Antisense transcripts with rice full-length cDNAs (original) (raw)
Short abstract
In this study, 687 sense-antisense transcript pairs from 32,127 full-length rice cDNA sequences were identified by aligning the cDNA sequences with rice genome sequences. The large number of pairs suggests that gene regulation by antisense transcripts occurs in plants and not only in animals.
Abstract
Background
Natural antisense transcripts control gene expression through post-transcriptional gene silencing by annealing to the complementary sequence of the sense transcript. Because many genome and mRNA sequences have become available recently, genome-wide searches for sense-antisense transcripts have been reported, but few plant sense-antisense transcript pairs have been studied. The Rice Full-Length cDNA Sequencing Project has enabled computational searching of a large number of plant sense-antisense transcript pairs.
Results
We identified sense-antisense transcript pairs from 32,127 full-length rice cDNA sequences produced by this project and public rice mRNA sequences by aligning the cDNA sequences with rice genome sequences. We discovered 687 bidirectional transcript pairs in rice, including sense-antisense transcript pairs. Both sense and antisense strands of 342 pairs (50%) showed homology to at least one expressed sequence tag other than that of the pair. Microarray analysis showed 82 pairs (32%) out of 258 pairs on the microarray were more highly expressed than the median expression intensity of 21,938 rice transcriptional units. Both sense and antisense strands of 594 pairs (86%) had coding potential.
Conclusions
The large number of plant sense-antisense transcript pairs suggests that gene regulation by antisense transcripts occurs in plants and not only in animals. On the basis of our results, experiments should be carried out to analyze the function of plant antisense transcripts.
Background
The transcripts of sense-antisense transcript pairs have complementary sequences. Natural antisense transcripts are transcripts of the opposite DNA strand to the sense strand, either at the same genomic locus as the sense strand (_cis_-encoded antisense transcripts) or at a different genomic locus (_trans_-encoded antisense transcripts). Antisense transcripts affect the expression of sense RNAs at all levels - transcription, RNA processing and transport, and RNA stability and translation - and thus may be involved in the control of development, adaptation to various stresses, and viral infection [1,2]. Genome imprinting [3-5] is sometimes triggered by antisense transcripts: 15% of imprinted genes are associated with antisense transcripts [6]. Antisense transcripts are also involved in methylation [7], X-chromosome inactivation [8-10], alternative splicing [11,12], RNA editing [13] and RNA interference [14].
Since the first examples of sense-antisense transcript pairs were reported in 1981 from human and mouse mitochondrial DNA [1,15,16], and overlapping sense and antisense transcripts were described in Drosophila [17], increasing numbers of endogenous antisense RNAs have been detected in numerous organisms: viruses, slime molds, insects, amphibians and birds, as well as in mammals (rats, mice, cows and humans) [1].
Genome sequences and sequences of mRNA transcripts of several species have been determined. These genome sequences enable us to search for sense-antisense mRNA candidates in the same loci in whole-genome sequences by aligning the mRNA sequences with genome sequences. Lehner et al. [18] performed a computational search for human sense-antisense candidates using mRNA sequences in public databases and identified 372 natural antisense transcripts. About the same time, Shendure and Church [19] found 217 sense-antisense candidates in public databases of mouse and human expressed sequence tags (ESTs) and detected 33 antisense transcripts by an orientation-specific reverse transcription (RT)-PCR assay (RT-PCR). In 2003, Yelin et al. [20] identified 2,667 sense-antisense transcripts from human expressed sequences in public databases and, using microarrays containing strand-specific oligonucleotide probes and northern blot analysis, confirmed that at least 1,600 sense-antisense candidates were transcribed from both DNA strands. In mouse, 2,481 mouse sense-antisense full-length cDNA pairs were identified from 60,770 mouse full-length cDNAs determined in our laboratory and mRNA sequences from public databases, and 4,511 sense-antisense transcripts among 4,962 candidates (2,481 pairs) were supported by at least one EST sequence [21].
Few sense-antisense transcript pairs have been reported in plants [2,22]. At present, no computational search for sense-antisense candidates from large numbers of plant mRNAs and whole-genome sequences has been reported (since the submission of this study, a large number of Arabidopsis antisense mRNAs have been reported [23]). To remedy this lack of data, in April 2000 we began a comprehensive Rice Full-Length cDNA Sequencing Project (RFLSP) [24]. We determined ESTs of cDNA clones and classified them to reduce redundant cDNAs and determine low-redundancy full-length cDNA sequences [25]. We obtained 32,127 low-redundancy Oryza sativa full-length cDNA sequences. In this study, we conducted an initial large-scale search for plant sense-antisense candidates on a large scale from these O. sativa full-length cDNA sequences and 1,687 O. sativa mRNAs in public databases.
