Beyond PSA: The next generation of prostate cancer biomarkers (original) (raw)
. Author manuscript; available in PMC: 2013 Oct 19.
Abstract
Since the introduction of serum prostate specific antigen (PSA) screening twenty-five years ago, prostate cancer diagnosis and management have been guided by this biomarker. Yet, PSA has proven controversial as a diagnostic assay due to its limitations. The next wave of prostate cancer biomarkers has emerged, introducing new assays in serum and urine that may supplement or, in time, replace PSA due to higher cancer specificity. This expanding universe of biomarkers has been facilitated, in large part, by new genomic technologies that have enabled an unbiased look at cancer biology. Such efforts have produced several notable success stories, moving biomarkers from the bench to the clinic rapidly. However, biomarker research has centered on disease diagnostics, rather than prognosis and prediction, which could work toward disease prevention—an important focus moving forward. We review the current state of prostate cancer biomarker research, including the PSA revolution, its impact on early prostate cancer detection, the recent advances in biomarker discovery, and the future efforts that promise to improve clinical management of this disease.
Introduction
The introduction of biomarkers for disease diagnosis and management has revolutionized the practice of oncology. Biomarkers are molecules whose detection or evaluation provides information about a disease beyond the standard clinical parameters that are routinely gathered by the clinician. Biomarkers can be proteins, metabolites, RNA transcripts, DNA, or epigenetic modifications of DNA, among other alterations. They can be detected through patient tissue samples, obtained either by biopsy or surgical resection, or non-invasively through the isolation of cells and/or molecules from bodily fluids, such as blood or urine. While increasing interest in biomarkers has spurred much research recently, controversies regarding what constitutes a robust biomarker and how to investigate biomarkers clinically remain, and these subjects will be addressed later in this review.
Broadly, there are seven common clinical roles for biomarkers (1), which address specific clinical questions when managing cancer patients or patients suspected to have a malignancy:
- Disease disposition: What is a patient’s risk of developing cancer in the future?
- Screening: Does earlier detection of patients with cancer decrease mortality?
- Diagnostic: Who has cancer? What is the grade of the cancer?
- Prognostic: What clinical outcome is most likely if therapy is not administered?
- Predictive: Which therapy is most appropriate?
- Monitoring: Was therapy effective? Did the patient’s disease recur?
- Pharmacogenomic: What is the risk for adverse reaction to the prescribed therapeutic dose?
A few successful examples of cancer biomarkers have emerged that illustrate these categories. For example, the commercially available OncotypeDx gene expression assay serves as prognostic biomarker to help predict breast cancer recurrence (2). Amplification of the human epidermal growth factor receptor 2 (HER2) oncogene (3), mutation in v-raf murine sarcoma viral oncogene homolog B1 (BRAF) (4), or the presence of a fusion between the echinoderm microtubule-associated protein-like 4 (EML4) gene and the anaplastic lymphoma kinase (ALK) gene (EML4-ALK) (5) are predictive biomarkers for breast cancer (HER2), melanoma (BRAF), or lung cancer (EML4-ALK) that help identify which patients will most likely benefit from targeted therapies against those genetic aberrations (6). Serum PSA is commonly used for monitoring disease progression following hormonal therapy of hormone-naïve prostate cancer (7).
The ideal biomarker for clinical use should have three major characteristics: 1) a safe and easy means of measurement, preferably non-invasively; 2) high sensitivity, specificity, and positive and negative predictive values for its intended outcome; and 3) improves decision-making abilities in conjunction with clinicopathological parameters. Although a biomarker that performs well in several of the aforementioned categories would be ideal, the reality is that multiple biomarkers will be likely required for cancer to fully cover screening, diagnosis, prognosis, and prediction.
PSA as a prostate cancer biomarker
Prostate cancer is the most common non-cutaneous cancer in men, with over 200,000 prostate cancer diagnoses per year in the United States. The lifetime risk for a U.S. male to develop prostate cancer is approximately 1 in 6, although the risk of dying from prostate cancer is only 1 in 35 (8). This discrepancy between prostate cancer incidence and lethality has led to widespread scrutiny of prostate cancer patient management, particularly for low-grade, low-stage disease (“indolent” disease) (9).
Unlike most solid tumors, prostate cancer management has long employed biomarkers. The first of these, prostatic acid phosphatase (PAP), was noted in the 1930s to be elevated in the serum of men with metastatic prostate cancer, and for nearly 50 years PAP was investigated as a clinical marker for disease progression (10).
In the 1980s, PAP was rapidly replaced by PSA, a secreted protein first studied in the late 1970s (11). PSA is encoded by the prostate-specific gene kallikrein 3 (KLK3), a member of the tissue kallikrein family, a gene family of serine proteases located on chromosome 19q13.4 that also includes KLK2 and KLK4 (12). Mature PSA is the result of two proteolytic cleavages of two inactive precursor peptides, pre-proenzyme PSA (pre-proPSA) and proPSA. In its final form, PSA is secreted into semen (12). Under normal conditions, only low levels of PSA can be detected in blood, and the increase of serum PSA found in prostate cancer can represent abnormalities in prostate gland architecture and vascularization, although the exact mechanism is unclear (7).
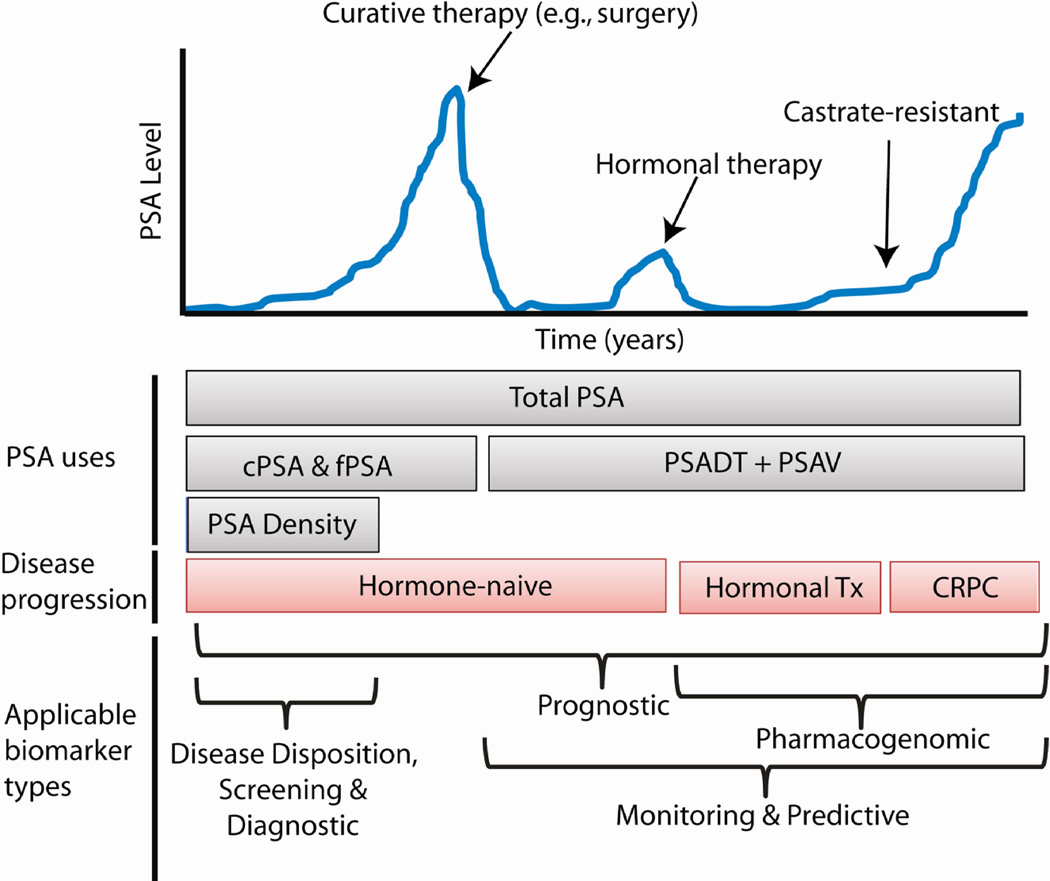
Initial reports suggested a role for PSA as a biomarker for monitoring the progression of patients already diagnosed with prostate cancer or for recurrence following curative therapy for organ-confined disease (Fig. 1). In a landmark study, Stamey et al. performed the first large-scale analysis of serum PSA as a prostate cancer biomarker in 1987, convincingly demonstrating that PSA was more sensitive than PAP for monitoring the disease (13). They showed that PSA level increased with advancing clinical stage and was useful for detecting disease recurrence after curative therapy (13).
Figure 1. PSA clinical course and biomarker uses.
In this model, PSA levels or increases suggest the presence of prostate cancer and can inform management decisions. Several types of PSA measurement can be employed, including total PSA, complexed and free PSA (cPSA and fPSA), PSA doubling time (PSADT) and velocity (PSAV), and PSA density. This cartoon plot illustrates the clinical course of some patients with recurrent prostate cancer, in which disease recurs following curative therapy. Hormonal therapy in this example leads to castrate-resistant prostate cancer (CRPC), in which the cancer becomes refractory to conventional hormonal therapies. The bottom segment of the plot indicates the type of biomarkers applicable for measurement for disease management.
