Variation in Consumption of Human Milk Oligosaccharides by Infant Gut-Associated Strains of Bifidobacterium breve (original) (raw)
Abstract
Human milk contains a high concentration of complex oligosaccharides that influence the composition of the intestinal microbiota in breast-fed infants. Previous studies have indicated that select species such as Bifidobacterium longum subsp. infantis and Bifidobacterium bifidum can utilize human milk oligosaccharides (HMO) in vitro as the sole carbon source, while the relatively few B. longum subsp. longum and Bifidobacterium breve isolates tested appear less adapted to these substrates. Considering the high frequency at which B. breve is isolated from breast-fed infant feces, we postulated that some B. breve strains can more vigorously consume HMO and thus are enriched in the breast-fed infant gastrointestinal tract. To examine this, a number of B. breve isolates from breast-fed infant feces were characterized for the presence of different glycosyl hydrolases that participate in HMO utilization, as well as by their ability to grow on HMO or specific HMO species such as lacto-_N_-tetraose (LNT) and fucosyllactose. All B. breve strains showed high levels of growth on LNT and lacto-_N_-neotetraose (LNnT), and, in general, growth on total HMO was moderate for most of the strains, with several strain differences. Growth and consumption of fucosylated HMO were strain dependent, mostly in isolates possessing a glycosyl hydrolase family 29 α-fucosidase. Glycoprofiling of the spent supernatant after HMO fermentation by select strains revealed that all B. breve strains can utilize sialylated HMO to a certain extent, especially sialyl-lacto-_N_-tetraose. Interestingly, this specific oligosaccharide was depleted before neutral LNT by strain SC95. In aggregate, this work indicates that the HMO consumption phenotype in B. breve is variable; however, some strains display specific adaptations to these substrates, enabling more vigorous consumption of fucosylated and sialylated HMO. These results provide a rationale for the predominance of this species in breast-fed infant feces and contribute to a more accurate picture of the ecology of the developing infant intestinal microbiota.
INTRODUCTION
In addition to essential nutrients such as lactose lipids and proteins, human milk contains a large concentration of oligosaccharides (1). Human milk oligosaccharides (HMO) are complex and diverse glycans. These molecules are composed of monomers of glucose (Glc), galactose (Gal), and _N_-acetylglucosamine (GlcNAc) and often contain fucose (Fuc) and/or _N_-acetylneuraminic acid (NeuAc), linked via several glycosidic bonds (2, 3). All HMO contain the disaccharide lactose at their reducing end. Lactose can be sialylated to form sialyllactose or fucosylated to form fucosyllactose. In more complex HMO, the lactose core is conjugated with repeats of lacto-_N_-biose I (Galβ1-3GlcNAc; LNB; type 1 chain) or _N_-acetyllactosamine (Galβ1-4GlcNAc; type 2 chain), producing molecules with a degree of polymerization larger than 4 (4). These core structures can be modified by fucose and sialic acid residues via different linkages, with fucosylated neutral oligosaccharides the most representative in human milk (5). Although a large number of different HMO structures have been determined, a few isomers can represent up to 70% of the total molecules (6).
Remarkably, the energetic value of HMO for the infant is minimal. Various researchers have shown that HMO are resistant to enzymatic hydrolysis from intestinal brush border membrane and pancreatic juices, and therefore the majority of these molecules transit the intestinal tract, reaching the colon intact (7–9). During their transit HMO are believed to prevent pathogen colonization by serving as decoy binding sites for epithelial glycans (10).
One of the more fascinating roles of HMO is their ability to influence the composition of the intestinal microbiota in the first years of life. While the microbial community in breast-fed infants is often dominated by the genus Bifidobacterium, formula-fed infants show increased bacterial diversity (11, 12). This suggests that both pro- and antimicrobial elements in breast milk account for these differences. A conceptual basis for coevolution between bifidobacteria and milk glycans is supported by recent definition of the molecular mechanisms by which these microbes catabolize HMO. In Bifidobacterium longum subsp. infantis (B. longum subsp. infantis) ATCC 15697 these mechanisms include oligosaccharide transporters and intracellular glycosyl hydrolases (GH) such as fucosidases, hexosaminidases, and sialidases (13).
Bifidobacterium breve, B. longum subsp. infantis, B. longum subsp. longum, Bifidobacterium catenulatum, Bifidobacterium pseudocatenulatum, and Bifidobacterium bifidum are the species most frequently detected in breast-fed infant feces (11, 14, 33). In general, B. breve and B. longum subsp. infantis are more exclusively found in infants, and B. longum subsp. longum and B. bifidum are found in both infants and adults. While several strains of B. bifidum and B. longum subsp. infantis have been shown to grow vigorously on HMO in vitro (15, 16), this phenotype has been largely unexplored for larger numbers of B. breve and B. longum subsp. longum isolates. Only one strain of B. breve, ATCC 15700, has been shown to utilize a far less diverse range of HMO isomers than B. longum subsp. infantis ATCC 15697 (17). Whether this pattern is representative of the B. breve species is not clear. To better address what factors explain the predominance of this species in the infant gut microbiota, we isolated and characterized a representative number of strains of B. breve and examined the molecular mechanisms for their consumption of milk oligosaccharides.
MATERIALS AND METHODS
Subjects.
Fecal samples were collected from 40 exclusively breast-fed term infants at 3 and 4 months of age (UC Davis Lactation Study). None of the infants enrolled in this study received antibiotic treatment, infant formula, or solid food before the time of sampling. Parents transferred their infant fecal samples into sterile plastic tubes and were instructed to immediately store the samples at −20°C until transported by study personnel. Fecal samples were transported to the laboratory on dry ice and stored at −80°C before processing. The University of California Davis Institutional Review Board approved all aspects of the study and informed consent was obtained from all subjects. This trial was registered on clinicaltrials.gov (identification number NCT01817127).
Microbial isolations.
For isolation of Bifidobacterium, 100 mg of each fecal sample was taken aseptically, transferred to a sterile tube, diluted 10-fold with 1% peptone water (Becton, Dickinson, Sparks, MD), and homogenized by vortexing. Ten-fold dilutions were prepared with 1% peptone water and inoculated on modified basal salts medium (BSM) agar (mBSM). Modified BSM agar was prepared by supplementing de Man-Rogosa-Sharpe (MRS) medium (Becton, Dickinson, Sparks, MD) with 15 g/liter agar, 500 mg/liter l-cysteine-HCl, 100 mg/liter mupirocin, 25 mg/liter kanamycin, 4.28 mg/liter polymyxin B, 25 mg/liter iodoacetate, 20 mg/liter nalidixic acid, and 25 mg/ml of 2,3,5-triphenyltetrazolium chloride (Sigma, St. Louis, MO). The plates were incubated for 48 h at 37°C in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) in an atmosphere containing 5% carbon dioxide, 5% hydrogen, and 90% nitrogen. Resulting colonies were streaked onto mBSM agar, and after two passages they were grown in MRS broth supplemented with 0.05% l-cysteine-HCl and stored at −80°C in 25% glycerol. Prior to each assay all bacteria strains were subcultured twice in MRS broth supplemented with 0.05% l-cysteine-HCl and incubated at 37°C for 18 h in an anaerobic chamber.
