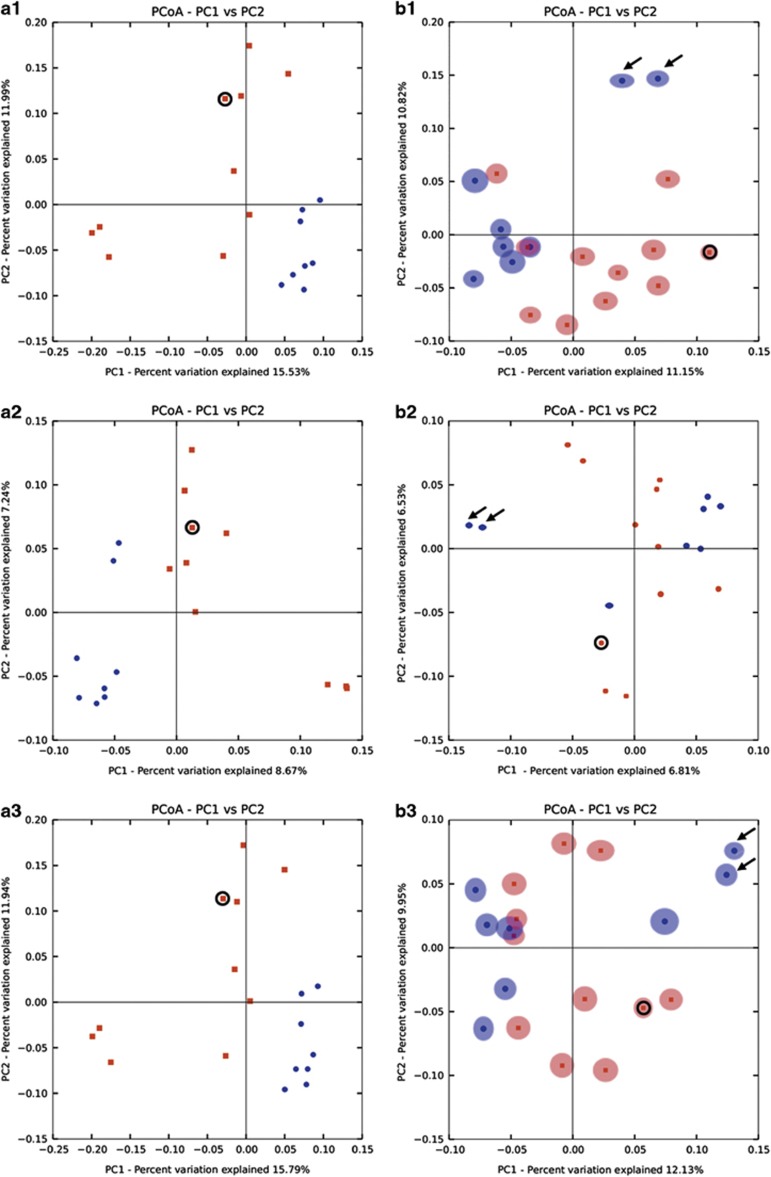
The diversity among samples was assessed with a Principal Coordinate Analysis of the microbiota and viral communities, which is represented in the two-dimensional plot in the Figure 6. For the microbiota, the plots show that the control group samples are less scattered than the CD ones, and the control samples are grouped separately from the CD samples, regardless of the dissimilarity/distance matrix used, suggesting the existence of two distinct clusters. This pattern is not as sharp for the viral communities because, although there appear to be two clusters with the CD group more scattered than the control group, the presence of exceptions involving samples from both groups blurs this distinction. For example, samples V5 and V7 (marked with arrows), which exhibit a much lower diversity than other control samples, appear to be more closely related to each other and separate from the remaining control samples.
Figure 6.
Diversity between samples. Jackknifed replicate Principal Coordinate Analysis (PCoA) two-dimensional plots obtained for the (a) microbiota, and for the (b) assembled reads virome using from the top to the bottom, the Bray–Curtis (a1 and b1), Canberra (a2 and b2), and Manhattan (a3 and b3) distance matrices. Only comparisons of P1 vs. P2 axes are shown. Red dots represent the samples from the CD group; blue dots represent the samples from the control group. The circle indicates the intestinal tissue sample. Black arrows indicate samples V5 and V7.
