Mutant p53 in Cancer: New Functions and Therapeutic Opportunities (original) (raw)
Abstract
Many different types of cancer show a high incidence of TP53 mutations, leading to the expression of mutant p53 proteins. There is growing evidence that these mutant p53s have both lost wild-type p53 tumor suppressor activity and gained functions that help to contribute to malignant progression. Understanding the functions of mutant p53 will help in the development of new therapeutic approaches that may be useful in a broad range of cancer types.
Main Text
p53 is one of the most intensively studied tumor suppressor proteins, with mutations that lead to loss of wild-type p53 activity frequently detected in many different tumor types. Perturbations in p53 signaling pathways are believed to be required for the development of most cancers, and there is evidence to suggest that restoration or reactivation of p53 function will have significant therapeutic benefit. For the first 10 years of investigation, p53 was considered to be the product of an oncogene, with many studies describing proliferative and transforming activities for p53. This mistake in the initial classification of p53 was the result of a simple error; the TP53 gene that had been cloned and used in the initial experiments encoded a mutant version of the wild-type gene. The tumor suppressor credentials of wild-type p53 are no longer in doubt, but the early studies provided a tantalizing hint of what has become an extremely active area of study—the suggestion that mutations in p53 can result in both loss of wild-type activity and gain of a novel transforming function. Moving in a circle in the past 30 years, we have come back around to considering that p53, albeit mutant versions of p53, can function as oncoproteins. In this review, we highlight recent progress in our understanding of how mutant p53 functions, discuss the avenues that are being explored to target mutant p53 tumors, and explore future directions for mutant p53 research.
TP53 is the most commonly mutated gene in human cancer (Kandoth et al., 2013). Alterations have been found in virtually every region of the protein (Leroy et al., 2013), but only a handful of the most frequently occurring mutations have been studied in depth for their contribution to cancer progression. In some cases, frameshift or nonsense mutations result in the loss of p53 protein expression, as seen with other tumor suppressors. However, more frequently, the tumor-associated alterations in p53 result in missense mutations, leading to the substitution of a single amino acid in the p53 protein that can be stably expressed in the tumor cell. These substitutions occur throughout the p53 protein, but most commonly cluster within the DNA binding region of p53, with six “hotspot” amino acids that are most frequently substituted. These mutations generally lead to a loss or diminution of the wild-type activity of p53, and because p53 normally acts as a tetramer, these mutant proteins may also function as dominant negative inhibitors over any remaining wild-type p53. Indeed, in a mouse model, the expression of mutant p53 has been shown to dampen (but not prevent) the therapeutic response to restoration of wild-type p53 (Wang et al., 2011). However, it is becoming clear that at least some of these mutant p53 proteins give rise to a more aggressive tumor profile, indicating that they have acquired novel functions in promoting tumorigenesis.
Gain of Function of Mutant p53
The concept that mutant p53 may show a neomorphic gain of function (GOF) was first suggested 20 years ago (Dittmer et al., 1993), when the introduction of mutant p53 into p53 null cells was shown to give rise to a new phenotype. Since then, a large number of publications have demonstrated many GOFs in numerous cell lines with a variety of p53 mutations, summarized in Table 1. The GOF acquired by mutant p53 is further supported by the finding that patients carrying a TP53 missense mutation (leading to expression of a mutant p53 protein) in the germline have a significantly earlier cancer onset than patients with mutations in TP53 that result in loss of p53 protein expression (Bougeard et al., 2008; Zerdoumi et al., 2013). Consistently, in vivo experiments showed