RNF168 ubiquitylates 53BP1 and controls its response to DNA double-strand breaks (original) (raw)
Significance
DNA damage-repair mechanisms are highly regulated, and posttranslational modifications are one means of such regulation. p53-binding protein 1 (53BP1) is important for the response to DNA double-strand breaks (DSBs). Here we report that its functions are regulated by RING finger 168 (RNF168), a DNA DSB-signaling protein that is mutated in the human radiosensitivity, immunodeficiency, dysmorphic features, and learning difficulties syndrome. We show that before their localization to DNA DSBs, RNF168 interacts with 53BP1 and modifies it through the addition of a chain of ubiquitin-polypeptides. We also show that Lysine 1268 of 53BP1 is important for its ubiquitin modification by RNF168 and that loss of this modification impairs 53BP1 recruitment to sites of DNA damage and restrains its functions in the repair of DNA damage and maintenance of genomic stability.
Keywords: ubiquitin, G2/M checkpoint, HR repair pathways, NHEJ repair pathway
Abstract
Defective signaling or repair of DNA double-strand breaks has been associated with developmental defects and human diseases. The E3 ligase RING finger 168 (RNF168), mutated in the human radiosensitivity, immunodeficiency, dysmorphic features, and learning difficulties syndrome, was shown to ubiquitylate H2A-type histones, and this ubiquitylation was proposed to facilitate the recruitment of p53-binding protein 1 (53BP1) to the sites of DNA double-strand breaks. In contrast to more upstream proteins signaling DNA double-strand breaks (e.g., RNF8), deficiency of RNF168 fully prevents both the initial recruitment to and retention of 53BP1 at sites of DNA damage; however, the mechanism for this difference has remained unclear. Here, we identify mechanisms that regulate 53BP1 recruitment to the sites of DNA double-strand breaks and provide evidence that RNF168 plays a central role in the regulation of 53BP1 functions. RNF168 mediates K63-linked ubiquitylation of 53BP1 which is required for the initial recruitment of 53BP1 to sites of DNA double-strand breaks and for its function in DNA damage repair, checkpoint activation, and genomic integrity. Our findings highlight the multistep roles of RNF168 in signaling DNA damage.
The DNA-damage response (DDR) is critical for genomic integrity (1) and is regulated by posttranslational modifications (PTMs) such as ubiquitylation of histones by the E3 ligases RING finger 8 (RNF8) and RING finger 168 (RNF168). Other PTMs important for DDR include dimethylation of histone H4 (H4K20me2), which allows p53-binding protein 1 (53BP1), a key mediator of DDR, to interact with chromatin.
Mutations of RNF168 have been associated with the human radiosensitivity, immunodeficiency, dysmorphic features, and learning difficulties (RIDDLE) syndrome, (2–4). RNF168 has an N-terminal RING finger domain, three ubiquitin (Ub)-binding domains (UBDs); two motif interacting with Ub (MIU) domains; and one Ub interacting motif (UIM)- and MIU-related (UMI) UBD (3, 5, 6). Current data support RNF168 function in DNA double-strand break (DSB) signaling downstream of H2A.X, mediator of DNA damage checkpoint 1 (MDC1), and RNF8 and indicate its requirement for 53BP1 recruitment to DSB sites (3, 5). Through its UBDs, RNF168 recognizes RNF8 ubiquitylated non-nucleosomal protein(s) at DSB-flanking sites, leading to its recruitment at these sites of DNA damage (7). With the Ub-conjugating enzyme UBC13, RNF168 initiates ubiquitylation of lysine (K) 13 or 15 of histones H2A and H2A.X, leading to the recruitment of DDR proteins, including 53BP1, to DSBs. Although H2A.X, MDC1, and RNF8 are important for the retention of 53BP1 at these DSBs, its initial and transient recruitment to DNA breaks still occurs in their absence (8–11). In contrast, deficiency of Rnf168 in mouse embryonic fibroblasts (MEFs) completely abolishes 53bp1 recruitment to DSB sites (12). Similar to _53bp1_−/− mice, but in contrast to _H2a.x_−/− mice (13), young Rnf168 −/− males are fertile (12). These data suggest that, in addition to its role in DSB signaling downstream of the H2A.X–MDC1–RNF8 axis, RNF168 also functions in DSB signaling independently of this pathway. Therefore, we postulated that RNF168 also might regulate DSB signaling through direct modulation of 53BP1 functions.
In the present study, we demonstrate that RNF168 associates with 53BP1 independently of the γ-H2A.X–MDC1–RNF8 signaling axis. RNF168 ubiquitylates 53BP1 before its localization to DSB sites, and this ubiquitylation is important for the initial recruitment of 53BP1 to DSB sites and its function in nonhomologous end joining (NHEJ) and activation of checkpoints. Collectively our data support a requirement for RNF168 at different stages of the DSB-signaling cascade and highlight the central role of RNF168 ubiquitylation of 53BP1 in maintaining genomic integrity.
Results
Rnf168 Interacts with 53bp1.