The intestinal sample microbiota, IC1 (circled), appears quite divergent from the other samples suggesting a distinct distribution, which reinforces the results from composition and abundance analyses (see above, Figure 3). The same plots for viruses are less conclusive.
All in all, the results suggest the existence of greater variation in the beta diversity in the case of the CD samples in both microbial and viral communities but lower diversity within samples, whereas in the control group there is more diversity within samples in general, but also more homogeneity between sample.
Comparison of sample clustering based on bacterial and viral composition and abundance
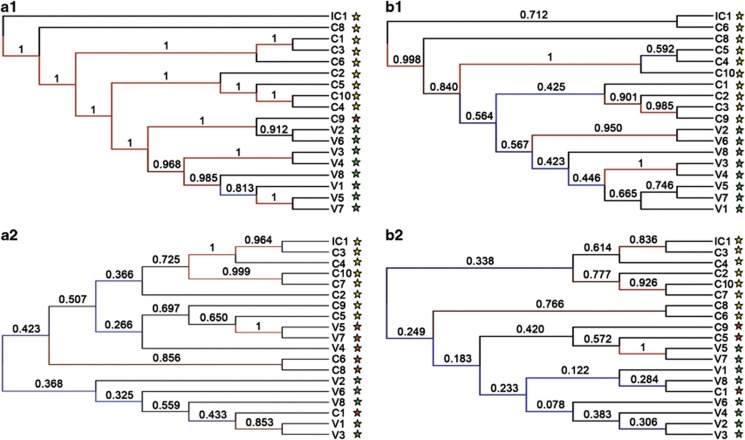
The cluster trees obtained with statistical jackknifing support for bacteria and viruses (see Figure 7) show differences in the way samples are clustered. Bacterial-based clustering shows a high statistical support (above 0.9) for most of the nodes. Two groups of samples (CD and control) are identified with one exception: sample C9 clusters within the control group when the Bray–Curtis matrix is used, but with the Canberra distance matrix the two groups of samples are separate.
Figure 7.
Cluster tree with jackknifing support for bacteria (a1) and for viruses (a2) obtained with Bray–Curtis dissimilarity matrix on the left, and with Canberra distance matrix (b1) and (b2) on the right. Jackknifing support values of the nodes are represented as well as the color scale for them. Yellow stars represent CD samples; green stars, control samples; and red stars, samples that are grouped within the other group. Ten thousand replicates were carried out.
Virus-based clustering results display lower values of statistical support for most nodes, due in part to the lower number of viral hits. The cluster tree shows that relationships among samples vary greatly depending on the assembly, the distance matrix and taxonomic level used, but overall the reconstructions differ from the bacterial-based clustering, displaying much lower resolution. Therefore, the CD and control samples can only partially be separated into two groups based on viral composition and abundance, and with low statistical support.
Comparison of the bacterial composition and abundance based on the 16S rDNA sequences and the bacterial hosts inferred from the bacteriophages
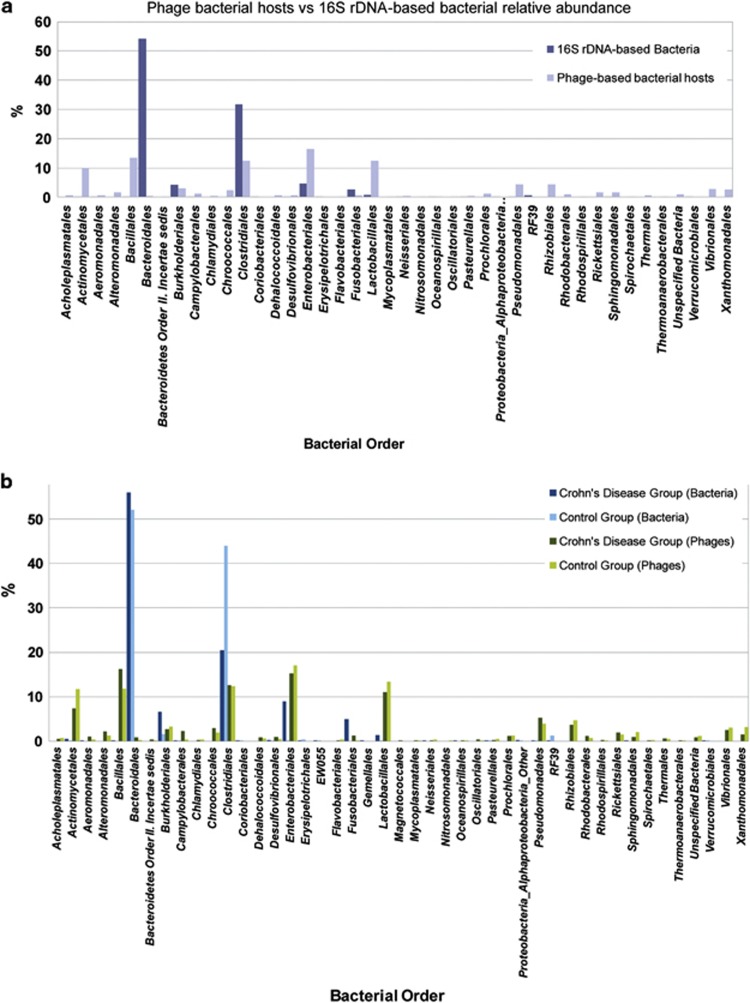
The comparison of the bacterial composition and abundance at the order level (see Figure 8a) identified two unrelated patterns of bacterial taxonomic distribution. One is derived from the direct sequencing of the 16S rDNA and reflects the predominance of Bacteroidales (54%) and Clostridiales (32%), and to a lesser extent of Enterobacteriales (4.7%), Burkholderiales (4.23%), and Fusobacteriales (2.57%), with a much lower presence of other orders. In the second, the list of bacterial orders is derived from the bacterial hosts of the bacteriophages present in the samples and differs considerably from that based on the 16S rDNA, as it reflects the presence of a greater range of potential bacterial hosts, with more bacterial orders represented. The most common orders (Enterobacteriales, Bacillales, Clostridiales, Lactobacillales, and Actinomycetales) are only marginally dominant (all of them within a range from 10 to 16.5%).
