ATP Binding and ATP Hydrolysis Play Distinct Roles in the Function of 26S Proteasome (original) (raw)
. Author manuscript; available in PMC: 2014 Mar 12.
Summary
The 26S proteasome degrades polyubiquitinated proteins by an energy-dependent mechanism. Here we define multiple roles for ATP in 26S proteasome function. ATP binding is necessary and sufficient for assembly of 26S proteasome from 20S proteasome and PA700/19S subcomplexes and for proteasome activation. Proteasome assembly and activation may require distinct ATP binding events. The 26S proteasome degrades nonubiquitylated, unstructured proteins without ATP hydrolysis, indicating that substrate translocation per se does not require the energy of hydrolysis. Nonubiquitylated folded proteins and certain polyubiquitylated folded proteins were refractory to proteolysis. The latter were deubiquitylated by an ATP-independent mechanism. Other folded as well as unstructured polyubiquitylated proteins required ATP hydrolysis for proteolysis and deubiquitylation. Thus, ATP hydrolysis is not used solely for substrate unfolding. These results indicate that 26S proteasome-catalyzed degradation of polyubiquitylated proteins involves mechanistic coupling of several processes and that such coupling imposes an energy requirement not apparent for any isolated process.
Introduction
The 26S proteasome is an energy-dependent molecular machine that degrades polyubiquitinated proteins (Voges et al., 1999; Coux et al., 1996; Pickart and Cohen, 2004). Proteasome-catalyzed proteolysis of ubiquitinmodified proteins plays a regulatory role in nearly every aspect of normal cellular function and in many diseases. The 2,400,000 dalton 26S proteasome is composed of two large multisubunit subcomplexes: a protease component, the 20S proteasome, and a regulatory component, PA700 (19S regulatory particle) (Voges et al., 1999; DeMartino and Slaughter, 1999; Baumeister et al., 1998). The 20S proteasome is a 700,000 dalton cylinder-shaped particle composed of four axially stacked heptameric rings (Baumeister et al., 1998). The inner rings contain two copies of each of three different protease subunits whose catalytic sites line an interior chamber (Bochtler et al., 1999). This topology sequesters the catalytic sites from substrates. Access of substrates to the catalytic sites is achieved via narrow 13 Å pores at either end of the 20S proteasome cylinder, thereby limiting substrates to short peptides and unfolded proteins. An additional restriction on substrate access to catalytic sites is imposed by occlusion of the pores by N-terminal peptide extensions of subunits of the outer proteasome rings (Groll et al., 1997). Thus, a 20S proteasome with occluded pores is catalytically inert. Proteasome activation is achieved by conformational changes that remove the occlusions, a process exerted by regulatory proteins that bind to the outer rings of the 20S proteasome (Whitby et al., 2001; Forster et al., 2005; Ortega et al., 2005; Rechsteiner and Hill, 2005), certain ionic conditions (Groll et al., 2001; McGuire et al., 1989; Wilk and Orlowski, 1983), certain hydrophobic peptides (Kisselev et al., 2002), and some intrinsically disordered protein substrates (Liu et al., 2003). Cells contain multiple proteasome regulators, but PA700/19S is the most prominent and physiologically best understood (DeMartino and Slaughter, 1999).
PA700 is a 700,000 dalton ATPase composed of 18 different gene products. PA700 is devoid of protease activity but contributes other functions essential for energy-dependent degradation of polyubiquitylated proteins by the 26S proteasome, including the following: (1) opening of the pores of the 20S proteasome upon binding (proteasome assembly and activation), (2) binding polyubiquitin chains of substrate proteins (substrate recognition), (3) destabilization of tertiary structure of folded substrate proteins (substrate unfolding), (4) movement of the unfolded polypeptide chain through the opened pore to the site of proteolysis in the interior chamber of the 20S proteasome (substrate translocation), and (5) catalytic removal of the polyubiquitin chain from the substrate (substrate deubiquitylation). 26S proteasome-catalyzed degradation requires continuous ATP hydrolysis, and each function listed above has been shown or suggested to require ATP (Ogura and Tanaka, 2003; Pickart and Cohen, 2004). PA700 contains six homologous AAA ATPase subunits arranged as a ring that abuts the outer rings of the 20S proteasome (Glickman et al., 1998a; Baumeister et al., 1998; Voges et al., 1999; Ferrell et al., 2000). This topology places them in a critical position to mediate aspects of proteasome function that require ATP. However, the precise mechanistic basis for the ATP dependence of proteolysis remains unclear.
To clarify the role of ATP in mediation of 26S proteasome function, we have examined the effect of ATP binding and hydrolysis on various features of 26S proteasome function. We show that ATP binding is necessary and sufficient for several aspects of 26S proteasome function, including assembly and activation. Additional ATP is not required for deubiquitylation of degradation-resistant substrates or for degradation of nonubiquitylated unstructured substrates. Surprisingly, however, ATP hydrolysis is required for degradation of both folded and unstructured polyubiquitylated proteins. Moreover, our results indicate that ATP hydrolysis-dependent degradation of polyubiquitylated proteins by the 26S proteasome is imposed by mechanistic coupling of several otherwise energy-independent steps.
Results
The 26S Proteasome Is Assembled from 20S Proteasome and PA700 Subcomplexes
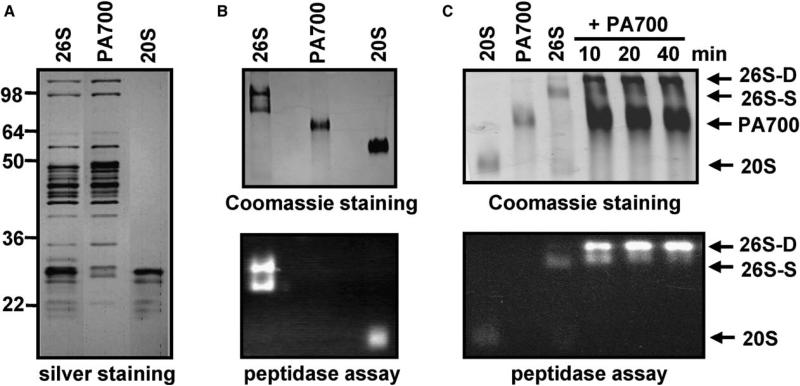
To investigate the role of ATP in 26S proteasome function, we developed purification schemes for intact 26S proteasome and for isolated 20S proteasome and PA700 subcomplexes. Optimal purification of intact 26S proteasome (described in the Supplemental Experimental Procedures available with this article online) required ATP in all buffers, suggesting that ATP stabilized 26S proteasome structure. Accordingly, efficient purification of 20S proteasome and PA700 subcomplexes was achieved in buffers lacking ATP. 20S proteasome and PA700 were revealed as single proteins, respectively, by native polyacrylamide gel electrophoresis; each displayed characteristic subunit patterns after SDS-PAGE (Figures 1A and 1B). Most preparations of 26S proteasome consisted of two bands on native polyacrylamide gels, corresponding to 26S proteasomes with either one or two copies of PA700 (Elasser et al., 2005). Each form was catalytically active, as demonstrated by gel overlay assays using a peptide substrate, Suc-Leu-Leu-Val-Tyr-AMC, to detect proteasome activity. The proportions of singly and doubly capped 26S proteasomes varied modestly from preparation to preparation. A highly enriched population of doubly capped 26S proteasomes could be generated from a population of mixed or predominantly singly capped proteasomes by incubation with excess PA700 in the presence of ATP (Figure 1C and see below).
Figure 1. Characterization of Purified 26S Proteasome from Bovine Blood Red Cells.
(A) 26S proteasome (960 ng), PA700 (400 ng), and 20S proteasome (260 ng) were subjected to SDS-PAGE and visualized by silver staining.
(B) 26S proteasome (3 mg), PA700 (3 mg), and 20S proteasome (3 mg) were subjected to 4% native PAGE and either stained with Coomassie blue (upper) or assayed for proteasome activity with an overlay assay (lower) using Suc-Leu-Leu-Val-Tyr-AMC as a fluorogenic substrate.
