Versatile and open software for comparing large genomes (original) (raw)
Short abstract
The newest version of MUMmer easily handles comparisons of large eukaryotic genomes at varying evolutionary distances, as demonstrated by applications to multiple genomes.
Abstract
The newest version of MUMmer easily handles comparisons of large eukaryotic genomes at varying evolutionary distances, as demonstrated by applications to multiple genomes. Two new graphical viewing tools provide alternative ways to analyze genome alignments. The new system is the first version of MUMmer to be released as open-source software. This allows other developers to contribute to the code base and freely redistribute the code. The MUMmer sources are available at http://www.tigr.org/software/mummer.
Background
Genome sequence comparison has been an important method for understanding gene function and genome evolution since the early days of gene sequencing. The pairwise sequence-comparison methods implemented in BLAST [1] and FASTA [2] have proved invaluable in discovering the evolutionary relationships and functions of thousands of proteins from hundreds of different species. The most commonly used application of these sequence-analysis programs is for comparing a single gene (either a DNA sequence or the protein translation of that sequence) to a large database of other genes. The results of such protein and nucleotide database searches have been used in recent years as the basis for assigning function to most of the newly discovered genes emerging from genome projects. In recent years, an important new sequence-analysis task has emerged: comparing an entire genome with another. Until 1999, each new genome published was so distant from all previous genomes that aligning them would not yield interesting results. With the publication of the second strain of Helicobacter pylori [3] in 1999, following the publication of the first strain [4] in 1997, the scientific world had its first chance to look at two complete bacterial genomes whose DNA sequences lined up very closely. Comparison of these genomes revealed an overall genomic structure that was very similar, but showed evidence of two large inversion events centered on the replication origin. The comparison also made it clear that a new type of bioinformatics program was needed, one that could efficiently compare two megabase-scale sequences, something that BLAST cannot do. In response to this need, TIGR released MUMmer 1.0, the first system that could perform genome comparisons of this scale [5]. The first two releases of MUMmer had over 1,600 site licensees, a number that has grown since moving to an open-source license in May 2003.
The number of pairs of closely related genomes has increased dramatically in recent years, with a corresponding increase in the number of scientific studies of genome structure and evolution, facilitated by new software that permits the comparisons of these genomes. As of mid-2003, there are more than 150 complete published genomes, with over 380 prokaryotic genome projects and 240 eukaryotic projects under way. Many of these involve species that are closely related to published genomes. The published databases already include 33 species for which at least one other closely related species has been sequenced; for a detailed list see [6]. More distantly related pairs of species, for example, Plasmodium falciparum and P. yoelii, fail to show DNA sequence similarity but do show large-scale similarity when their translated protein sequences are aligned, as described in earlier studies [7,8].
Related to the growing number of closely related species that have been sequenced is a rapid growth in the number of known species whose genomes are similar but have undergone significant rearrangement. The human and mouse genomes, for example, are both available in draft form, and the chromosomes of either species can be aligned with the other at the DNA level. Various lines of evidence in the past have pointed to massive genome rearrangements separating the species, and the latest analysis indicates that the mouse genome can be split into 217 large segments that can be rearranged to produce the same gene order as in the human genome [9]. This very large-scale similarity interrupted by rearrangements places additional demands on genome-comparison programs: essentially, one must produce all pairs of similar regions in the sequences (in form of local alignments), not merely a single 'best' or longest global alignment of the entire sequences.
In addition to the need for whole-genome alignment programs, another need has become evident recently - a means of reliably evaluating and comparing genome assemblies. The explosion of genome sequencing has brought with it an explosion in genome-assembly programs, with several new assemblers either under development or recently released [10-12]. Unlike the previous generation of assemblers (TIGR Assembler [13], phrap [14], and CAP3 [15]), these second-generation assemblers are designed to handle large eukaryotic genomes. Assembly of large genomes is a major technical challenge, and once an assembly has been produced, evaluating it can be almost as difficult. Debates over the relative quality of assemblies produced by different assemblers are ongoing, and whole-genome comparison algorithms represent a critical tool in these analyses. Different assemblies of the same data should be nearly 100% identical, making the comparison problem analogous to the problem of comparing closely related species. Assembly differences may represent errors in one of the algorithms, and are useful for providing insights into the strengths and weaknesses of different methods. The large-scale comparison problem also occurs for assemblies delivered by the same software but from different inputs; for example, assemblies at threefold (3×) coverage and sixfold (6×) coverage of the same genome. With larger eukaryotic projects, multiple assemblies are run at different stages of the project, and comparisons of the successive assemblies provide a map showing how to transfer any analyses (such as gene predictions) from one assembly to another.
