Repair of Strand Breaks by Homologous Recombination (original) (raw)
Breaks in both strands of DNA are caused by environmental agents (e.g., ionizing radiation) or errors in normal cellular processes (e.g., replication). Homologous recombination is one major pathway to repair such breaks.
Abstract
In this review, we discuss the repair of DNA double-strand breaks (DSBs) using a homologous DNA sequence (i.e., homologous recombination [HR]), focusing mainly on yeast and mammals. We provide a historical context for the current view of HR and describe how DSBs are processed during HR as well as interactions with other DSB repair pathways. We discuss the enzymology of the process, followed by studies on DSB repair in living cells. Whenever possible, we cite both original articles and reviews to aid the reader for further studies.
It was long recognized by radiation biologists that γ-irradiation was capable of causing DNA strand breaks. Geneticists posited that radiation-induced broken ends were treated differently from the ends of chromosomes (see Webb et al. 2013). Much of our early view of how HR is used in yeast to repair strand breaks emerged from studies of γ-irradiation-induced breaks and targeted integration during the 1970s and early 1980s. A model for the repair of γ-irradiation-induced breaks was proposed by Resnick and involved pairing of the broken ends with the homolog (Resnick 1976).
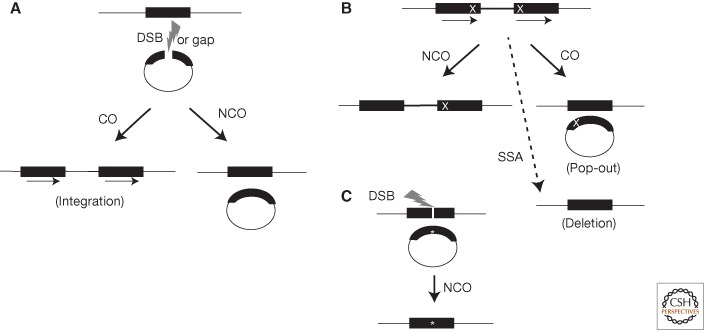
This model foreshadowed the repair of broken plasmid DNA (Orr-Weaver et al. 1981). Plasmid molecules containing yeast genes were linearized with restriction enzymes that cut once within a yeast gene and were introduced into yeast cells via transformation (Fig. 1A). By cutting these plasmids, the number of transformants compared to uncut circular DNA was increased 10- to 1000-fold. In addition, most of the transformants resulted from plasmid integration into the yeast genome. In plasmids that contained two yeast genes, the integration event was directed to the chromosomal copy of the gene that was cut on the plasmid. Gaps in the plasmid created by removing sequences from the yeast gene also directed integration into the homologous gene in the genome and were filled-in using the chromosomal sequence as a template (Orr-Weaver et al. 1981). The addition of yeast origins of replication to the plasmids allowed the detection of those events that restored the plasmid sequences unassociated with an integration event (noncrossover [NCO]) versus integration events associated with gap repair (crossover [CO]) (Fig. 1A) (Orr-Weaver and Szostak 1983). These observations showed that gene conversion of the gapped sequences occurred, which lead directly to the DSB repair model for HR (Szostak et al. 1983), which in its simplest form postulated a double Holliday junction (dHJ) recombination intermediate flanking the gap. Holliday junctions are named for the cross-strand exchange structure in Robin Holliday's model for genetic recombination (Holliday 1964).
Figure 1.
HR events. (A) Plasmid-by-chromosome HR initiated by a DSB or gap (lightning bolt) in the plasmid. The black bars represent the homology between the chromosome and plasmid. In this case, the chromosomal sequence is used as the donor template to repair the gap in the recipient plasmid. Crossovers (COs) lead to plasmid integration and the formation of direct repeats (indicated by arrows). Noncrossovers (NCOs) are also detected if the plasmid contains an origin of replication. (B) Recombination between direct repeats is frequently used to assay HR. Different mutations (X) are present in each repeat. HR associated with gene conversion of one mutation leads to restoration of one intact repeat (solid black bars), which is wild type. NCOs maintain the direct repeat configuration, whereas a CO leads to a plasmid “pop-out” event. Whereas bona fide HR events (i.e., those involving gene conversion) maintain both repeats, SSA (for details, see Fig. 2) leads to deletion of one repeat and the segment between the repeats. (C) DSB-induced gene targeting. Because nonhomologous integration of plasmids in mammalian cells is so efficient, creation of a site-specific DSB in the chromosome is used to induce HR. For the purposes of genome modification (*), the repair of this DSB can be from an incoming plasmid donor sequence, which can be circular (shown) or linear (not shown).
Discovery of a role for HR in DSB repair in mammalian cells lagged significantly behind that of yeast and other model organisms. In fact, cell culture and mouse studies had originally pointed investigators away from considering HR as having a vital role in mammalian cells. Analysis of the fate of integrated DNA following transfection or injection in cells and in transgenic mice showed nonhomologous sites of integration (Lacy et al. 1983; Smithies et al. 1985), unlike the uniform homologous integration of transformed DNA in yeast. More direct studies of DSBs in plasmid DNA showed efficient nonhomologous repair (i.e., nonhomologous end-joining [NHEJ]) (Wake et al. 1984; Chiruvella et al. 2013). Subsequently, antigen receptor rearrangement was determined to involve DSB repair by NHEJ (Roth et al. 1992), which piqued the interest of investigators in NHEJ, but dissuaded them from considering that HR played a significant role in DSB repair in mammalian cells. Furthermore, when sequence repeats (homology) were provided in the plasmids, repair was consistent with annealing of complementary strands (single-strand annealing [SSA]) (Lin et al. 1984) rather than the invasion of one homologous strand into the other, the defining step of HR (Fig. 2). These observations were important for understanding the deletions that occur during direct repeat recombination in fungi and mammalian cells (Rudin and Haber 1988; Liang et al. 1996).
Figure 2.
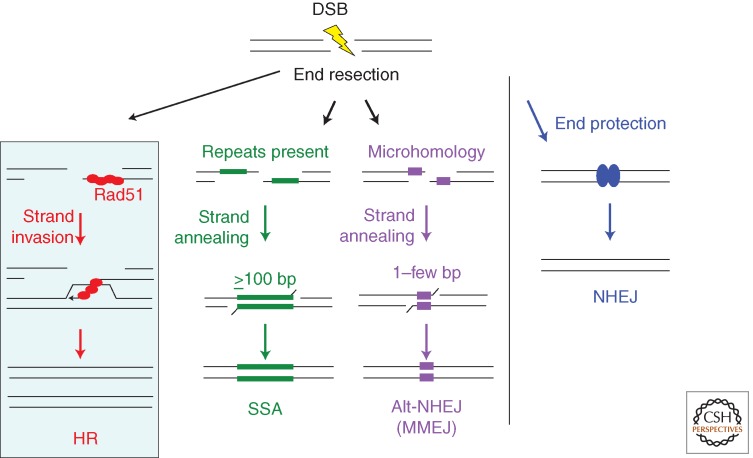
HR is only one pathway of DSB repair and can collaborate with and compete with other pathways to repair DSBs. An early determinant of DSB repair pathway choice is DNA end resection—the processing of DNA ends to generate 3′ single-strands, which is required for HR but inhibits canonical NHEJ in which DNA ends are protected with minimal processing before joining. The defining step of HR is strand invasion by Rad51 or a related recombinase (red balls). Rad51 forms a nucleoprotein filament on single-stranded DNA after end resection. End resection also provides an intermediate in nonconservative single-strand annealing (SSA) and alternative-NHEJ (alt-NHEJ) involving microhomology, as it gives rise to single strands, which can anneal at long (SSA) or short (alt-NHEJ) complementary sequences (green and purple bars, respectively). HR and SSA require more extensive end resection than alt-NHEJ, because minimal end resection will uncover microhomologies present near DNA ends. Alt-NHEJ involving microhomology is also termed microhomology-mediated NHEJ (MMEJ).