Figure 8.
16S rDNA and bacteriophage-based bacterial composition and abundance. Comparison of the composition and relative abundance of bacterial orders based on the 16S rDNA sequences and on the bacterial hosts of the bacteriophages, in the (a) overall data set and (b) with the distinction between the Crohn's disease and the control group. Only bacterial orders represented over 0.1% in at least one group are displayed.
When looking deeper into this taxonomic bacterial distribution by groups (see Figure 8b), some notable differences between the CD and control groups are observed in both cases. For example, bacterial composition based on the 16S rDNA reveals a difference in the percentage of Enterobacteriales (+8.78%), Burkholderiales (+4.97%), Fusobacteriales (+4.90%), Bacteroidales (+3.85%), and Lactobacillales (+1.25%) in favor of the CD group, which shows a significant decrease mainly of Clostridiales (−23.47%) and RF39 (−1.20%) compared with the control group. However, when comparing the ratios, orders that are overall represented in low percentages may show greater differences between groups than orders that account for the majority of the bacteria. For example, some orders only appear in the control group, such as Aeromonadales, Bacillales, Fusobacteriales, Gemellales, Neisseriales, Oceanospirillales, and Sphingomonadales, or are significantly more represented in that group, such as Actinomycetales (246.98 fold), or Enterobacteriales (99.75 fold), compared with only a 1.07-fold increase of Bacteroidales in this group. No bacterial order is found exclusively in the CD group, but five orders are better represented in this group: Erysipelotrichales (1.97 fold), Clostridiales (2.15 fold), Verrucomicrobiales (3.63 fold), Pasteurellales (7.55 fold), and especially RF39 (80.97 fold).
The comparison between groups based on the phage-hosting bacteria shows an uneven distribution when compared with their 16S rDNA data set counterparts. For example, bacteriophages whose bacterial hosts belong to the orders Bacteriodetes, Bacillales, Desulfovibrionales, Fusobacteriales, or Pseudomonadales are more abundant and Neisseriales, Oceanospirillales, or Pasteurellales less abundant in the CD group in both data sets. However, for other orders, comparing the two groups gives the opposite result between bacteria and bacterial hosts of the phages, for example, in the Actinomycetales, Burkholderiales, Clostridiales, Enterobacteriales, or Lactobacillales.
