Long Non-coding RNAs: Novel Targets for Nervous System Disease Diagnosis and Therapy (original) (raw)
Abstract
The human genome encodes tens of thousands of long non-coding RNAs (lncRNAs), a novel and important class of genes. Our knowledge of lncRNAs has grown exponentially since their discovery within the last decade. lncRNAs are expressed in a highly cell- and tissue-specific manner, and are particularly abundant within the nervous system. lncRNAs are subject to post-transcriptional processing and inter- and intra-cellular transport. lncRNAs act via a spectrum of molecular mechanisms leveraging their ability to engage in both sequence-specific and conformational interactions with diverse partners (DNA, RNA, and proteins). Because of their size, lncRNAs act in a modular fashion, bringing different macromolecules together within the three-dimensional context of the cell. lncRNAs thus coordinate the execution of transcriptional, post-transcriptional, and epigenetic processes and critical biological programs (growth and development, establishment of cell identity, and deployment of stress responses). Emerging data reveal that lncRNAs play vital roles in mediating the developmental complexity, cellular diversity, and activity-dependent plasticity that are hallmarks of brain. Corresponding studies implicate these factors in brain aging and the pathophysiology of brain disorders, through evolving paradigms including the following: (i) genetic variation in lncRNA genes causes disease and influences susceptibility; (ii) epigenetic deregulation of lncRNAs genes is associated with disease; (iii) genomic context links lncRNA genes to disease genes and pathways; and (iv) lncRNAs are otherwise interconnected with known pathogenic mechanisms. Hence, lncRNAs represent prime targets that can be exploited for diagnosing and treating nervous system diseases. Such clinical applications are in the early stages of development but are rapidly advancing because of existing expertise and technology platforms that are readily adaptable for these purposes.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-013-0199-0) contains supplementary material, which is available to authorized users.
Keywords: ANRIL, Long Non-Coding RNA, Non-Coding RNA, Microvesicle, NEAT2, Neurological Disease
Introduction
In the last decade, our understanding of the human nuclear and mitochondrial genomes has been revolutionized by technological innovations, including next-generation sequencing, and by major international consortia focused on characterizing functional genomic elements, such as the Encyclopedia of DNA Elements Project [1, 2]. One of the principal paradigm shifts is an appreciation of the existence of diverse classes of non-protein-coding RNAs (ncRNAs) with vital and wide-ranging biological roles [3]. Indeed, it is now clear that the genome contains two types of genes, those that code for proteins and those that are transcribed but lack open reading frames, and are thus not translated (i.e., ncRNA genes). Of note, this dichotomy is conceptual and not absolute, as some protein-coding RNAs have important functions at the transcript level and some ncRNAs can be translated.
ncRNAs are typically categorized based on salient features, including size, activity, and genomic context. ncRNA research is vigorously ongoing, with novel classes and subclasses of ncRNA genes, unique ncRNA transcripts, and their critical cellular functions being identified at a rapid pace. For example, microRNAs (miRNAs) are one well-characterized class of ncRNAs. These single-stranded transcripts engage in post-transcriptional regulation of target RNAs—either messenger RNAs (mRNAs) or other ncRNAs with complementary sequences that serve as miRNA binding sites—via RNA interference (RNAi) pathways. The physiological and pathophysiological roles of miRNAs have been the focus of intense scrutiny. However, interrogating long non-coding RNAs (lncRNAs)—the most novel, numerous, heterogeneous, and mechanistically and biologically complex class of ncRNAs—is now at the pinnacle of ncRNA research efforts [4–6].
A recent analysis published by Encyclopedia of DNA Elements identified more than 9,000 distinct lncRNA genes within the human genome [7]. The LNCipedia database (www.lncipedia.org) version 2.0 (accessed January 2013), which curates human lncRNA data from a number of different sources, contains entries for 17,513 lncRNA genes [8]. Other strategies for lncRNA gene discovery suggest that there are more than 50,000 in the human genome [9]. These numbers are astonishing because the widespread existence of lncRNAs has been recognized only very recently and also because the human genome only encodes approximately 20,000 protein-coding genes. These observations suggest that these previously unrecognized lncRNAs are essential players in human cell biology [4–6]. Many lines of evidence confirm that particular lncRNAs are, in fact, involved in seminal biological processes, including pluripotency, the establishment and maintenance of cell identity, and the activation of stress responses, such as DNA damage detection and repair and apoptosis. The lncRNAs that have been studied act via a remarkably diverse spectrum of molecular mechanisms, and play prominent roles in transcriptional, post-transcriptional, and epigenetic regulation. However, for the majority of lncRNAs, their biological significance has yet to be elucidated fully.
Some lncRNA genes are highly evolutionarily conserved, supporting their fundamental roles in cellular processes. Other lncRNA genes have evolved quite rapidly in a lineage-specific manner, suggesting that they served as substrates for evolutionary innovations, including the emergence of human brain structure and higher-order cognitive and behavioral functions [10]. Conspicuously, a large proportion of lncRNAs are preferentially expressed in the brains of humans and other species in specific temporal and spatial profiles [7, 11–14]. ncRNAs, as a whole, and lncRNAs, in particular, are extremely relevant within the nervous system, where they are implicated in mediating many aspects of brain evolution, development, homeostasis, and plasticity [10, 15]. An increasing array of defects in lncRNA genes (i.e., mutations) and lncRNA transcript expression, localization, and function are, correspondingly, being linked to the pathogenesis of neurological and psychiatric disorders.
In this review, we discuss our rapidly emerging knowledge of lncRNAs, including their roles in the nervous system, and provide our perspective on what we believe to be the significant and wholly untapped potential for targeting these novel factors to diagnose and treat neurological and psychiatric disorders.
Understanding the Biology of lncRNAs
A ncRNA is designated as being “long” if it is greater than 200 nucleotides. This definition reflects the size-based methodologies that are often used to isolate RNAs in the laboratory, but it belies the tremendous diversity and complexity of the lncRNA landscape that is now emerging [4–6]. lncRNA genes and associated lncRNA transcripts are quite heterogeneous in terms of their genomic context, size, regulation, life cycles, mechanisms of action, and functional repertoire.
Different classes of lncRNAs have been described based on genomic context, which refers to specific features of the locus encoding a particular lncRNA, such as the presence of protein-coding genes and their regulatory regions (e.g., promoters and enhancers), retrotransposons and other repetitive elements, and additional landmarks. A lncRNA gene found in a region that is free of other genes is referred to as a long intergenic/intervening ncRNA (lincRNA). More complex loci contain multiple genes, which can be arrayed in many different orientations relative to each other. These include transcripts derived from the same (i.e., sense) strand of DNA and those on opposite strands in a sense–antisense configuration. They can overlap partially, completely (with one gene contained entirely within another), or not at all (e.g., divergently transcribed sense–antisense pairs). In some cases, the actions of lncRNAs have been uncovered by analyzing their genomic contexts. For example, 70 % of protein-coding genes are associated with antisense lncRNAs, called natural antisense transcripts, which are thought to regulate the corresponding protein-coding genes through various mechanisms (see below).
Like their protein-coding counterparts, lncRNAs can be comprised of multiple exons and can be hundreds of kilobases in length (e.g., X active coating transcript) [16]. The expression of lncRNA genes can be modulated by transcriptional and epigenetic regulatory factors, including other classes of ncRNAs (i.e., miRNAs). lncRNA transcripts can be subject to post-transcriptional processing, intracellular and intercellular transport, association with RNA binding proteins, and myriad interactions with other molecules. Accordingly, lncRNAs harbor sequence motifs and other elements that govern, for example, their regulation (e.g., transcription factor binding sites and miRNA binding sites); post-transcriptional processing (e.g., splicing donor/acceptor sites, polyadenylation sites, and RNA editing sites); their folding into secondary and higher-order structures; and their functional relationships with other nucleic acids and proteins (e.g., binding domains).
Because of this cache of features, lncRNAs can serve as highly dynamic, modular, multimodal, and energy efficient machines for sensing and integrating information, and, in turn, for coordinating when and where cellular programs are executed with great precision [6]. Operationally, lncRNAs act within the nucleus primarily, but also in the cytoplasm, and are implicated in a broad range of processes involving multiple cellular ensembles—the genome, the transcriptome, and the proteome. lncRNAs have a diversity of molecular mechanisms of action that are still emerging, including roles in mediating local and higher-order epigenetic states, transcription and post-transcriptional processes, ncRNA regulatory network topologies and dynamics, and nuclear architecture (Table 1). These have been discussed in detail in many excellent review articles [4–6].
