Comprehensive genomic characterization defines human glioblastoma genes and core pathways (original) (raw)
Main
Cancer is a disease of genome alterations: DNA sequence changes, copy number aberrations, chromosomal rearrangements and modification in DNA methylation together drive the development and progression of human malignancies. With the complete sequencing of the human genome and continuing improvement of high-throughput genomic technologies, it is now feasible to contemplate comprehensive surveys of human cancer genomes. The Cancer Genome Atlas (TCGA) aims to catalogue and discover major cancer-causing genome alterations in large cohorts of human tumours through integrated multi-dimensional analyses.
The first cancer studied by TCGA is glioblastoma, the most common primary brain tumour in adults1. Primary glioblastoma, which comprises more than 90% of biopsied or resected cases, arises de novo without antecedent history of low-grade disease, whereas secondary glioblastoma progresses from previously diagnosed low-grade gliomas1. Patients with newly diagnosed glioblastoma have a median survival of approximately 1 year with generally poor responses to all therapeutic modalities2. Two decades of molecular studies have identified important genetic events in human glioblastomas, including the following: (1) dysregulation of growth factor signalling via amplification and mutational activation of receptor tyrosine kinase (RTK) genes; (2) activation of the phosphatidylinositol-3-OH kinase (PI(3)K) pathway; and (3) inactivation of the p53 and retinoblastoma tumour suppressor pathways1. Recent genome-wide profiling studies have also shown remarkable genomic heterogeneity among glioblastoma and the existence of molecular subclasses within glioblastoma that may, when fully defined, allow stratification of treatment3,4,5,6,7,8. Albeit fragmentary, such baseline knowledge of glioblastoma genetics sets the stage to explore whether novel insights can be gained from a more systematic examination of the glioblastoma genome.
As a public resource, all TCGA data are deposited at the Data Coordinating Center (DCC) for public access (http://cancergenome.nih.gov/). TCGA data are classified by data type (for example, clinical, mutations, gene expression) and data level to allow structured access to this resource with appropriate patient privacy protection. An overview of the data organization is provided in the Supplementary Methods, and a detailed description is available in the TCGA Data Primer (http://tcga-data.nci.nih.gov/docs/TCGA_Data_Primer.pdf).
Biospecimen collection
Retrospective biospecimen repositories were screened for newly diagnosed glioblastoma based on surgical pathology reports and clinical records (Supplementary Fig. 1). Samples were further selected for having matched peripheral blood as well as associated demographic, clinical and pathological data (Supplementary Table 1). Corresponding frozen tissues were reviewed at the Biospecimen Core Resource (BCR) to ensure a minimum of 80% tumour nuclei and a maximum of 50% necrosis (Supplementary Fig. 1). DNA and RNA extracted from qualified biospecimens were subjected to additional quality control measurements (Supplementary Methods) before distribution to TCGA centres for analyses (Supplementary Fig. 2).
After exclusion based on insufficient tumour content (n = 234) and suboptimal nucleic acid quality or quantity (n = 147), 206 of the 587 biospecimens screened (35%) were qualified for copy number, expression and DNA methylation analyses. Of these, 143 cases had matched normal peripheral blood DNAs and were therefore appropriate for re-sequencing. This cohort also included 21 post-treatment glioblastoma cases used for exploratory comparisons (Supplementary Table 1). Although it is possible that a small number of progressive secondary glioblastomas were among the remaining 185 cases of newly diagnosed glioblastomas, this cohort represents predominantly primary glioblastoma. Indeed, when compared with published cohorts, overall survival of the newly diagnosed glioblastoma cases in TCGA is similar to that reported in the literature (Supplementary Fig. 3, P = 0.2)9,10,11,12.
Genomic and transcriptional aberrations
Genomic copy number alterations (CNAs) were measured on three microarray platforms (Supplementary Methods) and analysed with multiple analytical algorithms13,14,15 (Supplementary Fig. 4 and Supplementary Tables 2–4). In addition to the well-known alterations3,13,14, we detected significantly recurrent focal alterations not previously reported in glioblastomas, such as homozygous deletions involving NF1 and PARK2, and amplifications of AKT3 (Fig. 1a and Supplementary Tables 2–4). Search for informative but infrequent CNAs also uncovered rare focal events, such as amplifications of FGFR2 and IRS2, and deletion of PTPRD (Supplementary Table 4). Abundance of protein-coding genes and non-coding microRNA was also measured by transcript-specific and exon-specific probes on multiple platforms (Supplementary Methods). The resulting integrated gene expression data set showed that ∼76% of genes within recurrent CNAs have expression patterns that correlate with copy number (Supplementary Table 2). In addition, single-nucleotide-polymorphism (SNP)-based analyses also catalogued copy-neutral loss of heterozygosity (LOH), with the most significant region being 17p, which contains TP53 (Supplementary Methods).
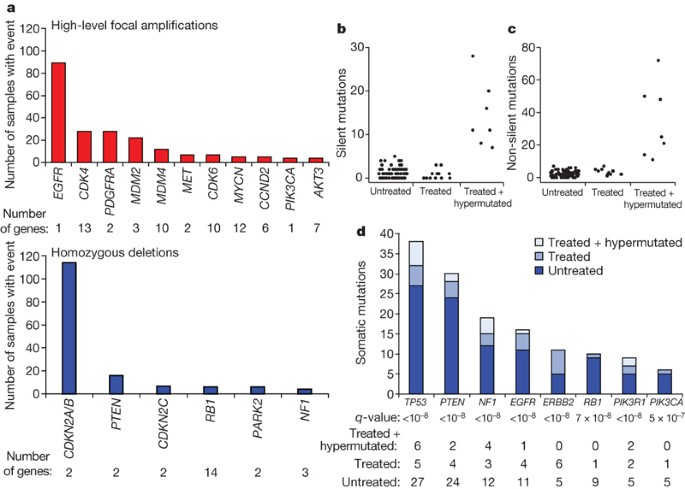
Figure 1: Significant copy number aberrations and pattern of somatic mutations.
a, Frequency and significance of focal high-level CNAs. Known and putative target genes are listed for each significant CNA, with ‘Number of genes’ denoting the total number of genes within each focal CNA boundary. b, c, Distribution of the number of silent (b) and non-silent (c) mutations across the 91 glioblastoma samples separated according to their treatment status, showing hypermutation in 7 out of the 19 treated samples. d, Significantly mutated genes in 91 glioblastomas. The eight genes attaining a false discovery rate <0.1 are displayed here. Somatic mutations occurring in untreated samples are in dark blue; those found in statistically non-hypermutated and hypermutated samples among the treated cohort are in respectively lighter shades of blue.
Patterns of somatic nucleotide alterations in glioblastoma
A total of 91 matched tumour–normal pairs (72 untreated and 19 treated cases) were selected from the 143 cases for detection of somatic mutations in 601 selected genes (Supplementary Table 5). The resulting sequences, totalling 97 million base pairs (1.1 ± 0.1 million bases per sample), uncovered 453 validated non-silent somatic mutations in 223 unique genes, 79 of which contained two or more events (Supplementary Table 6; see also http://tcga-data.nci.nih.gov/docs/somatic_mutations/tcga_mutations.htm). The background mutation rates differed markedly between untreated and treated glioblastomas, averaging 1.4 versus 5.8 somatic silent mutations per sample (98 among 72 untreated versus 111 among 19 treated, P < 10-21), respectively. This difference was predominantly driven by seven hypermutated samples, as determined by frequencies of both silent and non-silent mutations (Fig. 1b, c). Four of the seven hypermutated tumours were from patients previously treated with temozolomide and three were from patients treated with CCNU (lomustine) alone or in combination (Supplementary Table 1b). A hypermutator phenotype in glioblastoma has been described in three glioblastoma specimens with MSH6 mutations16,17, prompting us to perform a systematic analysis of the genes involved in mismatch repair (MMR). Indeed, six of the seven hypermutated samples harboured mutations in at least one of the MMR genes MLH1, MSH2, MSH6, or PMS2, as compared with only one sample among the eighty-four non-hypermutated samples (P = 7 × 10-8), suggesting a role of decreased DNA repair competency in these highly mutated samples derived from treated patients.