Subsequent studies shifted the focus of PSA towards early detection of prostate cancer. In 1986, the U.S. Food and Drug Administration (FDA) approved PSA as an adjunctive test to the DRE for the detection of prostate cancer in men over the age of 50. In 1991, Catalona and colleagues demonstrated that the combination of a serum PSA measurement of more than ≥4.0 ng/mL with other clinical findings, such as the results of a digital rectal exam (DRE), improved detection of prostate cancer in a prospective study of 1653 healthy men with no history of cancer (14). Numerous groups confirmed that PSA was useful as a diagnostic test for prostate cancer (15).
Impact of PSA on diagnosis and treatment: More harm than good?
Between 1985 and 1995, prostate cancer incidence doubled in the U.S., from approximately 55 to 110 cases per 100,000 men (16, 17). This dramatic increase was paralleled by an even more striking increase in invasive procedures for prostate cancer treatment: radical prostatectomy rates were nearly 6-fold higher in 1990 than in 1984 (18). These major shifts in the detection and treatment of prostate cancer have been attributed to the use of PSA as a diagnostic test, coupled with improvements in the safety of the radical prostatectomy procedure (19).
The introduction of PSA into the prostate cancer diagnostics community also led to its widespread use as a screening test among asymptomatic men. Subsequently, the proportion of men with metastatic prostate cancer at the time of diagnosis decreased dramatically, a major feat for the prostate cancer community that altered disease management (16). More men were being diagnosed with prostate cancer, with the majority having early-stage, clinically indolent disease. More men with benign conditions such as inflammation or hyperplasia were also being biopsied. PSA therefore enables the early detection of many latent prostate cancers, the majority of which may never have led to harm (16). This discrepancy between decreasing disease aggressiveness and increasing treatment has led to widespread criticism that prostate cancer is now an “overdiagnosed” and “overtreated” cancer. The majority of low-grade, low-stage tumors are unlikely to cause significant symptoms or mortality, and it is estimated that up to 50% of new prostate cancer diagnoses detect a tumor that was unlikely to surface clinically in the absence of PSA screening (9). A subsequent analysis by Draisma et al. suggested an overdiagnosis rate of 20 to 42% (20).
Moreover, treatment of indolent cancer may cause a patient more harm than good. Biopsies and prostate cancer treatments have been associated with psychological distress, loss of bodily function, pain, and suffering for patients (21). Side effects of radiotherapy and radical prostatectomy, including sexual dysfunction, urinary incontinence, and impaired bowel/rectal function, occur in large fractions of patients, adding to a patient’s distress (22). Rarely, treatment of prostate cancer directly contributes to a life-threatening adverse event (23).
Although mortality from prostate cancer has been decreasing since the mid-1990s, it is unclear to what extent PSA screening may be responsible. The two largest prospective screening trials to date—the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) in the U.S. and the European Randomized Study of Screening for Prostate Cancer (ERSPC) in Europe—failed to demonstrate a concordant benefit in overall patient survival from PSA screening (24, 25). At best, the ERSPC trial demonstrated that PSA screening decreased prostate cancer related mortality; however, in order to prevent 1 death from prostate cancer, a physician must screen 1410 men for serum PSA and treat 48 (25). In 2002 the U.S. Preventive Services Task Force (USPSTF) deemed the evidence to be insufficient to recommend routine use of PSA as a screening test among men younger than age 75 (http://www.uspreventiveservicestaskforce.org/uspstf/uspsprca.htm). The USPSTF reviewed the available evidence again in 2011 and, in a draft report, concluded that population benefit from PSA screening was inconclusive, recommending against PSA-based prostate cancer screening at any age (N.B. This draft is currently in the public forum for feedback) (26).
A biomarker with limitations
The diagnostic test performance characteristics of PSA are variable. First, its specificity and sensitivity ranges from 20 to 40% and 70 to 90%, respectively, depending on the PSA cutoff values used (e.g. 3 ng/mL vs 4 ng/mL) (27). The area-under-the-curve (AUC) metric of the receiver operating characteristic (ROC) curve is between 0.55 and 0.70 for the ability of PSA to identify patients with cancer, where a score of 1.0 is perfect discrimination and 0.5 is a coin-toss (27). One of the major reasons for such poor specificity is the fact that several non-cancerous events may elevate the level of PSA. Indeed, inflammation, infection, trauma, and benign prostatic hyperplasia (BPH) are more common causes of elevated serum PSA than cancer (7, 12, 27). BPH is known to be present in over 50% of men >50 years old, thus confounding PSA as a cancer biomarker (8, 12). Not surprising then, PSA-based screening for prostate cancer is plagued by false positives, resulting in a positive predictive value of only 25 to 40% (28). Conversely, approximately 15% of men with a low-level PSA (<4.0 ng/mL) have prostate cancer, and 15% of these display a high Gleason score (29, 30).
PSA derivatives
There have been numerous efforts to improve the performance of the PSA test, such as normalizing PSA to the size of the gland (the PSA “density”) (13, 31, 32) or monitoring the dynamics of PSA change in serum (the PSA velocity and doubling time) (33–37). In addition, assays measuring alternative molecular traits of PSA have also gained attention, including free and complexed PSA (fPSA and cPSA, respectively) (38–41), and isoforms of the PSA protein (proPSA, most commonly) (Fig. 1) (42, 43).
Among these, cPSA and fPSA have been considered adjunctive tests to total serum PSA rather than replacement assays (Fig. 1). cPSA measurements exploit the molecular interactions of PSA mainly with α-1-antichymotrypsin (ACT) in the blood (39). Conversely, fPSA measures the percentage of total serum PSA not bound to ACT. This %fPSA decreases in prostate cancer, making it useful in distinguishing men with BPH from men with cancer. A %fPSA of less than 25% has been shown to improve the sensitivity and specificity of a total PSA test and to reduce unnecessary biopsies (38, 41). %fPSA has thus gained FDA approval for use when patients have a total PSA in the 4 – 10 ng/mL “gray zone.” Furthermore, combined measurement of [−2] pro-PSA (a peptide precursor to mature PSA) with fPSA may help diagnose early prostate cancers with a PSA of 2 to 10 ng/mL (42, 43). fPSA has several drawbacks, such as the potential instability of the fPSA measurement if sample processing occurs after 24 hours of collection (44). The %fPSA may also increase following DRE or biopsy procedures (45), confounding its use in those settings.
PSA dynamics, namely PSA velocity (PSAV) and doubling time (PSADT), have prognostic value (46). PSAV is defined as the change in PSA concentration per year, with a high PSAV being strongly associated with prostate cancer and a 9-fold elevated risk of cancer-death following prostatectomy (33, 34, 47). PSADT is defined as the time necessary for the serum PSA level to double. PSADT is most commonly used to monitor disease progression following curative therapy for organ-confined disease, as an increasing PSA level following radiotherapy or prostatectomy indicates the presence of residual tumor cells. Numerous studies have demonstrated that a more rapid PSADT (<10 months) is associated with reduced survival (35, 36). In rare cases, disease may recur in the absence of an elevated PSA (48). Nevertheless, neither test has been shown to improve over a standard PSA measurement for prostate cancer screening (37). Taken together, measurement of PSA isoforms and dynamics have modestly improved care but are largely hindered by the same issues confounding PSA itself.
