HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses (original) (raw)
Abstract
The identification of recurrent somatic mutations in genes encoding epigenetic enzymes has provided a strong rationale for the development of compounds that target the epigenome for the treatment of cancer. This notion is supported by biochemical studies demonstrating aberrant recruitment of epigenetic enzymes such as histone deacetylases (HDACs) and histone methyltransferases to promoter regions through association with oncogenic fusion proteins such as PML-RAR_α_ and AML1-ETO. HDAC inhibitors (HDACi) are potent inducers of tumor cell apoptosis; however, it remains unclear why tumor cells are more sensitive to HDACi-induced cell death than normal cells. Herein, we assessed the biological and molecular responses of isogenic normal and transformed cells to the FDA-approved HDACi vorinostat and romidepsin. Both HDACi selectively killed cells of diverse tissue origin that had been transformed through the serial introduction of different oncogenes. Time-course microarray expression profiling revealed that normal and transformed cells transcriptionally responded to vorinostat treatment. Over 4200 genes responded differently to vorinostat in normal and transformed cells and gene ontology and pathway analyses identified a tumor-cell-selective pro-apoptotic gene-expression signature that consisted of BCL2 family genes. In particular, HDACi induced tumor-cell-selective upregulation of the pro-apoptotic gene BMF and downregulation of the pro-survival gene BCL2A1 encoding BFL-1. Maintenance of BFL-1 levels in transformed cells through forced expression conferred vorinostat resistance, indicating that specific and selective engagement of the intrinsic apoptotic pathway underlies the tumor-cell-selective apoptotic activities of these agents. The ability of HDACi to affect the growth and survival of tumor cells whilst leaving normal cells relatively unharmed is fundamental to their successful clinical application. This study provides new insight into the transcriptional effects of HDACi in human donor-matched normal and transformed cells, and implicates specific molecules and pathways in the tumor-selective cytotoxic activity of these compounds.
Keywords: epigenetics, apoptosis, tumor-selective, microarray
Deregulation of histone deacetylase (HDAC) activity in tumors, through over-expression of individual HDACs,1, 2, 3, 4, 5, 6, 7, 8 or through aberrant HDAC recruitment to promoter regions through association with oncogenic fusion proteins such as PML-RAR_α_9, 10 and AML1-ETO11, 12 has provided a strong rationale for the clinical application of HDAC inhibitors (HDACi). HDACi can induce cell-cycle arrest, differentiation and cell death in vitro and mediate tumor-cell-selective apoptosis at drug concentrations that leave normal cells relatively unharmed.13, 14, 15 We previously demonstrated that apoptotic sensitivity of tumor cells to HDACi in vitro correlated with therapeutic responsiveness in vivo.16, 17 However, it is not clear why tumor cells are relatively hypersensitive to HDACi-mediated apoptosis and normal cells are relatively resistant.
To assess the tumor-specific molecular and biological activities of HDACi, we used a defined model of human cell transformation in which normal cells had been converted to transformed (tumorigenic) states through the sequential introduction of genes encoding the SV40 large T and small t antigens, hTERT and H-RASG12V.18, 19 The HDACi vorinostat and romidepsin induced apoptosis in a tumor-cell-selective manner. Time-course gene expression profiling revealed that normal and transformed cells transcriptionally responded to HDACi treatment; however, 4200 genes responded differently to vorinostat treatment in normal and transformed cells. Gene ontology (GO) and pathway analyses identified a tumor-cell-selective pro-apoptotic gene-expression signature containing effectors of the intrinsic apoptotic pathway. Regulation of a pro-apoptotic _BCL2_-family gene signature correlated with the relative sensitivity of transformed cells to HDACi-mediated apoptosis. Furthermore, a role for HDACi-mediated repression of BCL2A1 in the induction of tumor cell apoptosis was confirmed, and we formally demonstrated that forced expression of BFL-1 encoded by BCL2A1 suppressed the apoptotic effects of vorinostat in transformed BJ fibroblasts. Collectively, these data enhance our understanding of the molecular consequences of HDAC inhibition, and provide a mechanistic basis for the tumor-selective biological effects of these agents.
Results
HDAC inhibitors selectively kill tumor cells
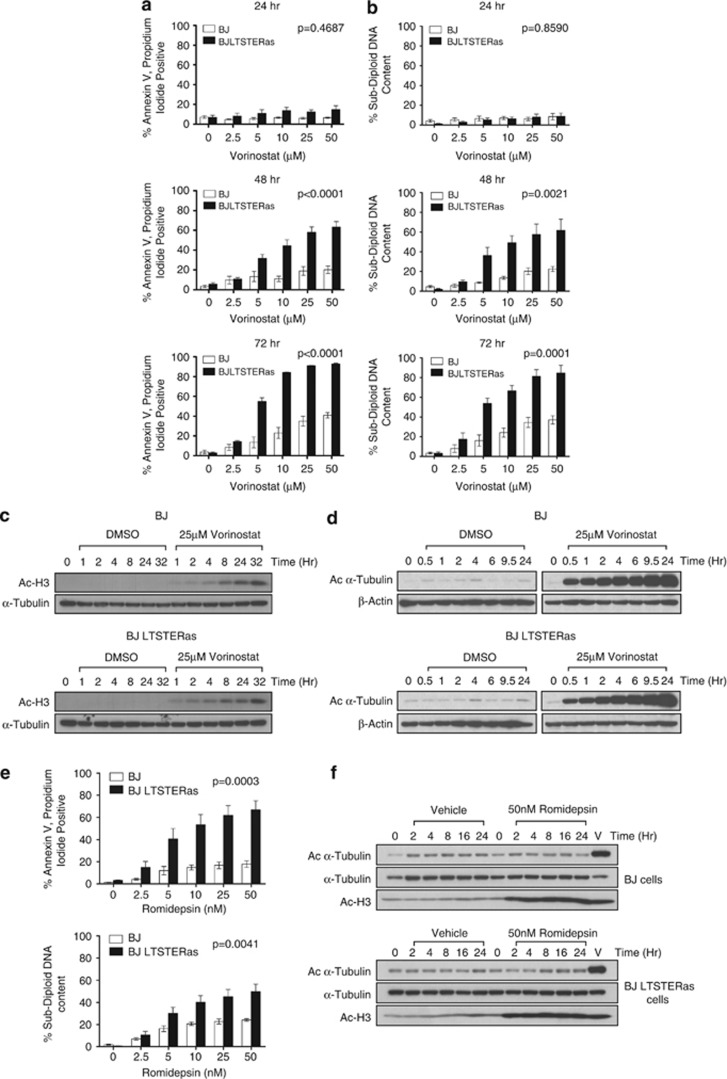
Matched normal (BJ) and transformed (BJ LTSTERas) fibroblasts were treated with vorinostat over 72 h, and cell death was analyzed (Figures 1a and b). Following 24 h vorinostat treatment, there was a marginal increase in death of transformed BJ LTSTERas fibroblasts that increased substantially following extended drug exposure. BJ LTSTERas fibroblasts were significantly more sensitive to vorinostat than BJ cells (Figures 1a and b). Vorinostat induced comparable time-dependent hyperacetylation of histone H3 (Figure 1c) and _α_-tubulin (Figure 1d) in both cell types, suggesting that the relative resistance of normal cells to vorinostat-mediated apoptosis was not due to a failure of vorinostat to inhibit HDACs in these cells. We next tested the sensitivity of BJ and BJ LTSTERas fibroblasts to romidepsin, an HDACi that selectively inhibits class I HDACs.20, 21, 22 BJ LTSTERas fibroblasts were more sensitive than BJ cells to romidepsin-induced apoptosis, even though there was equivalent acetylation of histone H3 in romidepsin-treated BJ and BJ LSTERas fibroblasts (Figures 1e and f). As _α_-tubulin is deacetylated by HDAC6 that is not effectively inhibited by romidepsin, romidepsin did not induce hyperacetylation of _α_-tubulin.
Figure 1.
Matched normal and tumor cells are differentially sensitive to vorinostat-mediated apoptosis. (a, b) BJ and BJ LTSTERas cells were treated with 0, 2.5, 5, 10, 25 or 50 _μ_M doses of vorinostat for 24, 48 or 72 h. Following treatment, all cells were harvested and (a) stained for annexin-V-APC binding and propidium iodide uptake, and analyzed by flow cytometry; (b) fixed with 50% (v/v) ethanol in PBS, stained with propidium iodide and analyzed by flow cytometry. The percentage of annexin-V-APC, propidium iodide double positive cells or cells with sub-diploid DNA content is shown in panels (a, b), respectively. Data are presented as mean±standard error of at least three independent experiments. _P_-values indicate statistical significance of cell type and dose response interactions as measured by two-way ANOVA, where P<0.05 is considered to be significant. (c, d) BJ (top panels) and BJ LTSTERas (bottom panels) cells were treated with 25 _μ_M vorinostat or DMSO equivalent for 0–32 h. Protein extracts were analyzed by western blotting for: (c) acetylation of histone H3 with a pan-acetylated Histone H3 antibody, expression of _α_-Tubulin was used as a loading control; (d) acetylation of _α_-Tubulin with an anti-acetylated tubulin antibody, expression of _β_-Actin served as loading control. Western blots shown are representative of two independent experiments. (e) BJ and BJ LTSTERas were treated with 0, 2.5, 5, 10, 25 and 50 nM concentrations of romidepsin for 48 h. Following treatment, all cells were harvested and stained for annexin-V-APC binding and propidium iodide uptake (top panel) and analyzed by flow cytometry or fixed with 50% (v/v) ethanol in PBS, stained with propidium iodide (bottom panel) and analyzed by flow cytometry. The percentage of annexin-V-APC, propidium iodide double positive cells or cells with sub-diploid DNA content is shown. Data are presented as mean±standard error of at least five independent experiments. The difference in apoptotic sensitivity of BJ and BJ LTSTERas (interaction between cell type and romidepsin dose) was statistically significant (P<0.05), as determined by two-way ANOVA. (f) BJ and BJ LTSTERas fibroblasts were treated with 50 nM romidepsin for 0–24 h or with 25 _μ_M vorinostat for 6 h (V). Immunoblots were probed with antibodies against acetylated histone H3 (AcH3), acetylated _α_-Tubulin (Ac-_α_-Tubulin) and total _α_-Tubulin to control for protein loading. Images are representative of two independent experiments
The sensitivity of donor-matched normal and transformed bone marrow endothelial cells (BMECs) to HDACi was also assessed. BMEC and BMEC LTSTERas were treated with vorinostat for 48 h and analyzed for annexin-V binding and propidium iodide uptake (Supplementary Figure 1A). Consistent with observations in the BJ fibroblast system, transformed BMEC were comparatively hypersensitive to vorinostat-mediated death.