By applying a statistical analysis of mutation significance18, we identified eight genes as significantly mutated (false discovery rate <10-3) (Fig. 2d and Supplementary Table 6). Interestingly, 27 TP53 mutations were detected in the 72 untreated glioblastomas (37.5%) and 11 mutations in the 19 treated samples (58%). All of those mutations clustered in the DNA binding domain, a well-known hotspot for p53 mutations in human cancers (Supplementary Fig. 5 and Supplementary Table 6). Given the predominance of primary glioblastoma among this newly diagnosed collection, that result unequivocally proves that p53 mutation is a common event in primary glioblastoma.
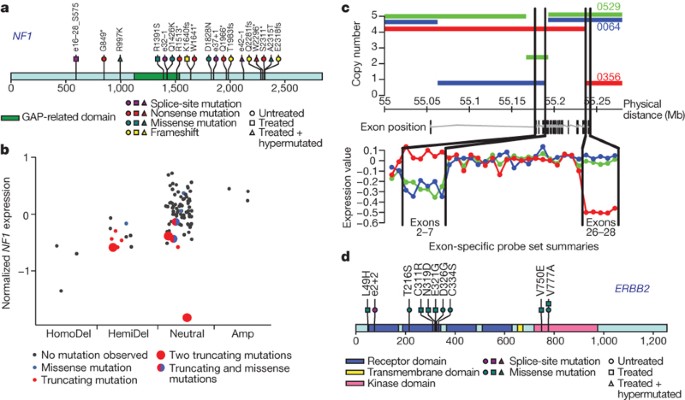
Figure 2: Mutations in NF1 tumour suppressor gene and EGFR family members.
a, NF1 somatic mutations in 91 glioblastoma tumours. Both missense mutations and truncating nonsense, frameshift and splice site mutations were observed. Splice positions are given in number of bases to the closest exon (e#) numbered according to the NF1 reference transcript in the Human Gene Mutation Database; positive indicates 3′ of exon, negative indicates 5′ of exon. Asterisk indicates a stop codon. fs, frameshift. b, Correlation of copy number and mutation status at the NF1 locus with level of expression (y axis). Mutation events predicted to result in fewer expressed copies (including deletion, nonsense, splice site and frameshift mutations) generally have lower observed expression. HomoDel, homozygous deletion; HemiDel, single-copy loss; Neutral, no change in copy number (presumed diploid); Amp, increased copy number. Copy number status of the NF1 locus in each sample was determined as described in the Supplementary Information. c, DNA copy number and mRNA expression profiles for TCGA samples TCGA-08-0356 (red), TCGA-02-0064 (blue) and TCGA-02-0529 (green) at the EGFR locus. The upper panel shows the segmented DNA copy number (based on Affymetrix SNP6.0 data) versus genomic coordinates on chromosome 7. The lower panel shows relative exon expression levels across the known EGFR exons from the Affymetrix Exon array ordered by genomic position, where relative expression is the median-centred difference in exon intensity and gene intensity. The EGFR gene model lies between the two plots. Black lines map the genomic positions of exons 2 through to 7 and 26 through to 28. Note that structural deletions cause the relatively lower expression of exons 2–7 in the green and blue samples and exons 26–28 in the red sample. d, ERBB2 somatic mutations in 91 glioblastoma tumours. Mutations cluster in the extracellular domain in both genes. Splice site mutation position is given in number of bases to the closest exon (e#); positive indicates 3′ of exon.
NF1 is a human glioblastoma suppressor gene
Although somatic mutations in NF1 have been reported in a small series of human glioblastoma tumours19, their role remains controversial20, despite strong genetic data in mouse model systems20,21,22. Here, 19 NF1 somatic mutations were identified in 13 samples (14% of 91), including 6 nonsense mutations, 4 splice site mutations, 5 missense changes and 4 frameshift insertions/deletions (indels) (Fig. 2a). Five of these mutations—R1391S (ref. 23), R1513* (ref. 24), e25 -1 and e29 +1 (ref. 25), and Q1966* (ref. 26)—have been reported as germline alterations in neurofibromatosis patients, and thus are probably inactivating. In addition, 30 heterozygous deletions in NF1 were observed among the entire interim sample set of 206 cases, 6 of which also harbour point mutation (Supplementary Tables 8 and 9). Some samples also exhibited loss of expression without evidence of genomic alteration (Fig. 2b). Overall, at least 47 of these 206 patient samples (23%) harboured somatic NF1 inactivating mutations or deletions, definitively addressing _NF1_’s relevance to sporadic human glioblastoma.
Prevalence of EGFR family activation
EGFR is frequently activated in primary glioblastomas. Variant III deletion of the extracellular domain (‘vIII mutant’)27 has been the most commonly described event, in addition to extracellular domain point mutations and cytoplasmic domain deletions28,29. Here, high-resolution genomic and exon-specific transcriptomic profiling readily detected vIII and carboxy-terminal deletions with correspondingly altered transcripts (Fig. 2c). Among the 91 glioblastoma cases with somatic mutation data, 22 harboured focal amplification of wild-type EGFR with no point mutation, 16 had point mutations in addition to focal amplification, and 3 had EGFR point mutations but no amplification (Supplementary Fig. 6 and Supplementary Table 9). Collectively, EGFR alterations were observed in 41 of the 91 sequenced samples.
ERBB2 mutation has previously been reported in only one glioblastoma tumour30. In the TCGA cohort, 11 somatic ERBB2 mutations in 7 of 91 samples were validated, including 3 in the kinase domain and 2 involving V777A, a site of recurrent missense and in-frame insertion mutations in lung, gastric and colon cancers31. The remaining eight mutations (including seven missense and one splice-site mutation) occurred in the extracellular domain of the protein, similar to somatic EGFR substitutions in glioblastoma (Fig. 2d). Unlike in breast cancers, focal amplifications of ERBB2 were not observed in glioblastomas.
Somatic mutations of the PI(3)K complex in human glioblastoma
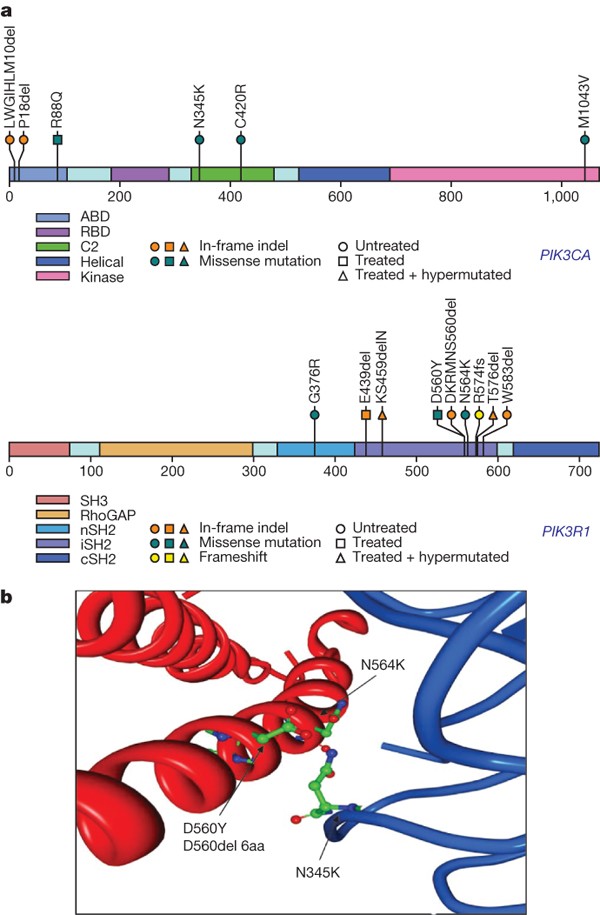
The PI(3)K complex is comprised of a catalytically active protein, p110α, encoded by PIK3CA, and a regulatory protein, p85α, encoded by PIK3R1. Frequent activating missense mutations of PIK3CA have been reported in multiple tumour types, including glioblastoma32,33. These mutations occur primarily in the adaptor binding domain (ABD) as well as the C2 helical and kinase domains34,35,36. Indeed, PIK3CA somatic nucleotide substitutions were detected in 6 of the 91 sequenced samples (Supplementary Table 6). Besides the four matching events already reported in the COSMIC database (http://www.sanger.ac.uk/genetics/CGP/cosmic/), two novel in-frame deletions were detected in the adaptor binding domain of PIK3CA (‘L10del’ and ‘P17del’). Those deletions may disrupt interactions between p110α and its regulatory subunit, p85α (ref. 37).