A third use for rapid, large-scale alignment programs has come up in our own applications. As part of our annotation 'pipeline' at TIGR, we routinely rebuild a database containing the results of all-against-all BLAST searches for all known proteins. Each time a new genome is added to the public archives, many thousands of searches need to be re-run to incorporate the newly sequenced genes. Because of the size of the archive, these additional searches take a relatively long time. A rapid method for identifying potential hits is used as a pre-screen as follows: for each new gene that is being added to the database, we use the high-speed method (MUMmer) to determine if it has any potential hits. If it does not, then it can be omitted from subsequent BLAST searches. If a new genome has a large number of novel proteins, this pre-screening step can substantially reduce the time required to search it against the database.
The new MUMmer system, version 3.0, addresses all of the above uses and more, including new graphical modules for viewing assembly comparisons and for looking at more distantly related species alignments. In addition, the implementations of all the fundamental search operations are now either optimal or nearly optimal, in the sense of running in time proportional to the sum of their input and output sizes. Other parts of the code have also been rewritten to improve their efficiency.
What may be the most significant change with MUMmer 3.0 is that it is now an open-source system. All code is publicly available without restriction on its use or redistribution, and we encourage others to add to the code base and distribute their own improvements. The modularity of the code base makes it easily extendable as well. Others can build on our matching algorithm, for example, and create their own clustering and extension steps.
Results
Since the development of MUMmer 1.0 in 1999, several other programs for large-scale genome comparison have been developed, for example, SSAHA [16], AVID [17], MGA [18], BLASTZ [19], and LAGAN [20] (see also [21] for a review). Most of these programs follow an anchor-based approach, which can be divided into three phases: computation of potential anchors; computation of a colinear sequence of non-overlapping potential anchors - these anchors form the basis of the alignment; and alignment of the gaps in between the anchors. The traditional methods to compute potential anchors, that is, maximal matches of some length l or longer, use a generate-and-test approach. In a first step, all matches of some fixed length k < l, called _k_-mers, are generated using a method based on hashing (adopted from [22]). Each such _k_-mer is checked to see whether it can be extended to a maximal exact match of length at least l. The extension is done by pairwise character comparisons, and thus the run time of this approach depends not only on the number of potential anchors, but also on their lengths. This can be illustrated by an example where all maximal matches of length 20 or larger between two different strains of Escherichia coli (strain K12, 4,639,221 base-pairs (bp) and strain O157:H7, 5,528,445 bp) are computed. With k = 10, a typical choice for k, the hashing approach first generates 4.99 × 107 _k_-mers and then performs 1.66 × 107 character comparisons to determine all 46,629 maximal matches of length 20 or larger. Thus, less than 0.1% of the generated _k_-mers are extended to maximal matches of the specified length. For this reason, the generate-and-test approach leads to long running times, if the sequences under consideration share long substrings.
Recognizing this disadvantage of the hashing approach, MUMmer 1.0 was the first software system to use suffix trees to find potential anchors for an alignment. Suffix trees have been studied for almost three decades in computer science (see [23] for an overview). A suffix tree is a data structure for representing all the substrings of a string, whether that string is a DNA sequence, a protein sequence, or plain text. Suffix trees have the following nice features which make them an important data structure for large-scale genome analysis: a suffix tree for a string S of length n can be represented in space proportional to n; fast algorithms have been designed that can construct a suffix tree in time proportional to n [24,25]; given the suffix tree of S and a query string Q of length m, there are algorithms to compute all unique maximal matches between S and Q of any specified minimum length (the potential anchors) in time proportional to m. All maximal matches, unique or not, can be found in near-optimal time. Note especially that, unlike the hashing approaches, the run-time of the suffix-tree algorithms does not depend on the length of the maximal matches.
Details of the suffix-tree algorithms incorporated in earlier versions of MUMmer have been described in [5,7]. Here we will focus on novel developments. MUMmer is among the fastest programs for large-scale alignment; one recent test reported times for MUMmer that ranged from 4 to 110 times faster than AVID, BLASTZ, and LAGAN [20]. At its default settings, MUMmer is less sensitive at detecting matches than these programs; however, we have added several command-line options to MUMmer 3.0 that permit the detection of much weaker matches than the system would find otherwise. Note that the modularity of MUMmer, and its availability as open-source code, means that others can now build a hybrid system using, for example, the suffix-tree matching algorithm in MUMmer and the match extension program code from LAGAN or AVID.