Finally, we determined whether the mechanisms by which normal cells were transformed affected the relative sensitivity of cells to vorinostat-induced apoptosis. Transformed BJ fibroblasts in which SV40 large T antigen was functionally replaced with HPV16 E6 and E7,23 were tested for vorinostat-sensitivity. BJ cells expressing hTERT, E6, E7, H-RASG12V and ST (BJ E6E7 TRST) were sensitive to vorinostat relative to normal BJ fibroblasts when measured over a 72-h time course (Supplementary Figure 1B). These data demonstrated that HDACis selectively killed transformed cells while mediating equivalent histone hyperacetylation in normal and transformed cells.
Vorinostat induces tumor-cell-selective gene expression changes
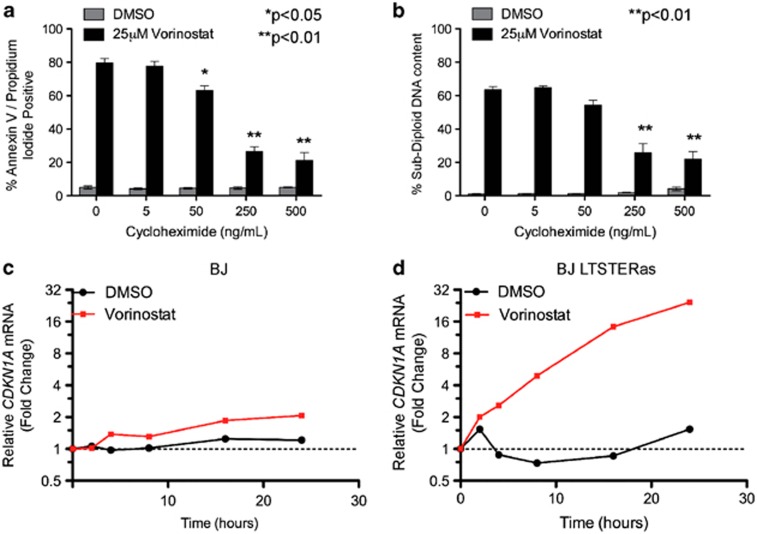
We hypothesized that tumor-cell-selective gene expression changes underpinned the differential sensitivity of isogenic tumor and normal cells to HDACi-induced apoptosis. To investigate whether de novo protein synthesis was required for HDACi-induced death, BJ LTSTERas fibroblasts were pre-treated for 1 h with cycloheximide (CHX) before the addition of vorinostat. CHX treatment significantly inhibited vorinostat-mediated apoptosis after 48 h of drug treatment (Figures 2a and b). Given the requirement of de novo protein expression for the induction of apoptosis by vorinostat, a time-course microarray study was conducted. An early (4 h) and intermediate (12 h) time point was selected for the microarray study on the basis of candidate quantitative real-time polymerase chain reaction (qRT-PCR) analyses of CDKN1A (Figures 2c and d), a gene commonly induced by HDACi.20, 24 CDKN1A mRNA was induced by vorinostat in BJ and BJ LTSTERas cells; however, the magnitude of induction was greater in transformed cells (Figures 2c and d). The abundance of CDKN1A mRNA in BJ and BJ LTSTERas cells after 24 h of vorinostat treatment was similar, as the threshold cycle (Ct) values relative to the control gene RPL32 were similar in both cell types (data not shown). The hyper-induction of CDKN1A in BJ LTSTERas fibroblasts over time reflects the lower basal expression in these cells (at time 0 h).
Figure 2.
Vorinostat-mediated apoptosis requires de novo protein synthesis. (a, b) BJ and BJ LTSTERas cells were pre-treated with 0, 5, 50, 250 and 500 ng/ml CHX to inhibit new protein synthesis and incubated with 25 _μ_M vorinostat for 48 h. Cells were harvested and analyzed for cell death by flow cytometry. (a) Cells were stained with annexin-V-APC and propidium iodide. The percentages of cells staining positively for both markers are shown. (b) Cells were fixed with 50% (v/v) ethanol in PBS, stained with propidium iodide and analyzed for DNA content. The percentages of cells with sub-2N DNA content are shown. All data are presented as mean±S.E.M. of three independent experiments. *P<0.05, **P<0.01 as determined by a two-tailed Student's _t_-test, comparing 25 _μ_M vorinostat treatments in the presence and absence of CHX. (c, d) BJ and BJ LTSTERas cells were treated with 25 _μ_M vorinostat (red) or DMSO (black) for 0, 2, 4, 8, 16 and 24 h. Expression of CDKN1A in (c) BJ and (d) BJ LTSTERas cells was analyzed by qRT-PCR. Messenger RNA levels were calculated relative to that of transcripts from the non-HDACi-regulated control gene RPL32. Fold expression changes at each time point are plotted relative to time 0 h (untreated cells)
Vorinostat-induced changes in gene expression in BJ and BJLSTRas cells
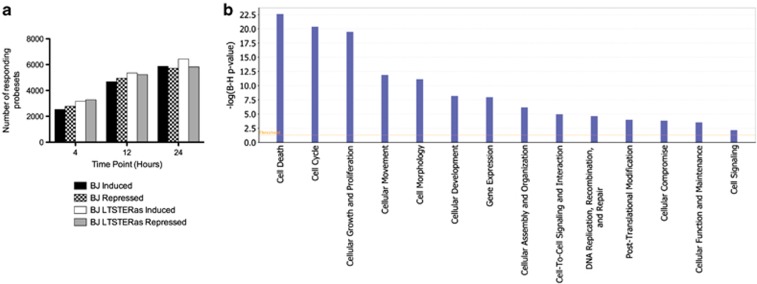
Affimetrix microarray assays showed that the number of vorinostat-regulated probe sets increased in both cell types as a function of time; however, the overall response in BJ LTSTERas cells consistently exceeded that of BJ cells by ∼1000 probe sets at every time point (Figure 3a). At the time of onset of apoptosis (24 h), the number of vorinostat-regulated probe sets in BJ and BJ LTSTERas cells corresponded to 21.1 and 23.0% of the total number of probe sets represented on the arrays, respectively. Collectively, these data indicated that both BJ and BJ LTSTERas fibroblasts were capable of mounting a transcriptional response to vorinostat although ∼1000 additional probe sets were significantly regulated by vorinostat in the transformed cells at every time point analyzed.
Figure 3.
Regulation of gene expression in BJ and BJ LTSTERas by vorinostat. (a) The number of probe sets that were significantly altered in BJ and BJ LTSTERas cells in response to treatment with vorinostat. Probe sets that were induced and repressed by at least 1.5-fold at any of the 4, 12 and 24 h time points relative to time 0 h are shown. (b) Genes that responded differently to vorinostat treatment in matched normal and transformed cells (contrast genes) were analyzed using the IPA tool. The associations of various molecular and cellular functions with contrast genes are plotted in decreasing order of statistical significance, according to −log2 (_P_-value). _P_-values were adjusted for multiple testing using the method of Benjamini and Hochberg. The orange line indicates the threshold for statistical significance (P<0.05)
The Connectivity Map (Cmap) tool25 was used to test whether the vorinostat signatures from BJ and BJ LTSTERas fibroblasts were similar to other drug-induced expression profiles. The Cmap compound with highest connectivity to the BJ and BJ LTSTERas profiles generated in this study was vorinostat (Tables 1 and 2). Two hydroxamic-acid based HDACi, trichostatin A (TSA) and scriptaid, also demonstrated high degrees of connectivity, which indicated that the probe sets regulated by vorinostat in BJ and BJ LTSTERas fibroblasts were similar to those regulated by different HDACi in diverse cell types.
Table 1. Connectivity map for vorinostat-responding genes in treated BJ cells.
| Rank | Cmap name | Mean | n | Enrichment | P-value | Specificity | % Non-null |
|---|---|---|---|---|---|---|---|
| 1 | Vorinostat | 0.762 | 12 | 0.977 | 0.00000 | 0.0151 | 100 |
| 2 | Trichostatin A | 0.695 | 182 | 0.922 | 0.00000 | 0.0047 | 96 |
| 3 | Resveratrol | 0.342 | 9 | 0.873 | 0.00000 | 0.0049 | 100 |
| 4 | Valproic acid | 0.238 | 57 | 0.488 | 0.00000 | 0.0000 | 68 |
| 5 | Sirolimus | 0.102 | 44 | 0.439 | 0.00000 | 0.1024 | 50 |
| 6 | LY-294002 | 0.194 | 61 | 0.421 | 0.00000 | 0.0940 | 67 |
| 7 | Scriptaid | 0.747 | 3 | 0.980 | 0.00004 | 0.0000 | 100 |
| 8 | Rifabutin | 0.695 | 3 | 0.973 | 0.00004 | 0.0000 | 100 |
| 9 | Prochlorperazine | 0.209 | 16 | 0.571 | 0.00004 | 0.0631 | 81 |
| 10 | Trifluoperazine | 0.204 | 16 | 0.549 | 0.00006 | 0.1442 | 68 |
| 11 | Thioridazine | 0.196 | 20 | 0.509 | 0.00008 | 0.2329 | 80 |
| 12 | Syrosingopine | 0.327 | 4 | 0.900 | 0.00010 | 0.0093 | 100 |
| 13 | CP-690334-01 | 0.275 | 8 | 0.685 | 0.00036 | 0.0121 | 75 |
| 14 | Loperamide | 0.235 | 6 | 0.756 | 0.00050 | 0.0152 | 100 |
| 15 | Parbendazole | −0.365 | 4 | −0.874 | 0.00054 | 0.0000 | 50 |
| 16 | Tanespimycin | 0.115 | 62 | 0.248 | 0.00064 | 0.5285 | 54 |
| 17 | Mefloquine | 0.291 | 5 | 0.795 | 0.00084 | 0.0588 | 100 |
| 18 | Fluphenazine | 0.166 | 18 | 0.441 | 0.00089 | 0.1814 | 66 |
| 19 | Securinine | −0.582 | 4 | −0.852 | 0.00092 | 0.0127 | 75 |
| 20 | MS-275 | 0.609 | 2 | 0.974 | 0.00111 | 0.0433 | 100 |
Table 2. Connectivity map for vorinostat-responding genes in treated BJ LTSTERas cells.