Functional analysis of the viromes
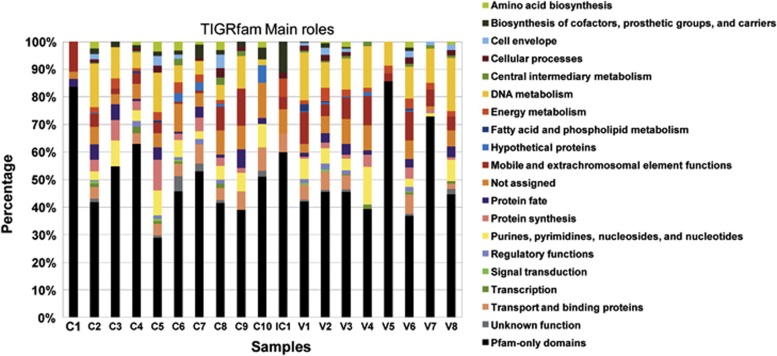
Figure 9 displays the TIGRfam functions found per sample grouped into higher functional categories, known as main roles. Contigs showing no homology with TIGRfam database (E value <10−3) but that were positive when compared with Pfam entries are listed in Supplementary Table S3. The latter comprise around half of the functional hits.
Figure 9.
Functional annotation of the assembled viral metagenomic sequences of each sample, grouped by TIGRfam main roles. An additional category: “Pfam-only domains”, which includes those reads without homology to the TIGRfam functional database but with hits against the Pfam database, is included. For more details on this category see Supplementary Table S3 in the Supplementary Materials and Methods.
Regarding the TIGRfam roles, those with greater prevalence among the samples, excluding the “not assigned” category, are proteins related to DNA metabolism (9.6%), followed by mobile and extrachromosomal element functions (5.9%), purines, pyrimidines, nucleosides, and nucleotides (5.1%), and transport and binding proteins (4.0%), whereas the less represented roles are signal transduction (0.2%), and fatty acid and phospholipid metabolism (0.3%). The two main roles, DNA metabolism and the mobile and extrachromosomal element functions, are typically associated with viral replication and structure, and are especially common in the control group (13.1% and 7.3%, respectively) compared with the CD group (7.1% and 4.9%, respectively).
DISCUSSION
Our report is the first metagenomic study to investigate the viral communities associated with a multifactorial chronic intestinal disease, namely CD. It also takes into account the microbial community composition, abundance, and diversity so that a comparison of the two communities can be established between the two sample groups under study.
Our analysis includes a study of the microbiota, already the subject of numerous investigations (reviewed for example by refs 2, 62, 63). These have previously reported the dysbiosis associated to the CD, including some of the features that we have also found, such a decrease in clostridia concentration (although not accompanied by a decrease in Bacteroidetes), as well as the relative abundance of members of the Enterobacteriaceae. However, unlike previous studies on inflammatory bowel disease, our efforts have been focused on the viral community associated with one particular form of this disease (CD), using a metagenomic approach combined with the massive sequencing tools.
We were able to retrieve more viral hits than previous approaches because of the extensive database we used, which comprises a comprehensive collection of four extant databases. This is despite the exclusion of environmental samples, unless they had been taxonomically assigned and appear in one of the databases used.
Similarly to bacteria, we observed a lower diversity in viral communities in CD samples compared with the control group. In addition, from our results we infer the existence of greater levels of variation within the CD group than within the control group, especially when analyzing the bacterial diversity, but also with viruses. We have also identified that more OTUs, in both viruses and bacteria, are underrepresented in the CD group samples compared with the control samples. However, the exceptions to this pattern, such as the case of viruses similar to members of the family Retroviridae, could be of interest for further investigation, particularly given the links between members of this family and immunodeficiency and the immune responses, key factors in CD.