Table 1.
Emerging functional roles for long non-coding RNAs
| Local and higher-order epigenetic states |
|---|
| Guide non-selective chromatin-modifying proteins to specific sites throughout the genome |
| Recruit chromatin-modifying proteins to modify local epigenetic environments |
| Recruit DNA methylation enzymes/factors to specific sites |
| Act as scaffolds for assembly of multiple chromatin-modifying proteins |
| Act as tethers for direct allele- and locus-specific control in cis |
| Promote chromatin looping |
| Promote formation of chromatin boundaries |
| Promote global gene network reprogramming |
| Transcription |
| Modulate the transcription of proximally located genes directly |
| Modulate the transcription of proximally located genes indirectly via effects on transcriptional co-regulators |
| Act as decoys for transcription factors |
| Compete with transcription factors for DNA binding sites |
| Influence the activity/subcellular localization of transcription factors |
| Interfere directly with RNA polymerase II-mediated initiation complex formation/elongation |
| Post-transcriptional processes |
| Modulate post-transcriptional RNA modifications (e.g., alternative splicing) |
| Modulate RNA stability |
| Modulate RNA trafficking |
| Modulate RNA quality control |
| Modulate mRNA translation |
| Modulate local mRNA translation in the synaptic compartment |
| Non-coding RNA network topologies and dynamics |
| Regulate other long and short non-coding RNAs at the epigenetic, transcriptional, and post-transcriptional level |
| Serve as hosts for the biogenesis of certain short non-coding RNAs |
| Act as microRNA sinks/sponges that bind to miRNAs, sequestering them away from their target mRNAs |
| Bind to microRNA binding sites in mRNAs, masking these sites and preventing miRNA-mediated regulatory events |
| Have dual roles as long non-coding RNA transcripts and as substrates for translation |
| Nuclear architecture |
| Act as scaffolds for the formation of subnuclear domains and their dynamic reorganization |
Briefly, in terms of the best characterized roles of lncRNAs in establishing local and higher-order epigenetic states, lncRNA transcripts can recruit histone modifying enzymes and chromatin remodeling complexes to an individual genomic locus, affecting local chromatin states associated with a particular gene or gene cluster, as in genomic imprinting. These local chromatin states can subsequently be propagated across an entire chromosome, as in X chromosome inactivation. lncRNA transcripts can also guide histone-modifying enzymes and chromatin-remodeling complexes to sites distributed throughout the entire genome promoting global epigenetic reprogramming, which occurs, for example, during growth and development. With respect to transcription, the process of lncRNA transcription (rather than the lncRNA transcript itself) can interfere with the recruitment of additional transcriptional machinery (i.e., RNA polymerase II) to the same locus, leading to the silencing of nearby genes [17]. lncRNA transcripts can tether transcription factors and other co-regulatory molecules to a specific genomic locus, affecting the transcription of genes located proximal to this site. Also, lncRNA transcripts can modulate post-transcriptional RNA processing, transport, stabilization, metabolism, and translation through a range of potential mechanisms, often through the modulation of ncRNA regulatory networks. For example, miRNA-mediated regulation of a particular mRNA can be mitigated by the presence of (i) a lncRNA that binds to the miRNA directly (i.e., a competing endogenous RNA that acts as a sink or sponge for miRNAs) or (ii) an antisense lncRNA that binds to the mRNA and masks the corresponding miRNA binding site. lncRNA transcripts can serve as precursors for the biogenesis of classes of short ncRNAs via cleavage of the host lncRNA. lncRNA transcripts can promote trafficking of proteins between the nucleus and cytoplasm and between other subcellular compartments. Lastly, lncRNA transcripts can serve as architectural components of subnuclear domains (e.g., speckles and paraspeckles). These dynamic structures assemble functionally related loci from different chromosomal regions, along with various enzymes, regulatory proteins, and other nuclear factors into hubs coupling transcription and specific post-transcriptional processes within the three-dimensional context of the nucleus. These domains promote the rapid and efficient execution of genomic programs, such as cell growth and the deployment of stress responses.
These highly versatile lncRNAs are generally expressed at lower levels than mRNAs but in a more cell type-selective manner. Accordingly, important physiological roles are increasingly being recognized for lncRNAs in particular tissues.
Uncovering Roles for lncRNAs in the Nervous System
lncRNAs are preferentially expressed in the nervous system compared with other organ systems. Data from animal models, as well as human tissues, reveals patterns that are regional; cell type-, subcellular compartment-, and developmental stage-specific; activity-dependent; and even sexually dimorphic [7, 10–15, 18–20]. An increasing number of studies are focused on interrogating the corresponding neurobiological roles played by these lncRNAs. The best-characterized functions are those linked to brain development, the establishment and maintenance of neural cell identity, plasticity, and the deployment of stress responses.
Brain Development
Several lines of evidence, including studies of lncRNA evolution, genomic features, expression and functional manipulation, strongly suggest that a significant subset of lncRNAs serve as mediators of brain development. An expanding inventory of different lncRNAs is expressed during brain development in species ranging from fly and zebrafish to mouse and human [11, 12, 21, 22]. Some of these lncRNAs are rapidly evolving in a lineage-specific manner and may have played a role in the emergence of human brain structure and function. For example, the highly accelerated region 1A/B lncRNAs (HAR1A/B) are transcribed from a Human Accelerated Region, which refers to regions of the genome subject to positive selection since our divergence from the great apes. HAR1A is expressed in Cajal-Retzius cells of the developing human neocortex during gestational weeks 7–19 in a pattern similar to that of reelin, a key protein that is responsible for orchestrating forebrain development [23]. In contrast, other lncRNAs are evolutionarily conserved. Some orthologous lncRNAs exhibit similar expression patterns in the embryonic and postnatal brains of birds, marsupials, and eutherian mammals, implying that they have fundamental roles in brain development [13]. Furthermore, a large proportion of lncRNAs expressed in the developing brain are associated, because of their genomic context, with protein-coding genes known to play roles in neural gene regulation and brain development. These observations, along with our current understanding of lncRNA mechanisms of action, support the conclusion that lncRNAs mediate brain development, at least in part, by modulating proximally-located neural developmental genes.
Additional data from functional manipulations of lncRNAs confirms the effect of lncRNAs on brain development. A study in zebrafish reported that knocking down the megamind lncRNA results, initially, in defects in ventricular morphology (i.e., midbrain ventricle expansion and forebrain ventricle contraction) and, subsequently, in small heads and eyes, enlarged ventricles (i.e., hydrocephalus), and loss of neurogenic differentiation-positive neurons in the retina and tectum [22]. Knock down of the cyrano lncRNA similarly produced small heads and eyes, defects in neural tube opening, loss of neurogenic differentiation-positive neurons in the retina and tectum, and enlarged nasal placodes. Another example is loss of the Dlx6 antisense RNA 1 (Dlx6as1) lncRNA in mouse, which leads to deregulated expression of transcription factors involved in the development of gamma-aminobutyric acid (GABA)-ergic interneurons [24]. In turn, loss of Dlx6as1 results in decreased numbers of GABAergic interneurons within the early postnatal hippocampus and dentate gyrus. This GABAergic interneuron population subsequently normalizes; however, abnormalities in excitability remain present throughout life, highlighting the relevance of Dlx6as1 in generating forebrain circuitry.
Establishment and Maintenance of Neural Cell Identity
lncRNAs also serve as critical regulators of cellular identity. There are examples from most, if not all, cell types that have been examined, including embryonic and other stem cells [25], neuronal and glial cells, muscle [26], heart [27], pancreas [28], blood [29], and fat [30], as well as other types of cells [31]. The cell-intrinsic, cell–cell, and environmental factors known to be involved in modulating cell identity (e.g., signaling pathways, cell cycle regulation, transcription factor codes, histone modifications and chromatin remodeling, and miRNA networks) interface with lncRNAs through the diverse mechanisms of action described above. Conversely, the expression and function of lncRNAs can also be subject to regulation by these factors.