| Rank | Cmap name | Mean | n | Enrichment | P-value | Specificity | % Non-null |
|---|---|---|---|---|---|---|---|
| 1 | Vorinostat | 0.816 | 12 | 0.978 | 0.00000 | 0.0151 | 100 |
| 2 | Trichostatin A | 0.703 | 182 | 0.932 | 0.00000 | 0.0000 | 96 |
| 3 | Resveratrol | 0.243 | 9 | 0.861 | 0.00000 | 0.0098 | 100 |
| 4 | CP-690334-01 | 0.358 | 8 | 0.802 | 0.00000 | 0.0121 | 87 |
| 5 | 15-Delta prostaglandin J2 | 0.202 | 15 | 0.696 | 0.00000 | 0.0447 | 86 |
| 6 | LY-294002 | 0.168 | 61 | 0.577 | 0.00000 | 0.0134 | 73 |
| 7 | Valproic acid | 0.171 | 57 | 0.482 | 0.00000 | 0.0000 | 61 |
| 8 | Scriptaid | 0.777 | 3 | 0.978 | 0.00004 | 0.0000 | 100 |
| 9 | Rifabutin | 0.650 | 3 | 0.973 | 0.00010 | 0.0000 | 100 |
| 10 | 5707885 | 0.282 | 4 | 0.899 | 0.00030 | 0.0052 | 100 |
| 11 | Thiostrepton | 0.247 | 4 | 0.880 | 0.00034 | 0.0392 | 100 |
| 12 | Phenoxybenzamine | 0.261 | 4 | 0.876 | 0.00046 | 0.2030 | 100 |
| 13 | Ciclopirox | 0.196 | 4 | 0.864 | 0.00101 | 0.0338 | 100 |
| 14 | Isotretinoin | 0.197 | 4 | 0.842 | 0.00050 | 0.0000 | 100 |
| 15 | Bacitracin | −0.410 | 3 | −0.913 | 0.00112 | 0.0121 | 100 |
| 16 | Sertaconazole | −0.249 | 4 | −0.834 | 0.00137 | 0.0000 | 75 |
| 17 | Mycophenolic acid | 0.252 | 3 | 0.910 | 0.00154 | 0.0452 | 100 |
| 18 | Syrosingopine | 0.260 | 4 | 0.822 | 0.00173 | 0.0326 | 100 |
| 19 | MG-262 | 0.314 | 3 | 0.902 | 0.00184 | 0.1676 | 100 |
| 20 | Fenoprofen | −0.187 | 6 | −0.683 | 0.00266 | 0.0255 | 50 |
Identification of genes that respond differently to vorinostat treatment in normal and transformed fibroblasts
We identified probe sets that responded differently to vorinostat treatment in matched normal and transformed cells (contrast probe sets) at any of the three time points (relative to time 0 h), as we hypothesized that contrast gene expression might underpin the different biological responses of donor-matched cells to vorinostat treatment. In total, 5959 contrast probe sets were identified and these were differently regulated by vorinostat in terms of either the direction (induction or repression) or the magnitude (degree of induction or repression) of the vorinostat response in BJ and BJ LTSTERas cells. Of the 5959 contrast probe sets, 2945 were equally expressed in untreated normal and tumor cells. However, 6226 of the 9240 probe sets with basal expression differences did not respond differently to vorinostat in the two cell types. These data indicated that the basal gene expression levels did not determine the capacity of genes to differently respond to vorinostat treatment in BJ and BJ LTSTERas fibroblasts. GO and pathway analyses were performed on the contrast gene list using the Ingenuity Pathways Analysis (IPA) tool (Figure 3b). ‘Cell Death' was the most significantly represented molecular and cellular function associated with contrast genes (false discovery rate (FDR)=3 × 10−23) while genes involved ‘cell-cycle' (FDR=5 × 10−21) and ‘cellular growth and proliferation' (FDR=4 × 10−20) were also significantly represented on the contrast list.
We further analyzed the contrast responses, in particular those genes that were selectively or hyper-induced in BJ LTSTERas cells compared with BJ cells, using GO analyses and the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool (Supplementary Tables 1–3). Genes involved in ‘induction of programmed cell death' (_P_=0.0130) and ‘apoptotic program' (_P_=0.0024) were over-represented in vorinostat-treated BJ LSTRas cells compared with BJ cells (Supplementary Tables 1–3). Many genes encoding components of the intrinsic and extrinsic apoptotic pathway were selectively induced in BJ LTSTERas cells. A candidate approach was subsequently undertaken to more thoroughly investigate the transcriptional regulation of apoptotic pathway components by vorinostat using the IPA program.
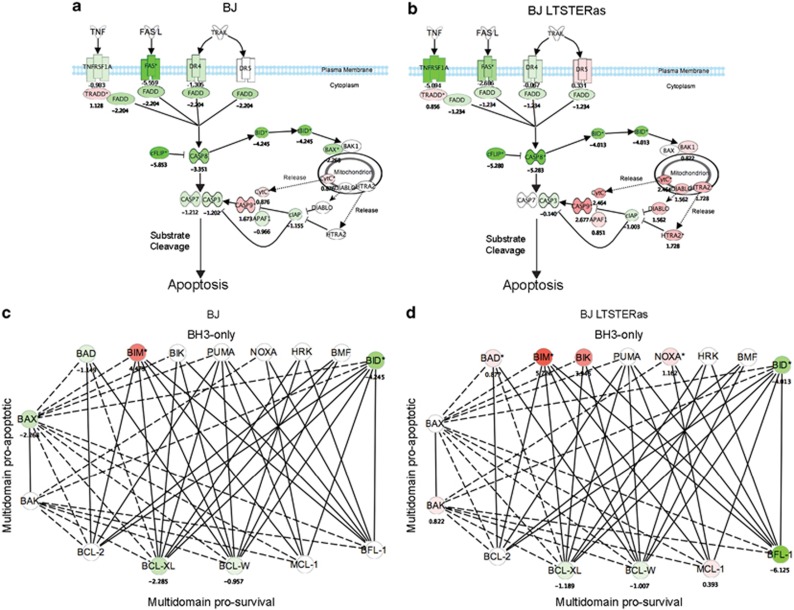
Vorinostat treatment resulted in decreased expression of TNFRSF6 (FAS), TNFRSF10A (DR4/TRAILR1) and TNFRSF1A (TNF_α_ receptor) in both normal and transformed fibroblasts (Figures 4a and b). Only a mild tumor-selective induction of TNFRSF10B (DR5/TRAILR2) was observed. We did not observe any transcriptional modulation of genes encoding death receptor ligands in response to vorinostat in either cell type (Figures 4a and b). Moreover, the genes encoding FADD (FADD), caspase 8 (CASP8) and BID were downregulated in BJ and BJ LTSTERas, suggesting that death receptor signaling was transcriptionally repressed by vorinostat in BJ and BJ LTSTERas fibroblasts. Collectively, the downregulation of genes encoding receptors, adaptors and proteolytic intermediaries in the death receptor pathway suggested that the extrinsic apoptotic pathway was unlikely to play a substantial role in promoting selective apoptosis of BJ LTSTERas fibroblasts.
Figure 4.
Ingenuity pathways analysis. Interaction map of the death receptor pathway was created using the ingenuity pathways analysis tool (a) BJ and (b) BJ LTSTERas fibroblasts. (c, d) An interaction map of the BCL-2 family of apoptotic regulators was created using the ingenuity pathways analysis tool, and overlaid with transcriptional responses to vorinostat in (c) BJ and (d) BJ LTSTERas fibroblasts. For (a–d), the direction and magnitude of vorinostat-mediated expression changes following 24 h of treatment, relative to time 0 h are indicated by pseudo-coloring. Induced (red), repressed (green) and non-responding probe sets (white) are shown. Log2 expression values of the strongest responding probe sets for each vorinostat-responding pathway component are shown. Interactions between BH3-only and pro-survival proteins are based upon Biacore analyses described in reference 26. Interactions between multi-domain proteins BAX/BAK and other BCL-2 family members were based on ingenuity pathway information
Vorinostat treatment caused the general transcriptional induction of the pro-apoptotic BH3-only protein encoding genes BAD, BCL2L11 (BIM), BIK, PMAIP1 (NOXA), the multi-domain pro-apoptotic gene BAK1 and genes encoding death effector components downstream of mitochondrial damage (DIABLO, APAF1, CASP9, HTRA2 and CYTC) in transformed fibroblasts. Moreover, the pro-survival genes BCL2A1, BCL2L1 (encoding BCL-XL) and BCL2L2 (encoding BCL-w) were concomitantly repressed in these cells (Figures 4c and d). Collectively, this transcriptional response is predicted to confer a strong pro-death signal in the tumor cells.