(C) Assembly of doubly capped 26S proteasome (26S-D) from singly capped 26S proteasome (26S-S) and PA700. Predominately singly capped 26S proteasome (40 nM) was incubated with purified PA700 (400 nM) in the presence of 200 μM ATP for the indicated times and subjected to 4% native PAGE. Gels were either stained with Coomassie blue (upper) or assayed for proteasome activity with a substrate overlay assay (lower). Migration positions of purified 20S and PA700 were established as references by electrophoresis of respective proteins.
ATP Binding Is Necessary and Sufficient for Assembly, Activation, and Stability of the 26S Proteasome
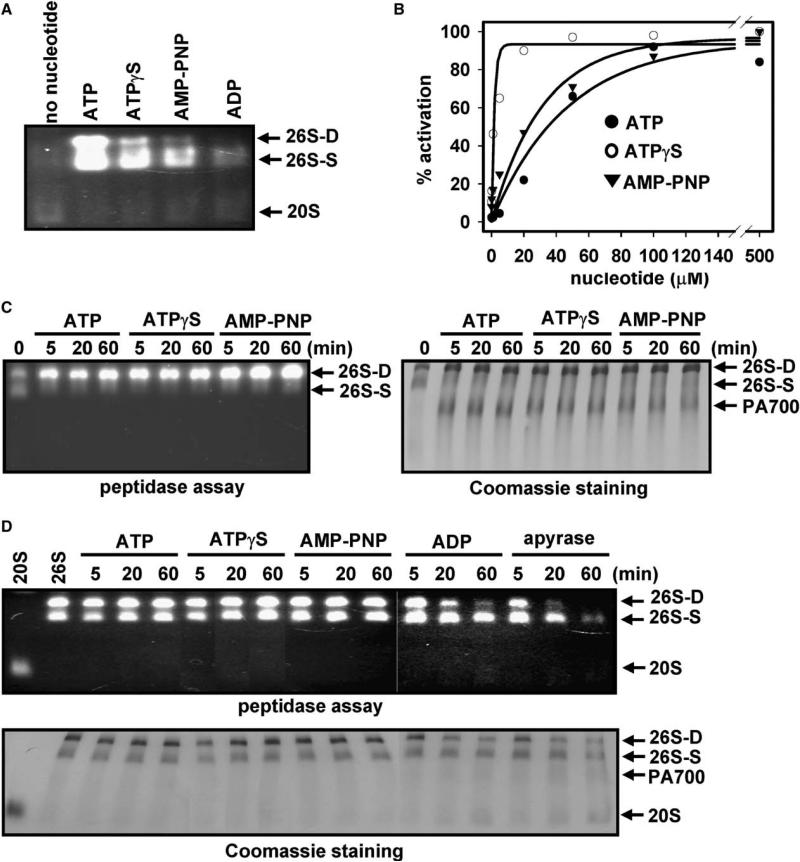
We and others previously demonstrated ATP-dependent in vitro assembly of 26S proteasome from purified 20S proteasome and PA700 (DeMartino et al., 1994; Ma et al., 1994; Hoffman et al., 1992). Assembly can be documented by native polyacrylamide gel electrophoresis or by glycerol density gradient centrifugation and is accompanied by a 20- to 50-fold increase in hydrolysis of Suc-Leu-Leu-Val-Tyr-AMC, as assessed by solution assays (Ma et al., 1994) or by substrate overlay assays in native gels (Figure 2A). Half-maximal proteasome activation was achieved at approximately 40 μM ATP (Figure 2B). The magnitude of ATP-dependent proteasome activation by PA700 was similar to that of ATP-independent proteasome activation by SDS or PA28 (Table S1). Increased proteasome activity against peptide substrates promoted by each of these agents likely results from opening of pores at the ends of the 20S proteasome.
Figure 2. ATP Binding Is Necessary and Sufficient for Assembly of 26S Proteasome from PA700 and 20S Proteasome and for 26S Proteasome Stability.
(A) ATP and nonhydrolyzable ATP analogs promote assembly and activation of the 26S proteasome. 20S proteasome (40 nM) was incubated with PA700 (320 nM) for 60 min with no nucleotide or in the presence of 1 mM ATP, ATPγS, AMP-PNP, or ADP. Samples were subjected to a native gel electrophoresis and assayed for proteasome activity with a substrate overlay assay by using Suc-Leu-Leu-Val-Tyr-AMC.
(B) Assembly reactions were as described in (A) at the indicated nucleotide concentrations. Soluble proteasome assays were conducted with Suc-Leu-Leu-Val-Tyr-AMC. Results are expressed as a percentage of maximal activation for each nucleotide. ATP, ATPγS, and AMP-PNP maximally activated proteasome activity by 19.8-, 10.1-, and 4.4-fold, respectively. No activation of 20S proteasome by PA700 was achieved in the absence of nucleotides.
(C) Nucleotide binding-dependent assembly of doubly capped 26S from singly capped 26S proteasome and PA700. Purified 26S proteasome (40 nM, approximately 60% doubly capped versus 40% singly capped) was incubated with PA700 (120 nM) in buffer A at 37°C for the indicated times. In reactions with ATPγS, AMP-PNP, or no ATP (2ATP), 26S proteasome was preincubated for 1 min at 30°C with apyrase (8 mU/μl) prior to the addition of ATPγS, AMP-PNP (1 mM), or buffer, respectively. Samples were subjected to native gel electrophoresis and assayed for proteasome activity using Suc-Leu-Leu-Val-Tyr-AMC (left panel) or stained with Coomassie blue (right panel).
(D) 26S proteasome (40 nM) was incubated at 37°C in the presence of 1 mM ATP or treated with 8 mU/μl apyrase for 1 min at 30°C prior to supplementation with 1 mM ATPγS, 1 mM AMP-PNP, 1 mM ADP, or buffer and incubated at 37°C. At the indicated times, samples were subjected to native PAGE and either assayed for proteasome activity by using Suc-Leu-Leu-Val-Tyr-AMC (upper) or stained with Coomassie blue (lower). Purified, nonincubated 20S and 26S proteasomes were electrophoresed and processed for reference.
To determine the mechanism of ATP-dependent assembly and activation of the 26S proteasome, we tested the relative ability of other nucleotides to mediate assembly and activation of the 26S proteasome from 20S and PA700 subcomplexes. Nonhydrolyzable ATP analogs ATPγS and AMP-PNP promoted proteasome assembly and activation, suggesting that ATP binding is sufficient for these processes. Although the extent of assembly and activation by each nonhydrolyzable nucleotide was less than that by ATP, half-maximal effects were achieved at similar or lower concentrations: 1 μM for ATPγS and 20 μM for AMP-PNP (Figure 2B). In each case, the extent of activation was proportional to the extent of assembly as determined by comparison of proteasome activity and amounts of 26S proteasome revealed by Coomassie blue staining after native gel electrophoresis (Figure 2 and data not shown). The effects of all nucleotides on assembly and activation required Mg2+. ADP was an insignificant activator and poorly supported assembly. Because certain ATPases, including the bacterial AAA ATPase ClpX (Burton et al., 2003), hydrolyze ATPγS at appreciable rates, we compared hydrolytic rates of ATP and ATPγS by 26S proteasome and isolated PA700. Although 26S proteasome and PA700 hydrolyzed ATP at similar rates, neither complex hydrolyzed ATPγS at detectable rates (Table S2). These results demonstrate that ATP binding, but not hydrolysis, is required for the assembly and activation of 26S proteasome. Similar features for nucleotide requirements were obtained for assembly of doubly capped 26S proteasome from a heterogeneous population of singly and doubly capped 26S proteasomes (Figure 2C). The later experiment, and others described below, required depletion of ATP from the buffer in which the 26S proteasome was purified and stored, followed by repletion with nucleotide of choice; ATP depletion was achieved efficiently by a brief preincubation with apyrase (see Experimental Procedures and Table S3). Incubation of 26S proteasome in the absence of any nucleotide promoted dissociation into 20S proteasome and PA700 subcomplexes, as shown below. Thus, ATP binding, but not hydrolysis, is also necessary and sufficient for assembly of doubly capped 26S proteasomes from singly capped proteasomes.