Additional features added to MUMmer 3.0 are a new Java viewer, DisplayMUMs; a new graphical output program to generate images in fig-format or PDF, showing the alignment of a set of contigs to a reference chromosome; and new options to find non-unique matches. These will be described below.
Optimized suffix-tree data structure and suffix-tree library
The most significant technical improvement in MUMmer 3.0 is a complete rewrite of the suffix-tree code, based on the compact suffix-tree representation of [26]. This representation was also used in the repeat analysis tool REPuter [27]. However, REPuter could only accommodate sequences up to 134 million bp (Mbp). For MUMmer 3.0 the implementation was improved so that it allows sequences up to 250 Mbp on a PC with 4 gigabytes (GB) of real memory, at the cost of a slightly larger space usage per base pair. For example, one can construct the suffix tree for human chromosome 2 (237.6 Mbp, the largest human chromosome) using 15.4 bytes per base-pair. For processing DNA sequences less than 134 Mbp in length, MUMmer can be compiled so that it uses only about 12.5 bytes per bp [26]. Since suffix trees for DNA sequences are typically larger than for protein sequences, the bytes per base-pair ratio is even better for the latter.
MUMmer now requires approximately 25% less memory than release 2.1 and it runs slightly faster. Compared to the initial release in 1999, the system is more than twice as fast and uses less than half the memory. As in MUMmer 2.1, release 3.0 streams the query sequence against the suffix tree of the reference sequence. Thus the total space requirement of MUMmer is the size of the suffix tree plus the size of the reference and the query sequences. Table 1 shows run times and memory requirements for MUMmer release 2.1 and 3.0, when computing maximal matches for different pairs of genomes or chromosomes.
Table 1.
Time and space requirements of MUMmer 2.1 and 3.0 when computing all exact matches of length 20 or longer for different pairs of sequences
| Reference genome | Query genome | MUMmer 2.1 | MUMmer 3.0 | ||||
|---|---|---|---|---|---|---|---|
| Species | Size (Mbp) | Species | Size (Mbp) | Time (sec) | Space (MB) | Time (sec) | Space (MB) |
| E. coli K12 | 4.6 | E. coli O157:H7 | 5.5 | 18 | 102 | 17 | 77 |
| A. fumigatus | 28.0 | A. nidulans | 30.1 | 128 | 578 | 120 | 459 |
| Saccharomyces cerevisiae | 13.0 | Schizosaccharomyces pombe | 13.8 | 51 | 261 | 47 | 204 |
| D. melanogaster (chromosome 2L) | 22.2 | D. pseudoobscura (all chromosomes) | 150.0 | 546 | 684 | 520 | 485 |
| Homo sapiens (chromosome 21) | 44.7 | Mus musculus (chromosome 16) | 99.2 | - | - | 430 | 759 |
While the previous versions of MUMmer implemented the main suffix tree construction and traversal algorithms in one monolithic program of 1,700 lines of code, the current version is based on a well-structured and well-documented software library. This provides data types for handling multiple DNA or protein sequences and their suffix trees. The library contains functions to construct the suffix tree and traverse it. In this way, a programmer who intends to modify or extend the code base can use the well-documented interfaces provided by the library, without the need to learn all of the low-level implementation details of the suffix tree.
With release 3.0, MUMmer now has the ability to run a multi-contig query against a multi-contig reference. Previously this was available by using the Nucmer package, but not directly within the core mummer program. In Table 1, for example, the genome sequence of Aspergillus fumigatus consisted (at the time of this study) of 19 scaffolds that were aligned to 248 contigs from A. nidulans. This comparison was handled by a simple call to the mummer program in release 3.0, but in release 2.1 the reference sequence needs to be first collapsed into a single contig and, after matching, the coordinates have to be re-mapped (by Nucmer) to the correct contig locations. Both releases handle multi-contig query files. Table 1 also shows the times for aligning the 22.2 Mbp chromosome 2L of the fruit fly Drosophila melanogaster to an interim assembly (before the genome project was complete) of D. pseudoobscura. In this case the query sequence, consisting of 4,653 scaffolds containing approximately 150 Mbp of sequence, was much longer than the reference. The program required 485 Mb of total memory, approximately 310 Mb for the suffix tree and the rest to hold the input sequences.