Additional B. breve strains were obtained from the Japanese Collection of Microorganisms (RIKEN BioResource Center, Japan), the American Type Culture Collection (Manassas, VA), and the University of California—Davis Viticulture and Enology Culture Collection (see Table S1 in the supplemental material).
Identification of bifidobacteria by 16S rRNA gene sequencing.
Genomic DNA was obtained from 1 ml of each culture and centrifuged for 5 min at 2,000 × g. The bacterial pellet was resuspended and incubated for 30 min at 37°C with enzymatic lysis buffer consisting of 20 mM Tris-Cl, pH 8.0, 2 mM sodium EDTA, 1.2% Triton X-100, and 40 mg/ml lysozyme (Sigma). After enzymatic lysis, bacterial DNA was isolated from the samples using a DNeasy tissue kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. DNA quality and yield were checked using a Nanodrop spectrophotometer (Wilmington, DE); the DNA was then stored at −20°C until further use. To identify the isolates at species level, the 16S rRNA gene was amplified by PCR using the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG) and 1492R (5′-TACGGTTACCTTGTTACGA) on an Applied Biosystems 2720m Thermal Cycler (Applied Biosystems, Mountain View, CA) (18). One microliter of extracted DNA was added to a 50-μl reaction mixture containing 50 pmol of primers, 500 μM each deoxynucleoside triphosphate (dNTP), 0.1 volume of 10× PCR buffer, 2.5 mM MgCl2, and 1 U of AmpliTaq Gold polymerase (Applied Biosystems). Amplification mixtures were subjected to 4 min of denaturation at 94°C, followed by 30 cycles of 94°C for 30 s, 55°C for 40 s, and 72°C for 1 min 30s, with a final extension period of 7 min at 72°C. The resulting amplicons were separated on a 1% agarose gel, followed by GelRed staining (Phenix Research Products, Candler, NC) and purification using a QIAquick PCR Purification Kit (Qiagen). Sequencing was performed on an ABI 3730 Capillary Electrophoresis Genetic Analyzer using BigDye Terminator chemistries at the University of California—Davis DNA Sequencing Facility. The sequences were analyzed using BioEdit, version 7.0 (http://www.mbio.ncsu.edu/BioEdit/BioEdit.html), and checked by nucleotide-nucleotide BLAST comparison at the NCBI database for species identification.
MLST of strains.
Multilocus sequence typing (MLST) analysis of B. breve isolates targeted intragenic regions of seven housekeeping genes (clpC, purF, gyrB, fusA, ileS, rplB, and rpoB) selected based on a previous study (19). PCR amplifications were prepared as described above, and cycling conditions were optimized for every primer set (see Table S2 in the supplemental material) and consisted of an initial denaturation at 95°C for 4 min, followed by 35 cycles of 95°C for 30 s, annealing at 60 to 67°C for 30 s, elongation at 72°C for 60 s, with a final extension at 72°C for 7 min and holding at 4°C. The PCR products were separated and sequenced as described above.
Sequencing data for all loci were edited using BioEdit, version 7.0, and aligned using CLUSTAL W (20). Phylogenetic analysis and concatenations of the sequenced loci were performed using the Molecular Evolutionary Genetic Analysis (MEGA) software, version 5 (http://megasoftware.net). Descriptive evolutionary analysis including mol% G+C content, number of polymorphic sites, nucleotide diversity (π/site), and average number of nucleotide differences (k) was performed using DnaSP, version 5.10 (see Table S3 in the supplemental material). Allelic sequences were assigned as described previously (21) (see Table S4). A minimum evolution tree of the concatenated loci was calculated using MEGA, version 5.0 (see Fig. S1).
Glycosyl hydrolase gene sequencing.
α-Fucosidases (Blon_2336, Blon_2335, Blon_0248/0426, and Blon_0346), α-sialidases (Blon_2348, and Blon_0646), and the β-hexosaminidase Blon_0459 protein sequences from B. longum subsp. infantis ATCC 15697 were aligned with homologous sequences from the GenBank database (accession numbers are shown in Table S5 in the supplemental material) using Bioedit, version 7.0, and degenerate primers were designed to amplify conserved regions (see Table S6). To further differentiate between Blon_0248 and Blon_0426, strains that were PCR positive for the fucosidase Blon_0248/0426 were also amplified with the primers designed previously (22) to amplify the complete gene in B. longum subsp. infantis ATCC 15697 (see Table S6). PCR amplifications were prepared as described above with 200 pmol of primers, and cycling conditions were optimized for every primer set (see Table S6); the PCR program consisted of an initial denaturation at 95°C for 4 min, followed by 35 cycles of 95°C for 30 s, annealing at 45 to 55°C for 30 s, elongation at 72°C for 60 s, with a final extension at 72°C for 7 min and holding at 4°C. The resulting amplicons were separated and sequenced as described above. B. longum subsp. infantis ATCC 15697 and Bifidobacterium animalis subsp. lactis JCM 10602 were used as positive- and negative-control strains, respectively.
Bifidobacterial growth in vitro on HMO.
The 24 B. breve strains listed in Table S1 in the supplemental material were tested for growth in the presence of seven different substrates: HMO (23), lacto-_N_-tetraose (LNT), lacto-_N_-neotetraose (LNnT), 2′-fucosyllactose (2FL), 3′-fucosyllactose (3FL) (Glycom, Denmark), 3′-sialyllactose (3SL), and 6′-sialyllactose (6SL) (GenChem Co., South Korea). B. longum subsp. infantis ATCC 15697 and B. animalis subsp. lactis JCM 10602 were included as positive and negative controls for growth on HMO, respectively. Two microliters of each resulting overnight culture was used to inoculate 200 μl of modified MRS medium (mMRS), devoid of glucose and supplemented with 2% (wt/vol) of each sterile-filtered substrate as the sole carbohydrate source, and another 2 μl was inoculated into mMRS medium without added sugar. The medium was supplemented with 0.05% (wt/vol) l-cysteine, and in all the cases the cultures in the wells of the microtiter plates were covered with 30 μl of sterile mineral oil to avoid evaporation. The incubations were carried out at 37°C in an anaerobic chamber (Coy Laboratory Products). Cell growth was monitored in real time by assessing optical density at 600 nm (OD600) using a BioTek PowerWave 340 plate reader (BioTek, Winoosky, VT) every 30 min, preceded by 15 s of shaking at variable speed. Two biological replicates and three technical replicates each were performed for every studied strain. Maximum OD and growth rates were calculated and expressed as the mean of all replicates with the respective standard deviation. These calculations were performed as previously described (24). The OD obtained for each strain grown on the different substrates was compared with the OD obtained in the absence of a sugar source. This difference in OD (ΔOD) was used as a parameter to evaluate each strain's ability for growth on the different substrates.