Results
Detection of bidirectional transcript pairs
We aligned the rice full-length cDNA sequences determined by the RFLSP and mRNA sequences from a public database with rice genome sequences. From the successfully aligned sequences, we selected those that overlapped with sequences on the other strand of the rice genome sequence as bidirectional transcript pairs. Then we classified these pairs according to the same system used to classify mouse bidirectional transcript pairs [21] to investigate the exon-intron structures of the pairs. First, the pairs were broadly divided into two categories according to whether the exons of the pairs overlapped - that is, whether mRNAs of the pairs included complementary regions of sequence. We termed pairs with a complementary region of sequence 'sense-antisense transcript pairs' and those without such a region 'non-antisense bidirectional transcript pairs'. Then the sense-antisense transcript pairs were divided into two subcategories, and the non-antisense bidirectional transcript pairs into three subcategories, based on the exon-intron structure and position (Figure 1). Most of the bidirectional transcript pairs were sense-antisense transcript pairs (categories 1 and 2; Figure 1). This distribution was also observed among mouse bidirectional transcript pairs [21]. However, more non-antisense bidirectional transcript pairs were in category 4 than in either category 3 or 5; this tendency was not observed in mouse. Figure 2 shows the distribution of sense-antisense transcript pairs according to the length of overlap (in base-pairs) of the exons. The numbers of pairs that mapped onto each chromosome are shown in Table 1. All chromosomes contained some pairs. However, the percentage of genes on each chromosome that include bidirectional transcript pairs cannot be calculated at present, because the rice genome sequence is not fully assembled, and the number of genes on each chromosome has not yet been estimated accurately.
Figure 1.
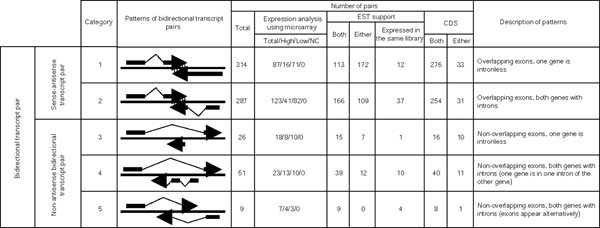
Bidirectional transcript pairs divided into five categories according to their patterns of exon-intron structure. The total numbers of bidirectional transcript pairs in each category were counted. The numbers of pairs detected by using a microarray are shown under 'Expression analysis using microarray'; in this column, 'Total' is the number of pairs put on the microarray, 'High' is the number of pairs in which both transcripts of the pairs were expressed at high intensity, 'Low' is the number of pairs in which the transcript of at least one member of the pair was expressed at low intensity, and 'NC' is the number of pairs in which neither transcript was expressed. The numbers of pairs in which sequences showed homology to at least one EST sequence are shown under 'EST support'. 'Both' indicates the number of pairs in which both sense-antisense strands showed homology to at least one EST sequence. 'Either' is the number of pairs in which either a sense or an antisense strand of the pairs showed homology to at least one EST sequence. 'CDS' is the number of pairs with coding potential. Here, 'Both' means that both sense-antisense strands of the pairs had coding potential, and 'Either' means that either the sense or the antisense strand of the pairs had coding potential. The numbers of pairs in which both sense and antisense transcripts were expressed in at least one identical library are listed under 'Expressed in the same library'.
Figure 2.
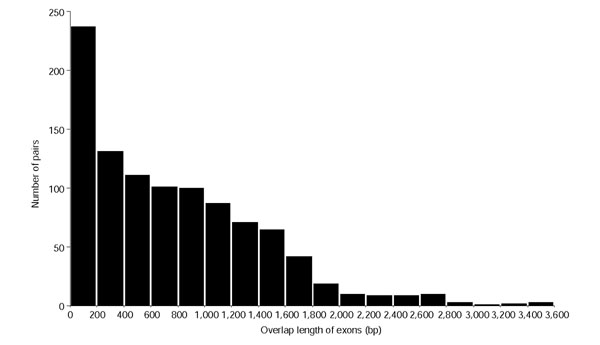
Distribution of overlap lengths of exons in sense-antisense transcript pairs. The number of pairs (y axis) is plotted against the overlap length (bp) of exons in each bidirectional transcript pair (x axis).
Table 1.
Numbers of bidirectional transcript pairs per chromosome and their lengths
| Chromosome | Number of sense-antisense transcript pairs | Number of non-antisense transcript pairs | Length of chromosome (Mb) |
|---|---|---|---|
| 1 | 215 | 30 | 51.4 |
| 2 | 130 | 8 | 43.8 |
| 3 | 162 | 52 | 47.3 |
| 4 | 109 | 11 | 36.6 |
| 5 | 62 | 17 | 33.8 |
| 6 | 63 | 24 | 35.4 |
| 7 | 54 | 7 | 33.1 |
| 8 | 64 | 7 | 33.6 |
| 9 | 45 | 8 | 27.2 |
| 10 | 43 | 4 | 23.7 |
| 11 | 46 | 34 | 33.6 |
| 12 | 112 | 59 | 31.2 |
Characterization of bidirectional transcript pairs
To confirm the reproducibility of the bidirectional transcript pairs, we used FASTA [26] to search for them against 32,718 rice 5'-EST sequences in the GenBank database, 91,425 rice 5'- and 175,642 rice 3'-EST sequences sequenced by the RFLSP. Among the 687 bidirectional transcript pairs identified, 642 showed homology to at least one EST sequence, not including the 5'- and 3'-EST sequences of the bidirectional transcript pairs themselves. Among these 642 pairs, 342 contained sequences homologous to at least one EST sequence on both strands of the pair; 300 contained such sequences on just one strand. The number of bidirectional transcript pairs in each category with EST support is shown in Figure 1 (the complete EST support dataset is available on our website [27]). The frequency distribution of EST matches on bidirectional transcript pair cDNA sequences is presented in Figure 3. Among 1,374 sequences of bidirectional transcript pairs, 931 sequences contain contiguous exons that are separated by a putative intron in the genomic sequence, and thus underwent splicing; therefore these pairs could not have been derived from contamination of the genome sequences.
Figure 3.
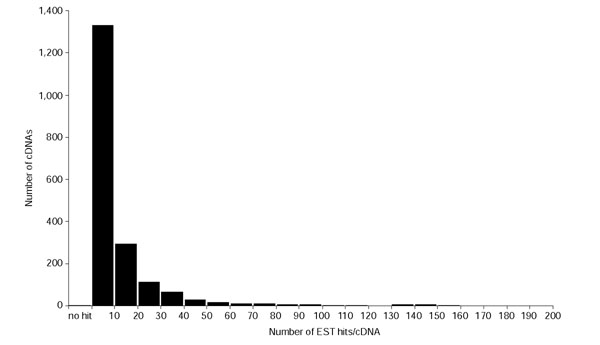
Frequency distribution of ESTs having homology to the bidirectional transcript pairs. The number of cDNAs (y axis) is plotted against the number of ESTs showing homology to each bidirectional transcript pair cDNA (x axis).
We also investigated the coding potential of the bidirectional transcript pairs. There is no computer program designed to predict coding sequence (CDS) regions in rice full-length cDNA or mRNA sequences, so we used the NCBI SEALS Wimklein program [28], which is based on the longest open reading frame (ORF) method, to deduce protein sequences from the sequences of the bidirectional transcript pairs. Among 687 bidirectional transcript pairs, 594 (86%) included a CDS region (at least 300 bp long) in both strands of the pair, and 86 (13%) included a CDS region in one of the strands (Figure 1). Thus, in contrast to mouse bidirectional transcript pairs [21], most rice bidirectional transcript pairs included a CDS region in one or both strands.
To study the library origins of the bidirectional transcript pairs, we also searched the 5'- and 3'-EST sequences from several kinds of rice full-length cDNA libraries constructed by the RFLSP against the sequences of the bidirectional transcript pairs using FASTA. The presence of EST sequences with ≥ 96% identity over 80% of overall length was the criterion used to identify the library origins of the matched bidirectional transcript pairs. Among the 687 bidirectional transcript pairs, 64 were expressed in a single library, as judged on the basis of EST support (Figure 1). The frequency of both members of the rice bidirectional transcript pairs in the same library (9.3%) was lower than that for mouse (23.2%) [21], perhaps because of the difference in the number of available EST sequences between rice and mouse. However, the results of the microarray experiments, presented in the next section, showed that 32% of the pairs on the microarray had high expression intensities on both strands of the pair. The distribution of the bidirectional transcripts in the 32,127 full-length rice cDNAs among the different libraries is shown on our website [27]. The library origins of all redundant bidirectional transcript pairs, including the 687 representative bidirectional transcript pairs, are also shown on our website.
Expression analysis of bidirectional transcript pairs
A single-strand oligo microarray has been designed on the basis of 21,938 rice full-length cDNA sequences, which were determined and selected as transcriptional units in RFLSP [24,29]. Oligo microarray measurements that were performed by Agilent Technologies were in good agreement with Q-PCR measurements [30]. We used the microarray to investigate the expression of the bidirectional transcripts. Among 687 nonredundant bidirectional transcript pairs, 258 pairs were aligned on the oligo microarray. We hybridized mRNAs derived from eight kinds of libraries - young leaf (YL), germinating seed (GS), mature leaf (ML), panicle (P), root (R), apical meristem (Ap), callus (Ca), and primary callus (Pc) - on the microarray. Figure 4 shows the distribution of the expression intensities of 21,938 mRNAs derived from YL and 956 mRNAs of the bidirectional transcripts derived from YL. The expression intensities of all mRNAs and of mRNAs of bidirectional transcripts were not clearly divided into low and high intensities, but were distributed broadly from low to high intensity and varied among libraries. We therefore normalized the expression intensities by subtracting the median expression intensity of 315 negative controls instead of the median of the 21,938 mRNAs from the intensities of the bidirectional transcripts. As a result, all bidirectional transcripts on the microarray were expressed with more than the median intensity of the negative controls. We divided the expression intensities into three categories: high (++), low (+) and not detected (-); we defined mRNAs that were expressed in greater amount than the median of the expression intensities of all 21,928 mRNAs on the microarray as having high expression, those lower than the median and higher than the median intensities of the negative controls as low, and those lower than the median as not detected. Among 258 nonredundant bidirectional transcript pairs on the microarray, in 82 (32%) both transcripts showed high expression intensity in at least one of the eight libraries, and in 176 (68%) at least one transcript showed low expression intensity in eight libraries. The numbers of bidirectional transcripts in each category showing each level of expression intensity are shown in Figure 1. The bidirectional transcripts in categories 1 and 3 included 122 and 23 intronless transcripts in each category; all intronless transcripts in nonredundant bidirectional transcript pairs were more highly expressed than the median of the negative controls; and 46 (38%) intronless transcripts in categories 1 and 16 (70%) in category 3 were expressed at high intensity. This shows that the intronless bidirectional transcripts are not the contamination of the genome sequences but mRNAs. The expression intensities of all bidirectional transcript pairs including redundant pairs on the microarray are available from our website [27].
Figure 4.
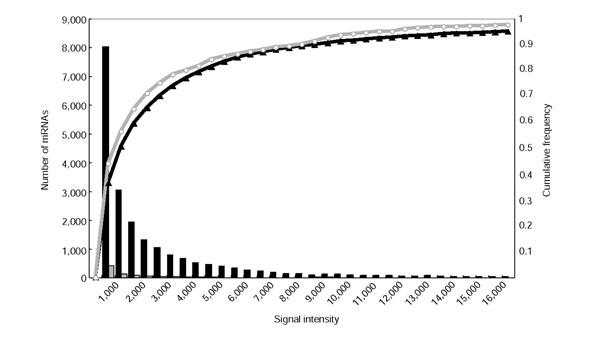
Cumulative frequency distribution of the expression intensities of 21,928 rice transcriptional units (black line) and 258 bidirectional transcript pairs on the microarray (gray line). Signal intensity (_x_-axis) is plotted against the number of mRNAs with that signal intensity (bars) and also against the normalized cumulative frequency (lines).
All bidirectional transcript pairs on the array, except for three pairs in category 1, seven pairs in category 2, three pairs in category 3 and two pairs in category 4, were expressed in all eight libraries at a higher level than the median expression intensity of the negative controls. However, some bidirectional transcripts were expressed at high intensity in some libraries and expressed at low intensity in the other libraries. Although microarrays can detect very low expression of mRNAs, we cannot determine the threshold of the expression intensity of mRNAs where they are activated. Thus, bidirectional transcripts expressed at low intensity in some libraries may not be functioning in the library.
Discussion
Although searches for large numbers of sense-antisense mRNA sequences in mammals have been reported [18,21], this is the first report of a search for a large number of sense-antisense pairs in plants. Through the RFLSP [24] and the rice genome project [31-34], it was possible to search for bidirectional transcript pairs in 32,127 rice full-length cDNA sequences by aligning them with rice genome sequences. As a result, we identified 687 bidirectional transcript pairs in rice. We had previously searched for mouse bidirectional transcript pairs by the same methods [21] and found 3,380 pairs, including 2,481 sense-antisense transcript pairs. The RFLSP produced 32,127 full-length rice cDNA sequences, estimated to consist of 20,447 transcriptional units [24], which we used in this study to search for rice bidirectional transcript pairs. The 60,770 mouse full-length cDNA sequences produced by the mouse full-length cDNA project were estimated to consist of 33,409 transcriptional units [35]. Even considering the difference in the number of transcriptional units between rice and mouse, the number of rice bidirectional transcript pairs identified is smaller than that of mouse. However, the mouse transcriptome was determined by producing close to 2 million sequences from strongly subtracted libraries [36], and this difference in coverage may partly account for the discrepancy. Taking into account the number of _trans_-encoded bidirectional transcript candidates or bidirectional transcript candidates from nonpolyadenylated RNAs, which were not included in this study [37], might increase the number of bidirectional transcripts in both mouse and rice.
Another difference between the mouse and rice bidirectional transcripts is that most of the rice bidirectional transcript pairs apparently include CDSs. About 86% of the rice bidirectional transcript pairs included CDSs on both strands, whereas about 27% of the mouse bidirectional transcripts included CDSs. To search for CDSs in rice bidirectional transcripts, we used the SEALS Wimklein computer program [30], which uses the longest-ORF method, because the available software for predicting CDS regions is not designed to detect rice cDNA or mRNA sequences. To evaluate the reliability of the longest-ORF method, we searched for CDSs from 32,127 rice full-length cDNA sequences and 60,770 mouse FANTOM2 sequences [35] using the SEALS Wimklein software. Among the 32,127 rice full-length cDNA sequences, 29,430 (91.6%) included CDSs, and among the 33,409 mouse FANTOM2 transcriptional units, 19,494 (58.3%) did. The frequency of mouse cDNA coding sequences was almost the same as that estimated by human curation (52.7%) [35]. Thus, the longest-ORF method should reliably detect CDS regions in rice full-length cDNA sequences. Furthermore, a study of CDS annotation of the mouse FANTOM2 sequences found that the longest-ORF method was particularly useful for predicting CDS regions in which no frameshift or stop codon errors were present [38]. In addition, an InterPro database search [39] showed that, among 32,071 rice full-length cDNA sequences, 21,702 coded for known protein-domain structures. However, the total number of rice full-length cDNA sequences is smaller than that of mouse, so if additional rice full-length sequences are determined, then the frequency of noncoding mRNAs may increase.
The expression and library origins of the bidirectional transcript pairs were checked by using 5'-EST sequences. Two million EST sequences are available for mouse, but only about 124,000 rice 5'-EST sequences produced by the RFLSP and in public databases are currently available. Among the 687 rice bidirectional transcript pairs, 342 (50%) matched at least one EST sequence on both strands, and 300 (44%) matched at least one EST sequence on one strand. These EST sequence matches do not include matches with the 5'- and 3'-EST sequences of the pair itself. When more rice EST sequences become available, the number of bidirectional transcript pairs with EST support on both strands may increase. The coexpression of the bidirectional transcript pairs was also studied by using EST sequences produced by the RFLSP. Recently, however, we used single-strand oligo microarrays to analyze the expression of bidirectional transcripts and detected more coexpression of the bidirectional transcript pairs, including intronless bidirectional transcripts, than predicted by the EST support. This result supported the idea that most intronless bidirectional transcripts were really expressed and were not merely contaminants of the genome sequences. Furthermore, because the genome sequences of each rice chromosome are not yet assembled, but consist of several hundred contigs, the exact number of genes or loci on each chromosome and the frequency of rice bidirectional transcript pairs in all rice genes cannot yet be estimated. The total number of rice genes has been roughly estimated by using gene-prediction software as 32,000-55,000 [31-34]. The availability of more rice EST sequences will also improve the detection of genes and loci in the rice genome.
Although far fewer antisense transcripts have been reported from plants compared with mammals and prokaryotes [2], the ability of some transgenes to silence the expression of homologous (chromosomal) loci was first observed in plants [40,41]. In these cases, introduced transgenes did not affect transcription of the target locus, but dramatically decreased the half-life of target RNAs. Such processes have been called post-transcriptional gene silencing (PTGS), but they were originally called 'co-suppression' in plants, 'RNA interference' in worms and flies, and 'quelling' in fungi. PTGS can be suppressed by several virus-encoded proteins and is closely related to RNA-mediated virus resistance and cross-protection in plants. Therefore, PTGS may represent a natural antiviral defense mechanism, and transgenes might be targeted because they, or their RNA, are perceived as viruses. PTGS may also represent a defense system against transposable elements that acts during plant development. PTGS in plants can be triggered effectively by double-stranded RNA (dsRNA), which in plants can be produced in two different ways. The first is the simultaneous expression of sense and antisense sequences (or of an RNA hairpin) corresponding to the desired target gene; the second is the simultaneous expression of a viral RNA replicase with a specific single-stranded RNA (ssRNA) that has been engineered to contain viral replication signals. Then the enzyme Dicer, or a Dicer-like enzyme, processes the dsRNA into small interfering RNAs (siRNA) of about 22 nucleotides each. The siRNAs are incorporated into multicomponent nucleases known as RNA-induced silencing complexes (RISCs). Each RISC then uses an unwound siRNA as a guide to the target mRNA [42-50]. Antisense transcripts may function through mechanisms such as PTGS. Recent studies on PTGS have revealed another function for dsRNA in plants: it can induce sequence-specific DNA methylation, known as RdDM [48]. Through the study of PTGS, breeders may become able to exploit dsRNA for crop improvement, and PTGS will also be useful in functional genomic studies.
Although PTGS is induced by mRNAs, not by proteins, as many as 86% of rice and 27% of mouse bidirectional transcript pairs are expected to include CDSs in both strands of the pair [21]. Among 20,447 rice transcript units, 18,807 (92%) are expected to include CDSs, and among 33,409 mouse transcript units, 19,494 (58.3%) include CDSs. Although the ratio of mouse noncoding mRNA sequences in all mouse transcripts is much higher than in rice, the numbers of protein-coding sequences are almost the same in rice and mouse: among full-length cDNAs sequenced so far in our projects; 594 pairs of rice and 519 pairs of mouse bidirectional transcripts include potential CDSs in both strands of each pair [21]. These bidirectional transcripts encoding protein sequences may function through mechanisms other than PTGS.
The existence of a large number of plant antisense transcripts is beginning to be revealed, and regulation of genes by antisense transcripts is more widespread than previously thought. The functional analyses of bidirectional transcript pairs presented here should contribute greatly to identifying the mechanisms of gene regulation in plants and to understanding the differences in these mechanisms between plants and animals.
Conclusions
We detected a large number of plant sense-antisense transcript pairs, which will be of key importance in analyses of the mechanisms and functions of plant post-transcriptional gene regulation. Further experimental analysis should reveal the function of these antisense transcripts.
Materials and methods
Search for bidirectional transcript pairs from rice full-length cDNA and mRNA sequences
We used 32,127 O. sativa full-length cDNA sequences and 1,687 O. sativa mRNA sequences (retrieved using the taxonomic ID number for O. sativa (japonica cultivar group) 39947 and including the words 'complete cds' in their definitions) from the GenBank database. We aligned those O. sativa cDNA and mRNA sequences with O. sativa genome sequences from the Institute for Genomic Research (TIGR) using BLAST [51] and Spidey [52] software. First, the cDNA and mRNA sequences were searched against the rice genome sequences using BLAST. The BLAST searches listed cDNA and mRNA sequences with equal or greater than 90% identity and a length of 50 bp or greater. Then the matched sequences were aligned with the rice genome sequences again using Spidey. From the Spidey results, cDNA and mRNA sequences with equal or greater than 96% identity over 50% or more of their overall lengths were selected as successfully mapped sequences. The criterion for overall length was set loosely ($50%) because the rice genome sequences have not yet been assembled as one contig per chromosome. When the same cDNA or mRNA sequence could align with several parts of the rice genome sequence, the position where it aligned with the longest region on the genome sequence was selected as the best-aligned position. Using the same method as was used in searches for mouse sense-antisense transcripts [21], we extracted sense-antisense transcript pairs and non-antisense bidirectional transcript pairs based on overlapping loci of the aligned cDNA and mRNA sequences. Then we grouped the sense-antisense pairs and non-antisense bidirectional transcript pairs into the five categories according to their exon-intron structures.
Search for EST sequences homologous to the bidirectional transcript pairs
To confirm that the bidirectional transcript pairs were derived from natural transcripts, we performed a FASTA search [26] for these bidirectional transcripts against 32,718 5'-EST sequences (retrieved using the taxonomic ID number for O. sativa (japonica cultivar group) 39947) in the GenBank database and 91,425 5'-EST sequences produced by the RFLSP. The EST sequences that matched each bidirectional transcript with equal or greater than 94% identity over 80% or more of the overall length were counted.
Library origins of the bidirectional transcript pairs
To analyze the library origins of the sequences of the bidirectional transcript pairs, we used the libraries of origin of not only the full-length sequences, but also of O. sativa EST sequences highly homologous to the full-length sequences. We searched the sequences against 91,425 5'-EST sequences and 175,642 3'-EST sequences using FASTA. The libraries in which we found 5'- and 3'-EST sequences with 94% or greater identity over 80% or more of the overall length were counted as libraries of origin of the matched sense or antisense sequences. Next, we counted the frequency of the libraries in which both sense and antisense clones were expressed simultaneously as follows. For each library, we counted the number of rice full-length cDNA clones expressed in the library and the number of sense and antisense clones expressed in the library and then calculated the ratio of the number of the sense and antisense clones to the number of rice full-length cDNA clones. We sorted the names of the libraries of origin according to each category of bidirectional transcript pairs by these ratios. The results are listed on our website [27]. We also counted the numbers of sense and antisense clones expressed in only one library, and these are also listed at the website [27].
Prediction of CDSs from the bidirectional transcript pairs
We used the NCBI SEALS Wimklein program [28], which is based on the longest-ORF method, to predict CDS regions from the sequences of the bidirectional transcript pairs. The optional parameters were set to
'-code = Standard -frames = all -mode = longest_orf -cutoff = 100'
to remove CDSs that coded for fewer than 100 amino acids (300 bp). The bidirectional transcript pairs including CDS regions with more than 100 amino acids were selected as pairs with coding potential.
Sample preparation for microarray analysis
Rice seeds (O. sativa L. cv Nipponbare) were supplied by Masahiro Yano of the National Institute of Agrobiological Sciences (NIAS), Japan. Rice seedlings were grown under hydroponic conditions at 28°C under a 16 h light/8 h dark cycle for 10 days. After germination, the young leaf (YL), root (R), and apical meristem (Ap) were harvested. The panicle (P) and mature leaf (ML) were cultivated in a pot at the Nagaoka University of Technology, Japan. Calluses (Ca) were induced from mature seeds placed on MS+2,4-D medium (10 ml of 0.2 mg/ml 2,4-dichlorophenoxyacetic acid in a medium made from 30 g sucrose added to 1,000 ml Murashige and Skoog vitamin mixture (Wako), the pH adjusted to 5.6 and 8 g agarose added) [53]. Seven days after the seeds were put on the medium, some of the primary calluses (Pc) were harvested and the other parts of the primary calluses were transferred onto new medium. Two weeks later the calluses were harvested. Total mRNA was prepared from each sample using a RNeasy kit (Qiagen, Valencia, CA), and the mRNAs purified by Oligotex-dT30 super mRNA Purification Kit (Takara Bio, Shiga, Japan).
Microarray analysis
We produced amplified cRNAs labeled with cyanine-3 CTP (Cy3) and cyanine-5 CTP (Cy5) from 200-500 ng mRNA using a Fluorescent Linear Amplification Kit (Agilent Technologies, Palo Alto, CA). We hybridized the fluorescent linear amplified cRNAs to custom-made _in situ_-synthesized 60-mer oligo microarrays containing 21,938 unique rice full-length transcripts (Agilent). Before hybridization, Cy3- and Cy5-labeled cRNAs were mixed and fragmented to an average size of 100-200 bases by using In situ Hybridization Kit Plus (Agilent) with incubation at 60°C for 30 min. Fragmented cRNAs were added to hybridization buffer, applied to the rice 22 K oligo microarray, and hybridized for 17 h at 60°C. The slides were washed and scanned on an Agilent Technologies G2565BA Microarray Scanner System. Scanned microarray images were processed with Feature Extraction 6.1.1 software (Agilent). To improve the accuracy of the expression data, we performed the experiment twice, labeling the same RNA templates in two separate reactions and analyzing four datasets per library. We used only data points that could be reproduced more than three times among four datasets per RNA template. Then we removed the results that the software had flagged as being due to corrupted spots from the data.
Acknowledgments
Acknowledgements
This work would not have been possible without the encouragement and support of Keiji Kainuma. We thank Itaru Nakayama and Hidenori Kiyosawa for their valuable comments and suggestions during the search for mouse antisense transcripts. We also thank the members of the RIKEN Genome Exploration Research Group Science Center for data preparation, Kimihisa Tasaki of Tochigi Prefectural Agricultural Experiment Station, Japan, Lee Jung-Sook of the National Institute of Agricultural Biotechnology, Korea, Koji Yamamoto of Nagaoka University of Technology, Japan, and Setsuko Kimura of the National Institute of Agricultural Science, Japan for sample preparations and microarray analysis. This study was supported by a research grant from the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science, and Technology of the Japanese Government to Y.H. and by ACT-JST (Research and Development for Applying Advanced Computational Science and Technology) of the Japan Science and Technology Corporation (JST) to Y.H. This work was also supported by a research grant for the Rice Genome Full-Length cDNA Library Construction Project from BRAIN (Bio-oriented Technology Research Advancement Institution) to Y.H.
References
- Vanhee-Brossollet C, Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene. 1998;211:1–9. doi: 10.1016/S0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- Terryn N, Rouze P. The sense of naturally transcribed antisense RNAs in plants. Trends Plant Sci. 2000;5:394–396. doi: 10.1016/S1360-1385(00)01696-4. [DOI] [PubMed] [Google Scholar]
- Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci USA. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, Mukai T, Niikawa N, Ogawa M, Wagstaff J, Kishino T. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet. 2003;12:837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Willard HF, Carrel L. Making sense (and antisense) of the X inactivation center. Proc Natl Acad Sci USA. 2001;98:10025–10027. doi: 10.1073/pnas.191380198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon BR, Chowdhury AK, Dunston JA, McIntosh I. Identification of TSIX, encoding an RNA antisense to human XIST, reveals differences from its murine counterpart: implications for X inactivation. Am J Hum Genet. 2001;69:951–960. doi: 10.1086/324022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe SH, Lazar MA. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J Biol Chem. 1991;266:22083–22086. [PubMed] [Google Scholar]
- Sureau A, Soret J, Guyon C, Gaillard C, Dumon S, Keller M, Crisanti P, Perbal B. Characterization of multiple alternative RNAs resulting from antisense transcription of the PR264/SC35 splicing factor gene. Nucleic Acids Res. 1997;25:4513–4522. doi: 10.1093/nar/25.22.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NT, Rohrbach JA, Zalewski BA, Byrkett CM, Vaughn JC. RNA editing and regulation of Drosophila 4f-rnp expression by sas-10 antisense readthrough mRNA transcripts. RNA. 2003;9:698–710. doi: 10.1261/rna.2120703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and organization of the mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Spencer CA, Gietz RD, Hodgetts RB. Overlapping transcription units in the dopa decarboxylase region of Drosophila. Nature. 1986;322:279–281. doi: 10.1038/322279a0. [DOI] [PubMed] [Google Scholar]
- Lehner B, Williams G, Campbell RD, Sanderson CM. Antisense transcripts in the human genome. Trends Genet. 2002;18:63–65. doi: 10.1016/S0168-9525(02)02598-2. [DOI] [PubMed] [Google Scholar]
- Shendure J, Church GM. Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biol. 2002;3:research0044.1–0044.14. doi: 10.1186/gb-2002-3-9-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- Kiyosawa H, Yamanaka I, Osato N, Kondo S, RIKEN GER Group and GSL Members. Hayashizaki Y. Antisense-transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant ncRNA database http://www.prl.msu.edu/PLANTncRNAs/database.html
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al. Collection, mapping, and annotation of 28,000 full-length cDNA clones from Japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Osato N, Itoh M, Konno H, Kondo S, Shibata K, Carninci P, Shiraki T, Shinagawa A, Arakawa T, Kikuchi S, et al. A computer-based method of selecting clones for a full-length cDNA project: simultaneous collection of negligibly redundant and variant cDNAs. Genome Res. 2002;12:1127–1134. doi: 10.1101/gr.75202. 10.1101/gr.75202. Article published online before print in June 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supplementary materials for this study http://cdna01.dna.affrc.go.jp/cDNA/Analysis/antisenseweb
- Walker DR, Koonin EV. SEALS: a system for easy analysis of lots of sequences. Proc Int Conf Intell Syst Mol Biol. 1997;5:333–339. [PubMed] [Google Scholar]
- Shimatani Z, Yazaki J, Kishimoto N, Hashimoto A, Nagata Y, Shimbo K, Fujii F, Taya T, Tonouchi M, Nelson C., et al. 22,000 single strand oligo-based microarray system for expression analysis of rice genes. Seventh International Congress of Plant Molecular Biology. 2003. Abstract.
- Carter MG, Hamatani T, Sharov AA, Carmack CE, Qian Y, Aiba K, Ko NT, Dudekula DB, Brzoska PM, Hwang SS, et al. In situ-synthesized novel microarray optimized for mouse stem cell and early developmental expression profiling. Genome Res. 2003;13:1011–1021. doi: 10.1101/gr.878903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Zhang Y, Hao P, Wang S, Fu G, Huang Y, Li Y, Zhu J, Liu Y, Hu X, et al. Sequence and analysis of rice chromosome 4. Nature. 2002;420:316–320. doi: 10.1038/nature01183. [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Matsumoto T, Yamamoto K, Sakata K, Baba T, Katayose Y, Wu J, Niimura Y, Cheng Z, Nagamura Y, et al. The genome sequence and structure of rice chromosome 1. Nature. 2002;420:312–316. doi: 10.1038/nature01184. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. i ndica). Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- FANTOM Consortium; RIKEN Genome Exploration Research Group Phase I & II Team Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Carninci P, Waki K, Shiraki T, Konno H, Shibata K, Itoh M, Aizawa K, Arakawa T, Ishii Y, Sasaki D, et al. Targeting a complex transcriptome: the construction of the mouse full-length cDNA encyclopedia. Genome Res. 2003;13:1273–1289. doi: 10.1101/gr.1119703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael GG. Antisense starts making more sense. Nat Biotechnol. 2003;21:371–372. doi: 10.1038/nbt0403-371. [DOI] [PubMed] [Google Scholar]
- Furuno M, Kasukawa T, Saito R, Adachi J, Suzuki H, Baldarelli R, Hayashizaki Y, Okazaki Y. CDS annotation in full-length cDNA sequence. Genome Res. 2003;13:1478–1487. doi: 10.1101/gr.1060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, et al. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 2003;31:315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Q, Wang HY, Jorgensen RA. Distinct patterns of pigment suppression are produced by allelic sense and antisense chalcone synthase transgenes in petunia flowers. Plant J. 1998;13:401–409. doi: 10.1046/j.1365-313X.1998.00038.x. [DOI] [Google Scholar]
- Jorgensen RA, Que Q, Stam M. Do unintended antisense transcripts contribute to sense cosuppression in plants? Trends Genet. 1999;15:11–12. doi: 10.1016/S0168-9525(98)01651-5. [DOI] [PubMed] [Google Scholar]
- Fire A. RNA-triggered gene silencing. Trends Genet. 1999;15:358–363. doi: 10.1016/S0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Beclin C, Fagard M. Post-transcriptional gene silencing in plants. J Cell Sci. 2001;114:3083–3091. doi: 10.1242/jcs.114.17.3083. [DOI] [PubMed] [Google Scholar]
- Brantl S. Antisense-RNA regulation and RNA interference. Biochim Biophys Acta. 2002;1575:15–25. doi: 10.1016/S0167-4781(02)00280-4. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. RNAi: Nature abhors a double-strand. Curr Opin Genet Dev. 2002;12:225–232. doi: 10.1016/S0959-437X(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Wang MB, Waterhouse PM. Application of gene silencing in plants. Curr Opin Plant Biol. 2002;5:146–150. doi: 10.1016/S1369-5266(02)00236-4. [DOI] [PubMed] [Google Scholar]
- Zamore PD. Ancient pathways programmed by small RNAs. Science. 2002;296:1265–1269. doi: 10.1126/science.1072457. [DOI] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Wheelan SJ, Church DM, Ostell JM. Spidey: a tool for mRNA-to-genomic alignments. Genome Res. 2001;11:1952–1957. doi: 10.1101/gr.195301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]