Unlike PIK3CA, PIK3R1 has rarely been reported as mutated in cancers. Among the five reported PIK3R1 nucleotide substitutions in cancers38,39, one was in a glioblastoma39. In our TCGA cohort, 9 PIK3R1 somatic mutations were detected among the 91 sequenced glioblastomas. None of them was in samples with PIK3CA mutations. Of the nine mutations, eight lay within the intervening SH2 (or iSH2) domain and four are 3-bp in-frame deletions (Fig. 3a and Supplementary Table 6). In accord with the crystal structure of PI(3)K, which identifies the D560 and N564 amino acid residues in p85α as contact points with the N345 amino acid residue in the C2 domain of p110α (ref. 37), the mutations detected in glioblastoma cluster around those three amino acid residues (Fig. 3b), including a N345K mutation in PIK3CA (previously reported in colon and breast cancers40) and two novel D560 mutations in PIK3R1 (D560Y and N564K). We also identified an 18-base-pair deletion spanning residues D560 to S565 (DKRMNS) in PIK3R1 (Fig. 3b) in addition to three other novel deletions (R574del, T576del and W583del) in proximity to the three key residues. We speculate that spatial constraints due to these deletions might prevent inhibitory contact of the p85α N-terminal SH2 (nSH2) domain with the helical domain of p110α, causing constitutive PI(3)K activity. Taken together, the pattern of clustering of the mutations around key residues defined by the crystal structure of PI(3)K strongly suggests that these novel PIK3R1 point mutations and indels disrupt the important C2–iSH2 interaction, relieving the inhibitory effect of p85α on p110α.
Figure 3: PIK3R1 and PIK3CA mutations in glioblastoma.
a, The locations of mutations found in TCGA tumours are indicated above the backbone. ABD, adaptor binding domain; RBD, Ras binding domain; C2, membrane-binding domain; nSH2, N-terminal SH2 domain; iSH2, inter-SH2 domain; cSH2, C-terminal SH2 domain. b, Four mutations found in the interaction interface of the C2 domain of p110α with iSH2 of p85α. Two residues of p85α, D560 and N564, are within hydrogen-bonding distance of the C2 residue of p110α, N345.
MGMT methylation and MMR in treated glioblastomas
Cancer-specific DNA methylation of CpG dinucleotides located in CpG islands within the promoters of 2,305 genes was measured relative to normal brain DNA (Supplementary Table 7 and Supplementary Methods). The promoter methylation status of MGMT, a DNA repair enzyme that removes alkyl groups from guanine residues41, is associated with glioblastoma sensitivity to alkylating agents42,43. Among the 91 sequenced cases, 19 samples were found to contain MGMT promoter methylation (including 13 of the 72 untreated cases and 6 of the 19 treated cases). When juxtaposed with somatic mutation data, an intriguing relationship between the hypermutator phenotype and MGMT methylation status emerged in the treated samples. Specifically, MGMT methylation was associated with a profound shift in the nucleotide substitution spectrum of treated glioblastomas (Fig. 4a). Among the treated samples lacking MGMT methylation (n = 13), 29% (29 out of 99) of the validated somatic mutations occurred as G˙C to A˙T transitions in CpG dinucleotides (characteristic of spontaneous deamination of methylated cytosines), and a comparable 23% (23 out of 99) of all mutations occurred as G˙C to A˙T transitions in non-CpG dinucleotides. In contrast, in the treated samples with MGMT methylation (n = 6), 81% of all mutations (146 out of 181) turned out to be of the G˙C to A˙T transition type in non-CpG dinucleotides whereas only 4% (8 out of 181) of all mutations were G˙C to A˙T transition mutations within CpGs. That pattern is consistent with a failure to repair alkylated guanine residues caused by treatment. In other words, MGMT methylation shifted the mutation spectrum of treated samples to a preponderance of G˙C to A˙T transition at non-CpG sites.
Figure 4: Pattern of somatic mutations, MGMT DNA methylation and MMR gene mutations in treated glioblastomas.
a, The mean number of validated somatic nucleotide substitutions per tumour for key sample groups is indicated on the y axis and denoted by the height of the bar histograms. Samples are grouped along the x axis according to treatment status of the patient (minus indicates untreated; plus indicates treated), DNA methylation status of MGMT (Meth, DNA methylated; minus, not methylated), and genetic status of MMR genes (minus, no genes mutated; Mut, one or more of the MLH1, MSH2, MSH6, or PMS2 genes mutated); the number below each bar indicates the number of samples in the group. Bars are colour-coded for types of nucleotide substitutions including G-to-A transitions at non-CpG sites (blue), G-to-A transitions at CpG sites (green), and other mutation types (grey). b, Bar histogram for mutation spectrum in the MMR genes as a function of treatment status and methylation status of MGMT. The colour code for substitution types is the same as in a.
Notably, the mutational spectra in the MMR genes themselves reflected MGMT methylation status and treatment consequences. All seven mutations in MMR genes found in six MGMT methylated, hypermutated (treated) tumours occurred as G˙C to A˙T mutations at non-CpG sites (Fig. 4b and Supplementary Table 6), whereas neither MMR mutation in non-methylated, hypermutated tumours was of this characteristic. Hence, these data show that MMR deficiency and MGMT methylation together, in the context of treatment, exert a powerful influence on the overall frequency and pattern of somatic point mutations in glioblastoma tumours, an observation of potential clinical importance.
Integrative analyses define glioblastoma core pathways
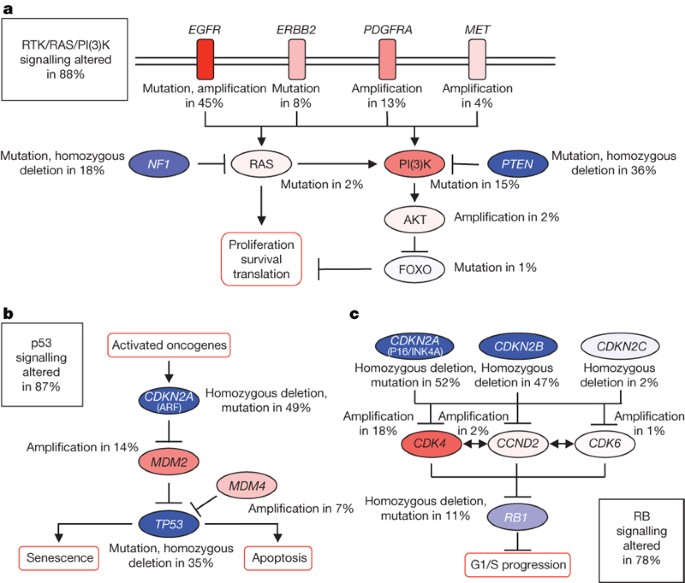
To begin to construct an integrated view of common genetic alterations in the glioblastoma genome, we mapped the unequivocal genetic alterations—validated somatic nucleotide substitutions, homozygous deletions and focal amplifications—onto major pathways implicated in glioblastoma1. That analysis identified a highly interconnected network of aberrations (Supplementary Figs 7 and 8), including three major pathways: RTK signalling, and the p53 and RB tumour suppressor pathways (Fig. 5).