The roles of lncRNAs in the establishment and maintenance of neural cell types were among the first to be discovered. Many lncRNAs exhibit specific patterns of expression in developing and mature neural cell types. These lncRNAs are often linked, because of their genomic context and because of correlating expression profiles with protein-coding genes known to play roles in neural lineage restriction, neuronal and glial lineage specification, maturation, and terminal differentiation. One study uncovered modules of lncRNAs that are dynamically regulated during GABAergic neurogenesis and progressive stages of oligodendrocyte (OL) lineage elaboration [32]. For example, it identified a lncRNA transcribed divergently, but sharing a promoter region with SRY-box containing gene 8 (Sox8), an important OL developmental transcription factor. This lncRNA, Sox8 Opposite Transcript (AK079380), exhibited a concordant expression profile with Sox8, implying a role in OL lineage specification and maturation. Other studies have similarly revealed lncRNAs with putative functions in OL lineage maturation; GABAergic, dopaminergic, and glutamatergic neurogenesis; and retinal cell fate specification utilizing different neural developmental cellular paradigms [33–37]. A significant number of these lncRNAs are implicated in recruiting chromatin-remodeling complexes that are known to regulate neural cell identity, such as RE1-silencing transcription factor (REST), corepressor for RE1-silencing transcription factor (CoREST), and polycomb repressive complex 2 (PRC2), to their genomic sites of action [38, 39].
Plasticity
lncRNAs are also implicated in modulating synapse formation and function. In particular, nuclear enriched abundant transcript 2 (NEAT2)/metastasis-associated lung adenocarcinoma transcript 1, brain cytoplasmic RNA 1, and testes-specific X-linked are all lincRNAs that regulate synaptogenesis, local protein synthesis at the synapse, and fear-conditioning and short-term hippocampal memory consolidation, respectively [40–42]. Other lncRNAs are transcribed from genomic loci encompassing protein-coding genes involved in synapse development and functioning. For example, loci for neurogranin and calcium/calmodulin-dependent protein kinase II inhibitor 1, genes that are critical for synaptic plasticity and long-term potentiation, also encode multiple antisense lncRNAs, which are dynamically expressed in the mouse brain [43]. Profiling lncRNAs in the nucleus accumbens of cocaine-conditioned mice, an animal model that exhibits aberrant plasticity, revealed hundreds of differentially expressed lncRNAs, including those associated with plasticity-related genes [44]. lncRNAs are similarly deregulated in relevant human neuropathological specimens from patients with alcohol and heroin addiction, as well as other disorders characterized by abnormal plasticity, such as autism and intractable epilepsy [45–48]. Many of these lncRNAs are linked with genes that have key roles in long-term potentiation, synaptic activity and memory, such as brain-derived neurotrophic factor (BDNF) and activity-regulated cytoskeleton-associated protein. In addition, thousands of neuronal cell-specific lncRNAs, designated enhancer RNAs (eRNAs), are transcribed from enhancer elements in a stimulus-dependent manner [49]. The expression levels of these eRNAs are positively correlated with those of proximally located genes, suggesting that eRNAs modulate activity-dependent neuronal gene transcription.
Deployment of Stress Responses
lncRNAs induced by stress can promote the execution of highly coordinated responses, such as global reductions in gene transcription and protein synthesis, and regulation of genes and gene networks that are protective against particular insults. Indeed, lncRNAs and associated factors are increasingly being implicated in mediating stress responses that are important in the nervous system, including those against heat shock, DNA damage, hypoxia, nutrient limitation, lipid-induced oxidation, endoplasmic reticulum stress, cellular transformation, and infection [50, 51]. For example, DNA damage provokes transcription of lncRNAs from the cyclin-dependent kinase inhibitor 1A (CDKN1A) gene promoter [52]. Expression of one of these lncRNAs, P21 associated ncRNA DNA damage activated, is p53-dependent, like that of the CDKN1A mRNA. However, it acts independently and inhibits apoptotic gene expression. Other lncRNAs are also induced by DNA damage in a p53-dependent manner [53]. One of these, which is encoded upstream of P21 associated ncRNA DNA damage activated, lincRNA-p21, acts as a repressor in the p53 pathway. Antisense non-coding RNA in the INK4 locus (ANRIL) is another lncRNA that is up-regulated by DNA damage, in an ataxia telangiectasia mutated signaling-dependent manner. In turn, ANRIL inhibits the expression of the CDKN2A/alternate open reading frame and CDKN2B genes during the late stage of the DNA damage response, facilitating the completion of DNA repair [54]. Similarly, ionizing radiation enhances the expression of lncRNAs encoded upstream of the cyclin D1 (CCND1) promoter that recruit regulatory factors, such as the RNA/DNA binding protein, translocated in liposarcoma—which is linked to amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD)—to the cyclin D1 promoter region ultimately leading to gene repression [55]. In addition, heat shock can induce lncRNAs, including Alu lncRNAs and satellite III repeat-derived lncRNAs, in humans, and B2 SINE RNA, in mice, that repress general mRNA transcription. Alu and B2 SINE RNA ncRNAs bind to RNA polymerase II and inhibit its functional interaction with promoter DNA [56, 57]. By contrast, satellite III ncRNAs promote the formation of nuclear stress bodies that rapidly effect changes to gene expression [58, 59]. Nuclear stress bodies consist of transcriptional regulators and RNA processing factors, including the heat shock transcription factor 1, which promotes transcription of cytoprotective genes. Further, heat shock RNA 1 is a lncRNA that is required to activate heat shock transcription factor 1 [60, 61].
lncRNAs can also mediate stress responses through changes in their nuclear-cytoplasmic localization. For example, a lncRNA transcribed antisense to ubiquitin carboxy-terminal hydrolase L1, a susceptibility gene for Parkinson’s disease and other forms of neurodegeneration, translocates from the nucleus to the cytoplasm in dopaminergic neurons when the mammalian target of rapamycin signaling pathway—a master regulator of growth, metabolism, autophagy, and other homeostatic processes—is inhibited [62]. Increased levels of this lncRNA in the cytoplasm promote translation of ubiquitin carboxy-terminal hydrolase mRNA, leading to increased levels of this neuroprotective protein. Cellular stress can also alter the subcellular localization of brain-specific repetitive RNA, a lncRNA transcribed from the Dlk1-Dio3 locus that is largely found in the nucleus [63]. Under cellular stress conditions, brain-specific repetitive RNA translocates selectively into cytoplasmic stress granules in hypothalamic neurons. Although its precise function within these RNA granules is unknown, it is likely that it also plays a role in translational control given this localization.
Functional manipulation experiments further suggest that stress-induced lncRNAs are important in nervous system disorders. For example, in Drosophila models of several different polyglutamine (polyQ) diseases, over expression of the heat shock RNA omega (hsrω) lncRNA increases polyQ-mediated cytotoxicity, whereas hsrω down-regulation reduces polyQ protein aggregation and mitigates neurodegeneration [51].
Brain Aging
Changes in the expression of genes mediating stress responses and mitochondrial functions underlie brain aging and are evolutionarily conserved [64–66]. By contrast, certain alterations, such as the coordinated down-regulation of neuronal genes during human brain aging, have emerged more recently during evolution. Unexpectedly, human brain aging seems to mirror brain development, with gene expression trajectories found in the later decades of life recapitulating patterns present in infancy. As a corollary, these observations support the conclusion that long ncRNAs with established and emerging roles in brain development are likely to be important in brain aging. Indeed, preliminary studies in animal models have revealed differential expression of lncRNAs in the aging brain [67]. In humans, the level of at least one neural lncRNA, BC200, is down regulated by 60 % in the cortex with aging [68]. Given its neurobiological role, decreased levels of BC200 likely influence age-associated changes in synaptic functioning. It is intriguing to speculate that lncRNAs—like miRNAs—may be responsible for brain aging, rather than simply being markers thereof [69].