Empirical evidence from the 26S proteasome purification procedure suggests that, in addition to its requirement for assembly, ATP retards dissociation of 26S proteasome into 20S proteasome and PA700 subcomplexes. To test directly the role of ATP on stability of the 26S proteasome, we incubated the purified complex in the presence or absence of ATP. Incubation of 26S proteasome at 37°C for several hours in the presence of ATP had no detectable effect on proteasome structure or activity (Figure 2D and data not shown). In contrast, a similar incubation after removal of ATP resulted in dissociation of the 26S proteasome into 20S proteasome and PA700 complexes (Figure 2D). ATPγS or AMP-PNP stabilized 26S proteasome structure and activity during the incubation, whereas ADP was a poor stabilizer. Thus, ATP binding, but not hydrolysis, is required for maintenance of 26S proteasome structure and catalytic activity.
Assembly and Activation of 26S Proteasome Appear to Require Distinct ATP-Binding Events
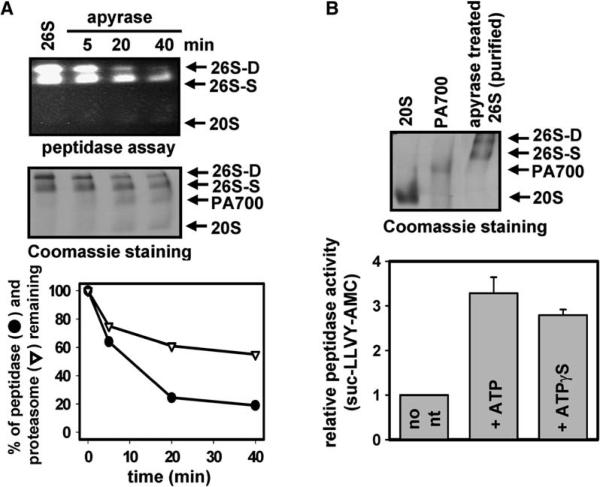
Although 26S proteasome assembly and activation were temporally indistinguishable, the time-dependent dissociation of 26S proteasome into component subcomplexes upon ATP removal resulted in loss of proteasome activity that was proportionately greater than the loss of 26S proteasome protein. For example, in a representative experiment shown in Figure 3A, a 40% loss in 26S proteasome protein (after 40 min) corresponded to a greater than 80% loss of proteasome activity. This result suggested that ATP binding could play distinct roles in 20S/PA700 association and proteasome activation. To test this possibility further, we isolated apyrase-treated 26S proteasome from dissociated 20S proteasome or PA700 by gel filtration chromatography and determined the effect of nucleotides on 26S proteasome activity. ATP and ATPγS activated hydrolysis of Suc-Leu-Leu-Val-Tyr-AMC by about 2- to 3-fold in various experiments (Figure 3B). These results indicate that ATP directly activates 26S proteasome. Thus, distinct ATP binding events appear to be responsible for 26S proteasome assembly/disassembly and activation.
Figure 3. 26S Proteasome Assembly and Activation Require Distinct ATP Binding Events.
(A) Depletion of ATP promotes dissociation of 26S proteasome and loss of activity. Purified 26S proteasome (40 nM) was incubated with 8 mU/μl apyrase at 37°C. At the indicated times, samples were subjected to native PAGE and assayed for proteasome activity using a gel overlay assay with Suc-Leu-Leu-Val-Tyr-AMC (upper panel), stained with Coomassie blue (middle panel), or assayed for proteasome activity using a soluble quantitative fluorescence assay with Suc-Leu-Leu-Val-Tyr-AMC (lower panel). The protein levels in the native gel were quantified by densitometry and plotted with the corresponding proteasome activity (lower panel).
(B) ATP binding promotes reactivation of ATP-deficient 26S proteasome. 26S proteasome was incubated for 60 min with apyrase as described above and subjected to Superose 6 gel filtration chromatography to separate the 26S proteasome from the dissociated 20S and PA700 subcomplexes. 26S proteasome isolated from the gel filtration column was subjected to native PAGE (upper panel) and to activity assays in the absence or presence of 0.5 mM ATP or 0.5 mM ATPγS (lower panel). Activity is expressed as arbitrary fluorescent units and represents the mean value of triplicate assays (±SEM). Similar results were obtained in three independent experiments.
ATP Hydrolysis Is Not Required for Degradation of Natively Disordered Proteins by the 26S Proteasome
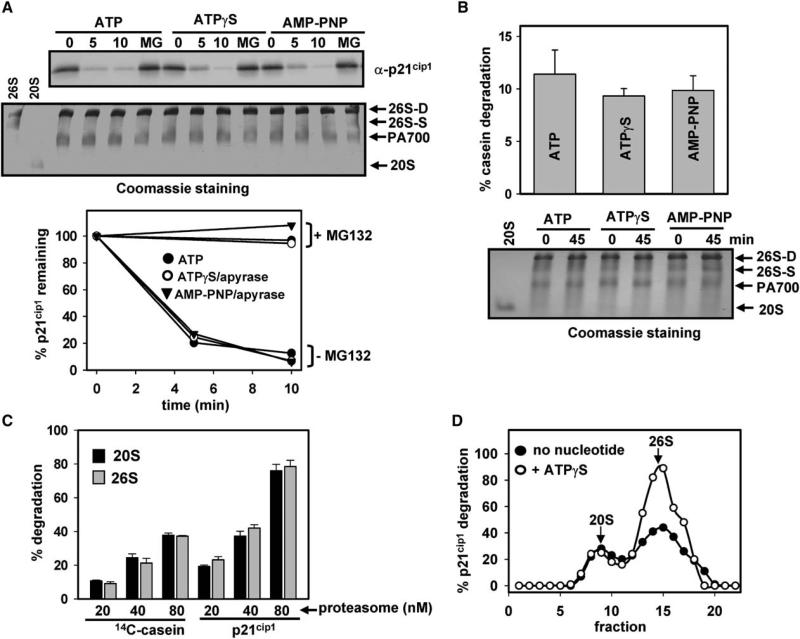
The 26S proteasome, as shown previously and in Figure 5, has the capacity to degrade certain natively disordered, nonubiquitinated proteins, such as p21cip1, casein, and α-synuclein, in the presence of ATP (Liu et al., 2003). Although ATP binding is sufficient to assemble and maintain 26S proteasome structure and to activate proteolysis of short peptides, we hypothesized that ATP hydrolysis would be required for translocation and degradation of larger protein substrates. To test this hypothesis, we examined the nucleotide requirement for the degradation of disordered protein substrates by the 26S proteasome. ATP was removed from doubly capped 26S proteasomes by apyrase, and the buffer was supplemented with ATPγS or AMP-PNP to maintain doubly capped 26S proteasome structure (Figures 4A and 4B). In contrast to our expectations, ATPγS or AMP-PNP supported 26S proteasome-catalyzed degradation of p21cip1 and casein at rates indistinguishable from those supported by ATP. To exclude the possibility that ATP hydrolysis-independent degradation of protein substrates was accomplished exclusively by small amounts of free 20S proteasome in these assays (estimated by native PAGE to be at least 20-fold lower than 26S proteasome, Figure 4), we conducted two additional experiments. First, we directly compared rates of p21cip1 and casein hydrolysis by doubly capped 26S proteasome and 20S proteasome. Equimolar concentrations of 20S and 26S proteasomes degraded these unstructured proteins at comparable rates (Figure 4C). Thus, proteolysis in assays containing predominantly doubly capped 26S proteasome cannot be explained by the exclusive action of 20S proteasome, and most degradation must be catalyzed by the 26S proteasome. Second, we subjected doubly capped, apyrase-treated 26S proteasome to glycerol density gradient centrifugation to separate 26S proteasome from 20S proteasome and then assayed gradient fractions for proteolysis of p21cip1 and casein in the presence and absence of ATPγS. Because the apyrase-treated 26S proteasome was isolated without ATP or ATPγS required for stabilization of its structure, the contribution of 20S proteasome to proteolysis was increased in this experiment due to partial dissociation of 26S proteasome (as compared to experiments in which apyrase-treated proteasomes were stabilized by immediate addition of nucleotides and subsequent centrifugation in nucleotide-containing buffer). Nevertheless, the results clearly demonstrate that 26S proteasome degrades p21cip1 (Figure 4D) and casein (data not shown) by an ATP hydrolysis-independent mechanism. Moreover, ATP binding appears to activate 26S proteasome-catalyzed degradation of proteins beyond its effect on 26S proteasome assembly (Figure 4) in a manner similar to its effect on the hydrolysis of peptide substrates described above (Figure 3). These results confirm that the contribution of 20S proteasome to proteolysis in assays containing doubly capped 26S proteasome is minimal and that 26S proteasome catalyzes degradation of unstructured proteins by an ATP hydrolysis-independent mechanism. In contrast to unstructured proteins, well-folded proteins, such as GFP, were not degraded by the 26S proteasome or 20S proteasome under any tested assay condition, including incubations extended to more than 4 hr. In sum, these surprising results indicate that the 26S proteasome degrades unstructured proteins by a mechanism that does not require ATP hydrolysis, and they suggest that such substrates reach the catalytic sites by passive diffusion through the opened pores of the proteasome.