Figure 5: Frequent genetic alterations in three critical signalling pathways.
a–c, Primary sequence alterations and significant copy number changes for components of the RTK/RAS/PI(3)K (a), p53 (b) and RB (c) signalling pathways are shown. Red indicates activating genetic alterations, with frequently altered genes showing deeper shades of red. Conversely, blue indicates inactivating alterations, with darker shades corresponding to a higher percentage of alteration. For each altered component of a particular pathway, the nature of the alteration and the percentage of tumours affected are indicated. Boxes contain the final percentages of glioblastomas with alterations in at least one known component gene of the designated pathway.
By copy number data alone, 66%, 70% and 59% of the 206 samples harboured somatic alterations of the RB, TP53 and RTK pathways, respectively (Supplementary Table 8). In the 91 samples for which there was also sequencing data, the frequencies of somatic alterations increased to 87%, 78% and 88%, respectively (Supplementary Table 9). There was a statistical tendency towards mutual exclusivity of alterations of components within each pathway (_P_-values of 9.3 × 10-10, 2.5 × 10-13 and 0.022, respectively, for the p53, RB and RTK pathways; Supplementary Table 10), consistent with the thesis that deregulation of one component in the pathway relieves the selective pressure for additional ones. However, we observed a greater than random chance (one-tailed, P = 0.0018) that a given sample harbours at least one aberrant gene from each of the three pathways (Supplementary Table 10). In fact, 74% harboured aberrations in all three pathways, a pattern suggesting that deregulation of the three pathways is a core requirement for glioblastoma pathogenesis.
Besides frequent deletions and mutations of the PTEN lipid phosphatase tumour suppressor gene, 86% of the glioblastoma samples harboured at least one genetic event in the core RTK/PI3K pathway (Fig. 5a). In addition to EGFR and ERBB2, PDGFRA (13%) and MET (4%) showed frequent aberrations (Supplementary Table 9). A total of 10 of the 91 sequenced samples have amplifications or point mutations in at least 2 of the 4 RTKs catalogued (EGFR, ERBB2, PDGFRA and MET; Supplementary Table 9), suggesting that genomic activation can be a mechanism for co-activated RTKs44.
Inactivation of the p53 pathway occurred in the form of ARF deletions (55%), amplifications of MDM2 (11%) and MDM4 (4%), in addition to mutations of p53 itself (Fig. 5b and Supplementary Table 8). Among 91 sequenced samples (Supplementary Table 9), genetic lesions in TP53 were mutually exclusive of those in MDM2 or MDM4 (odds ratios of 0.00 for both; P = 0.02 and 0.068, respectively; Supplementary Table 10), but not of those in ARF. In fact, 10 of the 32 tumours with TP53 mutations also had deleted ARF, suggesting that homozygous deletion of the CDKN2A locus (which encodes both p16INK4A and ARF) was at least in part driven by p16INK4A.
Among the 77% samples harbouring RB pathway aberrations (Fig. 5c), the most common event was deletion of the CDKN2A/CDKN2B locus on chromosome 9p21 (55% and 53%), followed by amplification of the CDK4 locus (14%) (Fig. 1a and Supplementary Tables 8 and 9). Although CNAs in the CDK/RB pathway members can co-occur in the same tumour14, all nine samples with RB1 nucleotide substitutions (Table S9) lacked CDKN2A/CDKN2B deletion or other CNAs in the pathway, suggesting that inactivation of RB1 by nucleotide substitution, in contrast to copy number loss, obviates the genetic pressure for activation of upstream cyclin/cyclin-dependent kinases.
Discussion
In establishing this pilot programme, TCGA has developed important principles in biospecimen banking and collection, and established the infrastructure that will serve similar efforts in the future. Although it ensured high-quality data, the stringent biospecimen selection criteria may have introduced a degree of bias because small samples and samples with high levels of necrosis were excluded. Nonetheless, the clinical parameters of this cohort are similar to other published cohorts (Supplementary Fig. 3 and Supplementary Table 1).
The integrated analyses of multi-dimensional genomic data from complementary technology platforms have proved informative. In addition to pinpointing deregulation of RB, p53 and RTK/RAS/PI(3)K pathways as obligatory events in most, and perhaps all, glioblastoma tumours, the patterns of mutations may also inform future therapeutic decisions. It would be reasonable to speculate that patients with deletions or inactivating mutations in CDKN2A or CDKN2C or patients with amplifications of CDK4/CDK6 would be candidates for treatment with CDK inhibitors, a strategy not likely to be effective in patients with RB1 mutation. Similarly, patients with PTEN deletions or activating mutations in PIK3CA or PIK3R1 might be expected to benefit from a PI(3)K or PDK1 inhibitor, whereas tumours in which the PI(3)K pathway is altered by AKT3 amplification might prove refractory to those modalities. The presence of genomic co-amplification reinforces the recent report of multiple phosphorylated (activated) RTKs in individual glioblastoma specimens44, suggesting a way to tailor anti-RTK therapeutic cocktails to specific patterns of RTK mutation. In addition, combination anti-RTK therapy might synergize with downstream inhibition of PI(3)K or cell cycle mediators. In contrast, glioblastomas with NF1 mutations might benefit from a RAF or MEK inhibitor as part of a combination, as shown for BRAF mutant cancers45.
One of the most important biomarkers for glioblastomas is the methylation status of MGMT, which predicts sensitivity to temozolomide42,43, an alkylating agent that is the current standard of care for glioblastoma patients. Integrative analysis of mutation, DNA methylation and clinical (treatment) data, albeit with small sample numbers, suggests a series of inter-related events that may have an impact on clinical response and outcome. Newly diagnosed glioblastomas with MGMT methylation respond well to treatment with alkylating agents, in part as a consequence of unrepaired alkylated guanine residues initiating cycles of futile mismatch repair, which can lead to cell death46,47,48. Therefore, treatment of _MGMT_-deficient glioblastomas with alkylating therapy introduces a strong selective pressure to lose mismatch repair function49. That conclusion is consistent with our observation that the mismatch repair genes themselves are mutated with characteristic C˙G to A˙T transitions at non-CpG sites resulting from unrepaired alkylated guanine residues. Thus, initial methylation of MGMT, in conjunction with treatment, may lead to both a shift in mutation spectrum affecting mutations at mismatch repair genes and selective pressure to lose mismatch repair function. In other words, our finding raises the possibility that patients who initially respond to the frontline therapy in use today may evolve not only treatment resistance, but also an MMR-defective hypermutator phenotype. If such a hypothesis is validated, one may speculate that selective strategies designed to target mismatch-repair-deficient cells50 would represent a rational upfront combination with alkylating agent that may prevent or minimize emergence of such resistance. Conversely, such a treatment-mediated mutator phenotype may enhance pathway mutations that can confer resistance to targeted therapies, thereby cautioning the combination of alkylating agents with targeted agents, as this may substantially increase the probability of developing resistance to such targeted drugs.