Despite the downregulation of BCL2L1 and BCL2L2, normal BJ fibroblasts failed to mount a substantial pro-apoptotic transcriptional response. BIK, PMAIP1 and BAK1 probe sets were unchanged following vorinostat treatment, and BAX and BAD were repressed. Although the maximally responding probe set for BCL2L11 was induced in BJ and BJ LTSTERas cells (therefore, BIM is pseudo-colored red in Figures 4c and d), three additional BCL2L11 probe sets were selectively induced in BJ LTSTERas cells (Supplementary Figure 2). Collectively, these data strongly support the hypothesis that the transformed cells were hypersensitive to vorinostat-mediated apoptosis due to acute transcriptional alterations in the balance of pro-death and pro-survival BCL2 family genes, whose protein products functionally converge to govern the integrity of the mitochondrial outer membrane.
Validation of BCL2 family gene expression by qRT-PCR
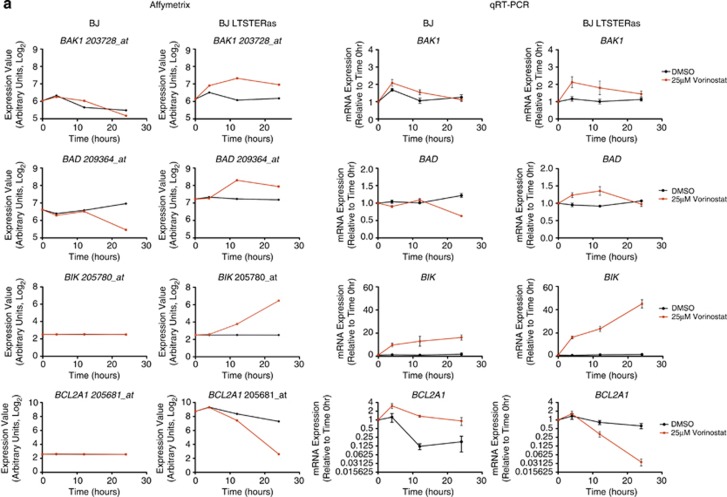
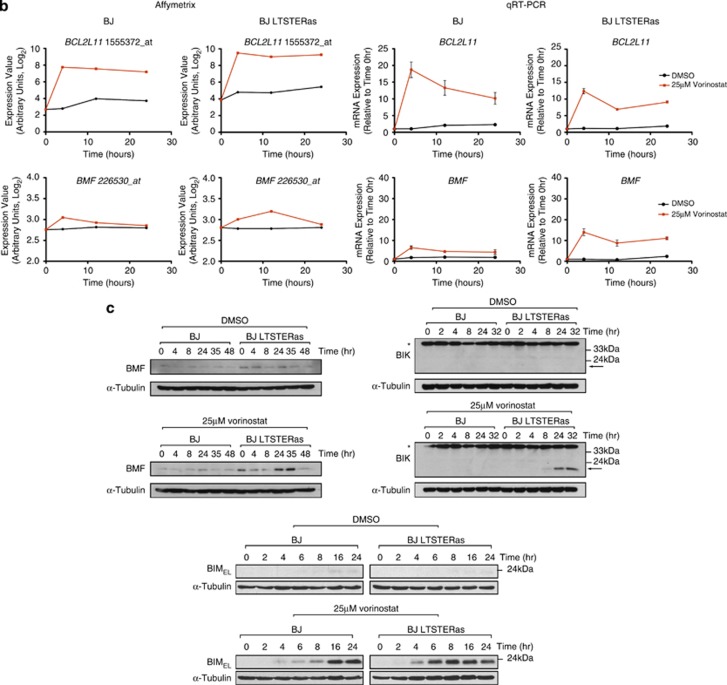
Expression of selected BCL2 family genes following vorinostat and dimethyl sulphoxide (DMSO) treatment was verified by qRT-PCR. qRT-PCR and microarray detection of vorinostat-induced transcriptional changes in BAD, BAK1, BIK, BCL2A1 were concordant (Figure 5a). BCL2L11 (encoding BIM) was transcriptionally induced in normal and transformed BJ fibroblasts following vorinostat treatment, and BCL2L11 induction at the 4 h time point (by qRT-PCR) was consistently greater in the normal cells (Figure 5b). Seven Affymetrix probe sets map to BCL2L11. Interestingly, three of these probe sets responded to vorinostat in both cell types and three probe sets were selectively induced in BJ LTSTERas fibroblasts in a time-dependent manner (Supplementary Figure 2). BMF was shown by us and others to be a primary HDACi response gene.27, 28, 29 In our microarray analyses, BMF was classified as a non-responding gene most likely due to the combined effects of low basal expression and the location of the sole probe set (226530_at) toward the 5′ end of the BMF transcripts. The expression of BMF using qRT-PCR demonstrated that the gene was hyper-induced by vorinostat in BJ LTSTERas fibroblasts (Figure 5b). Taken together, the qRT-PCR data supported the microarray-based observation of a tumor-cell-selective pro-apoptotic gene expression signature, which temporally preceded the onset of apoptosis in BJ LTSTERas cells.
Figure 5.
Validation of vorinostat-induced transcriptional responses by qRT-PCR. Transcriptional responses of BJ and BJ LTSTERas fibroblasts following DMSO (black) and 25 _μ_M vorinostat treatment (red) were validated by qRT-PCR using the same template RNA as that used in the microarray study. Log2 transformed expression intensities of HG U133 plus 2.0 probe sets corresponding to (a) BAK1, BAD, BIK and BCL2A1 and (b) BCL2L11 and BMF are shown. Gene names and probe set IDs are indicated in the title of each panel. Normalized probe set intensity values (_Y_-axis) are presented, which allows direct comparison of basal expression (time 0 h), and log2 transcriptional responses over time between cell types (BJ and BJ LTSTERas). Data represent the summarized probe set intensities from three independent microarray time course experiments. qRT-PCR verification of selected genes is shown. Transcript expression was calculated relative to the control gene RPL32 (that does not respond to vorinostat or DMSO treatment), and fold-change mRNA expression over time was calculated relative to the 0 h time point. Data are presented as the mean±S.E.M. of three independent experiments. For both Affymetrix and qRT-PCR results, data sets for BJ cells are in the left panels while data sets for BJ LTSTERas are in the right panels. (c) Whole cell protein extracts from BJ and BJ LTSTERas fibroblasts treated with DMSO or 25 _μ_M vorinostat for 0–48 h were separated by SDS-PAGE and analyzed for the expression of BMF, BIK and BIM by western blotting. The expression of _α_-Tubulin was analyzed to confirm equal protein loading in each lane. The asterisk (*) denotes a non-specific band in the BIK western blot, which acts as a positive control for activity of the BIK antibody. The arrow denotes the location of BIK. Images are representative of two independent experiments
Western blotting was performed to confirm that the vorinostat-mediated transcriptional changes corresponded to alterated protein expression. BIK and BMF levels increased within 24 h of vorinostat treatment in BJ LTSTERas fibroblasts (Figure 5c). Despite the slight transcriptional induction of BMF and BIK mRNA in normal cells, changes in BIK and BMF protein levels were undetectable following vorinostat treatment in this cell type. BIMEL protein levels were increased in BJ and BJ LTSTERas fibroblasts within 4 h of vorinostat treatment (Figure 5c); however, the time to maximal BIMEL induction differed between the two cell types (16 h in BJ fibroblasts versus 6 h in BJ LTSTERas fibroblasts).
Vorinostat-mediated expression changes are HDACi-specific
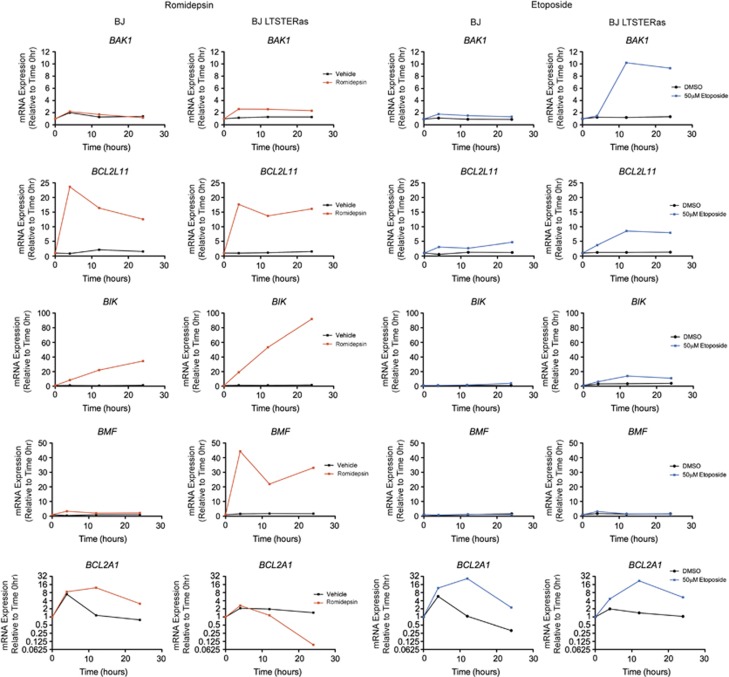
To investigate whether vorinostat-mediated changes in expression of Bcl-2-family genes were HDACi-specific, BJ and BJ LTSTERas cells were treated with equipotent concentrations of romidepsin or the DNA damaging agent etoposide and the expression of BAK1, BCL2L11, BIK, BMF and BCL2A1 was analyzed by qRT-PCR (Figure 6). Similar to the response observed with vorinostat treatment, romidepsin induced BCL2L11 in normal and transformed fibroblasts and induced BAK1 in the transformed cells. Furthermore, romidepsin treatment caused the hyper-induction of BIK and BMF, together with the marked repression of BCL2A1 selectively in transformed cells. By contrast, etoposide failed to substantially induce BMF or BIK. Furthermore, BCL2A1 was induced 22-fold (rather than repressed over time as observed with HDACi) in BJ LTSTERas fibroblasts by etoposide treatment. Etoposide was however clearly capable of modulating pro-apoptotic gene expression in matched fibroblasts, and caused the hyper-induction of BAK1 in BJ LTSTERas cells with a considerably higher magnitude than the changes induced by vorinostat or romidepsin. Collectively, these data strongly suggested that vorinostat-mediated alterations in the expression of BCL2 family genes such as BMF, BIK and BCL2A1 were HDACi-specific.