Non-unique maximal matches
Previous versions of MUMmer emphasized maximal unique matches (MUMs) as prospective anchors for an alignment. MUMs are unique in that they occur exactly once in each of the genomes. In some cases, the uniqueness constraint will prevent MUMmer from finding all matches for a repetitive substring. For example, if the reference genome has two exact copies of a particular string and the query has just one copy, then earlier versions of MUMmer would generally miss one of the matching copies, depending on the surrounding sequence. To overcome this problem, the new MUMmer system can find all maximal matches - including non-unique ones - between two input sequences simply by providing a command-line option to the program mummer. Other command-line options allow the user to produce MUMs that are unique in both the query and the reference sequence or MUMs that are unique only in the reference sequence.
Although the algorithm to produce all maximal matches is more complicated than the algorithm to produce unique maximal matches, it still runs in nearly optimal time, where optimal time would be proportional to the sum of the sizes of the input strings and the number of matches found. The run times to produce any of the three types of maximal matches are generally similar. Note, however, that when the program is directed to find all non-unique matches, including short ones, the size of the output can be extremely large, and the time to create the output file will be the dominant part of the computation.
Distant matches
One of the criticisms that has been leveled at MUMmer 1.0 was that it only finds exact matches, whereas in practice we often want to find approximate matches, that is, matches between sequences that are less than 100% identical. We addressed this concern in release 2.1, with the introduction of the Nucmer and Promer packages built on top of MUMmer. These have been substantially improved in the 3.0 release, and now exhibit performance only marginally slower than the basic search itself. The speed-up of Nucmer and Promer compared to release 2.1 is approximately 10-fold.
Both Nucmer and Promer produce a collection of local alignments using the algorithm described below. The difference between the two programs is that Nucmer constructs nucleotide alignments between two sets of DNA sequences, while Promer constructs amino acid alignments. Each set of sequences is a collection of one or more sequences from the same genome, for example, a collection of contigs produced by a genome assembler. Promer first translates both reference and query in all six frames, finds all matches in the amino acid sequences, and then maps the matches back to the original DNA coordinate system. For the extension step below, Promer uses a standard amino acid substitution matrix (BLOSUM62 is the default) to score mismatches.
The Nucmer/Promer alignment algorithm is as follows. First, both programs run MUMmer to find all exact matches longer than a specified length l, measured in nucleotides for Nucmer and amino acids for Promer. Second, the matches are clustered in preparation for extending them. Two matches are joined into the same cluster if they are separated by no more than g nucleotides (Nucmer) or amino acids (Promer). Then from each cluster, the maximum-length colinear chain of matches is extracted and processed further if the combined length of its matches is at least c nucleotides/amino acids. (Note that a chain can consist of a single matching region if l >c.) The parameters l, g, and c can all be set on the command line. The chain matches are then extended using an implementation of the Smith-Waterman dynamic programming algorithm [28], which is applied to the regions between the exact matches and also to the boundaries of the chains, which may be extended outward. This 'match and extend' step in the algorithm is essentially the same as that used by FASTA [29], BLAST [30], and many other sequence-alignment programs.
When two species are very similar, such as the two isolates of the Bacillus anthracis Ames strain sequenced at TIGR [31-33], then MUMmer is ideally suited for aligning the genomes. In that comparison of anthrax isolates, only four single-nucleotide differences separated the two 5.3 Mbp main chromosomes from one another. Similarly, in our comparison of a clinical isolate of Mycobacterium tuberculosis to a laboratory strain [31], MUMmer quickly found the approximately 1,100 SNPs and a handful of IS elements that distinguished the strains. However, when the species being compared are more distant, Nucmer and Promer provide much more detailed and more useful alignments than MUMmer alone. In the examples described below, we show how each of the programs described here may be run for genomes at different evolutionary distances
Fly versus fly
The 130 Mbp genome of D. melanogaster is largely complete, with the six main chromosome arms containing only a few gaps. Recently, the Human Genome Sequencing Center at Baylor College of Medicine completed the shotgun sequencing of D. pseudoobscura, a closely related species with a genome of approximately the same size. These two species are close enough that almost all genes are shared, and exons show a high level of sequence identity. However, they are sufficiently distant that intergenic regions and introns do not align well, and there have been hundreds of large-scale chromosomal rearrangements since the species diverged. Thus, one cannot simply align each chromosome arm to its counterpart. Complicating matters further, the D. pseudoobscura shotgun assembly consists of thousands of scaffolds and contigs. To facilitate comparison, the first computational task is to align all the scaffolds to each of the D. melanogaster arms. (The comprehensive analysis of D. pseudoobscura, organized by the sequencing center scientists and their collaborators, will appear in a future paper. The description here is primarily intended to illustrate the use and capabilities of Nucmer.)
