Type I interferons down regulate myeloid cell IFNGR by inducing recruitment of an Egr3/Nab1 complex that silences ifngr1 transcription (original) (raw)
. Author manuscript; available in PMC: 2014 Sep 15.
Published in final edited form as: J Immunol. 2013 Aug 9;191(6):3384–3392. doi: 10.4049/jimmunol.1203510
Abstract
The ability of type I interferons (IFNs) to increase susceptibility to certain bacterial infections correlates with down regulation of myeloid cell surface IFNGR, the receptor for the type II IFN (IFNγ), and reduced myeloid cell responsiveness to IFNγ. Here, we show that the rapid reductions in mouse and human myeloid cell surface IFNGR1 expression that occur in response to type I IFN treatment reflect a rapid silencing of new ifngr1 transcription by repressive transcriptional regulators. Treatment of macrophages with IFNβ reduced cellular abundance of ifngr1 transcripts as rapidly and effectively as actinomycin D treatment. IFNβ treatment also significantly reduced the amounts of activated RNA polymerase II (pol II) and acetylated histones H3 and H4 at the ifngr1 promoter, and the activity of an _ifngr1_-luc reporter construct in macrophages. The suppression of _ifngr1_-luc activity required an intact early growth response factor (Egr)-binding site in the proximal ifngr1 promoter. Three Egr proteins and two Egr/NGFI-A binding (Nab) proteins were found to be expressed in bone macrophages, but only Egr3 and Nab1 were recruited to the ifngr1 promoter upon IFNβ stimulation. Knockdown of Nab1 in a macrophage cell line prevented down regulation of IFNGR1 and prevented the loss of acetylated histones from the ifngr1 promoter. These data suggest that type I IFN stimulation induces a rapid recruitment of a repressive Egr3/Nab1 complex that silences transcription from the ifngr1 promoter. This mechanism of gene silencing may contribute to the anti-inflammatory effects of type I IFNs.
Keywords: Myeloid cell, Inflammation, Interferon, IFNGR, Transcription, Nab, Egr
Introduction
Type I IFNs (e.g. IFNs α and β) were originally recognized for their ability to induce an antiviral state following autocrine or paracrine signaling in fibroblasts and other cell types (1,2). Consequently, responses to type I IFNs are required for optimal host resistance in diverse viral infection models, and considerable effort has focused on understanding how these cytokines stimulate protective immune responses(1-4). However, in apparent contrast to the protective effects of type I IFNs during viral infections, these cytokines have also been found to suppress or negatively regulate inflammatory and immune responses in other disease settings. For example, responsiveness to type I IFNs reduces host resistance to a number of bacterial infections (5-7). The mechanisms responsible for these negative effects of type I IFNs have been unclear.
The modulation of host cell gene expression by type I IFNs requires binding of these cytokines to a ubiquitously expressed cell surface receptor, IFNAR (1,2). Such binding elicits signals that alter expression of hundreds of target genes (1-3,8,9). There is considerable information on how type I IFNs positively regulate gene expression, but the mechanisms they use to negatively regulate gene expression have been less clear. Recently, it was shown that the ability of type I IFNs to negatively regulate expression of certain induced genes involves recruitment of repressive epigenetic factors, such as methyltransferases (10), or histone deacetylases (HDACs) (11,12). However, it is not known if similar mechanisms contribute to the ability of type I IFNs to silence expression of basally transcribed genes.
Prior studies by our lab and others established that type I IFNs suppress myeloid cell activation during infections by certain intracellular bacteria, including Mycobacterium tuberculosis and Listeria monocytogenes(13,14). The induction of macrophage activation in these and other infections involves type II IFN, IFNγ, and plays an essential role in host resistance (6,7,15-17). Cellular responsiveness to IFNγ requires cell surface expression of a heterodimeric receptor, IFNGR, which is comprised of the IFNGR1 and IFNGR2 subunits. Our prior findings using L. monocytogenes infection showed that the reduced activation of myeloid cells corresponded with type I IFN-dependent reductions in myeloid cell surface IFNGR1 and IFNGR2, which correlated with reduced abundance of ifngr1 (but not ifngr2) transcripts (14). These findings suggest that the abundance of ifngr1 transcripts regulates cell surface IFNGR levels in mouse myeloid cells. Here, we sought to investigate how type I IFNs negatively regulate myeloid cell ifngr1 expression.
The silencing of basally transcribed genes often involves recruitment of repressive transcription factors to the target gene promoter. The early growth response (Egr) family of transcription factors comprises four members (Egr1, Egr2, Egr3, and Egr4). DNA-binding domains in these Egr proteins are formed by three zinc-finger motifs that bind to the consensus sequence CGCCCCCGC (18). Egr proteins were originally recognized for their role in the genetic regulation of cell growth and differentiation in response to extracellular stimuli, particularly in the context of the nervous system (19). They are now also known to promote expression of a diverse group of genes, including several with important immunological functions (20-24). Egr family members can also repress the transcription of certain target genes, particularly in response to external stimuli such as cytokines (25,26). The mechanisms for gene repression include interference with transcriptional activators such as Sp1 (25-29) and TATA binding protein (TBP) (30,31), and recruitment of a family of Egr corepressors known as NGFI-A binding proteins (Nab) (32,33). Egr1, Egr2, and Egr3 proteins (but not Egr4) contain a repression domain (R1) that binds to the highly conserved NCD1 domain present in both Nab family members, Nab1 and Nab2 (34-38). Nab proteins are unable to bind to DNA alone (38), and thus suppress transcription upon recruitment to a DNA-bound Egr family member (32,33,38). The repressive Egr-Nab complexes often silence or maintain repression of gene expression by recruiting factors that can induce epigenetic gene silencing, such as HDACs (33,39).
Here, we showed that type I IFN treatment rapidly silences ifngr1 transcription in mouse and human macrophages, but not T cells, and describe a mechanism contributing to this silencing. We identified putative Egr binding sites in the mouse and human ifngr1 promoters and showed that a proximal Egr site is required for silencing of ifngr1 transcription in mouse myeloid cells treated with IFNβ. Chromatin immunoprecipitation analysis further indicated that type I IFNs induce rapid recruitment of Egr3 to a region of the ifngr1 promoter containing this proximal Egr binding site in myeloid but not T cells. Recruitment of Egr3 correlated with reductions in activated RNA polymerase II (pol II) and preceded recruitment of Nab1. Nab1 recruitment coincided with and was required for deacetylation of the ifngr1 promoter and down regulation of cell surface IFNGR. These data demonstrate involvement of a Egr3/Nab1 complex in the silencing of ifngr1 transcription and down regulation of IFNGR by type I IFNs. Putative Egr binding sites were also identified in the promoters of other constitutively expressed genes known to be repressed by type I IFNs, suggesting Egr3 and Nab1 may play a general role in negative regulation of myeloid cell gene expression.
Materials and Methods
Mice
C57BL/6 mice were obtained from Jackson laboratory. IFNAR-/- crossed to C57BL/6 (Jackson Laboratory, Bar Harbor, ME) for >10 generations were previously described (14). Mice were housed in the National Jewish Health Biological Resource Center. The National Jewish Health Institutional Animal Care and Use Committee approved all studies.
Cell Culture and IFNβ Treatment
To culture BMDMs, cells were flushed from the femurs, tibias, and fibulas of mice and cultured for 6 d in BM macrophage media (DMEM supplemented with 10% FBS, 1% sodium pyruvate, 1% L-glutamine, 1% penicillin/streptomycin, 2-mercaptoethanol, plus 10% L-cell conditioned media). Media components were Gibco (Life Technologies, Carlsbad, CA). Fresh media was added at day 3 and BMDMs were used for experiments on day 7. RAW264.7 murine macrophage cells and EL4 murine T cells were cultured in DM10 media (DMEM supplemented with 10% FBS, 1% sodium pyruvate, 1% L-glutamine, 1% penicillin/streptomycin). THP-1 cells were cultured in suspension with RP10 media (RPMI supplemented with 10% FBS, 1% sodium pyruvate, 1% L-glutamine, 1% penicillin/streptomycin). 24 hours prior to experimentation, THP-1 cells were stimulated with 0.1μg/mL phorbol 12-myristate 13-acetate (P-8139; Sigma-Aldrich, St. Louis, MO) to obtain adherent cells. To obtain human peripheral blood mononuclear cells (hPBMCs), de-identified blood from donors was collected in heparin-containing vacuum tubes, and white blood cells were separated from whole blood by Ficoll-Paque gradient (Histopaque-1077; Sigma-Aldrich). Isolated cells were incubated overnight in 6-well culture plates in DMEM supplemented with human serum. BMDMs, RAW264.7, and EL4 cells were treated at various time points with 100 U/mL murine IFNβ (PBL Interferon Source, Piscataway, NJ). THP-1 cells and hPBMCs were treated at various time points with 100 U/mL human IFNβ (PBL Interferon Source).