Figure 6.
Regulation of BCL2 family gene expression by the HDACi romidepsin and the DNA damaging agent etoposide. BJ (left panels) and BJ LTSTERas (right panels) fibroblasts were treated for 0, 4, 12 and 24 h with 50 nM romidepsin (red), 50 _μ_M etoposide (blue) and analyzed for the expression of BAK1, BCL2L11, BIK, BMF and BCL2A1 by qRT-PCR. Expression of each gene was calculated relative to the non-romidepsin- and non-etoposide-responding control gene RPL32 and fold-change expression over time was calculated relative to time 0 h. Gene expression changes resulting from vehicle treatment (black) are also shown. Data are presented as the mean expression fold change from two independent time course experiments
Forced expression of BCL2A1 protects BJ LTSTERas fibroblasts from vorinostat-mediated apoptosis
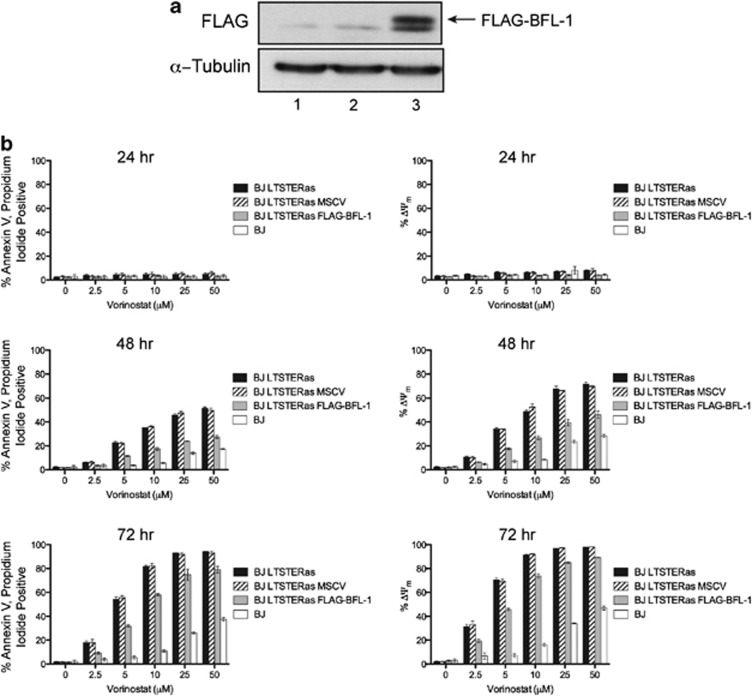
BCL2A1 was highly expressed in untreated BJ LTSTERas fibroblasts and demonstrated the strongest vorinostat response of all BCL2 family genes with the pro-survival gene being strongly repressed in a tumor-cell-selective manner following HDACi treatment (refer to Figure 5a). BCL2A1 (encoding BFL-1) was over-expressed in BJ LTSTERas fibroblasts to ascertain whether restoration of BCL2A1 expression in BJ LTSTERas fibroblasts could protect cells from vorinostat-mediated death and to formally demonstrate that vorinostat-induced BJ LTSTERas cell death proceeds through induction of the intrinsic apoptotic cascade. BJ LTSTERas cells were transduced with an MSCV-based retroviral construct encoding FLAG-tagged human BFL-1 (FLAG–BFL-1), and expression of the exogenous FLAG–BFL-1 protein in GFP-positive cells was confirmed by western blot (Figure 7a).
Figure 7.
Forced expression of BFL-1 protects BJ LTSTERas fibroblasts from vorinostat-mediated apoptosis. (a) Proteins were extracted from exponentially growing BJ LTSTERas fibroblasts, and MSCV and MSCV FLAG–BFL-1 transduced variants. Proteins were separated on 10% polyacrylamide gels by SDS-PAGE and analyzed for the expression of FLAG–BFL-1 by western blotting with an anti-FLAG antibody. The expression of _α_-Tubulin levels served as a control for protein loading. (b) BJ, BJ LTSTERas, BJ LTSTERas MSCV and BJ LTSTERas FLAG–BFL-1 cells were incubated with 0, 2.5, 5, 10, 25 and 50 _μ_M vorinostat for 24, 48 and 72 h. Cells were harvested and stained with annexin-V-APC and propidium iodide as an apoptosis readout or stained for loss of mitochondrial outer membrane potential (MOMP, ΔΨm) using the mitochondrial dye tetramethylrhodamine ethyl ester (TMRE). Cells were then analyzed by flow cytometry. The percentage of annexin-V-APC, propidium iodide double positive cells are shown in the left panels while cells that had lost the ability to bind TMRE (ΔΨm) are in the right panels. Data are presented as mean±S.E.M. of three independent experiments
The sensitivity of BJ LTSTERas cells transduced with MSCV empty vector or FLAG–BFL-1 to vorinostat-mediated apoptosis was evaluated alongside parental BJ LTSTERas and normal BJ fibroblasts. Expression of FLAG–BFL-1 significantly protected BJ LTSTERas fibroblasts from vorinostat-mediated apoptosis at the 48 and 72 h time points, relative to parental and MSCV transduced BJ LTSTERas fibroblasts (Figure 7b, P<0.01, as measured by two-way ANOVA at 48 and 72 h). Collectively, these data showed that protection of cells at the level of mitochondrial integrity (through over-expression of a pro-survival BCL2 family gene) significantly attenuated the induction of vorinostat-mediated apoptosis in transformed fibroblasts, and therefore formally demonstrated a role for the intrinsic apoptotic cascade in the death of BJ LTSTERas fibroblasts.
Discussion
HDACi are being extensively used in the clinic as anti-cancer agents given their manageable toxicity profiles and broad anti-tumor activities.30 While tumor-cell-selective killing by HDACi has been observed,13, 31, 32, 33, 34 exactly why transformed cells are selectively hypersensitive to HDACi remains unclear. We aimed to determine whether donor-matched normal and transformed cells demonstrate the same or different gene expression profiles following HDACi treatment. Time-course microarray expression profiling revealed that normal and transformed cells transcriptionally responded to HDACi treatment, and that vorinostat signatures resembled other HDACi signatures in the Cmap database. Therefore, it appears that diverse HDACi modulate the expression of a common set of genes, irrespective of cellular phenotype or the tissues from which cells were originally isolated, and that a restricted set of genes are particularly amenable to transcriptional modulation following HDACi treatment. Many genes responded differently to vorinostat treatment in normal and transformed cells and a tumor-cell-selective pro-apoptotic gene-expression signature was identified. A role for HDACi-mediated repression of BCL2A1 in the induction of tumor cell apoptosis was confirmed, and we formally demonstrated that the intrinsic apoptotic pathway was required for the cytotoxic activities of HDACi in transformed BJ fibroblasts.
A link between tumor-cell-selective gene induction and cell death following HDACi treatment was previously proposed.35, 36 Kobayashi et al. analyzed romidepsin-mediated transcriptional responses in non-donor-matched normal melanocytes and melanoma cell lines and noted that RAP1A was selectively induced in melanoma cells, and was important for the cytotoxic effect of romidepsin in these cells. By contrast, we found RAP1A to be more highly induced in normal BJ fibroblasts than in BJ LTSTERas fibroblasts (data not shown), suggesting that any contribution of RAP1A towards HDACi-mediated cytotoxicity is likely to be cell-type-dependent. Noh et al.36 investigated the transcriptional responses of non-donor-matched normal bronchial epithelium and lung cancer cells following TSA treatment. That study showed TSA mediated tumor-selective induction of CDKN1A and tumor-selective repression of ERBB2,36 both of which were also tumor-cell-selective events in our microarray study. Chiba et al.37 reported on the transcriptional responses of non-donor-matched hepatocytes and hepatocellular carcinoma cells. In agreement with our study, an approximately equal number of genes were activated and repressed in normal cells following HDACi treatment, although in our study a considerably higher percentage of total genes on the array were significantly modulated by vorinostat.38
Previous studies had proposed that altered ROS production and differential expression of ROS scavengers governs the differential sensitivity of normal and transformed cells to HDACi-mediated apoptosis.31, 32, 33 We did not observe any vorinostat-mediated transcriptional alteration in thioredoxin gene (TXN) expression in normal or transformed cells. Moreover, the gene encoding TBP-2 (TXNIP) was equivalently induced with vorinostat treatment in both cell types. Therefore, vorinostat-mediated transcriptional modulation of TXN and TXNIP was unlikely to be a direct determinant of normal BJ fibroblast resistance, or transformed BJ fibroblast sensitivity to vorinostat-mediated apoptosis.
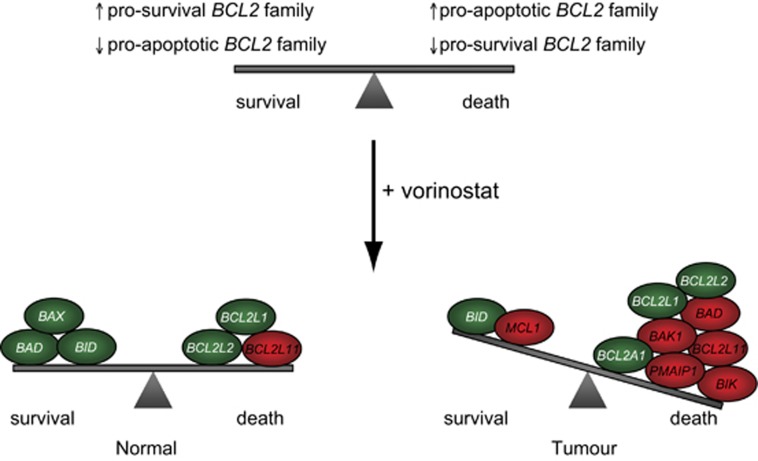
The most notable finding from our candidate and GO studies was the direction in which pro- and anti-apoptotic genes were transcriptionally altered in the normal and transformed cells. Vorinostat-treated BJ LTSTERas fibroblasts elicited a strong pro-apoptotic transcriptional response, consisting of the induction of pro-apoptotic _BCL2_-family genes and the concomitant repression of pro-survival genes. Although BCL2L1 and BCL2L2 were repressed in normal cells following vorinostat treatment, other pro-apoptotic genes such as BAX, BAD and BID were also downregulated. Collectively, these data supported a model in which a tumor-cell-selective alteration in the balance between pro- and anti-apoptotic molecules following HDACi treatment ultimately determines the apoptotic sensitivity of cells exposed to these agents (Figure 8).