We ran the Nucmer program with a minimum match length of 25, which was adequate to capture virtually all matching exons. Because matching genes are much longer, we required cluster chains to contain at least 100 matching nucleotides. To account for long introns and to allow the program to cluster together multiple genes, we allowed the gap between exact matches to be as long as 3,000 bp. At the time of our analysis (before completion of the sequencing project), the D. pseudoobscura assembly contained 4,653 scaffolds spanning 150 Mbp. We ran Nucmer separately to align the full set of scaffolds to each D. melanogaster chromosome arm. Using these settings, the program takes about 6 minutes per arm and uses approximately 490 Mb of memory on a 2.8 GHz desktop Pentium 4 PC running Linux.
Fly versus mosquito
When the two species are more distantly related, the only means of detecting large-scale similarity is through comparisons on the amino acid level. One example of this phenomenon arose during our comparison of the genomes of the malaria mosquito, Anopheles gambiae, and the fruit fly D. melanogaster. Because Anopheles was the second insect genome to be sequenced, the only available species for comparison was fruit fly. Our detailed analysis, done jointly with colleagues at the European Molecular Biology Laboratory in Heidelberg, was based on a combination of BLAST and MUMmer analysis [34]. These two species diverged about 250 million years ago, and they have an average protein sequence identity of 56%, less than that shared between humans and pufferfish. Although the two insects have the same number of chromosomes, the Anopheles genome is approximately twice as large, and the gene order has been almost completely shuffled, as our alignments revealed. Only small, but numerous, regions of 'microsynteny' remain: we reported 948 regions, the largest containing 8 genes in Anopheles and 31 in Drosophila. An interesting finding, though, was that despite extensive shuffling, each chromosome arm had a clear predominance of homologs on a single arm in the other species, indicating that intrachromosome gene shuffling was the primary force affecting gene order (see Figure 7 of [34]).
Fungus versus fungus
In a current application, we are using both Nucmer and Promer to compare two related fungal genomes, Aspergillus fumigatus (a human pathogen) and A. nidulans (a non-pathogenic model organism). Shotgun sequencing of these two genomes has been completed, and A. fumigatus is in the process of being completely finished; that is, all gaps are being closed. (A. fumigatus is a joint sequencing project of TIGR and The Sanger Institute, while A. nidulans is being sequenced at the Whitehead/MIT Genome Center.) At the time of our most recent comparison, the A. fumigatus genome had progressed to the point where it was assembled into 19 scaffolds spanning 28 Mbp, and the A. nidulans genome was assembled into 238 contigs spanning 30 Mbp. For this comparison, we first ran Nucmer and found that most of the two genomes mapped onto one another quite clearly: there are sufficient matches to reveal large segments of similarity in a simple dot plot. There has been extensive rearrangement of the chromosomes, but large-scale synteny is still present. For example, the largest contig (A1058) in A. fumigatus, at 2.9 Mbp, representing an essentially complete chromosome, maps onto five different scaffolds in A. nidulans. If one looks only at the Nucmer alignment of the largest of these, a 2.1 Mbp scaffold containing 10 contigs, it appears to be rearranged into multiple segments, but the matches are so scattered that it is difficult to tell how many segments there are (Figure 1, left-hand side).
Figure 1.
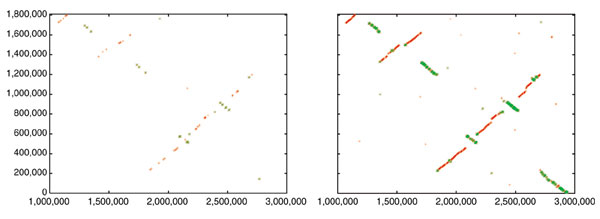
Dot-plot alignments of a 2.9 Mbp chromosome of A. fumigatus (_x_-axis) to a 2.1 Mbp scaffold of A. nidulans (_y_-axis). Left: nucleotide-based alignment with Nucmer. Right: amino-acid-based alignment with Promer. Aligned segments are represented as dots or lines, up to 3,000 bp long in the Nucmer alignment and up to 9,500 bp in the Promer alignment. These alignments were generated by the mummerplot script and the Unix program gnuplot.