The next generation of biomarkers: -omics in prostate cancer
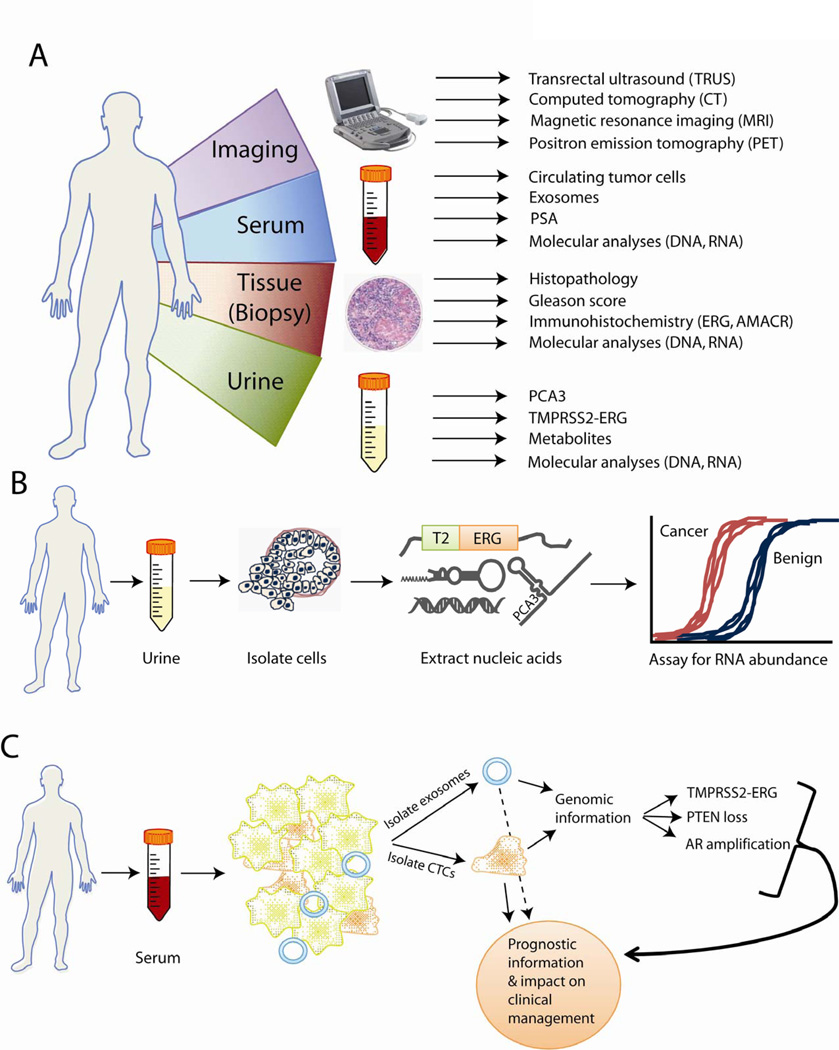
The 20 years since the widespread adoption of PSA have witnessed a remarkable maturation of genomic technologies, such as microarrays and whole-genome sequencing (49). These advances in DNA sequence and RNA transcriptome profiling have enabled detailed dissections of cancer biology at a level previously unattainable (49, 50). As a result, the world of biomarker research has shifted to use these “-omics” methods, populating the prostate cancer literature with discoveries based on profiling prostate tumors for aberrations in either DNA, RNA, or epigenetic DNA methylation states. Here, we will focus on the discovery and characterization of emerging urine assays for prostate cancer, including PCA3 and the TMPRSS2-ERG gene fusion, although the biomarker research also includes advances in tissue and imaging-based tools as well (Fig. 2A).
Figure 2. Advances in prostate cancer biomarker uses.
(A) The emerging clinical paradigm for prostate cancer biomarkers, including the combined application of imaging biomarkers and biomarkers found in serum, urine, and tissue. (B) Recent advances in molecular biology have enabled the robust detection of transcriptomic, proteomic, and genomic biomarkers in patient urine. PCA3 and TMPRSS2-ERG screening lend increased specificity for detecting cancer, resulting in fewer false positive test results. (C) Promising avenues of biomarker research are the isolation of circulating tumor cells (CTCs) and exosomes from patient serum. Molecular analysis of CTCs and exosomes for common genetic aberrations may further provide predictive information for prostate cancer.
PCA3
The most prominent biomarker emerging as a non-PSA-based diagnostic test for prostate cancer is prostate cancer antigen 3 (PCA3). PCA3 is a long noncoding RNA (lncRNA) that has been shown to be elevated in >90% of prostate cancer tissues, but not normal or BPH tissues—an important distinction to serum PSA (51, 52). The high sensitivity and specificity of PCA3 in tissues led to studies of PCA3 as a non-invasive biomarker, where numerous assays have been developed to detect the RNA transcript in patient urine samples, which contain cells shed from the prostate during urination (Fig. 2B). Over the past decade, several iterations of PCA3 urine tests have emerged (53), and currently a clinical-grade assay based on transcription-mediated amplification is available (53).
Urine PCA3 measurements have consistently added to the diagnostic information obtained from the PSA test, with higher AUC values of 0.66 to 0.72 (compared to 0.54 to 0.63 for serum PSA alone) (54). A particularly important attribute of PCA3 is the fact that, unlike PSA, urine PCA3 levels are independent of prostate size (55). Sensitivities for urine PCA3 levels range from 47 to 69%, with most between 58 and 69%, although it is difficult to compare the studies directly because of different analysis platforms, different criteria for enrolling patients (serum PSA elevated to varying levels), and relatively small patient cohorts (several hundred men) (54). While PCA3 has been established as a robust biomarker despite these variations, the differences in methodology illustrate the challenges of biomarker research and development even for a highly sensitive tissue biomarker such as PCA3. In addition, combining a serum PSA value with a urine PCA3 analysis improves both measures, with the combination AUC of 0.71 to 0.75 (56). In 2012, PCA3 was approved by the FDA as a diagnostic test for prostate cancer in the setting of a prior negative prostate biopsy.
TMPRSS2-ERG
A gene fusion product arising from a translocation of the androgen-induced transmembrane protease, serine 2 (TMPRSS2) gene with the transcription factor v-ets erythroblastosis virus E26 oncogene homolog (ERG) is one of the most common genetic events in prostate cancer, present in approximately 50% of all cases and accounting for 90% of prostate cancer fusions (57). TMPRSS2-ERG fusions are specific for prostate cancer, and can even be detected in precursor lesions, such as prostate intraepithelial neoplasia (PIN), if these lesions are proximal to, or contiguous with, regions of cancer (58).
The detection of TMPRSS2-ERG RNA in patient urine has also been investigated (59, 60) (Fig. 2B). Yet, TMPRSS2-ERG is absent in approximately 50% of cancers; therefore, its ideal usage lies in multiplexed assays with other biomarkers. To this end, Hessels et al. measured urinary TMPRSS2-ERG in conjunction with PCA3 and found that the sensitivity of the combined test was 0.73—better than either test alone (59). Similarly, a large study of more than 1300 men demonstrated recently that combined measurement of PCA3 and TMPRSS2-ERG in urine outperformed serum PSA for prostate cancer diagnosis (AUC = 0.71 — 0.77 for TMPRSS2-ERG + PCA3; AUC = 0.61 for PSA), thus adding to available clinical information in the Prostate Cancer Prevention Trial (PCPT) risk estimates for predicting cancer (60).
There has been some debate as to whether the presence of a TMPRSS2-ERG fusion is itself a prognostic biomarker when detected in tissues. While several groups have reported an association between TMPRSS2-ERG and aggressive prostate cancer (60–62), others have not observed this association (63, 64). One complication to these studies has been heterogeneity in the patient populations studied and the clinical outcomes evaluated. Interestingly, quantitative levels of TMPRSS2-ERG detected in urine, however, appear to be associated with clinically significant prostate cancer based on Epstein criteria, which stratifies disease aggressiveness using PSA density and characteristics of the patient’s biopsy (Gleason score as well as the percent tumor observed in the biopsy core and the number of cores with tumor) (60).
Limitations of PCA3 and TMPRSS2-ERG assays
As with any assay, there are limitations to the PCA3 and TMPRSS2-ERG tests. First, these tests are currently adjunctive to PSA, and head-to-head trials to determine whether these tests perform well in the absence of PSA screening are lacking. Secondly, urine expression of PCA3 or TMPRSS2-ERG is determined relative to urine PSA mRNA (59, 60), because PSA transcript abundance indicates the relative yield of prostate cells in the urine sediment. Thus, if the PSA transcript level is too low, the tests are uninformative.
AMACR
Another biomarker nominated by RNA expression profiling is the enzyme alpha-methylacyl-CoA racemase (AMACR), which has demonstrated high sensitivities and specificities, each >90% when tested as a diagnostic biomarker on prostate biopsy tissue (65). Low AMACR expression in prostate cancer has also been correlated with metastasis and biochemical recurrence (66). However, AMACR is not specific to prostate cancer, and is also not suitable for non-invasive detection in urine (67), rendering it most useful as a tissue biomarker when prostate biopsy cores yield ambiguous pathological results.
Germline prostate cancer risk loci
In addition to profiling urine RNA and DNA, genomic analyses have recently uncovered several single nucleotide polymorphisms (SNPs) associated with prostate cancer. These loci may be germline indications of an individual’s risk for developing prostate cancer. To date, more than 50 SNPs have been proposed as putative risk loci for prostate cancer, of which ~30 have been validated in multiple studies (68). Although each individual SNP is likely to contribute a minor degree to disease risk (thus making individual assays ineffective) combining multiple SNPs may yield more informative results. In a retrospective cohort of 2893 prostate cancer patients and 1781 control patients, Zheng et al. defined a core set of 5 disease-associated SNPs that were then combined with family history to predict risk (up to tenfold) for developing prostate cancer (69). Recently, rare SNP variants in HOXB13, an AR cofactor, have recently been implicated in familial predisposition to early-onset prostate cancer as well, although these variants occur at low prevalence in the general prostate cancer population (<1%) (70).
Other “-omic” biomarkers
Other studies have used high-throughput proteomics and metabolomics platforms to elucidate signatures of serum proteins and urine metabolites in prostate cancer. One major advantage of profiling the human serum proteome is the vast dynamic range—greater than 10 logs (1010)—over which serum proteins can be accurately detected (71). Rosenzweig et al. used mass spectrometry to nominate serum proteins signatures for predicting prostate cancer biochemical recurrence (72). Mass spectrometry has also been used to identify candidate serum proteins that may indicate response to chemotherapy (73). Profiling of culture media from prostate cancer cell lines for secreted proteins has also identified several potentially diagnostic proteins (74). Similarly, a study of urine metabolites in prostate cancer patients lead to the identification of a series of metabolites elevated in aggressive forms of prostate cancer, including sarcosine, a metabolite of glycine (75).