Figure 8.
Model for the induction of tumor-selective apoptosis by vorinostat. The tumor-selective induction of apoptosis following vorinostat treatment may be due to acute alterations in the expression of pro-survival and pro-apoptotic BCL2 family genes. The vorinostat-mediated transcription changes observed in BJ LTSTERas cells are hypothesized to tip the balance of pro-survival and pro-apoptotic gene expression in a direction that favors cell death. Although pro-survival and pro-apoptotic expression changes were observed in normal cells, the net change of expression in terms of ability to ‘tip the balance' is relatively neutral. Genes are pseudo-colored according to the direction of gene expression change at the 24-h time point, relative to time 0 h: induced (red) and repressed (green), and are placed on the balance according to whether the expression change is expected to confer a pro-death or pro-survival response
Previous studies have shown that HDACi can induce pro-death transcriptional signatures in human tumor lines.39, 40, 41 The regulation of BCL2 family genes by HDACi in normal cells has to our knowledge not previously been investigated, making this study the first to characterize a vorinostat-mediated BCL2 family transcriptional signature in donor-matched normal and transformed cells. The vorinostat-regulated BCL2 family genes identified herein (e.g., BMF, BCL2L11, PMAIP1, BCL2L1) have also been shown in independent studies to impinge upon HDACi-mediated apoptosis in tumor cells,16, 27, 42, 43, 44, 45 suggesting that tumor-cell-selective regulation of pro-apoptotic genes might be a common mechanism employed by HDACi to exert their tumor-selective cytotoxic effect.
BCL2L11 and BMF were rapidly induced early by vorinostat, indicating that they may be primary HDACi-response genes as has been reported in the literature for BMF.27 Despite the early transcriptional induction, an increase in BMF protein (selectively in tumor cells) was detected considerably later (24 h), and was consistent with BMF being a possible mediator of apoptosis in BJ LTSTERas fibroblasts. Previous reports suggested that elevated BIM expression alone (through deregulated E2F-1 activity) by HDACi was sufficient to induce apoptosis in E1A-expressing human fibroblasts.43, 44 Moreover, BIM was reportedly important for the induction of apoptosis by HDACi in leukemic cell lines and primary cells from patients with chronic lymphocytic leukemia.42 We did observe vorinostat- and romidepsin-mediated induction of BCL2L11 mRNA by microarray and qRT-PCR in normal and transformed cells. Moreover, BIMEL protein levels were elevated in both normal and transformed cells well before the onset of apoptosis (BIM induction was observed at 4 h). These data were, therefore, inconsistent with BIM being the critical mediator of tumor-cell-selective apoptosis in the BJ fibroblast system and support the notion that BIM might instead cooperate with the many other HDACi-induced BH3-only proteins (such as BMF and BIK) to induce apoptosis in sensitive cells.
Our data suggest that pro-death gene expression changes are likely to be important for the tumor-selective effects of HDACi; however, it is not yet known whether the HDACi signatures are driven by histone hyperacetylation in the promoters of specific loci and/or if gene modulation is due to the altered activities of transcription regulators. A recent report demonstrated a correlation between vorinotat-mediated induction of BMF, NOXA and BCL2L11 and histone H4 hyperacetylation within the promoter regions of these genes following HDACi treatment.42 Whether such a correlation between HDACi-mediated selective induction of gene expression and promoter histone acetylation occurs on a global scale and is the basis for the tumor cell-specific effects of these anti-cancer agents remains to be determined.
Materials and Methods
Cell culture
BJ and BJ LTSTERas cells (kindly provided by Professor William Hahn, Dan Farber Cancer Institute, Boston, MA, USA) were grown in media containing four parts HEPES buffered DMEM, pH 7.4 (SAFC Biosciences, Brooklyn, Australia) to one part Medium 199 (Sigma, St. Louis, MO, USA), supplemented with 15% (v/v) fetal bovine serum (FBS; Commonwealth Serum Laboratories, Parkville, Australia), 1 × GlutaMAX (Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin and 100 _μ_g/ml streptomycin (Life Technologies, Inc., Grand Island, NY, USA). BMECs were cultured in Medium 199 (Biowhittaker, Walkersville, MD, USA) supplemented with 20% FBS, 3% human serum, 20 _μ_g/ml endothelial cell growth factor (Biomedical Technologies, Stroughton, MA, USA), 90 _μ_g/ml sodium heparin (Sigma), 2 mM ℒ-Glutamine, 80 U/ml penicillin and 80 _μ_g/ml streptomycin. The establishment of transformed BMEC (previously referred to as BMSVhTERT-4Nras) was previously described.19 Transformed BMEC were maintained in Iscove's Modified Dulbecco's Medium (IMDM; Life Technologies) with 20% FBS. All cells were maintained at 37 °C in a humidified incubator containing 5% CO2.
Chemical compounds
Vorinostat (100 mM) and CHX (5, 50, 500 _μ_g/ml) stock solutions were made in DMSO (Sigma) and stored at −20 °C. Romidepsin was dissolved in 80% (v/v) 1,2 propanediol, 20% (v/v) ethanol by the Peter MacCallum Cancer Centre Pharmacy, and diluted to a 1 mM stock concentration and stored at −20 °C. Stocks were further diluted to working concentrations with growth media immediately before use. Cells were incubated with 0–50 _μ_M vorinostat for 0−72 h, 0–50 nM romidepsin for 0–48 h and 50 _μ_M etoposide for 0–48 h to induce cell death and protein hyperacetylation. To inhibit new protein synthesis, cells were pre-treated (prior to the addition of HDACi) with 5, 50, 250 or 500 ng/ml CHX for 1 h, and CHX was left in media throughout the HDACi treatment time course.
In vitro cell death analysis
BJ fibroblasts (5 × 104 cells) were seeded into wells of six-well plates (Greiner Bio-One, Stonehouse, UK). BMEC (1 × 105 cells) were seeded into 0.1% gelatin coated six-well plates. Cells were allowed to adhere overnight. Immediately prior to each experiment, spent media was replaced with fresh media and drugs/vehicles were added to working concentrations from stock solutions. At nominated time points, all floating and adherent cells (detached by trypsinization) were collected, washed with PBS, stained for viability markers and analyzed by flow cytometry. Mitochondrial outer-membrane permeability was measured with tetramethyl-rhodamine ethyl ester (TMRE—Molecular Probes, Eugene, OR, USA)46 and DNA content determined by propidium iodide staining as previously described.16, 17 Cell viability was measured by propidium iodide uptake and Annexin-V-APC staining. Briefly, cells were resuspended in annexin-V binding buffer (10 mM Hepes pH 7.4, 140 mM NaCl, 5 mM CaCl2) containing 1/200 dilution of annexin-V-APC (BD Biosciences, San Jose, CA, USA) and 1/100 dilution of PI citrate buffer. Cells were analyzed using LSRII and FACSCanto II flow cytometers (BD Biosciences).
Western blotting
Cells (1 × 106) were seeded onto 10-cm plates and incubated in the presence of 25 _μ_M vorinostat or DMSO. Cells were harvested by scraping into ice-cold PBS, washed once with PBS and lysed as described below. For analysis of BMF, BIK and BFL-1 cells were lysed in ice-cold whole-cell lysis buffer (150 mM NaCl, 50 mM Tris-HCl pH 8.0, 0.1 mM EDTA, 0.1 mM EGTA, 0.5% Triton X-100, 10% glycerol) supplemented with protease inhibitors (1 _μ_g/ml Aprotinin, 1 _μ_g/ml Leupeptin, 1 _μ_g/ml Pepstatin and 2 mM PMSF) for 1 h, and the extracts cleared by centrifugation. For the analysis of acetylated proteins and BIM, cells were lysed on ice for 5 min with SDS lysis buffer (250 mM NaCl, 50 mM Tris-HCl pH 7.5, 5 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, 20% glycerol, 1 _μ_g/ml Aprotinin, 1 _μ_g/ml Leupeptin, 1 _μ_g/ml Pepstatin and 2 mM PMSF), sonicated twice for 15 s and extracts cleared by centrifugation. Proteins were separated on SDS-polyacrylamide gels and electroblotted onto Immobilon-P membranes (Millipore, Bedford, MA, USA). The following primary antibodies were used: anti-Acetylated tubulin (Clone 6-11 B-1, Sigma), anti-acetylated H3 (Upstate, Bellerica, MA, USA), anti-_α_-tubulin (Clone B-5-1-2, Sigma) anti-_β_-Actin (Clone AC-74, Sigma), anti-BMF (Abcam, Cambridge, UK), anti-BIM (Stressgen, Farmingdale, NY, USA), anti-BIK (Santa Cruz, Dallas, TX, USA) and anti-FLAG (M2, Sigma). Membranes were incubated overnight with primary antibodies at 4 °C, followed by short-term incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (DAKO) at room temperature. Immunoreactive bands were visualized by enhanced chemiluminescence (GE Healthcare, Little Chalfont, UK).