The syntenic alignment is much more clearly visible, however, if we use Promer instead. The simplest summary is just the number of bases included in the alignments: if we look at the Nucmer alignment between the scaffolds, the total number of matching bases is 81 kbp. In contrast, the Promer alignment covers 1.87 Mbp of A1058, beginning at nucleotide position 1,000,000 and continuing to the end of the chromosome. A graphical illustration is shown in Figure 1, which displays both the Promer and Nucmer alignments between the 2.1 Mbp scaffold from A. nidulans and scaffold A1058 of A. fumigatus. As the figure makes clear, the amino-acid-based alignment covers much more of the sequence of both species, and is therefore much more useful for determining homologous relationships between genes and chromosomal relationships.
Human versus human
One of the most challenging computational tasks one can perform today is the cross-comparison of mammalian genomes. The human and mouse genomes are sufficiently complete that much ongoing research is based on mappings between these two species. As shown in Table 1, MUMmer 3.0 can compare human and mouse chromosomes in a matter of minutes. The table shows the time (7 minutes 10 seconds, on a 2.4 GHz Pentium processor) required to align mouse chromosome 16 (Mm16) to human chromosome 21 (Hs21). These two were chosen because nearly all of Hs21 maps to one end of Mm16; in fact, researchers have developed a mouse model of Down syndrome that has an extra copy of this part of Mm16.
We ran a benchmark test of MUMmer 3.0 in which we compared the human genome (version of 3 January 2003, downloaded from GenBank) to itself by computing all maximal matches of length at least 300 between each chromosome and all the others. The resulting 631,975 matches allow one to identify both large- and small-scale interchromosomal duplications. Note that the run-times reported in [6] are only for the match-finding part of MUMmer. The time for processing clusters and performing alignments in the gaps between matches are omitted as these vary widely depending on the parameters used.
For this test, we needed a maximum of about 4 GB of memory. As we did not have a PC available with this amount of memory, we used a Sun-Sparc computer running the Solaris operating system, with 64 GB of memory and a 950 MHz processor.
We ran the alignment as follows. Each human chromosome was used as a reference, and the rest of the genome was used as a query and streamed against it. To avoid duplication, we only included chromosomes in the query if they had not already been compared; thus we first used chromosome 1 as a reference, and streamed the other 23 chromosomes against it. Then we used chromosome 2 as a reference, and streamed chromosomes 3-22, X, and Y against that, and so on.
The total length of all human chromosomes for this test was 2,839 Mbp. The time required to build all the suffix trees was 4.7 hours. The space requirement for the suffix tree was remarkably constant, with about 15.5 bytes per base-pair (with only one exception). The total query time was 101.5 hours, and memory usage never exceeded 3.9 GB (see [6] for details). Thus, in approximately 4.5 days on a single processor, we matched the human genome against itself. This could easily be divided up among multiple computers, with each chromosome handled separately, bringing the time down to just 11 hours.
Graphical viewers
Because the text-format output of MUMmer 3.0 is often voluminous, we have developed two graphical viewers, one for the purpose of comparing two genome assemblies or near-identical sequences, and the other for comparing more distantly related genomes, such as two distinct species. The first viewer, DisplayMUMs, is an open-source, platform-independent Java program. It has been tested on a variety of Unix/Linux platforms and also runs on Apple Macintosh (OS X) or Microsoft Windows computers. The program, which takes as input the results of running MUMmer, allows the user to align and view the results of two different assemblies of the same or very closely related genomes and to tile one set of contigs onto the other. This provides a powerful graphical front end for assembly comparison, a function that is frequently used in the process of assembling and finishing genomes. It allows a user to visualize the tiling of sequence reads onto an assembly in order to understand why contigs might not have properly merged together. Alternatively, one can compare the output of different genome assemblers on the same data, a task that can be quite bewildering when the genome is large and the assemblers disagree.