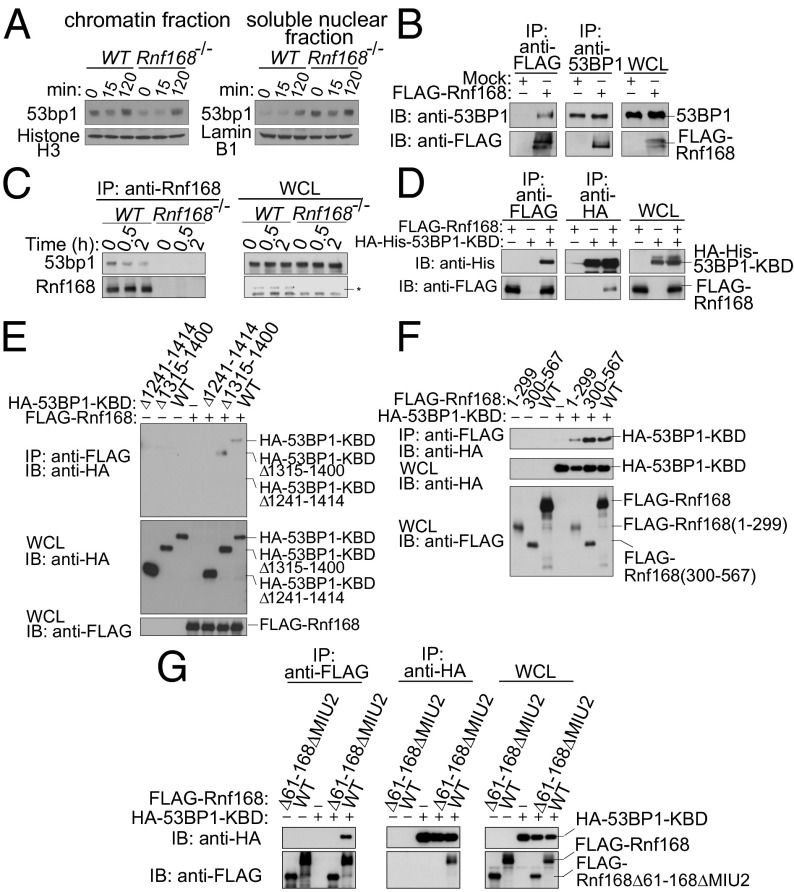
Cells deficient for Rnf168 fail to recruit or retain 53bp1 at sites of DNA damage (2, 3, 12). To examine the mechanisms that mediate Rnf168 regulation of 53bp1 recruitment to DSB sites, we tested whether Rnf168 contributes to 53bp1’s association with chromatin. We observed that 53bp1 was bound to chromatin in untreated (UT) WT thymocytes and was enriched in chromatin-rich insoluble fractions after irradiation (IR) (Fig. 1_A_). Although deficiency of H2a.x or Rnf8 does not affect 53bp1’s affinity for chromatin (14, 15), the levels of 53bp1 in chromatin-enriched insoluble fractions were reduced in Rnf168 −/− thymocytes compared with controls, both under UT conditions and 15 min post-IR (Fig. 1_A_). Because 53BP1 associates with chromatin through its binding to H4K20me2 (16, 17), we examined the level of this histone mark in UT or irradiated WT and Rnf168 −/− primary MEFs. Our data indicated no defects in H4K20me2 levels in the absence of Rnf168 (Fig. S1_A_). These data suggested that Rnf168, unlike H2a.x and Rnf8, is important for 53bp1’s association with chromatin both under UT conditions and in response to DNA damage and that H4K20me2 is not a limiting factor for 53bp1 recruitment to DSBs in Rnf168-deficient cells. Next we examined whether Rnf168 interacts physically with 53bp1. Immunoprecipitation (IP) and immunoblot (IB) analysis using HEK293T cells expressing exogenous Rnf168 revealed its interaction with 53BP1 (Fig. 1_B_). Endogenous Rnf168 and 53bp1 also were found to interact in WT thymocytes (Fig. 1 and Fig. S1_B_). 53BP1 contains a kinetochore-binding domain (KBD; amino acids 1235–1616) that includes a Tudor domain (Fig. S1_C_) essential for 53BP1 binding to chromatin (14, 18). Thus, we assessed whether 53BP1-KBD may interact with Rnf168. The amino acid sequence of the KBD domain of 53BP1 is highly conserved between humans and mice (92.9% identity) (Fig. S1_D_). Therefore, we used human 53BP1-KBD for these analyses and found that this domain was sufficient to mediate 53BP1 interaction with Rnf168 (Fig. 1_D_). By coexpressing a series of deletion mutants of HA-tagged 53BP1-KBD (Fig. S1_C_) with FLAG-Rnf168 in HEK293T cells and using IP and IB analysis, we identified the minimal amino acid residues (1241–1315) required for 53BP1’s binding to Rnf168 (Fig. 1_E_). These 53BP1 amino acid residues are 100% conserved between humans and mice (Fig. S1_D_). To determine Rnf168 region(s) that bind 53BP1, we performed an in vivo binding assay using FLAG-tagged truncated forms of Rnf168 (Fig. S2_A_). We transiently expressed each FLAG-tagged Rnf168 with HA-tagged 53BP1-KBD in HEK293T cells. 53BP1-KBD interacted with both the truncated and full-length forms of Rnf168 (Fig. 1_F_). We next performed in vivo binding assay using HA-tagged 53BP1-KBD and FLAG-tagged Rnf168ΔRING, Rnf168ΔMIU1, Rnf168ΔMIU2, Rnf168ΔMIU1/2, Rnf168(61–168), Rnf168Δ61–168Δ194–299, and full-length Rnf168. All Rnf168 mutants interacted with 53BP1-KBD (Fig. S2 A and B). These results suggest that multiple regions of Rnf168 and 53BP1 might be involved in their interaction. To examine further the region(s) of Rnf168 that mediate its binding to 53BP1, we performed an in vivo binding assay using HA-tagged 53BP1-KBD and FLAG-tagged Rnf168 truncated mutants [Rnf168(1–299), Rnf168(1–299)Δ61–168], and full-length Rnf168. Interestingly, additional deletion of residues 61–168 in Rnf168(1–299) prevented its interaction with 53BP1-KBD (Fig. S2 A_–_C). Moreover, we performed a similar in vivo binding assay using HA-tagged 53BP1-KBD, FLAG-tagged full-length Rnf168, and FLAG-tagged Rnf168 deletion mutants including Rnf168(300–567), Rnf168(300–567)ΔMIU2, and Rnf168ΔMIU2 (Fig. S2_A_). Our data indicated that, in contrast to Rnf168(300–567) (Fig. 1_F_), Rnf168(300–567)ΔMIU2 failed to bind 53BP1-KBD (Fig. S2_D_). An in vivo binding assay using HA-tagged 53BP1-KBD, FLAG-tagged Rnf168Δ61–168ΔMIU2, and full-length Rnf168 also indicated the inability of Rnf168Δ61–168ΔMIU2 to interact with 53BP1-KBD (Fig. 1_G_ and Fig. S2_A_). These results suggest that the KBD of 53BP1 binds at multiple sites of Rnf168 including its C-terminal MIU2 domain and its N-terminal amino acid residues 61–168 that overlap with its coiled-coil domain.
Fig. 1.
53bp1 interacts with Rnf168. (A) WT and Rnf168 −/− thymocytes were either UT or irradiated (5 Gy), and chromatin and nuclear fractions were isolated post-IR. (B) HEK293T cells were transfected with FLAG-Rnf168 or empty vector, and IP and IB were performed. Three per cent of the whole-cell lysate (WCL) used for IP was subjected to IB analysis. (C) Thymocytes were either UT or harvested at different time points post-IR (2 Gy). IP was performed with anti-Rnf168 antibody, and IB was carried out as indicated. The asterisk indicates nonspecific bands. (D) HEK293T cells were transfected with FLAG-Rnf168 and HA-His–tagged truncated 53BP1 (53BP1-KBD) expression vectors as indicated. Anti-FLAG IP and anti-HA IB are shown. (E) FLAG-Rnf168, HA-tagged 53BP1-KBD, or its truncated forms were transfected into HEK293T cells as indicated, and anti-FLAG IP were subjected to IB analysis. (F and G) HA-53BP1-KBD, FLAG-Rnf168 (WT), and its truncated forms were transfected into HEK293T cells, and IP and IB were performed. Representative Immunoblots of three independent experiments are shown.