Further relationships may exist between brain aging and lncRNA biology, potentially associated with stem cell aging. The phenomenon of stem cell aging refers to a diminished capacity for self-renewal, proliferation, and differentiation, and activation of senescence and apoptosis pathways, leading, in turn, to impairments in tissue homeostasis and repair [70]. lncRNAs seem to regulate the maintenance of stem cell pools and are integrated with genomic loci encoding factors involved in senescence and lifespan, such as insulin-like growth factor 2 (i.e., H19) and CDKN2A/B and alternate open reading frame (i.e., ANRIL) [71, 72]. Other lncRNAs, such as Alu RNAs, may similarly play roles in brain aging through the modulation of transcriptional and epigenetic regulatory programs, genomic stability, and DNA damage and other stress responses [73, 74].
Establishing Paradigms for lncRNA-Related Pathology
Given the important biological roles played by lncRNAs, it is not surprising that these factors are also implicated in mediating disease processes. They are linked to common disease states including, most prominently, cancer and nervous system disorders, as well as immunological, metabolic, vascular, and other pathologies [75]. These include, but are not limited to, susceptibility to viral and bacterial pathogens; human immunodeficiency virus replication; type 2 diabetes; hemolysis, elevated liver enzymes, and low platelets syndrome; brachydactyly and other developmental defects; and cancer risk, progression, and metastasis. An evolving literature is focused on better understanding and interpreting this lncRNA-related pathology, and several distinct (but non-mutually exclusive) paradigms are emerging. Here, we highlight examples within this framework that are pertinent to the nervous system (Fig. 1; Table 2).
Fig. 1.
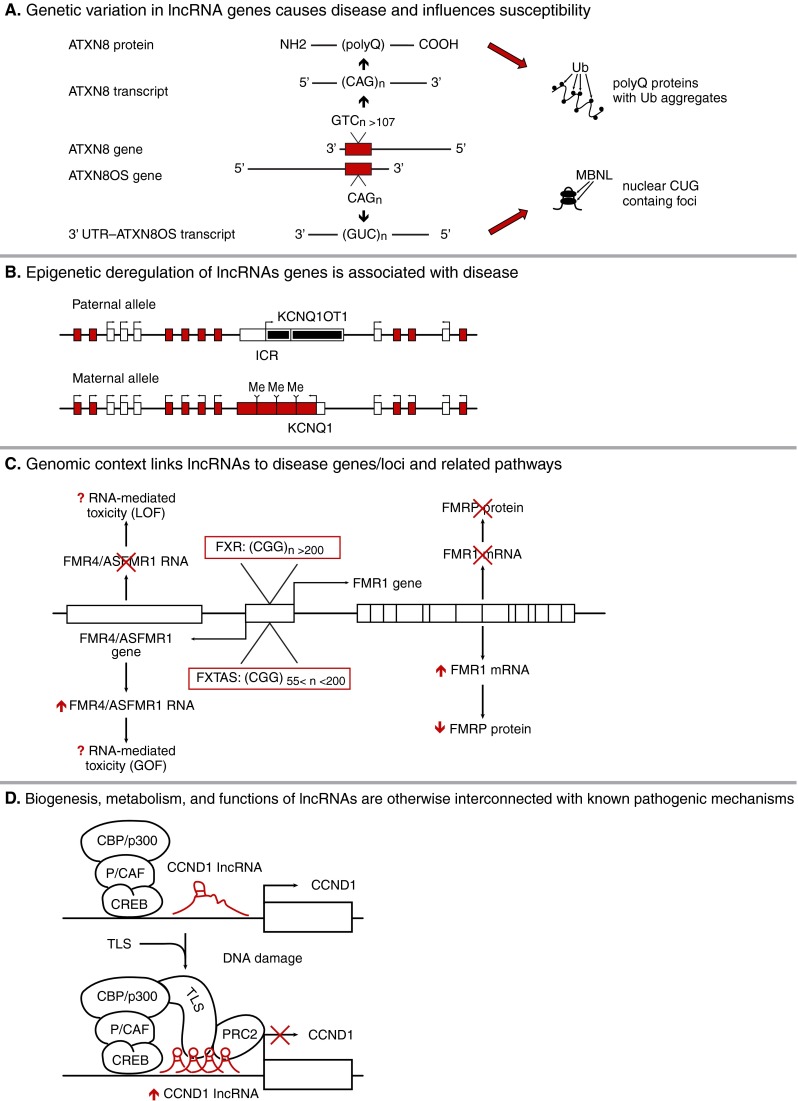
(A) Bidirectional CTG/CAG expansion at the spinocerebellar ataxia, type 8 locus gives rise to CUG expansion RNAs from the long non-coding ataxin 8 opposite strand (ATXN8OS) gene and polyglutamine expansion proteins produced by ataxin 8 (ATXN8) CAG transcripts expressed in the opposite direction, resulting in accumulation of ribonuclear inclusions that co-localize with muscleblind-like (MBNL) and polyglutamine proteins with ubiquitin (Ub) aggregates, respectively. (B) Mutations in the potassium voltage-gated channel KQT-like subfamily member1 (KCNQ1) imprinted region are associated with Beckwith-Wiedemann and Russell-Silver syndromes, and imprinting is regulated by epigenetic modification of the long non-coding RNA, KCNQ overlapping transcript 1 (KCNQ1OT1). The imprinting control region (ICR) for KCNQ1 contains the promoter for KCNQ1OT1: on the paternal allele, the ICR is unmethylated allowing KCNQ1OT1 expression that silences the paternal allele of linked genes in cis; on the maternal allele, KCNQ1OT1 expression is repressed by ICR methylation and the adjacent imprinted genes are expressed. Red boxes with arrows denote active maternal allele. (C) The fragile-X mental retardation locus contains an expanded CGG repeat of variable length in the 5’ untranslated region (UTR) of the fragile-X mental retardation 1 (FMR1) gene resulting in either a developmental disorder [fragile-X mental retardation (FXR)] or a neurodegenerative disorder [fragile-X tremor ataxia syndrome (FXTAS)] depending on full or pre-mutation length expansion repeats and the associated absence of FMR1 mRNA and fragile-X mental retardation protein (FMRP) protein or increased levels of FMR1 mRNA and reduced levels of FMRP protein, respectively. A long non-coding antisense RNA [fragile-X mental retardation 4/antisense FMR1 (FMR4/ASFMR1)] is also generated from this locus, possibly from a bidirectional promoter associated with the FMR1 5’ UTR, and exhibits parallel changes in RNA expression profiles to those seen in FXR or FXTAS; this may contribute to unique forms of RNA-mediated disease toxicity. (D) The expression of the cyclin D1 (CCND1) gene is positively regulated by a transcription factor complex [cAMP response element-binding protein (CREB)-binding protein (CBP)/p300, p300/CBP associated factor (P/CAF), CREB] present within the promoter region that also contains a CCND1 long non-coding RNA. Following DNA damage, CCND1 mRNA expression is repressed by induction of CCND1 long non-coding RNAs in cis that recruit the RNA binding protein, translocated in liposarcoma (TLS), which inhibits the histone acetyltransferase activity of CBP/p300 and also recruits the repressive polycomb repressive complex 2 (PRC2) complex. GOF = gain-of-function; LOF = loss-of-function; Me = methylated; polyQ = polyglutamine
Table 2.