The power of TCGA to produce unprecedented multi-dimensional data sets using statistically robust numbers of samples sets the stage for a new era in the discovery of new cancer interventions. The integrative analyses leading to the formulation of an unanticipated hypothesis on a potential mechanism of resistance highlights precisely the value and power of such project design, demonstrating how unbiased and systematic cancer genome analyses of large sample cohorts can lead to important discoveries.
Methods Summary
Biospecimens were screened from retrospective banks of tissue source sites under appropriate Institutional Review Board approvals for newly diagnosed glioblastoma with minimal 80% tumour cell percentage. RNA and DNA extracted from qualified specimens were distributed to TCGA centres for analysis. Whole-genome-amplified genomic DNA samples from tumours and normal samples were sequenced by the Sanger method. Mutations were called, verified using a second genotyping platform, and systematically analysed to identify significantly mutated genes after correcting for the background mutation rate for nucleotide type and the sequence coverage of each gene. DNA copy number analyses were performed using the Agilent 244K, Affymetrix SNP6.0 and Illumina 550K DNA copy number platforms. Sample-specific and recurrent copy number changes were identified using various algorithms (GISTIC, GTS, RAE). Messenger RNA and microRNA (miRNA) expression profiles were generated using Affymetrix U133A, Affymetrix Exon 1.0 ST, custom Agilent 244K, and Agilent miRNA array platforms. mRNA expression profiles were integrated into a single estimate of relative gene expression for each gene in each sample. Methylation at CpG dinucleotides was measured using the Illumina GoldenGate assay. All data for DNA sequence alterations, copy number, mRNA expression, miRNA expression and CpG methylation were deposited in standard common formats in the TCGA DCC at http://cancergenome.nih.gov/dataportal/. All archives submitted to DCC were validated to ensure a common document structure and to ensure proper use of identifying information.
Change history
17 September 2008
The final version of this paper, published online on 17 September 2008, replaces an earlier 'non final proof' version published online on 4 September 2008, and incorporates minor changes made in proof.
References
- Furnari, F. B. et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 21, 2683–2710 (2007)
Article CAS PubMed Google Scholar - Mischel, P. S. & Cloughesy, T. F. Targeted molecular therapy of GBM. Brain Pathol. 13, 52–61 (2003)
Article PubMed Google Scholar - Mischel, P. S., Nelson, S. F. & Cloughesy, T. F. Molecular analysis of glioblastoma: pathway profiling and its implications for patient therapy. Cancer Biol. Ther. 2, 242–247 (2003)
Article CAS PubMed Google Scholar - Phillips, H. S. et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006)
Article CAS PubMed Google Scholar - Maher, E. A. et al. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res. 66, 11502–11513 (2006)
Article CAS PubMed Google Scholar - Lee, J. et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 (2006)
Article CAS PubMed Google Scholar - Diehn, M. et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc. Natl Acad. Sci. USA 105, 5213–5218 (2008)
Article CAS ADS PubMed PubMed Central Google Scholar - Liang, Y. et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc. Natl Acad. Sci. USA 102, 5814–5819 (2005)
Article CAS ADS PubMed PubMed Central Google Scholar - Freije, W. A. et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 64, 6503–6510 (2004)
Article CAS PubMed Google Scholar - Murat, A. et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J. Clin. Oncol. 26, 3015–3024 (2008)
Article CAS PubMed Google Scholar - Nutt, C. L. et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 63, 1602–1607 (2003)
CAS PubMed Google Scholar - Sun, L. et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 9, 287–300 (2006)
Article CAS PubMed Google Scholar - Beroukhim, R. et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl Acad. Sci. USA 104, 20007–20012 (2007)
Article CAS ADS PubMed PubMed Central Google Scholar - Wiedemeyer, R. et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell 13, 355–364 (2008)
Article CAS PubMed PubMed Central Google Scholar - Taylor, B. S. et al. Functional copy-number alterations in cancer. PLoS ONE (in the press)
- Hunter, C. et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 66, 3987–3991 (2006)
Article CAS PubMed PubMed Central Google Scholar - Cahill, D. P. et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin. Cancer Res. 13, 2038–2045 (2007)
Article CAS PubMed PubMed Central Google Scholar - Getz, G. et al. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science 317, 1500 (2007)
Article CAS ADS PubMed Google Scholar - Thiel, G. et al. Somatic mutations in the neurofibromatosis 1 gene in gliomas and primitive neuroectodermal tumours. Anticancer Res. 15, 2495–2499 (1995)
CAS PubMed Google Scholar - Zhu, Y. et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell 8, 119–130 (2005)
Article CAS PubMed PubMed Central Google Scholar - Reilly, K. M., Loisel, D. A., Bronson, R. T., McLaughlin, M. E. & Jacks, T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nature Genet. 26, 109–113 (2000)
Article CAS PubMed Google Scholar - Kwon, C. H. et al. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 68, 3286–3294 (2008)
Article CAS PubMed PubMed Central Google Scholar - Upadhyaya, M. et al. Mutational and functional analysis of the neurofibromatosis type 1 (NF1) gene. Hum. Genet. 99, 88–92 (1996)
Article Google Scholar - Side, L. et al. Homozygous inactivation of the NF1 gene in bone marrow cells from children with neurofibromatosis type 1 and malignant myeloid disorders. N. Engl. J. Med. 336, 1713–1720 (1997)
Article CAS PubMed Google Scholar - Fahsold, R. et al. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am. J. Hum. Genet. 66, 790–818 (2000)
Article CAS PubMed PubMed Central Google Scholar - Messiaen, L. M. et al. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum. Mutat. 15, 541–555 (2000)
Article CAS PubMed Google Scholar - Humphrey, P. A. et al. Anti-synthetic peptide antibody reacting at the fusion junction of deletion-mutant epidermal growth factor receptors in human glioblastoma. Proc. Natl Acad. Sci. USA 87, 4207–4211 (1990)
Article CAS ADS PubMed PubMed Central Google Scholar - Lee, J. C. et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 3, e485 (2006)
Article PubMed PubMed Central Google Scholar - Ekstrand, A. J., Sugawa, N., James, C. D. & Collins, V. P. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc. Natl Acad. Sci. USA 89, 4309–4313 (1992)
Article CAS ADS PubMed PubMed Central Google Scholar - Stephens, P. et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature 431, 525–526 (2004)