Further refinements of genomic technologies, such as next generation transcriptome sequencing (RNA-Seq), promise to uncover additional biomarkers in an unbiased manner, including tissue-specific non-coding RNAs similar to PCA3 (50). With throughputs of thousands of molecules simultaneously, advances in computational biology and informatics will continue to be integral to fish out cancer-specific indicators in a sea of “-omics” data. In some cases, these high-throughput approaches can be used to define patient-specific biomarkers that may further the concept of personalized medicine (76). To this end, Roychowdhury et al. recently employed several kinds of sequencing approaches to comprehensively define the genomic and transcriptomic aberrations in metastatic prostate cancer patients in a clinically-relevant time-frame of <4 weeks post-biopsy (77). Further developments in personalized sequencing efforts may enable biomarker discovery in a patient-specific manner and impact disease management.
Circulating tumor cells
One area of expanding investigation is circulating tumor cells (CTCs). CTCs are found in the bloodstream and are particularly prevalent in locally aggressive or metastatic disease. CTCs can be both a biomarker for cancer detection and a source of molecular information, such as TMPRSS2-ERG, androgen receptor (AR) and phosphatase and tensin homolog (PTEN) copy number status (Fig. 2C) (78). In support of CTCs as a predictive biomarker, several groups have demonstrated that an increased abundance of CTCs in the blood of castration-resistant prostate cancer patients predicted worse overall survival (79, 80). However, detecting CTCs and extracting molecular information is currently labor-intensive and expensive, and it is yet unknown to what extent CTC abundance in blood represents aggressive disease undergoing hematogenous spread versus cells that have simply dislodged from the main tumor bulk into the bloodstream.
Exosomes
A similar effort has recently focused on developing assays to detect prostate-derived exosomes (also called prostatosomes). Exosomes are small vesicles (50 – 150 nm in diameter) generated from internalized parts of the cellular membrane that are subsequently secreted into the blood, semen, or urine (Fig. 2C) (81). Prostate cancer patients exhibit increased numbers of exosomes in their serum compared to men with no disease, and elevated levels of exosomes may also correlate with increasing Gleason score (82). Prostate cancer RNA biomarkers, including PCA3 and TMPRSS2-ERG, can also be detected in urine-derived exosomes from prostate cancer patients (83). Although these efforts remain mainly research-oriented at this time, they provide promising future directions for biomarker research.
The role of biomarkers in prostate cancer diagnosis
PSA has persisted in clinical practice owing in large part to the public’s demand for prostate cancer screening. Indeed, PSA remains an inexpensive and sensitive biomarker for disease detection and monitoring progression and recurrence following curative therapy of local disease (7, 34). Furthermore, because PSA screening is so common, the clinical evaluation of new biomarkers has only occurred in patient populations previously screened for PSA. Thus, future iterations of prostate cancer biomarkers will most likely retain PSA as a primary clinical tool in conjunction with other tests, unless new biomarkers are shown to be superior to PSA in head-to-head comparisons. In this regard, new biomarker assays will likely complement PSA-based detection of prostate cancer (Fig. 2A, B).
A common theme in prostate cancer biomarker development is the desirability of non-invasive assays to replace biopsy as the diagnostic “gold standard”. Biopsy procedures are associated with increased risk of adverse events, such as bleeding and sepsis, owing to their invasive nature. Studies have routinely shown that biopsies are associated with a 15 – 20% false negative rate (84, 85), perhaps owing to inefficient sampling, where normal tissue is biopsied in addition to diseased tissue. Non-invasive biomarkers in serum and urine have the potential to improve the standard tissue biopsy procedure, although they cannot provide direct histopathological or spatiotemporal information. As such, supplementing PSA measurements with urine biomarker analyses may become standard practice in the near future.
Finally, these developments also need to be considered in conjunction with tissue biomarkers and imaging technologies, such as transrectal ultrasound (TRUS), computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET) (Fig. 2A). Indeed, the role of imaging is crucial to patient management for visualizing and staging both localized prostate cancers and metastatic disease, especially in the bone.
Future directions of biomarker discovery in prostate cancer
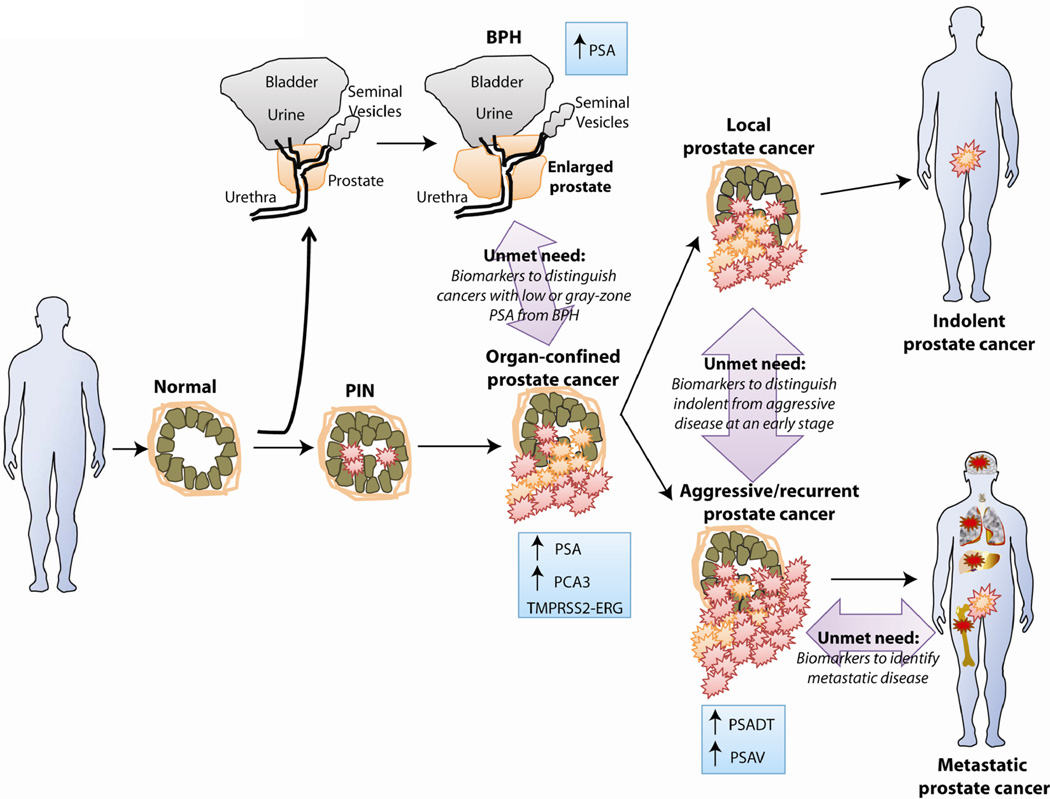
The most critical biomarker studies will focus on biomarker candidates that address the current gaps in prostate cancer biomarker development, including prognostic and predictive biomarkers (Fig. 3). The utility of PSA as a diagnostic biomarker for prostate cancer is limited by the fact that only about 3% of PSA-screened men with prostate cancer have lethal disease, thus leading to overtreatment of indolent disease (9). Development of new biomarkers that only identify more prostate cancer cases does not address this discrepancy. It follows, then, that the identification and validation of novel biomarkers to “rule out” lethal prostate cancer at the point of screening is the greatest unmet clinical need, as this may reduce unnecessary interventions that may cause more harm than good.
Figure 3. Future challenges for prostate cancer biomarker research.
Current clinical practice relies on PSA to help diagnose prostate cancer. New prostate cancer biomarkers should be targeted to addressing unmet clinical needs in prostate cancer management, including indicators for disease with low PSA values (<10ng/mL), prognostic markers to distinguish indolent from aggressive disease, and biomarkers for metastatic cancer.
One approach to identifying predictive biomarkers is to focus on genomic disease signatures, such as loss of the PTEN tumor suppressor or gain of ETS transcription factor gene fusions, which influence the biological characteristics of an individual cancer. For example, PTEN loss activates the phosphoinositide 3-kinase (PI3K) pathway, which inhibits AR signaling and causes resistance to AR-based therapies (86). Treatment of _PTEN_-null mouse tumors with combined pharmacologic inhibition of PI3K and AR signaling has led to tumor regression (86). Clinically, PTEN deletion is associated with poor outcome and hormone-refractory disease in prostate cancer (87). Therefore, PTEN deletion may be both prognostic and predictive of response to therapy. Similarly, TMPRSS2-ERG fusions may predict for tumor sensitivity to poly (ADP-ribose) polymerase 1 (PARP1) inhibition (88), and may add prognostic information detailing more aggressive disease, especially in conjunction with PTEN deletion (61, 89).