DisplayMUMs creates a stand-alone display, illustrated in Figure 2. It contains three main areas. The upper area can show a variety of types of information, including zoomed-in nucleotide alignments. The central panel shows a summary of the alignment, with the reference shown as a gray bar. The matches of the queries to the reference are shown as green (forward) and red (reverse) rectangles, with gaps indicated in gray. A second gray bar shows the gaps in blue, which may seem redundant but is useful when the scale is zoomed out; for example, if the sequence has only one small gap and the scale shows 1 Mbp, then the small gap will be invisible in the upper bar but will still be visible on the lower bar. The lower panel shows the tiling of all the query sequences on the reference, with red and green colors indicating the forward and reverse matching substrings. As Figure 2 shows, some sequences might match for only a small portion of their length, while others will match across their entire length. DisplayMUMs has many other features, including mouse-over and searching functions, all of which are documented in the software. As this example makes clear, its primary purpose is to improve the utility of MUMmer for genome-assembly analysis.
Figure 2.
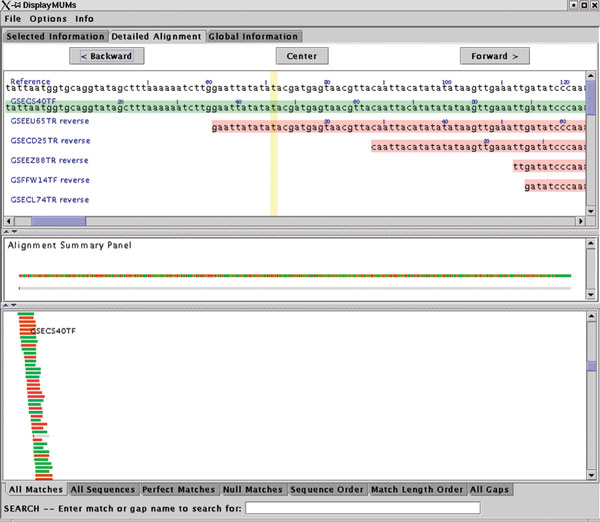
Sample display from DisplayMUMs, showing whole-genome alignment of individual shotgun reads (query sequences) to a contig from the Staphylococcus epidermidis genome. The display illustrates how exact matches of the tiling reads can be seen against the contig consensus. Green and red colors in the query sequences indicate alignment on the forward and reverse strands, respectively.
The second viewer, MapView, creates a picture of the mapping between two species based on Nucmer or Promer output. The motivation for creating this viewer was the rapidly increasing number of genome projects that are undertaken to enhance our understanding of another, already completed genome. In these projects, the second genome may have only faint DNA sequence similarity to the first, and in some cases the similarity may be detectable only through protein sequence alignments, such as those produced by Promer. A good example of such a project is the recent effort to sequence D. pseudoobscura mentioned above. The primary motivation for this project is to improve the annotation of D. melanogaster, and MUMmer is one of the tools being used to map the newly assembled D. pseudoobscura onto it. Because the reference genome is well annotated, we included in the viewer the option to display the locations of the genes (and their identifiers) along with the mapping at either the DNA or amino acid sequence level. A snapshot of this alignment by MapView is in Figure 3, which makes it clear that the amino acid conservation between these two species closely matches the annotated exon structure. This viewer can be used to highlight areas of a genome where exons might have been missed in previous analyses.
Figure 3.
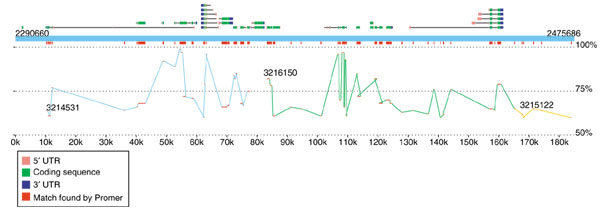
Sample display created by the MapView program, showing a 185 kbp slice of D. melanogaster chromosome 2L and its alignment to D. pseudoobscura. The alignment, generated by Promer, shows all regions of conserved amino acid sequence. The blue rectangle spanning the figure represents the reference (D. melanogaster), with annotated genes shown above it. Alternative splice variants of the same gene are stacked vertically. Exons are shown as boxes, with intervening introns connecting them. The 5' and 3' UTRs are colored pink and blue to indicate the gene's direction of translation. Promer matches are shown twice, once just below the reference genome, where all matches are collapsed into red boxes, and in a larger display showing the separate matches within each contig, where the contigs are colored differently to indicate contig boundaries. The vertical position of the matches indicates their percent identity, ranging from 50% at the bottom of the display to 100% just below the red rectangles.