Glycoprofiling.
Bacterial cultures in mMRS medium with 2% HMO were collected at the end of the exponential phase and centrifuged at 12,000 × g for 30 min. In the case of B. breve SC95, the samples were collected at three different points in the growth curve, i.e., OD600 values of approximately 0.2, 0.5, and 0.8. At least two biological replicates were performed in triplicates. Supernatants were filtered using a multiscreen 96-well, 0.22-μm-pore-size filtration plate (Millipore, Billerica, MA) prior to storage at −80°C. Remaining oligosaccharides were recovered from the supernatants (25 μl) and reduced to their alditol forms with 1 M NaBH4 at 65°C for 1.5 h. Each replicate was desalted by solid-phase extraction (SPE) on graphitized carbon cartridges. Salts were removed with 6 ml of deionized water, and oligosaccharides were eluted with 20% (vol/vol) acetonitrile in water and with 40% acetonitrile in 0.01% trifluoroacetic acid (vol/vol). SPE fractions were combined and dried under vacuum. Samples were reconstituted in 100 μl of deionized water and diluted 50-fold for liquid chromatography-mass spectrometry (LC-MS) analysis.
An Agilent high-performance liquid chromatography–chip–time of flight (HPLC-chip/TOF) mass spectrometer equipped with a capillary pump for sample loading and a nano-pump for chromatographic separation was used for HMO analysis. Separation was performed on a microfluidic chip equipped with enrichment and nano-LC analytical columns, both packed with porous graphitized carbon, using a previously described method (3, 25). Briefly, HMO were separated by a 65-min gradient using a binary solvent system consisting of 3% acetonitrile-water in 0.1% formic acid (vol/vol) (solvent A) and 90% acetonitrile-water in 0.1% formic acid (vol/vol) (solvent B). HMO were analyzed in positive ion mode, with a mass range between 300 and 2,000 m/z. Agilent's Masshunter software was used for data acquisition and data analysis (version B.03.01).
HMO monosaccharide composition was determined using accurate mass within a ±20 ppm mass error of theoretically calculated masses. Specific structures were assigned to HMO peaks by matching the reproducible retention times to those reported in previously annotated HMO libraries (2, 25). Percent consumption was calculated using a label-free method, employing the uninoculated HMO pool as an external standard. Total HMO consumption was calculated with respect to the uninoculated control by normalizing the summed abundance of all identified oligosaccharide spectra in ion counts in the bacterial supernatant to that of the control using the following equation:
| [1−(Σi=1n API bacteria sampleΣi=1n API uninoculated control)]×100 | (1) |
|---|
where API is absolute peak intensity and n is the number of identified HMO. The consumption of individual HMO species was quantified in the same manner, in which the absolute peak intensity of a specific HMO structure was normalized to the peak intensity of the corresponding structure in the uninoculated control.
Gene expression analysis.
The full nucleotide sequences of the genes encoding a GH95 and a GH29 α-fucosidase in the strain B. breve SC95 generated were used to design qPCR primers using the primer-BLAST tool at NCBI (see Table S6 in the supplemental material). For relative quantification, the rnpA gene was used as described by O'Connell Motherway et al. (26). B. breve SC95 was grown as described above in mMRS medium supplemented with either 2% lactose, 2% HMO, or 2% 2FL in a microplate reader, and cultures were taken at mid-exponential phase (OD of 0.6 to 1.0). Samples were immediately pelleted at 12,000 × g for 1 min and stored in RNA Later (Ambion). RNA extraction, cDNA conversion, and quantitative PCR (qPCR) were performed as described by Garrido et al. (27).
RESULTS
Isolation and identification of Bifidobacterium from breast-fed infant feces.
In order to obtain a representative number of B. breve strains, we collected fecal samples from 40 breast-fed term infants. Diluted fecal samples were grown on mBSM medium, and six colonies from the highest dilution were isolated. We obtained around 500 isolates that were identified by 16S rRNA gene sequence analysis, and a total of 461 isolates were identified as Bifidobacterium. Seven species of bifidobacteria were detected, and the species B. longum was the most represented (297 isolates), followed by B. breve with 77 isolates (Table 1).
Table 1.
Distribution of isolates of bifidobacteria from breast-fed infants identified by 16S rRNA gene sequencing
| Species | No. of isolates identified |
|---|---|
| B. longum | 297 |
| B. breve | 77 |
| B. pseudocatenulatum | 45 |
| B. bifidum | 22 |
| B. dentium | 8 |
| B. adolescentis | 7 |
| B. animalis | 5 |
We further investigated the identity of the B. breve isolates at the strain level by MLST, as described elsewhere (19). Our analysis also included nine strains from culture collections (see Table S1 in the supplemental material). A total of 172 single nucleotide polymorphisms (SNPs) were found in seven loci, and they generated between 8 rplB and 65 purF polymorphic sites (see Table S3). Twenty different allelic profiles were identified in the 86 B. breve isolates analyzed (see Table S4). Taking a conservative approach, strains isolated from the unrelated infants that shared identical MLST profiles were considered separate isolates in our study. This resulted in a library of 24 strains of B. breve (see Table S1), for which a consensus phylogenetic tree of the concatenated MLST data is shown in Fig. S1 in the supplemental material.
Glycosyl hydrolase genes in B. breve isolates.
In order to study the possible adaptations of the B. breve isolates for growth on HMO, we first determined the presence of three key GH classes required to cleave HMO into its constituent monosaccharides. The β-galactosidase activity was not examined because it is widespread in the Bifidobacterium genus. The genome of B. breve UCC2003 (26) contains an α-fucosidase, an α-sialidase, and a β-hexosaminidase with significant homology to cognate enzymes in B. longum subsp. infantis ATCC 15697. No homology was found to the same glycosyl hydrolases in B. bifidum genomes. Based on this, we used degenerate primers to look for genes encoding these GH in the assembled B. breve strains (Table 2). All of the B. breve strains possessed a gene homologous to β-hexosaminidase Blon_0459 in B. longum subsp. infantis (27) and an α-fucosidase similar to Blon_2335 in B. longum subsp. infantis ATCC 15697 (22), and all strains excepting JCM 7020 possessed an α-sialidase, related to Blon_0646 in B. longum subsp. infantis (28). Moreover, five strains possessed a second α-fucosidase, homologous to Blon_0248 in B. longum subsp. infantis ATCC 15697 (22), belonging to GH family 29 (GH29) (Table 2; see also Fig. S2 in the supplemental material).
