A transcriptional network in polycystic kidney disease (original) (raw)
Abstract
Mutations in cystic kidney disease genes represent a major genetic cause of end-stage renal disease. However, the molecular cascades controlling the expression of these genes are still poorly understood. Hepatocyte Nuclear Factor 1β (HNF1β) is a homeoprotein predominantly expressed in renal, pancreatic and hepatic epithelia. We report here that mice with renal-specific inactivation of _HNF1_β develop polycystic kidney disease. We show that renal cyst formation is accompanied by a drastic defect in the transcriptional activation of Umod, Pkhd1 and Pkd2 genes, whose mutations are responsible for distinct cystic kidney syndromes. In vivo chromatin immunoprecipitation experiments demonstrated that HNF1β binds to several DNA elements in murine Umod, Pkhd1, Pkd2 and Tg737/Polaris genomic sequences. Our results uncover a direct transcriptional hierarchy between _HNF1_β and cystic disease genes. Interestingly, most of the identified HNF1β target gene products colocalize to the primary cilium, a crucial organelle that plays an important role in controlling the proliferation of tubular cells. This may explain the increased proliferation of cystic cells in MODY5 patients carrying autosomal dominant mutations in _HNF1_β.
Keywords: cilium, cysts, HNF1β, MODY5, proliferation
Introduction
The development of the kidney depends on reciprocal interactions between the ureteric bud and the metanephric mesenchyme. Factors secreted by the ureteric bud induce the aggregation of the mesenchyme and subsequent conversion into the epithelium. The induced mesenchyme in turn sends signals to the ureteric bud to promote its branching. This reciprocal induction proceeds in an ordered manner to give rise to the collecting duct system and simultaneously to the nephrons (see Vainio and Lin, 2002).
Failure of the differentiation program during nephrogenesis can lead to diverse pathological conditions that can perturb renal structure and function. Mutations in cystic kidney disease genes constitute the most common genetic cause of chronic renal failure in both children and adults (see Igarashi and Somlo, 2002). Despite their genetic, clinical and histopathological heterogeneity, these renal diseases result in a similar outcome: dilation of tubules and cystogenesis. These observations have led to the hypothesis that the gene defects underlying these disorders might disrupt a common pathway. However, the mechanisms of cyst formation are still not completely elucidated. They are postulated to involve dysfunctions in the control of cell proliferation, apoptosis, cell polarity, the localization of ion channels, fluid secretion and cell–cell adhesion or cell–matrix interactions (see Igarashi and Somlo, 2002). During the past decade, positional cloning strategies have identified several genes responsible for cystic kidney diseases. Genes mutated in autosomal dominant polycystic kidney disease (ADPKD) (PKD1 and PKD2) (Hughes et al, 1995; Mochizuki et al, 1996), autosomal recessive polycystic kidney disease (ARPKD) (PKHD1) (Ward et al, 2002), nephronophthisis (NPHP1 to 4) (Hildebrandt et al, 1997; Otto et al, 2002; Olbrich et al, 2003; Otto et al, 2003) and medullary cystic kidney disease (MCKD) type 2 (UMOD/THP) (Hart et al, 2002) have been identified. Recently, ADPKD, ARPKD and nephronophthisis gene products have been localized in the primary cilium, a solitary non-motile organelle that extends from the apical surface of renal epithelial cells (see Watnick and Germino, 2003). _Pkd1_- and _Pkd2_-encoded proteins may be part of a mechanosensitive Ca2+-signaling pathway in renal cilia that inhibits renal tubular cell growth (Nauli et al, 2003). If ciliary structure or function is perturbed, tubular cell growth may continue, leading to cyst formation. The role of the proteins involved in ARPKD and nephronophthisis in the cilium is still not clear. Polaris, a protein involved in a mouse polycystic kidney disease model, is also localized in the cilium, and plays a role in the assembly of this organelle (Pazour et al, 2000). Mutations in the human UMOD gene are responsible for MCKD type 2, but the mechanisms underlying the cyst formation in this disease also remain elusive.
Relatively little is known about the transcriptional networks that control epithelial differentiation of renal tubules. Hepatocyte Nuclear Factor 1β (HNF1β) is a homeodomain-containing transcription factor that binds DNA and transactivates transcription as homodimer, or heterodimers with the closely related factor, HNF1α (see Cereghini, 1996). Both proteins were initially described as liver-enriched transcription factors, but their expression pattern is not restricted to hepatocytes (Blumenfeld et al, 1991; see Cereghini, 1996). In particular, these proteins are expressed in the tubular epithelial cells of the kidney (Lazzaro et al, 1992; Pontoglio et al, 1996). HNF1β is expressed throughout the entire nephron, from proximal tubules to collecting ducts, whereas HNF1α expression is restricted to proximal tubules. Interestingly, HNF1β is also highly expressed in other tubular epithelia such as biliary and pancreatic ducts (Coffinier et al, 1999a).
Autosomal dominant mutations in the _HNF1_β gene are associated with a particular form of diabetes mellitus called Maturity Onset Diabetes of the Young type 5 (MODY5) (Horikawa et al, 1997). Affected patients also present diverse forms of non-diabetic nephropathies and, with less penetrant occurrence, genital malformations and liver dysfunction (Nishigori et al, 1998; Lindner et al, 1999; Bingham et al, 2000). The most common and penetrant trait of these patients is the presence of renal cysts and diabetes (RCAD). The renal manifestations of the disease also include cystic renal dysplasia, probably due to a defective nephrogenesis, oligomeganephronia and hypoplastic Glomerulocystic kidney disease (GCKD) (Bingham et al, 2001). In zebrafish, mutations of _HNF1_β affect pronephros development leading to cystic pronephric tubules (Sun and Hopkins, 2001). The expression of HNF1β mutants in Xenopus laevis affects the normal development of pronephric structures (Bohn et al, 2003). Furthermore, _HNF1_β plays an essential role during mouse early embryogenesis. _HNF1_β-null mice die around embryonic day 7 (E7) due to a defect in visceral endoderm differentiation (Barbacci et al, 1999; Coffinier et al, 1999b). To assess the roles of HNF1β during late embryogenesis, we made use of a Cre-LoxP strategy. We demonstrate here that kidney-specific inactivation of _HNF1_β leads to a polycystic phenotype. We found that the expression of several genes involved in cystic diseases is severely affected in the conditionally inactivated mice. By chromatin immunoprecipitation, we show that three of them (Umod, Pkhd1 and Pkd2) are directly controlled by HNF1β. These results describe a direct transcriptional hierarchy between HNF1β and these genes, and establish the role of this transcription factor in regulating the terminal differentiation of renal tubular cells.
Results
Kidney-specific inactivation of the HNF1β gene
The early lethality of HNF1β-deficient embryos prevented the analysis of its function at later developmental stages. To circumvent this problem, we inactivated the _HNF1_β gene using the Cre-LoxP strategy. To restrict the inactivation to renal cells, we made use of a KspCre transgenic mouse strain that expresses the Cre recombinase under the control of the Ksp-cadherin (Cadherin 16) promoter. This transgene drives efficient Cre expression and Cre-mediated recombination in collecting ducts, loops of Henle and only occasionally (less than 5%) in proximal tubular cells (Shao et al, 2002).
Inactivation of the _HNF1_β gene was achieved by generating mice carrying a homozygous floxed _HNF1_β gene (Coffinier et al, 2002) along with the KspCre transgene (KspCre; HNF1_β_flox/flox). In a parallel way, we also generated compound heterozygous animals carrying a null _HNF1_β lacZ knock-in (Coffinier et al, 1999b) and a floxed allele along with the KspCre transgene (KspCre; HNF1_β_flox/LacZ). These two genotypes involved a renal-specific inactivation of _HNF1_β and gave rise to the same phenotype. For these reasons, we described these animals as ‘mutants'. Conversely, littermates carrying either (KspCre; HNF1_β_flox/+) or (HNF1_β_flox/flox) or (HNF1_β_flox/+) or (HNF1_β_flox/LacZ) were phenotypically indistinguishable from the wild-type mice. For this reason, we termed these animals collectively as ‘controls'.
Kidney-specific inactivation of HNF1β leads to postnatal lethality and renal failure
Pups carrying all possible genotypes were born in normal Mendelian ratios (_N_=159, _P_>0.05). No lethality was observed up to postnatal day 8 (P8). Between days P10 and P21, 75% of mutants died (_N_=8). Generally, mutant animals were growth retarded by P8. At this stage, the average weight of mutant pups was 11% lower than that of controls (_N_control=75, _N_mutant=22, P<0.05). Growth retardation increased with age, and mutants that survived weaning exhibited an approximately 25% decrease in body weight.
To investigate renal function, we determined seric urea and creatinine concentrations. Measurements were performed at different ages (P8 and on the few surviving animals at P17 and P30) (Figure 1). Both urea and creatinine levels were increased in mutant mice at all time points. These data indicated that mutants suffered, and died from, a severe impairment of renal function. Most measurements required euthanasia of the pups, therefore these data do not represent an average time-course progression of renal disease.
Figure 1.
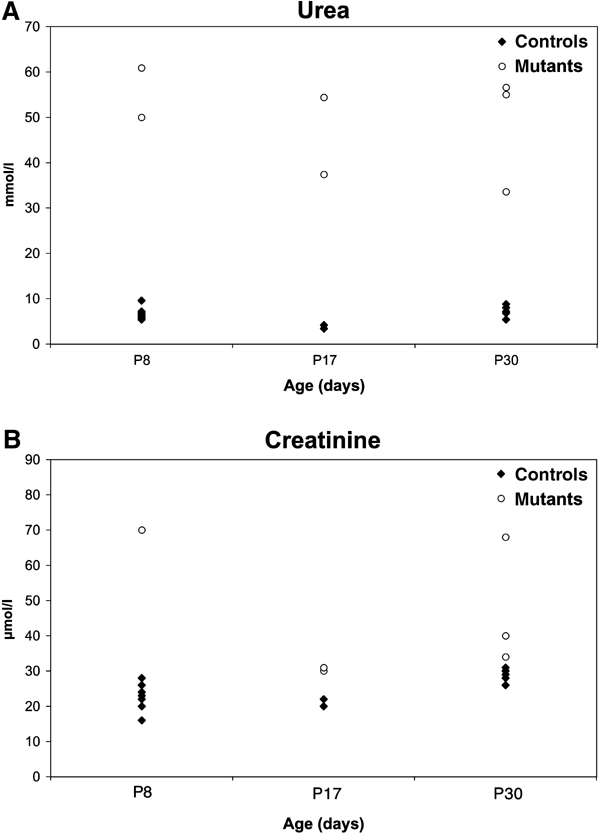
Impaired renal function in mutant mice. Serum concentrations of urea (A) and creatinine (B) at P8, P17 and P30.
Inactivation of HNF1β results in a renal polycystic phenotype
We observed bilateral ureteral dilation, also involving the renal pelvis, in most mutants at P8 (92%, _N_=25) (Figure 2). No signs of ureter obstruction were seen. As HNF1β is expressed in the ureteral epithelium, and as KspCre had driven recombination in the precursors of these cells (data not shown), we concluded that this abnormality was a direct consequence of _HNF1_β inactivation in ureter-epithelial cells.
Figure 2.
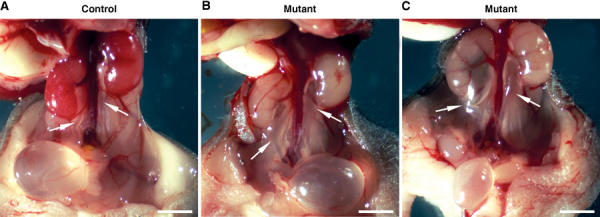
_HNF1_β inactivation results in bilateral ureteral dilation. Macroscopic view of urinary tract in control (A) and mutant (B, C) mice at P8. Some mutants exhibit a mild ureter dilation (arrows) (compare B versus A), while others present a more severe phenotype (compare C versus A). Mutant kidneys had a normal size, but were paler than controls. Scale bars: 0.25 cm.
The kidneys of mutants were much paler than the controls but were apparently normally sized at P8 (Figure 2). Gross abnormalities of mature nephrons suggested that terminal morphogenesis did not occur normally at P1. In particular, several tubules located in the medullary region of mutant kidneys were dilated and contained more cells than in controls (Figure 3A and B). Histological sections at P8 revealed that the kidneys of all mutant mice exhibited numerous cysts (Figure 3C and D). These cysts were mostly located in the medulla, but a few could also be observed in the cortex. The average size of cysts increased with age (Figure 3D and F). It is noteworthy that dilation of the urinary space in some glomeruli could be clearly observed at all stages (Figure 3F).
Figure 3.
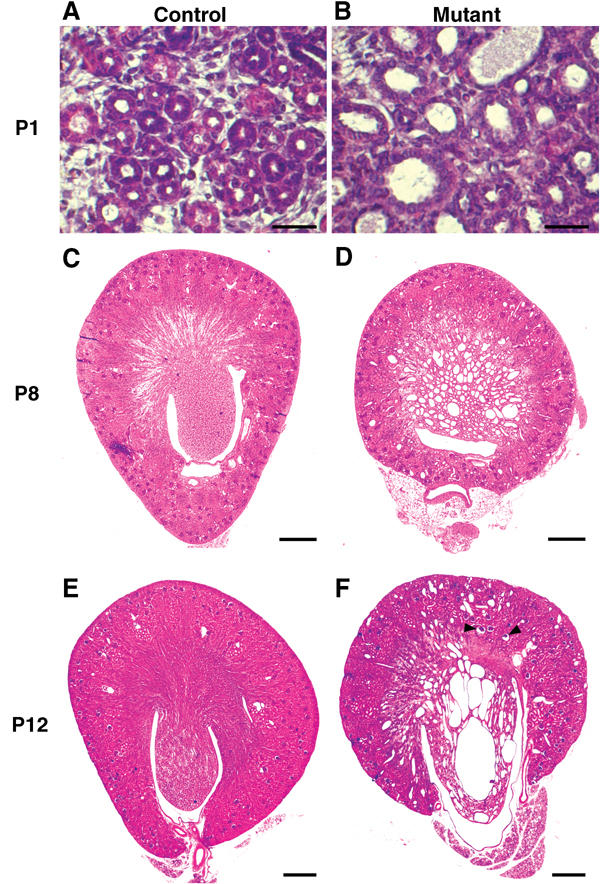
Inactivation of _HNF1_β in kidney cells leads to polycystic kidney disease. Hematoxylin–eosin-stained kidney sections from control and mutant mice at P1 (A, B), P8 (C, D) and P12 (E, F). Tubular dilations were visible in the medullary region of mutants at P1, with an increased number of nuclei per tubule section compared to controls (B versus A). In mutants, the medulla was completely disrupted by large cysts by P8 (D). The size of cysts increased with age (F). A few glomerular cysts were observed in the deep cortex (F, arrowheads). Scale bars: A, B: 100 μm, C–F: 400 μm.
Renal cysts are lined with Cre recombined HNF1β −/− cells
The KspCre transgene drives efficient recombination in the renal medulla. Histological abnormalities in _HNF1_β-inactivated mice mainly affected this region, suggesting that the renal cysts were a direct consequence of _HNF1_β inactivation. To confirm this hypothesis, we monitored the recombination status of cystic cells using the ROSA26R reporter allele (Soriano, 1999). We generated mice with kidney-specific inactivated _HNF1_β combined with the ROSA26R reporter allele (KspCre; HNF1_β_flox/flox; ROSA26R). This allele contains a lacZ gene whose activity is induced by the Cre-driven recombination (Soriano, 1999). These mice had the same ureteral and cystic phenotype as KspCre; HNF1_β_flox/lacZ animals (Figure 4B and 5F and data not shown). X-gal stained kidney sections revealed that the renal cysts were lined with β-galactosidase-positive cells (Figure 4B). As expected, control mice (KspCre; HNF1_β_flox/+; ROSA26R) showed β-gal-positive cells lining most medullary segments of the kidney tubules without any cyst formation (Figure 4A). These data indicate that all cystic cells underwent Cre-mediated recombination or were derived from recombined progenitor cells.
Figure 4.
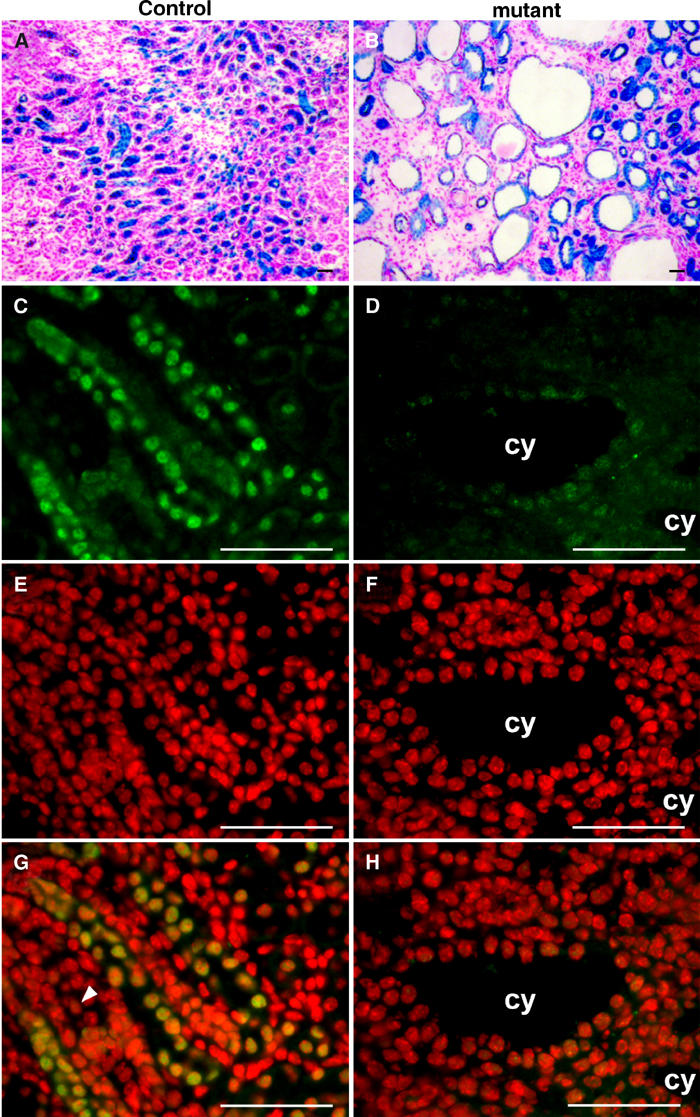
Cystic cells underwent recombination and lack HNF1β expression. (A, B) X-gal staining on kidney sections of control KspCre; HNF1_β_flox/+; ROSA26R (A) and mutant KspCre; HNF1_β_flox/flox; ROSA26R (B) at P14. β-gal activity is an indicator of Cre-driven recombination on the ROSA26R locus. (A) The medulla of control mice showed recombination in the tubular epithelium. (B) In mutants, all cysts were lined with recombined cells. (C–H) Kidney sections of control HNF1_β_flox/lacZ and mutant KspCre; HNF1_β_flox/lacZ at P8. (C, D) HNF1β staining (fluorescein/green). (E, F) Nuclear staining of the same section (DAPI/red). (G, H) Merging. (G) In controls, HNF1β is expressed in tubular but not in mesenchymal cells (arrowhead). (H) Mutants showed no expression of HNF1β in cysts. cy: cyst. Scale bars: 75 μm.
Figure 5.
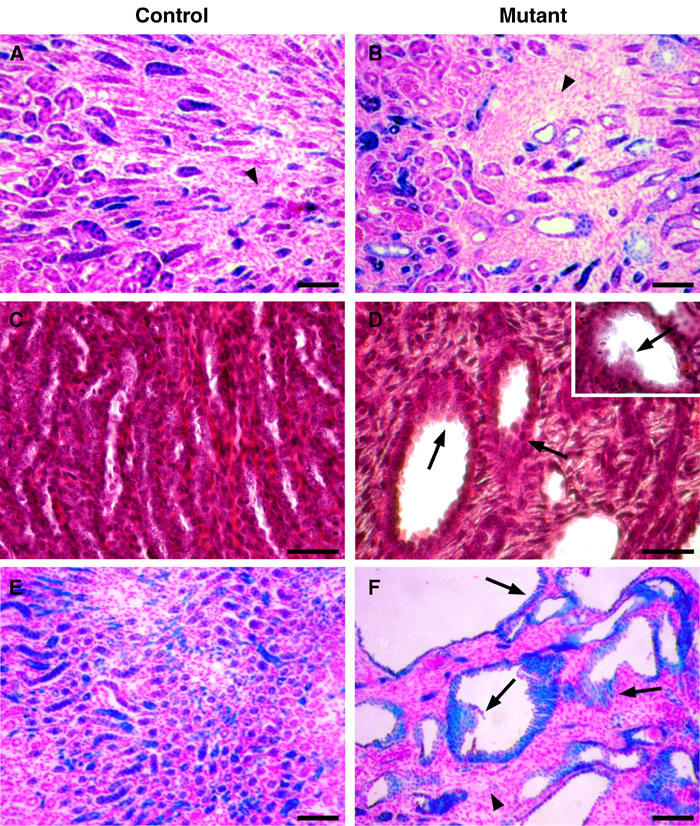
_HNF1_β inactivation leads to multilayered tubular epithelia. (A, B) X-gal staining on kidney sections of control HNF1_β_flox/lacZ (A) and mutant KspCre; HNF1_β_flox/lacZ (B) at P8. β-gal activity is an indicator of the endogenous _HNF1_β promoter activity. (A) _HNF1_β was expressed in all tubular cells but not in mesenchyme (arrowhead). (B) Mesenchyme is β-gal negative (arrowhead), indicating that it was not programmed to express _HNF1_β. (C, D) Hematoxylin–eosin-stained sections of control (C) and mutant (D) at P8. Multilayered tubular epithelial cells were observed (arrows), as well as polyps growing into the cyst lumen (D and insert). (E, F) X-gal staining on kidney sections of control KspCre; HNF1_β_flox/+; ROSA26R (E) and mutant KspCre; HNF1_β_flox/flox; ROSA26R (F) at P14. β-gal activity is an indicator of Cre-driven recombination. (E) KspCre-driven recombination was seen in a large proportion of medullary tubular cells. (F) Several cysts lack the typical monolayered epithelial structure (arrows). All epithelial cells are β-gal-positive, demonstrating that they underwent Cre recombination. The mesenchyme in mutants was not affected by recombination (arrowhead). Scale bars: 200 μm.
To further prove that Cre activity had inactivated the endogenous _HNF1_β gene, we performed immunofluorescent detection using a novel polyclonal antibody (HNF1β-NC-AR) (Figure 4C–H). This analysis revealed that the HNF1β protein was indeed absent from cystic epithelium (Figure 4D and H), whereas its expression was conserved in proximal tubules (data not shown). Thus, renal cysts were lined with recombined HNF1β-null cells.
To establish the cellular origin of cysts, we analyzed specific markers of collecting ducts and Henle's loops. Most cysts expressed Aquaporin2 (Aqp2) protein, a collecting duct cell marker (Figure 7G). Some cysts expressed the sodium–potassium-chloride cotransporter isoform 2 (Slc12a1), a specific marker of thick ascending limb of Henle's loop (Figure 7C and K) (see Delpire et al, 1996). Taken together, these data indicate that the cysts were mainly derived from two nephron segments where KspCre is expressed.
Figure 7.
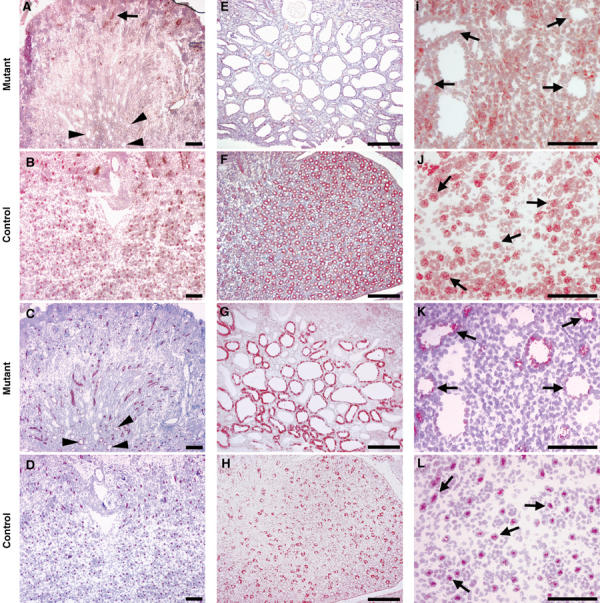
_HNF1_β inactivation leads to a strong decrease of Umod, Pkhd1 and Pkd2 protein levels in cystic cells. Immunostainings for uromodulin (A, B), Pkhd1 (E, F), Pkd2 (I, J), Slc12a1 (C, D, K, L), and Aqp2 (G, H) in mutant and control mice at P8. (A–D) Uromodulin expression pattern. In mutants, no uromodulin staining was detected in the medullary cysts (compare A versus B). Some cysts were clearly shown to be thick ascending limb (TAL)-derived (see the arrowheaded positive staining with α-Slc12a1 antibody in C). Arrowheads in A indicate the position of the corresponding cysts in the adjacent section. In controls, both uromodulin and Slc12a1 proteins colocalized (compare adjacent sections B and D, respectively). In mutants, only few cortical nondilated TAL segments expressed uromodulin (see arrow in A). (E–H) Pkhd1 expression pattern. In mutants, Pkhd1 expression was strongly decreased in the vast majority of medullary cystic cells (compare E versus F). Pkhd1 downregulation is not due to the absence of collecting duct cells, as most cysts still expressed aquaporin2 (compare G versus E). (I–L) Pkd2 expression pattern. Pkd2 expression is absent in cystic epithelia (compare I versus J). Again, some cysts were clearly shown to be TAL-derived (see the arrowed positive staining with α-Slc12a1 antibody in K). Arrows in I indicate the position of the corresponding cysts, in the adjacent section. In control animals, both Pkd2 and Slc12a1 proteins colocalized (compare arrows in serial sections B and D, respectively). Scale bars: 300 μm.
Analysis of kidneys from P8 animals also showed that mesenchymal cells were present in both control and mutant mice, but they were more abundant in mutants (Figure 5A and B). From P12 onwards, the presence of mesenchymal cells was visible in mutants, but not in controls at the same age (Figure 3E and F). We investigated whether these mesenchymal cells expressed _HNF1_β. We took advantage of the _HNF1_β lacZ allele in heterozygous control mice (HNF1_β_flox/lacZ). Mesenchymal cells were β-gal-negative, indicating that they do not express _HNF1_β (Figure 5A). In addition, immunofluorescence experiments showed that these cells do not express the HNF1β protein (Figure 4G). Similarly, mesenchymal cells present in mutants (KspCre; HNF1_β_flox/lacZ) were also β-gal-negative, indicating that these cells were not programmed to express the endogenous _HNF1_β gene (Figure 5B). In a parallel manner, we investigated whether these mesenchymal cells had undergone KspCre-driven recombination using the KspCre; HNF1_β_flox/flox; ROSA26R mice. X-gal staining of kidney sections from these animals showed that all mesenchymal cells were β-gal-negative, whereas cells lining cysts were β-gal-positive (Figure 5F). Thus, these mesenchymal cells did not express KspCre and were not derived from cells where the KspCre had been expressed. Both experiments demonstrate that the increase in mesenchymal cells in mutant kidneys is a secondary consequence of _HNF1_β inactivation.
Increased proliferation in mutant renal cysts is cell-autonomous
Increased proliferation is one of the major mechanisms underlying cyst formation in cystic kidney diseases. Notwithstanding this propensity to proliferate, renal cystic cells are usually organized in a monolayered epithelium (see Igarashi and Somlo, 2002). This is actually the case for most of the cysts that we found in mutants at P8. However, about 15% of cysts showed multilayered epithelia at this stage (Figure 5D). Similar to what has been observed in human polycystic kidney disease (see Bernstein et al, 1987), some polyps developed into the cyst lumen (insert in Figure 5D). Using the ROSA26R reporter allele, we have investigated the lineage of these proliferating cells. Interestingly, in mutant KspCre; HNF1_β_flox/flox; ROSA26R animals, all cells in multilayered structures were positively stained with X-gal, indicating that all of them had recombined (Figure 5F). Therefore, the presence of multilayered cysts is probably due to a cell-autonomous defect. This observation indicates that _HNF1_β inactivation in tubular epithelial cells results in increased proliferation.
Mutant mice show specific transcriptional defects of several cystic disease genes
To unravel the molecular mechanisms underlying cyst formation in our mouse model, we monitored the expression levels of genes known to be involved in cystogenesis in mice and humans. We performed quantitative RT–PCR experiments using kidney RNA from mutant and control animals. We found that the expression level of Umod (Uromodulin/Tamm–Horsfall glycoprotein) was decreased 10-fold in mutant kidney. This result could be due to a direct defective transcriptional activation of the Umod gene by HNF1β. Alternatively, this deficiency could be due to the absence of cells that normally express this gene (Bachmann et al, 1990) because of a developmental or differentiation defect. The grossly normal expression of a marker of the thick ascending limb of Henle's loops (TAL), the sodium–potassium-chloride cotransporter isoform 2 (Slc12a1/Nkcc2) (Figure 6), argued against this possibility. In addition, when the expression of Umod was normalized for the expression of Slc12a1, downregulation of Umod expression was still calculated to be 8.5-fold (Figure 6). This normalization allowed us to demonstrate that the Umod transcriptional defect is not linked to the absence of TAL cells in mutant mice.
Figure 6.
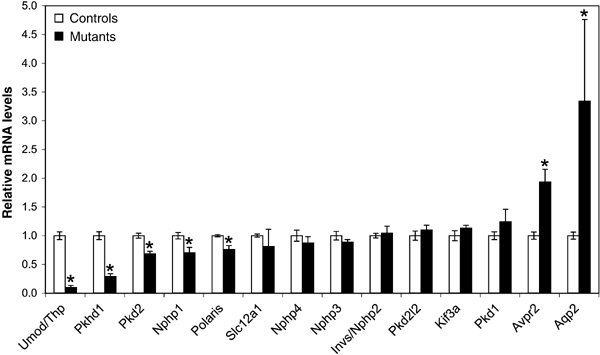
Defective transcriptional activation of cystic disease genes in mutant mice. Quantitative RT–PCR analysis of cystic disease genes and cell-specific markers. Umod/Thp, Pkhd1, Pkd2, Nphp1 and Tg737/Polaris were downregulated in mutant kidneys. Expression levels in mutants are indicated relative to controls. Results were normalized with β_-2 microglobulin_ expression level (except for Umod, which was normalized to Slc12a1 expression). Significant differences between mutants and controls (P<0.05) are indicated (*). Error bars represent standard error of the mean (_N_control=5, _N_mutant=3).
Interestingly, two other key genes of cyst formation, Pkhd1 and Pkd2, were also downregulated (3.4- and 1.5-fold, respectively). In contrast, Pkd1 and Pkd2l2 (a _Pkd2_-related gene) (Guo et al, 2000) expression levels were normal. Among the genes involved in nephronophthisis, only Nphp1 was significantly downregulated in mutant mice (1.4-fold), whereas Invs/Nphp2, Nphp3 and Nphp4 were normally expressed (Figure 6).
We also investigated the expression of two genes involved in cystogenesis in mice, Tg737/Polaris (Moyer et al, 1994) and Kif3a (Lin et al, 2003). The former was mildly, but significantly, dowregulated (1.3-fold) in mutant mice, whereas Kif3a was normally expressed (Figure 6).
We analyzed the expression of Aqp2 and arginine-vasopressin-receptor type 2 (Avpr2). These two genes are expressed in collecting ducts, a structure that contributes to cyst formation. Similar to what has been described in different polycystic rodent models (Gattone et al, 2003), both of these genes were upregulated in mutant kidneys (Figure 6).
Our results show that renal _HNF1_β inactivation results in a reduced expression of several genes known to play crucial roles in cystogenesis. In fact, at least five of these genes (Umod, Pkhd1, Pkd2, Nphp1 and Tg737/Polaris) were specifically affected by _HNF1_β inactivation.
Umod-, Pkhd1- and _Pkd2_-encoded proteins are dramatically downregulated in cystic epithelia
In order to assess the expression of Umod, Pkhd1 and Pkd2 proteins in HNF1β−/− cystic cells, we carried out immunohistochemical stainings. Uromodulin is known to be exclusively expressed in the cortical and medullary TAL (Bachmann et al, 1990). Our results show that in mutant kidney, the expression of this protein is drastically reduced in the medulla (Figure 7A), a compartment where a large fraction of the positive cells is normally detected (compare Figure 7A versus B). A residual expression of uromodulin is detected in rare non-dilated segments of cortical TAL (arrow, Figure 7A). As previously mentioned, the lack of this marker could underly a defective differentiation of TAL. However, our results have shown that another marker of TAL, the Slc12a1 protein (see Delpire et al, 1996), is normally expressed in mutant kidneys (compare Figure 7C versus D). In addition, serial histological sections have shown that the same TAL-derived cysts, positively stained for Slc12a1, were negative for uromodulin (compare arrowheads in Figure 7A and C). In a similar way, polyductin (Pkhd1) expression was dramatically reduced in cystic epithelia (compare Figure 7E versus F). As this protein is known to be predominantly expressed in collecting ducts (Ward et al, 2003), we assessed the presence of this cell type in the cystic epithelium by the detection of Aqp2, a specific marker of collecting ducts. Our results showed that this cell type was indeed present, as a large proportion of cysts was positive for Aqp2 (compare Figure 7G versus H). Notwithstanding a high background signal detected in mesenchymal cells, we showed that the expression of polycystin2 (Pkd2) was absent in cystic cells (compare arrowed epithelia in Figure 7I versus 7J). Again, since Pkd2 in the medulla is predominantly expressed in TAL (Foggensteiner et al, 2000), we monitored the expression of Slc12a1 in adjacent sections and showed that corresponding cysts expressed this marker but not Pkd2 (see arrows in Figure 7I versus K). Remarkably, Pkd2 was normally expressed throughout the whole cortex (Supplementary Figure 1). In this region of the kidney, KspCre is not expressed and therefore _HNF1_β is not deleted. As a result of this residual cortical expression, RT–PCR carried out on whole kidney RNAs might strongly underestimate the actual downregulation of Pkd2 expression. Taken together, these results clearly indicate that uromodulin, Pkhd1 and Pkd2 are drastically downregulated in HNF1β-deficient cells.
An important fraction of Pkhd1 and Pkd2 proteins is localized to the primary cilium of tubular cells (Pazour et al, 2002; Ward et al, 2003). To assess the morphological integrity of this organelle, we performed immunostaining of acetylated tubulin, a specific component of the primary cilium. We found that mutant cells had normal cilia (Supplementary Figure 2). Moreover, morphometric analysis showed that the cilium length, specifically measured in collecting duct cells (costained with α-Aqp2 antibodies), was normal.
HNF1_β_ binds to cis-acting control elements in several genes involved in cystogenesis
To investigate whether the affected genes were directly controlled by HNF1β we made use of the chromatin immunoprecipitation (ChIP) technique (Orlando et al, 1997; Hecht et al, 1996). This technique can monitor direct in vivo interactions between proteins, such as transcription factors, and their chromatin DNA target sites. To this end, we first identified the position of putative HNF1 binding sites with an in silico approach based on a previously described position-weight matrix (Tronche et al, 1997, MP and SG, unpublished results). Subsequently, we tested whether these sites were actually bound in vivo. The ChIP assays were performed with novel antibodies raised against HNF1β and α proteins (see Supplementary Method). Western blot analyses proved that both antibodies were highly specific and did not cross-react (data not shown). In the following, all given site positions will be based on the Ensembl Mouse release 15.30, which is built around the NCBI 30 composite assembly (see Supplementary Table IA).
Our results showed that the two sites detected close to the Umod gene (at −1.1 and −0.58 kb with respect to the putative transcriptional start site) were significantly bound by HNF1β in vivo (Figure 8). Given the very large size of the Pkhd1 gene (spanning more than 550 kb), we only considered 15 sites identified in a window of 100 kb around the transcriptional start site. Six of them showed a significant interaction with HNF1β in vivo. Interestingly, strongly enriched sites were located either in the proximal promoter (within ∼200 bp) or much further upstream (−35 kb). All _in vivo-_enriched HNF1 sites in this gene occurred as doublets with a maximal interdistance of 400 bp. The in silico search identified four intronic HNF1 sites in the Pkd2 gene and a doublet downstream of the 3′ end of the gene (at around +51 kb). Among the four intronic sites, only one was not enriched in vivo, whereas the 3′ doublet showed the highest enrichment (∼six-fold). The in silico search revealed four potential HNF1 binding sites around the Tg737/Polaris gene. In this case only the closest site (−6.64 kb) showed a weak, but significant, enrichment for HNF1β. Nphp1 was the only candidate gene for which we could not observe any in vivo interaction, notwithstanding the in silico prediction of 2 sites. In order to assess the specificity of our results, we also monitored the potential binding of HNF1β to Pkd1, another cystic disease gene, whose expression is not altered in our mice. We could predict a single HNF1 binding site positioned at +4 kb, but this site was not bound in vivo by HNF1β or HNF1α (Figure 8).
Figure 8.
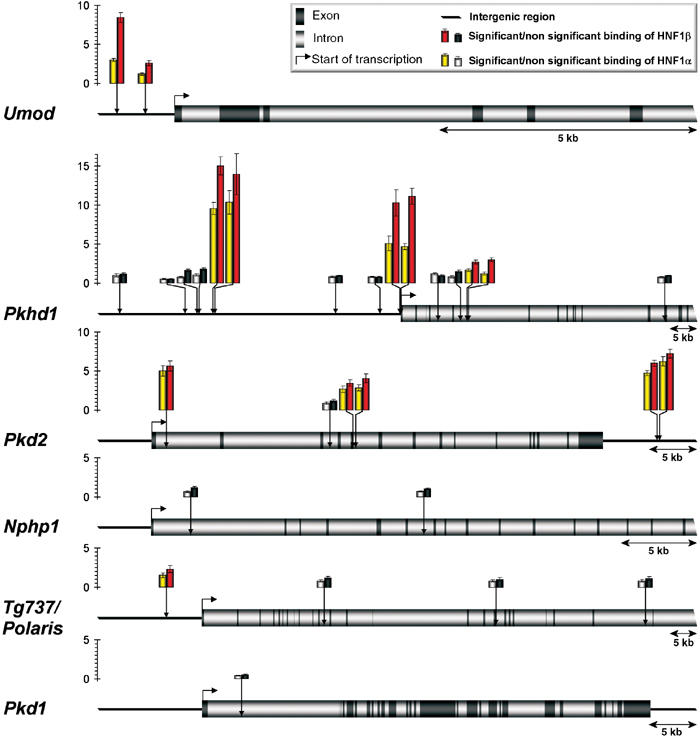
In vivo binding of HNF1 proteins to their chromatin target sites in cystic kidney disease genes. Predicted in silico HNF1 binding sites (vertical arrows) in Umod, Pkhd1, Pkd2, Nphp1, Tg737/Polaris and Pkd1 genes were tested in ChIP experiments for in vivo HNF1α and β binding. The relative enrichment for each DNA fragment upon immunoprecipitation of HNF1α or β is illustrated as histograms. Colored bars represent HNF1 binding sites with enrichments significantly higher than background (gray bars). PCR experiments were performed in triplicate and the standard errors of these quantifications are shown as error bars.
In vitro experiments have shown that HNF1α and HNF1β bind to their target sites with similar affinities, either as homo- or heterodimers (De Simone et al, 1991; Rey-Campos et al, 1991). Our results indicated that the enrichment obtained with the α-HNF1α-specific antibody was systematically weaker than that of HNF1β. This observation could be explained by the fact that HNF1β has a more widespread expression pattern than HNF1α in the kidney. However, the ratio of enrichment in the Umod gene was particularly striking. In this case, HNF1α binding was much lower, compared to that of HNF1β. Interestingly, the Umod gene is the only gene among the analyzed candidates that is not coexpressed with HNF1α. This observation suggests that HNF1α is unable to bind these sites efficiently in the proximal tubular cell, a cell type that does not express the Umod gene.
In summary, our ChIP experiments demonstrate that HNF1β clearly interacts with several HNF1 binding sites in the Umod, Pkhd1 and Pkd2 genes. These cystic disease genes can be considered as direct transcriptional targets of HNF1β. Moreover, interaction with Tg737/Polaris could also be shown, but to a weaker extent.
Discussion
Cystic kidney diseases represent a major cause of end-stage renal failure. Several genes have recently been identified to be involved in these disorders. However, little is known about the transcriptional networks that regulate the expression of these genes. HNF1β plays a crucial role in the development of extraembryonic visceral endoderm and in the morphogenesis of hepatic bile ducts. Here we show that mice lacking HNF1β specifically in the kidney suffer from severe polycystic kidney disease.
HNF1_β_ and cystic kidney diseases
Our results show that the lack of HNF1β is directly responsible for cyst formation via a cell-autonomous mechanism. In humans, heterozygous mutations of _HNF1_β are associated with the Renal Cysts and Diabetes (RCAD) syndrome. This syndrome combines a form of type II diabetes called maturity onset diabetes of the young type 5 (MODY5) with a non-diabetic renal disease (Horikawa et al, 1997; Nishigori et al, 1998; Lindner et al, 1999; Bingham et al, 2000, 2001). The manifestations of renal dysfunction are variable, ranging from isolated cysts to multicystic kidneys. Little is known about the molecular mechanisms underlying this phenotype. Here we show that HNF1β directly controls the transcription of several genes expressed in tubular epithelial cells, namely Umod, Pkhd1 and Pkd2. In addition, HNF1β might contribute to the transcriptional activation of Tg737/Polaris. All these genes are known to be mutated in distinct cystic kidney diseases: MCKD type 2 (UMOD) (Hart et al, 2002), ARPKD (PKHD1) (Ward et al, 2002), ADPKD (PKD2) (Mochizuki et al, 1996) and a murine model of ARPKD (Tg737/Polaris) (Moyer et al, 1994). Strikingly, although a defect in any of these genes in humans is sufficient to elicit renal cyst formation, we observed a concomitant downregulation of a specific subset of cystic disease genes in mutant mice. The combined defects in the expression of these genes could be the origin of the massive cyst formation observed in our mouse model. These results reveal a transcriptional hierarchy between _HNF1_β and different cystic disease genes, and could explain why carriers of autosomal dominant mutations in _HNF1_β develop renal cysts.
Interestingly, MODY5 patients do not develop a strict polycystic disease. They actually present a smaller number of cysts in comparison to ADPKD or ARPKD patients. This difference could be explained by the fact that the human disease occurs in heterozygous carriers, whereas our model involves homozygous _HNF1_β deletion. In order to understand dominant inheritance in humans, it would be interesting to test whether renal cysts in MODY5 patients develop upon loss of the residual wild-type _HNF1_β allele. This ‘two-hit' mechanism has already been described for genes involved in ADPKD (Pkd1 and Pkd2) (Watnick et al, 1998) and for tumor suppressor genes.
Polyductin (Pkhd1) and polycystin2 (Pkd2) proteins are preferentially localized in a specialized cellular organelle: the primary cilium (Pazour et al, 2002; Ward et al, 2003). Several studies suggest a link between a defect in cilium assembly, or function, and renal cyst formation (see Watnick and Germino, 2003). We show that HNF1β is not necessary for cilium formation. However, our data demonstrate that the genetic program driven by HNF1β involves the expression of genes that play a crucial role in cilium function.
HNF1_β_ inactivation leads to increased proliferation of epithelial cells
The presence of renal cysts and the appearance of multilayered epithelia in mutant mice strongly suggest that the inactivation of _HNF1_β leads to an increased rate of cellular proliferation. Mice with liver-specific _HNF1_β inactivation have dysplastic epithelia in both the gallbladder and in the few remaining intrahepatic bile ducts. Multilayered epithelial structures, reminiscent of those observed upon kidney-specific inactivation, are present in the biliary tract of these animals (Coffinier et al, 2002). Recently, HNF1α was shown to be involved in the control of cell proliferation. Indeed, biallelic inactivation of _HNF1_α is frequently observed in liver adenomas and in some hepatocellular carcinomas (Bluteau et al, 2002). For this reason, _HNF1_α can be considered as a tumor suppressor gene. We postulate that _HNF1_β could have the same properties, and that the increased proliferation of HNF1_β_−/− cells upon kidney- and liver-specific inactivation of _HNF1_β is the result of defective regulation of cell cycle in the absence of HNF1β. To validate this hypothesis, the incidence of biallelic inactivation of _HNF1_β in renal cancers and cholangiocarcinomas should be assessed. The antiproliferative effects of HNF1β could be mediated by the biological function of the Pkd1/Pkd2 complex, which has been recently shown to control p21 Cip1 transcription via the JAK/STAT pathway (Bhunia et al, 2002). In addition, Pkd2 has been shown to inhibit the Pkd1-dependent G protein activation (Delmas et al, 2002). However, we cannot rule out that defective expression of additional _HNF1_β target genes could result in a nonfully differentiated state that, in turn, could render growth arrest less efficient.
HNF1_β_ chromatin targets
We show that the defective expression of the cystic disease genes Umod, Pkhd1 and Pkd2 directly correlates with the presence of several HNF1 binding sites that are bound by HNF1β in vivo. This strongly suggests that HNF1β directly controls the expression of these genes. Interestingly, the weak interaction of HNF1β with its target site in Tg737/Polaris gene is reflected by its marginally decreased transcription in the mutant kidney.
The binding of HNF1β was not only restricted to proximal promoter sites but also involved far upstream and downstream sites. In the case of Pkhd1, HNF1 binding sites were identified at 35 kb upstream of the transcriptional start site. These sites occur as doublet and are conserved in both human and mouse genomes (data not shown). Interestingly, this doublet is particularly enriched, suggesting a strong interaction with HNF1β. Similarly, doublets of HNF1 binding sites in the Umod and Pkd2 genes were also strongly bound, whereas isolated binding sites were weakly bound or not bound at all. These data could suggest that HNF1β occupancy is enhanced by the presence of closely spaced sites on chromatin. Alternatively, a higher relative enrichment could also be explained by the increased probability for a DNA fragment encompassing two HNF1 sites to be immunoprecipitated.
The liver-specific inactivation of _HNF1_β leads to defective morphogenesis of intrahepatic bile ducts (IHBD) (ductal plate malformation) (Coffinier et al, 2002). Interestingly, polycystin2/Pkd2 and polyductin/Pkhd1 proteins are coexpressed with HNF1β in biliary epithelial cells (Ong et al, 1999; Ward et al, 2003). HNF1β could drive the expression of these genes in both renal tubules and bile ducts. Notably, patients with ARPKD/PKHD1 suffer from ductal plate malformations giving rise to biliary dysgenesis, whereas patients with ADPKD/PKD2 develop biliary cysts (see Igarashi and Somlo, 2002). Although these patients do not strictly phenocopy the hepatic HNF1β deficiency, concomitant transcriptional defects affecting both Pkhd1 and Pkd2 genes could explain why IHBD fail to differentiate in HNF1_β_−/− livers.
Our study provides novel insights into the molecular mechanisms underlying HNF1β function in normal kidney and in cystic disorders. Our results demonstrate that HNF1β is a critical regulator of a genetic cascade that is essential for controlling the proliferation and differentiation of renal tubular epithelial cells.
Materials and methods
Animals
Kidney-specific inactivation of _HNF1_β was obtained using a Cre-LoxP strategy. KspCre (Shao et al), HNFsβlacz/+(Coffinier et al, 1999b) HNFsβflox/flox (Coffinier et al, 2002) and ROSA26R (Soriano) mice were previously described. All animals received humane care, and the institutional review committee approved the study protocol.
Biochemical analysis
Serum urea and creatinine levels were determined by the Centre d'Explorations Fonctionnelles Intégré (Faculté de Médecine X. Bichat).
Production of HNF1_α_ and β antibodies
We constructed GST fusion polypeptides of HNF1 α and β by excluding regions of high homology between both proteins using gene splicing by overlap extension PCR (SOE PCR) (Horton, 1995). All SOE PCRs were carried out with corresponding rat cDNAs. See Supplementary method 1 for more details.
Immunohistochemical procedures
Kidney cryosections were fixed in 4% PFA and treated as previously described (Foggensteiner et al, 2000) with the following antibodies: rabbit α-HNF1β-NC-AR, 1:150; rabbit α-NKCC2 (a gift from M Knepper), 1:1000; rabbit α-Pkd2 (a gift from R Sandford), 1:200; α-THP (Biogenesis), 1:50. Aqp2 (rabbit α-Aqp2 antibody (a gift from M Knepper), 1:400) and pkhd1 (mouse α-Pkhd1 antibody (a gift from C Ward), 1:200) were detected on paraffin sections according to Ward et al, (2003). X-gal stainings were performed as described (Pontoglio et al, 1996). For histological analysis, paraffin sections were stained with hematoxylin and eosin.
Quantitative RT–PCR
cDNAs were generated from total kidney RNAs using the Invitrogen Superscript II reagents. Quantitative RT–PCR was carried out on an ABI PRISM® 7000 sequence detection system using SYBR® green. Primers were designed using PrimerExpress 2.0® software (Applied Biosystems). β_-2 microglobulin_ was used as the control gene for normalization. All genes have been tested in P8 control (_N_=5) and mutant (_N_=3) mice. For primer sequences, see Supplementary Table II.
Chromatin immunoprecipitation (ChIP)
Nuclei were prepared from pooled kidneys (Cereghini et al, 1987). Chromatin preparation and ChIP were performed as described in Supplementary method 2. Three independent ChIP experiments were performed with α-HNF1α-NC-AR or α-HNF1β-NC-AR antibodies, and corresponding eluates were pooled. Primers were designed using PrimerExpress 2.0® software. For primer sequences, see Supplementary Table IB. Quantification of precipitated DNA fragments was carried out on an ABI PRISM® 7000 sequence detection system using SYBR® green in triplicate. Relative fold in vivo enrichment of DNA fragments was calculated using the following formula: (ChIPtarget/ChIPnormalizer)/(inputtarget/inputnormalizer). As normalizer, we used a DNA fragment lacking any HNF1 site, located in the first intron of aortic smooth muscle alpha-actin 2 gene.
Note added in proof
Supporting our results, HNF1β has been shown to regulate the activity of the pkhd1 promoter in cultured cells (Hiesberger T, Bai Y, ShaoX, McNally BT, Sinclair AM, Tian X, Somlo S, Igarashi P (2004) J Clin Invest (in press).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Table I
Supplementary Table II
Supplementary Method 1
Supplementary Method 2
Acknowledgments
We are grateful to F Bourgade and A Doyen for technical assistance and advice, to C Ward for the Pkhd1 antibody, to M Knepper for the Slc12a1 and Aqp2 antibodies and to R Sandford for the Pkd2 antibody. We thank F Terzi for helpful discussion and J Weitzman for critical reading of the manuscript. LG was supported by a fellowship from the French Ministry of Research and the Pierre et Marie Curie University. EF is a fellow of La Ligue Nationale Contre le Cancer. AR is supported by Boehringer Ingelheim Fonds. This work was supported by HFSP RGP0024/2001-M, La Ligue contre le Cancer, Genzyme Corp. and (NIH grant number R01 DK42921 and P50 DK57328) to PI.
References
- Bachmann S, Metzger R, Bunnemann B (1990) Tamm-Horsfall protein-mRNA synthesis is localized to the thick ascending limb of Henle's loop in rat kidney. Histochemistry 94: 517–523 [DOI] [PubMed] [Google Scholar]
- Barbacci E, Reber M, Ott MO, Breillat C, Huetz F, Cereghini S (1999) Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development 126: 4795–4805 [DOI] [PubMed] [Google Scholar]
- Bernstein J, Evan AP, Gardner KD Jr. (1987) Epithelial hyperplasia in human polycystic kidney diseases. Its role in pathogenesis and risk of neoplasia. Am J Pathol 129: 92–101 [PMC free article] [PubMed] [Google Scholar]
- Bhunia AK, Piontek K, Boletta A, Liu L, Qian F, Xu PN, Germino FJ, Germino GG (2002) PKD1 induces p21(waf1) and regulation of the cell cycle via direct activation of the JAK-STAT signaling pathway in a process requiring PKD2. Cell 109: 157–168 [DOI] [PubMed] [Google Scholar]
- Bingham C, Bulman MP, Ellard S, Allen LI, Lipkin GW, Hoff WG, Woolf AS, Rizzoni G, Novelli G, Nicholls AJ, Hattersley AT (2001) Mutations in the hepatocyte nuclear factor-1beta gene are associated with familial hypoplastic glomerulocystic kidney disease. Am J Hum Genet 68: 219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham C, Ellard S, Allen L, Bulman M, Shepherd M, FraylingT, Berry PJ, Clark PM, Lindner T, Bell GI, Ryffel GU, Nicholls AJ, Hattersley AT (2000) Abnormal nephron development associated with a frameshift mutation in the transcription factor hepatocyte nuclear factor-1 beta. Kidney Int 57: 898–907 [DOI] [PubMed] [Google Scholar]
- Blumenfeld M, Maury M, Chouard T, Yaniv M, Condamine H (1991) Hepatic nuclear factor 1 (HNF1) shows a wider distribution than products of its known target genes in developing mouse. Development 113: 589–599 [DOI] [PubMed] [Google Scholar]
- Bluteau O, Jeannot E, Bioulac-Sage P, Marques JM, Blanc JF, Bui H, Beaudoin JC, Franco D, Balabaud C, Laurent-Puig P, Zucman-Rossi J (2002) Bi-allelic inactivation of TCF1 in hepatic adenomas. Nat Genet 32: 312–315 [DOI] [PubMed] [Google Scholar]
- Bohn S, Thomas H, Turan G, Ellard S, Bingham C, Hattersley AT, Ryffel GU (2003) Distinct molecular and morphogenetic properties of mutations in the human HNF1beta gene that lead to defective kidney development. J Am Soc Nephrol 14: 2033–2041 [DOI] [PubMed] [Google Scholar]
- Cereghini S (1996) Liver-enriched transcription factors and hepatocyte differentiation. FASEB J 10: 267–282 [PubMed] [Google Scholar]
- Cereghini S, Raymondjean M, Carranca AG, Herbomel P, Yaniv M (1987) Factors involved in control of tissue-specific expression of albumin gene. Cell 50: 627–638 [DOI] [PubMed] [Google Scholar]
- Coffinier C, Barra J, Babinet C, Yaniv M (1999a) Expression of the vHNF1/HNF1beta homeoprotein gene during mouse organogenesis. Mech Dev 89: 211–213 [DOI] [PubMed] [Google Scholar]
- Coffinier C, Gresh L, Fiette L, Tronche F, Schutz G, Babinet C, Pontoglio M, Yaniv M, Barra J (2002) Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1beta. Development 129: 1829–1838 [DOI] [PubMed] [Google Scholar]
- Coffinier C, Thepot D, Babinet C, Yaniv M, Barra J (1999b) Essential role for the homeoprotein vHNF1/HNF1beta in visceral endoderm differentiation. Development 126: 4785–4794 [DOI] [PubMed] [Google Scholar]
- De Simone V, De Magistris L, Lazzaro D, Gerstner J, Monaci P, Nicosia A, Cortese R (1991) LFB3, a heterodimer-forming homeoprotein of the LFB1 family, is expressed in specialized epithelia. EMBO J 10: 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas P, Nomura H, Li X, Lakkis M, Luo Y, Segal Y, Fernandez-Fernandez JM, Harris P, Frischauf AM, Brown DA, Zhou J (2002) Constitutive activation of G-proteins by polycystin-1 is antagonized by polycystin-2. J Biol Chem 277: 11276–11283 [DOI] [PubMed] [Google Scholar]
- Delpire E, Kaplan MR, Plotkin MD, Hebert SC (1996) The Na-(K)-Cl cotransporter family in the mammalian kidney: molecular identification and function(s). Nephrol Dial Transplant 11: 1967–1973 [DOI] [PubMed] [Google Scholar]
- Foggensteiner L, Bevan AP, Thomas R, Coleman N, Boulter C, Bradley J, Ibraghimov-Beskrovnaya O, Klinger K, Sandford R (2000) Cellular and subcellular distribution of polycystin-2, the protein product of the PKD2 gene. J Am Soc Nephrol 11: 814–827 [DOI] [PubMed] [Google Scholar]
- Gattone VH, Wang X, Harris PC, Torres VE (2003) Inhibition of renal cystic disease development and progression by a vasopressin V2 receptor antagonist. Nat Med 9: 1323–1326 [DOI] [PubMed] [Google Scholar]
- Guo L, Schreiber TH, Weremowicz S, Morton CC, Lee C, Zhou J (2000) Identification and characterization of a novel polycystin family member, polycystin-L2, in mouse and human: sequence, expression, alternative splicing, and chromosomal localization. Genomics 64: 241–251 [DOI] [PubMed] [Google Scholar]
- Hart TC, Gorry MC, Hart PS, Woodard AS, Shihabi Z, Sandhu J, Shirts B, Xu L, Zhu H, Barmada MM, Bleyer AJ (2002) Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J Med Genet 39: 882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M (1996) Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96 [DOI] [PubMed] [Google Scholar]
- Hildebrandt F, Otto E, Rensing C, Nothwang HG, Vollmer M, Adolphs J, Hanusch H, Brandis M (1997) A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat Genet 17: 149–153 [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn BN, Lindner T, Yamagata K, Ogata M, Tomonaga O, Kuroki H, Kasahara T, Iwamoto Y, Bell GI (1997) Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nat Genet 17: 384–385 [DOI] [PubMed] [Google Scholar]
- Horton RM (1995) PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol Biotechnol 3: 93–99 [DOI] [PubMed] [Google Scholar]
- Hughes J, Ward CJ, Peral B, Aspinwall R, Clark K, San Millan JL, Gamble V, Harris PC (1995) The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat Genet 10: 151–160 [DOI] [PubMed] [Google Scholar]
- Igarashi P, Somlo S (2002) Genetics and pathogenesis of polycystic kidney disease. J Am Soc Nephrol 13: 2384–2398 [DOI] [PubMed] [Google Scholar]
- Lazzaro D, De Simone V, De Magistris L, Lehtonen E, Cortese R (1992) LFB1 and LFB3 homeoproteins are sequentially expressed during kidney development. Development 114: 469–479 [DOI] [PubMed] [Google Scholar]
- Lin F, Hiesberger T, Cordes K, SinclairAM, Goldstein LS, Somlo S, Igarashi P (2003) Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc Natl Acad Sci USA 100: 5286–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner TH, Njolstad PR, Horikawa Y, Bostad L, Bell GI, Sovik O (1999) A novel syndrome of diabetes mellitus, renal dysfunction and genital malformation associated with a partial deletion of the pseudo-POU domain of hepatocyte nuclear factor-1beta. Hum Mol Genet 8: 2001–2008 [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Wu G, Hayashi T, Xenophontos SL, Veldhuisen B, Saris JJ, Reynolds DM, Cai Y, Gabow PA, Pierides A, Kimberling WJ, Breuning MH, Deltas CC, Peters DJ, Somlo S (1996) PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272: 1339–1342 [DOI] [PubMed] [Google Scholar]
- Moyer JH, Lee-Tischler MJ, Kwon HY, Schrick JJ, Avner ED, Sweeney WE, Godfrey VL, Cacheiro NL, Wilkinson JE, Woychik RP (1994) Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science 264: 1329–1333 [DOI] [PubMed] [Google Scholar]
- Nauli SM, Alenghat FJ, LuoY, Williams E, Vassilev P, Li X, Elia AE, Lu W, Brown EM, Quinn SJ, Ingber DE, Zhou J (2003) Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet 33: 129–137 [DOI] [PubMed] [Google Scholar]
- Nishigori H, Yamada S, Kohama T, Tomura H, Sho K, Horikawa Y, Bell GI, Takeuchi T, Takeda J (1998) Frameshift mutation, A263fsinsGG, in the hepatocyte nuclear factor-1beta gene associated with diabetes and renal dysfunction. Diabetes 47: 1354–1355 [DOI] [PubMed] [Google Scholar]
- Olbrich H, Fliegauf M, Hoefele J, Kispert A, Otto E, Volz A, Wolf MT, Sasmaz G, Trauer U, Reinhardt R, Sudbrak R, Antignac C, Gretz N, Walz G, Schermer B, Benzing T, Hildebrandt F, Omran H (2003) Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat Genet 34: 455–459 [DOI] [PubMed] [Google Scholar]
- Ong AC, Ward CJ, Butler RJ, Biddolph S, Bowker C, Torra R, Pei Y, Harris PC (1999) Coordinate expression of the autosomal dominant polycystic kidney disease proteins, polycystin-2 and polycystin-1, in normal and cystic tissue. Am J Pathol 154: 1721–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R (1997) Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11: 205–214 [DOI] [PubMed] [Google Scholar]
- Otto E, Hoefele J, Ruf R, Mueller AM, Hiller KS, Wolf MT, Schuermann MJ, Becker A, Birkenhager R, Sudbrak R, Hennies HC, Nurnberg P, Hildebrandt F (2002) A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am J Hum Genet 71: 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto EA, Schermer B, Obara T, O'Toole JF, Hiller KS, Mueller AM, Ruf RG, Hoefele J, Beekmann F, Landau D, Foreman JW, Goodship JA, Strachan T, Kispert A, Wolf MT, Gagnadoux MF, Nivet H, Antignac C, Walz G, Drummond IA, Benzing T, Hildebrandt F (2003) Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left–right axis determination. Nat Genet 34: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG (2000) Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol 151: 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB (2002) Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr Biol 12: R378–380 [DOI] [PubMed] [Google Scholar]
- Pontoglio M, Barra J, Hadchouel M, Doyen A, Kress C, Bach JP, Babinet C, Yaniv M (1996) Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell 84: 575–585 [DOI] [PubMed] [Google Scholar]
- Rey-Campos J, Chouard T, Yaniv M, Cereghini S (1991) vHNF1 is a homeoprotein that activates transcription and forms heterodimers with HNF1. EMBO J 10: 1445–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Somlo S, Igarashi P (2002) Epithelial-specific Cre/lox recombination in the developing kidney and genitourinary tract. J Am Soc Nephrol 13: 1837–1846 [DOI] [PubMed] [Google Scholar]
- Soriano P (1999) Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21: 70–71 [DOI] [PubMed] [Google Scholar]
- Sun Z, Hopkins N (2001) vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev 15: 3217–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Ringeisen F, Blumenfeld M, Yaniv M, Pontoglio M (1997) Analysis of the distribution of binding sites for a tissue-specific transcription factor in the vertebrate genome. J Mol Biol 266: 231–245 [DOI] [PubMed] [Google Scholar]
- Vainio S, Lin Y (2002) Coordinating early kidney development: lessons from gene targeting. Nat Rev Genet 3: 533–543 [DOI] [PubMed] [Google Scholar]
- Ward CJ, Hogan MC, Rossetti S, Walker D, Sneddon T, Wang X, Kubly V, Cunningham JM, Bacallao R, Ishibashi M, Milliner DS, Torres VE, Harris PC (2002) The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat Genet 30: 259–269 [DOI] [PubMed] [Google Scholar]
- Ward CJ, Yuan D, Masyuk TV, Wang X, Punyashthiti R, Whelan S, Bacallao R, Torra R, LaRusso NF, Torres VE, Harris PC (2003) Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum Mol Genet 12: 2703–2710 [DOI] [PubMed] [Google Scholar]
- Watnick T, Germino G (2003) From cilia to cyst. Nat Genet 34: 355–356 [DOI] [PubMed] [Google Scholar]
- Watnick TJ, Torres VE, Gandolph MA, Qian F, Onuchic LF, Klinger KW, Landes G, Germino GG (1998) Somatic mutation in individual liver cysts supports a two-hit model of cystogenesis in autosomal dominant polycystic kidney disease. Mol Cell 2: 247–251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Table I
Supplementary Table II
Supplementary Method 1
Supplementary Method 2