Quantitative real-time polymerase chain reaction
RNA was prepared from vehicle- and drug-treated cells (1 × 106) using 1 ml TRIzol reagent according to the manufacturer's instructions (Invitrogen). RNA was resuspended in diethyl pyrocarbonate (DEPC)-treated, autoclaved MQH2O and first-strand cDNA was subsequently prepared using a random primer technique according to the manufacturer's instructions (Promega, Madison, WI, USA). Quantitative real-time PCR reactions were performed in triplicate using the SYBR green detection method in a 384-well plate format. Amplification and fluorescence detection was performed in a 7900 HT fast real-time PCR system with SDS v2.3 software (Applied Biosystems, Grand Island, NY, USA). Initial denaturation of template and enzyme activation was performed at 95 °C for 15 min followed by 40 cycles of 95 °C for 30 s, 65 °C for 30 s. Dissociation protocols were always performed for each qRT-PCR reaction to ensure the presence of a single ‘major' amplification product per well. An average Ct value for the three technical replicates was calculated for each sample. Fold-change expression of ‘test' mRNA was determined relative to RPL32 expression using the 2−ΔΔCt method. Expression fold-change over time was plotted relative to mRNA expression at the 0 h time point.
QRT-PCR primer sequences (5′–3′)
RPL32
Forward TTCCTGGTCCACAACGTCAAG
Reverse TTGTGAGCGATCTCGGCAC
CDKN1A
Forward GGCGGCAGACCAGCATGACAGATT
Reverse GCAGGGGGCGGCCAGGGTAT
BCL2A1 (BFL-1)
Forward CGTAGACACTGCCAGAACACTA
Reverse GGGCAATTTGCTGTCGTAGA
BAK1
Forward ACACTTGCTCCCAACCCATTCA
Reverse AACCAAGTCCTAAGCCCTGT
BIK
Forward ATCTTGATGGAGACCCTCCTGT
Reverse TCACTGCCCTCCATGCATT
BMF
Forward ACCAGCCAGGAAGACAAAGCT
Reverse CGATAGCCAGCATTGCCATAA
BAD
Forward AGTGACGAGTTTGTGGACTCCT
Reverse AGCTTTGCCGCATCTGCGTT
BCL2L11 (BIM)
Reverse GTTGCTTTGCCATTTGGTCT
Forward CCACCACTTGATTCTTGCAG
Microarray hybridizations
Total RNA (2 _μ_g) was prepared for hybridization to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays according to the manufacturer's recommendations. Poly-A RNA controls from Bacillus subtilis (lys, phe, thr and dap) were diluted in Poly-A control dilution buffer (serial dilutions of 1 : 20, 1 : 50 and 1 : 25) and 2 _μ_l of the third serial dilution was spiked into each RNA sample prior to cDNA synthesis. RNA was reverse transcribed using a one-cycle cDNA synthesis kit (Affymetrix, Santa Clara, CA, USA) and all annealing, amplification and incubation steps were performed in accordance with the manufacturer's instructions. Double-stranded cDNA was purified using a GeneChip sample cleanup module according to the manufacturer's instructions (Affymetrix) and cDNA was eluted from columns with 14 _μ_l cDNA elution buffer. Biotin-labeled cRNA was synthesized from cDNA using a GeneChip IVT labeling kit (Affymetrix) in a total reaction volume of 40 _μ_l. cRNA was purified and concentrated using the GeneChip sample cleanup module. Full-length cRNA (20 _μ_g) was fragmented to 35–200 bp lengths by incubation with cRNA fragmentation reagent for 35 min at 94 °C. Fragmented cRNA (15 _μ_g) was added to a hybridization mix according to Affymetrix instructions and hybridized to Affymetrix HG U133 plus 2.0 probe arrays at 45 °C in an Affymetrix Hybridization oven 640 with 60 r.p.m. rotation for 16 h. Array washes and three-stage streptavidin phycoerythrin (SAPE) staining was performed according to the manufacturer's instructions on a GeneChip fluidics station 450 (Affymetrix). HG U133 plus 2.0 arrays were individually scanned using an Affymetrix GeneChip 3000 scanner with GeneChip Operating Software (GCOS, Affymetrix).
Microarray differential expression analyses
Intensities from the Affymetrix arrays were background corrected, normalized and summarized using the GC-Robust Multi-Array Analysis (GC-RMA) algorithm.47 Probe sets were defined by the default Affymetrix HG U133 plus 2.0 chip definition file. Box-plots of expression intensities and RNA degradation plots confirmed good consistent quality of the samples and the hybridizations. Microarray data are available through the Gene Expression Omnibus, GEO accession number GSE43010.
Differential expression analysis of the normalized log2-expression values was performed using the Bioconductor software package limma.48 All the arrays were analyzed together using a linear model. Empirical Bayes moderated _t_-tests were used to assess differential expression between the BJ and BJLTSTERas fibroblasts and between untreated and treated cells.48, 49, 50 Specifically, baseline differences between the cell lines were identified by comparing untreated BJ and to BJ LTSTERas fibroblasts at time 0 h. Probe sets responding to DMSO or vorinostat were identified by comparing each of the 4, 12 or 24 h time points back to untreated cells, and this was done separately for BJ and BJ LTSTERas fibroblasts. For all comparisons, probe sets were considered to be significantly differentially expressed if the fold-change exceeded 1.5 (i.e., 50% difference in expression) and the FDR was <0.05.51
Probe sets responding differently to vorinostat treatment in BJ LTSTERas fibroblasts as compared with BJ cells (i.e., contrast genes), were identified using empirical Bayes moderated F-tests.49, 50 F-tests allowed us to combine information across the three treatment time points, looking for probe-sets with differing responses at any of the times. First we used an F-test to find probe sets whose gene expression changes (from 0 h to any of 4, 12 or 24 h vorinostat treatment) were different in BJ fibroblasts compared with BJ LTSTERas fibroblasts. This test is analogous to testing for interaction in a two-way ANOVA. Second, we subtracted from this list any probe set whose expression response to vorinostat could not be distinguished from that to DMSO (the vehicle control). To do this, we used an F-test to find probe sets whose response to vorinostat relative to DMSO was different between BJ fibroblasts and BJ LTSTERas fibroblasts at any of the paired time points. Only probe sets detected by both the above F-tests were retained in the contrast list. The final contrast list contained therefore only bona fide vorinostat-responding genes. For all F-tests, probe sets were considered statistically significant if the FDR was <0.05 and at least one comparison involved in the test achieved a 1.5-fold change.
GO analyses
Gene lists were tested for over-represented GO terms using the DAVID website.52 The Affymetrix HG U133 Plus 2.0 Array served as the background gene list. Only GO terms at specificity levels 4 or 5 within the directed acyclic graph hierarchy defined by the GO Consortium were included.53 The minimum hit threshold was set to 3. Over-represented GO terms were ordered by the EASE score _P_-value, a conservative modification of Fisher's Exact test designed to de-prioritise annotation terms containing very small numbers of genes.54
Ingenuity pathway analyses
Gene lists and log2 expression fold-changes were uploaded into the IPA tool (Ingenuity Systems, www.ingenuity.com). When more than one probe set mapped to the same gene, the probe set with the largest fold-change was used. Pathways and interaction maps were constructed using the path designer function, with expression changes overlayed onto the pathways. Gene lists were also analyzed in IPA for over-represented cellular and molecular functions and for association with disease. _P_-values were adjusted by the Benjamini and Hochberg algorithm to control the FDR at <5%.
Connectivity map analyses
Probe sets with statistically significantly (F-test) responses to vorinostat at any time were ranked by their expression fold-change at 24 h versus 0 h, and this was performed separately for BJ and BJ LTSTERas cells. The 500 most induced and 500 most repressed probe sets were uploaded into the Connectivity Map (Cmap) v02 (http://www.broad.mit.edu/cmap). Cmap ranked drug compounds with gene expression signatures similar to the uploaded gene lists on the basis of connectivity _P_-values and enrichment scores.25 Cmap required Affymetrix HG U133A probe set identifiers, so the identifiers from our HG U133 Plus 2.0 Arrays were mapped back to equivalent HG 133A probe sets using the NetAffx analysis center (http://www.affymetrix.com/analysis/index.affx).
Retroviral transduction of BJ LTSTERas fibroblasts
BFL-1 cDNA was cloned into the MSCV-IRES-GFP retroviral vector and transduction of BJ LSTERas cells was performed as previously described.16
Acknowledgments
We thank Ben Martin, Christopher Clarke and Richard Pearson for constructive criticism and helpful advice, and William Hahn for normal and transformed cell lines. RWJ is a Principal Research Fellow of the National Health and Medical Research Council of Australia (NHMRC) and supported by NHMRC Program and Project Grants, the Susan G Komen Breast Cancer Foundation, Cancer Council Victoria, The Victorian Cancer Agency, The Leukemia Foundation of Australia and the Victorian Breast Cancer Research Consortium. JEB was supported by The Cancer Research Institute Predoctoral Emphasis Pathway in Tumor Immunology. KLM was supported by funds from NHMRC, Cancer Institute NSW and NSW Cancer Council. GKS is a Senior Research Fellowship of the NHMRC. GKS and WS are supported by NHMRC Program Grant 490037.