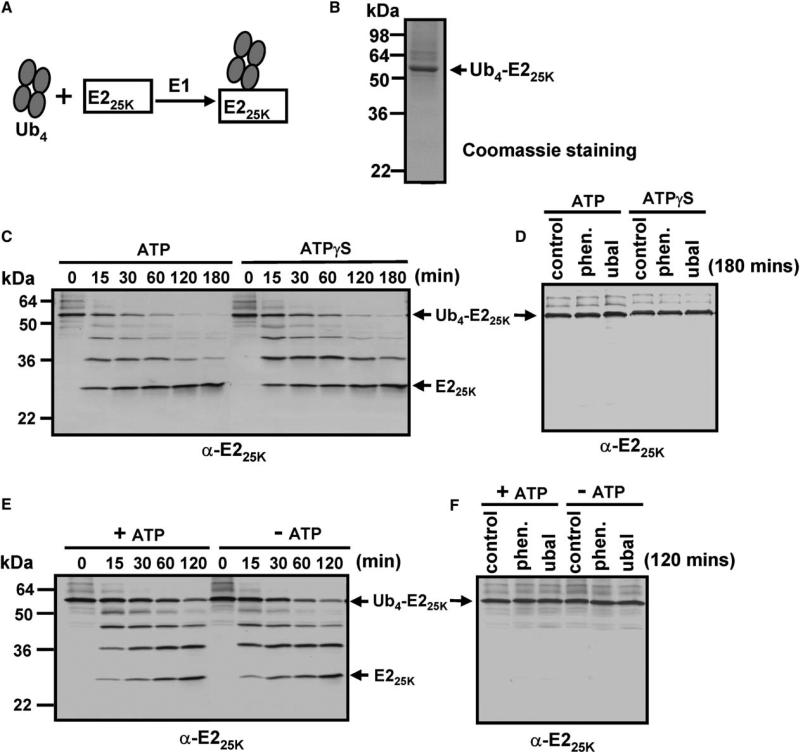
Figure 5. Polyubiquitin Chain Engagement and Deubiquitylation Do Not Require ATP Hydrolysis.
(A) Scheme for preparation of Ub4-E225K.
(B) Ub4-E225K was purified by Superdex 200 chromatography; Ub4-E225K (2 μg) was subjected to 10% SDS-PAGE and visualized by Coomassie staining.
(C–F) Deubiquitylation of Ub4-E225K by the 26S proteasome and PA700. Deubiquitylation was monitored by blotting with anti-E225K. (C) Ub4-E225K (120 nM) and purified 26S proteasome (80 nM) were incubated at 37°C for indicated times in the presence of 1 mM ATP or 1 mM ATPγS after depletion of ATP by apyrase. (D) Reactions as in (C) after 180 min of incubation with no 26S proteasome (control) or 26S proteasome after 10 min preincubation with 2.5 mM 1,10-phenanthroline (1, 10 phen) or 2 μM ubiquitin aldehyde (Ubal). (E and F) Deubiquitylation of Ub4-E225K by PA700. Reactions are as in (C) and (D) except with 160 nM PA700 without (–) or with (+) 1 mM ATP.
Figure 4. ATP Hydrolysis Is Not Required for Degradation of Nonubiquitylated Unstructured Proteins by the 26S Proteasome.
(A) Degradation of p21cip1 by doubly capped 26S proteasome. Doubly capped 26S proteasome was prepared in the presence of 1 mM ATP, ATPγS, or AMP-PNP. p21cip1 (1.5 μM) was incubated with doubly capped 26S proteasome (40 nM) at 37°C for the indicated times. Lanes denoted MG indicate that proteasomes were preincubated with 100 μM MG132 prior to addition of substrate and initiation of assay. Samples were either subjected to western blotting against p21cip1 antibody (upper gel) or evaluated for integrity of the 26S proteasome during degradation by Coomassie staining after native PAGE (lower gel). Purified 26S proteasome (containing both doubly capped and singly capped proteasome) and 20S proteasome were also electrophoresed as references for unassembled complexes. p21cip1 was quantified by densitometry after western blotting (lower). Similar results were obtained in three independent experiments.
(B) Degradation of [methyl-14C] casein by purified 26S proteasome. Reactions contained 40 nM doubly capped 26S proteasome and 0.2 mg/ml of [methyl-14C] casein in the presence of 1 mM ATP, ATPγS, or AMP-PNP. Proteolysis is expressed as the percentage of casein degraded and represents mean values of triplicate assays (±SEM). Samples before and after the reaction were also subjected to native PAGE to evaluate the integrity of the 26S proteasome (lower gel).
(C) Degradation of p21cip1 by 20S proteasome and 26S proteasome. p21cip1 was incubated with indicated concentrations of 20S proteasome or doubly capped 26S proteasome for 8 min at 37°C. Residual p21cip1 was quantified by densitometry after western blotting. Proteolysis is expressed as the percent of initial p21cip1 degraded. Values represent means of triplicate assays (±SEM).
(D) 26S proteasome does not require ATP hydrolysis to degrade unstructured proteins. ATP was removed from doubly capped 26S proteasome by apyrase. The sample was subjected to glycerol density gradient centrifugation in ATP-deficient buffer. Fraction 1 (12.5% glycerol), fraction 22 (40% glycerol). Samples from fractions were assayed for degradation of p21cip1 or [methyl-14C] casein (data not shown) as described above in the presence or absence of 0.5 mM ATPγ S. Arrows denote sedimentation positions of purified untreated 20S proteasome and 26S proteasomes in separate control experiments.