In our study, bacteriophages are directly inferred from the metagenomic samples. However, in analyzing the bacterial composition it must be noted that two different comparisons are carried out: one was inferred directly from bacterial 16S sequences, whereas the other used an indirect inference of the potential bacterial hosts from the bacteriophages detected in the samples, which does not necessarily correlate with, or even reflect, the actual bacterial composition and abundance in a particular environment, in the same way that the composition of predators does not necessarily allow the inference of the composition and abundance of their prey. In addition, the range of potential bacterial hosts for a bacteriophage can sometimes be very narrow, but other bacteriophages may predate a wider range of bacteria, which can further distort this picture. Furthermore, we have shown that bacterial inference based on the sampling of viruses cannot replace or even complement the analyses of the microbiome, due to the bias in the characterized bacteriophages that hinders any chance to consider them representative of the bacterial communities. For example, the databases have a bias toward bacteriophages of more well-studied bacterial orders because of their health or economic importance, such as Enterobacteriales, Actinomycetales, or Lactobacillales. Conversely, bacteriophages from largely predominant gut bacteria, such as Bacteroidales and Clostridiales, are underrepresented in the virome samples because many of their hosts remain poorly characterized despite their abundance in the human gut, probably due to the fact that many of them are uncultivable bacteria. These discrepancies may also explain the disparate clustering of the samples based on viruses, which does not match the one based on bacteria. There is also less support for clustering when based on viruses, resulting in more ambiguous and variable results.
There are a series of methodological considerations to be taken into account when analyzing the viral community composition and abundance, and the derived taxonomic and functional analyses. Thus, the assembly of the reads into contigs has an impact in the distribution of the viral hits, reducing the absolute number of OTUs in terms of composition. OTU abundance is also affected by a reduction of the relative number of viruses represented by higher numbers of reads, which are therefore more likely to be redundant, as they produce a reduced number of contigs after assembly. In contrast, viruses represented by a low number of individual reads are less prone to be assembled into contigs and so tend to increase their relative OTU frequency after assembly. Analyses carried out in our group have demonstrated that the read assembly significantly increases the performance of the functional analyses (data not shown), making it preferable to assemble into contigs for the functional analysis.
One point of caution is that the identification of viral hits prior to their taxonomic assignment relies on the blast search, which allows identification of the “most similar viruses” in the database. This does not necessarily imply that the viruses present in the public databases are the actual ones present in the sample. Also, slight variations in the blast results can result in a different taxonomic assignment of the E value-based best hits, and therefore variations in the viral distribution. This can result from the assembly of reads into contigs, for example, which can change the best hits in the blast results.
Another noteworthy issue would be the heterogeneity in the number of reads obtained per sample, which makes comparison between samples a more difficult process. Thus, reaching the most homogeneous possible number of reads would be desirable.
Finally, even though we are able to retrieve more viral hits than with the existing pipelines, such as MetaVir, most of the reads remain still unknown. So far, we can only state that we detect candidates related to those viruses available in the public databases, but we cannot rule out the possibility that other viruses, possibly more relevant to understanding the etiology and progression of the CD, may be “hidden” within the uncharacterized reads. It is necessary to expand extant viral databases and other tools to identity viruses not only by homology search but also by means that are independent of sequence.
Study Highlights
Acknowledgments
We thank Sébastien Varlez, Adriana Cordova, Pau Esparza, Sara Ferrando, and Inés Moret for assistance with various aspects of the work presented here. We are especially grateful to Dr C. Graham Clark for his helpful comments and language editing of this manuscript.
Guarantor of the article: Andrés Moya, PhD.
Specific author contributions: Pilar Nos, Belén Beltrán, and Vicente Pérez-Brocal collected the samples. Andrés Moya, Vicente Pérez-Brocal conceived, and designed the experiments. Vicente Pérez-Brocal, Rodrigo García-López, and Jorge Vázquez-Castellanos performed the experiments and analyzed the data. Andrés Moya and Amparo Latorre contributed reagents/materials/analysis tools. Vicente Pérez-Brocal wrote the paper.
Financial support: This work was supported by grants SAF-2009-13032-C02-01 from the Spanish Ministry of Science and Innovation (MICINN) and SAF-2012-31187 from the Ministry of Economy and Competitiveness (MECO) to Andrés Moya. Vicente Pérez-Brocal has a research contract from the Instituto de Salud Carlos III (ISCIII).