Table 2.
Presence of glycosyl hydrolases and growth on different HMO by B. breve strains
| Strain | Glycosyl hydrolase and family_a_ | Bacterial growth on_b_: | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| α-Fucosidase | α-Sialidase (GH33) | β-Hexosaminidase (GH20) | HMO | LNT | LNnT | 2FL | 3FL | 3SL | 6SL | ||
| GH95 | GH29 | ||||||||||
| UCC2003 | + | − | + | + | + | +++ | +++ | − | + | − | − |
| ATCC15700 | + | − | + | + | + | +++ | +++ | − | − | − | − |
| ATCC15698 | + | − | + | + | ++ | +++ | +++ | − | − | − | − |
| ATCC15701 | + | − | + | + | +++ | +++ | +++ | − | − | − | − |
| JCM7017 | + | − | + | + | ++ | +++ | +++ | − | − | − | − |
| JCM7019 | + | − | + | + | ++ | +++ | +++ | + | + | − | − |
| JCM7020 | + | + | − | + | ++ | +++ | +++ | − | − | − | − |
| S-17c | + | − | + | + | + | +++ | +++ | − | + | − | − |
| S-46 | + | − | + | + | ++ | +++ | +++ | − | + | − | − |
| SC81 | + | − | + | + | ++ | +++ | +++ | − | − | − | − |
| SC95 | + | + | + | + | +++ | +++ | +++ | +++ | + | − | − |
| SC139 | + | − | + | + | ++ | +++ | +++ | − | − | − | − |
| SC154 | + | + | + | + | +++ | +++ | +++ | − | − | − | − |
| SC500 | + | − | + | + | ++ | +++ | +++ | − | − | − | − |
| SC506 | + | + | + | + | ++ | +++ | +++ | − | − | − | − |
| SC508 | + | − | + | + | + | +++ | +++ | − | − | − | − |
| SC522 | + | − | + | + | ++ | +++ | +++ | − | + | − | − |
| SC559 | + | − | + | + | ++ | +++ | +++ | − | − | − | − |
| SC567 | + | − | + | + | ++ | +++ | +++ | − | − | − | − |
| SC568 | + | + | + | + | ++ | +++ | +++ | +++ | + | − | − |
| SC573 | + | − | + | + | + | +++ | +++ | − | + | − | − |
| SC580 | + | − | + | + | ++ | +++ | +++ | − | + | − | − |
| SC670 | + | − | + | + | + | +++ | +++ | − | − | − | − |
| KA179 | + | − | + | + | ++ | +++ | +++ | + | − | + | + |
| ATCC15697 | + | + | + | + | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| JCM10602 | − | − | − | − | − | − | − | − | − | − | − |
Growth of B. breve on pooled and individual HMO.
The panel of B. breve strains was examined for growth in vitro on total HMO isolated from breast milk as the sole carbon source (23) or on individual milk oligosaccharides that represent the chemical diversity of HMO, such as neutral nonfucosylated HMO (LNT and LNnT), fucosylated HMO (2FL and 3FL), and sialylated HMO (3SL and 6SL). Growth behavior on HMO (shown in Fig. S3 in the supplemental material) and maximum OD values obtained were parameters to arbitrarily classify this panel in three groups (Table 2; see also Table S7).
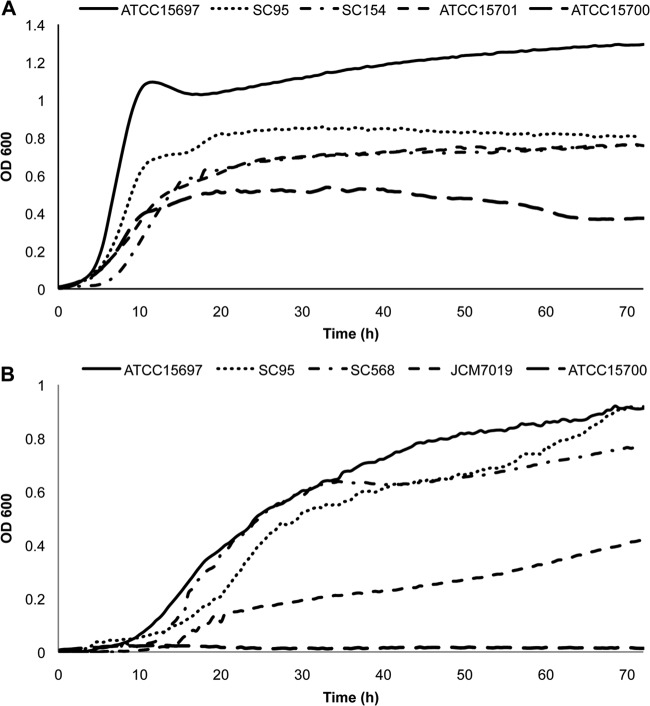
In general, moderate growth on HMO was witnessed for all the strains (Table 2; see also Fig. S3 and Table S7 in the supplemental material), with strain-level differences among the isolates to utilize HMO (Table 2). Interestingly, three strains (SC95, SC154, and ATCC 15701) exhibited remarkable growth on HMO compared to the type strain B. breve ATCC 15700 but still lower overall growth and a lower growth rate than B. longum subsp. infantis ATCC 15697 (Fig. 1A; see also Table S7).
Fig 1.
Growth of B. breve on HMO. B. breve isolates were inoculated on mMRS medium supplemented with 2% (wt/vol) HMO (A) and 2FL (B). B. longum subsp. infantis ATCC 15697 was included as a high-growth control. Fermentations were carried out in triplicate.
All B. breve strains grew on LNT and LNnT to high cell densities and at levels comparable to those of B. longum subsp. infantis ATCC 15697 (Table 2; see also Fig. S4 and S5 in the supplemental material). Interestingly, a few strains were able to grow on fucosylated HMO (Fig. 1B and Table 2). The isolates SC95 and SC568 grew well on 2FL and to a similar extent as B. longum subsp. infantis ATCC 15697. Using 3FL as the sole carbon source, only the strains SC95 and JCM 7019 grew, albeit minimally (Table 2; see also Fig. S6). Finally, growth on 3SL and 6SL was only observed for one strain, KA179 (Table 2).
Glycoprofiling of the HMO consumption by B. breve isolates.