Non-mutually exclusive paradigms for characterizing how long non-coding RNAs (lncRNAs) are intimately connected with neurological and psychiatric diseases with examples
| I. Genetic variation in lncRNA genes causes disease and influences susceptibility | ||
|---|---|---|
| Alzheimer’s disease | ANRIL | [78, 108–112] |
| Atherosclerosis | ||
| Diabetes | ||
| Glaucoma | ||
| Glioma | ||
| Intracranial aneurysm | ||
| Plexiform neurofibroma | ||
| Stroke | ||
| Progressive encephalopathy with severe infantile anorexia | SLC7A2-IT1A/B | [77] |
| Intellectual and developmental disability | LINC00299 | [76] |
| Prader-Willi syndrome | IPW | [113, 114] |
| Spinocerebellar ataxia type 8 | ATXN8OS | [115, 116] |
| West syndrome | BX118339 | [117] |
| II. Epigenetic deregulation of lncRNAs genes is associated with disease | ||
| Beckwith-Wiedemann syndrome | KCNQ1OT1 | [118] |
| Facioscapulohumeral muscular dystrophy | DBE-T | [81] |
| Glioma | H19 | [119, 120] |
| Medulloblastoma | ||
| Meningioma | ||
| Neural tube defects | ||
| Glioma | MEG3 | [121–124] |
| Medulloblastoma | ||
| Meningioma | ||
| Neuroblastoma | ||
| Pituitary adenoma | ||
| Prader-Willi syndrome | MKRN3-AS1 | [125] |
| III. Genomic context links lncRNAs to disease genes/loci and related pathways | ||
| Alzheimer's disease | BACE1-AS | [126, 127] |
| Inclusion body myositis | ||
| Angelman syndrome | UBE3A-ATS | [128] |
| Autism spectrum disorder | DISC2 | [129] |
| Bipolar disorder | ||
| Major depression | ||
| Schizophrenia | ||
| Fragile X | FMR4/ASFMR | [130, 131] |
| Fragile X-associated tremor and ataxia syndrome | ||
| Neurodevelopmental syndromes associated with the SOX2 locus | SOX2OT | [132] |
| Parkinson’s disease | UCH1LAS | [62] |
| Spinocerebellar ataxia type 7 | SCAANT1 | [133] |
| IV. Biogenesis, metabolism, and functions of lncRNAs are otherwise interconnected with known pathogenic mechanisms | ||
| Diffuse cerebral hypomyelination with cerebellar atrophy and hypoplasia of the corpus callosum | RNA polymerase III-dependent lncRNAs | [83] |
| TDP-43-associated pathological states | NEAT1/2 | [84] |
| TLS-associated pathological states | CCND1 promoter-derived lncRNAs | [55] |
| Huntington’s disease | REST/CoREST-regulated lncRNAs | [134] |
| Macular degeneration | Alu lncRNAs | [135] |
| p53-associated pathological states | PANDA, lincRNA-p21 | [52, 53] |
It is clear that genetic variation in lncRNA genes can directly cause disease and influence susceptibility. A recent study identified a severely intellectually and developmentally disabled 16-year-old woman with a 46, XX, t(2;11)(p25.1;p15.1)dn balanced translocation that selectively disrupts the LINC00299 gene, providing compelling evidence linking this genomic locus to the disease phenotype [76]. This lncRNA is ubiquitously expressed—most prominently in brain. Another study similarly reported that progressive encephalopathy with severe infantile anorexia (i.e., Ravine encephalopathy) is mediated by a point mutation in the SLC7A2-IT1 lncRNA gene locus within a primate-specific retroelement. Two lncRNAs, SLC7A2-IT1A and SLC7A2-IT1B, are transcribed from this locus and expressed in brain. In vitro knock down of these lncRNAs leads to neuronal apoptosis, consistent with the human neuropathological findings [77].
Another intriguing example is the ANRIL lncRNA gene on chromosome 9p21, which has been described as a major unexpected hotspot in genome-wide association studies for human diseases, with strong association signals for vascular disorders and cancer [78]. Although this locus encompasses protein-coding genes with tumor suppressor functions and other roles in regulating cell cycle, apoptosis, senescence, and aging, polymorphisms in ANRIL are preferentially associated with disease risk. For example, rs6475606, rs564398 and rs496892, rs1063192, and rs2151280 modify susceptibility to intracranial aneurysm, large-vessel ischemic stroke, glioma, and plexiform neurofibroma, respectively. Other correlations between lncRNA polymorphisms and disease are actively being investigated [79]. One study analyzed data from the Alzheimer Disease (AD) Neuroimaging Initiative and reported that the rs7990916:T>C polymorphism in the brain-specific, rapidly evolving lincRNA, TCONS_00021856/linc-SLITRK5-11, is associated with regional cortical gray matter volumes in normal controls and in patients with mild cognitive impairment and AD [80]. These examples offer one explanation for interpreting genome-wide association studies disease association signals in non-coding genomic regions.
Epigenetic deregulation of lncRNAs genes can also be associated with disease. For example, in facioscapulohumeral muscular dystrophy, the pathogenic D4Z4 binding element transcript lncRNA is transcribed from the facioscapulohumeral muscular dystrophy locus on chromosome 4q35. This results from disruption of the repressive profiles of DNA methylation, histone modification, and higher-order chromatin usually present at this locus in healthy people [81]. These epigenetic abnormalities arise because of disease-causing deletions of the D4Z4 polymorphic tandem repeat sequence located on chromosome 4q35, which reduces the number of repeats to less than 11. In turn, D4Z4 binding element transcript promotes further epigenetic and transcriptional changes at this locus by recruiting the Trithorax group protein, ASH1L, which catalyzes histone 3, lysine 36 dimethylation, and histone 3, lysine 4 trimethylation, and activates pathogenic 4q35 gene transcription.
In addition, genomic context can link lncRNAs to disease genes/loci and related pathways. In particular, antisense lncRNAs are associated with many disease-related factors, including those mediating common neurodevelopmental, neurodegenerative, neuro-oncological, and psychiatric disorders. For example, BACE1 antisense RNA (BACE1-AS) is a lncRNA transcribed antisense to the β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) gene, which is important for amyloid metabolism and implicated in the pathogenesis of AD. BACE1-AS stabilizes the BACE1 mRNA by forming a duplex with the BACE1 mRNA and masking the miR-485-5p miRNA binding site [82]. BACE1-AS activity is influenced by stress and its levels are elevated in brain tissues from AD patients, supporting its significance in AD pathophysiology.
Moreover, the biogenesis, metabolism, and functions of lncRNAs can otherwise be interconnected with known pathogenic mechanisms. For example, a study of patients with an inherited leukoencephalopathy (i.e., diffuse cerebral hypomyelination with cerebellar atrophy and hypoplasia of the corpus callosum) identified mutations in subunits of RNA polymerase III as the cause of this syndrome [83]. It suggested that the underlying pathogenic mechanism is deregulation of Pol III-transcribed ncRNAs, which include non-polyadenylated lncRNAs and those that can be cleaved into short ncRNAs. Furthermore, TAR DNA binding protein (TDP-43) and TLS are DNA/RNA binding proteins involved in transcriptional and post-transcriptional regulation, and in the pathogenesis of ALS and FTLD. Notably, both of these factors bind to numerous lncRNAs, such as NEAT1 and NEAT2, and these profiles are perturbed in FTLD and ALS [84, 85].
Lastly, increasing evidence from animal models and patient-derived tissues reveals that many lncRNAs are differentially expressed in a spectrum of neurological and psychiatric disorders. These include, as examples, lncRNA signatures in neuropathological specimens from patients with AD [86], addiction to alcohol [46], addiction to heroin [45], autism [47], gliomas [87, 88], Huntington’s disease [89], and pituitary adenomas [90]. Importantly, these abnormal expression patterns are not only found in brain but they can also be present in more accessible fluids and tissues. The contributions of these peripheral lncRNA profiles to central disease pathophysiology are largely unknown.
Membrane-bound exosomes and microvesicles that can circulate between cells within the brain and between cells in the brain and those in the periphery (and vice versa) are emerging as carriers for lncRNAs [91–93]. It has been suggested that these extracellular vehicles carry the bulk of RNA circulating in the periphery, and they are very relevant to disease. In cancers, such as glioblastoma, for example, these factors modulate immune responses and the tumor microenvironment, and promote angiogenesis and tumor invasiveness [91, 92].
Developing Clinical Applications Targeting lncRNAs
Because of the rapid pace of discoveries linking lncRNAs to different pathologies, this field has been christened a potential “goldmine” for identifying novel targets to diagnose and treat human diseases [94–97]. Many of these lncRNA targets have yet to be validated, and the development of clinical applications exploiting lncRNAs remains in its early stages. Nevertheless, our growing appreciation for the biology of lncRNAs, their roles in the nervous system, their seemingly ubiquitous deregulation in neurological and psychiatric diseases, and the magnitude of unmet clinical needs related to these disorders suggest that diagnostic and therapeutic strategies targeting lncRNAs offer mechanistic and also potentially effective approaches for distinguishing and mitigating disease pathology that are complementary to, and likely more precise than, existing ones. Clinical applications can even be propelled into development in a prompt and relatively cost-effective manner because tools for interrogating lncRNAs and for modulating them can be adapted, at least in part, from existing technology platforms designed for different, but related, purposes, such as targeting aberrant mRNAs and miRNAs. Salient examples include the use of next-generation sequencing, molecular imaging, microfluidics, microvesicles, nanotechnologies, genome editing, antisense oligonucleotides (ASOs), and RNAi.