Design, interpretation, and challenges for future prostate cancer biomarkers
One of the important lessons learned from the popularization of PSA as a screening test is that biomarker development requires a priori deliberation of the intended role. Initially developed to aid the monitoring of prostate cancer recurrence, widespread uptake of the PSA test to screening even of asymptomatic men has resulted in the net overdiagnosis and overtreatment of indolent disease. As a result, clinicians must now endeavor to educate their patients about the limitations of the PSA test and also inform patients that treating prostate cancer is not always beneficial to the patient. Hindsight begs the question: “What is the best path to validate a new biomarker for clinical application?” The National Cancer Institute’s Early Detection Research Network (EDRN) has headed a response to this question with their unique biomarker discovery and validation infrastructure as well as their standardized prospective-specimen-collection, retrospective-blinded-evaluation (PRoBE) approach to biomarker validation (90).
Design
A general model for biomarker development, led by efforts from the EDRN, consists of five phases (Table 1): 1) biomarker discovery; 2) clinical assay development; 3) retrospective studies to clarify target populations; 4) prospective screening studies to determine efficacy; 5) analysis of biomarker impact in terms of cost-effectiveness and patient compliance (90, 91). The problem with biomarker development often lies not in the general framework outlined above, but poor adherence to it. Perhaps the largest shortcoming of many failed biomarker trials is that independent groups have been unable to generate concordant results; nonetheless, biomarker development proceeds (92).
Table 1.
Questions and challenges in biomarker discovery and validation
| Based on refs. 99 & 100 |
|---|
| Phase 1: Biomarker Disovery |
| Is the biomarker disease-specific? |
| Is the biomarker tissue-specific? |
| If normally expressed in another tissue, non-invasive detection may be confounded. |
| What is the dynamic range of biomarker expression? |
| What is the absolute level of biomarker expression? |
| Low-expressed biomarkers are often less reliable. |
| Phase 2: Clinical Assay Development |
| Is the assay non-invasive? |
| Is the assay reproducible? |
| Is the assay easy or difficult to perform? |
| Difficult assays have a lower likelihood of clinical utility. |
| Do biomarker assay results correlate with other confounding factors? |
| Patient age, gender, ethnicity, etc, could be confounding. |
| Phase 3: Retrospective Studies |
| What endpoints will be measured? |
| Is the cohort size adequate to evaluate the desired endpoints? |
| Are concordant results obtained from multiple independent cohorts? |
| Lack of reproducibility halts development of many initial biomarkers. |
| Do cohorts accurately reflect the target patient population? |
| Are the control and experimental cohorts matched uniformly? |
| Biases in cohort design account for many false-positive biomarker reports. |
| Does the biomarker improve upon existing clinical tests? |
| If the novel biomarker does not offer an improvement, further development may be halted. |
| Phase 4: Prospective Studies |
| Are the control and experimental cohorts matched uniformly? |
| Biases in cohort design account for many false-positive biomarker reports. |
| What endpoints will be measured? |
| The biomarker may be primarily diagnostic, prognostic, or predictive in nature. |
| What performance criteria will be measured? |
| Biomarker efficacy can be in terms of sensitivity/specificity, postive/negative predictive values,etc. |
| What clinical patient population will be used? |
| Defining the appropriate clinical context is an essential aspect often overlooked. |
| Is large-scale implementation of the biomarker feasible? |
| A feasibility analysis suggests whether the test could be used widely. |
| How does the biomarker compare to other clinical tests? |
| Phase 5: Analysis of Biomarker Impact |
| What statistical test is most appropriate? |
| What type of cross-validation is appropriate? |
| Does the analysis overfit the data? |
| Overfitting is a common problem with many analyses. |
| Is the biomarker protocol conducive to usage? |
| Acquisition of the biomarker should be simple and robust. |
| Is the biomarker cost-effective? |
| Were there issues with compliance to the biomarker? |
A major implication of this framework is that the time required from the initial discovery and retrospective studies to clinical adoption of a biomarker is often lengthy. In effect, the framework describes an adapted version of phase I/II/III clinical trials, where the idea is to establish sequential levels of evidence—from discovery to retrospective to prospective studies— that show utility of the biomarker. Ultimately, biomarker studies for prostate cancer are unlikely to be evaluated in terms of overall patient survival or progression-free survival, as these metrics may take decades to evaluate for a novel biomarker. The only practical means to potentially assess such endpoints is to create large repositories for a range of tissues on the basis of ongoing screening and therapeutic trials. Such repositories would enable large-scale evaluation of new biomarkers in the clinical trial setting in a relatively more rapid time frame (years as opposed to decades).
Interpretation
Statistical interpretation is a core consideration in biomarker studies. Any interpretation must first determine that the study is designed with sufficient power to evaluate the desired endpoints (Table 1). Then, a classic biomarker analysis evaluates the sensitivity and specificity, often using an ROC curve analysis. However, Shaw et al. argued that standard ROC curves are not always appropriate analyses, especially in the context of prostate cancer screening (93). The issue derives from the fact that new prostate cancer biomarkers are generally combined with PSA to identify improvements in the combined test over PSA. In this case, studies become biased when they cannot evaluate the performance of the secondary marker in the absence of the first marker—i.e., if a patient is PSA-negative. Therefore, a “relative” ROC (rROC) curve may be more appropriate, in which the relative true- and false-positive rates—but not their absolute true- and false-positive rates—are evaluated (93). Promising new prostate cancer biomarkers should therefore be evaluated both in combination with PSA and independent of PSA screening status. For this latter option, one approach is to move the research out of the urologist’s office and into the primary care setting, where men could be screened for a promising novel biomarker prior to receiving PSA testing and a DRE. Another option would be to design trials that include biopsies of men with an abnormal measurement of a new biomarker even if the PSA and DRE results suggest only a low risk for cancer.
Monitoring sensitivity and specificity is standard practice in biomarker studies, but this may not be sufficient to evaluate efficacy. These metrics measure the proportion of individuals, either positive or negative for the test, that have been detected accurately. But, in fact, positive and negative predictive values are more clinically informative statistics. These two metrics report a confidence for the relative value of a positive or negative test result. Even with a reasonable sensitivity and specificity, a test may actually have a low positive predictive value. Herein lies another fundamental problem with PSA: even when the sensitivity is set reasonably high, the resulting positive predictive value is only ~25% (e.g. when a 4 ng/mL cut-off value is applied) (28).
Challenges and common errors
Biomarker studies are often fraught with systematic errors in the design and execution (Table 1) (94), which has resulted in widespread failure of initially “promising” biomarker trials (92, 95). In the literature, there are five common errors that render many biomarker studies ineffectual: lack of a robust assay protocol for reproducibility; biased comparison groups in the study (case vs. controls); unclear or inappropriate clinical role of the biomarker; underpowered study size; and inappropriate statistical analyses, including overfitting of data. These errors can be made at any stage of the biomarker development process. But most frequently they occur in preclinical stages and the weakness of the biomarker is later revealed in larger trials (92).
Of these, the lack of a clear clinical role and inappropriate statistical methods are particularly germane to our discussion. First, the clinical role of newly discovered biomarkers is often only vaguely defined—if at all—leading to poorly executed clinical studies (91, 96, 97). A biomarker, by definition, is employed for only a specific patient population for a specific clinical purpose (e.g. prognosis). Extension of such a biomarker beyond its intended context is unlikely to result in positive results. PSA screening trials were commissioned decades after PSA was introduced into clinical practice as a screening test, only to conclude that PSA screening offers negligible benefit at the population level (24, 25). Perhaps the best way to avoid such complications in biomarker development is to clearly define a specific context(s) for a candidate biomarker through rigorous retrospective evaluation of the biomarker in clinically-annotated tissue repositories.
Second, the statistical analysis of biomarker trials is challenging, and there is a concern that biomarker studies too often suffer from overfitting the data for an individual dataset (90–92, 94). This will lead to positive results for a single trial that are unable to be reproduced independently. In this regard, cross-validation of the statistical analysis is an important, but only partial solution, as it still does not employ an independent set of samples. A biomarker is not considered “validated” until independent research groups have rigorously demonstrated concordant results in independent trials.
Another issue that plagues biomarker studies is that substantial bias is introduced through selective reporting of data. This “non-reporting” bias tends to mask negative reports, whereas published articles may be more positive (90, 98). For instance, in the cancer literature, Kyzas et al. demonstrated that published articles showed a significant association of p53 mutations with clinical outcome in head and neck squamous cell cancers, whereas unpublished data or data located in large, unwieldy supplemental files were markedly less positive (94). This issue of transparency may be best addressed during the peer review process, where journals can promote thorough evaluation of submitted manuscripts by providing longer periods of time for review of manuscripts with large amounts of supplementary material and specifically ask reviewers for comments on the integrity of the supplementary material.
Conclusions
The era of PSA testing in prostate cancer has imparted lasting changes in the way we think about prostate cancer biology and clinical management. Although it is natural for patients to want to know if they have prostate cancer, the high prevalence of latent cancers detected by PSA-based screening singularly argues for the use of adjunctive biomarkers that better refine disease risk. Moreover, prostate cancer biomarkers should be evaluated in terms of their intended use and clinical context: reduction in unnecessary biopsies, reduction in unnecessary prostatectomies/radiotherapy, stratification of organ-confined tumors (curable by surgery), ability to monitor progression during watchful waiting, detection of micrometastatic disease below the limit of detection for imaging modalities, or reduction in overall mortality. A more rational approach to biomarker discovery, combined with modern molecular science and bioinformatics, will eventually allow clinicians to better diagnose and target treatment for those patients who are most likely to benefit.