Flow Cytometry
BMDMs and adherent cell lines were lifted from culture dishes with cold PBS. Adherent hPBMCs were lifted from culture dishes with cold PBS and added to non-adherent cells. Murine Fc receptors were blocked before staining using supernatant from hybridoma 2.4G2 (rat anti-CD16/32), and human Fc receptors were blocked using pooled human serum in PBS. To detect murine IFNGR1, cells were stained with biotinylated antibodies to IFNGR1/CD119 (BD Biosciences, San Jose, CA), followed by streptavidin-APC secondary antibody (eBiosciences, San Diego, CA). To detect human IFNGR1, cells were stained with biotinylated antibodies to IFNGR1/CD119 (Caltag Labs; Life Technologies, Carlsbad, CA), followed by strepavidin-APC secondary. Primary human T cells and monocytes were detected using CD3-FITC and CD14-PE antibodies (eBiosciences) respectively. To detect MHCI, EL4 cells were stained with anti-H2Db- -FITC (eBiosciences). All antibodies were diluted in surface staining buffer (PBS/1%BSA/0.01% NaN3). The mean fluorescence intensities (MFIs) for each of three treated samples per time point were normalized to mean MFI for three untreated samples using the following formula: Relative Surface Staining = (MFI treated)/(MFI untreated). For statistical analyses, we pooled the relative MFI values from three separate experiments each using at least three control and three treated samples.
Real-time Quantitative PCR
Preparation and analysis of samples for quantitative RT-PCR (qPCR) was described previously (14). Briefly, 9×106 BMM or RAW264.7 cells were distributed into 3 wells of a 6-well plate (Cell Star®; Sigma-Aldrich), for each treatment time point. Cells were pooled from 3 wells and the RNA was isolated using the RNeasy kit (QIAGEN, Valencia, CA). Complementary DNA synthesis was conducted with 1μg of total RNA using Oligo(dT) primers (Promega, Madison, WI). Commercial oligonucleotide primer sets from Applied Biosystems (Life Technologies, Carlsbad, CA) were used to quantify mouse ifngr1 and human ifngr1 transcripts, and the primer set, sense 5′ – GCACTGGGTGGAATGAGACTATTG – 3′ and antisense 5′ – GACCTGTCAGTTGATGCCTCAGAA – 3′, was used to quantify mouse IFNβ transcripts. Real-time quantitative PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) and an ABI PRISM 7300 Sequence detector. The equation used to determine relative transcript abundance = 2(x002C6)(-1*((IFNGR1 Ct – Mean GAPDH Ct) – (Mean Unstimulated dCT))). Transcript half-life calculated using the equation: half-life=(elapsed time*log2)/(log (beginning amount/ending amount)), and STD determined by pooling values from three separate experiments.
Chromatin Immunoprecipitation
The chromatin immunoprecipitation (ChIP) experiments were performed according to the protocol provided for the Active Motif ChIP Express kit (Active Motif, Carlsbad, CA). Briefly, after treatment with IFNβ, RAW 264.7, EL4, and BMDMs were cross-linked with 1% methanol-free formaldehyde for 7 minutes at room temperature. Fixed cells (7×106 in 300μL) were resuspended in kit lysis buffer plus protease inhibitors and incubated at 30 minutes at 4°C. Cell nuclei were pelleted and resuspended in 300 μL of kit shearing buffer plus protease inhibitors. A Covaris S2 sonicator was used to shear the samples, using a 27-cycle treatment. To ensure that the shearing process consistently produced fragments of 200-500bp, 5 μL of all supernatants were run on a 1% agarose gel (Fig. S3A). An additional 10 μL of supernatant was saved for use as total input DNA. All samples were stored at -80°C until use. Immunoprecipitations were performed with protein G magnetic beads overnight at 4°C, using an estimated 7 μg of sheared chromatin and antibodies specific for pS5-RNA pol II (ab5131; Abcam, Cambridge, MA), acetyl-Histone H3 (39139, Active Motif), pan-acetyl-Histone H4 (39925, Active Motif), Total Histone H3 (ab1791; Abcam) Egr1 (#4153, Cell Signaling, Danvers, MA), Egr2 (PRB-236P; Covance, Princeton, NJ), Egr3 (ab75461, Abcam), and Nab1 (NBP1-71838; Novus, Littleton, CO). Equal amounts of chromatin were also immunoprecipitated using equivalent amounts of a control IgG antibody (ab46540, Abcam). Following immunoprecipitation, beads were washed, and the immune complexes were eluted with kit elution buffer. Reverse cross-linking buffer was added to each eluted supernatant at 1:1, and the samples and input DNA were heated for 1 hour at 95°C. After treatment with 10μg/mL proteinase K for 1 hour at 37°C, samples were purified using QIAGEN PCR purification kit, then used for qPCR. The promoter primer sequences used to analyze chromatin immunoprecipitations for pS5-RNA pol II, acetylated histone H3, total histone H3, and acetylated histone H4 were sense 5′-GCAATTGTGTCCCTCGCGCAGGAATGGGCC-3′ and antisense 5′-GCTCGTCAAAGCTCCACTCCCGACC-3′. The primer sequences used to analyze all Egr and Nab immunoprecipitations were sense 5′-CCTCAGGCTAGTCCACCCCTTCTCC-3′ and antisense 5′-GGAGGCGTGTCTTGGCGGG-3′. Real-time quantitative PCR was performed using an Absolute QPCR SYBR Green PCR ROX Mix (Thermo, Waltham, MA) and an ABI PRISM 7900 Sequence detector. The equation used for determining percent input = 2(x002C6)(Input Ct – IP Ct). The equation used for determining Fold Enrichment Over Isotype = 2(x002C6)(-1*((IP Ct – Input Ct) – (Isotype Ct – Input Ct))).
Luciferase Constructs
The wildtype ifngr1 promoter luciferase reporter construct (IFNGR1pr-luc) was created by amplification of the proximal portion of the ifngr1 promoter from -2320 to +1 using C57BL/6 genomic DNA as template. Primers were: sense 5′-GGGGTACCAGACTAAGCCAATCCTGCCCCACC-3′ and antisense 5′-CAGTCTCCACAGGGAGCGCGTCCTGAGCTCGG-3′. KpnI and XhoI restriction sites were included in the primers and used to clone the amplified sequence into the multiple cloning site of pGL3-Basic (Promega). The hygromycin resistance gene from pGL4.15[Hygro] (Promega) was subsequently cloned into the BamHI and SalI sites of pGL3 to generate IFNGR1pr-luc. For mutagenesis, the Stratagene QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) was used. The primer set for mutagenesis of the Egr site in mEgr-luc was: sense 5′-CCAACTTAGTGAAACTTCTCAGTACACGCAGCG-3′ and antisense 5′-GGTTGAATCACTTTGAAGAGTCATGTGCGTCGC-3′. The two independent IFNGR1pr-luc and mEgr-luc cell lines used for experiments were obtained from two separate transfections.
Creation of Putative Transcription Factor Binding Maps
To construct a putative transcription factor binding site map of the mouse and human ifngr1 promoters, the DNA sequences for the first 2320 base pairs were entered into the TFSearch program (40). Results were limited to consensus sequences with a score of 85.0 or better according to the program's algorithm. The presence and location of these binding sites were confirmed using TESS (41) and ENCODE (42).
Reporter Cell Lines and Luciferase Assay
To make stable cell lines, 5×106 RAW 264.7 cells were transfected with 1μg of linearized IFNGR1pr-luc or mEgr-luc plasmid by electroporation (250 V, 950 μF) using a BioRad electroporator. Following electroporation, the cells were resuspended in 10 mL of DM10 and plated at 100 μL/well in a 96-well plate. The transfected cells were selected for using 250 μg/mL hygromycin. To determine the luciferase activity, expanded cell lines were plated at 1×105 in a 24-well plate and stimulated with 100 U/mL of IFNβ at various time points. Following stimulation, lysates were harvested using lysis buffer from the Promega Luciferase Assay System. Luminescence was measured with a Synergy 2 plate reader (BioTek, Winooski, VT).
Western Blots
BMDM or RAW cell cultured monolayers were lysed in 100 μL of 1× SDS-PAGE buffer (0.0625 M Tris-Cl, pH 6.8/2% SDS/10% glycerol/5% 2-ME/0.01% Bromophenol Blue) containing HALT® protease inhibitors (Thermo) at 1× concentration. Cell lysates were scraped from plates and frozen at -20°C. The lysate volume corresponded to 3.5×105 cells for all Egr family members and 2×105 cell equivalents per lane for all other proteins. Proteins were transferred to nitrocellulose and probed with antibodies to total STAT1 (#9172; Cell Signaling), STAT1pY701 (#9167; Cell Signaling), Egr1 (#4153; Cell Signaling), Egr2 (PRB-236P; Covance), Egr3 (ab75461; Abcam), Nab1 (NBP1-71838; Novus), and Nab2 (sc-22815; Santa Cruz Biotechnology, Santa Cruz, CA). As loading control, blots were stripped and re-probed with an anti-Actin antibody (MAB1501; Millipore, Billerica, MA).