Glossary
HDAC
histone deacetylase
BMEC
bone marrow endothelial cell
HDACi
histone deacetylase inhibitor
CHX
cycloheximide
PI
propidium iodide
Dr. Johnstone's work has been funded by a research grant from Novartis.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
Supplementary Information
References
- Song J, Noh JH, Lee JH, Eun JW, Ahn YM, Kim SY, et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS. 2005;113:264–268. doi: 10.1111/j.1600-0463.2005.apm_04.x. [DOI] [PubMed] [Google Scholar]
- Choi JH, Kwon HJ, Yoon BI, Kim JH, Han SU, Joo HJ, et al. Expression profile of histone deacetylase 1 in gastric cancer tissues. Jpn J Cancer Res. 2001;92:1300–1304. doi: 10.1111/j.1349-7006.2001.tb02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusche CA, Wulfing P, Kersting C, Vloet A, Bocker W, Kiesel L, et al. Histone deacetylase-1 and -3 protein expression in human breast cancer: a tissue microarray analysis. Breast Cancer Res Treat. 2005;90:15–23. doi: 10.1007/s10549-004-1668-2. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yamashita H, Toyama T, Sugiura H, Omoto Y, Ando Y, et al. HDAC6 expression is correlated with better survival in breast cancer. Clin Cancer Res. 2004;10:6962–6968. doi: 10.1158/1078-0432.CCR-04-0455. [DOI] [PubMed] [Google Scholar]
- Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- Zhu P, Martin E, Mengwasser J, Schlag P, Janssen KP, Göttlicher M. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell. 2004;5:455–463. doi: 10.1016/s1535-6108(04)00114-x. [DOI] [PubMed] [Google Scholar]
- Wilson AJ, Byun DS, Popova N, Murray LB, L'Italien K, Sowa Y, et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- Spurling CC, Godman CA, Noonan EJ, Rasmussen TP, Rosenberg DW, Giardina C. HDAC3 overexpression and colon cancer cell proliferation and differentiation. Mol Carcinog. 2008;47:137–147. doi: 10.1002/mc.20373. [DOI] [PubMed] [Google Scholar]
- Wu WS, Vallian S, Seto E, Yang WM, Edmondson D, Roth S, et al. The growth suppressor PML represses transcription by functionally and physically interacting with histone deacetylases. Mol Cell Biol. 2001;21:2259–2268. doi: 10.1128/MCB.21.7.2259-2268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- Ferrara FF, Fazi F, Bianchini A, Padula F, Gelmetti V, Minucci S, et al. Histone deacetylase-targeted treatment restores retinoic acid signaling and differentiation in acute myeloid leukemia. Cancer Res. 2001;61:2–7. [PubMed] [Google Scholar]
- Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinga A, Monestiroli S, Ronzoni S, Gelmetti V, Marchesi F, Viale A, et al. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat Med. 2005;11:71–76. doi: 10.1038/nm1160. [DOI] [PubMed] [Google Scholar]
- Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- Lindemann RK, Newbold A, Whitecross KF, Cluse LA, Frew AJ, Ellis L, et al. Analysis of the apoptotic and therapeutic activities of histone deacetylase inhibitors by using a mouse model of B cell lymphoma. Proc Natl Acad Sci USA. 2007;104:8071–8076. doi: 10.1073/pnas.0702294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold A, Lindemann RK, Cluse LA, Whitecross KF, Dear AE, Johnstone RW. Characterisation of the novel apoptotic and therapeutic activities of the histone deacetylase inhibitor romidepsin. Mol Cancer Ther. 2008;7:1066–1079. doi: 10.1158/1535-7163.MCT-07-2256. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- MacKenzie KL, Franco S, Naiyer AJ, May C, Sadelain M, Rafii S, et al. Multiple stages of malignant transformation of human endothelial cells modelled by co-expression of telomerase reverse transcriptase, SV40 T antigen and oncogenic N-ras. Oncogene. 2002;21:4200–4211. doi: 10.1038/sj.onc.1205425. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Grant C, Rahman F, Piekarz R, Peer C, Frye R, Robey RW, et al. Romidepsin: a new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumors. Expert Rev Anticancer Ther. 2010;10:997–1008. doi: 10.1586/era.10.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson M, Johnstone RW, Prince HM. Histone deacetylase inhibitors: potential targets responsible for their anti-cancer effect. Invest New Drugs. 2010;28 (Suppl 1:S3–20. doi: 10.1007/s10637-010-9596-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, et al. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol Cell Biol. 2002;22:2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Adachi M, Kawamura R, Imai K. Bmf is a possible mediator in histone deacetylase inhibitors FK228 and CBHA-induced apoptosis. Cell Death Differ. 2006;13:129–140. doi: 10.1038/sj.cdd.4401686. [DOI] [PubMed] [Google Scholar]
- Labi V, Grespi F, Baumgartner F, Villunger A. Targeting the Bcl-2-regulated apoptosis pathway by BH3 mimetics: a breakthrough in anticancer therapy. Cell Death Differ. 2008;15:977–987. doi: 10.1038/cdd.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmans AP, Alsop AE, Bots M, Cluse LA, Williams SP, Banks KM, et al. Deciphering the molecular events necessary for synergistic tumor cell apoptosis mediated by the histone deacetylase inhibitor vorinostat and the BH3 mimetic ABT-737. Cancer Res. 2011;71:3603–3615. doi: 10.1158/0008-5472.CAN-10-3289. [DOI] [PubMed] [Google Scholar]
- Rasheed W, Bishton M, Johnstone RW, Prince HM. Histone deacetylase inhibitors in lymphoma and solid malignancies. Expert Rev Anticancer Ther. 2008;8:413–432. doi: 10.1586/14737140.8.3.413. [DOI] [PubMed] [Google Scholar]
- Butler LM, Zhou X, Xu WS, Scher HI, Rifkind RA, Marks PA, et al. The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc Natl Acad Sci USA. 2002;99:11700–11705. doi: 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerstedt JS, Sowa Y, Xu WS, Shao Y, Dokmanovic M, Perez G, et al. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:673–678. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Ngo L, Perez G, Dokmanovic M, Marks PA. Intrinsic apoptotic and thioredoxin pathways in human prostate cancer cell response to histone deacetylase inhibitor. Proc Natl Acad Sci USA. 2006;103:15540–15545. doi: 10.1073/pnas.0607518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Varghese DS, Gillam MC, Peyton M, Modi B, Schiltz RL, et al. Differential response of cancer cells to HDAC inhibitors trichostatin A and depsipeptide. Br J Cancer. 2012;106:116–125. doi: 10.1038/bjc.2011.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Ohtsuki M, Murakami T, Kobayashi T, Sutheesophon K, Kitayama H, et al. Histone deacetylase inhibitor FK228 suppresses the Ras-MAP kinase signaling pathway by upregulating Rap1 and induces apoptosis in malignant melanoma. Oncogene. 2006;25:512–524. doi: 10.1038/sj.onc.1209072. [DOI] [PubMed] [Google Scholar]
- Noh EJ, Jang ER, Jeong G, Lee YM, Min CK, Lee JS. Methyl CpG-binding domain protein 3 mediates cancer-selective cytotoxicity by histone deacetylase inhibitors via differential transcriptional reprogramming in lung cancer cells. Cancer Res. 2005;65:11400–11410. doi: 10.1158/0008-5472.CAN-05-1092. [DOI] [PubMed] [Google Scholar]
- Chiba T, Yokosuka O, Fukai K, Kojima H, Tada M, Arai M, et al. Cell growth inhibition and gene expression induced by the histone deacetylase inhibitor, trichostatin A, on human hepatoma cells. Oncology. 2004;66:481–491. doi: 10.1159/000079503. [DOI] [PubMed] [Google Scholar]
- Chiba T, Yokosuka O, Arai M, Tada M, Fukai K, Imazeki F, et al. Identification of genes up-regulated by histone deacetylase inhibition with cDNA microarray and exploration of epigenetic alterations on hepatoma cells. J Hepatol. 2004;41:436–445. doi: 10.1016/j.jhep.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci USA. 2004;101:540–545. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart MJ, Smyth G, Van Laar R, Bowtell D, Richon V, Marks PA, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PS, Barbi S, Donadelli M, Costanzo C, Bassi C, Palmieri M, et al. Gene expression profiling after treatment with the histone deacetylase inhibitor trichostatin A reveals altered expression of both pro- and anti-apoptotic genes in pancreatic adenocarcinoma cells. Biochim Biophys Acta. 2004;1693:167–176. doi: 10.1016/j.bbamcr.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Inoue S, Riley J, Gant TW, Dyer MJ, Cohen GM. Apoptosis induced by histone deacetylase inhibitors in leukemic cells is mediated by Bim and Noxa. Leukemia. 2007;21:1773–1782. doi: 10.1038/sj.leu.2404760. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tan J, Zhuang L, Jiang X, Liu ET, Yu Q. Inhibitors of histone deacetylases target the Rb-E2F1 pathway for apoptosis induction through activation of proapoptotic protein Bim. Proc Natl Acad Sci USA. 2005;102:16090–16095. doi: 10.1073/pnas.0505585102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Zhuang L, Jiang X, Yang KK, Karuturi KM, Yu Q. Apoptosis signal-regulating kinase 1 is a direct target of E2F1 and contributes to histone deacetylase inhibitor- induced apoptosis through positive feedback regulation of E2F1 apoptotic activity. J Biol Chem. 2006;281:10508–10515. doi: 10.1074/jbc.M512719200. [DOI] [PubMed] [Google Scholar]
- Xargay-Torrent S, Lopez-Guerra M, Saborit-Villarroya I, Rosich L, Campo E, Roué G, et al. Vorinostat-induced apoptosis in mantle cell lymphoma is mediated by acetylation of proapoptotic BH3-only gene promoters. Clin Cancer Res. 2011;17:3956–3968. doi: 10.1158/1078-0432.CCR-10-3412. [DOI] [PubMed] [Google Scholar]
- Peart MJ, Tainton KM, Ruefli AA, Dear AE, Sedelies KA, O'Reilly LA, et al. Novel mechanisms of apoptosis induced by histone deacetylase inhibitors. Cancer Res. 2003;63:4460–4471. [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- Smyth GK.Limma: linear models for microarray dataIn: Gentleman R, Carey V, Dudoit S, Irizarry R, H W, (eds).Bioinformatics and Computational Biology Solutions using R and Bioconductor Springer: New York; 397–420.2005 [Google Scholar]
- Smyth GK.Linear models and empirical bayes methods for assessing differential expression in microarray experiments Stat Appl Genet Mol Biol 20043Article3. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Ritchie M, Thorne N, Wettenhall J. limma: Linear Models for Microarray Data Users Guide. 2005.
- Benjamini Y, Hochberg Y. Controlling false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc B. 1995;57:289–300. [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information