Article CAS ADS PubMed Google Scholar - Bamford, S. et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 91, 355–358 (2004)
Article CAS PubMed PubMed Central Google Scholar - Samuels, Y. et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554 (2004)
Article CAS PubMed Google Scholar - Gallia, G. L. et al. PIK3CA gene mutations in pediatric and adult glioblastoma multiforme. Mol. Cancer Res. 4, 709–714 (2006)
Article CAS PubMed Google Scholar - Bader, A. G., Kang, S., Zhao, L. & Vogt, P. K. Oncogenic PI3K deregulates transcription and translation. Nature Rev. Cancer 5, 921–929 (2005)
Article CAS Google Scholar - Bader, A. G., Kang, S. & Vogt, P. K. Cancer-specific mutations in PIK3CA are oncogenic in vivo. . Proc. Natl Acad. Sci. USA 103, 1475–1479 (2006)
Article CAS ADS PubMed PubMed Central Google Scholar - Liu, Z. & Roberts, T. M. Human tumor mutants in the p110α subunit of PI3K. Cell Cycle 5, 675–677 (2006)
Article CAS PubMed Google Scholar - Huang, C. H. et al. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science 318, 1744–1748 (2007)
Article CAS ADS PubMed Google Scholar - Philp, A. J. et al. The phosphatidylinositol 3′-kinase p85α gene is an oncogene in human ovarian and colon tumors. Cancer Res. 61, 7426–7429 (2001)
CAS PubMed Google Scholar - Mizoguchi, M., Nutt, C. L., Mohapatra, G. & Louis, D. N. Genetic alterations of phosphoinositide 3-kinase subunit genes in human glioblastomas. Brain Pathol. 14, 372–377 (2004)
Article CAS PubMed Google Scholar - Zhang, H. et al. Comprehensive analysis of oncogenic effects of PIK3CA mutations in human mammary epithelial cells. Breast Cancer Res. Treat. doi: 10.1007/s10549-007-9847-6 (2007)
- Pegg, A. E., Dolan, M. E. & Moschel, R. C. Structure, function, and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog. Nucleic Acid Res. Mol. Biol. 51, 167–223 (1995)
Article CAS PubMed Google Scholar - Esteller, M. et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 343, 1350–1354 (2000)
Article CAS PubMed Google Scholar - Hegi, M. E. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 352, 997–1003 (2005)
Article CAS PubMed Google Scholar - Stommel, J. M. et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318, 287–290 (2007)
Article CAS ADS PubMed Google Scholar - Solit, D. B. et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature 439, 358–362 (2006)
Article CAS ADS PubMed Google Scholar - Drablos, F. et al. Alkylation damage in DNA and RNA-repair mechanisms and medical significance. DNA Repair 3, 1389–1407 (2004)
Article CAS PubMed Google Scholar - Hirose, Y., Kreklau, E. L., Erickson, L. C., Berger, M. S. & Pieper, R. O. Delayed repletion of O6-methylguanine-DNA methyltransferase resulting in failure to protect the human glioblastoma cell line SF767 from temozolomide-induced cytotoxicity. J. Neurosurg. 98, 591–598 (2003)
Article CAS PubMed Google Scholar - Kaina, B., Christmann, M., Naumann, S. & Roos, W. P. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair 6, 1079–1099 (2007)
Article CAS PubMed Google Scholar - Casorelli, I., Russo, M. T. & Bignami, M. Role of mismatch repair and MGMT in response to anticancer therapies. Anticancer Agents Med. Chem. 8, 368–380 (2008)
Article CAS PubMed Google Scholar - Yang, J. L., Qu, X. J., Yu, Y., Kohn, E. C. & Friedlander, M. L. Selective sensitivity to carboxyamidotriazole by human tumor cell lines with DNA mismatch repair deficiency. Int. J. Cancer 123, 258–263 (2008)
Article CAS PubMed Google Scholar
Acknowledgements
We thank the members of TCGA’s External Scientific Committee and the Glioblastoma Disease Working Group (http://cancergenome.nih.gov/components) for discussions; D. N. Louis for discussions; A. Mirick, J. Melone and C. Collins for administrative coordination of TCGA activities; and L. Gaffney for graphic art. This work was supported by the following grants from the United States National Institutes of Health: U54HG003067, U54HG003079, U54HG003273, U24CA126543, U24CA126544, U24CA126546, U24CA126551, U24CA126554, U24CA126561 and U24CA126563.
Author Contributions The TCGA research network contributed collectively to this study. Biospecimens were provided by the tissue source sites and processed by the Biospecimen Core Resource. Data generation and analyses were performed by the genome sequencing centres and cancer genome characterization centres. All data were released through the Data Coordinating Center. Project activities were coordinated by the NCI and NHGRI project teams. The following TCGA investigators contributed substantively to the writing of this manuscript. Leaders: L. Chin and M. Meyerson. Neuropathology: K. Aldape, D. Bigner, T. Mikkelsen and S. VandenBerg. Databases: A. Kahn. Biospecimen analysis: R. Penny, M. L. Ferguson and D. S. Gerhard. Copy number: G. Getz, C. Brennan, B. S. Taylor, W. Winckler, P. Park and M. Ladanyi. Gene expression: K. A. Hoadley, R. G. W. Verhaak, D. N. Hayes and P. Spellman. LOH: D. Absher and B. A. Weir. Sequencing: G. Getz, L. Ding, D. Wheeler, M. S. Lawrence, K. Cibulskis, E. Mardis, Jinghui Zhang and R. K. Wilson. TP53: L. Donehower and D. A. Wheeler. NF1: W. Winckler, L. Ding and Jinghui Zhang. EGFR: E. Purdom and W. Winckler. ERBB2: W. Winckler. PIK3R1: L. Ding, J. Wallis and E. Mardis. DNA methylation: P. W. Laird, J. G. Herman, L. Ding, D. J. Weisenberger and S. B. Baylin. Pathway analysis: N. Schultz, L. Donehower, D. A. Wheeler, Jun Yao, R. Wiedemeyer, J. Weinstein and C. Sander. General: S. B. Baylin, R. A. Gibbs, J. Gray, R. Kucherlapati, M. Ladanyi, E. S. Lander, R. M. Myers, C. M. Perou, J. Weinstein and R. K. Wilson.
Author information
Authors and Affiliations
- Department of Pathology,,
Roger McLendon & Darrell Bigner - Department of Surgery, Duke University Medical Center, Durham, North Carolina 27710, USA.,
Allan Friedman - Department of Neurosurgery,,
Erwin G. Van Meir, Gena M. Mastrogianakis & Jeffrey J. Olson - Department of Hematology and Medical Oncology,,
Erwin G. Van Meir & Jeffrey J. Olson - Winship Cancer Institute,,
Erwin G. Van Meir, Daniel J. Brat & Jeffrey J. Olson - Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, Georgia 30322, USA.,
Daniel J. Brat - Department of Neurological Surgery,,
Tom Mikkelsen - Department of Pathology, Henry Ford Hospital, Detroit, Michigan 48202, USA.,
Norman Lehman - Department of Pathology,,
Ken Aldape - Department of Neuro-Oncology,,
W. K. Alfred Yung - Department of Neurosurgery, University of Texas M.D. Anderson Cancer Center, Houston, Texas 77030, USA.,
Oliver Bogler - Department of Pathology,,
Scott VandenBerg - Department of Neurosurgery, University of California San Francisco, San Francisco, California 94143, USA.,
Mitchel Berger & Michael Prados - Human Genome Sequencing Center, Baylor College of Medicine, Houston, Texas 77030, USA.,
Donna Muzny, Margaret Morgan, Steve Scherer, Aniko Sabo, Lynn Nazareth, Lora Lewis, Otis Hall, Yiming Zhu, Yanru Ren, Omar Alvi, Jiqiang Yao, Alicia Hawes, Shalini Jhangiani, Gerald Fowler, Anthony San Lucas, Christie Kovar, Andrew Cree, Huyen Dinh, Jireh Santibanez, Vandita Joshi, Manuel L. Gonzalez-Garay, Christopher A. Miller, Aleksandar Milosavljevic, David A. Wheeler & Richard A. Gibbs - Graduate Program in Structural and Computational Biology and Molecular Biophysics,,
Christopher A. Miller & Aleksandar Milosavljevic - Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas 77030, USA.,
Aleksandar Milosavljevic - Department of Molecular Virology and Microbiology, Human Genome Sequencing Center, Baylor College of Medicine, Houston, Texas 77030, USA.,
Larry Donehower - The Eli and Edythe L. Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, Massachusetts 02142, USA.,
Kristian Cibulskis, Carrie Sougnez, Tim Fennell, Scott Mahan, Jane Wilkinson, Liuda Ziaugra, Robert Onofrio, Toby Bloom, Rob Nicol, Kristin Ardlie, Jennifer Baldwin, Stacey Gabriel, Eric S. Lander, Gad Getz, Wendy Winckler, Roel G. W. Verhaak, Michael S. Lawrence, Michael O’Kelly, Jim Robinson, Gabriele Alexe, Rameen Beroukhim, Scott Carter, Derek Chiang, Josh Gould, Supriya Gupta, Josh Korn, Craig Mermel, Jill Mesirov, Stefano Monti, Huy Nguyen, Melissa Parkin, Michael Reich, Nicolas Stransky, Barbara A. Weir, Levi Garraway, Todd Golub & Matthew Meyerson - Department of Biology, Institute of Massachusetts Institute of Technology, Cambridge, Massachusetts 02142, USA.,
Eric S. Lander - Department of Systems Biology, Harvard University, Boston, Massachusetts 02115, USA.,
Eric S. Lander - Department of Genetics, The Genome Center at Washington University, Washington University School of Medicine, St Louis, Missouri 63108, USA.,
Li Ding, Robert S. Fulton, Michael D. McLellan, John Wallis, David E. Larson, Xiaoqi Shi, Rachel Abbott, Lucinda Fulton, Ken Chen, Daniel C. Koboldt, Michael C. Wendl, Rick Meyer, Yuzhu Tang, Ling Lin, John R. Osborne, Brian H. Dunford-Shore, Tracie L. Miner, Kim Delehaunty, Chris Markovic, Gary Swift, William Courtney, Craig Pohl, Scott Abbott, Amy Hawkins, Shin Leong, Carrie Haipek, Heather Schmidt, Maddy Wiechert, Tammi Vickery, Sacha Scott, David J. Dooling, Asif Chinwalla, George M. Weinstock, Elaine R. Mardis & Richard K. Wilson - Department of Medical Oncology,,
Wendy Winckler, Roel G. W. Verhaak, Rameen Beroukhim, Derek Chiang, Craig Mermel, Barbara A. Weir, Levi Garraway, Todd Golub, Matthew Meyerson, Lynda Chin & W. Ruprecht Wiedemeyer - Center for Cancer Genome Discovery,,
Wendy Winckler, Roel G. W. Verhaak, Rameen Beroukhim, Barbara A. Weir, Levi Garraway, Todd Golub & Matthew Meyerson - Center for Applied Cancer Science of the Belfer Institute for Innovative Cancer Science, Dana-Farber Cancer Institute, Boston, Massachusetts 02115, USA.,
Lynda Chin, Alexei Protopopov, Jianhua Zhang, Ilana Perna, Georgia Ren, Jun Yao & Yonghong Xiao - Department of Dermatology, Harvard Medical School, Boston, Massachusetts 02115, USA.,
Lynda Chin - Harvard Medical School-Partners HealthCare Center for Genetics and Genomics, Boston, Massachusetts 02115, USA.,
Sandy Aronson, Narayan Sathiamoorthy, Hyunsoo Kim, Isaac S. Kohane, Peter J. Park & Raju Kucherlapati - Informatics Program,,
Sek Won Kong, Isaac S. Kohane & Peter J. Park - Department of Cardiology, Children’s Hospital, Boston, Massachusetts 02115, USA.,
Sek Won Kong - Center for Biomedical Informatics,,
Isaac S. Kohane & Peter J. Park - Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA.,
Jon Seidman - USC Epigenome Center, University of Southern California, Los Angeles, California 90089, USA.,
Peter W. Laird, Daniel J. Weisenberger, Fei Pan & David Van Den Berg - Biometry and Clinical Trials Division,,
Leslie Cope - Cancer Biology Division, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University, Baltimore, Maryland 21231, USA.,
James G. Herman, Joo Mi Yi, Kornel E. Schuebel & Stephen B. Baylin - Department of Molecular Biotechnology, Faculty of Bioscience and Engineering, Ghent University, Ghent B-9000, Belgium.,
Leander Van Neste - HudsonAlpha Institute for Biotechnology, Huntsville, Alabama 35806, USA.,
Devin M. Absher & Richard M. Myers - Department of Human Genetics, University of Michigan, Ann Arbor, Michigan 48109, USA.,
Jun Z. Li - Department of Genetics,,
Audrey Southwick, Shannon Brady, Amita Aggarwal, Tisha Chung & Gavin Sherlock - Department of Urology, Stanford University School of Medicine, Stanford, California 94305, USA.,
James D. Brooks - Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, California 94720, USA.,
Paul T. Spellman, Lakshmi R. Jakkula, Anna V. Lapuk, Henry Marr, Shannon Dorton, Ju Han, Amrita Ray, Steffen Durinck, Nicholas J. Wang, Gerald V. Fontenay, John G. Conboy, Bahram Parvin, Heidi S. Feiler & Joe W. Gray - Department of Statistics,,
Elizabeth Purdom, Victoria Wang & Terence P. Speed - Department of Molecular and Cellular Biology, University of California at Berkeley, Berkeley, California 95720, USA.,
Yoon Gi Choi, Karen Vranizan, Vivian Peng, Eric Van Name & John Ngai - Walter and Eliza Hall Institute, Parkville, Victoria 3052, Australia.,
Mark Robinson & Terence P. Speed - Department of Neurosurgery,,
Cameron Brennan - Computational Biology Center, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, USA.,
Nicholas D. Socci, Barry S. Taylor, Alex Lash, Nikolaus Schultz, Boris Reva, Yevgeniy Antipin, Alexey Stukalov, Benjamin Gross, Ethan Cerami, Wei Qing Wang & Chris Sander - Department of Epidemiology and Biostatistics, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, USA.,
Adam Olshen, Li-Xuan Qin & Venkatraman E. Seshan - Department of Physiology and Biophysics, Weill Cornell Graduate School of Medical Sciences, New York, New York 10065, USA.,
Barry S. Taylor - Genomics Core Laboratory, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, USA.,
Liliana Villafania, Magali Cavatore & Agnes Viale - Department of Pathology, Human Oncology and Pathogenesis Program, Memorial Sloan-Kettering Cancer Center, New York, New York 10065, USA.,
Laetitia Borsu, William Gerald & Marc Ladanyi - Department of Genetics,,
Charles M. Perou & Katherine A. Hoadley - Department of Pathology and Laboratory Medicine,,
Charles M. Perou & Michael D. Topal - Department of Internal Medicine, Division of Medical Oncology, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.,
D. Neil Hayes & Yuan Qi - Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.,
Michael D. Topal, Sai Balu, Yan Shi & Junyuan Wu - International Genomics Consortium, Phoenix, Arizona 85004, USA.,
Robert Penny, Troy Shelton, Elizabeth Lenkiewicz, Scott Morris, Debbie Beasley & Sheri Sanders - Computational Biology Division, Translational Genomics Research Institute, Phoenix, Arizona 85004, USA.,
Michael Bittner - SRA International, Fairfax, Virginia 22033, USA.,