Diagnostic Strategies
Diagnostic strategies can be envisioned that focus on analyzing lncRNA genes, the epigenetic status of lncRNA gene loci, and lncRNA transcript expression and function.
One approach that can be implemented immediately is examining annotated lncRNA genes in tissues from patients with nervous system disorders. As we have highlighted, variation in lncRNAs genes may cause disease or influence susceptibility. Given the emerging roles of lncRNAs in regulating brain development and neural cell identity, this methodology may be particularly relevant for uncovering certain forms of intellectual and developmental disability and somatic genomic alterations in lncRNAs that are associated with brain tumors and conceivably indicative of sensitivities to specific drugs. Importantly, lncRNA sequence information is already embedded within whole genome sequencing data sets that are encountered not only in research laboratories but also increasingly commonly in clinical settings.
Another strategy that can be implemented immediately is probing the epigenetic status of lncRNA gene loci. A variety of tools and techniques have been developed in the last decade to profile DNA methylation, histone modification, and chromatin states present at an individual gene locus or throughout the entire genome. These can be used not only to interrogate protein-coding gene loci but also lncRNA gene loci, as the catalog of disease-associated epigenetic abnormalities in lncRNA gene loci continues to expand. As a corollary, these measurements could serve as biomarkers for drug screening assays and, clinically, for monitoring disease activity and response to therapies.
An additional approach is evaluating lncRNA transcript biogenesis, processing, transport, degradation, and function via expression profiling, molecular imaging and other methods. Levels of lncRNA transcripts and even their profiles of post-transcriptional modifications can be quantified with widely available nucleic acid detection techniques such as polymerase chain reaction, microarray analysis, and RNA sequencing. Levels of lncRNAs that are trafficked intercellularly can also be determined, specifically, by isolating microvesicles in blood, cerebrospinal fluid, and other biological fluids, and examining their contents [98]. The emergence of more advanced technologies with the capacity to analyze single cells, single molecules, and very small specimen volumes promises to simplify sample preparation, enhance sensitivity and specificity, and lower costs. These innovations are likely to further promote the adoption of lncRNA expression analysis in the clinical arena. Moreover, novel molecular imaging probes, such as oligonucleotide molecular beacons and quantum dot nanoparticles, are already enabling the visualization of lncRNAs, potentially even for real-time in vivo imaging. These can be developed for clinical applications, for example, to better define tumor margins with lncRNA molecular markers during surgery. Proof of principle for this approach is provided by a recent study that reported using a lncRNA-specific peptide nucleic acid-based molecular beacon as a diagnostic probe for detecting a disease-associated lncRNA in vitro, ex vivo, and in situ (i.e., in biopsy material from human tissue) [99].
Practical and effective strategies for systemically investigating more complex networks between lncRNAs and other nervous system disease-related DNA, RNA, and protein molecules, including the molecular substrates mediating these relationships at the primary sequence or secondary and tertiary structure levels, are yet to emerge, but these would, obviously, have value for a range of pre-clinical and clinical applications, including screening for, and the development of, precision medicines targeting these interactions.
Therapeutic Strategies
Therapeutic strategies can be envisioned that focus on targeting lncRNA genes, the epigenetic status of lncRNA gene loci, and lncRNA transcript expression and functions.
An interesting approach is editing lncRNA genes with effects that could include disrupting regulatory, structural, or functional motifs. Emerging technologies for genome editing with engineered nucleases, such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases, and oligonucleotide-directed methods represent state-of-the-art gene therapy techniques that can be used to target lncRNAs. For example, one study reported that the NEAT2 lncRNA could be silenced in human cells very effectively with ZFNs designed to insert RNA destabilizing elements into its gene locus biallelically [100]. Although this was a preliminary in vitro illustration, ZFN-based therapies are being studied in clinical trials for neurological diseases, including an adoptive T cell approach for glioblastoma (NCT01082926). These observations suggest that this strategy is amenable to further development, particularly for cellular therapies. In addition to integrating destabilizing elements, other sequences such as post-transcriptional regulatory elements (e.g., miRNA binding sites and triple helix stabilization motifs), transport signals, and zipcode-like sequences for loading into microvesicles can potentially be edited.
Another approach is to modulate the epigenetic state of lncRNA gene loci with the plethora of agents currently being developed to target readers, writers, and erasers of epigenetic marks. In fact, epigenetic enzymes, such as histone deacetylases, are involved in disease context-specific regulation of lncRNAs [101]; and drugs targeting these enzymes clearly influence the expression of lncRNAs [32] and, therefore, have the potential to mitigate disease processes. Related strategies for modulating lncRNAs indirectly include targeting factors and signaling pathways that regulate lncRNA expression, such as REST, CoREST, and p53.
Another strategy is modulating lncRNAs at the post-transcriptional level by targeting their metabolism and functional interactions. These techniques leverage the considerable expertise that exists in developing oligonucleotide therapeutics, such as ASOs, RNAi, and related platforms and chemistries for enhancing specificity, stability, and penetration of the blood–brain barrier (e.g., locked nucleic acids and nanodelivery systems). One particularly intriguing example is the antagoNAT modified ASO technology, which is designed to target natural antisense lncRNAs and, thus, to up-regulate corresponding protein-coding genes [102]. For example, administering an antagoNAT targeting the BDNF-AS lncRNA leads to an increase in the level of BDNF protein in neuronal cells. ASOs targeting other lncRNAs have also been reported and show benefits in animal models of diseases, such as cancer. For example, a recent study demonstrated that an ASO inhibiting the NEAT2 lncRNA in a mouse lung cancer xenograft model prevented tumor metastasis [103]. These observations are intriguing because the development of ASO therapeutics is undergoing a renaissance. Indeed, mipomersen (Kynamro; Isis Pharmaceuticals, Carlsbad, CA) was recently approved, and emerging data from preclinical and later-stage clinical trials for additional candidate ASO drugs, including those for neurological disease indications (e.g., HD, spinal muscular atrophy, myotonic dystrophy type 1, and Duchenne muscular dystrophy) seem promising. In addition, oligonucleotides and small molecules can, potentially, be engineered to modulate lncRNA transcript activity by interfering with their sequence-specific and structural interactions with other molecules [i.e., DNA, RNA, and proteins (e.g., chromatin remodeling complexes)]. Targeting interactions between lncRNAs and chromatin remodeling complexes has been preliminarily reported for activation of PRC2-regulated genes [104]. Relationships between lncRNAs and REST, CoREST, and other transcriptional and epigenetic factors could similarly be targeted with small molecules, oligonucleotides, and peptides. Moreover, lncRNA–miRNA, lncRNA–mRNA, and miRNA–mRNA circuitries could be modulated by approaches that change binding site stoichiometry (e.g., introducing sponges and masks) and affinities, effectively changing the topologies and functions of these networks.
Ultimately, as more organizing principles for lncRNA sequence, structure, and function relationships emerge, computational and functional assays will also be enabled to repurpose known drugs and identify compounds for modulating these novel molecular targets.
Perspectives
Our understanding of human genomic organization, regulation, and activity has been revolutionized by the recent discovery of tens of thousands of lncRNA genes and associated transcripts. The study of these factors is revealing that they have important roles in mediating brain development and function, and in promoting neurological and psychiatric disease pathogenesis. Many future studies are necessary to better define the actions of specific lncRNAs and to uncover how exactly they contribute to disease mechanisms. Nevertheless, previously existing and more recently developed technologies are already promising to deliver innovative tools for diagnosis and treatment by exploiting known lncRNA targets. For example, high-profile scientists have established commercial entities [i.e., CURNA (now OPKO-CURNA) and RaNA Therapeutics] focused explicitly on designing and developing oligonucleotide therapeutics targeting lncRNAs for nervous system (and additional) disorders. Others with experience in interrelated oligonuceotide platforms (e.g., ASO, RNAi, and miRNA) or with other approaches have also expressed an interest in pursuing lncRNA targets as well. However, significant challenges do exist, including those linked to crossing the blood–brain barrier, cell type- and subcellular compartment-specific delivery, modulating low abundance transcripts, and minimizing off-target effects. One interesting strategy that has been proposed to overcome the dilemma of delivery is to utilize exosomes, loaded with designer oligonucleotide cargo, as a delivery vehicle, which can traverse from the periphery into the brain [105–107].