Acknowledgements
We thank S. Roychowdhury and members of the Chinnaiyan lab for helpful discussions and comments on this manuscript. We further acknowledge the numerous labs, authors, and publications that we were unable to cite in this review due to space restrictions.
Funding: This work was supported by the Early Detection Research Network (EDRN) grant U01 CA 11275 (A.M.C. and M.A.R.), the EDRN grant 5U01 CA113913 (J.T.W.), the Department of Defense grants PC100171 (A.M.C.) and PC094290 (J.R.P), NIH Prostate Specialized Program of Research Excellence grant P50CA69568 (to A.M.C.). A.M.C. is supported by the Doris Duke Charitable Foundation Clinical Scientist Award, the Prostate Cancer Foundation, the American Cancer Society, and the Howard Hughes Medical Institute. J.R.P. is a Fellow of the University of Michigan Medical Scientist Training Program. A.M.C. is a Taubman Scholar of the University of Michigan.
Footnotes
Competing interests: A.M.C. serves as an advisor to Gen-Probe, Inc., who has developed diagnostic tests using PCA3 and TMPRSS2-ERG. The University of Michigan has licensed the development of TMPRSS2-ERG-based prostate cancer diagnostic assays to Gen-Probe and A.M.C. and M.A.R. are named as co-inventors. Gen-Probe was not involved in the writing or approval of this manuscript.
References and notes
- 1.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 2.Oakman C, Santarpia L, Di Leo A. Breast cancer assessment tools and optimizing adjuvant therapy. Nat Rev Clin Oncol. 2010;7:725–732. doi: 10.1038/nrclinonc.2010.170. [DOI] [PubMed] [Google Scholar]
- 3.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 4.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O’Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.La Thangue NB, Kerr DJ. Predictive biomarkers: a paradigm shift towards personalized cancer medicine. Nat Rev Clin Oncol. 2011 doi: 10.1038/nrclinonc.2011.121. [DOI] [PubMed] [Google Scholar]
- 7.Lilja H, Ulmert D, Vickers AJ. Prostate-specific antigen and prostate cancer: prediction detection and monitoring. Nat Rev Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 8.Wolf AM, Wender RC, Etzioni RB, Thompson IM, D’Amico AV, Volk RJ, Brooks DD, Dash C, Guessous I, Andrews K, DeSantis C, Smith RA. American Cancer Society guideline for the early detection of prostate cancer: update 2010. CA Cancer J Clin. 2010;60:70–98. doi: 10.3322/caac.20066. [DOI] [PubMed] [Google Scholar]
- 9.Etzioni R, Cha R, Feuer EJ, Davidov O. Asymptomatic incidence and duration of prostate cancer. Am J Epidemiol. 1998;148:775–785. doi: 10.1093/oxfordjournals.aje.a009698. [DOI] [PubMed] [Google Scholar]
- 10.Lowe FC, Trauzzi SJ. Prostatic acid phosphatase in 1993. Its limited clinical utility. Urol Clin North Am. 1993;20:589–595. [PubMed] [Google Scholar]
- 11.Ercole CJ, Lange PH, Mathisen M, Chiou RK, Reddy PK, Vessella RL. Prostatic specific antigen and prostatic acid phosphatase in the monitoring and staging of patients with prostatic cancer. J Urol. 1987;138:1181–1184. doi: 10.1016/s0022-5347(17)43543-9. [DOI] [PubMed] [Google Scholar]
- 12.Balk SP, Ko YJ, Bubley GJ. Biology of prostate-specific antigen. J Clin Oncol. 2003;21:383–391. doi: 10.1200/JCO.2003.02.083. [DOI] [PubMed] [Google Scholar]
- 13.Stamey TA, Yang N, Hay AR, McNeal JE, Freiha FS, Redwine E. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N Engl J Med. 1987;317:909–916. doi: 10.1056/NEJM198710083171501. [DOI] [PubMed] [Google Scholar]
- 14.Catalona WJ, Smith DS, Ratliff TL, Dodds KM, Coplen DE, Yuan JJ, Petros JA, Andriole GL. Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N Engl J Med. 1991;324:1156–1161. doi: 10.1056/NEJM199104253241702. [DOI] [PubMed] [Google Scholar]
- 15.Parkes C, Wald NJ, Murphy P, George L, Watt HC, Kirby R, Knekt P, Helzlsouer KJ, Tuomilehto J. Prospective observational study to assess value of prostate specific antigen as screening test for prostate cancer. BMJ. 1995;311:1340–1343. doi: 10.1136/bmj.311.7016.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potosky AL, Feuer EJ, Levin DL. Impact of screening on incidence and mortality of prostate cancer in the United States. Epidemiol Rev. 2001;23:181–186. doi: 10.1093/oxfordjournals.epirev.a000787. [DOI] [PubMed] [Google Scholar]
- 17.Shibata A, Ma J, Whittemore AS. Prostate cancer incidence and mortality in the United States and the United Kingdom. J Natl Cancer Inst. 1998;90:1230–1231. doi: 10.1093/jnci/90.16.1230. [DOI] [PubMed] [Google Scholar]
- 18.Lu-Yao GL, McLerran D, Wasson J, Wennberg JE. An assessment of radical prostatectomy. Time trends, geographic variation, and outcomes. The Prostate Patient Outcomes Research Team. JAMA. 1993;269:2633–2636. doi: 10.1001/jama.269.20.2633. [DOI] [PubMed] [Google Scholar]
- 19.Walsh PC. The discovery of the cavernous nerves and development of nerve sparing radical retropubic prostatectomy. J Urol. 2007;177:1632–1635. doi: 10.1016/j.juro.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 20.Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, Gulati R, Feuer E, de Koning H. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101:374–383. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fowler FJ, Jr, Barry MJ, Walker-Corkery B, Caubet JF, Bates DW, Lee JM, Hauser A, McNaughton-Collins M. The impact of a suspicious prostate biopsy on patients’ psychological, socio-behavioral, and medical care outcomes. J Gen Intern Med. 2006;21:715–721. doi: 10.1111/j.1525-1497.2006.00464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanda MG, Dunn RL, Michalski J, Sandler HM, Northouse L, Hembroff L, Lin X, Greenfield TK, Litwin MS, Saigal CS, Mahadevan A, Klein E, Kibel A, Pisters LL, Kuban D, Kaplan I, Wood D, Ciezki J, Shah N, Wei JT. Quality of life and satisfaction with outcome among prostate-cancer survivors. N Engl J Med. 2008;358:1250–1261. doi: 10.1056/NEJMoa074311. [DOI] [PubMed] [Google Scholar]
- 23.Nanda A, Chen MH, Braccioforte MH, Moran BJ, D’Amico AV. Hormonal therapy use for prostate cancer and mortality in men with coronary artery disease-induced congestive heart failure or myocardial infarction. JAMA. 2009;302:866–873. doi: 10.1001/jama.2009.1137. [DOI] [PubMed] [Google Scholar]
- 24.Andriole GL, Crawford ED, Grubb RL, 3rd, Buys SS, Chia D, Church TR, Fouad MN, Gelmann EP, Kvale PA, Reding DJ, Weissfeld JL, Yokochi LA, O’Brien B, Clapp JD, Rathmell JM, Riley TL, Hayes RB, Kramer BS, Izmirlian G, Miller AB, Pinsky PF, Prorok PC, Gohagan JK, Berg CD. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–1319. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, Denis LJ, Recker F, Berenguer A, Maattanen L, Bangma CH, Aus G, Villers A, Rebillard X, van der Kwast T, Blijenberg BG, Moss SM, de Koning HJ, Auvinen A. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 26.Chou R, Croswell JM, Dana T, Bougatsos C, Blazina I, Fu R, Gleitsmann K, Koenig HC, Lam C, Maltz A, Rugge JB, Lin K. Screening for Prostate Cancer: A Review of the Evidence for the U.S. Preventive Services Task Force. Ann Intern Med. 2011 doi: 10.7326/0003-4819-155-11-201112060-00375. [DOI] [PubMed] [Google Scholar]
- 27.Brawer MK. Prostate-specific antigen: current status. CA Cancer J Clin. 1999;49:264–281. doi: 10.3322/canjclin.49.5.264. [DOI] [PubMed] [Google Scholar]
- 28.Schroder FH, Carter HB, Wolters T, van den Bergh RC, Gosselaar C, Bangma CH, Roobol MJ. Early detection of prostate cancer in 2007. Part 1: PSA and PSA kinetics. Eur Urol. 2008;53:468–477. doi: 10.1016/j.eururo.2007.10.047. [DOI] [PubMed] [Google Scholar]
- 29.Lucia MS, Darke AK, Goodman PJ, La Rosa FG, Parnes HL, Ford LG, Coltman CA, Jr, Thompson IM. Pathologic characteristics of cancers detected in The Prostate Cancer Prevention Trial: implications for prostate cancer detection and chemoprevention. Cancer Prev Res (Phila) 2008;1:167–173. doi: 10.1158/1940-6207.CAPR-08-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, Minasian LM, Ford LG, Lippman SM, Crawford ED, Crowley JJ, Coltman CA., Jr Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med. 2004;350:2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 31.Carter HB, Sauvageot J, Walsh PC, Epstein JI. Prospective evaluation of men with stage T1C adenocarcinoma of the prostate. J Urol. 1997;157:2206–2209. [PMC free article] [PubMed] [Google Scholar]