Rnf168 Forms a Protein Complex with 53bp1 and Mdc1.
Previous studies suggested interaction of 53BP1 and MDC1 in undamaged cells (19–21). The 53BP1 region that mediates its binding to tandem breast cancer 1 (BRCA1) C-terminal domains of MDC1 is located at residues 1288–1409 and thus overlaps with the minimal 53BP1 binding region (residues 1241–1315) to Rnf168 (Fig. S1_C_). We therefore examined whether Rnf168, 53bp1, and Mdc1 interact. We first determined whether Rnf168 binds MDC1 using HEK293T cells expressing FLAG-tagged Rnf168, HA-MDC1, or both. IP and IB experiments demonstrated interaction of Rnf168 with MDC1 (Fig. S3_A_). Further analyses demonstrated that interaction of endogenous 53bp1 and Mdc1 was enhanced in the presence of Rnf168 (Fig. S3_B_). Similarly, interaction of exogenous 53BP1 and MDC1 was enhanced in the presence of exogenous Rnf168 protein (Fig. S3_C_). These data suggest that RNF168 interacts with 53BP1 and MDC1 and that its presence facilitates the formation of a protein complex that includes these three proteins.
Rnf168 Mediates 53bp1 Ubiquitylation.
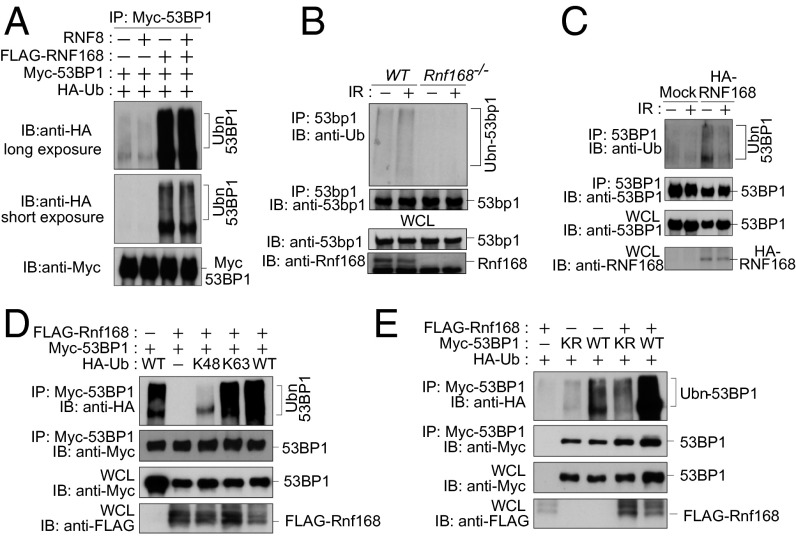
Based on the interaction of Rnf168 and 53bp1 and the Ub ligase function of Rnf168 (3, 5), we hypothesized that 53bp1 may represent a substrate for Rnf168. Indeed intracellular ubiquitylation assays indicated that Rnf168 mediates polyubiquitylation of 53BP1 (Fig. 2_A_ and Fig. S4_A_). The smear of ubiquitylated 53BP1 became stronger when both exogenous Rnf168 and Ub were coexpressed with full-length 53BP1 (Fig. 2_A_). Next we examined the importance of the RING finger of Rnf168 in mediating 53BP1 ubiquitylation. We observed reduced 53BP1 ubiquitylation in the presence of the Rnf168C21S (CS) RING finger mutant compared with WT Rnf168 (Fig. S4_B_), suggesting the importance of the RNF168 RING finger domain for 53BP1 ubiquitylation. We also examined whether RNF8 interacts and ubiquitylates 53BP1. Unlike RNF168, RNF8 failed to interact with 53BP1 (Fig. S4_C_), and overexpression of RNF8 was unable to mediate 53BP1 ubiquitylation or to affect its ubiquitylation by RNF168 (Fig. 2_A_). We next examined ubiquitylation of endogenous 53bp1 using WT and Rnf168 −/− splenocytes. When 53bp1 was immunoprecipitated from WT splenocytes, we observed smears with slower mobility, which also could be detected by antibodies specific for Ub (Fig. 2_B_). In addition examination of WT splenocytes indicated that the level 53bp1 ubiquitylation was increased 15 min post- IR (Fig. 2_B_), a time point where 53bp1 irradiation-induced foci (IRIF) already are formed, but it was reduced 2 h post-IR (Fig. S4_D_). In contrast to WT cells, Rnf168 −/− splenocytes displayed no significant 53bp1 ubiquitylation under UT or IR conditions (Fig. 2_B_), suggesting that ubiquitylation of endogenous 53bp1 is Rnf168 dependent. Ubiquitylation of 53BP1 under UT or IR conditions also was defective in human fibroblasts from the RNF168-deficient RIDDLE patient 15–9BI (Fig. 2_C_) (2). Complementation of these RIDDLE cells with RNF168 restored 53BP1 ubiquitylation (Fig. 2_C_). In contrast to Rnf168, loss of endogenous Rnf8 had no effect on 53bp1 ubiquitylation (Fig. S4_E_). Taken together, these results indicate that Rnf168, but not Rnf8, mediates 53bp1 ubiquitylation.
Fig. 2.
RNF168 mediates 53BP1 ubiquitylation. (A) HEK293T cells were transfected with the indicated expression vectors, and IP and IB were performed as indicated. (B) Splenocytes either UT or 15 min post-IR were examined by IP/IB as indicated. (C) RNF168-deficient fibroblasts from a RIDDLE patient complemented with empty vector (Mock) or HA-tagged RNF168 were either UT or were collected 2 h post-IR (5 Gy). IP and IB analyses were performed as in B. (D and E) HEK293T cells were transfected with the indicated vectors, and IP/IB analyses were performed. KR, Myc-53BP1-K1268R; Ubn, ubiquitylated proteins.
K1268 of 53BP1 Is Important for Its Ubiquitylation by Rnf168.
Although our data indicate that Rnf168 interacts with 53bp1 and mediates its polyubiquitylation, previous studies reported that, in response to DNA damage, 53BP1 is monoubiquitylated on K1268 by RAD18 and that this ubiquitylation is important for 53BP1 retention at DSBs (22). When examined under the same conditions, Rad18 −/− MEFs, in contrast to Rnf168 −/− MEFs (12), showed no significant defects in the initial recruitment or retention of 53bp1 to DSB sites (Fig. S5 A_–_C). We also examined tightly associated 53bp1 IRIF, which are resistant to salt extraction, because previously they were reported to be defective in Rad18 −/− MEFs (22). These 53bp1 IRIFs were significantly reduced in Rad18 −/− MEFs and were completely lacking in Rnf168 −/− MEFs (Fig. S5_D_). We next examined the effect of Rad18 loss on 53bp1 polyubiquitylation and IRIF of Ub. Our data indicated similar levels of 53bp1 polyubiquitylation in WT and _Rad18_−/− MEFs under unstressed or irradiated conditions (Fig. S5_E_). In response to IR, WT and _Rad18_−/− cells showed similar levels of Ub foci formation (Fig. S5_F_). These results suggest that, in contrast to Rnf168, loss of Rad18 had no effect on 53bp1 polyubiquitylation or Ub IRIF at DSB sites.
Polyubiquitin linkages via K48 and K63 are used frequently for polyubiquitylation (23); therefore we used Ub in which all lysines except K48 or K63 are mutated to arginine and examined the Ub-linkage type used for RNF168 ubiquitylation of 53BP1. We observed that Rnf168 mediates K63-linked but not K48-linked ubiquitylation of 53BP1 (Fig. 2_D_). We next examined 53BP1 sites ubiquitylated by Rnf168 and assessed whether K1268 of 53BP1, known to be ubiquitylated by RAD18 (22), also is targeted for ubiquitylation by Rnf168. Ubiquitylation of full-length 53BP1 by Rnf168 was reduced significantly by the substitution of its K1268 residue to arginine (R) (53BP1-K1268R) (Fig. 2_E_). These data indicate that RNF168 mediates K63- but not K48-linked ubiquitylation of 53BP1 and that K1268 of 53BP1 is important for this ubiquitylation.
Interaction of Rnf168 with 53bp1 and Its E3 Ligase Activity Are Required for the Initial Recruitment and Retention of 53bp1 to DNA Damage Sites.
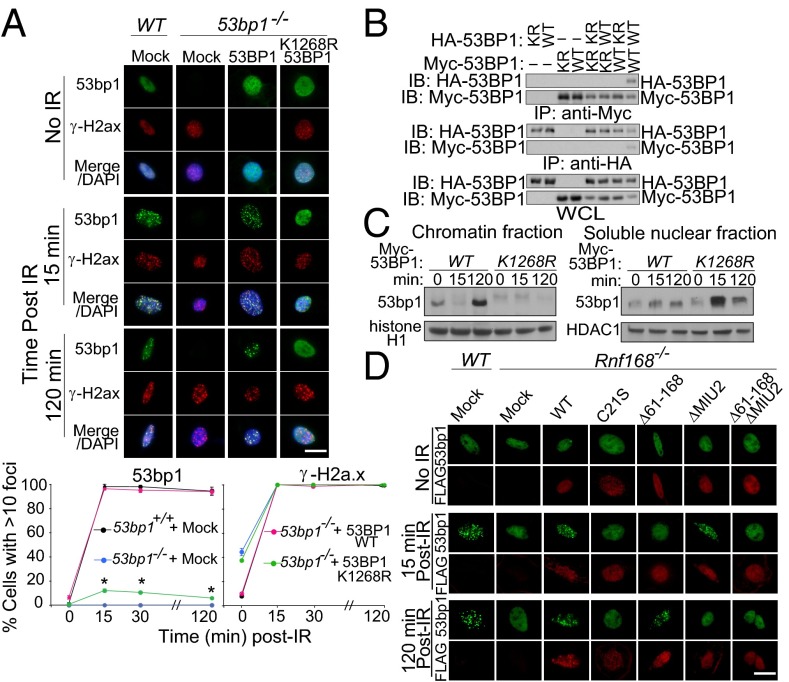
To address the functional significance of 53BP1 ubiquitylation by RNF168, we first examined whether loss of ubiquitylation of 53BP1 on K1268 affects its recruitment to DSB sites. Although complementation of 53bp1 −/− MEFs with WT 53BP1 fully restored IR-induced 53BP1 foci, the 53BP1 K1268R mutant expressed in 53bp1 −/− MEFs failed to form transient or stable foci in response to IR (Fig. 3_A_ and Fig. S6_A_). Because 53BP1 oligomerization is important for its function (24), and K1268 lies within its oligomerization domain, we examined whether the K1268R mutation affects 53BP1 oligomerization (Fig. 3_B_). Full-length 53BP1-WT and 53BP1-K1268R proteins bearing either Myc or HA tags were ectopically expressed in HEK293T cells and were subjected to IP and IB analysis. IP of HA-53BP1-WT and of Myc-53BP1-WT brought down Myc-53BP1-WT and HA-53BP1-WT proteins, respectively. However, these IP of HA-53BP1-WT and Myc-53BP1-WT failed to bring down tagged 53BP1-K1268R protein. In addition, IP of Myc-53BP1-K1268R with anti-Myc antibody failed to bring down HA-tagged 53BP1-K1268R or HA-tagged 53BP1-WT proteins. These results also were confirmed by the failure of HA-53BP1-K1268R IP to bring down Myc-tagged 53BP1-K1268R or Myc-tagged 53BP1-WT proteins. The observed oligomerization of 53BP1-WT but not 53BP1-K1268R suggests that ubiquitylation of 53BP1 on K1268 may play an important role in its oligomerization. We next addressed whether 53BP1 ubiquitylation on K1268 alters its affinity for chromatin. The effect of K1268 mutation on the subcellular distribution of 53BP1 was examined using 53bp1 −/− MEFs reconstituted with either WT or mutated (K1268R) Myc-tagged 53BP1 (Fig. 3_C_). As expected, irradiation of 53bp1 −/− cells reconstituted with WT 53BP1 resulted in enrichment of 53BP1 in chromatin-rich insoluble fractions. In contrast, irradiated 53bp1 −/− cells reconstituted with 53BP1-K1268R displayed reduced levels of 53BP1 mutant proteins in their chromatin-enriched insoluble fractions. These results suggest that ubiquitylation of 53BP1 on K1268 alters its affinity for chromatin. RNF8 and RNF168 ubiquitylation of the histone demethylase jumonji domain-containing (JMJD) protein-2A (JMJD2A) was reported to be necessary for 53BP1 IRIF, and combined knockdown of JMJD2A and JMJD2B was shown to restore 53BP1 IRIF in the absence of either RNF8 or RNF168 (25). Therefore, we used 53bp1 −/− MEFs complemented with Myc-tagged 53BP1-K1268R and examined whether knockdown of JMJD2A and JMJD2B could restore the recruitment of 53BP1-K1268R to DSB-flanking sites. Our data indicate that combined knockdown of JMJD2A and JMJD2B cannot restore IRIF for 53BP1-K1268R (Fig. S6 B and C).
Fig. 3.
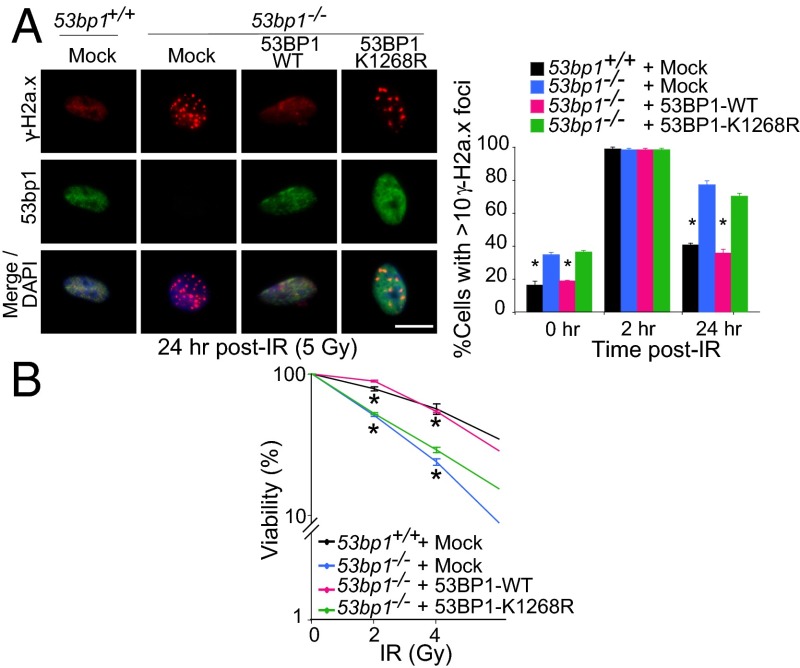
RNF168 is required for the initial recruitment and retention of 53BP1 to DNA damage sites. (A) (Upper) WT and 53bp1 −/− MEFs stably infected with the indicated retroviruses were stained with anti-53bp1, anti–γ-H2a.x, and DAPI. (Lower) The number of cells with >10 53bp1 foci. Data are presented as the mean ± SEM. n = 3. (Scale bar: 20 μm.) *P < 0.05. (B) WT or K1268R (KR) HEK293T cells were transfected with Myc- or HA-tagged 53BP1, and IP and IB were performed. (C) 53bp1 −/− MEFs reconstituted with Myc-tagged 53BP1-WT or 53BP1-K1268R were either UT or were irradiated (5 Gy), and chromatin and nuclear fractions were examined by IB. (D) WT and Rnf168 −/− MEFs reconstituted with Mock or Rnf168 (WT or mutants) were fixed post-IR (5 Gy) and processed for immunofluorescence. Representative images of three experiments are shown. Scale bar: 20 μM.
Rnf168 amino acid residues 61–168 and its MIU2 domain were responsible for its interaction with 53BP1 (Fig. 1_F_ and Fig. S2 C and D). Therefore we examined the effect of their loss on 53bp1 IRIF (Fig. 3_D_ and Fig. S6 D and E). Unlike exogenous WT Rnf168, which fully rescued 53bp1 IRIF in Rnf168 −/− MEFs, complementation of these MEFs with either Rnf168C21S or Rnf168Δ61–168ΔMIU2, which lacks domains important for Rnf168-53bp1 interaction, failed to restore the initial recruitment or retention of 53bp1 at DSB sites. Expression of Rnf168Δ61–168 in Rnf168 −/− MEFs delayed the initial formation of 53bp1 IRIF, whereas expression of Rnf168ΔMIU2 in these cells failed to rescue the retention of 53bp1 at DSB sites. The ability of 53bp1 to form transient foci in Rnf168 −/− MEFs expressing Rnf168ΔMIU2, a mutant that cannot localize to sites of DNA damage, indicates that in the absence of Rnf168 recruitment to DSB sites, 53BP1 can bind chromatin and form transient foci; however, these IRIF cannot be retained. We next investigated whether ubiquitylation of 53BP1 occurs before or after its localization to sites of DNA damage. Using an intracellular ubiquitylation assay, we examined 53BP1 ubiquitylation in the presence of WT Rnf168 or its mutant Rnf168ΔMIU2. We found that, despite the lack of its recruitment to DSB-flanking sites, the Rnf168ΔMIU2 mutant retained its ability to ubiquitylate 53BP1 to a level similar to that of WT Rnf168 (Fig. S6_F_). Therefore, Rnf168 ubiquitylation of 53BP1 is important for the initial recruitment of 53bp1 to DSB sites, and Rnf168 mediates this ubiquitylation independently of its recruitment to DSBs. These results suggest that 53bp1 interaction with Rnf168 and its ubiquitylation by this E3 ligase are important for its recruitment to DSB sites. Furthermore, our data also indicate that Rnf168 ubiquitylates 53bp1 before its localization to sites of DNA damage.
Important Role for 53bp1 Ubiquitylation for G2/M Checkpoint, NHEJ Repair, and Genomic Integrity.
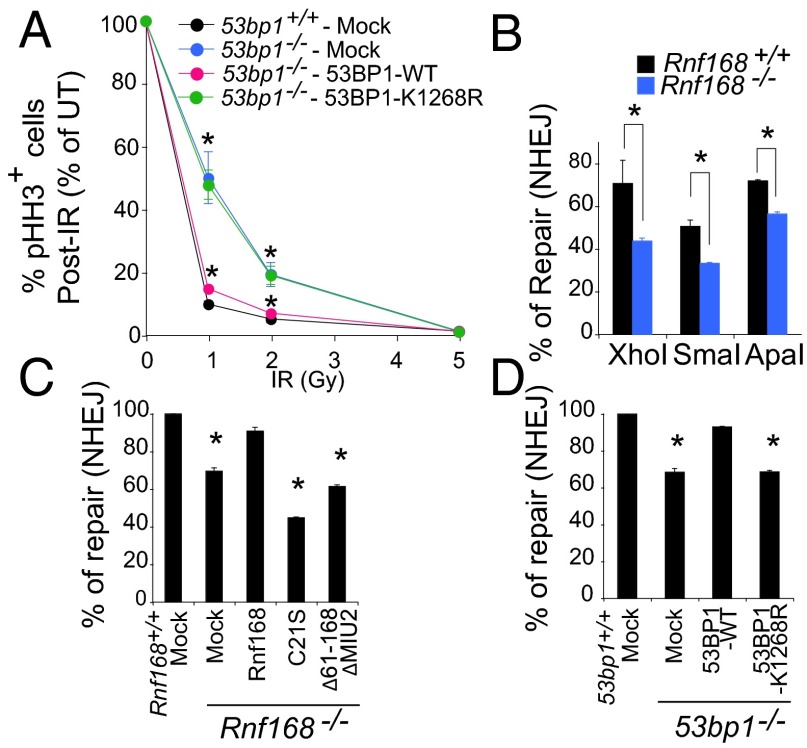
Cell-cycle checkpoints and DDR pathways are critical for the response to damaged DNA (26). Because Rnf168 −/− and 53bp1 −/− cells display a defective G2/M checkpoint (12, 27), we investigated the effect of 53BP1 ubiquitylation on the activation of this checkpoint. Although 53bp1 −/− MEFs displayed a pronounced defect at the G2/M checkpoint, their complementation with 53BP1-WT fully rescued this defect (Fig. 4_A_ and Fig. S7_A_). In contrast, 53BP1-K1268R failed to rescue the defective G2/M checkpoint of 53bp1 −/− MEFs (Fig. 4_A_), suggesting the importance of 53BP1 ubiquitylation for this checkpoint. 53BP1 also plays important roles in NHEJ (13, 28, 29). Although recently 53BP1 has been reported to be important for the choice between the NHEJ and homologous recombination (HR) repair pathways (30–32), previous studies demonstrated that 53BP1 is not required for HR (33) and IRIF for Rad51, a recombinase integral for HR (34, 35). We therefore examined the effect of Rnf168 deficiency on DSB repair using reporter plasmids for NHEJ and HR (36–38). Although the loss of Rnf168 in MEFs impaired NHEJ; it did not significantly affect HR (Fig. 4_B_ and Fig. S7_B_). Moreover, the number of Rad51 IRIF did not differ in WT and Rnf168 −/− MEFs (Fig. S7_C_). Consistent with their defective retention of 53bp1 at DSB sites, Rnf8 −/− MEFs displayed reduced NHEJ efficiency (Fig. S7_B_). We next examined NHEJ in WT and in Rnf168 −/− MEFs complemented with mutated Rnf168 proteins (Rnf168C21S and Rnf168Δ61–168ΔMIU2) that are deficient in the E3 ligase activity or lack regions critical for the Rnf168 interaction with 53BP1. Unlike WT Rnf168, Rnf168 mutants failed to rescue impaired NHEJ of Rnf168 −/− MEFs (Fig. 4_C_ and Fig. S7). Similarly, we observed that, in contrast to WT 53BP1, 53BP1-K1268R protein failed to rescue NHEJ in 53bp1 −/− MEFs (Fig. 4_D_ and Fig. S7_E_). These data support the importance of Rnf168 in NHEJ and indicate that K1268 of 53BP1, a target for Rnf168 ubiquitylation, is critical for 53BP1 function in NHEJ.
Fig. 4.
53BP1 ubiquitylation is important for G2/M checkpoint activation and NHEJ. (A) G2/M checkpoint analysis. MEFs either UT or irradiated (2 Gy) were stained with anti-pHH3 antibody and propidium iodide, and the percentage of mitotic cells was determined by FACS. (B_–_D) Efficiency of NHEJ in WT and Rnf168 −/− MEFs using the pCMS end reporter. (C and D) NHEJ was examined as in B. Each panel shows the results of three experiments. Data are presented as the mean ± SEM. *P < 0.05.
Delayed rejoining of DSBs and sustained DDR can correlate with the persistence of γ-H2a.x foci (39). Therefore we examined spontaneous γ-H2a.x foci in WT, 53bp1 −/−, and reconstituted 53bp1 −/− MEFs. Consistent with their defective DSB repair, 53bp1 −/− MEFs displayed elevated levels of γ-H2a.x foci under UT conditions and 24 h post-IR (Fig. 5_A_ and Fig. S8). In contrast to 53bp1 −/− MEFs complemented with 53BP1-WT, 53bp1 −/− MEFs complemented with 53BP1-K1268R retained their elevated number of spontaneous γ-H2a.x foci. 53bp1 −/− MEFs expressing exogenous 53BP1-WT or 53BP1-K1268R displayed similar numbers of γ-H2a.x foci 2 h post-IR. However, at 24 h post-IR, 53bp1 −/− MEFs complemented with 53BP1-WT displayed a lower number of γ-H2a.x foci, similar to WT MEFs, whereas complementation of these cells with 53BP1-K1268R had no effect on their elevated level of γ-H2a.x foci (Fig. 5_A_ and Fig. S8). To examine further the physiological relevance of 53BP1 ubiquitylation, clonogenic assays were performed using UT or IR-treated WT and 53bp1 −/− MEFs reconstituted with empty vector, 53BP1-WT, or 53BP1-K1268R. In contrast to 53BP1-WT, which restored cell survival of irradiated 53bp1 −/− MEFs to a level similar to WT MEFs, expression of 53BP1-K1268R in 53bp1 −/− MEFs failed to rescue their increased radiosensitivity (Fig. 5_B_). Collectively, these results suggest that ubiquitylation of 53BP1 on K1268 is critical for its functions in DDR and genomic integrity.
Fig. 5.
53BP1 ubiquitylation is Important for genomic integrity. (A) Mock-infected WT and 53bp1 −/− MEFs and stably reconstituted 53bp1 −/− MEFs with mock or Myc-tagged 53BP1 (WT or 53BP1-K1268R) were fixed 24 h post-IR (5 Gy) and processed for immunofluorescence. The histogram data are presented as mean ± SEM. *P < 0.05. (B) A clonogenic assay was performed to examine the radiosensitivity of 53bp1 +/+ and 53bp1 −/− MEFs complemented with WT or mutant (K1268R) 53BP1. Data shown are representative of three independent experiments. Graphs show mean ± SEM. *P < 0.05.
Discussion
In this work we have identified a mechanism for RNF168 regulation of DSB signaling and repair. We show that RNF168 interacts with 53BP1 and facilitates the formation of a protein complex containing RNF168, 53BP1, and MDC1. Moreover, we show that RNF168 mediates K63-linked polyubiquitylation of 53BP1 and that Rnf168 ubiquitylation of 53bp1 takes place before their localization to sites of DNA damage. Our data also indicate that the K1268 residue of 53BP1 is important for its ubiquitylation by RNF168 and indicate that this ubiquitylation has a critical role in the initial recruitment of 53BP1 to DSB sites and in its function in DDR and genomic stability.
53BP1 is important for class switch recombination (CSR), V(D)J recombination, NHEJ of dysfunctional telomeres, and genomic stability (13, 14, 29, 40). Through its Tudor domain, 53BP1 associates with H4K20me2, even in the absence of DNA damage, and both its Tudor and oligomerization domains are required for its efficient focus formation and the protection from DNA end resection in response to genotoxic stress (14, 16, 17). Initial recruitment and subsequent retention of 53BP1 to DSB sites are mechanistically distinct processes that are not fully understood. The initial and transient recruitment to DSB sites of DDR proteins, including 53BP1 and BRCA1, depends on the MRE11–RAD50–NBS1 complex and was proposed to set up for DNA repair (8). Although the loss of the γ-H2A.X and MDC1 interaction does not affect the initial recruitment of NBS1, 53BP1, and BRCA1, it impairs their retention at DSB-flanking sites (8, 9, 41, 42). 53BP1 recruitment to DSB sites also is regulated by RNF8 and RNF168 ubiquitylation of histones and other proteins at these sites of DNA damage (7, 43, 44). RNF8 and RNF168 also have been shown to facilitate 53BP1 binding to H4K20me2 at DSB sites (45). A recent study also suggested a role for RNF8 and RNF168 ubiquitylation and proteasomal degradation of JMJD2A in exposing H4K20me2 and facilitating 53BP1 recruitment to DSB sites (25). Although current data support the function of RNF168 in the DSB-signaling cascade downstream of RNF8, the requirement for this E3 ligase, but not RNF8 (3, 8, 11, 12), for the initial recruitment of 53BP1 to DSB sites suggests that RNF168 also may function independently of RNF8 in the DSB-signaling cascade.
We report that the loss of RNF168 attenuates the initial loading of 53BP1 to chromatin, suggesting that it has a role in modulating 53BP1’s affinity for chromatin. We also report the interaction of RNF168 with 53BP1. 53BP1 interacts with MDC1 in unstressed cells, and this interaction was reported to be required for its retention to DSB sites (19–21). Our data indicate that, in addition to 53BP1, RNF168 also interacts with MDC1 in UT cells and that the loss of RNF168 attenuates 53BP1–MDC1 interactions. RNF168’s role in facilitating 53BP1 interaction with MDC1 is consistent with its importance in regulating 53BP1 recruitment to sites of DNA damage. We also provide evidence that RNF168 mediates K63-linked ubiquitylation of 53BP1. Although RNF168 ubiquitylation of 53BP1 increased at early time points after DNA damage, the level of this ubiquitylation was lower when examined 2 h post-IR. Although the mechanism for this reduced polyubiquitylation at late time points post-IR remains to be investigated, it may involve deubiquitylases that serve to restrain 53BP1 ubiquitylation in response to DNA damage.
Our data also indicate that K1268R substitution in 53BP1 markedly reduces its ubiquitylation by RNF168. These data suggest that, although RNF168 likely ubiquitylates 53BP1 at multiple residues, the K1268 residue remains critical for this ubiquitylation. We also report that the initial recruitment of 53BP1 to DSB sites is significantly impaired by the presence of the RNF168 deletion mutant (Rnf168Δ61–168ΔMIU2), which fails to interact with 53BP1, and by the K1268R substitution of 53BP1, which restrains its ubiquitylation by RNF168. Using full-length 53BP1, we demonstrate that mutation of K1268 that lies within 53BP1 oligomerization domain impairs oligomerization of 53BP1. We also show that this mutation reduces 53BP1’s affinity for chromatin, highlighting the importance of 53BP1 ubiquitylation on K1268 for its recruitment to DSB sites. Thus, initial recruitment of 53BP1 to sites of DNA damage is dependent not only on its interaction with RNF168 but also on its ubiquitylation by this E3 ligase. Our data also indicate that the RNF168ΔMIU2 mutant, which cannot be recruited to the DSB-flanking site, retains its ability to ubiquitylate 53BP1. Thus, RNF168 polyubiquitylation of 53BP1 does not require its localization to DSB sites. Furthermore, our data demonstrate the importance of 53BP1 ubiquitylation on K1268 for G2/M checkpoint activation, NHEJ repair of DSBs, and radiosensitivity. Consistent with RNF8-independent initial recruitment of 53BP1 to DSBs (8, 11), RNF8 failed to interact with 53BP1 or mediate its ubiquitylation. In addition, no additive effect on 53BP1 ubiquitylation was observed in the presence of both RNF8 and RNF168. These data suggest RNF168-dependent but RNF8-independent ubiquitylation of 53BP1. This difference in 53BP1 ubiquitylation by these two E3 ligases may contribute to their differential requirement for the initial 53BP1 IRIF.
In addition to RNF8 and RNF168, the E3 ligase RAD18 also is involved in DSB signaling (22, 46). Although RNF168 interacts with 53BP1 in UT cells, RAD18 interacts with 53BP1 in an IR-dependent manner (22). In addition, in contrast to RNF168 polyubiquitylation of 53BP1 in UT cells, RAD18 monoubiquitylates 53BP1 in response to DNA damage and enhances its retention at DSBs (22). Although the loss of RNF168 impairs 53BP1 polyubiquitylation and suppresses its transient recruitment and retention at DSBs, as is consistent with previous studies (22), we observed no defect in 53BP1 polyubiquitylation or its initial recruitment to DSB sites in Rad18 −/− cells. Thus, although 53BP1 is ubiquitylated by two E3 ligases under different conditions, its RNF168 polyubiquitylation primes its initial recruitment to DSBs, whereas its RAD18 monoubiquitylation is important only for enforcing its retention to sites of DNA damage.
Through its ubiquitylation of 53BP1 and H2A-type histones, RNF168 plays a central role in DSB signaling. RNF168 ubiquitylates 53BP1 without requiring their localization to DSBs, and this 53BP1 ubiquitylation is required for its initial localization to sites of damage. Once Rnf168 is recruited to DSB sites, its ubiquitylation of H2A on K15 provides an epitope for 53BP1 interaction with chromatin, allowing its retention at these breaks (44). In patients with the RIDDLE syndrome, the defect of these ubiquitylation events mediated by RNF168 is likely to cause the complete absence of 53BP1 IRIF associated with this syndrome (2–5). The multistep requirement for RNF168 in the DSB-signaling cascade, its polyubiquitylation of 53BP1, and the importance of this ubiquitylation for the regulation of 53BP1 functions highlight the critical role this E3 ligase plays in DNA-damage signaling and repair and in maintaining genomic integrity.
Materials and Methods
Cell Cultures.
MEFs and HEK293T cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Wincent). Splenocytes, thymocytes, and lymphocytes were cultured in RPMI1640 (Gibco) with 10% (vol/vol) FBS. Rnf168 +/+ and Rnf168 −/− MEFs (12) were generated using standard procedures. WT, 53bp1 −/−, and Rad18 −/− MEFs were used (22, 27). RIDDLE 15–9BI cells and 15–9BI cells complemented with WT RNF168 were described previously (2).
Intracellular Ubiquitylation Assay.
The calcium phosphate method was used to transfect HEK293T cells with expression vectors encoding FLAG-tagged WT or mutated mouse Rnf168, FLAG-RNF8, HA-tagged WT or mutated 53BP1, HA-His–53BP1-KBD, WT or mutated Myc-53BP1, and WT or mutated HA-Ub [Addgene: pRK5-HA-Ub-WT (ID 17608), pRK5-HA-Ub-K48 (ID 17605), pRK5-HA-Ub-K63 (ID 17606)] or Myc-Ub as indicated. After 48 h, cells were lysed in modified RIPA buffer, and IP was performed using anti-HA or anti-Myc antibody and protein A-Sepharose.
Supplementary Material
Supporting Information
Acknowledgments
We thank V. Stambolic, D. Durocher, P. Cheung, and L. Salmena for helpful discussions and D. Durocher, J. Chen, R. Schiestl, S. Tateishi, V. Gorbunova, T. Atsumi, S. Hatakeyama, and K. Iwabuchi for providing reagents. This work was supported by the Canadian Institutes of Health Research and the Canadian Breast Cancer Foundation. G.S.S. was supported by Cancer Research Senior Fellowship C17183/A13030.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Panier S, Durocher D. Push back to respond better: Regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol. 2013;14(10):661–672. doi: 10.1038/nrm3659. [DOI] [PubMed] [Google Scholar]
- 2.Stewart GS, et al. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci USA. 2007;104(43):16910–16915. doi: 10.1073/pnas.0708408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136(3):420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 4.Devgan SS, et al. Homozygous deficiency of ubiquitin-ligase ring-finger protein RNF168 mimics the radiosensitivity syndrome of ataxia-telangiectasia. Cell Death Differ. 2011;18(9):1500–1506. doi: 10.1038/cdd.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 6.Pinato S, Gatti M, Scandiuzzi C, Confalonieri S, Penengo L. UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol Cell Biol. 2011;31(1):118–126. doi: 10.1128/MCB.00818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattiroli F, et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150(6):1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Yuan J, Chen J. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. J Biol Chem. 2010;285(2):1097–1104. doi: 10.1074/jbc.M109.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celeste A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5(7):675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 10.Bekker-Jensen S, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173(2):195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, et al. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med. 2010;207(5):983–997. doi: 10.1084/jem.20092437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bohgaki T, et al. Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet. 2011;7(4):e1001381. doi: 10.1371/journal.pgen.1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Difilippantonio S, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456(7221):529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bothmer A, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42(3):319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santos MA, et al. Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J Exp Med. 2010;207(5):973–981. doi: 10.1084/jem.20092308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127(7):1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pei H, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470(7332):124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwabuchi K, et al. Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J Biol Chem. 2003;278(38):36487–36495. doi: 10.1074/jbc.M304066200. [DOI] [PubMed] [Google Scholar]
- 19.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421(6926):961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Stern DF. NFBD1/MDC1 regulates ionizing radiation-induced focus formation by DNA checkpoint signaling and repair factors. FASEB J. 2003;17(13):1842–1848. doi: 10.1096/fj.03-0310com. [DOI] [PubMed] [Google Scholar]
- 21.Eliezer Y, Argaman L, Rhie A, Doherty AJ, Goldberg M. The direct interaction between 53BP1 and MDC1 is required for the recruitment of 53BP1 to sites of damage. J Biol Chem. 2009;284(1):426–435. doi: 10.1074/jbc.M807375200. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe K, et al. RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Res. 2009;37(7):2176–2193. doi: 10.1093/nar/gkp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6(10):776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 24.Zgheib O, Pataky K, Brugger J, Halazonetis TD. An oligomerized 53BP1 tudor domain suffices for recognition of DNA double-strand breaks. Mol Cell Biol. 2009;29(4):1050–1058. doi: 10.1128/MCB.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallette FA, et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31(8):1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432(7015):316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 27.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23(7):2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie A, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28(6):1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456(7221):524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bothmer A, et al. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207(4):855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17(6):688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165(4):459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orsburn B, et al. Differential requirement for H2AX and 53BP1 in organismal development and genome maintenance in the absence of poly(ADP)ribosyl polymerase 1. Mol Cell Biol. 2010;30(10):2341–2352. doi: 10.1128/MCB.00091-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298(5597):1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 36.Secretan MB, et al. Effect of Ku86 and DNA-PKcs deficiency on non-homologous end-joining and homologous recombination using a transient transfection assay. Mutat Res. 2004;554(1-2):351–364. doi: 10.1016/j.mrfmmm.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 37.Mao Z, Jiang Y, Liu X, Seluanov A, Gorbunova V. DNA repair by homologous recombination, but not by nonhomologous end joining, is elevated in breast cancer cells. Neoplasia. 2009;11(7):683–691. doi: 10.1593/neo.09312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Löbrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7(11):861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 40.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5(5):481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 41.Lukas C, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23(13):2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21(2):187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 43.Bekker-Jensen S, Mailand N. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 2011;585(18):2914–2919. doi: 10.1016/j.febslet.2011.05.056. [DOI] [PubMed] [Google Scholar]
- 44.Fradet-Turcotte A, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499(7456):50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Acs K, et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol. 2011;18(12):1345–1350. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- 46.Huang J, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11(5):592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information