Based on their growth phenotypes and ability to utilize certain glycans, six strains of B. breve were selected to examine the consumption of 22 different oligosaccharides during growth on total HMO. This included strains SC95, SC154, SC568, SC580, ATCC 15701, and JCM7019, as well as B. longum subsp. infantis ATCC 15697 and B. breve ATCC 15700 as positive and negative controls, respectively. The supernatant was collected at the end of the exponential phase during growth on HMO, and remaining oligosaccharides were purified and reduced and later detected and quantified by nano-HPLC-chip/TOF MS. Identification of specific oligosaccharides and isomers in this study was aided by two oligosaccharide structure libraries (2, 3). The oligosaccharides quantified include the most abundant neutral and sialylated HMO (6), and Table 3 shows their names, masses, chemical structures, and degree of polymerization (DP).
Table 3.
Names, structures, and masses of human milk oligosaccharides glycoprofiled_a_
| HMO type | Name | Abbreviation or designation | Mass (m/z) | Structure | DP_b_ |
|---|---|---|---|---|---|
| Neutral | |||||
| Nonfucosylated | Lacto-_N_-tetraose/lacto-_N_-neotetraose | LNT/LNnT | 709 | Galβ1-3GlcNacβ1-3Galβ1-4Glc/Galβ1-4GlcNacβ1-3Galβ1-4Glc | 4 |
| Lacto-_N_-hexaose | LNH | 1,074 | Galβ1-3GlcNacβ1-3[Galβ1-4GlcNacβ1-6]Galβ1-4Glc | 6 | |
| Lacto-_N_-neohexaose | LNnH | 1,074 | Galβ1-4GlcNacβ1-3[Galβ1-4GlcNacβ1-6]Galβ1-4Glc | 6 | |
| _para_-Lacto-_N_-hexaose | p-LNH | 1,074 | Galβ1-3GlcNacβ1-3Galβ1-4GlcNacβ1-3Galβ1-4Glc | 6 | |
| Fucosylated | 2′-Fucosyllactose | 2FL | 490 | Fucα1-2Galβ1-4Glc | 3 |
| Lacto-_N_-fucopentaose | LNFPI | 855 | Fucα1-2Galβ1-3GlcNacβ1-3Galβ1-4Glc | 5 | |
| LNFPII | 855 | Galβ1-3[Fucα1-4]GlcNacβ1-3Galβ1-4Glc | 5 | ||
| Monofucosyllacto-_N_-hexaose | MFLNHI | 1,220 | Fucα1-2Galβ1-3GlcNacβ1-3[Galβ1-4GlcNacβ1-6]Galβ1-4Glc | 7 | |
| Difucosyllacto-_N_-hexaose | DFLNH-A | 1,366 | Fucα1-2Galβ1-3GlcNacβ1-3[Galβ1-4[Fucα1-4]GlcNacβ1-6]Galβ1-4Glc | 8 | |
| DFLNH-B | 1,366 | Galβ1-3[Fucα1-4]GlcNacβ1-3[Galβ1-4[Fucα1-3]GlcNacβ1-6]Galβ1-4Glc | 8 | ||
| DFLNH-C | 1,366 | Fucα1-2Galβ1-3[Fucα1-4]GlcNacβ1-3[Galβ1-4GlcNacβ1-6]Galβ1-4Glc | 8 | ||
| DFpLNHII | 1,366 | Galβ1-3[Fucα1-4]GlcNacβ1-3Galβ1-4[Fucα1-3]GlcNacβ1-3Galβ1-4Glc | 8 | ||
| Acidic sialylated | 3-Sialylactose | 3SL | 635 | NeuAcα2-3Galβ1-4Glc | 3 |
| Monosialyllacto-_N_-tetraose | LSTa | 1,000 | NeuAcα2-3Galβ1-3GlcNacβ1-3Galβ1-4Glc | 5 | |
| LSTb | 1,000 | Galβ1-3[NeuAcα2-6]GlcNacβ1-3Galβ1-4Glc | 5 | ||
| Monosialyllacto-_N_-neotetraose | LSTc | 1,000 | NeuAcα2-6Galβ1-4GlcNacβ1-3Galβ1-4Glc | 5 | |
| Monofucosylmonosialyllacto-_N_-tetraose | F-LSTc | 1,146 | NeuAcα2-6Galβ1-4GlcNacβ1-3Galβ1-4[Fucα1-3]Glc | 5 | |
| Monosialyllacto-_N_-hexaose | S-LNH | 1,365 | Galβ1-3GlcNacβ1-3[[NeuAcα2-6]Galβ1-4GlcNacβ1-6]Galβ1-4Glc | 7 | |
| 4021a | 1,365 | Galβ1-3[NeuAcα2-6]GlcNacβ1-3[Galβ1-4GlcNacβ1-6]Galβ1-4Glc | 7 | ||
| Monosialyllacto-_N_-neohexaose | S-LNnHII | 1,365 | [NeuAcα2-6]Galβ1-4GlcNacβ1-3[Galβ1-4GlcNacβ1-6]Galβ1-4Glc | 7 | |
| Monofucosylmonosialyllacto-_N_-hexaose | 4121a/4121b | 1,511 | [NeuAcα2-6]Galβ1-4[Fucα1-3]GlcNacβ1-3Galβ1-4GlcNacβ1-3Galβ1-4Glc/[NeuAcα2-6]Galβ1-4GlcNacβ1-3Galβ1-4[Fucα1-3]GlcNacβ1-3Galβ1-4Glc | 8 | |
| FS-LNHIII | 1,511 | Galβ1-3[Fucα1-4]GlcNacβ1-3[[NeuAcα2-6]Galβ1-4GlcNacβ1-6]Galβ1-4Glc | 8 | ||
| 4121 unknown | 1,511 | Not defined | 8 |
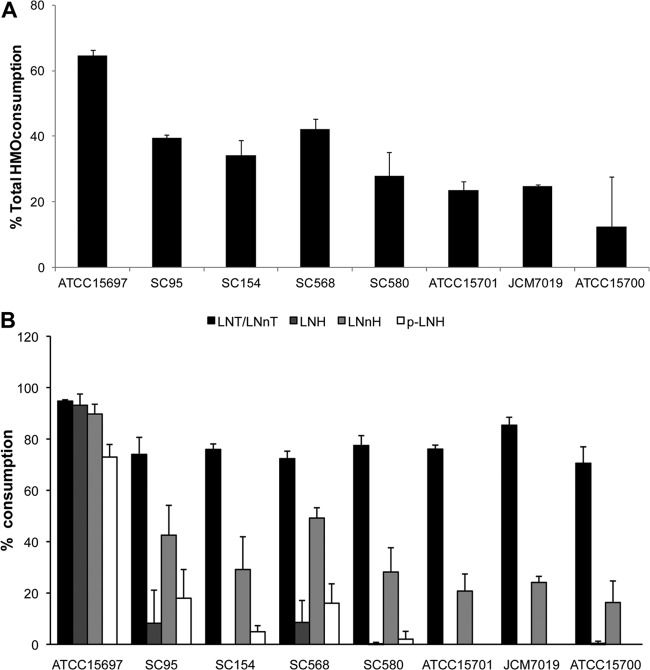
Among the six strains selected, total consumption of HMO ranged between 23 and 42%. These values are lower than for B. longum subsp. infantis ATCC 15697 (64% consumption) but clearly higher than for B. breve ATCC 15700 (Fig. 2A), the only B. breve strain glycoprofiled to date (17, 29, 30). Figure 3B presents the consumption of neutral nonfucosylated HMO in more detail, showing that consumption patterns among the B. breve strains were similar. All strains were able to deplete LNT/LNnT from the culture medium to a high extent as previously witnessed (17). Among three major hexaoses found in HMO, a preference for lacto-_N_-neohexaose (LNnH) was observed over lacto-_N_-hexaose (LNH) and _para_-lacto-_N_-hexaose (p-LNH).