- 32.Kundu SD, Roehl KA, Yu X, Antenor JA, Suarez BK, Catalona WJ. Prostate specific antigen density correlates with features of prostate cancer aggressiveness. J Urol. 2007;177:505–509. doi: 10.1016/j.juro.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 33.Carter HB, Ferrucci L, Kettermann A, Landis P, Wright EJ, Epstein JI, Trock BJ, Metter EJ. Detection of life-threatening prostate cancer with prostate-specific antigen velocity during a window of curability. J Natl Cancer Inst. 2006;98:1521–1527. doi: 10.1093/jnci/djj410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Amico AV, Chen MH, Roehl KA, Catalona WJ. Preoperative PSA velocity and the risk of death from prostate cancer after radical prostatectomy. N Engl J Med. 2004;351:125–135. doi: 10.1056/NEJMoa032975. [DOI] [PubMed] [Google Scholar]
- 35.Makarov DV, Humphreys EB, Mangold LA, Carducci MA, Partin AW, Eisenberger MA, Walsh PC, Trock BJ. The natural history of men treated with deferred androgen deprivation therapy in whom metastatic prostate cancer developed following radical prostatectomy. J Urol. 2008;179:156–161. doi: 10.1016/j.juro.2007.08.133. discussion 161–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281:1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 37.Ulmert D, Serio AM, O’Brien MF, Becker C, Eastham JA, Scardino PT, Bjork T, Berglund G, Vickers AJ, Lilja H. Long-term prediction of prostate cancer: prostate-specific antigen (PSA) velocity is predictive but does not improve the predictive accuracy of a single PSA measurement 15 years or more before cancer diagnosis in alarge, representative, unscreened population. J Clin Oncol. 2008;26:835–841. doi: 10.1200/JCO.2007.13.1490. [DOI] [PubMed] [Google Scholar]
- 38.Catalona WJ, Partin AW, Slawin KM, Brawer MK, Flanigan RC, Patel A, Richie JP, deKernion JB, Walsh PC, Scardino PT, Lange PH, Subong EN, Parson RE, Gasior GH, Loveland KG, Southwick PC. Use of the percentage of free prostate-specific antigen to enhance differentiation of prostate cancer from benign prostatic disease: a prospective multicenter clinical trial. JAMA. 1998;279:1542–1547. doi: 10.1001/jama.279.19.1542. [DOI] [PubMed] [Google Scholar]
- 39.Lilja H, Christensson A, Dahlen U, Matikainen MT, Nilsson O, Pettersson K, Lovgren T. Prostate-specific antigen in serum occurs predominantly in complex with alpha 1-antichymotrypsin. Clin Chem. 1991;37:1618–1625. [PubMed] [Google Scholar]
- 40.Partin AW, Brawer MK, Bartsch G, Horninger W, Taneja SS, Lepor H, Babaian R, Childs SJ, Stamey T, Fritsche HA, Sokoll L, Chan DW, Thiel RP, Cheli CD. Complexed prostate specific antigen improves specificity for prostate cancer detection: results of a prospective multicenter clinical trial. J Urol. 2003;170:1787–1791. doi: 10.1097/01.ju.0000092695.55705.dd. [DOI] [PubMed] [Google Scholar]
- 41.Prestigiacomo AF, Lilja H, Pettersson K, Wolfert RL, Stamey TA. A comparison of the free fraction of serum prostate specific antigen in men with benign and cancerous prostates: the best case scenario. J Urol. 1996;156:350–354. doi: 10.1097/00005392-199608000-00004. [DOI] [PubMed] [Google Scholar]
- 42.Catalona WJ, Bartsch G, Rittenhouse HG, Evans CL, Linton HJ, Horninger W, Klocker H, Mikolajczyk SD. Serum pro-prostate specific antigen preferentially detects aggressive prostate cancers in men with 2 to 4 ng/ml prostate specific antigen. J Urol. 2004;171:2239–2244. doi: 10.1097/01.ju.0000127737.94221.3e. [DOI] [PubMed] [Google Scholar]
- 43.Sokoll LJ, Chan DW, Mikolajczyk SD, Rittenhouse HG, Evans CL, Linton HJ, Mangold LA, Mohr P, Bartsch G, Klocker H, Horninger W, Partin AW. Proenzyme psa for the early detection of prostate cancer in the 2.5–4.0 ng/ml total psa range: preliminary analysis. Urology. 2003;61:274–276. doi: 10.1016/s0090-4295(02)02398-1. [DOI] [PubMed] [Google Scholar]
- 44.Piironen T, Pettersson K, Suonpaa M, Stenman UH, Oesterling JE, Lovgren T, Lilja H. In vitro stability of free prostate-specific antigen (PSA) and prostate-specific antigen (PSA) complexed to alpha 1-antichymotrypsin in blood samples. Urology. 1996;48:81–87. doi: 10.1016/s0090-4295(96)00616-4. [DOI] [PubMed] [Google Scholar]
- 45.Ornstein DK, Rao GS, Smith DS, Ratliff TL, Basler JW, Catalona WJ. Effect of digital rectal examination and needle biopsy on serum total and percentage of free prostate specific antigen levels. J Urol. 1997;157:195–198. [PubMed] [Google Scholar]
- 46.Heidenreich A, Aus G, Bolla M, Joniau S, Matveev VB, Schmid HP, Zattoni F. EAU guidelines on prostate cancer. Eur Urol. 2008;53:68–80. doi: 10.1016/j.eururo.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Carter HB, Morrell CH, Pearson JD, Brant LJ, Plato CC, Metter EJ, Chan DW, Fozard JL, Walsh PC. Estimation of prostatic growth using serial prostate-specific antigen measurements in men with and without prostate disease. Cancer Res. 1992;52:3323–3328. [PubMed] [Google Scholar]
- 48.Leibovici D, Spiess PE, Agarwal PK, Tu SM, Pettaway CA, Hitzhusen K, Millikan RE, Pisters LL. Prostate cancer progression in the presence of undetectable or low serum prostate-specific antigen level. Cancer. 2007;109:198–204. doi: 10.1002/cncr.22372. [DOI] [PubMed] [Google Scholar]
- 49.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, Onofrio R, Carter SL, Park K, Habegger L, Ambrogio L, Fennell T, Parkin M, Saksena G, Voet D, Ramos AH, Pugh TJ, Wilkinson J, Fisher S, Winckler W, Mahan S, Ardlie K, Baldwin J, Simons JW, Kitabayashi N, MacDonald TY, Kantoff PW, Chin L, Gabriel SB, Gerstein MB, Golub TR, Meyerson M, Tewari A, Lander ES, Getz G, Rubin MA, Garraway LA. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM. Chinnaiyan, Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N, Isaacs WB. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 52.de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, Swinkels DW, Schalken JA. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–2698. [PubMed] [Google Scholar]
- 53.Hessels D, Schalken JA. The use of PCA3 in the diagnosis of prostate cancer. Nat Rev Urol. 2009;6:255–261. doi: 10.1038/nrurol.2009.40. [DOI] [PubMed] [Google Scholar]
- 54.Roobol MJ, Schroder FH, van Leeuwen P, Wolters T, van den Bergh RC, van Leenders GJ, Hessels D. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. Eur Urol. 2010;58:475–481. doi: 10.1016/j.eururo.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 55.Haese A, de la Taille A, van Poppel H, Marberger M, Stenzl A, Mulders PF, Huland H, Abbou CC, Remzi M, Tinzl M, Feyerabend S, Stillebroer AB, van Gils MP, Schalken JA. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol. 2008;54:1081–1088. doi: 10.1016/j.eururo.2008.06.071. [DOI] [PubMed] [Google Scholar]
- 56.Wang R, Chinnaiyan AM, Dunn RL, Wojno KJ, Wei JT. Rational approach to implementation of prostate cancer antigen 3 into clinical care. Cancer. 2009;115:3879–3886. doi: 10.1002/cncr.24447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prensner JR, Chinnaiyan AM. Oncogenic gene fusions in epithelial carcinomas. Curr Opin Genet Dev. 2009;19:82–91. doi: 10.1016/j.gde.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, Palanisamy N, Tomlins SA, Chinnaiyan AM, Shah RB. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22:1083–1093. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hessels D, Smit FP, Verhaegh GW, Witjes JA, Cornel EB, Schalken JA. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin Cancer Res. 2007;13:5103–5108. doi: 10.1158/1078-0432.CCR-07-0700. [DOI] [PubMed] [Google Scholar]