Potential competing interests: None.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Supplementary Material
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3
References
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, et al. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Wegrzyn G, Thomas MS. Modulation of the susceptibility of intestinal bacteria to bacteriophages in response to Ag43 phase variation—a hypothesis. Med Sci Monit. 2002;8:HY15–HY18. [PubMed] [Google Scholar]
- Kasman LM. Barriers to coliphage infection of commensal intestinal flora of laboratory mice. Virol J. 2005;2:34. doi: 10.1186/1743-422X-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górski A, Weber-Dabrowska B. The potential role of endogenous bacteriophages in controlling invading pathogens. Cell Mol Life Sci. 2005;62:511–519. doi: 10.1007/s00018-004-4403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Sozzi T, Turroni F, et al. The impact of bacteriophages on probiotic bacteria and gut microbiota diversity. Genes Nutr. 2011;6:205–207. doi: 10.1007/s12263-010-0188-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BM, Waldor MK.Mobile genetic elements and bacteria pathogenesisIn Craig NL Gragie R Gellert M Lambowitz AM (eds)Mobile DNA II ASM Press: Washington, DC, USA; 20021040–1055. [Google Scholar]
- Breitbart M, Salamon P, Andresen B, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Felts B, Kelley S, et al. Diversity and population structure of a nearshore marine sediment viral community. Proc Biol Sci. 2004;271:565–574. doi: 10.1098/rspb.2003.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Angly FE, Felts B, Breitbart M, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bench SR, Hanson TE, Williamson KE, et al. Metagenomic characterization of Chesapeake Bay virioplankton. Appl Environ Microbiol. 2007;73:7629–7641. doi: 10.1128/AEM.00938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld T, Patterson M, Richardson PM, et al. Assembly of viral metagenomes from Yellowstone hot springs. Appl Environ Microbiol. 2008;74:4164–4174. doi: 10.1128/AEM.02598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SJ, Rusch DB, Yooseph S, et al. The Sorcerer II global ocean sampling expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One. 2008;3:e1456. doi: 10.1371/journal.pone.0001456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bueno A, Tamames J, Velázquez D, et al. High diversity of the viral community from an Antarctic lake. Science. 2009;326:858–861. doi: 10.1126/science.1179287. [DOI] [PubMed] [Google Scholar]
- Tamaki H, Zhang R, Angly FE, et al. Metagenomic analysis of DNA viruses in a wastewater treatment plant in tropical climate. Environ Microbiol. 2011;14:441–452. doi: 10.1111/j.1462-2920.2011.02630.x. [DOI] [PubMed] [Google Scholar]
- Cann AJ, Fandrich SE, Heaphy S. Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes. 2005;30:151–156. doi: 10.1007/s11262-004-5624-3. [DOI] [PubMed] [Google Scholar]
- Vega Thurber RL, Barotta KL, Halla D, et al. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc Natl Acad Sci USA. 2008;105:18413–18418. doi: 10.1073/pnas.0808985105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee B, Freeborough MJ, Maree HJ, et al. Deep sequencing analysis of viruses infecting grapevines: Virome of a vineyard. Virology. 2010;400:157–163. doi: 10.1016/j.virol.2010.01.023. [DOI] [PubMed] [Google Scholar]
- Li L, Victoria JG, Wang C, et al. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol. 2010;84:6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shan T, Wang C, et al. The fecal viral flora of California sea lions. J Virol. 2011;85:9909–9917. doi: 10.1128/JVI.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T, Li L, Simmonds P, et al. The fecal virome of pigs on a high-density farm. J Virol. 2011;85:11697–11708. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Rohwer F. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques. 2005;39:729–736. doi: 10.2144/000112019. [DOI] [PubMed] [Google Scholar]
- Allander T, Tammi MT, Eriksson M, et al. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D, Furlan M, Haynes M, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cochetiere MF, Durand T, Lalande V, et al. Effect of antibiotic therapy on human fecal microbiota and the relation to the development of Clostridium difficile. Microb Ecol. 2008;56:395–402. doi: 10.1007/s00248-007-9356-5. [DOI] [PubMed] [Google Scholar]
- Othman M, Aguero R, Lin HC. Alterations in intestinal microbial flora and human disease. Curr Opin Gastroenterol. 2008;24:1–16. doi: 10.1097/MOG.0b013e3282f2b0d7. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, et al. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman EF, Iwasaki A. Genome-virome interactions: examining the role of common viral infections in complex disease. Nat Rev Microbiol. 2011;9:254–264. doi: 10.1038/nrmicro2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus EV, Jr., Sandborn WJ. Epidemiology of inflammatory bowel disease. Gastroenterol Clin North Am. 2002;31:1–20. doi: 10.1016/s0889-8553(01)00002-4. [DOI] [PubMed] [Google Scholar]
- Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- Halme L, Paavola-Sakki P, Turunen U, et al. Family and twin studies in inflammatory bowel disease. World J Gastroenterol. 2006;12:3668–3672. doi: 10.3748/wjg.v12.i23.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekbom A. Twin studies in IBD and other disorders. J Pediatr Gastroenterol Nutr. 2008;46 (Suppl. 1:E9. doi: 10.1097/01.mpg.0000313821.86218.db. [DOI] [PubMed] [Google Scholar]
- Phillpotts RJ, Hermon-Taylor J, Brooke BN. Virus isolation studies in Crohn's disease: a negative report. Gut. 1979;20:1057–1062. doi: 10.1136/gut.20.12.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillpotts RJ, Hermon-Taylor J, Teich NM, et al. A search for persistent virus infection in Crohn's disease. Gut. 1980;21:202–207. doi: 10.1136/gut.21.3.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura HH, Estes MK, Graham DY. Search for evidence of a viral aetiology for inflammatory bowel disease. Gut. 1984;25:347–355. doi: 10.1136/gut.25.4.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kruiningen HJ, Poulin M, Garmendia AE, et al. Search for evidence of recurring or persistent viruses in Crohn's disease. APMIS. 2007;115:962–968. doi: 10.1111/j.1600-0463.2007.apm_564.x. [DOI] [PubMed] [Google Scholar]
- Sura R, Gavrilov B, Flamand L, et al. Human herpesvirus-6 in patients with Crohn's disease. APMIS. 2010;118:394–400. doi: 10.1111/j.1600-0463.2010.02613.x. [DOI] [PubMed] [Google Scholar]
- Lepage P, Colombet J, Marteau P, et al. Dysbiosis in inflammatory bowel disease: a role for bacteriophages. Gut. 2008;57:424–425. doi: 10.1136/gut.2007.134668. [DOI] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard VM, Cadwell K. Viruses, autophagy genes, and Crohn's disease. Viruses. 2011;3:1281–1311. doi: 10.3390/v3071281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Haynes M, Kelley S, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Zhang T, Breitbart M, Lee WH, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:0108–0118. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Park EJ, Roh SW, et al. Diversity and abundance of single-stranded DNA viruses in human feces. Appl Environ Microbiol. 2011;77:8062–8070. doi: 10.1128/AEM.06331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Sinha R, Chen J, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner SR, Allred AF, Tarr PI, et al. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Yang CS, Sakon N, et al. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One. 2009;4:e4219. doi: 10.1371/journal.pone.0004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria JG, Kapoor A, Li L, et al. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R, Lima-Mendez G, Toussaint A. ACLAME: A CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res. 2010;38 (database issue:D57–D61. doi: 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S, Faubladier M, Paulhe N, et al. Metavir: a web server dedicated to virome analysis. Bioinformatics. 2011;27:3074–3075. doi: 10.1093/bioinformatics/btr519. [DOI] [PubMed] [Google Scholar]
- Angly FE, Willner D, Prieto-Davó A, et al. The GAAS metagenomic yool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput Biol. 2009;5:e1000593. doi: 10.1371/journal.pcbi.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria; 2009. [Google Scholar]
- Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Chen J, Li W, et al. Community cyberinfrastructure for advanced microbial ecology research and analysis: the CAMERA resource. Nucleic Acids Res. 2011;2010:D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comito D, Romano C. Dysbiosis in the pathogenesis of pediatric inflammatory bowel diseases. Int J Inflam. 2012;2012:687143. doi: 10.1155/2012/687143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalingam NA, Lynch SV. Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2012;18:968–984. doi: 10.1002/ibd.21866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Supplementary Table 2
Supplementary Table 3