ATP Hydrolysis Is Not Required for Deubiquitylation of Nondegradable Polyubiquitylated Proteins by the 26S Proteasome
Despite its capacity to degrade certain disordered, nonubiquitinylated proteins, the 26S proteasome's main physiological substrates are proteins modified with polyubiquitin chains. Commonly accepted but incompletely documented models of 26S proteasome function for degradation of such substrates posit that ATP hydrolysis is linked to substrate unfolding, translocation, and deubiquitylation, three functions required for passage of substrates through the narrow opened pores of the proteasome and transit to the catalytic sites. The results presented above exclude translocation of a polypeptide chain as a process obligatorily linked to ATP hydrolysis. To determine the role of ATP hydrolysis on substrate unfolding and deubiquitylation, we prepared K48-linked polyubiquitylated forms of four folded proteins: Ub5-GFP, Ubn-MDM2, Ub4-E225K, and Ub4-Ubch10. These substrates had different susceptibilities to proteolysis by the 26S proteasome. For example, Ub5-GFP and Ub4-E225K were refractory to degradation but were deubiquitylated to unmodified proteins that remained resistant to proteolysis even after extended (>4 hr) incubation (Figure 5 and Figure S1). Deubiquitylation of these substrates was independent of ATP hydrolysis and was catalyzed by both 26S proteasome and isolated PA700. Moreover, PA700, whose stability was unaffected in the absence of ATP, deubiquitylated these substrates at indistinguishable rates in the presence or absence of ATP, ATPγ S, or AMP-PNP (Figure 5, Figure S1, and data not shown). These results demonstrate that deubiquitylation per se requires neither ATP binding nor ATP hydrolysis.
Deubiquitylation of nondegradable substrates was inhibited by ubiquitin aldehyde and by 1,10-phenanthroline (Figures 5D and 5F and Figure S1) but was unaffected by proteasome inhibitors such as MG132 and epoxomicin (data not shown). GFP was not degraded when deubiquitylation was inhibited by either ubiquitin aldehyde or 1,10-phenanthroline (Figure S1) or when it was fused to ubiquitin by a noncleavable peptide bond at residue 76 (Ub-V76-GFP), as described previously (Guterman and Glickman, 2004) (data not shown).
ATP Hydrolysis Is Utilized to Couple Processes Required for Degradation of Folded and Unstructured Polyubiquitinated Proteins by the 26S Proteasome
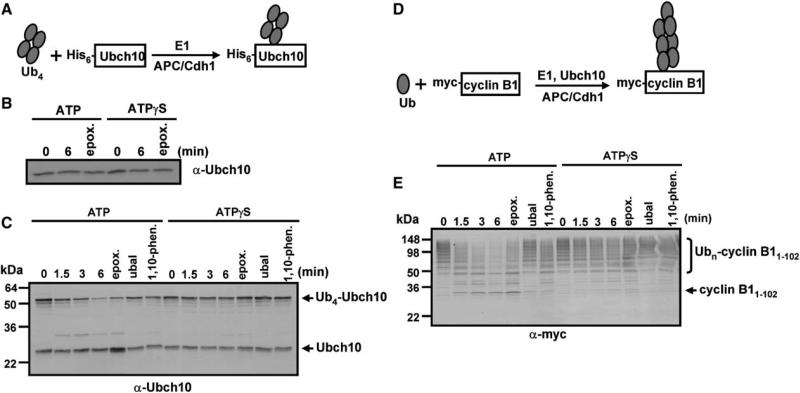
In contrast to Ub5-GFP and Ub4-E225K, other polyubiquitylated, folded proteins such as Ub4-Ubch10 and Ubn-MDM2 were degraded rapidly by the 26S proteasome (Figure 6 and data not shown). Proteolysis required both polyubiquitin modification and ATP hydrolysis because nonubiquitylated forms of these proteins were not degraded and degradation of Ub4-Ubch10 and ubiquitylated Ubch10 and Ubn-MDM2 was not supported by ATPγS or AMP-PNP (Figures 6B and 6C, and data not shown). Degradation was documented by the time-dependent disappearance of Ub4-Ubch10 without concurrent accumulation of nonubiquitylated Ubch10 and was blocked by proteasome inhibitors such as epoxomicin and MG132 (Figure 6C). Because the results described in previous sections demonstrated that neither substrate translocation nor substrate deubiquitylation requires ATP hydrolysis, these results could indicate that ATP hydrolysis is utilized mainly for substrate unfolding as a requisite for degradation. To test this possibility, we examined the degradation of polyubiquitylated myccyclin B11–102 (Figures 6D and 6E). This N-terminal fragment of cyclin B1 has been documented by NMR to be structurally disordered (Z.T. and H.Y., unpublished data). We reasoned that ATP hydrolysis would not be required for proteolysis of this substrate if ATP hydrolysis were used principally for substrate unfolding. However, Ubn-cyclin B11–102 was degraded much more rapidly in the presence of ATP than in the presence of ATPγS (Figure 6E). Therefore, these and other results described below reveal a more complex role for ATP hydrolysis in proteolysis than can be explained by substrate unfolding alone. For example, inhibitors of 26S proteasome deubiquitylating activities, such as 1,10-phenanthroline and ubiquitin aldehyde, inhibited both deubiquitylation and proteolysis, as judged by the stabilization of Ub4-Ubch10 (Figures 6C and 6E and data not shown). These results show that proteolysis is coupled to substrate deubiquitylation and are consistent with previous results (Verma et al., 2002; Yao and Cohen, 2002). Moreover, deubiquitylation of both Ub4-Ubch10 and Ubncyclin B11–102 required ATP hydrolysis, as revealed by the accumulation of deubiquitylated substrate by epoxomicin-inhibited proteasome in the presence of ATP, but not in the presence of ATPγS or AMP-PNP (Figures 6C and 6E). These results demonstrate a close mechanistic coupling among multiple processes required for proteolysis of ubiquitylated proteins. They also suggest that some individual processes, which in isolation are independent of ATP hydrolysis, become dependent on ATP hydrolysis when they are mechanistically linked (Figure 7).
Figure 6. ATP Hydrolysis Is Required for Degradation of a Polyubiquitylated Protein.
(A) Scheme for synthesis of Ub4-Ubch10.
(B) Nonubiquitinated Ubch10 is not degraded by the 26S proteasome. Reactions contained Ubch10 (100 nM) and 26S proteasome (30 nM) in the presence of 1 mM ATP or ATPγS. In reactions containing epoxomicin, proteasome was preincubated with 100 μM epoxomicin for 10 min prior to initiation, and reactions were stopped at the last time point.
(C) ATP hydrolysis-dependent degradation of Ub4-Ubch10 by the 26S proteasome. Doubly capped 26S proteasome (30 nM) was incubated with Ub4-Ubch10 (100 nM) with 1 mM ATP or 1 mM ATPγS for the indicated times. Ubch10 was detected by western blotting. Lanes denoted Ubal, 1,10-phen, and Epox indicate that proteasome was preincubated with either 2 μM ubiquitin aldehyde, 2.5 mM 1,10-phenanthroline, or 100 μM epoxomicin, respectively, for 10 min prior to initiation of reaction. Ubch10 was monitored by western blotting with an anti-Ubch10 antibody.
(D) Scheme for synthesis of Ubn-cyclin B11–102.
(E) ATP hydrolysis-dependent degradation of Ubn-cyclin B11–102. Degradation of Ubn-cyclin B11–102 (100 nM) was assessed by western blotting with an anti-myc antibody in reactions analogous to (C). Reactions contained 20 nM 26S proteasome in the presence of 1 mM ATP or ATPγS.
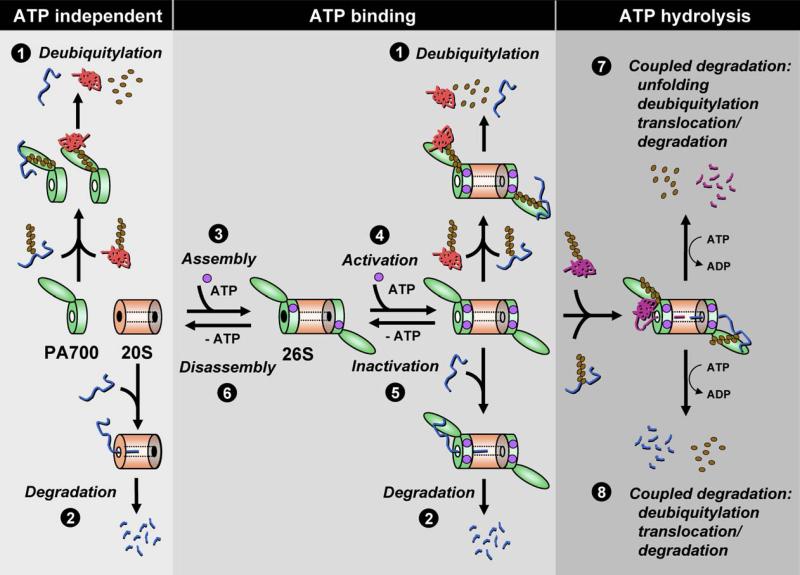
Figure 7. A Model for the Roles of ATP in 26S Proteasome Function.