Fig 2.
Glycoprofiling of HMO consumption by select B. breve strains. (A) Total utilization of HMO. Consumption is calculated as the percent difference in HMO between the start and the end of exponential phase. (B) Glycoprofiles of the consumption of neutral nonfucosylated HMO by seven B. breve strains. B. longum subsp. infantis ATCC 15697 was included as a positive control.
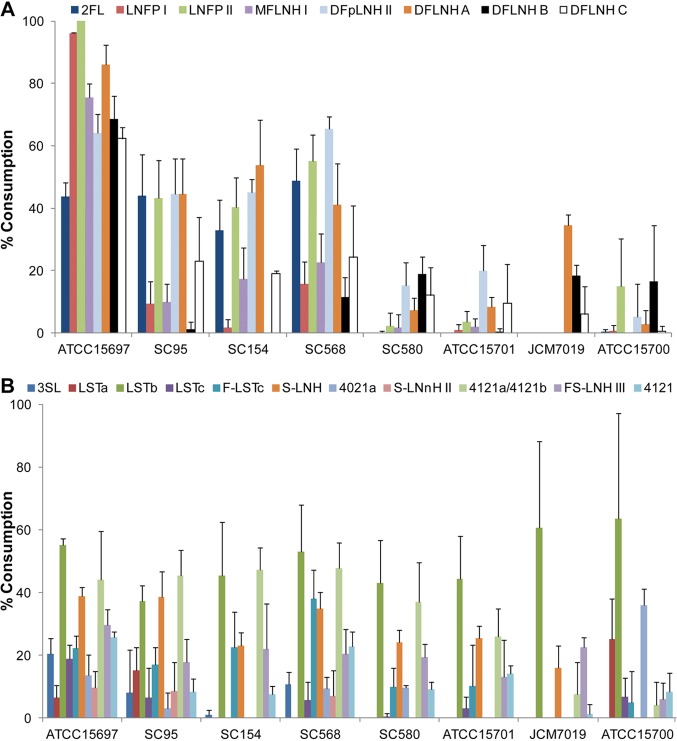
Fig 3.
Glycoprofiling of the consumption of fucosylated and acidic HMO by select B. breve strains. Consumption of eight fucosylated HMO (A) and 11 sialylated HMO (B) was calculated. B. longum subsp. infantis ATCC 15697 was included as a positive control. HMO consumption is represented as the percent difference in HMO between the start and the end of the exponential phase.
In general, the ability of B. breve to metabolize fucosylated HMO was lower than that of B. longum subsp. infantis, which showed high consumption levels for all the HMO tested (Fig. 3A). However, strains SC95, SC154, and SC568 showed a significant consumption of monofucosylated 2FL and lacto-_N_-fucopentaose (LNFPII) and difucosyllacto-_N_-hexaoses (DFpLNHII and DFLNH-A) (Fig. 3A and Table 3). Interestingly, even though 2FL cannot support the growth in vitro of strain SC154 (Table 2), this strain utilizes larger fucosylated HMO.
We observed that growth on fucosylated HMO was more prominent in strains possessing an additional GH29 α-fucosidase (Fig. 3A and Table 2). To determine if this gene is involved, we evaluated the relative gene expression of the fucosidases in strain SC95. Growth on 2FL as the sole carbon source upregulated 40-fold the expression of the GH29 fucosidase gene (Fig. 4), while expression of the GH95 fucosidase was not altered, suggesting that the presence of the GH29 fucosidase endows these strains with the ability to consume fucosylated oligosaccharides. In contrast, growth on total HMO did not affect the expression of these genes.
Fig 4.
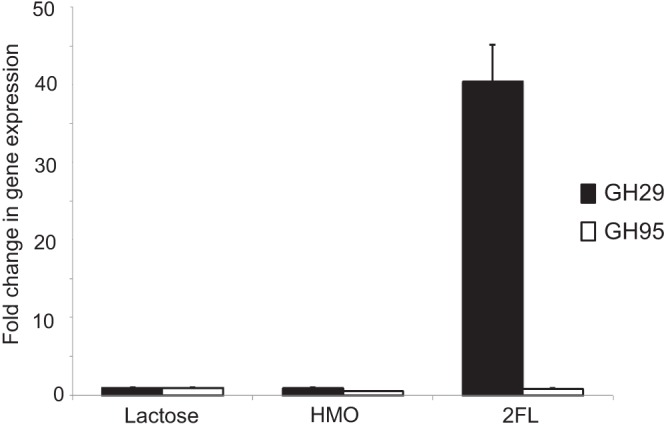
Fold in change expression for genes encoding α-fucosidases from GH families 95 and 29 in B. breve SC95 during mid-exponential-phase growth on HMO and 2FL. Growth on lactose was used as a control.
Acidic HMO represent approximately 15% of total HMO, and in this study we also screened the consumption of 11 sialylated HMO in the spent supernatants of these strains during growth on total HMO (Fig. 3B). The levels of consumption were very similar among the strains tested and comparable to those of B. longum subsp. infantis. In particular sialyl-LNT b (LSTb), sialyl-lacto-_N_-hexaose (S-LNH), and mass 4121a/4121b were preferentially utilized by B. breve (Fig. 3B).
Finally, to elucidate possible substrate preferences in a B. breve strain with high HMO consumption, we monitored the consumption of nine representative oligosaccharides at different points during the fermentation of HMO by strain SC95 (Fig. 5). These nine HMO correspond to neutral and acidic HMO that were consumed at levels higher than 40%. Remarkably, glycoprofiling of early exponential growth revealed that acidic HMO disappeared from the medium first, together with LNnH. In contrast, LNT seems to be metabolized during the mid-exponential phase and, as observed previously, is substantially depleted at the end of the growth on HMO. We also observed that while monofucosylated HMO such as 2FL and LNFPII are depleted from the medium at the mid-exponential phase and not later, difucosylated HMO appear to be steadily consumed during the three growth phases (Fig. 5).
Fig 5.
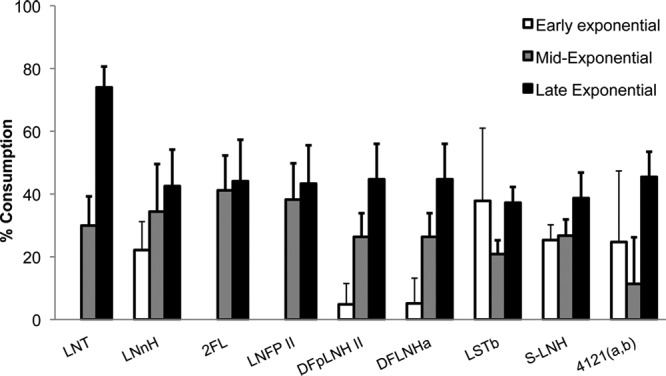
Temporal glycoprofile of the consumption of select neutral and acidic HMO by B. breve SC95 at different stages in the exponential phase.
DISCUSSION
B. breve is one of the most representative species of bifidobacteria found in the infant intestinal microbiota. Unfortunately, the factors that contribute to this persistence in infants are not well understood. We have previously determined that some B. breve strains are equipped with enzymes that release _N_-glycans from host glycoproteins, and some strains can use _N_-glycosylated proteins as the sole carbon source (31). This suggested that glycoproteins could play a prebiotic role for B. breve. In order to determine whether free HMO also contribute to the persistence of B. breve in the infant intestinal microbiota, we evaluated a significant number of B. breve strains for growth on and consumption of HMO.
The dominance of B. breve in the gut community has been supported by several studies, especially in breast-fed infants (11, 14), where this species together with B. longum subsp. longum and B. longum subsp. infantis can largely outnumber other microorganisms (32, 33). It can be considered that breast milk itself is also another habitat for this species, which, in addition to the vaginal and fecal microbiota of the mother, contributes to intestinal colonization of the infant (34). Some strains of this species are currently studied for their probiotic properties, such as production of conjugated linoleic acids or important immunomodulatory activities (26, 35).
Since the predominance of bifidobacteria in breast-fed infants can be attributed in part to milk components such as HMO, the utilization of these substrates in vitro and in vivo is an important reflection of the adaptations of intestinal microorganisms to the environmental conditions prevalent in the infant gut. HMO utilization has been shown for a few Bacteroides species and certain infant gut-associated bifidobacteria, especially strains of B. longum subsp. infantis and B. bifidum (16, 30). Previous studies have addressed this phenotype only for the type strain of B. breve, ATCC 15700 (JCM 1192), indicating that this microorganism has a limited ability to consume HMO (17, 29, 30). In this study, we have expanded these observations and concluded that several infant-associated strains of B. breve can readily utilize HMO, consuming larger amounts of total HMO than the type strain ATCC 15700. The HMO consumption in B. breve is, however, moderate compared to that of B. longum subsp. infantis ATCC 15697 (Fig. 2A).
Mass spectrometry-based analysis of the HMO remaining after growth provides a detailed representation of the preferences of these strains for different oligosaccharide subsets present in the HMO pool. For example, all strains showed vigorous growth on LNT and LNnT, and the molecular mass representing both species (709 m/z) was the most consumed in pooled HMO. The utilization of LNnT is interesting since this oligosaccharide is not readily fermented by all species of Bifidobacterium found in the infant gut (36). Moreover, growth on LNnT was shown to enable B. longum subsp. infantis to outcompete Bacteroides fragilis in a mouse model (37).
HMO with a mass of 1,074 m/z represent approximately 10% of the total (6) and includes three neutral isomers, LNH, LNnH, and p-LNH (Table 3). Interestingly, LNnH is the most abundant of the three isomers (3), and it was selectively consumed by B. breve compared to the other two isomers. This indicates structure-based preferences in HMO consumption in B. breve (Fig. 2B and Table 3).
Strain-dependent differences were more evident in the growth of B. breve on fucosylated HMO. Fucosidase activity has not been described previously in B. breve, and while all the strains studied possessed a gene encoding a GH95 α-fucosidase, we observed that the presence of a second α-fucosidase from GH29 in isolates SC95, SC568, and SC154 correlated with their consistent growth and consumption of fucosylated HMO (Fig. 3A and Table 2). Some strains with this additional GH29 α-fucosidase, however, did not grow on 2FL and 3FL. We hypothesize that these smaller HMO are imported by different transport mechanisms. For example, in B. longum subsp. infantis, 2FL and larger fucosylated HMO are likely imported by different solute binding proteins (38).
Remarkably, all B. breve strains seem to consume acidic HMO to a significant extent, and an α-sialidase was identified. All strains glycoprofiled showed a preferential consumption of select acidic HMO such as LSTb and monosialyllacto-_N_-hexaose (S-LNH), but not smaller HMO, which might additionally explain why growth on 3SL and 6SL was negligible (Table 2).
B. longum subsp. infantis ATCC 15697 is currently a model to study genetic adaptations to growth on human milk glycans (39). Several observations here indicate that the mechanisms of HMO consumption in B. breve are very similar to those of B. longum subsp. infantis, with a preference to import intact oligosaccharides followed by intracellular degradation, rather than the extracellular degradation observed in B. bifidum (13). For example, B. breve strain ATCC 15700 can quickly deplete LNT from the spent medium, and the absence of intermediate monosaccharides indicates that this strain imports this substrate (40). Moreover, the GH genes identified in this study lacked signal peptides, suggesting intracellular localization. Finally, the GH gene sequences obtained are homologous to previously characterized enzymes in B. longum subsp. infantis, including β-hexosaminidase Blon_0459, two α-fucosidases (Blon_2335 and Blon_0248), and an α-sialidase (Blon_0646), suggesting a common origin. Further studies will address in more detail the molecular mechanisms that B. breve deploys to utilize HMO.
In conclusion, these results provide a rationale for the predominance of B. breve in the infant intestinal microbiota, improving our understanding about the ecology of this unique environment. The genetic variation of these strains and the strain-dependent character of the HMO utilization are factors to consider in probiotic and prebiotic studies. Finally, we consider that better characterization of the diversity and physiology of beneficial strains of bifidobacteria and more selective substrates that allow their implantation in the intestine could be useful in the near future to design more selective synbiotic preparations.
Supplementary Material
Supplemental material
ACKNOWLEDGMENTS
This work was supported by the University of California Discovery Grant Program, the California Dairy Research Foundation, National Institutes of Health awards R01AT007079, R01HD065122, and R01HD061923, and the Bill and Melinda Gates Foundation. S.R.-M. was supported by the Ministry of Education and Science of Spain and University of Extremadura, Spain, and D.A.G. was supported in part by a scholarship from the National Milk Producers Federation.
We thank Karen Kalanetra for her support during this study.
Footnotes
Published ahead of print 26 July 2013
REFERENCES
- 1.Petherick A. 2010. Development: mother's milk: a rich opportunity. Nature 468:S5–S7 [DOI] [PubMed] [Google Scholar]
- 2.Wu S, Grimm R, German JB, Lebrilla CB. 2011. Annotation and structural analysis of sialylated human milk oligosaccharides. J. Proteome Res. 10:856–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu S, Tao N, German JB, Grimm R, Lebrilla CB. 2010. Development of an annotated library of neutral human milk oligosaccharides. J. Proteome Res. 9:4138–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bode L, Jantscher-Krenn E. 2012. Structure-function relationships of human milk oligosaccharides. Adv. Nutr. 3:383S–91S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Leoz MLA, Gaerlan SC, Strum JS, Dimapasoc LM, Mirmiran M, Tancredi DJ, Smilowitz JT, Kalanetra KM, Mills DA, German JB, Lebrilla CB, Underwood MA. 2012. Lacto-_N_-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J. Proteome Res. 11:4662–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ninonuevo MR, Park Y, Yin H, Zhang J, Ward RE, Clowers BH, German JB, Freeman SL, Killeen K, Grimm R, Lebrilla CB. 2006. A strategy for annotating the human milk glycome. J. Agric. Food Chem. 54:7471–7480 [DOI] [PubMed] [Google Scholar]
- 7.Chaturvedi P, Warren CD, Altaye M, Morrow AL, Ruiz-Palacios G, Pickering LK, Newburg DS. 2001. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology 11:365–372 [DOI] [PubMed] [Google Scholar]
- 8.Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. 2000. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am. J. Clin. Nutr. 71:1589–1596 [DOI] [PubMed] [Google Scholar]
- 9.Gnoth MJ, Kunz C, Kinne-Saffran E, Rudloff S. 2000. Human milk oligosaccharides are minimally digested in vitro. J. Nutr. 130:3014–3020 [DOI] [PubMed] [Google Scholar]
- 10.Newburg DS, Ruiz-Palacios GM, Morrow AL. 2005. Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 25:37–58 [DOI] [PubMed] [Google Scholar]
- 11.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. 2010. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156:3329–3341 [DOI] [PubMed] [Google Scholar]
- 12.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrido D, Barile D, Mills DA. 2012. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr. 3:415S–21S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avershina E, Storro O, Oien T, Johnsen R, Wilson R, Egeland T, Rudi K. 2013. Bifidobacterial succession and correlation networks in a large unselected cohort of mothers and their children. Appl. Environ. Microbiol. 79:497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. 2010. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl. Environ. Microbiol. 76:7373–7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U. S. A. 107:19514–19519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strum JS, Kim J, Wu S, De Leoz ML, Peacock K, Grimm R, German JB, Mills DA, Lebrilla CB. 2012. Identification and accurate quantitation of biological oligosaccharide mixtures. Anal. Chem. 84:7793–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deletoile A, Passet V, Aires J, Chambaud I, Butel MJ, Smokvina T, Brisse S. 2010. Species delineation and clonal diversity in four Bifidobacterium species as revealed by multilocus sequencing. Res. Microbiol. 161:82–90 [DOI] [PubMed] [Google Scholar]
- 20.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai H, Rodriguez BT, Zhang W, Broadbent JR, Steele JL. 2007. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology 153:2655–2665 [DOI] [PubMed] [Google Scholar]
- 22.Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom HJ, Joachimiak A, Lebrilla CB, Mills DA. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl. Environ. Microbiol. 78:795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. 2006. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl. Environ. Microbiol. 72:4497–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breidt F, Romick TL, Fleming HP. 1994. A rapid method for the determination of bacterial growth kinetics J. Rapid Methods Autom. Microbiol. 3:59–68 [Google Scholar]
- 25.Totten SM, Zivkovic AM, Wu S, Ngyuen U, Freeman SL, Ruhaak LR, Darboe MK, German JB, Prentice AM, Lebrilla CB. 2012. Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. J. Proteome Res. 11:6124–6133 [DOI] [PubMed] [Google Scholar]
- 26.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U. S. A. 108:11217–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrido D, Ruiz-Moyano S, Mills DA. 2012. Release and utilization of _N_-acetyl-d-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp.infantis. Anaerobe 18:430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sela DA, Li Y, Lerno L, Wu S, Marcobal AM, German JB, Chen X, Lebrilla CB, Mills DA. 2011. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J. Biol. Chem. 286:11909–11918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. 2007. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J. Agric. Food Chem. 55:8914–8919 [DOI] [PubMed] [Google Scholar]
- 30.Locascio RG, Ninonuevo MR, Kronewitter SR, Freeman SL, German JB, Lebrilla CB, Mills DA. 2009. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb. Biotechnol. 2:333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrido D, Nwosu C, Ruiz-Moyano S, Aldredge D, German JB, Lebrilla CB, Mills DA. 2012. Endo-beta-_N_-acetylglucosaminidases from infant gut-associated bifidobacteria release complex _N_-glycans from human milk glycoproteins. Mol. Cell. Proteomics 11:775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikami K, Takahashi H, Kimura M, Isozaki M, Izuchi K, Shibata R, Sudo N, Matsumoto H, Koga Y. 2009. Influence of maternal bifidobacteria on the establishment of bifidobacteria colonizing the gut in infants. Pediatr. Res. 65:669–674 [DOI] [PubMed] [Google Scholar]
- 33.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. 10.1371/journal.pone.0036957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin R, Jimenez E, Heilig H, Fernandez L, Marin ML, Zoetendal EG, Rodriguez JM. 2009. Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR. Appl. Environ. Microbiol. 75:965–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeon SG, Kayama H, Ueda Y, Takahashi T, Asahara T, Tsuji H, Tsuji NM, Kiyono H, Ma JS, Kusu T, Okumura R, Hara H, Yoshida H, Yamamoto M, Nomoto K, Takeda K. 2012. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 8:e1002714. 10.1371/journal.ppat.1002714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miwa M, Horimoto T, Kiyohara M, Katayama T, Kitaoka M, Ashida H, Yamamoto K. 2010. Cooperation of beta-galactosidase and beta-_N_-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology 20:1402–1409 [DOI] [PubMed] [Google Scholar]
- 37.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL. 2011. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10:507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrido D, Kim JH, German JB, Raybould HE, Mills DA. 2011. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One 6:e17315. 10.1371/journal.pone.0017315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrido D, Dallas DC, Mills DA. 2013. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology 159:649–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. 2011. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 286:34583–34592 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material