Stable Knockdown of Egr and Nab Family Members
Sigma Aldrich MISSION® shRNA constructs Egr3 (TRCN# 96139), Nab1 (TRCN# 96134), and Nab2 (TRCN# 96349) were obtained from the University of Colorado Cancer Center (University of Colorado-Anschutz Medical Campus, Aurora, CO). To make stable cell lines, each construct was linearized and 1μg was used for transfection of 5×106 RAW 264.7 cells. Transfected cells were selected using 5 μg/mL Puromycin. The two independent Nab1-KD and Nab2-KD cell lines used for experiments were obtained from two separate transfections.
Results
Type I IFNs decrease surface expression of IFNGR1 on mouse and human macrophages
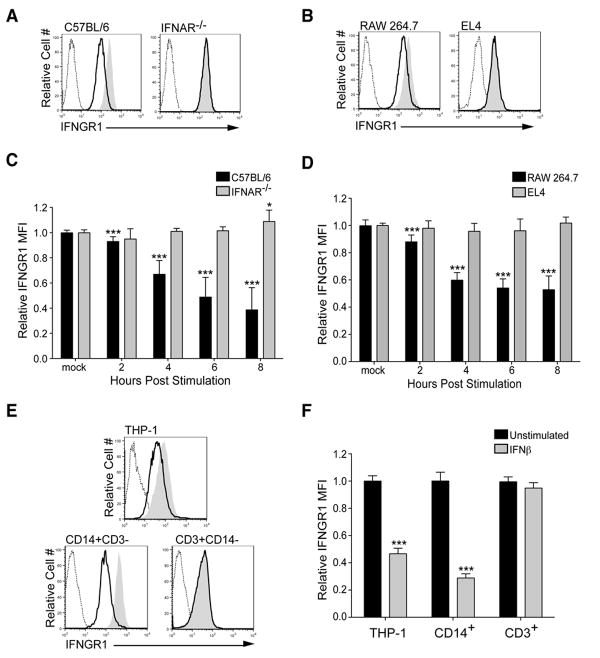
Consistent with our previous studies (14), we observed that cell surface IFNGR1 staining was reduced at 6 h after IFNβ treatment (100 U/mL) in bone marrow-derived macrophages (BMDMs) from wt C57BL/6, but not IFNAR-/-, mice (Fig. 1A). The intensity of IFNGR1 staining on cells from IFNGR1-/- mice was similar to that of the secondary reagent alone, demonstrating specificity of the stain (Fig. S1A). The ability of IFNβ treatment to reduce IFNGR1 staining was also observed with RAW264.7 macrophages (Fig. 1B). However, down regulation of IFNGR1 was not seen in mouse EL4 T cells (Fig. 1B), despite the fact that they responded to IFNβ as judged by upregulation of MHC class I (Fig. S1B), and phosphorylation of STAT1 (Fig. S1C). Increasing the concentration of IFNβ used for stimulation did not further reduce IFNGR1 down regulation for either BMDMs or RAW 264.7 cells (Fig. S1D). We also observed similar reductions in IFNGR1 staining when BMDMs were treated with IFNα (Fig. S1E). To facilitate statistical comparisons of cell surface IFNGR1 staining across multiple experiments and time points, the mean fluorescence intensities (MFI) of staining on IFNβ-treated cells were normalized to those of mock-treated control cells. When normalized data from three independent experiments were plotted and analyzed, the reduction in cell surface IFNGR1 staining in both wt BMDM and RAW264.7 cells was found to be significant as early as 2 h post-stimulation (hps) with IFNβ (Fig. 1C,D). These data indicate that both primary mouse macrophages and RAW264.7 cells respond to type I IFNs by rapidly down regulating cell surface IFNGR1, though the reduction in IFNGR1 staining seen with RAW264.7 cells (40-50%) was consistently less than that seen with wt C57BL/6 BMDMs (55-65%).
Figure 1. Type I IFNs reduce cell surface IFNGR1 on mouse and human macrophages.
Cells were treated or not with 100 U/mL of IFNβ, then stained to quantify cell surface IFNGR1. Histograms (A, B, E) illustrate reduced cell surface IFNGR1 staining typically seen on macrophages after 6 h of IFNβ treatment (black lines). Control histograms included unstimulated cells (gray shading) and cells stained with secondary reagent alone (dashed line). Graphs (C, D, F) depict mean fluorescence intensity (MFI) values normalized to those of the respective untreated cells (relative MFI=MFI from treated sample/average of MFI from untreated sample). Cells used included (A, C) live-gated C57BL/6 and B6.IFNAR-/- mouse BMDMs, (B, D) mouse RAW 264.7 macrophages and EL4 thymoma cells, and (E, F) THP-1 cells and gated CD14+ or CD3+ human PBMCs. Each bar in the graphs represents the mean ± STD of the pooled values for each condition from a total of at least three independent experiments. Statistical significance was determined using an unpaired t-test with * ≤ 0.05, **≤ 0.01, *** ≤ 0.001.
To determine whether human myeloid cells also down regulate IFNGR1 in response to type I IFNs, cell surface IFNGR1 staining was evaluated in human THP-1 macrophage-like cells, and in primary peripheral blood mononuclear cells (PBMCs). Similar to mouse myeloid cells, IFNGR1 staining was selectively and significantly reduced in THP-1 cells and CD3-CD14+, but not CD3+CD14-, PBMCs at 6 h post-stimulation (hps) with 100 U/mL of human IFNβ (Fig. 1E,F; gating shown in Fig. S1F). The similar effect of type I IFNs on IFNGR expression in mouse and human myeloid cells is consistent with the known ability of type I IFNs to suppress activation of both mouse and human macrophages (43,44), and the ability of IFNβ treatment to ameliorate mouse and human neuroinflammatory diseases (45). In addition, the lack of IFNGR1 down regulation in mouse EL4 T cells (Fig. 1B,D), mouse splenic CD3+ T cells (14), and human CD3+ PBMCs (Fig. 1E,F) suggests that this conserved response to type I IFNs primarily occurs in myeloid cells.
Type I IFNs silence transcription of ifngr1
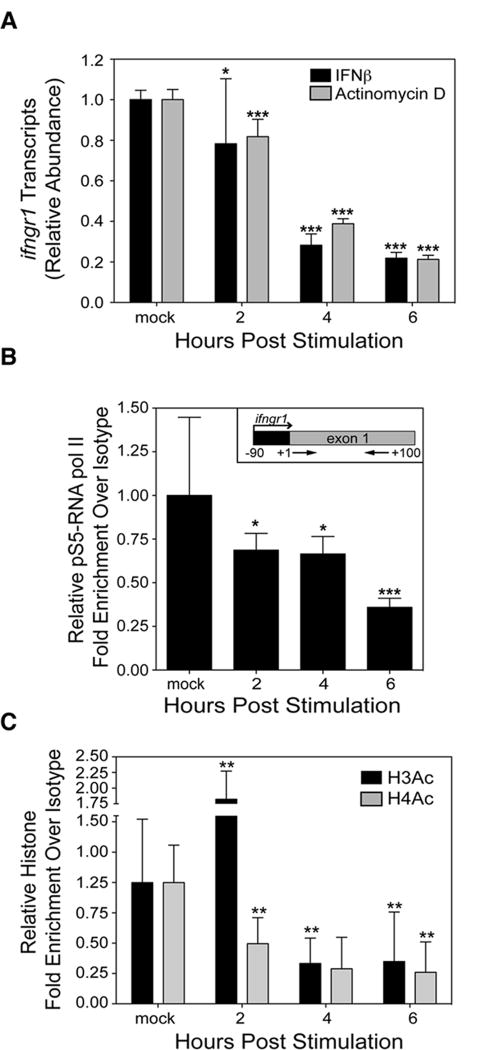
The observed reductions in cell surface IFNGR1 staining in mouse macrophages correlated with similar magnitude reductions in the abundance of ifngr1 transcripts as measured by quantitative real-time RT-PCR (qPCR) (Figs. 2A and S2A). The effects of IFNβ on ifngr1 transcript abundance were significant as early as 2 hps. A similarly rapid decrease in transcript abundance was seen upon chemical inhibition of the transcription machinery using inhibitory concentrations of Actinomycin D (Fig. 2A). The Actinomycin D concentration used (1μg/mL) did not affect BMDM viability, but was sufficient to block de novo expression of IFNβ in BMDM treated with poly I:C (Fig S2B). Based on the changes in transcript abundance over time, we calculated the half-life of ifngr1 mRNA in the BMDM to be 2.74 h (± 0.257 STD) following IFNβ treatment, and 2.70 h (± 0.169 STD) following Actinomycin D treatment. These results suggest that IFNβ treatment rapidly and fully silences de novo transcription of ifngr1.
Figure 2. Type I IFNs repress transcription of_ifngr1_.
(A) Total RNA was isolated from BMDMs treated or not with 100 U/mL of IFNβ or 1μg/mL of Actinomycin D. Relative change in normalized expression between treated and untreated BMDMs was quantified by qRT-PCR. Graph depicts ifngr1 transcript abundance values normalized to those of the respective untreated cells (relative ifngr1 transcript abundance=transcript abundance from treated sample/average abundance from untreated sample). ChIP assays for (B) pS5-RNA pol II and (C) H3Ac and H4Ac in BMDMs treated or not with 100 U/mL IFNβ. Primers that amplify 100 base pairs within exon 1 of ifngr1 were used to quantify promoter-associated immunoprecipitated chromatin (B-inset). Graphs depict fold enrichment over isotype values normalized to those of the respective untreated cells (relative fold enrichment over isotype=fold enrichment from treated sample/average fold enrichment from untreated sample). Each bar in the graphs represents the mean ± STD of the pooled values for each condition from a total of at least three independent experiments. Statistical significance was determined using an unpaired t-test with * ≤ 0.05, **≤ 0.01, *** ≤ 0.001. Representative data showing shearing efficiency and percent input graphs for each of these ChIP assays are in Fig. S3.
As an independent method to confirm whether IFNβ treatment silenced new ifngr1 transcription, we used chromatin immunoprecipitation (ChIP) to evaluate accumulation of active RNA pol II to a 100 base pair region adjacent to the predicted transcriptional initiation start site (TSS) of the ifngr1 gene (Fig. 2B, inset). Chromatin isolated from BMDMs 0, 2, 4, and 6 hps with IFNβ was immunoprecipitated with an antibody specific for a phosphorylated isoform of the RNA pol II complex (pS5-RNA pol II). This antibody specifically recognizes RNA pol II phosphorylated at serine 5 within the C-terminal domain heptapeptide repeat, a modification that is necessary for the initiation of transcription (46). The relative abundance of promoter DNA immunoprecipitated with anti-pS5-RNA pol II was determined using qPCR (Fig. 2B). The data showed that IFNβ treatment significantly reduced association of activated RNA pol II with this region of the ifngr1 promoter by 2 hps. Consistent with the conclusion that IFNβ prevents new ifngr1 transcription, the association of activated RNA pol II and the ifngr1 promoter remained low for at least 6 hps.
The silencing of ifngr1 transcription suggested that type I IFN stimulation might alter the epigenetic structure of the ifngr1 promoter. Epigenetic changes associated with transcriptionally inactive condensed chromatin include deacetylation of lysine residues in histones H3 and H4 (47,48). We used antibodies specific for lysine N-acetylation of histones H3 (H3Ac) and H4 (H4Ac) to conduct ChIP assays on BMDMs stimulated with IFNβ for 0, 2, 4, and 6 hrs. Type I IFN stimulation induced a transient increase in H3Ac signal at 2 hps, presumably reflecting rapid changes in the structure of the promoter. However, starting at 4 hps the stimulation significantly reduced acetylation of histones H3 and H4 at the ifngr1 promoter (Fig. 2C). Together, the pS5-RNA pol II, H3Ac, and H4Ac ChIP results indicate that stimulation of primary macrophages and cell lines with type I IFNs induces a rapid and sustained silencing of ifngr1 transcription.
An Egr site is required for silencing of transcription from the proximal ifngr1 promoter
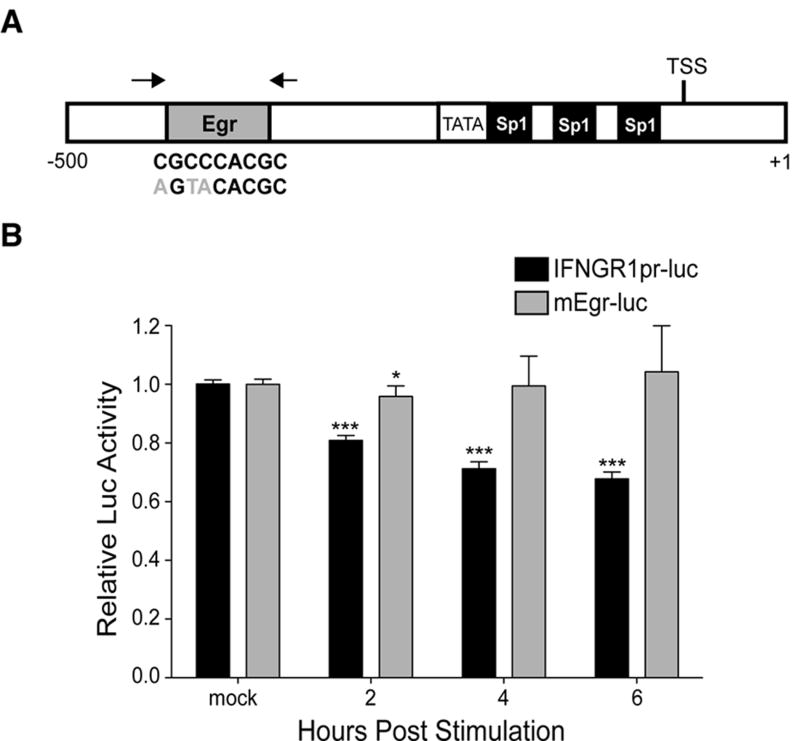
To determine whether the proximal ifngr1 promoter was responsive to transcriptional repression by type I IFNs, we stably transfected RAW 264.7 macrophages with a luciferase construct containing the 2,320 base pair region from the proximal mouse ifngr1 promoter (IFNGR1pr-luc). The type I IFN-responsive region of the mouse promoter contained three putative Sp1 binding sites adjacent to a TATA box, and a putative Egr binding site near the ifngr1 transcriptional start site (Fig. 3A). Putative binding sites for Sp1 and Egr1 were also present in the proximal region of the human ifngr1 promoter (Fig. S4A). A previous study implicated Sp1 as a positive regulator of ifngr1 transcription (49), and others suggested that Egr family members repress Sp1-dependent transcriptional activity (26-29). We thus engineered a mutated version of IFNGR1pr-luc with three point mutations at the putative Egr-binding site (Fig. 3A). The selected mutations were previously shown to disrupt the ability of DNA to bind Egr1 protein (50). The IFNGR1pr-luc reporter cells experienced a rapid and significant decrease in luciferase activity following treatment with IFNβ, demonstrating a decrease in ifngr1 promoter activity (Fig 3B). In contrast, the reporter activity in RAW 264.7 cells stably transfected with the mutated construct (mEgr-luc) were not affected by treatment with IFNβ. These data suggested that the presence of a functional Egr binding site is essential for the silencing of transcription from the proximal ifngr1 promoter.
Figure 3. An Egr site the proximal ifngr1 promoter confers sensitivity to transcriptional silencing by type I IFNs.
(A) Schematic of proximal region of mouse ifngr1 promoter. Black letters below Egr site represent DNA binding consensus sequence, and gray letters specify three inserted point mutations. Black arrows denote Egr3 and Nab1 primers used in ChIP assays. (B) Wildtype (IFNGR1pr-luc) and mutated (mEgr-luc) ifngr1 promoter constructs were stably transfected to generate RAW 264.7 luciferase reporter cells. Cells were stimulated or not with 100 U/mL of IFNβ. Graph depicts luciferase activity values normalized to those of the respective untreated cells (relative luc activity=luc activity from treated sample/average of luc activity from untreated sample). Values were derived from three separate experiments using at least 2 independently transfected IFNGR1pr-luc or mEgr-luc cell lines. Each bar in the graph represents the mean ± STD, and statistical significance was determined using an unpaired t-test with * ≤ 0.05, **≤ 0.01, *** ≤ 0.001.
Egr3 and Nab1 are recruited to the proximal Egr site of the ifngr1 promoter in response to type I IFN stimulation
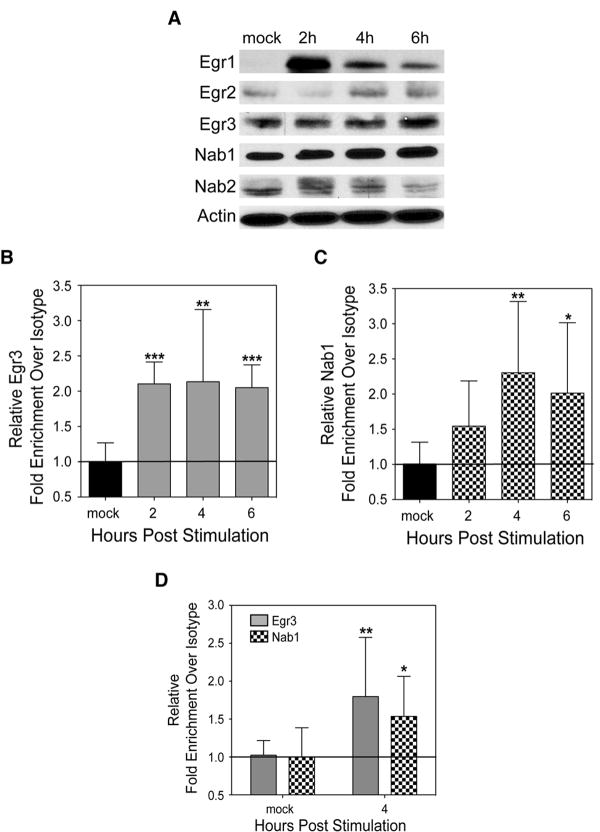
Previous studies reported expression of Egr1, Egr2, and Egr3 mRNAs in BMDMs (51). We thus used immunoblots to investigate expression of these Egr proteins in our BMDMs, and whether IFNβ treatment affected such expression (Fig. 4A). Egr1 protein was present at low or undetectable levels in the unstimulated cells, but was strongly induced within 2 hps with IFNβ. Egr2 protein was present in unstimulated cells and at late times post-stimulation, but appeared to undergo a transient depletion at 2 hps. Egr3 was readily detected in unstimulated cells and its expression was unaffected by type I IFN stimulation. We next used ChIP to ask whether Egr1, 2, or 3 associated with the proximal Egr site in the ifngr1 promoter before or after type I IFN stimulation of BMDMs. An initial percent input analysis revealed that Egr3 was the primary Egr family member recruited to the ifngr1 promoter following type I IFN stimulation (Fig. S4B). Moreover, the kinetics of Egr3 recruitment to the proximal Egr site (Fig. 4B) correlated well with loss of activated RNA pol II recruitment to the ifngr1 transcriptional start site (Fig. 2B), and reduced ifngr1 transcript abundance (Fig. 2A). These events all preceded the reduced cell surface IFNGR1 staining in IFNβ-treated BMDMs (Fig. 1C).
Figure 4. Egr3 and Nab1 are recruited to the proximal Egr site of the_ifngr1_promoter in response to type I IFN stimulation.
(A) Representative Western Blots of Egr1, Egr2, Egr3, Nab1, and Nab2 to determine protein expression in BMDMs stimulated or not with 100 U/mL IFNβ. Anti-actin antibody was used to determine that equivalent protein concentrations were loaded in each lane. ChIP assay for (B) Egr3 and (C) Nab1 in BMDMs or (D) in RAW 264.7 cells stimulated or not with 100 U/mL of IFNβ. Primers denoted in Fig. 3A were used for all qPCR analysis. Graphs depict fold enrichment over isotype values normalized to those of the respective untreated cells, calculated as in Fig 2B. Each bar in the graphs represents the mean ± STD of pooled values from a total of at least three independent experiments. Statistical significance was determined using an unpaired t-test with * ≤ 0.05, **≤ 0.01, *** ≤ 0.001.
Nab proteins are known to interact with DNA-bound Egr family members as corepressors of target gene expression (32,33). We thus asked whether Egr3 binding to the ifngr1 promoter might recruit Nab proteins to mediate silencing of ifngr1. Both Nab1 and Nab2 showed detectable levels of protein expression in unstimulated BMDMs (Fig. 4A). Stimulation with IFNβ modestly reduced Nab2 expression at late times (6 hps), but did not alter Nab1 protein amounts. ChIP assays were thus used to evaluate whether Nab1 was recruited to the ifngr1 promoter. By 4 hps, Nab1 was detected at a region of the ifngr1 promoter containing the Egr site (Figs. 4C; S4C). This result established that IFNβ treatment of BMDMs stimulates the recruitment of Nab1 subsequent to recruitment of Egr3 (Fig 4B), and at a time when there is deacetylation of the ifngr1 promoter (Fig 2C). Recruitment of Egr3 and Nab1 to the ifngr1 promoter also occurred in RAW 264.7 cells following stimulation with IFNβ for 4 hrs (Fig. 4D). However, Egr3 was not recruited to the ifngr1 promoter in EL4 cells (Fig. S4D), suggesting that these events selectively occur in myeloid cells as they silence ifngr1 transcription.
Nab1 knockdown prevents ifngr1 promoter deacetylation and down regulation of IFNGR1 in response to type I IFN stimulation
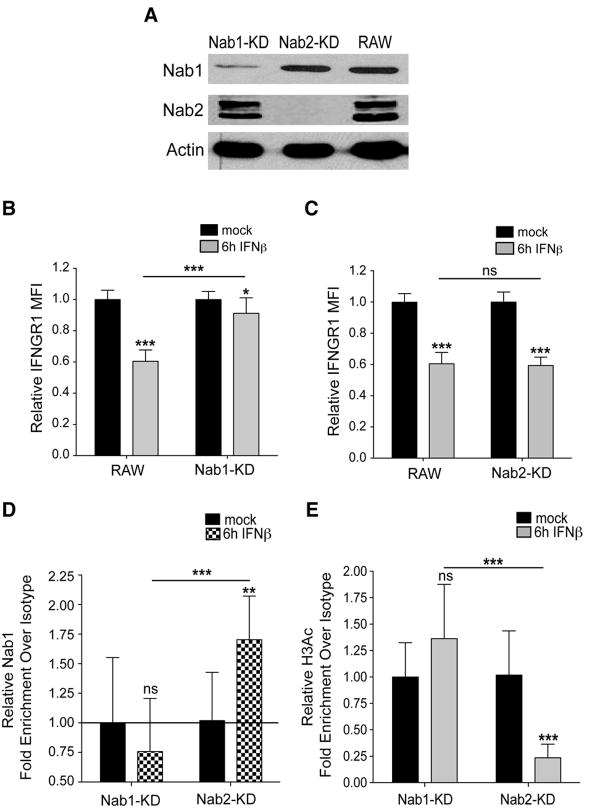
Repeated attempts to knockdown Egr3 expression in RAW264.7 cells failed, suggesting this factor may be important for macrophage viability. However, we were successful in generating knockdowns of Nab1 and Nab2. Independent, stably transfected cell lines were developed using constructs encoding shRNAs that targeted the nab1 and nab2 genes. These lines respectively showed large decreases in Nab1 (Nab1-KD) and almost complete elimination of Nab2 (Nab2-KD) protein expression when compared to control RAW 264.7 cells (Fig. 5A). In response to treatment with IFNβ, cell surface IFNGR1 staining was barely reduced in the Nab1-KD cells (Fig. 5B), whereas IFNGR1 staining in the Nab2-KD cells was reduced to a similar extent as seen in control RAW 264.7 cells (Fig. 5C). These data established that Nab1, but not Nab2, plays a role in the suppression of IFNGR1 expression.
Figure 5. Knockdown of Nab1 prevents down regulation of IFNGR1 and deacetylation of_ifngr1_ promoter by type I IFNs.
(A) Representative immunoblots to determine expression of Nab1 and Nab2 proteins in RAW 264.7 cell lines with or without stable knockdown of Nab1 (Nab1-KD) or Nab2 (Nab2-KD). Anti-actin antibody was used to confirm equivalent protein loading. Control RAW264.7 and (B) Nab1-KD or (C) Nab2-KD cell lines were treated or not with 100 U/mL of IFNβ then stained to quantify cell surface IFNGR1. ChIP assays for (D) Nab1 and (E) H3Ac in Nab1-KD or Nab2-KD cell lines stimulated or not for 6 h with 100 U/mL of IFNβ. Primers denoted in Fig.3A were used for Nab1 ChIP qPCR analysis, and primers denoted in Fig. 2B were used for H3Ac qPCR analysis. Graphs depict MFI (B, C) or fold enrichment over isotype (D, E) values normalized to those of the respective untreated cells, calculated as in Fig. 1C and Fig. 2B, respectively. Each bar in the graphs represents the mean ± STD of the pooled values for each condition from a total of at least three independent experiments. Statistical significance was determined using an unpaired t-test with * ≤ 0.05, **≤ 0.01, *** ≤ 0.001.
Nab proteins were previously shown to interact with complexes that actively deacetylate histones to silence gene expression (33,39). We thus asked whether reducing the recruitment of Nab1 to the ifngr1 promoter could prevent the histone deacetylation associated with silencing of ifngr1 transcription. ChIP for Nab1 and H3Ac was performed on Nab1-KD and Nab2-KD cell lines. In contrast to Nab2-KD cells, recruitment of Nab1 to the infgr1 promoter was not detectable in the Nab1-KD cells after 6 h of stimulation with IFNβ (Fig. 5D). This finding indicated that the knockdown efficiency in the Nab1-KD cells was adequate to prevent detectable Nab1 recruitment and that Nab2 knockdown did not prevent this recruitment. Furthermore, this lack of Nab1 recruitment was associated with a failure of IFNβ stimulation to induce deacetylation of histone H3 in Nab1-KD, but not Nab2-KD, macrophages (Fig. 5E). Thus, Nab1 recruitment is necessary for the histone deacetylation associated with silencing of the ifngr1 promoter in response to type I IFN stimulation.
Discussion
Although IFNβ has been widely used in treatment of multiple sclerosis and is known to impair resistance to certain bacterial infections, the mechanisms through which type I IFNs suppress inflammatory responses remain poorly understood. Our findings here provide further insight into this mystery by showing how type I IFNs down regulate expression of the IFNGR. Reduced IFNGR expression is known to correlate with reduced myeloid cell responsiveness to the pro-inflammatory cytokine IFNγ (14), and thus may itself contribute to the anti-inflammatory effects of type I IFNs. In addition, the mechanism suggested by our findings here may also contribute to the silencing of other genes involved in the pro-inflammatory responses of myeloid cells.
Transcripts for ifngr1 and ifngr2 are constitutively present in a variety of tissues, and functional IFNGRs are constitutively present at the surface of diverse cell types (2,52,53). However, variations in ifngr1 and ifngr2 expression occur in lymphoid and myeloid cells. Early studies established that the expression of IFNGR2 is selectively down regulated in Th1-type T cells (54,55), presumably to protect them from killing by the IFNγ they produce. IFNGR1 is not down regulated in T cells, but we recently reported that both IFNGR1 and IFNGR2 are lost from the cell surface of myeloid and B cells responding to type I IFNs (14). The loss of cell surface IFNGR in myeloid cells, which was associated with reduced abundance of ifngr1 (but not ifngr2) transcripts, correlated with reduced responsiveness to IFNγ and increased susceptibility to infection by the bacterium L. monocytogenes(14). This previous work indicated that type I IFN stimulation reduces both cell surface IFNGR1 and total cellular IFNGR1 in myeloid cells exposed to type I IFN (14). Moreover, the loss of cell surface IFNGR1 occurred with a half-life of ∼4h, matching the previously measured half-life of IFNGR1 protein (56). Silencing of ifngr1 transcription and the consequent reduction in cell surface IFNGR1 has also been observed in breast cancer cells, which enables the tumor cells to evade killing by IFNγ (57). These findings together argue that constitutive transcription of ifngr1 is necessary to maintain cell surface expression of the IFNGR, and that silencing of new ifngr1 transcription is sufficient to reduce sensitivity of myeloid and other non-lymphoid cell types to IFNγ.
Regarding the mechanism of ifngr1 silencing in myeloid cells, our studies here showed that an Egr binding region in the proximal ifngr1 promoter is responsive to type I IFNs. The type I IFN-responsive Egr site in the proximal ifngr1 promoter is near sites for binding of the Sp1 transcription factor. Sp1 promotes transcriptional activation through direct interaction with factors important for assembly of the RNA pol II complex (58), and has been shown to promote basal transcription of human ifngr1(49). It has also previously been established that Egr family members can interfere with Sp1 binding (26-28), and/or assembly of the RNA pol II transcriptional complex (30,31). Hence, we speculate that the binding of Egr3 may initially impair new transcription of ifngr1 by blocking Sp1 binding to the promoter. Consistent with this model, decreased binding of Sp1 to the ifngr1 promoter was associated with decreased transcription of ifngr1 in THP-1 cells infected with M. tuberculosis(59), and in breast cancer cells (57).
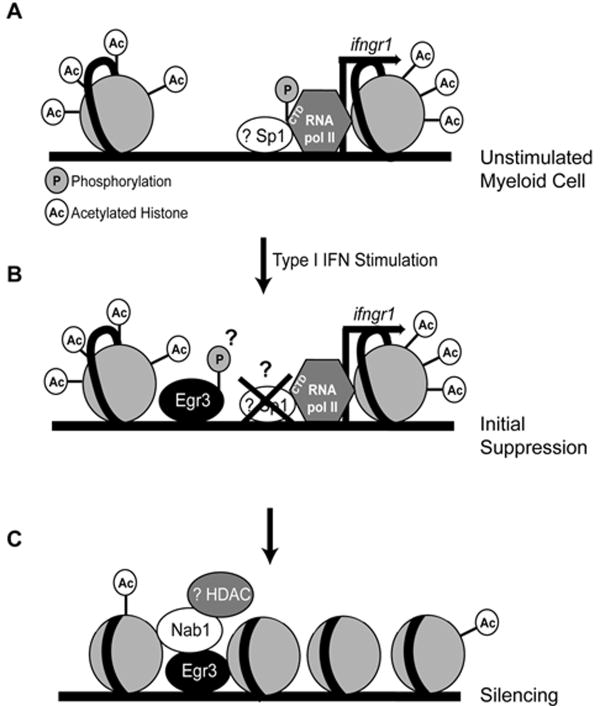
Prolonged silencing of active promoters often involves recruitment and activity of histone deacetylase enzymes (HDACs) by DNA-bound repressors (60). We found that binding of Egr3 to the ifngr1 promoter preceded recruitment of Nab1 and loss of acetylated histones H3 and H4. Nab family members have been shown to repress gene transcription by interacting with HDACs (33,39), and our data reveal that Nab1 knockdown largely prevents deacetylation of histone H3 at the ifngr1 promoter and IFNGR1 down regulation. Thus, we speculate that the prolonged and complete silencing of ifngr1 in myeloid cells involves the formation of an Egr3/Nab1 complex that fosters the recruitment of HDACs to deacetylate the ifngr1 promoter (Fig. 6).
Figure 6. Model for silencing of ifngr1 transcription by type I IFNs.
Schematic of observed and hypothesized (indicated by ?) events that mediate silencing of ifngr1 transcription by type I IFNs. Model represents factors present on the ifngr1 promoter in (A) unstimulated BMDMs, and following stimulation with type I IFNs for (B) 2 h or (C) 4-6 h.
Microarray analysis has shown that numerous basally active genes are rapidly repressed by type I IFN in BMDMs (9). The affected genes include several that code for proteins known to be associated with an activated macrophage state, such as ICAM-1, IL-1β, Jagged-1, and IL-12p40. The promoters for these four genes, and numerous other genes repressed by type I IFNs in myeloid cells, contain Egr sites. Thus, the down regulation of these genes may utilize a similar mechanism to that proposed here for ifngr1. In addition, such down regulation could arguably have anti-inflammatory effects. For example, the induction of Jagged-1 has recently been shown to be important for enhancing the responsiveness to IFNγ (61), suggesting that its down-regulation by type I IFN may synergize with the suppressive effects of IFNGR down regulation. Likewise, reductions in IL-12p40 would be expected to reduce the induction of IFNγ (and IL-17) production by T cells. Interestingly, the ifngr1 silencing in breast cancer cells (which is not known to require type I IFNs) was previously shown to involve the transcription factor AP-2α (57), rather than Egr3. A search of the ImmGen database showed that AP-2α is not expressed in immune cells (62), and our data here indicated that IFNβ stimulation failed to stimulate recruitment of Egr3 to the ifngr1 promoter in EL4 T cells. Thus, Egr3 appears to selectively participate in the silencing of ifngr1 transcription that is triggered by type I IFNs in myeloid cells. The observation that there are distinct mechanisms to permit repression of ifngr1 or ifngr2 transcription in myeloid, lymphoid, and other cell types underscores the biological importance of regulating responsiveness to IFNγ, and raises the possibility of therapies that selectively increase or reduce such responsiveness in specific cell types.
It remains unclear how stimulation with type I IFNs induces the rapid recruitment of Egr3/Nab1 to the ifngr1 promoter and why only these family members are recruited. As shown here, we failed to observe significant induction of Egr3 or Nab1 expression in response to type I IFN stimulation. We have also failed to see increased nuclear localization of Egr3 in stimulated macrophages (not shown). We thus hypothesize that recruitment of Egr3 is triggered by phosphorylation or other post-translational modification of Egr3 or an associated factor. Multiple studies have demonstrated post-translational modifications that alter the activity of Egr1 in response to exogenous stimuli (63-66). In addition, one study suggests that different modifications to the phosphorylation state of Egr1 alter whether this factor acts as an inducer or repressor of transcription at the same promoter (65). Phosphorylation or other modifications of Egr1 and 2 might also conceivably prevent their recruitment to the proximal ifngr1 promoter. Further investigation is needed to resolve these issues and determine more precisely how type I IFN stimulation regulates the association of Egr3 with the ifngr1 promoter.
Polymorphisms in the ifngr1 promoter are known to correlate with susceptibility to diverse diseases such as Leishmaniasis, tuberculosis, leprosy, and hepatitis infections (67-70). Suppression of ifngr1 transcription by type I IFNs also correlates with impaired macrophage activation, and compromised resistance to diverse bacterial infections (5-7). Yet, surprisingly little is known about the transcriptional regulation of ifngr1. Our efforts here have provided new insight into the mechanisms for silencing of ifngr1 expression by type I IFNs and thus the regulation of IFNGR expression in myeloid cells. These findings may also prove relevant for silencing of other myeloid cell gene expression by type I IFNs. Ultimately, such efforts may reveal new strategies for improving host resistance to a variety of infectious diseases.
Supplementary Material
1
2
3
4
5
Footnotes
1
Supported by the National Institutes of Health research grants R56AI65638, RO1AI065638, R21AI102264, and R21AI103782 to LLL. BPOC received support from the Walter Scott Foundation. SJK received support from a Cancer Research Institute Training Grant. Core facilities used in these studies received support from P01AI22296 and P30CA046934.
References
- 1.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nature reviews Drug discovery. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annual review of biochemistry. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Hwang SY, Hertzog PJ, Holland KA, Sumarsono SH, Tymms MJ, Hamilton JA, Whitty G, Bertoncello I, Kola I. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 5.Decker T, Muller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nature reviews Immunology. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- 6.Kearney S, Delgado C, Lenz LL. Differential effects of type I and II interferons on myeloid cells and resistance to intracellular bacterial infections. Immunologic research, 2012/09/18 Ed. 2012 doi: 10.1007/s12026-012-8362-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rayamajhi M, Humann J, Kearney S, Hill KK, Lenz LL. Antagonistic crosstalk between type I and II interferons and increased host susceptibility to bacterial infections. Virulence. 2010;1:418–422. doi: 10.4161/viru.1.5.12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decker T, Stockinger S, Karaghiosoff M, Muller M, Kovarik P. IFNs and STATs in innate immunity to microorganisms. The Journal of clinical investigation. 2002;109:1271–1277. doi: 10.1172/JCI15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ea CK, Hao S, Yeo KS, Baltimore D. EHMT1 Protein Binds to Nuclear Factor-kappaB p50 and Represses Gene Expression. The Journal of biological chemistry. 2012;287:31207–31217. doi: 10.1074/jbc.M112.365601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Icardi L, Lievens S, Mori R, Piessevaux J, De Cauwer L, De Bosscher K, Tavernier J. Opposed regulation of type I IFN-induced STAT3 and ISGF3 transcriptional activities by histone deacetylases (HDACS) 1 and 2. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2012;26:240–249. doi: 10.1096/fj.11-191122. [DOI] [PubMed] [Google Scholar]
- 12.Mittelstadt ML, Patel RC. AP-1 mediated transcriptional repression of matrix metalloproteinase-9 by recruitment of histone deacetylase 1 in response to interferon beta. PloS one. 2012;7:e42152. doi: 10.1371/journal.pone.0042152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonelli LR, Gigliotti Rothfuchs A, Goncalves R, Roffe E, Cheever AW, Bafica A, Salazar AM, Feng CG, Sher A. Intranasal Poly-IC treatment exacerbates tuberculosis in mice through the pulmonary recruitment of a pathogen-permissive monocyte/macrophage population. The Journal of clinical investigation. 2010;120:1674–1682. doi: 10.1172/JCI40817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rayamajhi M, Humann J, Penheiter K, Andreasen K, Lenz LL. Induction of IFN-alphabeta enables Listeria monocytogenes to suppress macrophage activation by IFN-gamma. The Journal of experimental medicine. 2010;207:327–337. doi: 10.1084/jem.20091746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper AM, Dalton DK, Stewart TA, Griffin JP, Russell DG, Orme IM. Disseminated tuberculosis in interferon gamma gene-disrupted mice. The Journal of experimental medicine. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dighe AS, Campbell D, Hsieh CS, Clarke S, Greaves DR, Gordon S, Murphy KM, Schreiber RD. Tissue-specific targeting of cytokine unresponsiveness in transgenic mice. Immunity. 1995;3:657–666. doi: 10.1016/1074-7613(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 17.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. The Journal of experimental medicine. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swirnoff AH, Milbrandt J. DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Molecular and cellular biology. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Donovan KJ, Tourtellotte WG, Millbrandt J, Baraban JM. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends in neurosciences. 1999;22:167–173. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- 20.Abdel-Latif MM, Windle HJ, Fitzgerald KA, Ang YS, Eidhin DN, Li-Weber M, Sabra K, Kelleher D. Helicobacter pylori activates the early growth response 1 protein in gastric epithelial cells. Infection and immunity. 2004;72:3549–3560. doi: 10.1128/IAI.72.6.3549-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cullen EM, Brazil JC, O'Connor CM. Mature human neutrophils constitutively express the transcription factor EGR-1. Molecular immunology. 2010;47:1701–1709. doi: 10.1016/j.molimm.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Droin NM, Pinkoski MJ, Dejardin E, Green DR. Egr family members regulate nonlymphoid expression of Fas ligand, TRAIL, and tumor necrosis factor during immune responses. Molecular and cellular biology. 2003;23:7638–7647. doi: 10.1128/MCB.23.21.7638-7647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Miao T, Sebastian M, Bhullar P, Ghaffari E, Liu M, Symonds AL, Wang P. The Transcription Factors Egr2 and Egr3 Are Essential for the Control of Inflammation and Antigen-Induced Proliferation of B and T Cells. Immunity. 2012 doi: 10.1016/j.immuni.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi L, Kishore R, McMullen MR, Nagy LE. Lipopolysaccharide stimulation of ERK1/2 increases TNF-alpha production via Egr-1. American journal of physiology Cell physiology. 2002;282:C1205–1211. doi: 10.1152/ajpcell.00511.2001. [DOI] [PubMed] [Google Scholar]
- 25.Nebbaki SS, El Mansouri FE, Afif H, Kapoor M, Benderdour M, Duval N, Pelletier JP, Martel-Pelletier J, Fahmi H. Egr-1 contributes to IL-1-mediated down-regulation of peroxisome proliferator-activated receptor gamma expression in human osteoarthritic chondrocytes. Arthritis research & therapy. 2012;14:R69. doi: 10.1186/ar3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan L, Peng H, Osaki M, Choy BK, Auron PE, Sandell LJ, Goldring MB. Egr-1 mediates transcriptional repression of COL2A1 promoter activity by interleukin-1beta. The Journal of biological chemistry. 2003;278:17688–17700. doi: 10.1074/jbc.M301676200. [DOI] [PubMed] [Google Scholar]
- 27.Bahouth SW, Beauchamp MJ, Vu KN. Reciprocal regulation of beta(1)-adrenergic receptor gene transcription by Sp1 and early growth response gene 1: induction of EGR-1 inhibits the expression of the beta(1)-adrenergic receptor gene. Molecular pharmacology. 2002;61:379–390. doi: 10.1124/mol.61.2.379. [DOI] [PubMed] [Google Scholar]
- 28.Huang RP, Fan Y, Ni Z, Mercola D, Adamson ED. Reciprocal modulation between Sp1 and Egr-1. Journal of cellular biochemistry. 1997;66:489–499. [PubMed] [Google Scholar]
- 29.Khachigian LM, Williams AJ, Collins T. Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth factor A-chain promoter in cultured vascular endothelial cells. The Journal of biological chemistry. 1995;270:27679–27686. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 30.Chiang SY, Welch JJ, Rauscher FJ, 3rd, Beerman TA. Effect of DNA-binding drugs on early growth response factor-1 and TATA box-binding protein complex formation with the herpes simplex virus latency promoter. The Journal of biological chemistry. 1996;271:23999–24004. doi: 10.1074/jbc.271.39.23999. [DOI] [PubMed] [Google Scholar]
- 31.Tatarowicz WA, Martin CE, Pekosz AS, Madden SL, Rauscher FJ, 3rd, Chiang SY, Beerman TA, Fraser NW. Repression of the HSV-1 latency-associated transcript (LAT) promoter by the early growth response (EGR) proteins: involvement of a binding site immediately downstream of the TATA box. Journal of neurovirology. 1997;3:212–224. doi: 10.3109/13550289709018296. [DOI] [PubMed] [Google Scholar]
- 32.Wang C, Dostanic S, Servant N, Chalifour LE. Egr-1 negatively regulates expression of the sodium-calcium exchanger-1 in cardiomyocytes in vitro and in vivo. Cardiovascular research. 2005;65:187–194. doi: 10.1016/j.cardiores.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 33.Mager GM, Ward RM, Srinivasan R, Jang SW, Wrabetz L, Svaren J. Active gene repression by the Egr2. NAB complex during peripheral nerve myelination The Journal of biological chemistry. 2008;283:18187–18197. doi: 10.1074/jbc.M803330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gashler AL, Swaminathan S, Sukhatme VP. A novel repression module, an extensive activation domain, and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Molecular and cellular biology. 1993;13:4556–4571. doi: 10.1128/mcb.13.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russo MW, Matheny C, Milbrandt J. Transcriptional activity of the zinc finger protein NGFI-A is influenced by its interaction with a cellular factor. Molecular and cellular biology. 1993;13:6858–6865. doi: 10.1128/mcb.13.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Molecular and cellular biology. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russo MW, Sevetson BR, Milbrandt J. Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swirnoff AH, Apel ED, Svaren J, Sevetson BR, Zimonjic DB, Popescu NC, Milbrandt J. Nab1, a corepressor of NGFI-A (Egr-1), contains an active transcriptional repression domain. Molecular and cellular biology. 1998;18:512–524. doi: 10.1128/mcb.18.1.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasan R, Mager GM, Ward RM, Mayer J, Svaren J. NAB2 represses transcription by interacting with the CHD4 subunit of the nucleosome remodeling and deacetylase (NuRD) complex. The Journal of biological chemistry. 2006;281:15129–15137. doi: 10.1074/jbc.M600775200. [DOI] [PubMed] [Google Scholar]
- 40.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic acids research. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schug J. Using TESS to Predict Transcription Factor Binding Sites in DNA Sequences. In: Baxevanis AD, editor. Current Protocols in Bioinformatics. J. Wiley and Sons; 2003. [DOI] [PubMed] [Google Scholar]
- 42.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling PD, Warren MK, Vogel SN. Antagonistic effect of interferon-beta on the interferon-gamma-induced expression of Ia antigen in murine macrophages. J Immunol. 1985;135:1857–1863. [PubMed] [Google Scholar]
- 44.Yoshida R, Murray HW, Nathan CF. Agonist and antagonist effects of interferon alpha and beta on activation of human macrophages Two classes of interferon gamma receptors and blockade of the high-affinity sites by interferon alpha or beta. The Journal of experimental medicine. 1988;167:1171–1185. doi: 10.1084/jem.167.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prinz M, Schmidt H, Mildner A, Knobeloch KP, Hanisch UK, Raasch J, Merkler D, Detje C, Gutcher I, Mages J, Lang R, Martin R, Gold R, Becher B, Bruck W, Kalinke U. Distinct and nonredundant in vivo functions of IFNAR on myeloid cells limit autoimmunity in the central nervous system. Immunity. 2008;28:675–686. doi: 10.1016/j.immuni.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 46.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes & development. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 47.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 48.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 49.Sakamoto S, Taniguchi T. Identification of a phorbol ester-responsive element in the interferon-gamma receptor 1 chain gene. The Journal of biological chemistry. 2001;276:37237–37241. doi: 10.1074/jbc.M105543200. [DOI] [PubMed] [Google Scholar]
- 50.Halvorson LM, Ito M, Jameson JL, Chin WW. Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone beta-subunit gene expression. The Journal of biological chemistry. 1998;273:14712–14720. doi: 10.1074/jbc.273.24.14712. [DOI] [PubMed] [Google Scholar]
- 51.Carter JH, Tourtellotte WG. Early growth response transcriptional regulators are dispensable for macrophage differentiation. J Immunol. 2007;178:3038–3047. doi: 10.4049/jimmunol.178.5.3038. [DOI] [PubMed] [Google Scholar]
- 52.Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annual review of immunology. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 53.Farrar MA, Schreiber RD. The molecular cell biology of interferon-gamma and its receptor. Annual review of immunology. 1993;11:571–611. doi: 10.1146/annurev.iy.11.040193.003035. [DOI] [PubMed] [Google Scholar]
- 54.Bach EA, Szabo SJ, Dighe AS, Ashkenazi A, Aguet M, Murphy KM, Schreiber RD. Ligand-induced autoregulation of IFN-gamma receptor beta chain expression in T helper cell subsets. Science. 1995;270:1215–1218. doi: 10.1126/science.270.5239.1215. [DOI] [PubMed] [Google Scholar]
- 55.Pernis A, Gupta S, Gollob KJ, Garfein E, Coffman RL, Schindler C, Rothman P. Lack of interferon gamma receptor beta chain and the prevention of interferon gamma signaling in TH1 cells. Science. 1995;269:245–247. doi: 10.1126/science.7618088. [DOI] [PubMed] [Google Scholar]
- 56.Shah KM, Stewart SE, Wei W, Woodman CB, O'Neil JD, Dawson CW, Young LS. The EBV-encoded latent membrane proteins, LMP2A and LMP2B, limit the actions of interferon by targeting interferon receptors for degradation. Oncogene. 2009;28:3903–3914. doi: 10.1038/onc.2009.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen C, Guo L, Shi M, Hu M, Hu M, Yu M, Wang T, Song L, Shen B, Qian L, Guo N. Modulation of IFN-gamma receptor 1 expression by AP-2alpha influences IFN-gamma sensitivity of cancer cells. The American journal of pathology. 2012;180:661–671. doi: 10.1016/j.ajpath.2011.10.040. [DOI] [PubMed] [Google Scholar]
- 58.Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2004;82:460–471. doi: 10.1139/o04-045. [DOI] [PubMed] [Google Scholar]
- 59.Singhal A, Jaiswal A, Arora VK, Prasad HK. Modulation of gamma interferon receptor 1 by Mycobacterium tuberculosis: a potential immune response evasive mechanism. Infection and immunity. 2007;75:2500–2510. doi: 10.1128/IAI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays : news and reviews in molecular, cellular and developmental biology. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 61.Monsalve E, Perez MA, Rubio A, Ruiz-Hidalgo MJ, Baladron V, Garcia-Ramirez JJ, Gomez JC, Laborda J, Diaz-Guerra MJ. Notch-1 up-regulation and signaling following macrophage activation modulates gene expression patterns known to affect antigen-presenting capacity and cytotoxic activity. J Immunol. 2006;176:5362–5373. doi: 10.4049/jimmunol.176.9.5362. [DOI] [PubMed] [Google Scholar]
- 62.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nature immunology. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 63.Huang RP, Fan Y, deBelle I, Ni Z, Matheny W, Adamson ED. Egr-1 inhibits apoptosis during the UV response: correlation of cell survival with Egr-1 phosphorylation. Cell death and differentiation. 1998;5:96–106. doi: 10.1038/sj.cdd.4400322. [DOI] [PubMed] [Google Scholar]
- 64.Jain N, Mahendran R, Philp R, Guy GR, Tan YH, Cao X. Casein kinase II associates with Egr-1 and acts as a negative modulator of its DNA binding and transcription activities in NIH 3T3 cells. The Journal of biological chemistry. 1996;271:13530–13536. doi: 10.1074/jbc.271.23.13530. [DOI] [PubMed] [Google Scholar]
- 65.Yu J, de Belle I, Liang H, Adamson ED. Coactivating factors p300 and CBP are transcriptionally crossregulated by Egr1 in prostate cells, leading to divergent responses. Molecular cell. 2004;15:83–94. doi: 10.1016/j.molcel.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 66.Yu J, Zhang SS, Saito K, Williams S, Arimura Y, Ma Y, Ke Y, Baron V, Mercola D, Feng GS, Adamson E, Mustelin T. PTEN regulation by Akt-EGR1-ARF-PTEN axis. The EMBO journal. 2009;28:21–33. doi: 10.1038/emboj.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cooke GS, Campbell SJ, Sillah J, Gustafson P, Bah B, Sirugo G, Bennett S, McAdam KP, Sow O, Lienhardt C, Hill AV. Polymorphism within the interferon-gamma/receptor complex is associated with pulmonary tuberculosis. American journal of respiratory and critical care medicine. 2006;174:339–343. doi: 10.1164/rccm.200601-088OC. [DOI] [PubMed] [Google Scholar]
- 68.Velayati AA, Farnia P, Khalizadeh S, Farahbod AM, Hasanzadh M, Sheikolslam MF. Interferon-gamma receptor-1 gene promoter polymorphisms and susceptibility to leprosy in children of a single family. The American journal of tropical medicine and hygiene. 2011;84:627–629. doi: 10.4269/ajtmh.2011.10-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou J, Chen DQ, Poon VK, Zeng Y, Ng F, Lu L, Huang JD, Yuen KY, Zheng BJ. A regulatory polymorphism in interferon-gamma receptor 1 promoter is associated with the susceptibility to chronic hepatitis B virus infection. Immunogenetics. 2009;61:423–430. doi: 10.1007/s00251-009-0377-8. [DOI] [PubMed] [Google Scholar]
- 70.Salih MA, Ibrahim ME, Blackwell JM, Miller EN, Khalil EA, ElHassan AM, Musa AM, Mohamed HS. IFNG and IFNGR1 gene polymorphisms and susceptibility to post-kala-azar dermal leishmaniasis in Sudan. Genes and immunity. 2007;8:75–78. doi: 10.1038/sj.gene.6364353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5