The 26S proteasome is composed of the 20S proteasome and the PA700 regulatory complex. Purified PA700 is an ATPase but deubiquitylates polybubiquitylated proteins by an ATP-independent process (1). The 20S proteasome can degrade certain unstructured proteins that promote gating (2). Assembly of the 26S proteasome from 20S proteasome and PA700 requires ATP binding, but not hydrolysis (3). Activation of the 26S proteasome may require a separate ATP binding event (4). Removal of ATP promotes inactivation (5) and disassembly (6) of the 26S proteasome. The 26S proteasome deubiquitylates, by an ATP-independent mechanism, some polyubiquitylated proteins that are refractory to degradation (1). The 26S proteasome degrades certain unstructured, nonubiquitylated proteins without ATP hydrolysis (2). The 26S proteasome degrades both folded and unstructured polyubiquitylated proteins by a process that requires ATP hydrolysis, and it couples mechanisms required for degradation including substrate unfolding (when needed), deubiquitylation, translocation, and degradation (7 and 8).
Discussion
Previous work has established that ATP hydrolysis is required for the overall degradation of polyubiquitylated proteins by the 26S proteasome. The 26S proteasome is an ATPase by virtue of six homologous AAA ATPase subunits that compose the base of the PA700-regulatory complex (Hoffman and Rechsteiner, 1996; DeMartino et al., 1994; Glickman et al., 1998b). However, the relative functions of these different ATPase subunits during proteolysis remain poorly defined. Biochemical and genetic data indicate that they play at least some discrete, nonredundant functions in both proteolytic and nonproteolytic roles of the proteasome (Rubin et al., 1998; Köhler et al., 2001; Gonzalez et al., 2002). Our studies have revealed several distinct roles for ATP in 26S proteasome function and have distinguished between requirements for ATP binding and ATP hydrolysis.
ATP binding is sufficient to promote assembly of the 26S proteasome from 20S proteasome and PA700 regulator (Figure 7). The simplest model for this effect is that ATP binding to one or more PA700 ATPase subunits increases affinity of PA700 for 20S proteasome. Because assembly occurs in vitro with highly purified preparations of 20S proteasome and PA700, our data indicate that no additional assembly factors are required. Previous reports have described extraproteasomal proteins that promote assembly and/or stability of 26S proteasome. For example, Hsp90, an ATP-regulated chaperone, was shown to promote assembly and to maintain stability of the 26S proteasome in yeast (Imai et al., 2003). We were unable to detect Hsp90 in our preparations of 26S proteasome, 20S proteasome, or PA700, and geldanamycin, an Hsp90 inhibitor, had no effect on any of the assembly or disassembly experiments reported here (data not shown). Ecm29 was identified as a stoichiometric binding partner for yeast 26S proteasome and proposed to enhance 26S proteasome stability by binding to both 20S and PA700 subcomplexes (Leggett et al., 2002). Ecm29 also binds to mammalian 26S proteasome (Gorbea et al., 2004) (G. Adams and G.N.D., unpublished data). However, none of our purified proteins contained detectable amounts of ecm29, and purified ecm29 had no effect on 26S proteasome in our in vitro assembly reactions (G. Adams and G.N.D., unpublished data). Thus, although these or other extraproteasomal proteins might affect features of ATP-dependent assembly and stability of the 26S proteasome under different conditions, we conclude that mammalian proteasome subcomplexes are sufficient for 26S proteasome assembly.
Once assembled, the 26S proteasome requires bound ATP for structural stability and dissociates into 20S proteasome and PA700 subcomplexes upon removal of ATP. In the presence of ATP or nonhydrolyzable ATP analogs, the 26S proteasome survived prolonged incubations in the absence or presence of degradable polyubiquitylated and nonpolyubiqutiylated substrates. These findings strongly suggest that the 26S proteasome is relatively stable during proteolysis.
ATP binding to PA700 and concomitant 26S proteasome assembly is necessary to increase hydrolysis of short synthetic peptide substrates, an effect that probably reflects PA700-induced opening of pores at the ends of the 20S proteasome. We were unable to temporally separate assembly from activation. This finding is consistent with a model in which ATP-dependent binding of PA700 to the 20S proteasome is necessary and sufficient for activation. However, other results suggest that distinct ATP binding events differentially regulate proteasome assembly and activation (Figure 7). For example, upon removal of ATP, the rate of 26S proteasome disassembly was slower than the rate of loss of proteasome activity. Moreover, ATP stimulated activity of 26S proteasome isolated in the absence of ATP, indicating that ATP can directly activate an already assembled 26S proteasome. These latter findings are consistent with results from several previous studies (Hoffman et al., 1992), including one in which a yeast 26S proteasome harboring a point mutation in the ATP-binding domain of the ATPase subunit Rpt2 has low catalytic activity against short peptides (Köhler et al., 2001; Rubin et al., 1998). Because this mutant proteasome is assembled normally, ATP binding at other sites might be sufficient for assembly, whereas ATP binding at Rpt2 has a separate role in activation. Distinct functions have been demonstrated for individual subunits of other multimeric AAA ATPases (Jeruzalmi et al., 2001; Zalk and Shoshan-Barmatz, 2003), and elegant studies of the bacterial ClpX ATPase have revealed that even the identical subunits of this homohexameric complex behave in nonequivalent fashion with respect to nucleotide binding, hydrolysis, and substrate interaction (Hersch et al., 2005; Martin et al., 2005). Although it is not evident that all multimeric AAA ATPases conform to a uniform mechanistic scheme, such studies reinforce the likelihood that individual subunits of PA700 will have nonequivalent functions. A major challenge for future work will be to determine the relative roles of the six PA700 AAA subunits in proteasome function.
The 26S proteasome exhibits preference for polyubiquitinated forms of most substrates but also can degrade certain structurally disordered, nonubiquitylated proteins. We utilized such substrates to distinguish among possible requirements for ATP hydrolysis during the degradation of polyubiquitylated proteins. Such requirements include the binding of the polyubiquitin chain to PA700 receptor subunits, deubiquitylation, substrate unfolding, and substrate translocation to the catalytic sites. Our data show that ATP hydrolysis was not required for degradation of nonubiquitylated unstructured proteins such as p21, casein, and α-synuclein. These results indicate that, as with short peptides, certain long unstructured polypeptides transit to the catalytic sites by a passive process and establish that translocation per se need not be coupled to ATP hydrolysis. While this manuscript was under review, Smith et al. reported that ATP hydrolysis was not required for either binding of archaeal 20S proteasome to the PAN ATPase complex or subsequent degradation of unstructured proteins (Smith et al., 2005), thereby clarifying a previous report that these features required ATP hydrolysis (Benarouodj et al., 2003). These results strongly support the conclusion that translocation of unstructured, nonubiquitinated proteins into the 20S degradation chamber does not require ATP hydrolysis (Figure 7), and they provide an example of how this process differs between the proteasome and Clp systems.