Ari Kahn, Robert Sfeir, Jessica Chen, David Nassau, Larry Feng & Erin Hickey - Center For Biomedical Informatics and Information Technology, National Cancer Institute, Rockville, Maryland 20852, USA.,
Jinghui Zhang, Carl Schaefer, Subhashree Madhavan & Kenneth H. Buetow - Department of Bioinformatics and Computational Biology, M.D. Anderson Cancer Center, Houston, Texas 77030, USA.,
John N. Weinstein - National Cancer Institute, National Institutes of Health, Bethesda, Maryland 20892, USA.,
Anna Barker, Daniela S. Gerhard, Joseph Vockley, Carolyn Compton, Jim Vaught & Peter Fielding - MLF Consulting, Arlington, Massachusetts 02474, USA.,
Martin L. Ferguson - National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland 20892, USA.,
Francis Collins, Peter Good, Mark Guyer, Brad Ozenberger, Jane Peterson & Elizabeth Thomson
Consortia
The Cancer Genome Atlas Research Network
Tissue source sites: Duke University Medical School
- Roger McLendon
- , Allan Friedman
- & Darrell Bigner
Emory University
- Erwin G. Van Meir
- , Daniel J. Brat
- , Gena M. Mastrogianakis
- & Jeffrey J. Olson
Henry Ford Hospital
- Tom Mikkelsen
- & Norman Lehman
MD Anderson Cancer Center
- Ken Aldape
- , W. K. Alfred Yung
- & Oliver Bogler
University of California San Francisco
- Scott VandenBerg
- , Mitchel Berger
- & Michael Prados
Genome sequencing centres: Baylor College of Medicine
- Donna Muzny
- , Margaret Morgan
- , Steve Scherer
- , Aniko Sabo
- , Lynn Nazareth
- , Lora Lewis
- , Otis Hall
- , Yiming Zhu
- , Yanru Ren
- , Omar Alvi
- , Jiqiang Yao
- , Alicia Hawes
- , Shalini Jhangiani
- , Gerald Fowler
- , Anthony San Lucas
- , Christie Kovar
- , Andrew Cree
- , Huyen Dinh
- , Jireh Santibanez
- , Vandita Joshi
- , Manuel L. Gonzalez-Garay
- , Christopher A. Miller
- , Aleksandar Milosavljevic
- , Larry Donehower
- , David A. Wheeler
- & Richard A. Gibbs
Broad Institute of MIT and Harvard
- Kristian Cibulskis
- , Carrie Sougnez
- , Tim Fennell
- , Scott Mahan
- , Jane Wilkinson
- , Liuda Ziaugra
- , Robert Onofrio
- , Toby Bloom
- , Rob Nicol
- , Kristin Ardlie
- , Jennifer Baldwin
- , Stacey Gabriel
- & Eric S. Lander
Washington University in St Louis
- Li Ding
- , Robert S. Fulton
- , Michael D. McLellan
- , John Wallis
- , David E. Larson
- , Xiaoqi Shi
- , Rachel Abbott
- , Lucinda Fulton
- , Ken Chen
- , Daniel C. Koboldt
- , Michael C. Wendl
- , Rick Meyer
- , Yuzhu Tang
- , Ling Lin
- , John R. Osborne
- , Brian H. Dunford-Shore
- , Tracie L. Miner
- , Kim Delehaunty
- , Chris Markovic
- , Gary Swift
- , William Courtney
- , Craig Pohl
- , Scott Abbott
- , Amy Hawkins
- , Shin Leong
- , Carrie Haipek
- , Heather Schmidt
- , Maddy Wiechert
- , Tammi Vickery
- , Sacha Scott
- , David J. Dooling
- , Asif Chinwalla
- , George M. Weinstock
- , Elaine R. Mardis
- & Richard K. Wilson
Cancer genome characterization centres: Broad Institute/Dana-Farber Cancer Institute
- Gad Getz
- , Wendy Winckler
- , Roel G. W. Verhaak
- , Michael S. Lawrence
- , Michael O’Kelly
- , Jim Robinson
- , Gabriele Alexe
- , Rameen Beroukhim
- , Scott Carter
- , Derek Chiang
- , Josh Gould
- , Supriya Gupta
- , Josh Korn
- , Craig Mermel
- , Jill Mesirov
- , Stefano Monti
- , Huy Nguyen
- , Melissa Parkin
- , Michael Reich
- , Nicolas Stransky
- , Barbara A. Weir
- , Levi Garraway
- , Todd Golub
- & Matthew Meyerson
Harvard Medical School/Dana-Farber Cancer Institute
- Lynda Chin
- , Alexei Protopopov
- , Jianhua Zhang
- , Ilana Perna
- , Sandy Aronson
- , Narayan Sathiamoorthy
- , Georgia Ren
- , Jun Yao
- , W. Ruprecht Wiedemeyer
- , Hyunsoo Kim
- , Sek Won Kong
- , Yonghong Xiao
- , Isaac S. Kohane
- , Jon Seidman
- , Peter J. Park
- & Raju Kucherlapati
Johns Hopkins/University of Southern California
- Peter W. Laird
- , Leslie Cope
- , James G. Herman
- , Daniel J. Weisenberger
- , Fei Pan
- , David Van Den Berg
- , Leander Van Neste
- , Joo Mi Yi
- , Kornel E. Schuebel
- & Stephen B. Baylin
HudsonAlpha Institute/Stanford University
- Devin M. Absher
- , Jun Z. Li
- , Audrey Southwick
- , Shannon Brady
- , Amita Aggarwal
- , Tisha Chung
- , Gavin Sherlock
- , James D. Brooks
- & Richard M. Myers
Lawrence Berkeley National Laboratory
- Paul T. Spellman
- , Elizabeth Purdom
- , Lakshmi R. Jakkula
- , Anna V. Lapuk
- , Henry Marr
- , Shannon Dorton
- , Yoon Gi Choi
- , Ju Han
- , Amrita Ray
- , Victoria Wang
- , Steffen Durinck
- , Mark Robinson
- , Nicholas J. Wang
- , Karen Vranizan
- , Vivian Peng
- , Eric Van Name
- , Gerald V. Fontenay
- , John Ngai
- , John G. Conboy
- , Bahram Parvin
- , Heidi S. Feiler
- , Terence P. Speed
- & Joe W. Gray
Memorial Sloan-Kettering Cancer Center
- Cameron Brennan
- , Nicholas D. Socci
- , Adam Olshen
- , Barry S. Taylor
- , Alex Lash
- , Nikolaus Schultz
- , Boris Reva
- , Yevgeniy Antipin
- , Alexey Stukalov
- , Benjamin Gross
- , Ethan Cerami
- , Wei Qing Wang
- , Li-Xuan Qin
- , Venkatraman E. Seshan
- , Liliana Villafania
- , Magali Cavatore
- , Laetitia Borsu
- , Agnes Viale
- , William Gerald
- , Chris Sander
- & Marc Ladanyi
University of North Carolina, Chapel Hill
- Charles M. Perou
- , D. Neil Hayes
- , Michael D. Topal
- , Katherine A. Hoadley
- , Yuan Qi
- , Sai Balu
- , Yan Shi
- & Junyuan Wu
Biospecimen Core Resource
- Robert Penny
- , Michael Bittner
- , Troy Shelton
- , Elizabeth Lenkiewicz
- , Scott Morris
- , Debbie Beasley
- & Sheri Sanders
Data Coordinating Center
- Ari Kahn
- , Robert Sfeir
- , Jessica Chen
- , David Nassau
- , Larry Feng
- , Erin Hickey
- , Jinghui Zhang
- & John N. Weinstein
Project teams: National Cancer Institute
- Anna Barker
- , Daniela S. Gerhard
- , Joseph Vockley
- , Carolyn Compton
- , Jim Vaught
- , Peter Fielding
- , Martin L. Ferguson
- , Carl Schaefer
- , Subhashree Madhavan
- & Kenneth H. Buetow
National Human Genome Research Institute
- Francis Collins
- , Peter Good
- , Mark Guyer
- , Brad Ozenberger
- , Jane Peterson
- & Elizabeth Thomson
Corresponding authors
Correspondence toMatthew Meyerson or Lynda Chin.
Additional information
A list of participants and affiliations appears at the end of the paper.
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1-S8 and Legends, Supplementary Table Legends (Supplementary Tables S1-S11 in accompanying Supplementary Excel file), Supplementary Methods and Supplementary References. Minor errors in Supplementary Fig. 2, part C of the Supplementary Methods and in the legend of Supplementary Methods Fig. 4 were corrected on 19th March 2009. (PDF 2259 kb)
Supplementary Tables
This file contains Supplementary Tables S1-S11 (Supplementary Table Legends in accompanying Supplementary PDF). (XLS 1429 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways.Nature 455, 1061–1068 (2008). https://doi.org/10.1038/nature07385
- Received: 28 July 2008
- Accepted: 01 September 2008
- Published: 04 September 2008
- Issue Date: 23 October 2008
- DOI: https://doi.org/10.1038/nature07385