- 60.Tomlins SA, Aubin SM, Siddiqui J, Lonigro RJ, Sefton-Miller L, Miick S, Williamsen S, Hodge P, Meinke J, Blase A, Penabella Y, Day JR, Varambally R, Han B, Wood D, Wang L, Sanda MG, Rubin MA, Rhodes DR, Hollenbeck B, Sakamoto K, Silberstein JL, Fradet Y, Amberson JB, Meyers S, Palanisamy N, Rittenhouse H, Wei JT, Groskopf J, Chinnaiyan AM. Urine TMPRSS2:ERG Fusion Transcript Stratifies Prostate Cancer Risk in Men with Elevated Serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, Berney D, Foster CS, Fletcher A, Gerald WL, Moller H, Reuter V, De Bono JS, Scardino P, Cuzick J, Cooper CS. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, Hoshida Y, Mosquera JM, Pawitan Y, Lee C, Adami HO, Mucci LA, Kantoff PW, Andersson SO, Chinnaiyan AM, Johansson JE, Rubin MA. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 63.Fine SW, Gopalan A, Leversha MA, Al-Ahmadie HA, Tickoo SK, Zhou Q, Satagopan JM, Scardino PT, Gerald WL, Reuter VE. TMPRSS2-ERG gene fusion is associated with low Gleason scores and not with high-grade morphological features. Mod Pathol. 2010;23:1325–1333. doi: 10.1038/modpathol.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gopalan A, Leversha MA, Satagopan JM, Zhou Q, Al-Ahmadie HA, Fine SW, Eastham JA, Scardino PT, Scher HI, Tickoo SK, Reuter VE, Gerald WL. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–1406. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, Barrette TR, Sanda MG, Pienta KJ, Ghosh D, Chinnaiyan AM. alpha-Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287:1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 66.Rubin MA, Bismar TA, Andren O, Mucci L, Kim R, Shen R, Ghosh D, Wei JT, Chinnaiyan AM, Adami HO, Kantoff PW, Johansson JE. Decreased alpha-methylacyl CoA racemase expression in localized prostate cancer is associated with an increased rate of biochemical recurrence and cancer-specific death. Cancer Epidemiol Biomarkers Prev. 2005;14:1424–1432. doi: 10.1158/1055-9965.EPI-04-0801. [DOI] [PubMed] [Google Scholar]
- 67.Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, Lonigro RJ, Tsodikov A, Wei JT, Tomlins SA, Chinnaiyan AM. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu H, Wang B, Han C. Meta-analysis of genome-wide and replication association studies on prostate cancer. Prostate. 2011;71:209–224. doi: 10.1002/pros.21235. [DOI] [PubMed] [Google Scholar]
- 69.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, Adami HO, Hsu FC, Zhu Y, Balter K, Kader AK, Turner AR, Liu W, Bleecker ER, Meyers DA, Duggan D, Carpten JD, Chang BL, Isaacs WB, Xu J, Gronberg H. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 70.Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, Wiley KE, Isaacs SD, Johng D, Wang Y, Bizon C, Yan G, Gielzak M, Partin AW, Shanmugam V, Izatt T, Sinari S, Craig DW, Zheng SL, Walsh PC, Montie JE, Xu J, Carpten JD, Isaacs WB, Cooney KA. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 72.Rosenzweig CN, Zhang Z, Sun X, Sokoll LJ, Osborne K, Partin AW, Chan DW. Predicting prostate cancer biochemical recurrence using a panel of serum proteomic biomarkers. J Urol. 2009;181:1407–1414. doi: 10.1016/j.juro.2008.10.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao L, Lee BY, Brown DA, Molloy MP, Marx GM, Pavlakis N, Boyer MJ, Stockler MR, Kaplan W, Breit SN, Sutherland RL, Henshall SM, Horvath LG. Identification of candidate biomarkers of therapeutic response to docetaxel by proteomic profiling. Cancer Res. 2009;69:7696–7703. doi: 10.1158/0008-5472.CAN-08-4901. [DOI] [PubMed] [Google Scholar]
- 74.Sardana G, Jung K, Stephan C, Diamandis EP. Proteomic analysis of conditioned media from the PC3, LNCaP, and 22Rv1 prostate cancer cell lines: discovery and validation of candidate prostate cancer biomarkers. J Proteome Res. 2008;7:3329–3338. doi: 10.1021/pr8003216. [DOI] [PubMed] [Google Scholar]
- 75.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 76.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, Kinzler KW, Vogelstein B, Diaz LA, Jr, Velculescu VE. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roychowdhury S, Iyer MK, Robinson DR, Lonigro RJ, Wu YM, Cao X, Kalyana-Sundaram S, Sam L, Balbin OA, Quist MJ, Barrette T, Everett J, Siddiqui J, Kunju LP, Navone N, Araujo JC, Troncoso P, Logothetis CJ, Innis JW, Smith DC, Lao CD, Kim SY, Roberts JS, Gruber SB, Pienta KJ, Talpaz M, Chinnaiyan AM. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A’Hern R, Levink R, Coumans F, Moreira J, Riisnaes R, Oommen NB, Hawche G, Jameson C, Thompson E, Sipkema R, Carden CP, Parker C, Dearnaley D, Kaye SB, Cooper CS, Molina A, Cox ME, Terstappen LW, de Bono JS. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–2918. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 79.Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, Lilja H, Schwartz L, Larson S, Fleisher M, Scher HI. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 80.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 81.Duijvesz D, Luider T, Bangma CH, Jenster G. Exosomes as biomarker treasure chests for prostate cancer. Eur Urol. 2011;59:823–831. doi: 10.1016/j.eururo.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 82.Tavoosidana G, Ronquist G, Darmanis S, Yan J, Carlsson L, Wu D, Conze T, Ek P, Semjonow A, Eltze E, Larsson A, Landegren UD, Kamali-Moghaddam M. Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Proc Natl Acad Sci U S A. 2011;108:8809–8814. doi: 10.1073/pnas.1019330108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, Widmark A. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moussa AS, Jones JS, Yu C, Fareed K, Kattan MW. Development and validation of a nomogram for predicting a positive repeat prostate biopsy in patients with a previous negative biopsy session in the era of extended prostate sampling. BJU Int. 2010;106:1309–1314. doi: 10.1111/j.1464-410X.2010.09362.x. [DOI] [PubMed] [Google Scholar]
- 85.Wolters T, van der Kwast TH, Vissers CJ, Bangma CH, Roobol M, Schroder FH, van Leenders GJ. False-negative prostate needle biopsies: frequency, histopathologic features, and follow-up. Am J Surg Pathol. 2010;34:35–43. doi: 10.1097/PAS.0b013e3181c3ece9. [DOI] [PubMed] [Google Scholar]
- 86.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, Scardino PT, Rosen N, Sawyers CL. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sircar K, Yoshimoto M, Monzon FA, Koumakpayi IH, Katz RL, Khanna A, Alvarez K, Chen G, Darnel AD, Aprikian AG, Saad F, Bismar TA, Squire JA. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol. 2009;218:505–513. doi: 10.1002/path.2559. [DOI] [PubMed] [Google Scholar]
- 88.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H, Yu J, Palanisamy N, Siddiqui J, Yan W, Cao X, Mehra R, Sabolch A, Basrur V, Lonigro RJ, Yang J, Tomlins SA, Maher CA, Elenitoba-Johnson KS, Hussain M, Navone NM, Pienta KJ, Varambally S, Feng FY, Chinnaiyan AM. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yoshimoto M, Joshua AM, Cunha IW, Coudry RA, Fonseca FP, Ludkovski O, Zielenska M, Soares FA, Squire JA. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451–1460. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 90.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–1438. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ransohoff DF. How to improve reliability and efficiency of research about molecular markers: roles of phases, guidelines, and study design. J Clin Epidemiol. 2007;60:1205–1219. doi: 10.1016/j.jclinepi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 92.Diamandis EP. Cancer biomarkers: can we turn recent failures into success? J Natl Cancer Inst. 2010;102:1462–1467. doi: 10.1093/jnci/djq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shaw PA, Pepe MS, Alonzo TA, Etzioni R. Methods for Assessing Improvement in Specificity when a Biomarker is Combined with a Standard Screening Test. Stat Biopharm Res. 2009;1:18–25. doi: 10.1198/sbr.2009.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kyzas PA, Loizou KT, Ioannidis JP. Selective reporting biases in cancer prognostic factor studies. J Natl Cancer Inst. 2005;97:1043–1055. doi: 10.1093/jnci/dji184. [DOI] [PubMed] [Google Scholar]
- 95.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 96.Freidlin B, McShane LM, Korn EL. Randomized clinical trials with biomarkers: design issues. J Natl Cancer Inst. 2010;102:152–160. doi: 10.1093/jnci/djp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumor MARKer prognostic studies (REMARK) Nat Clin Pract Oncol. 2005;2:416–422. [PubMed] [Google Scholar]
- 98.Andre F, McShane LM, Michiels S, Ransohoff DF, Altman DG, Reis-Filho JS, Hayes DF, Pusztai L. Biomarker studies: a call for a comprehensive biomarker study registry. Nat Rev Clin Oncol. 2011;8:171–176. doi: 10.1038/nrclinonc.2011.4. [DOI] [PubMed] [Google Scholar]