The features of 26S proteasome-catalyzed degradation of folded proteins differed appreciably from those of degradation of disordered proteins. No folded protein we tested was degraded unless it was modified by a polyubiquitin chain. This requirement reflects the established role of polyubiquitin as an important determinant for interactions between folded substrates and the 26S proteasome (Thrower et al., 2001). Although a polyubiquitin chain was necessary for degradation of folded proteins, it was not sufficient. Some ubiquitylated proteins were rendered refractory to degradation by ATP hydrolysis-independent deubiquitylation that proceeded faster than proteolysis, suggesting that these two processes are not obligately coupled. Deubiquitylation also was catalyzed by isolated PA700 in the absence of ATP, showing that deubiquitylation per se requires neither ATP binding nor ATP hydrolysis (Figure 7). The nearly complete inhibition of deubiquitylation by either ubiquitin aldehyde or 1,10-phenanthroline was unexpected based on previous reports (Verma et al., 2002; Yao and Cohen, 2002). These results suggest that the functions of the subunits responsible for this activity are linked. Although the precise explanation for this effect is unclear, we note that our proteasome preparations contain two ubiquitin aldehyde-sensitive subunits, Uch37 and Usp14, in addition to the 1,10-phenanthroline-sensitive S13/Rpn11 subunit, and therefore have a different composition of deubiquitylating proteins than yeast proteasomes. High concentrations of 1,10-phenanthroline, such as those used here, have been reported to inhibit deubiquitylation by nonspecific mechanisms, possibly by destabilizing proteasome structure (Guterman and Glickman, 2004; Yao and Cohen, 2002). However, we found no effect of 2.5 mM 1,10-phenanthroline on 26S proteasome structure (as determined by native PAGE) or function (as determined by assays of degradation of peptide or non-ubiquitylated protein substrates) (X.L. and G.N.D., unpublished data).
Other folded and unstructured polyubiquitylated proteins were susceptible to both deubiquitylation and degradation in a process that was ATP hydrolysis dependent and mechanistically coupled. The basis for distinction between degradation-susceptible and refractory polyubiquitylated substrates is unclear but could be the presence or absence, respectively, of sites on the substrate capable of interacting directly with PA700. Such sites might be localized unstructured regions of otherwise globally folded proteins that serve as points of engagement by the proteasome after initial binding of the polyubiquitin chain. Proteins that lack these sites may be deubiquitylated before becoming engaged in proteolysis. Previous work supports the view that both polyubiquitin modification and structural features of the protein contribute to susceptibility for proteolysis (Prakash et al., 2004). In this regard, we noted that the degradation of some unstructured, polyubiquitylated proteins occurred in two phases. In addition to the rapid, coupled, ATP hydrolysis-dependent process described above (Figure 6), a fraction of such substrates was deubiquitylated completely prior to proteolysis; degradation of the resulting unmodified protein occurred during subsequent incubation by an ATP hydrolysis-independent mechanism similar to that of unstructured proteins that had never been ubiquitylated (C. Liu, X.L., and G.N.D., unpublished data).
Finally, our data show that several, and perhaps most, processes essential for degradation of susceptible polyubiquitylated proteins are mechanistically coupled. Remarkably, the coupled action of these processes imposes a mutual dependence on ATP hydrolysis that is not manifested for an isolated process. The detailed mechanism of such coupling and its relationship to ATP hydrolysis are unknown. However, conformational changes in the ATPase subunits during ATP hydrolysis might be transmitted to other sites responsible for binding and deubiquitylation of the polyubiquitin chain and for binding, unfolding, and translocation of the protein substrate. In this manner, the close apposition of these sites could make the processes they support mutually dependent on one another and therefore on ATP hydrolysis.
Experimental Procedures
Reagents
Human E1, ubiquitin aldehyde, and epoxomicin were purchased from Boston Biochemical. Bovine ubiquitin, 1,10-phenanthroline, apyrase, ATP, ATPγ S, AMP-PNP, and ADP were purchased from Sigma. Succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcourmarin (AMC) was from Bachem. Antibodies were purchased against the following proteins: Ubch10, E225K (Boston Biochemical), p21Cip1 (Santa Cruz), GFP (Covance), and myc (Covance). Ubch5a was a generous gift of Dr. James Chen (UT Southwestern).
Recombinant Protein Expression and Purification
Cdc27-conjugated Affi-Prep Protein A beads were prepared as described previously (Rape and Kirschner, 2004). Cdh1 was purified from sf9 cells (Rape and Kirschner, 2004; Tang and Yu, 2004). His-p21cip1 and α-synuclein were prepared as described previously (Liu et al., 2003). GST-E2 25K, His6-Ubch10, and His6-Yuh1 were expressed in E. coli and purified. Linear fusion proteins of Ub(G76)-GFP-His6 and Ub(V76)-GFP-His6 were expressed in E. coli and purified. Myc-cyclin B11–102 was prepared as described previously (Tang and Yu, 2004).
Proteasome Purification
Latent 20S proteasome and PA700 were purified from bovine red blood cells, as described previously (DeMartino et al., 1996; McGuire et al., 1989). 26S proteasome was purified from bovine red bloods as described in Supplemental Data.
Measurement of Proteasome Activity against Flurogenic Peptide Substrates
Proteasome activity was assessed by measuring hydrolysis of Suc-Leu-Leu-Val-Tyr-AMC in either a soluble assay, as described previously (Ma et al., 1994) except that fluorescence of AMC was monitored in a 96-well plate reader (Bio-TEK), or by substrate overlay assays in native polyacrylamide gels (Elasser et al., 2005; Hoffman et al., 1992). Degradation of [methyl-14C]-casein was measured as described previously (McGuire et al., 1989). Degradation of other proteins was assessed as described either previously (Liu et al., 2003) or below.
Measurement of ATPase Activity
ATPase activity of 26S proteasome and isolated PA700 was measured as described previously (Van Veldoven and Mannaerts, 1987).
Native Polyacrylamide Gel Electrophoresis
PAGE was conducted with 4% acrylamide gels containing 0.5 mM ATP and 2.5 mM MgCl2 (Elasser et al., 2005). Gels were either stained for protein with Coomassie blue or overlayed with Suc-Leu-Leu-Val-Tyr-AMC (50 μM) for assessment of proteasome activity.
Assembly of 26S Proteasome from Subcomplexes
Assembly of 26S proteasome from purified 20S proteasome and PA700 was carried out as described previously (DeMartino et al., 1996) in buffer A (20 mM Tris [pH 7.1], 5 mM MgCl2, 1 mM ATP, 0.5 mM DTT).
SDS-PAGE and Immunoblotting
Proteins were separated by 10% or 12.5% SDS-PAGE, visualized by silver staining or Coomassie blue staining, or transferred into nitrocellulose and were blotted with appropriate antibodies. Immunoblotting was conducted with alkaline phosphatase-conjugated immunoreactions using appropriate secondary antibodies.
Preparation of Polyubiquitylated Proteins
Polyubiquitylated proteins were prepared by several methods. K48-linked tetraubiquitin (Ub4) was synthesized as described previously (Piotrowsi et al., 1997) and conjugated to Ub(G76)-GFP, E225K, or Ubch10. Ub5-GFP was synthesized in buffer A containing 100 nM human E1, 15 μM GST-E225K, 25 μM Ub4, and 2.5 μM Ub(G76)-GFP. After 3 hr at 37°C, Ub5-GFP was isolated with Ni2+-NTA resin (Qiagene); bound proteins were eluted with buffer C (20 mM Tris HCl [pH 7.1], 10% glycerol) containing 120 mM imidazole. Ub4-E225K was synthesized with a similar method, except that E225K replaced GST-E225K. Proteins were further purified with Superose 6 equilibrated in buffer C containing 150 mM NaCl. To obtain Ub4-Ubch10, the APC/C E3 complex was immunoprecipitated from extracts of interphase Xenopus oocytes by protein A-conjugated CDC27 antibody (Rape and Kirschner, 2004). The ubiquitylation reaction contained 100 nM E1, 4 μM His-Ubch10, 160 μl APC/C complex immunoprecipitated from 2 ml Xenopus extract, 10 μM Ub4, 0.5 mM β-mercaptoethanol, and an ATP regenerating system in XB buffer (10 mM HEPES [pH7.7], 100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2, 50 mM sucrose). After 1 hr at room temperature, Ub4-Ubch10 was purified with Ni-NTA resin; protein was eluted with 120 mM immidazole. Polyubiquitin chains of undefined lengths were conjugated to myc-cyclin B1(1–102). Myc-cyclin B1(1–102) was ubiquitylated as described previously (Tang and Yu, 2004).