The MapView program can produce output in three formats: fig (for viewing with the Unix xfig program), PostScript, or PDF. The most flexible format, fig, allows for unlimited scrolling and zooming, and for export to a wide range of additional formats. This makes it easy to view the mapping between a large collection of contigs and a large chromosome.
Conclusions
As the examples above show, the capabilities of MUMmer 3.0 enable a researcher to compare virtually any two genomes, or collections of genomic sequences, using computers widely available today. Bacterial genomes and relatively small eukaryotes can be aligned on a standard desktop computer, while larger genomes may require larger, server-class machines. With the state of the art representation of the suffix-tree data structure, the memory usage of MUMmer 3.0 is close to the minimum possible, while retaining optimal or near-optimal worst-case run time, depending on the match algorithm used. The additional features in MUMmer 3.0 allow one to find non-unique and non-exact matches, greatly enhancing the flexibility of the system. Finally, by making the system open source, we hope to encourage others to expand upon and improve the code base, which is freely available to all.
Acknowledgments
Acknowledgements
S.L.S. and A.L.D. were supported in part by NIH grant R01-LM06845, A.P. by NIH grant N01-AI-15447, C.A. by NSF grant MCB0114792, and M.S. by the University of Virginia Biotechnology Training Program (NIH grant T32 GM08715). S.K. was partly supported by the Deutsche Forschungsgemeinschaft (DFG-grant Ku 1257/1-3).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–80. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–47. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Kasif S, Fleischmann RD, Peterson J, White O, Salzberg SL. Alignment of whole genomes. Nucleic Acids Res. 1999;27:2369–76. doi: 10.1093/nar/27.11.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUMmer: comparative applications http://www.tigr.org/software/mummer/applications.html
- Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, Ermolaeva MD, Allen JE, Selengut JD, Koo HL, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419:512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- Batzoglou S, Jaffe DB, Stanley K, Butler J, Gnerre S, Mauceli E, Berger B, Mesirov JP, Lander ES. ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullikin JC, Ning Z. The phusion assembler. Genome Res. 2003;13:81–90. doi: 10.1101/gr.731003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G, White O, Adams M, Kerlavage AR. TIGR Assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci Technol. 1995;1:9–19. [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N, Dubchak I, Pachter L. AVID: a global alignment program. Genome Res. 2003;13:97–102. doi: 10.1101/gr.789803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Kurtz S, Ohlebusch E. Efficient multiple genome alignment. Bioinformatics. 2002;18 Suppl 1:S312–S320. doi: 10.1093/bioinformatics/18.suppl_1.s312. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain P, Kurtz S, Ohlebusch E, Slezak T. An applications-focused review of comparative genomics tools: capabilities, limitations and future challenges. Brief Bioinform. 2003;4:105–123. doi: 10.1093/bib/4.2.105. [DOI] [PubMed] [Google Scholar]
- Dumas JP, Ninio J. Efficient algorithms for folding and comparing nucleic acid sequences. Nucleic Acids Res. 1982;10:197–206. doi: 10.1093/nar/10.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusfield D. Algorithms on Strings, Trees, and sequences: Computer Science and Computational Biology. New York: Cambridge University Press; 1997. [Google Scholar]
- Weiner P. In Proc 14th IEEE Symp Switching and Automata Theory. Iowa City: University of Iowa; 1973. Linear pattern matching algorithms. pp. 1–11. [Google Scholar]
- McCreight EM. A space-economical suffix tree construction algorithm. J Assoc Comp Mach. 1976;23:262–272. doi: 10.1145/321941.321946. [DOI] [Google Scholar]
- Kurtz S. Reducing the space requirement of suffix trees. Software Practice Experience. 1999;29:1149–1171. doi: 10.1002/(SICI)1097-024X(199911)29:13<1149::AID-SPE274>3.0.CO;2-O. [DOI] [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Pearson WR. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, Salzberg SL, Pop M, Shumway M, Umayam L, Jiang L, Holtzapple E, Busch JD, Smith KL, Schupp JM, et al. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science. 2002;296:2028–2033. doi: 10.1126/science.1071837. [DOI] [PubMed] [Google Scholar]
- Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, Gill SR, et al. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, von Mering C, Letunic I, Torrents D, Suyama M, Copley RR, Christophides GK, Thomasova D, Holt RA, Subramanian GM, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]