The publication of DSB repair studies in yeast (Orr-Weaver and Szostak 1983) and the proposal of the DSB repair model (Orr-Weaver et al. 1981; Szostak et al. 1983) prompted direct investigation into a role for HR in mammalian cells. Initial studies were based on plasmid-by-chromosome recombination (Fig. 1A), in which a double-strand gap was introduced into the plasmid that could be repaired from the chromosome, leading to the production of intact virus (Jasin et al. 1985). These experiments showed a high frequency of DSB repair by HR. As much as 25% of the successfully transfected cells produced wild-type virus soon after plasmid transfection and about 10% of the initially gapped plasmids appeared to have undergone HR. Although designed for a virus readout, these experiments provided clear proof of DSB repair by HR in mammalian cells.
The product of these plasmid-by-chromosome HR experiments was essentially an NCO. An alternative outcome predicted by the original DSB repair model is a CO, which could not be detected in the viral system. Thus, the approach was revised to target the same locus but with a promoterless selectable marker gene substituting for viral sequences (Jasin and Berg 1988). DSB repair leading to plasmid integration (CO) was observed (Fig. 1A), further showing the recombinogenic nature of DSBs in mammalian cells, and led to efficient selection for the marker, now a frequently used approach. The enrichment of homologous integration events with a DSB was estimated to be 100-fold. Despite the large effect, however, these CO events were notably significantly less frequent than NCO events. The choice of CO and NCO outcomes in HR will be discussed below.
DIRECT REPEAT RECOMBINATION
In addition to plasmid repair studies, direct repeat recombination assays were developed in yeast, taking advantage of plasmids integrated at their homologous chromosomal site (Jackson and Fink 1981; Klein and Petes 1981). The integrations were often designed to create direct repeats with two different mutations (Fig. 1B). Selecting for recombination between the direct repeats leads to prototrophs that maintain the plasmid and these are called gene convertants. Prototrophs associated with plasmid loss are called “pop-outs.” Depending on the configuration of the alleles, triplications can also arise. Variations on these assays have formed the basis of many recombination studies in fungi and mammalian cells (Klein 1995; Liang et al. 1996; Lambert et al. 1999) including the now widely used GFP repeats (Pierce et al. 1999).
SITE-SPECIFIC DSBs
Molecular insights into DSB repair required the development of systems to introduce site-specific DSBs in the genome. Expression of the HO and I-_Sce_I endonucleases have been used to create site-specific DSBs in yeast (Rudin and Haber 1988) and mammalian cells (Rouet et al. 1994b) and later in other species including flies (Bellaiche et al. 1999; Rong et al. 2002), plants (Puchta et al. 1993, 1996), and bacteria (Ponder et al. 2005). The HO endonuclease was discovered studying mating type switching in budding yeast, which is initiated by a DSB (Strathern et al. 1982; Kostriken et al. 1983; Haber 2012). The HO endonuclease targets the active mating locus, creates a site-specific DSB, and then one of the two silent copies elsewhere on the chromosome is used as a template to switch mating type information. Studies of this highly regulated process led to the notion that the processed ends of a DSB could undergo “ectopic” recombination. The model of plasmid gap repair was used to explain the gene conversion of the silent locus into the MAT cassette (Orr-Weaver et al. 1981). Subsequent experiments in Schizosaccharomyces pombe (Arcangioli and de Lahondes 2000), where mating type is initiated by a nick that is converted to a DSB, and later in Saccharomyces cerevisiae (Ira et al. 2006) showed that the DNA synthesis during mating type switching is conservative.
Studies of mitochondrial genetics in budding yeast led to the discovery of an endonuclease-initiated HR event that involves cleavage of an 18-base-pair recognition site (Colleaux et al. 1988). The site is present only in mitochondrial genomes that do not contain the ORF encoding the endonuclease, because the ORF interrupts the recognition site (Dujon 1989). When mitochondria, which contain the ORF, fuse with ones that do not, the endonuclease cleaves the site in the ORF-minus genomes, initiating DSB gap repair. The ORF is subsequently copied into the broken site, formally a gene conversion event, which renders the site no longer sensitive to the endonuclease. The endonuclease responsible for this site-specific DSB is I-_Sce_I and the ORF encoding it was codon optimized for protein expression in E. coli (Colleaux et al. 1986), allowing expression vectors to be developed for a variety of cell types, including yeast (Plessis et al. 1992) and mammalian cells (Rouet et al. 1994a).
Many studies of DSB repair have centered on site-specific breaks introduced by the HO and I-_Sce_I endonucleases. Various assays have been designed to assess how different cell types and different mutations affect the process of DSB repair. Direct repeat assays with the DSB cut site inserted in different places are used to measure HR (here defined as a gene-conversion event), SSA, and/or NHEJ. Importantly, the efficiency of cutting by HO and I-_Sce_I is high enough in yeast cells to physically monitor DSB repair, leading to mechanistic insights into this process and the genetic control of discrete steps.
GENOME ENGINEERING BY REPAIR OF CHROMOSOME DSBs
DSB-induced plasmid recombination experiments were performed contemporaneously with the burgeoning field of gene targeting in mammalian cells for the purposes of modifying genes at their endogenous loci (Capecchi 1989). The finding that DSBs in plasmids induced recombination with the chromosome suggested that homologous DSB repair could be co-opted in mammalian cells for genome engineering. However, two limitations were immediately noted: the relatively low frequency of CO events (Jasin and Berg 1988), and that formally the plasmid, not the chromosome, is the one modified because it carries the DSB (Jasin et al. 1985). Thus, what was required to efficiently modify the genome by HR was a DSB in the chromosome, something that was unnecessary in yeast because homologous integration of plasmids was already much more efficient than nonhomologous events. This requirement could be met by expressing I-_Sce_I (Rouet et al. 1994a) and introducing its target site into the mammalian genome (Rouet et al. 1994b). I-_Sce_I was used for this purpose because its target site was well defined and long enough so that its expression was not expected to be toxic to cells even with complex genomes (Rouet et al. 1994a). DSBs in the mammalian genome are repaired by HR using this system (Fig. 1C), such that homologous gene targeting was increased several orders of magnitude (Rouet et al. 1994b). DSBs were also efficiently repaired by NHEJ in this system, with an estimate of approximately twice the level of NHEJ as HR (Rouet et al. 1994b). However, any such estimates comparing HR and NHEJ frequencies should be used with caution, given that the most common HR and NHEJ events (precise repair from the sister chromatid and religation of the site, respectively) are not scored in assays that require modification of the DSB site to be detected.
DSB repair studies with the I-_Sce_I endonuclease form the basis of current genome engineering approaches in mammalian cells. Critical for the success of these approaches is an endonuclease directed to cleave the locus to be modified. Nucleases using an array of zinc finger DNA-binding domains fused to a _Fok_I endonuclease domain were the first designed to target an endogenous mammalian locus (Urnov et al. 2005), similar to an approach developed for Drosophila (Bibikova et al. 2003). Nuclease design was greatly facilitated by the discovery of the simple DNA recognition code of TAL effector proteins from pathogenic bacteria, which invade plants (Boch et al. 2009; Moscou and Bogdanove 2009) and even more so recently by the discovery of an RNA-guided nuclease in bacterial adaptive immunity termed CRISPR/Cas9 (Gasiunas et al. 2012; Jinek et al. 2012). These “designer” nucleases are being used for genome engineering in a variety of organisms in addition to mammalian cells, including zebrafish (Hwang et al. 2013) and livestock (Carlson et al. 2012).
SINGLE-STRAND BREAKS AND HR
Single-strand breaks and gaps, like DSBs, can also be repaired by HR, as first described in E. coli (Michel et al. 2007). In the fission yeast Schizosaccharomyces pombe, the mating type switching system is likely induced from a site-specific nick or gap left on one chromosome, which in the next round of DNA synthesis becomes a DSB that is used to initiate the switch (Arcangioli and de Lahondes 2000). Interestingly, this DSB repair is conservative, likely to avoid COs that would result in deletion of the silent cassettes. The mechanism responsible for generating the nick at the fission yeast mating type locus is still unknown. Evidence that single-strand nicks or gaps are likewise repaired by HR in mammalian cells comes not only from studies with drugs like camptothecin but also from the use of site-specific nickases, such as meganucleases (Davis and Maizels 2011), zinc finger nucleases (Kim et al. 2012; Ramirez et al. 2012; Wang et al. 2012), CRISPR/Cas9 (Cong et al. 2013; Mali et al. 2013), and corrupted Rag proteins (Lee et al. 2004). For purposes of genome engineering, nickases have the advantage that single-strand breaks are not repaired by error-prone NHEJ, although the induction of HR is usually much less than that from DSBs.
MEIOSIS
The connection between mitotic HR repair and meiosis was made clear when DNA DSBs were identified as initiators of meiotic recombination (Sun et al. 1989). The search for the enzyme responsible for creating meiotic DSBs revealed a conserved gene, SPO11 (Bergerat et al. 1997; Keeney et al. 1997). SPO11, first identified as being defective in budding yeast sporulation (Esposito and Esposito 1969), which is the end product of successful meiosis, is homologous to the archael type VIA topoisomerase subunit with functional homologs identified in many eukaryotes (Keeney 2008), including mouse (Baudat et al. 2000; Romanienko and Camerini-Otero 2000). When introducing a DSB, Spo11 protein remains covalently bound to the ends before it is removed during endonucleolytic processing (Neale et al. 2005; Garcia et al. 2011). In organisms such as yeast and mouse, HR promotes pairing of chromosome homologs in addition to providing the physical connections between chromosome homologs (COs) critical for proper disjunction during meiosis I (Davis et al. 2001; Tesse et al. 2003; Henderson and Keeney 2004; Kauppi et al. 2013).
STEPS OF DSB REPAIR BY HR
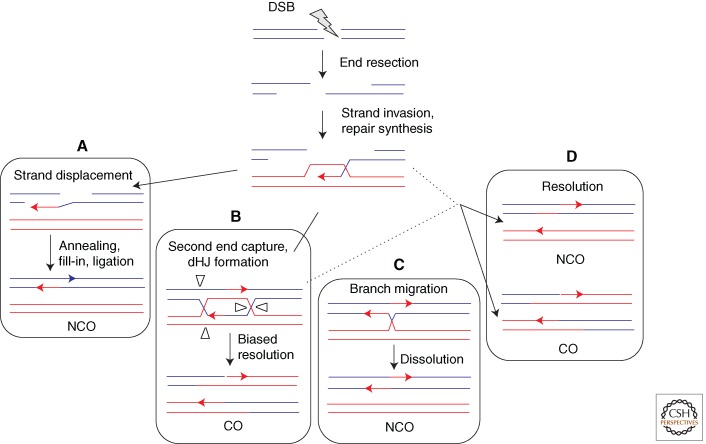
The defining step of HR is the invasion of 3′ single-stranded DNA into a homologous duplex (Figs. 2 and 3). Single-stranded DNA generated by resection of the ends of a DSB provides a substrate for assembly of the Rad51 filaments needed for strand invasion; moreover, the invading 3′ end provides a primer for repair synthesis templated by the intact duplex. The strand invasion intermediate (D loop) can be resolved in a number of different ways, ultimately leading to an NCO or CO.
Figure 3.
HR of a resected DSB after strand invasion and repair synthesis showing independent and interrelated steps for resolution. (A) Synthesis-dependent strand annealing involves displacement by DNA helicase(s) of the newly synthesized strand. The two ends can anneal by their shared complementarity. Synthesis, nuclease, and ligase reactions finish the event. (B) Double Holliday junctions (dHJs) are formed when the D loop captures the second end. In meiotic cells, dHJ resolution is biased to give COs. (C) dHJs can be dissolved without crossover by the action of the BLM(Sgs1)/TOP3α/RMI1 complex. (D) Resolution of intermediates that escape the BLM(Sgs1) complex may occur from the action of different resolvases (e.g., MUS81/EME1[Mms4], GEN1[Yen1], etc.).
Initiating Homologous Recombination: End Resection and the Control of Pathway Choice
As a key early step in HR reactions, the generation of 3′ single-stranded DNA by resection of the 5′ end acts as a control point for DSB repair pathway choice (Kass and Jasin 2010; Symington and Gautier 2011). End resection was observed in budding yeast by physical analysis of the fate of DSBs in vivo (Pâques and Haber 1999). HO endonuclease-generated DSBs undergo resection at their 5′ ends while maintaining relatively stable 3′ ends. Identifying the role of factors involved in resection proved challenging largely because of redundancy, but a breakthrough came from two laboratories in 2008 (Mimitou and Symington 2008; Zhu et al. 2008). Initial processing of the ends involves Mre11/Rad50/Xrs2, termed MRX (MRN in mammals), and the Sae2 protein. In mammalian cells, the functional counterpart of Sae2 is CtIP, as evidenced by the dependence on CtIP for recruitment of RPA to nuclear foci and for ATR activation (Sartori et al. 2007). Of note, the breast cancer suppressor BRCA1 interacts with MRN (Zhong et al. 1999) and CtIP (Yu et al. 1998) and importantly promotes both HR and SSA (Moynahan et al. 1999; Stark et al. 2004), as does CtIP (Sartori et al. 2007; Bennardo et al. 2008), suggesting that BRCA1 may also be involved in the initial step of end resection (Stark et al. 2004). More extensive resection involves the 5′-3′ exonuclease Exo1 or the combined helicase/nuclease activities of Sgs1/Dna2 (Mimitou and Symington 2008; Zhu et al. 2008). Roles for the mammalian counterparts EXO1 and BLM, respectively, is supported by in vitro end resection assays (Nimonkar et al. 2011).
Although programmed DSBs are channeled into specific repair pathways (e.g., meiotic DSBs into HR and immune system DSBs into NHEJ), DSBs from nucleases or radiation can be repaired by either HR or NHEJ pathways (Rouet et al. 1994b; Liang et al. 1998; Rothkamm et al. 2003). HR itself is regulated and is promoted during the S/G2 phase of the cell cycle (Ira et al. 2004; Escribano-Diaz et al. 2013). It is important to note that HR occurs within a cellular milieu in which NHEJ is also active, especially in mammalian cells where NHEJ is a prominent pathway throughout the cell cycle (Rothkamm et al. 2003). As such, end resection is a major determinant of whether canonical NHEJ or HR is used at DSBs. As shown by direct molecular analysis, resection is increased in yeast mutated in the canonical NHEJ proteins Ku and Dnl4/Lif1 (LIG4/XRCC4 in mammalian cells), with a greater effect in the Ku mutant (Lee et al. 1998; Zhang et al. 2007). Because Ku binds DNA ends, it presumably physically blocks access of the end resection machinery, whereas Dnl4/Lif1 may act to stabilize Ku binding to DNA ends. In cases in which the HR machinery successfully competes with Ku, MRX (MRN) and Sae2 (CtIP) may clip Ku from DNA ends, similar to its action to remove Spo11 from 5′ ends during meiotic recombination (Garcia et al. 2011).
Consistent with an increase in a resected intermediate, mutation of Ku or LIG4/XRCC4 in mammalian cells increases HR, with mutation of Ku showing the more profound effect (Pierce et al. 2001). Mutation of DNA-PKcs, which does not have a yeast homolog, also increases HR, again less profoundly than Ku (Pierce et al. 2001; Allen et al. 2002). Interestingly, an increase in HR in NHEJ mutants occurs whether the DSB is introduced by I-_Sce_I endonuclease, which generates a 3′ overhang, or the RAG recombinase, which generates a hairpin-capped end (Pierce et al. 2001; Weinstock and Jasin 2006). Like HR, SSA is increased with loss of Ku or LIG4/XRCC4 (Stark et al. 2004; Bennardo et al. 2009), in agreement with increased end resection in these mutants.
In addition to HR and SSA, a third DSB repair pathway initiates with end resection—that of microhomology-mediated NHEJ (MMEJ), also termed alternative NHEJ (alt-NHEJ) to distinguish it from canonical NHEJ (Fig. 2). Like HR and SSA, alt-NHEJ at microhomology is suppressed by canonical NHEJ components like Ku and LIG4/XRCC4, but it can also operate in the presence of the canonical pathway (Weinstock et al. 2007; Simsek and Jasin 2010; Simsek et al. 2011; Escribano-Diaz et al. 2013). On the other hand, end resection factors like CtIP promote alt-NHEJ, as well as HR and SSA (Bennardo et al. 2008; Simsek et al. 2011; Zhang and Jasin 2011).
Besides canonical NHEJ proteins like Ku and LIG4, the DNA damage response protein 53BP1 has a role in NHEJ (Nakamura et al. 2006; Stavnezer et al. 2008), and as with canonical NHEJ factors, 53BP1 disruption is associated with increased HR (Nakamura et al. 2006; Xie et al. 2007). In 2009, a surprising discovery was made showing that 53bp1 mutation rescues the lethality associated with Brca1 mutation in mice (Cao et al. 2009). Disruption of canonical NHEJ factors like Ku, however, does not similarly rescue (Bunting et al. 2012). Evidence indicates that 53bp1 disruption restores HR in BRCA1-deficient cells, likely by relieving 53BP1 inhibition of end resection at DSBs (Bouwman et al. 2010; Bunting et al. 2010). The yeast 53BP1 ortholog, Rad9, also regulates DNA end resection (Lazzaro et al. 2008). Thus, 53BP1 and BRCA1 may act antagonistically on end resection to control DSB repair pathway choice.
Likely a number of other proteins will be identified that regulate end resection, given the central role of end resection in DSB repair pathway choice. Several laboratories have recently described the RIF1 protein as an effector of DNA end resection control by 53BP1 (Chapman et al. 2013; Di Virgilio et al. 2013; Escribano-Diaz et al. 2013; Feng et al. 2013; Zimmermann et al. 2013). Thus, like disruption of 53BP1, loss of RIF1 restores end resection in BRCA1-depleted cells, as measured by RPA accumulation on chromatin and Rad51 focus formation. Genetic assays support this interpretation because HR, SSA, and alt-NHEJ are all increased by RIF1 depletion (Gunn and Stark 2012; Escribano-Diaz et al. 2013).
Rad51 Assembly and Strand Invasion: Another Decision Point
Rad51, a DNA-dependent ATPase that forms nucleoprotein filaments with DNA, is a homolog of the bacterial RecA protein (Shinohara et al. 1992, 1993; Cox 2007). The mechanism of strand exchange was illuminated by crystal structures of RecA filaments in complex with single- and double-stranded DNA (Chen et al. 2008), and is likely relevant to how Rad51 proteins function. Single-stranded DNA within the RecA filament has a repeating unit of three nucleotides, which maintains a B-form structure, while between every triplet the DNA is significantly stretched. The triplets in single-stranded DNA can pair through canonical Watson-Crick hydrogen bonds with complementary triplets in homologous duplex DNA, which is crucial for a stable interaction, as RecA itself has few contacts with the complementary DNA to stabilize the interaction. ATP hydrolysis promotes dissociation of the newly formed heteroduplex DNA and the displaced single strand.
Replication protein A (RPA) binds avidly to single-stranded DNA and effectively competes with Rad51, such that a number of proteins termed mediators are necessary to displace RPA to promote Rad51 binding (San Filippo et al. 2008). Critical mediators are Rad52 in yeast and BRCA2 in mammalian cells, as Rad51 recruitment to DSBs, and hence HR, are substantially impaired when these proteins are disrupted (Sugawara et al. 2003; Lisby et al. 2004; San Filippo et al. 2008; Moynahan and Jasin 2010). Formation of a Rad51 filament directs repair into an HR pathway and suppresses SSA. Thus, disruption of either BRCA2 or Rad51 leads to increased SSA while reducing HR (Moynahan et al. 2001; Tutt et al. 2001; Stark et al. 2004). The opposing effects on HR and SSA with Rad51 or BRCA2 disruption contrast with BRCA1 or CtIP disruption, in which both pathways are reduced, and RIF1 disruption, in which both pathways are increased. Where checked, alt-NHEJ is affected similarly to SSA. Overall, these results are consistent with an upstream role in HR for BRCA1/CtIP in promoting end resection, which is suppressed by 53BP1/RIF1, but a downstream role in HR for Rad51/BRCA2 in strand invasion. Importantly, Rad51 filament formation suppresses the potentially mutagenic pathways of SSA and alt-NHEJ, while promoting more precise HR.
Completing HR: Many Avenues
The extended strand invasion intermediate has many potential fates, ultimately leading to either an NCO or CO (Fig. 3) (Klein and Symington 2012). This “choice” of fate can have critical consequences. In meiotic cells, at least one DSB per chromosome pair must be resolved as a CO to ensure proper segregation of homologous chromosomes at the first meiotic division (Baker et al. 1976). However, in mitotic cells, COs between homologous chromosomes can lead to loss of heterozygosity of the segment of chromosome distal to the CO, an important mechanism for initiation of some types of tumors, whereas COs between repeats can lead to copy number variation, such as from duplications or deletions (Moynahan and Jasin 2010). Nonetheless, both COs and NCOs are observed in mitotic and meiotic cells, even if the balance is weighted differently for each (Cole et al. 2010; LaRocque et al. 2011).
In a primary pathway in mitotic cells to generate NCOs, the newly synthesized DNA strand dissociates from the D loop to anneal to the other DNA end, which has been termed synthesis-dependent strand annealing (Fig. 3A) (Nassif et al. 1994; Ferguson and Holloman 1996). In contrast, a key pathway in meiotic cells involves “capture” of the other DNA end by the D loop to form a dHJ; although originally postulated to be resolved as either a CO or NCO (Orr-Weaver and Szostak 1983), dHJ resolution appears to be biased to the formation of COs (Allers and Lichten 2001) by the combined activity of a MutL complex and Exo1 (Fig. 3B) (Zakharyevich et al. 2012). However, dHJs that form in mitotic cells can also be “dissolved” by the branch migration and topoisomerase activity of the BLM (Sgs1)/TOP3α/RMI1 complex (Fig. 3C) (Wu and Hickson 2003). Intermediates that escape the action of the BLM complex can potentially be resolved by several different resolvases, including MUS81/EME1(Mms4), GEN1(Yen1), and SLX1/SLX4 (Ho et al. 2010; Wechsler et al. 2011; De Muyt et al. 2012; Zakharyevich et al. 2012), the choice of which may be cell-cycle regulated (Fig. 3D) (Matos et al. 2011). Further complicating this already complex picture, Sgs1 regulates the formation of both COs and NCOs in meiosis (De Muyt et al. 2012; Klein and Symington 2012; Zakharyevich et al. 2012). Whether BLM performs a similarly complex role in mammals is still an open question.
CELL BIOLOGY OF DSB REPAIR IN LIVING CELLS
Immunohistochemistry provides a view of where the repair and recombination proteins are localized in the cell after DNA damage. For example, the colocalization of RAD51 and the breast cancer susceptibility genes, BRCA1 and BRCA2, in nuclear foci was important for linking them in a common pathway (Scully et al. 1997; Chen et al. 1998). As a result of the development of fluorescent tags, it has been shown that many repair proteins involved in HR form parts of dynamic giga-Dalton assemblies that contain many hundreds to thousands of copies of the different proteins (Lisby and Rothstein 2004). The proteins flow into and out of these complexes as shown by fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) analyses in mammalian cells (Essers et al. 2002). In addition, cells constantly assemble and disassemble these complexes as part of a quality control mechanism designed to monitor appropriate sites for HR activity (Kanaar et al. 2008). At this time, the completion of recombination events cannot be detected in individual cells and therefore one must be cautious in the interpretation of what the appearance of a focus represents—an attempt to recombine or a successful event. In addition, not all HR events may lead to detectable foci (Lisby and Rothstein 2004).
Choreography of the DNA Damage Response
DNA damage can be introduced into chromosomes via γ-irradiation, clastogens, laser microirradiation (in mammalian cells), or by using site-specific endonucleases. Precise tagging of chromosomal regions can be achieved by inserting multiple tandem arrays of bacterial operators into the chromosome and expression of fluorescently tagged repressor proteins allows the visualization of the specific chromosomal sites (Robinett et al. 1996). Addition of a meganuclease cut site adjacent to the array allows the monitoring of the proteins that are recruited to a site-specific DSB (Lisby et al. 2004). By combining tagged proteins and sites with mutants in the repair process and using time-lapse photography after induction of the break, many aspects of the choreography of the DNA damage response have been revealed (Lisby et al. 2004).
In budding yeast, repair centers are formed after DSBs, which are accompanied by the recruitment of checkpoint and repair proteins (Lisby and Rothstein 2004). One of the first proteins to form a focus after induction of DNA damage by γ-irradiation is the Mre11 protein, part of the Mre11/Rad50/Xrs2 (MRX) complex. It is also the first protein recruited to a site-specific DSB (Lisby et al. 2004). Resection of endonuclease-induced DSBs only occurs in S or G2 and is controlled by cyclin-dependent kinases (Ira et al. 2004). On the other hand, DSBs induced by γ-irradiation can be resected at any stage of the cell cycle (Barlow et al. 2008). The absence of the Mre11 protein blocks the appearance of Tel1 (the yeast ATM homolog) in foci, but interestingly does not block the appearance of many other downstream proteins including Rpa1, members of the yeast 9-1-1 complex (Ddc1, Rad17, and Mec3), Rad53 (a yeast CHK2 homolog), and the recombination protein, Rad52 (Lisby et al. 2004). Recruitment of budding yeast Rad9 (a 53BP1 homolog) and Rad53 depend on both Tel1 and Mec1 (the ATR homolog), whereas the downstream recombination protein, Rad52, does not (Lisby et al. 2004). The entire cascade of recombination proteins in yeast depends first on the recruitment of RPA to single-stranded DNA, which in turn recruits Rad52, which is necessary to recruit the Rad51 recombinase (Lisby et al. 2004).
In mammalian cells, like in yeast, recombination proteins are recruited to DSBs and many of these steps are conserved (Wyman and Kanaar 2006). For example, the mammalian Mre11/Rad50/Nbs1 (MRN) complex is one of the first sensors of DSBs followed by the downstream checkpoint proteins of the 9-1-1 complex (Rad9, Rad1, and Hus1) (Petrini and Stracker 2003). Other steps use slightly different players. For example, Rad52 in mammalian cells does not have as strong a recombination phenotype as in yeast (Essers et al. 2002; Stark et al. 2004) and it is the BRCA2 protein that is necessary to recruit Rad51 to IR-induced foci for HR (Yuan et al. 1999; Moynahan et al. 2001). However, knocking down both Rad52 and BRCA2 leads to synthetic lethality pointing to a role for Rad52 in mammalian cells (Feng et al. 2011). In addition, the Rad51 paralogues, RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3, are also required to recruit Rad51 to foci (Bishop et al. 1998; Takata et al. 2000; Rodrigue et al. 2006), whereas in budding yeast, the Rad51 paralogues, Rad55 and Rad57 are not (Lisby et al. 2004), although Rad51 foci are dimmer in a rad57 mutant (Fung et al. 2009). The Rad54 protein in mammalian cells plays a role both early and late in the recruitment of Rad51 to foci, the early role not being ATPase dependent (Agarwal et al. 2011).
In mammalian cells there are many more proteins found at laser-induced DSBs (Bekker-Jensen and Mailand 2010). In addition, the studies in mammalian cells have defined microcompartments with single-stranded DNA (RPA localization) as well as surrounding chromatin flanking the DSB after irradiation (Bekker-Jensen et al. 2006). Some recombination proteins, like the MRN complex and BRCA1, are in both compartments (Bekker-Jensen et al. 2006). However, there is much protein recruitment activity in the adjacent surrounding chromatin (Polo and Jackson 2011). For example, after a DSB, PARP activity is detected by the appearance of poly(ADP ribose) at a DSB (D'Amours et al. 1999; Luijsterburg and van Attikum 2012). Its accumulation and H2AX phosphorylation (γH2AX) signal the recruitment of MDC1 (Stucki et al. 2005), which in turn recruits 53BP1 (Luijsterburg and van Attikum 2012) and then the E3 ubiquitin ligases, RNF8, RNF168, and BRCA1 (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Wang and Elledge 2007; Doil et al. 2009; Stewart et al. 2009). These later proteins are necessary for the recruitment of the SUMO ligases PIAS1 and PIAS4, which help stabilize some of the ubiquitin conjugants (Galanty et al. 2009; Morris et al. 2009). It is thought that all of these processes aid in both the assembly and disassembly of recombination foci. Understanding their regulation will help us gain insights into their various functions during DSB repair.
INCREASED CHROMOSOME MOBILITY AFTER DNA DAMAGE
Recently, studies have focused on the movement of chromosomal loci before and after DNA damage. Many support the notion that DSBs cause increased mobility within the nucleus. For example, in yeast, cells that contain two double-strand breaks often form only a single repair focus, suggesting that the broken chromosomes move to a repair center (Lisby et al. 2003b). Furthermore, DSBs that are unable to be repaired move to the yeast nuclear periphery (Nagai et al. 2008; Oza et al. 2009). On the other hand, chromosome mobility in mammalian cells does not increase in G1 cells after induction of a single DSB, however mobility does increase slightly in the absence of Ku80 (Soutoglou et al. 2007). Different results were seen after creating DSBs in HeLa cells with α-particles (Jakob et al. 2009). On the other hand, γH2AX foci are more mobile favoring the view that distant DSBs can be juxtaposed (Aten et al. 2004). Increased chromosome movement of uncapped telomeres in mouse cells has recently been associated with the 53BP1 repair protein (Dimitrova et al. 2008). In addition, 53BP1 plays a role in long-range VDJ joining reactions, underlining the importance of properly regulating chromosome mobility (Difilippantonio et al. 2008). All of these results highlight the role of the DNA damage response and of repair proteins in the higher mobility of chromosomal loci.
High-resolution 4D tracking studies in the mid-1990s showed that chromosomal loci in budding yeast show confined diffusion exploring approximately 3% of the nuclear volume (Marshall et al. 1997). Recent work in haploid and diploid yeast cells has shown that after DNA damage, there is an increase in chromosome mobility (Hajjoul et al. 2009; Dion et al. 2012; Mine-Hattab and Rothstein 2012). Because the confinement radius of the broken chromosome more than doubles from that seen in the absence of DSBs, it means that it explores a more than 10 times larger nuclear volume. The increased mobility is general, as the dynamics of the unbroken chromosomes also increase depending on the number of DSBs as revealed by studies with γ-irradiation at doses delivering from between ∼4 DSBs and ∼20 DSBs per nucleus (Mine-Hattab and Rothstein 2012). In diploid cells, the pairing of the homologous chromosomes can be observed in real time, and once started it takes approximately 20 min before the loci separate again (Mine-Hattab and Rothstein 2012). This separation is associated with the disassembly of the repair center. Importantly, the single repair center associated with a DSB necessarily has the two ends held together and this tethering is at least partially dependent on MRX and Sae2 (Lisby et al. 2003a; Kaye et al. 2004; Lobachev et al. 2004; Clerici et al. 2005). Thus, the two ends of a DSB most often form a single Mre11 or Rad52 focus, suggesting that the homology search that occurs after a break has both ends ready for strand invasion at the homologous chromosomal locus. In summary, all of these studies suggest that the increase in chromosomal mobility likely facilitates the search for homology.
Combining genetics and cell biological studies have revealed some of the key players in regulating increased chromosome mobility. For example, in both haploid and diploid yeast cells, increased DNA dynamics depend on the Rad51 recombinase, which is similar to the observation that Rad51 is important for the movement of an unrepaired DSB to the nuclear periphery in haploid yeast cells (Oza et al. 2009; Dion et al. 2012; Mine-Hattab and Rothstein 2012). Similarly, in Drosophila, Rad51 is important for the increased mobility of heterochromatic DNA after induction of DSBs (Chiolo et al. 2011). Increased chromosome mobility in haploid yeast also depends on another recombination protein, Rad54, as well as on the checkpoint proteins, Rad9 and Mec1 (Dion et al. 2012). In addition, the pairing between homologous MAT and HML loci on chromosome III in haploid yeast cells seen after an HO-induced DSB at the MAT locus depends on two recombination and repair helicase proteins, Sgs1 and Srs2 (Houston and Broach 2006). Finally, delaying the appearance of single-stranded DNA in the absence of Sae2, a protein involved very early in the processing of DNA ends, also delays increased chromosome mobility (Mine-Hattab and Rothstein 2012). All of these studies point to the regulation of chromosome movements in response to DNA damage. It is interesting to note that increased mobility is not associated with a higher coefficient of diffusion, suggesting that global changes in the properties of chromatin after damage (Kruhlak et al. 2006; Ziv et al. 2006) may affect the movement of chromosomal loci.
PERSPECTIVES
Over the years, much progress has been made concerning the repair of DSBs; however, many questions remain in the field. How does the cell “know” what to do when confronted with broken DNA? What is the mechanism governing increased chromosome mobility? To what is the circuitry responding when it triggers the repair event? Clearly the state of the chromatin is used to both interpret and act as the scaffold to build repair centers. What are all of the proteins doing in the gigadalton complexes that form around broken DNA? Why do so many mediator proteins seem to be required to load Rad51 onto single-stranded DNA? How is the chromosomal end that is bound by Rad51 organized in this complex to find its partner? It is likely that concentrating the proteins in centers help ensure that biochemical reactions proceed rapidly and efficiently. They also play an important role in coordinating the reactions at the ends.
Finally, it is notable that HR proteins act as tumor suppressors (Walsh and King 2007; Turnbull and Rahman 2008). Furthermore, agents that cause strand breaks are tumorigenic. Thus, understanding DSB repair processes and the factors involved in them continue to be a priority. Still, the relationship between HR and tumor suppression remains to be elucidated. It is unclear whether genomewide instability is sufficient to promote tumorigenesis or whether a subset of loci is particularly vulnerable to lead to transformation. Synthetic lethality approaches are being exploited for therapy of HR-deficient tumors (Brough et al. 2011). Presumably, synthetic lethal screens will continue to be fruitful for developing therapeutic approaches and also to provide insight into the relationship of HR with other cellular pathways.
ACKNOWLEDGMENTS
We thank Lorraine Symington and Peter Thorpe for comments on the manuscript. M.J. also thanks Paige Arnold for her assistance. This work is supported by NIH NIGMS grants GM54668 and GM105421 to M.J. and GM50237 and GM67055 to R.R.
REFERENCES
*Reference is also in this collection.
- Agarwal S, van Cappellen WA, Guenole A, Eppink B, Linsen SE, Meijering E, Houtsmuller A, Kanaar R, Essers J 2011. ATP-dependent and independent functions of Rad54 in genome maintenance. J Cell Biol 192: 735–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C, Kurimasa A, Brenneman MA, Chen DJ, Nickoloff JA 2002. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc Natl Acad Sci 99: 3758–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers T, Lichten M 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106: 47–57 [DOI] [PubMed] [Google Scholar]
- Arcangioli B, de Lahondes R 2000. Fission yeast switches mating type by a replication-recombination coupled process. EMBO J 19: 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303: 92–95 [DOI] [PubMed] [Google Scholar]
- Baker BS, Carpenter AT, Esposito MS, Esposito RE, Sandler L 1976. The genetic control of meiosis. Ann Rev Genet 10: 53–134 [DOI] [PubMed] [Google Scholar]
- Barlow JH, Lisby M, Rothstein R 2008. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell 30: 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6: 989–998 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Mailand N 2010. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst) 9: 1219–1228 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol 173: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaiche Y, Mogila V, Perrimon N 1999. I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila. Genetics 152: 1037–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM 2008. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4: e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Gunn A, Cheng A, Hasty P, Stark JM 2009. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet 5: e1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417 [DOI] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, Shinohara A 1998. Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem 273: 21482–21488 [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 17: 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brough R, Frankum JR, Costa-Cabral S, Lord CJ, Ashworth A 2011. Searching for synthetic lethality in cancer. Curr Opin Genet Dev 21: 34–41 [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, et al. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell 46: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, et al. 2009. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell 35: 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR 1989. Altering the genome by homologous recombination. Science 244: 1288–1292 [DOI] [PubMed] [Google Scholar]
- Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC 2012. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci 109: 17382–17387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49: 858–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell 2: 317. [DOI] [PubMed] [Google Scholar]
- Chen Z, Yang H, Pavletich NP 2008. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453: 484–489 [DOI] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH 2011. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chiruvella KK, Liang Z, Wilson TE 2013. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol 5: a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP 2005. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem 280: 38631–38638 [DOI] [PubMed] [Google Scholar]
- Cole F, Keeney S, Jasin M 2010. Evolutionary conservation of meiotic DSB proteins: More than just Spo11. Genes Dev 24: 1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleaux L, d'Auriol L, Betermier M, Cottarel G, Jacquier A, Galibert F, Dujon B 1986. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell 44: 521–533 [DOI] [PubMed] [Google Scholar]
- Colleaux L, d'Auriol L, Gailbert F, Dujon B 1988. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci 85: 6022–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM 2007. Motoring along with the bacterial RecA protein. Nat Rev Mol Cell Biol 8: 127–138 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342: 249–268 [PMC free article] [PubMed] [Google Scholar]
- Davis L, Maizels N 2011. DNA nicks promote efficient and safe targeted gene correction. PloS ONE 6: e23981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Barbera M, McDonnell A, McIntyre K, Sternglanz R, Jin Q, Loidl J, Engebrecht J 2001. The Saccharomyces cerevisiae MUM2 gene interacts with the DNA replication machinery and is required for meiotic levels of double strand breaks. Genetics 157: 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell 46: 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 2008. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM 2012. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol 14: 502–509 [DOI] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, et al. 2013. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Dujon B 1989. Group I introns as mobile genetic elements: facts and mechanistic speculations—A review. Gene 82: 91–114 [DOI] [PubMed] [Google Scholar]
- Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, et al. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell 49: 872–883 [DOI] [PubMed] [Google Scholar]
- Esposito MS, Esposito RE 1969. The genetic control of sporulation in Saccharomyces. I: The isolation of temperature-sensitive sporulation-deficient mutants. Genetics 61: 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, Houtsmuller AB, van Veelen L, Paulusma C, Nigg AL, Pastink A, Vermeulen W, Hoeijmakers JH, Kanaar R 2002. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J 21: 2030–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN 2011. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci 108: 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Fong KW, Wang J, Wang W, Chen J 2013. RIF1 Counteracts BRCA1-mediated end resection during DNA repair. J Biol Chem 288: 11135–11143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DO, Holloman WK 1996. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci 93: 5419–5424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung CW, Mozlin AM, Symington LS 2009. Suppression of the double-strand-break-repair defect of the Saccharomyces cerevisiae rad57 mutant. Genetics 181: 1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP 2009. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462: 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SE, Gray S, Neale MJ 2011. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479: 241–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci 109: E2579–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn A, Stark JM 2012. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol Biol 920: 379–391 [DOI] [PubMed] [Google Scholar]
- Haber JE 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjoul H, Kocanova S, Lassadi I, Bystricky K, Bancaud A 2009. Lab-on-Chip for fast 3D particle tracking in living cells. Lab Chip 9: 3054–3058 [DOI] [PubMed] [Google Scholar]
- Henderson KA, Keeney S 2004. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc Natl Acad Sci 101: 4519–4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Mazón G, Lam AF, Symington LS 2010. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell 40: 988–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R 1964. A mechanism for gene conversion in fungi. Gen Res 5: 282–304 [DOI] [PubMed] [Google Scholar]
- Houston PL, Broach JR 2006. The dynamics of homologous pairing during mating type interconversion in budding yeast. PLoS Genet 2: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotech 31: 227–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Satory D, Haber JE 2006. Conservative inheritance of newly synthesized DNA in double-strand break-induced gene conversion. Mol Cell Biol 26: 9424–9429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JA, Fink GR 1981. Gene conversion between duplicated genetic elements in yeast. Nature 292: 306–311 [DOI] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Durante M, Taucher-Scholz G 2009. Live cell microscopy analysis of radiation-induced DNA double-strand break motion. Proc Natl Acad Sci 106: 3172–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Berg P 1988. Homologous integration in mammalian cells without target gene selction. Genes Dev 2: 1353–1363 [DOI] [PubMed] [Google Scholar]
- Jasin M, deVilliers J, Weber F, Schaffner W 1985. High frequency of homologous recombination in mammalian cells between endogenous and introduced SV40 genomes. Cell 43: 695–703 [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Wyman C, Rothstein R 2008. Quality control of DNA break metabolism: In the “end,” it's a good thing. EMBO J 27: 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass EM, Jasin M 2010. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett 584: 3703–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Lange J, Baudat F, Jasin M, Keeney S 2013. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev 27: 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP 2004. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol 14: 2096–2106 [DOI] [PubMed] [Google Scholar]
- Keeney S 2008. Spo11 and the formation of DNA double-strand breaks in meiosis. In Recombination and meiosis (ed. Egel R, Lankenau D-H), pp. 81–123 Springer-Verlag, Berlin: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS 2012. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res 22: 1327–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL 1995. Genetic control of intrachromosomal recombination. Bioessays 17: 147–159 [DOI] [PubMed] [Google Scholar]
- Klein HL, Petes TD 1981. Intrachromosomal gene conversion in yeast. Nature 289: 144–148 [DOI] [PubMed] [Google Scholar]
- Klein HL, Symington LS 2012. Sgs1—The maestro of recombination. Cell 149: 257–259 [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostriken R, Strathern JN, Klar AJ, Hicks JB, Heffron F 1983. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell 35: 167–174 [DOI] [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, Bazett-Jones DP, Nussenzweig A 2006. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol 172: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy E, Roberts S, Evans EP, Burtenshaw MD, Costantini FD 1983. A foreign β-globin gene in transgenic mice: Integration at abnormal chromosomal positions and expression in inappropriate tissues. Cell 34: 343–358 [DOI] [PubMed] [Google Scholar]
- Lambert S, Saintigny Y, Delacote F, Amiot F, Chaput B, Lecomte M, Huck S, Bertrand P, Lopez BS 1999. Analysis of intrachromosomal homologous recombination in mammalian cell, using tandem repeat sequences. Mutat Res 433: 159–168 [DOI] [PubMed] [Google Scholar]
- LaRocque JR, Stark JM, Oh J, Bojilova E, Yusa K, Horie K, Takeda J, Jasin M 2011. Interhomolog recombination and loss of heterozygosity in wild-type and Bloom syndrome helicase (BLM)-deficient mammalian cells. Proc Natl Acad Sci 108: 11971–11976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M 2008. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J 27: 1502–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409 [DOI] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Salus SS, Roth DB 2004. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell 117: 171–184 [DOI] [PubMed] [Google Scholar]
- Liang F, Romanienko PJ, Weaver DT, Jeggo PA, Jasin M 1996. Chromosomal double-strand break repair in Ku80-deficient cells. Proc Natl Acad Sci 93: 8929–8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Han M, Romanienko PJ, Jasin M 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci 95: 5172–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FL, Sperle K, Sternberg N 1984. Model for homologous recombination during transfer of DNA into mouse L cells: Role for DNA ends in the recombination process. Mol Cell Biol 4: 1020–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R 2004. DNA damage checkpoint and repair centers. Curr Opin Cell Biol 16: 328–334 [DOI] [PubMed] [Google Scholar]
- Lisby M, Antunez de Mayolo A, Mortensen UH, Rothstein R 2003a. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle 2: 479–483 [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R 2003b. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol 5: 572–577 [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R 2004. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K 2004. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol 14: 2107–2112 [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, van Attikum H 2012. Close encounters of the RNF8th kind: When chromatin meets DNA repair. Curr Opin Cell Biol 24: 439–447 [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol 7: 930–939 [DOI] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, West SC 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Baharoglu Z, Lestini R 2007. Genetics of recombination in the model bacterium Escherichia coli. In Molecular genetics of recombination, pp. 1–26 Springer, New York [Google Scholar]
- Mimitou EP, Symington LS 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine-Hattab J, Rothstein R 2012. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol 14: 510–517 [DOI] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462: 886–890 [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ 2009. A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M 1999. Brca1 controls homology-directed DNA repair. Mol Cell 4: 511–518 [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M 2001. BRCA2 Is required for homology-directed repair of chromosomal breaks. Mol Cell 7: 263–272 [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Sakai W, Kawamoto T, Bree RT, Lowndes NF, Takeda S, Taniguchi Y 2006. Genetic dissection of vertebrate 53BP1: A major role in non-homologous end joining of DNA double strand breaks. DNA Repair (Amst) 5: 741–749 [DOI] [PubMed] [Google Scholar]
- Nassif N, Penney J, Pal S, Engels WR, Gloor GB 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol 14: 1613–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436: 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW 1983. Yeast recombination: The association between double-strand gap repair and crossing-over. Proc Natl Acad Sci 80: 4417–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ 1981. Yeast transformation: A model system for the study of recombination. Proc Natl Acad Sci 78: 6354–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev 23: 912–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F, Haber JE 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini JH, Stracker TH 2003. The cellular response to DNA double-strand breaks: Defining the sensors and mediators. Trends Cell Biol 13: 458–462 [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13: 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 15: 3237–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessis A, Perrin A, Haber JE, Dujon B 1992. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics 130: 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Jackson SP 2011. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev 25: 409–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder RG, Fonville NC, Rosenberg SM 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell 19: 791–804 [DOI] [PubMed] [Google Scholar]
- Puchta H, Dujon B, Hohn B 1993. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res 21: 5034–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H, Dujon B, Hohn B 1996. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci 93: 5055–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CL, Certo MT, Mussolino C, Goodwin MJ, Cradick TJ, McCaffrey AP, Cathomen T, Scharenberg AM, Joung JK 2012. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res 40: 5560–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick MA 1976. The repair of double-strand breaks in DNA; a model involving recombination. J Theoret Biol 59: 97–106 [DOI] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135: 1685–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue A, Lafrance M, Gauthier MC, McDonald D, Hendzel M, West SC, Jasin M, Masson JY 2006. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J 25: 222–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6: 975–987 [DOI] [PubMed] [Google Scholar]
- Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Olivera BM, Brodsky M, Rubin GM, Golic KG 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev 16: 1568–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth DB, Nakajima PB, Menetski JP, Bosma MJ, Gellert M 1992. V(D)J recombination in mouse thymocytes: Double-strand breaks near T cell receptor delta rearrangement signals. Cell 69: 41–53 [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Kruger I, Thompson LH, Lobrich M 2003. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol 23: 5706–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M 1994a. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci 91: 6064–6068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M 1994b. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14: 8096–8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N, Haber JE 1988. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol 8: 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H 2008. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257 [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM 1997. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88: 265–275 [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T 1992. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69: 457–470 [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T 1993. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet 4: 239–243 [DOI] [PubMed] [Google Scholar]
- Simsek D, Jasin M 2010. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4/ligase IV during chromosomal translocation formation. Nat Struct Mol Biol 17: 410–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, et al. 2011. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet 7: e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS 1985. Insertion of DNA sequences into the human chromosomal ß-globin locus by homologous recombination. Nature 317: 230–234 [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T 2007. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 9: 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M 2004. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol 24: 9305–9316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE 2008. Mechanism and regulation of class switch recombination. Annu Rev Immunol 26: 261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Strathern JN, Klar AJ, Hicks JB, Abraham JA, Ivy JM, Nasmyth KA, McGill C 1982. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31: 183–192 [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell 12: 209–219 [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Schultes NP, Szostak JW 1989. Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338: 87–90 [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271 [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW 1983. The double-strand-break repair model for recombination. Cell 33: 25–35 [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, Swagemakers SM, Kanaar R, Thompson LH, Takeda S 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol Cell Biol 20: 6476–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesse S, Storlazzi A, Kleckner N, Gargano S, Zickler D 2003. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci 100: 12865–12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C, Rahman N 2008. Genetic predisposition to breast cancer: Past, present, and future. Annu Rev Genomics Hum Genet 9: 321–345 [DOI] [PubMed] [Google Scholar]
- Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, Griffin C, Thacker J, Ashworth A 2001. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J 20: 4704–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC 2005. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651 [DOI] [PubMed] [Google Scholar]
- Wake CT, Gudewicz T, Porter T, White A, Wilson JH 1984. How damaged is the biologically active subpopulation of transfected DNA? Mol Cell Biol 4: 387–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, King MC 2007. Ten genes for inherited breast cancer. Cancer Cell 11: 103–105 [DOI] [PubMed] [Google Scholar]
- Wang B, Elledge SJ 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci 104: 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Friedman G, Doyon Y, Wang NS, Li CJ, Miller JC, Hua KL, Yan JJ, Babiarz JE, Gregory PD, et al. 2012. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res 22: 1316–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Webb CJ, Wu Y, Zakian VA 2013. DNA repair at telomeres: Keeping the ends intact. Cold Spring Harb Perspect Biol 5: a012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler T, Newman S, West SC 2011. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature 471: 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Jasin M 2006. Alternative pathways for the repair of RAG-induced DNA breaks. Mol Cell Biol 26: 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Brunet E, Jasin M 2007. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol 9: 978–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R 2006. DNA double-strand break repair: All's well that ends well. Annu Rev Genet 40: 363–383 [DOI] [PubMed] [Google Scholar]
- Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, et al. 2007. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell 28: 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R 1998. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem 273: 25388–25392 [DOI] [PubMed] [Google Scholar]
- Yuan SS, Lee SY, Chen G, Song M, Tomlinson GE, Lee EY 1999. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res 59: 3547–3551 [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jasin M 2011. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol 18: 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE 2007. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol 14: 639–646 [DOI] [PubMed] [Google Scholar]
- Zhong Q, Chen CF, Li S, Chen Y, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH 1999. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science 285: 747–750 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T 2013. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 339: 700–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM-and KAP-1 dependent pathway. Nat Cell Biol 8: 870–876 [DOI] [PubMed] [Google Scholar]