Electronic supplementary material
Acknowledgments
We regret that space constraints have prevented the citation of many relevant and important references. M.F.M. is supported by grants from the National Institutes of Health (NS071571, HD071593, MH66290), as well as by the F.M. Kirby, Alpern Family, Mildred and Bernard H. Kayden, and Roslyn and Leslie Goldstein Foundations.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mercer TR, Neph S, Dinger ME, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 4.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 6.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volders PJ, Helsens K, Wang X, et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2013;41:D246–251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Managadze D, Lobkovsky AE, Wolf YI, Shabalina SA, Rogozin IB, Koonin EV. The vast, conserved mammalian lincRNome. PLoS Comput Biol. 2013;9:e1002917. doi: 10.1371/journal.pcbi.1002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipovich L, Tarca AL, Cai J, et al. Developmental Changes in the transcriptome of human cerebral cortex tissue: long noncoding rna transcripts. Cereb Cortex 2013 Feb 1 [Epub ahead of print]. [DOI] [PubMed]
- 12.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chodroff RA, Goodstadt L, Sirey TM, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belgard TG, Marques AC, Oliver PL, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallot C, Huret C, Lesecque Y, et al. XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nat Genet. 2013;45:239–241. doi: 10.1038/ng.2530. [DOI] [PubMed] [Google Scholar]
- 17.Latos PA, Pauler FM, Koerner MV, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 18.Taguchi S, Iwami M, Kiya T. Identification and characterization of a novel nuclear noncoding RNA, Fben-1, which is preferentially expressed in the higher brain center of the female silkworm moth, Bombyx mori. Neurosci Lett. 2011;496:176–180. doi: 10.1016/j.neulet.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Reinius B, Shi C, Hengshuo L, et al. Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics. 2010;11:614. doi: 10.1186/1471-2164-11-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qureshi IA, Mehler MF. Genetic and epigenetic underpinnings of sex differences in the brain and in neurological and psychiatric disease susceptibility. Prog Brain Res. 2010;186:77–95. doi: 10.1016/B978-0-444-53630-3.00006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young RS, Marques AC, Tibbit C, et al. Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol Evol. 2012;4:427–442. doi: 10.1093/gbe/evs020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pollard KS, Salama SR, Lambert N, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 24.Bond AM, Vangompel MJ, Sametsky EA, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klattenhoff CA, Scheuermann JC, Surface LE, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moran I, Akerman I, van de Bunt M, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun L, Goff LA, Trapnell C, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kretz M, Siprashvili Z, Chu C, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercer TR, Qureshi IA, Gokhan S, et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin M, Pedrosa E, Shah A, et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS ONE. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol. 2010;10:49. doi: 10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernard D, Prasanth KV, Tripathi V, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin D, Pestova TV, Hellen CU, Tiedge H. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol. 2008;28:3008–3019. doi: 10.1128/MCB.01800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anguera MC, Ma W, Clift D, Namekawa S, Kelleher RJ, 3rd, Lee JT. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011;7:e1002248. doi: 10.1371/journal.pgen.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling KH, Hewitt CA, Beissbarth T, et al. Spatiotemporal regulation of multiple overlapping sense and novel natural antisense transcripts at the Nrgn and Camk2n1 gene loci during mouse cerebral corticogenesis. Cereb Cortex. 2011;21:683–697. doi: 10.1093/cercor/bhq141. [DOI] [PubMed] [Google Scholar]
- 44.Bu Q, Hu Z, Chen F, et al. Transcriptome analysis of long non-coding RNAs of the nucleus accumbens in cocaine-conditioned mice. J Neurochem. 2012;123:790–799. doi: 10.1111/jnc.12006. [DOI] [PubMed] [Google Scholar]
- 45.Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J Neurochem. 2011;116:459–466. doi: 10.1111/j.1471-4159.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kryger R, Fan L, Wilce PA, Jaquet V. MALAT-1, a non protein-coding RNA is upregulated in the cerebellum, hippocampus and brain stem of human alcoholics. Alcohol. 2012;46:629–634. doi: 10.1016/j.alcohol.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 47.Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. J Mol Neurosci. 2013;49:589–593. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lipovich L, Dachet F, Cai J, et al. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics. 2012;192:1133–1148. doi: 10.1534/genetics.112.145128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim TK, Hemberg M, Gray JM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.St Laurent G, 3rd, Faghihi MA, Wahlestedt C. Non-coding RNA transcripts: sensors of neuronal stress, modulators of synaptic plasticity, and agents of change in the onset of Alzheimer's disease. Neurosci Lett. 2009;466:81–88. doi: 10.1016/j.neulet.2009.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakhotia SC. Long non-coding RNAs coordinate cellular responses to stress. Wiley Interdiscip Rev RNA. 2012;3:779–796. doi: 10.1002/wrna.1135. [DOI] [PubMed] [Google Scholar]
- 52.Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wan G, Mathur R, Hu X, et al. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. 2013;25:1086–1095. doi: 10.1016/j.cellsig.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mariner PD, Walters RD, Espinoza CA, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 57.Yakovchuk P, Goodrich JA, Kugel JF. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc Natl Acad Sci U S A. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valgardsdottir R, Chiodi I, Giordano M, Cobianchi F, Riva S, Biamonti G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol Biol Cell. 2005;16:2597–2604. doi: 10.1091/mbc.E04-12-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biamonti G, Vourc'h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol. 2010;2:a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 61.Shamovsky I, Nudler E. Isolation and characterization of the heat shock RNA 1. Methods Mol Biol. 2009;540:265–279. doi: 10.1007/978-1-59745-558-9_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carrieri C, Cimatti L, Biagioli M, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 63.Royo H, Basyuk E, Marty V, Marques M, Bertrand E, Cavaille J. Bsr, a nuclear-retained RNA with monoallelic expression. Mol Biol Cell. 2007;18:2817–2827. doi: 10.1091/mbc.E06-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–535. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kang HJ, Kawasawa YI, Cheng F, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Colantuoni C, Lipska BK, Ye T, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wood SH, Craig T, Li Y, Merry B, de Magalhaes JP. Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome. Age (Dordr) 2013;35:763–776. doi: 10.1007/s11357-012-9410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104:10679–10684. doi: 10.1073/pnas.0701532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu N, Landreh M, Cao K, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482:519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones DL, Rando TA. Emerging models and paradigms for stem cell ageing. Nat Cell Biol. 2011;13:506–512. doi: 10.1038/ncb0511-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 72.Ratajczak MZ. Igf2-H19, an imprinted tandem gene, is an important regulator of embryonic development, a guardian of proliferation of adult pluripotent stem cells, a regulator of longevity, and a 'passkey' to cancerogenesis. Folia Histochem Cytobiol. 2012;50:171–179. doi: 10.5603/fhc.2012.0026. [DOI] [PubMed] [Google Scholar]
- 73.Wang J, Geesman GJ, Hostikka SL, et al. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle. 2011;10:3016–3030. doi: 10.4161/cc.10.17.17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qureshi IA, Mehler MF. Alu transcription: A rheostat for stem cell aging? Cell Cycle. 2011;10:3820–3821. doi: 10.4161/cc.10.22.18194. [DOI] [PubMed] [Google Scholar]