Measurement of Proteolysis and Deubiquitylation of Protein Substrates by the Proteasome
Degradation and deubiquitylation reactions were conducted with purified proteins described above. In some experiments, purified 26S proteasome was preincubated at 37°C for 30 min in buffer A with PA700 to promote assembly of doubly capped proteasomes. For assays with ATPγS, AMP-PNP, or ADP 26S proteasome was treated with apyrase (8 mU/μl) for 1 min at 30°C. Measurements of ATP concentration showed it to be less than 1 nM after treatment. ATPγS or AMP-PNP was added to the proteasome for the indicated experiment. Degradation assays contained 20 mM Tris-HCl (pH 7.1) at 37°C, 125 mM NaCl, 5 mM MgCl2, 1 mM ATP or other nucleotide, 5% glycerol, 40 nM 26S proteasome, and substrates, After incubation at 37°C for indicated times, reactions were either terminated by SDS-PAGE sample buffer or subjected to native PAGE.
Supplementary Material
Supplement
Acknowledgments
We thank Cecile Pickart for generous advice regarding preparation of tetraubiquitin. This work was supported by a grant from the National Institutes of Health (DK46181 to G.N.D. and DK49835 to P.J.T.), the Muscular Dystrophy Association (to G.N.D.), and the Parkinson's Disease Foundation (to P.J.T.).
Footnotes
References
- Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Benarouodj N, Zwickl P, Seemüller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol. Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. The proteasome. Annu. Rev. Biophys. Biomol. Struct. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- Burton RE, Baker TA, Sauer RT. Energy-dependent degradation: linkage between ClpX-catalyzed nucleotide hydrolysis and protein-substrate processing. Protein Sci. 2003;12:893–902. doi: 10.1110/ps.0237603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- DeMartino GN, Slaughter CA. The proteasome, a novel protease regulated by multiple mechanisms. J. Biol. Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- DeMartino GN, Moomaw CR, Zagnitko OP, Proske RJ, Ma C-P, Afendis SJ, Swaffield JC, Slaughter CA. PA700, an ATP-dependent activator of the 20S proteasome, is an ATPase containing multiple members of a nucleotide-binding protein family. J. Biol. Chem. 1994;269:20878–20884. [PubMed] [Google Scholar]
- DeMartino GN, Proske RJ, Moomaw CR, Strong AA, Song X, Hisamatsu H, Tanaka K, Slaughter CA. Identification, purification, and characterization of a PA700-dependent activator of the proteasome. J. Biol. Chem. 1996;271:3112–3118. doi: 10.1074/jbc.271.6.3112. [DOI] [PubMed] [Google Scholar]
- Elasser S, Schmitt M, Finley D. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 2005;398:353–363. doi: 10.1016/S0076-6879(05)98029-4. [DOI] [PubMed] [Google Scholar]
- Ferrell K, Wilkinson CRM, Dubiel W, Gordon C. Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem. Sci. 2000;25:83–88. doi: 10.1016/s0968-0004(99)01529-7. [DOI] [PubMed] [Google Scholar]
- Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9Å structure of a proteasome-11S activator complex and implication for proteasome-PAN/PA700 interactions. Mol. Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998a;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fried VA, Fischer JE, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 1998b;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2002;286:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- Gorbea C, Teter K, Holmes RK, Rechsteiner M. Characterisation of mammalian ecm29, a 26S proteasome-associated protein that localizes to the nucleus and membrane vesicles. J. Biol. Chem. 2004;279:54849–54861. doi: 10.1074/jbc.M410444200. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of the 20S proteasome from yeast at 2.4Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MN, Finley D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2001;11:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- Guterman A, Glickman MH. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the protea-some. J. Biol. Chem. 2004;279:1729–1738. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- Hersch GL, Burton RE, Bolon DN, Baker TA, Sauer RT. Asymmetric interactions of ATP with the AAA+ ClpX 6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Hoffman L, Rechsteiner M. Nucleotidase activities of the 26S proteasome and its regulatory complex. J. Biol. Chem. 1996;271:32538–32545. doi: 10.1074/jbc.271.51.32538. [DOI] [PubMed] [Google Scholar]
- Hoffman L, Pratt G, Rechsteiner M. Multiple forms of the 20S multicatalytic and the 26S ubiquitin-ATP-dependent pro-teases from rabbit reticulocyte lysate. J. Biol. Chem. 1992;267:22362–22368. [PubMed] [Google Scholar]
- Imai J, Maruya M, Yashiroda H, Yahara I, Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003;22:3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, O'Donnell M, Kuriyan J. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001;106:417–428. [PubMed] [Google Scholar]
- Kisselev AF, Kaganovich D, Goldberg AL. Binding of hydrophobic peptides to several non-catalytic sites promotes peptide hydrolysis by all active sites of 20S proteasomes. J. Biol. Chem. 2002;277:22260–22270. doi: 10.1074/jbc.M112360200. [DOI] [PubMed] [Google Scholar]
- Köhler A, Cascio P, Leggett DS, Woo KM, Goldberg AL, Finley D. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell. 2001;7:1143–1152. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Waltz T, Ploeugh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell. 2002;10:498–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Liu C-W, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C-P, Vu JH, Proske RJ, Slaughter CA, DeMartino GN. Identification, purification, and characterization of a high-molecular weight, ATP-dependent activator (PA700) of the 20S proteasome. J. Biol. Chem. 1994;269:3539–3547. [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- McGuire MJ, McCullough ML, Croall DE, DeMartino GN. The high molecular weight multicatalytic proteinase, macro-pain, exists in a latent form in human erythrocytes. Biochim. Biophys. Acta. 1989;995:181–186. doi: 10.1016/0167-4838(89)90078-2. [DOI] [PubMed] [Google Scholar]
- Ogura T, Tanaka K. Dissecting various ATP-dependent steps involved in proteasomal degradation. Mol. Cell. 2003;11:3–5. doi: 10.1016/s1097-2765(03)00004-2. [DOI] [PubMed] [Google Scholar]
- Ortega J, Heymann JB, Kajava AV, Ustrell V, Rechsteiner M, Steven AC. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J. Mol. Biol. 2005;346:1221–1227. doi: 10.1016/j.jmb.2004.12.049. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Piotrowsi J, Beal R, Hoffman L, Wilkinson KD, Cohen RE, Pickart CM. Inhibition of the 26S proteasome by polyubiquitin chains synthesized to have defined lengths. J. Biol. Chem. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432:588–595. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Rubin DM, Glickman MH, Larsen CN, Druvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Kafri G, Cheng Y, Ng D, Wala T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Tang Z, Yu H. Functional analysis of the spindle checkpoint proteins using an in vitro ubiquitination assay. Methods Mol. Biol. 2004;281:227–242. doi: 10.1385/1-59259-811-0:227. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2001;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veldoven PP, Mannaerts GP. Inorganic and organic phosphate measurements in the nanomolar range. Anal. Biochem. 1987;161:45–48. doi: 10.1016/0003-2697(87)90649-x. [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR, Koonin EV, Deshaies RJ. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S protea-some: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2001;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- Wilk S, Orlowski M. Evidence that pituitary cation-sensitive neutral endopeptidase is a multicatalytic protease complex. J. Neurochem. 1983;40:842–849. doi: 10.1111/j.1471-4159.1983.tb08056.x. [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Zalk R, Shoshan-Barmatz V. ATP-binding sites in brain p97/VCP (valosin-containing protein), a multifunctional AAA ATPase. Biochem. J. 2003;374:473–480. doi: 10.1042/BJ20030219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement