Mutational heterogeneity in cancer and the search for new cancer genes (original) (raw)
. Author manuscript; available in PMC: 2014 Feb 10.
Published in final edited form as: Nature. 2013 Jun 16;499(7457):214–218. doi: 10.1038/nature12213
Abstract
Major international projects are now underway aimed at creating a comprehensive catalog of all genes responsible for the initiation and progression of cancer. These studies involve sequencing of matched tumor–normal samples followed by mathematical analysis to identify those genes in which mutations occur more frequently than expected by random chance. Here, we describe a fundamental problem with cancer genome studies: as the sample size increases, the list of putatively significant genes produced by current analytical methods burgeons into the hundreds. The list includes many implausible genes (such as those encoding olfactory receptors and the muscle protein titin), suggesting extensive false positive findings that overshadow true driver events. Here, we show that this problem stems largely from mutational heterogeneity and provide a novel analytical methodology, MutSigCV, for resolving the problem. We apply MutSigCV to exome sequences from 3,083 tumor-normal pairs and discover extraordinary variation in (i) mutation frequency and spectrum within cancer types, which shed light on mutational processes and disease etiology, and (ii) mutation frequency across the genome, which is strongly correlated with DNA replication timing and also with transcriptional activity. By incorporating mutational heterogeneity into the analyses, MutSigCV is able to eliminate most of the apparent artefactual findings and allow true cancer genes to rise to attention.
Recent cancer genome studies have led to the identification of scores of cancer genes, in glioblastoma1, ovarian2, colorectal3, lung4, head-and-neck5, multiple myeloma6, chronic lymphocytic leukemia7, diffuse large B-cell lymphoma8,9, and many other cancers. Studies are now underway through The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) and the International Cancer Genome Consortium (ICGC) (http://www.icgc.org/) to create a comprehensive catalog of significantly mutated genes across all major cancer types.
The expectation has been that larger sample sizes will increase the power both to detect true cancer driver genes (sensitivity) and to distinguish them from the background of random mutations (specificity). Alarmingly, recent results appear to show the opposite phenomenon: with large sample sizes, the list of apparently significant cancer genes grew rapidly and implausibly. For example, when we applied current analytical methods to whole-exome sequence data from 178 tumor-normal pairs of lung squamous cell carcinoma10, a total of 450 genes (Supplementary Table S1, Supplementary Method S2) were found to be mutated at a significant frequency (false-discovery rate q < 0.1). While the list contains some genes known to be associated with cancer, many of the genes seem highly suspicious based on their biological function or genomic properties. Almost a quarter (101/450) of the putative significant genes encode olfactory receptors. The list is also highly enriched for genes encoding extremely large proteins, including more than one-fifth of the 83 genes encoding proteins with >4,000 amino acids (p<10−11, Fisher’s exact test). These include the two longest human proteins, the muscle protein titin (36,800 amino acids) and the membrane-associated mucin _MUC16_ (14,500 amino acids), as well as another mucin (_MUC4_), cardiac ryanodine receptors (_RYR2_, _RYR3_), cytoskeletal dyneins (_DNAH5_, _DNAH11_), and the neuronal synaptic vesicle protein piccolo (_PCLO_). The prominence of these genes is not simply the consequence of their long coding regions, because the statistical tests already account for the larger target size. Furthermore, the list also contains genes with very long introns, including one-sixth of the 73 genes spanning a genomic region of >1Mb (p<10−6), such as those encoding cub- and-sushi-domain proteins (CSMD1, CSMD3), and many neuronal proteins, such as the neurexins NRXN1, NRXN4 (CNTNAP2), CNTNAP4, and CNTNAP5, the neural adhesion molecule CNTN5, and the Parkinson protein PARK2. When we performed similar analyses for several other cancer types with many samples, we similarly obtained large lists including many of the same genes (data not shown).
After recognizing the problem of apparent false-positive findings, we reviewed the published literature and found that some of these potentially spurious genes have already cropped up in recently published cancer genome studies, for example: LRP1B in glioblastoma (GBM)2 and lung adenocarcinoma1,4; CSMD3 in ovarian cancer2; PCLO in diffuse large B-cell lymphoma (DLBCL)9; MUC16 in lung squamous carcinoma11, breast cancer12 and DLBCL8; MUC4 in melanoma13; olfactory receptor OR2L13 in GBM14; and TTN in breast cancer12 and other tumor types15. We therefore set out to understand the source of the problem.
Analytical approaches in wide use today1-9,13-16 identify as significantly mutated those genes harboring more mutations than expected given the average background mutation frequency for the cancer type. These methods employ a handful of parameters: an average overall mutation frequency for a cancer type and a few parameters about the relative frequencies of different categories of mutations (small insertions/deletions and transitions vs. transversions at CpG dinucleotides, other C:G basepairs and A:T basepairs). Average values of these parameters are typically estimated from the samples under study. Various efforts, by us and others, have recently began to incorporate sample-specific mutation rates into the analysis.3,9
We hypothesized that the problem might be due to heterogeneity in the mutational processes in cancer. While it is obvious that assuming an average mutation frequency that is too low will lead to spuriously significant findings, it is less well appreciated that using the correct average rate but failing to account for heterogeneity in the mutational process can also wreak havoc. To illustrate this point, we compared two simple scenarios both sharing the same average mutation frequency: (a) constant frequency of 10 mutations per megabase (10/Mb) across all genes, versus (b) frequencies of 4/Mb, 8/Mb and 20/Mb in 25%, 50% and 25% of genes, respectively (Supplementary Figure S1). If one analyzes the second case under the erroneous assumption of a constant rate, many of the highly mutable genes will falsely be declared to be cancer genes. Notably, the problem grows with sample size: because the threshold for statistical significance decreases with sample size, modest deviations due to an erroneous model are declared significant. For the same reason, the problem is also more pronounced in tumor types with higher mutation rates. Heterogeneity in mutation frequencies across patients can also lead to inaccurate results, including the potential to produce both false-positive, as described above, and false-negative results if the baseline frequency is overestimated.
We therefore set out to study heterogeneity in mutation rates, in a data set of 3,083 tumor/normal pairs across 27 tumor types, with 2,957 having whole-exome sequence and 126 having whole-genome sequence (Supplementary Table S2). Approximately 92% of the samples were sequenced at the Broad Institute and thus were processed using a uniform experimental and analytical pipeline (see Methods). In this data set, an average of 30 Mb of coding sequence per sample was covered to adequate depth for mutation detection, yielding a total of 373,909 nonsilent coding mutations or an average of 4.0/Mb per sample (median of 44 nonsilent coding mutations per sample, or 1.5/Mb).
We analyzed three types of heterogeneity, with the aim of achieving more accurate detection of cancer genes.
(i) Heterogeneity across patients with a given cancer type
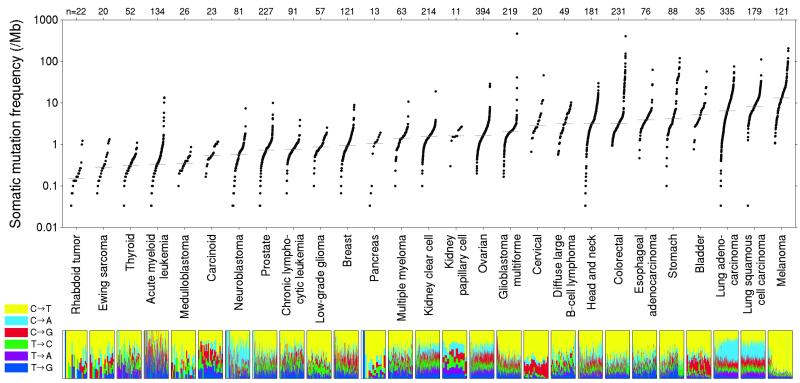
Analysis of the 27 cancer types revealed that the median frequency of non-synonymous mutations varied by more than 1000-fold across cancer types (Figure 1). About half of the variation in mutation frequencies (measured on a logarithmic scale) can be explained by tissue type of origin. Pediatric cancers showed frequencies as low as 0.1/Mb (approximately one change across the entire exome), while at the opposite extreme, melanoma and lung cancer exceeded 100/Mb. The high mutation frequencies are in some cases attributable to extensive exposure to well known carcinogens, such as UV radiation in the case of melanoma and tobacco smoke in the case of lung cancers.
Figure 1.
Somatic mutation frequencies observed in exomes from 3,083 tumor-normal pairs. Each dot corresponds to a tumor-normal pair, with vertical position indicating the total frequency of somatic mutations in the exome. Tumor types are ordered by their median somatic mutation frequency, with the lowest frequencies (left) found in hematological and pediatric tumors, and the highest (right) in tumors induced by carcinogens such as tobacco smoke and UV light. Mutation frequencies vary more than 1000-fold between lowest and highest mutation rates across cancer and also within several tumor types. The lower panel shows the relative proportions of the six different possible base-pair substitutions, as indicated in the legend on the left. (See also Supplementary Table S2.)
More surprisingly, mutation frequencies varied dramatically across patients within a cancer type. In melanoma and lung cancer, the frequency ranged across 0.1 - 100/Mb. Despite the low median frequency in AML (0.37/Mb), the patient-specific frequencies similarly spanned three orders of magnitude 0.01 - 10/Mb. Variation may in some cases be due to key biological factors, such as melanomas not attributed to UV exposure or on unexposed skin, colon cancers with or without mismatch repair defects3, or head and neck tumors with viral or non-viral origin5 (Supplementary Figure S2).
(ii) Heterogeneity in mutational spectrum
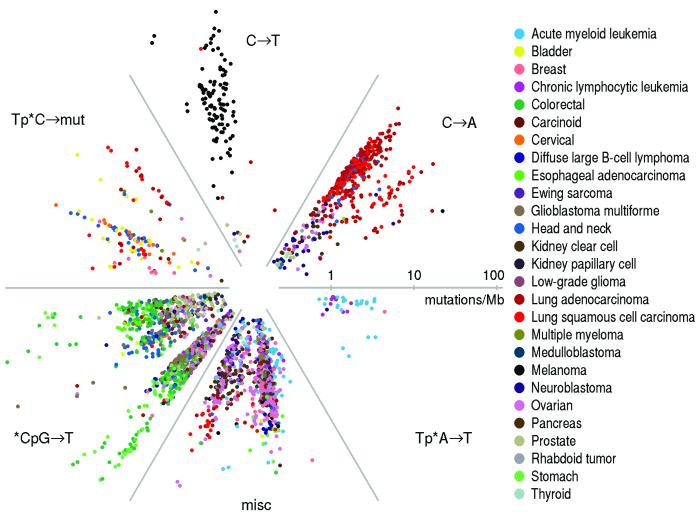
In addition to total mutation frequency, we examined the mutational spectrum in each tumor. Starting with all 96 possible mutations (12 mutations at a base times 16 possible flanking bases then collapsed by strand symmetry), we used non-negative matrix factorization to reduce the dimensionality, with each spectrum represented as a linear combination of six basic spectra (Methods). We represented the mutational spectrum of each tumor on a circular plot, with distance from the origin representing total mutation rate and angle representing the relative contribution of the six basic spectra (Figure 2). This representation reveals natural groupings with respect to mutational spectrum.
Figure 2.
Radial spectrum plot of the 2,892 tumor samples having at least 10 coding mutations. The angular space is compartmentalized into the six different factors discovered by NMF (see Methods). The distance from the center represents the total mutation frequency. Different tumor types segregate into different compartments based on their mutation spectra. Notable examples are: lung adenocarcinoma and lung squamous carcinoma (red; 2 o’clock position), melanoma (black; 12 o’clock position), stomach, esophageal and colorectal cancer (various shades of green; 8 o’clock position), samples harboring mutations of the HPV or APOBEC signature (bladder, cervical and head and neck cancer, marked in yellow, orange, and blue respectively; 10 o’clock position), and AML and CLL samples sharing the Tp*A→T signature, 4 o’clock position. (See also Supplementary Table S3.)
Lung cancers, for example, (red cluster at 2 o’clock position), share a mutational spectrum dominated by C→A mutations, consistent with their exposure to the polycyclic aromatic hydrocarbons in tobacco smoke17. Melanoma (black cluster at 12 o’clock) shows a distinct pattern reflecting the frequent C→T mutations caused by misrepair of UV-induced covalent bonds between adjacent pyrimidines18. Gastrointenstinal tumors (esophageal, colororectal, and gastric, corresponding to green cluster at 8 o’clock) show extremely high frequencies of transition mutations at CpG dinucleotides, which may reflect higher methylation levels in these tumor types3.
Interestingly, there is a multifarious cluster at the 10 o’clock position corresponding to cervical, head-and-neck, and bladder tumors, all sharing frequent mutations at C’s in the context TpC that change the C to either T or G or (less often) A. This pattern is characteristic of mutations caused by the APOBEC family of cytidine deaminases, innate immunity enzymes restricting propagation of retroviruses and retrotransposons19,20. Some APOBECs can be induced by certain classes of viruses21. Cervical cancer is known to be caused in over 90% of cases by the human papillomavirus (HPV)22. Recent studies have also implicated HPV in head-and-neck cancers5. The similar mutational spectrum in bladder cancer may indicate a viral etiology in a significant subset of this tumor type; a potential role of HPV in bladder cancer is a subject of active investigation23. This cluster also contains sporadic examples of breast tumors (consistent with a recent report12), as well as some tumors from lung and other tissues. Recent work19,20 has shown that the TpC mutations tend to occur in proximity to one another, consistent with the activity of APOBEC enzymes in damaged long single-strand DNA regions. One last minor cluster (4 o’clock position) consists of samples dominated by A→T mutations in the context TpA. This cluster contains mostly leukemia samples (AML and CLL), as well as one breast sample and one neuroblastoma sample.
In summary, the rich variation in mutational spectrum across tumors underscores the problems with using an overly simplistic model of the average mutational process for a tumor type and failing to account for heterogeneity within a tumor type.
(iii) Heterogeneity across the genome
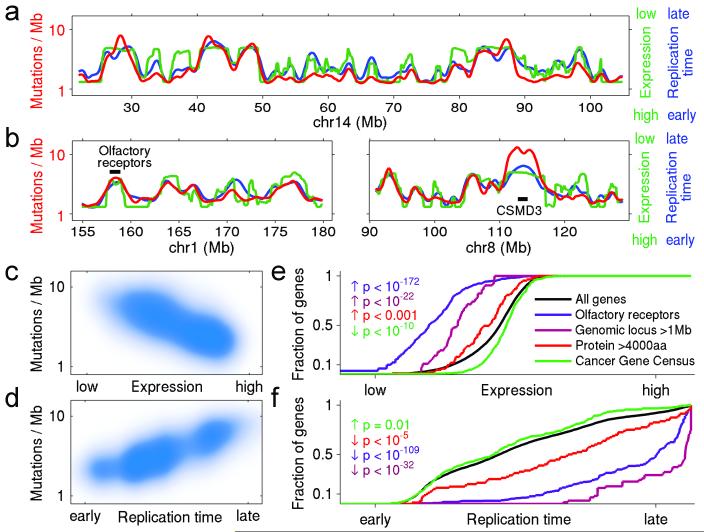
Of all the kinds of heterogeneity in mutational processes, the most important effect turns out to be regional heterogeneity across the genome. By examining whole-genome sequence from 126 tumor-normal pairs across ten tumor types, we found striking variation in mutation frequency across the genome, with differences exceeding 5-fold (Figure 3a,b); the profile of the genomic variation was similar across and within tumor types (Figure S3). Recent studies have noted regional variation in cancer mutation rates and begun to explore correlations with genomic features6,17,18,24.
Figure 3.
Mutation rate varies widely across the genome and correlates with DNA replication time and expression level. (a,b) Mutation rate, replication time, and expression level plotted across selected regions of the genome. Red shows total noncoding mutation rate calculated from whole-genome sequences of 126 samples (excluding exons). Blue shows replication time27. Green shows average expression level across 91 cell lines in the Cancer Cell Line Encyclopedia (CCLE), determined by RNA sequencing. (Note that low expression is at the top of the scale and high expression at the bottom, in order to emphasize the mutual correlations with the other variables). Shown are (a) entire chromosome 14 and (b) portions of chromosomes 1 and 8, with the locations of two specific loci: a cluster of 16 olfactory receptors on chr1 and the gene CSMD3 on chr8. These two loci have very high mutation rates, late replication times, and low expression levels. (The local mutation rate at CSMD3 is even higher than predicted from replication time and expression, suggesting contributions from additional factors, perhaps locally increased DNA breakage: the locus is a known fragile site). (c,d) Correlation of mutation rate with expression level and replication time, for all 100 Kb windows across the genome. (e,f) Cumulative distribution of various gene families as a function of expression level and replication time. Olfactory receptor genes, genes encoding long proteins (>4,000aa) and genes spanning large genomic loci (>1Mb) are significantly enriched towards lower expression and later replication. In contrast, known cancer genes (as listed in the Cancer Gene Census) trend toward slightly higher expression and earlier replication. (See also Supplementary Figure S9 and Supplementary Tables S4, S5, S6.)
We focused on two factors that were especially powerful in explaining mutational heterogeneity. The first factor is gene expression level. It is known that the germline mutation rate is somewhat lower in genes that are highly expressed in the germline18, due to a process termed transcription-coupled repair25. With the whole-genome and whole-exome data analyzed here, we found a strong correlation between somatic mutation frequency in cancers and gene expression level (averaged across many cell lines, with similar results for expression in matched normal tissue) (Figure 3a,b; Supplementary Figure S3; Supplementary Tables S4, S5). The average mutation rate is ~2.9-fold higher than the bottom percentile than in the top percentile. While statistically highly significant, this effect is insufficient to fully explain regional variation in mutation levels. The second important factor is the replication time of a DNA region during the cell cycle. Recent studies have reported that germline mutation rates are correlated with DNA replication time26-28: late-replicating regions have much higher mutation rates, possibly due to depletion of the pool of free nucleotides26. With the whole-genome and whole-exome data here, we see a striking correlation between somatic mutation frequency in cancers and DNA replication timing (as measured in HeLa cells27) (Figure 3a,b), with similar results for blood cell lines28 (Figure S3). The average mutation rate is ~2.9-fold higher in the latest- versus earliest-replicating percentile, and ~2.1-fold difference between the latest- and earliest-replicating decile.
These two features explain most of the suspicious entries on the putative cancer gene lists. Olfactory receptor genes, for example, have low expression (p<10−172, Kolmogorov-Smirnoff test, Figure 3e), are strikingly late in replication timing (p<10−109, Figure 3f), and show a high regional noncoding mutation rate (p<10−81), which accounts for the high frequency of somatic mutations in their coding regions. Large genes are similarly low-expressed and late-replicating (Figure 3e,f), including the genes cited in the lung cancer example above, such as titin and the ryanodine receptors. Importantly, these results undermine the evidence supporting several recent reports – such as the suggestion that CSMD3 is a cancer gene in ovarian cancer2. As an independent test, we confirmed that these two genomic features correlated strongly with the overall frequency of silent substitutions in coding regions and mutations in introns (Figure 3c,d; Supplementary Table S6). We note, however, that silent substitutions alone provide inadequate data to correct mutation frequencies on a gene-by-gene basis in most tumor types and for most genes, due to the sparsity of the data and the resulting uncertainty in estimated rates.
Using the observations above, we developed a new integrated approach to identify significantly mutated genes in cancer. The method (MutSigCV) corrects for variation by employing (i) patient-specific mutation frequency and spectrum, and (ii) gene-specific background mutation rates incorporating expression level and replication time (Supplementary Methods 3). MutSigCV is freely available for noncommercial use (http://www.broadinstitute.org/cancer/cga/mutsig).
When we applied MutSigCV to the lung cancer example above, the list of significantly mutated genes shrank from 450 to 11 genes. Most of the genes in this shorter list have been previously reported to be mutated in squamous cell lung cancer (TP53, KEAP1, NFE2L2, CDKN2A, PIK3CA, PTEN, RB111,16) or other tumor types (MLL2, NOTCH1, FBXW7). An additional novel gene in the list, HLA-A,suggests that mutations in immune-related genes may help tumors evade immune surveillance, a finding that requires follow-up experimental work. These significantly mutated genes are discussed in the TCGA lung squamous publication10, in which we applied our novel methodology.
With the ability to eliminate many obviously suspicious genes, it is now feasible to start analyzing large cancer collections, including combined data sets across many cancer types.
We note that other forms of heterogeneity in tumors merit further investigation. These include the co-occurrence of many mutations in proximity to each other (“kataegis”19 or “clustered mutations”20) (see Supplementary Figure S10) and transcription-coupled repair (see Supplementary Figure S11). In addition, heterogeneity across cancer cells within a tumor, reflecting the evolutionary process of a tumor, will be crucial to fully understand.29
Our results make clear that the accurate identification of new cancer genes will require accurate accounting of mutational processes. While MutSigCV resolves the most serious current problems, the ultimate solution will likely involve using empirically observed local mutation rates obtained from massive amounts of whole-genome sequencing.
Methods Summary
All samples were obtained under institutional IRB approval and with documented informed consent. A complete list of samples is given in Table S2. Whole-exome capture libraries were constructed and sequenced on Illumina HiSeq flowcells to average coverage of 118x. Whole-genome sequencing was done with the Illumina GA-II or Illumina HiSeq sequencer, achieving an average of ~30X coverage depth. Reads were aligned to the reference human genome build hg19 using an implementation of the Burrows-Wheeler Aligner, and a BAM file was produced for each tumor and normal sample using the Picard pipeline6. The Firehose pipeline was used to manage input and output files and submit analyses for execution. The MuTect30 and Indelocator (Sivachenko, A. et al., manuscript in preparation) algorithms were used to identify somatic single-nucleotide variants (SSNVs) and short somatic insertions and deletions, respectively. Mutation spectra were analyzed using non-negative matrix factorization (NMF). Significantly mutated genes were identified using MutSigCV, which estimates the background mutation rate (BMR) for each gene-patient-category combination based on the observed silent mutations in the gene and noncoding mutations in the surrounding regions. Because in most cases these data are too sparse to obtain accurate estimates, we increased accuracy by pooling data from other genes with similar properties (e.g. replication time, expression level). Significance levels (p-values) were determined by testing whether the observed mutations in a gene significantly exceed the expected counts based on the background model. False Discovery Rates (q-values) were then calculated, and genes with q≤0.1 were reported as significantly mutated. Full methods details are listed in Supplementary Information.
Supplementary Material
1
2
3
Acknowledgements
This work was conducted as part of The Cancer Genome Atlas (TCGA), a project of the National Cancer Institute (NCI) and National Human Genome Research Institute (NHGRI). This work was conducted as part of the Slim Initiative for Genomic Medicine (SIGMA), a joint U.S.-Mexico project founded by the Carlos Slim Health Institute. Support to DAG and SAR was through the Intramural Research Program of the NIEHS (NIH, DHHS) project ES065073 (PI Michael Resnick).
Footnotes
To whom correspondence should be addressed.
Author Contributions GG, ESL, SS, DAG, TRG, MM, LAG, AJB, KS, JAB, CWMR, SBG, CJW, SAM, JMZ and AHM conceived the project and provided leadership. CSo, LA, EN, ES, MLC, DA, WW, and KA provided project management. WW, KA, TF, RO, and MP planned and carried out DNA sequencing and genetic analysis. TF, DV, GS, MN, DD, PL, LL, and RJ developed and engineered software to support the project. MSL, PS, PP, GVK, KC, AS, SLC, CSt, CHM, SAR, AKi, PSH, AM, YD, LZ, AHR, TJP, NS, EH, JK, MI, BH, EH, SB, AMD, JL, DAL, CJW, JMZ, AHM, AKo, SAM, JM, BC, AJB, and DAG analyzed the data and contributed to scientific discussions. MSL, PS, PP, ESL, and GG wrote the paper.
Declaration of competing financial interests A patent application has been filed relating to this work.
References
- 1.TCGA Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.TCGA Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.TCGA Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature. 2012 doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–84. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.TCGA Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012 doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata T, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105:13568–73. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–73. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 17.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–90. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nik-Zainal S, et al. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts SA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–35. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vartanian JP, Guetard D, Henry M, Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–3. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- 22.Walboomers JM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–9. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 23.Jimenez-Pacheco A, Exposito-Ruiz M, Arrabal-Polo MA, Lopez-Luque AJ. Meta-analysis of studies analyzing the role of human papillomavirus in the development of bladder carcinoma. Korean J Urol. 2012;53:240–7. doi: 10.4111/kju.2012.53.4.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgkinson A, Eyre-Walker A. Variation in the mutation rate across mammalian genomes. Nat Rev Genet. 2011;12:756–66. doi: 10.1038/nrg3098. [DOI] [PubMed] [Google Scholar]
- 25.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 26.Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–5. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CL, et al. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res. 2010;20:447–57. doi: 10.1101/gr.098947.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koren A, et al. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am J Hum Genet. 2012;91:1033–40. doi: 10.1016/j.ajhg.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landau DA, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–26. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013 doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3