- 75.Chen G, Wang Z, Wang D, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Talkowski ME, Maussion G, Crapper L, et al. Disruption of a large intergenic noncoding RNA in subjects with neurodevelopmental disabilities. Am J Hum Genet. 2012;91:1128–1134. doi: 10.1016/j.ajhg.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cartault F, Munier P, Benko E, et al. Mutation in a primate-conserved retrotransposon reveals a noncoding RNA as a mediator of infantile encephalopathy. Proc Natl Acad Sci U S A. 2012;109:4980–4985. doi: 10.1073/pnas.1111596109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 79.Ning S, Wang P, Ye J, et al. A global map for dissecting phenotypic variants in human lincRNAs. Eur J Hum Genet. 2013 Mar 6 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 80.Chen G, Qiu C, Zhang Q, Liu B, Cui Q. Genome-wide analysis of human SNPs at long intergenic noncoding RNAs. Hum Mutat. 2013;34:338–344. doi: 10.1002/humu.22239. [DOI] [PubMed] [Google Scholar]
- 81.Cabianca DS, Casa V, Bodega B, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Faghihi MA, Zhang M, Huang J, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saitsu H, Osaka H, Sasaki M, et al. Mutations in POLR3A and POLR3B encoding RNA polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am J Hum Genet. 2011;89:644–651. doi: 10.1016/j.ajhg.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tollervey JR, Curk T, Rogelj B, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lagier-Tourenne C, Polymenidou M, Hutt KR, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Massone S, Vassallo I, Fiorino G, et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis. 2011;41:308–317. doi: 10.1016/j.nbd.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 87.Han L, Zhang K, Shi Z, et al. LncRNA pro fi le of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40:2004–2012. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 88.Zhang X, Sun S, Pu JK, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 89.Johnson R. Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol Dis. 2012;46:245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 90.Mezzomo LC, Gonzales PH, Pesce FG, et al. Expression of cell growth negative regulators MEG3 and GADD45gamma is lost in most sporadic human pituitary adenomas. Pituitary. 2012;15:420–427. doi: 10.1007/s11102-011-0340-1. [DOI] [PubMed] [Google Scholar]
- 91.Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28:1551–1558. doi: 10.3892/or.2012.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sanchez Y, Huarte M. Long non-coding RNAs: challenges for diagnosis and therapies. Nucleic Acid Ther. 2013;23:15–20. doi: 10.1089/nat.2012.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weinberg MS, Morris KV. Long non-coding RNA targeting and transcriptional de-repression. Nucleic Acid Ther. 2013;23:9–14. doi: 10.1089/nat.2012.0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bhartiya D, Kapoor S, Jalali S, et al. Conceptual approaches for lncRNA drug discovery and future strategies. Expert Opin Drug Discov. 2012;7:503–513. doi: 10.1517/17460441.2012.682055. [DOI] [PubMed] [Google Scholar]
- 97.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–165. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 98.Shao H, Chung J, Balaj L, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kam Y, Rubinstein A, Naik S, et al. Detection of a long non-coding RNA (CCAT1) in living cells and human adenocarcinoma of colon tissues using FIT-PNA molecular beacons. Cancer Lett 2013 Feb 14 [Epub ahead of print]. [DOI] [PubMed]
- 100.Gutschner T, Baas M, Diederichs S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011;21:1944–1954. doi: 10.1101/gr.122358.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang F, Huo XS, Yuan SX, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 102.Modarresi F, Faghihi MA, Lopez-Toledano MA, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci U S A. 2010;107:22196–22201. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lakhal S, Wood MJ. Exosome nanotechnology: an emerging paradigm shift in drug delivery: exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. Bioessays. 2011;33:737–741. doi: 10.1002/bies.201100076. [DOI] [PubMed] [Google Scholar]
- 106.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 107.Katakowski M, Buller B, Zheng X, et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013;335:201–204. doi: 10.1016/j.canlet.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang W, Chen Y, Liu P, et al. Variants on chromosome 9p21.3 correlated with ANRIL expression contribute to stroke risk and recurrence in a large prospective stroke population. Stroke. 2012;43:14–21. doi: 10.1161/STROKEAHA.111.625442. [DOI] [PubMed] [Google Scholar]
- 109.Pasmant E, Sabbagh A, Masliah-Planchon J, et al. Role of noncoding RNA ANRIL in genesis of plexiform neurofibromas in neurofibromatosis type 1. J Natl Cancer Inst. 2011;103:1713–1722. doi: 10.1093/jnci/djr416. [DOI] [PubMed] [Google Scholar]
- 110.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6:e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zuchner S, Gilbert JR, Martin ER, et al. Linkage and association study of late-onset Alzheimer disease families linked to 9p21.3. Ann Hum Genet. 2008;72:725–731. doi: 10.1111/j.1469-1809.2008.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wevrick R, Kerns JA, Francke U. Identification of a novel paternally expressed gene in the Prader-Willi syndrome region. Hum Mol Genet. 1994;3:1877–1882. doi: 10.1093/hmg/3.10.1877. [DOI] [PubMed] [Google Scholar]
- 114.Maina EN, Webb T, Soni S, et al. Analysis of candidate imprinted genes in PWS subjects with atypical genetics: a possible inactivating mutation in the SNURF/SNRPN minimal promoter. J Hum Genet. 2007;52:297–307. doi: 10.1007/s10038-007-0109-6. [DOI] [PubMed] [Google Scholar]
- 115.Moseley ML, Zu T, Ikeda Y, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 116.Daughters RS, Tuttle DL, Gao W, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vandeweyer G, Van der Aa N, Ceulemans B, van Bon BW, Rooms L, Kooy RF. A de novo balanced t(2;6)(p15;p22.3) in a patient with West Syndrome disrupts a lnc-RNA. Epilepsy Res. 2012;99:346–349. doi: 10.1016/j.eplepsyres.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 118.Higashimoto K, Urano T, Sugiura K, et al. Loss of CpG methylation is strongly correlated with loss of histone H3 lysine 9 methylation at DMR-LIT1 in patients with Beckwith-Wiedemann syndrome. Am J Hum Genet. 2003;73:948–956. doi: 10.1086/378595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Albrecht S, Waha A, Koch A, Kraus JA, Goodyer CG, Pietsch T. Variable imprinting of H19 and IGF2 in fetal cerebellum and medulloblastoma. J Neuropathol Exp Neurol. 1996;55:1270–1276. doi: 10.1097/00005072-199612000-00011. [DOI] [PubMed] [Google Scholar]
- 120.Muller S, Zirkel D, Westphal M, Zumkeller W. Genomic imprinting of IGF2 and H19 in human meningiomas. Eur J Cancer. 2000;36:651–655. doi: 10.1016/s0959-8049(99)00328-7. [DOI] [PubMed] [Google Scholar]
- 121.Balik V, Srovnal J, Sulla I, et al. MEG3: a novel long noncoding potentially tumour-suppressing RNA in meningiomas. J Neurooncol. 2013;112:1–8. doi: 10.1007/s11060-012-1038-6. [DOI] [PubMed] [Google Scholar]
- 122.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 123.Cheunsuchon P, Zhou Y, Zhang X, et al. Silencing of the imprinted DLK1-MEG3 locus in human clinically nonfunctioning pituitary adenomas. Am J Pathol. 2011;179:2120–2130. doi: 10.1016/j.ajpath.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jong MT, Gray TA, Ji Y, et al. A novel imprinted gene, encoding a RING zinc-finger protein, and overlapping antisense transcript in the Prader-Willi syndrome critical region. Hum Mol Genet. 1999;8:783–793. doi: 10.1093/hmg/8.5.783. [DOI] [PubMed] [Google Scholar]
- 126.Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nogalska A, Engel WK, Askanas V. Increased BACE1 mRNA and noncoding BACE1-antisense transcript in sporadic inclusion-body myositis muscle fibers—possibly caused by endoplasmic reticulum stress. Neurosci Lett. 2010;474:140–143. doi: 10.1016/j.neulet.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Le Meur E, Watrin F, Landers M, Sturny R, Lalande M, Muscatelli F. Dynamic developmental regulation of the large non-coding RNA associated with the mouse 7C imprinted chromosomal region. Dev Biol. 2005;286:587–600. doi: 10.1016/j.ydbio.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 129.Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci. 2011;12:707–722. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS ONE. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ladd PD, Smith LE, Rabaia NA, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 132.Amaral PP, Neyt C, Wilkins SJ, et al. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009;15:2013–2027. doi: 10.1261/rna.1705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sopher BL, Ladd PD, Pineda VV, et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron. 2011;70:1071–1084. doi: 10.1016/j.neuron.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Johnson R, Richter N, Jauch R, et al. The Human Accelerated Region 1 noncoding RNA is repressed by REST in Huntington's disease. Physiol Genomics 2010 Feb 23 [Epub ahead of print]. [DOI] [PubMed]
- 135.Kaneko H, Dridi S, Tarallo V, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.