that mice expressing mutant p53 display a tumor profile that is more aggressive and metastatic than p53 null or p53 wild-type mice (Doyle et al., 2010; Lang et al., 2004; Morton et al., 2010; Olive et al., 2004), although some tissue specificity of this effect has been suggested by further studies showing that introduction of similar p53 mutations in the lung did not reveal any detectable GOF activity over p53 loss (Jackson et al., 2005). Nevertheless, numerous in vitro and xenograft models have confirmed the ability of mutant p53s to drive enhanced invasion and motility, with evidence that mutant p53 can enhance signaling through receptors such as transforming growth factor β (TGF-β) receptor, epidermal growth factor receptor, and MET (Adorno et al., 2009; Grugan et al., 2013; Muller et al., 2009, 2012; Sauer et al., 2010; Wang et al., 2013a). In part, these responses reflect an ability of mutant p53 to promote integrin/RCP driven recycling (Muller et al., 2009, 2012) or increase the expression of growth factor receptors (Sauer et al., 2010; Wang et al., 2013a). Although mutant p53s have generally lost the ability to bind consensus p53 DNA binding regions in target gene promoters, their activity appears to reflect an ability to regulate gene expression directly (Weisz et al., 2007), although cytoplasmic and mitochondrial activities of mutant p53 in regulating apoptosis and autophagy have also been described (Chee et al., 2013; Frank et al., 2011; Morselli et al., 2008). Whereas various different mutant p53s can bind directly to DNA with some degree of selectivity (Brázdová et al., 2013; Göhler et al., 2005; Quante et al., 2012) and may thereby directly control the transcription of some genes (Weisz et al., 2007), there is increasing evidence that an indirect effect on gene expression through binding to other transcription factors underlies the novel activities of mutant p53s. For example, several studies have revealed a role for TAp63, a p53 family protein and transcription factor, which interacts with mutant but not wild-type p53 (Gaiddon et al., 2001; Strano et al., 2002). By inhibiting TAp63, mutant p53 can regulate a pro-invasive transcription program that includes regulation of the expression of Dicer, DEPDC1, Cyclin G2, and Sharp1 (Adorno et al., 2009; Girardini et al., 2011). The Dicer regulation by mutant p53 may be of particular importance, because several miRNAs that can in turn regulate genes involved in invasion have been described to be regulated by mutant p53, although this may not always involve TAp63 or Dicer inhibition (Dong et al., 2012; Neilsen et al., 2012; Tucci et al., 2012; Wang et al., 2013a).
Table 1.
The Different GOF Roles of Mutant p53 in Cells
| Mutation | Cell Line | Mutant p53 Expression | Reference |
|---|---|---|---|
| Invasion | |||
| R172H (human R175H), 175H | PDAC | endogenous (also stable/transient) | Muller et al., 2012 |
| R175H | KLE | endogenous (also stable/transient) | Dong et al., 2009 |
| R175H, R273H, R248Q, R280K, | H1299 | stable/transient | Adorno et al., 2009; Coffill et al., 2012; Muller et al., 2009; Noll et al., 2012; Yoshikawa et al., 2010 |
| G266E | MDA MB435 | endogenous | Yeudall et al., 2012 |
| R273H | A431 | endogenous | Muller et al., 2009 |
| R280K | MDA MB231 | endogenous | Coffill et al., 2012; Girardini et al., 2011; Muller et al., 2009 |
| Increased (Altered) Migrationa | |||
| R172H | MEF | endogenous | Adorno et al., 2009 |
| R175H, H179L, R248Q, R273H, D281G | H1299 | stable/transient | Adorno et al., 2009; Muller et al., 2009, 2012; Noll et al., 2012; Yeudall et al., 2012 |
| R175H, R248Q | HEC-50 | stable/transient | Dong et al., 2012 |
| R248Q | HEC-1 | endogenous | Dong et al., 2012 |
| R248W | HCT116−/− | endogenous | Muller et al., 2012 |
| R249S | KNS-62 | endogenous | Vaughan et al., 2012b |
| R267P | H1437 | endogenous | Vaughan et al., 2012b |
| R273H | HT29, A431, U373, SNB19 | endogenous | Huang et al., 2013; Muller et al., 2012 |
| R280K | MDA MB231 | endogenous | Adorno et al., 2009; Girardini et al., 2011; Li et al., 2011a |
| Proliferation, Propagation of Cell Cycle | |||
| P278S | ABC1 | endogenous | Vaughan et al., 2012a |
| R172H (human R175H) | MEF | endogenous | Lang et al., 2004 |
| R175H | SK-BR3, VMRC | endogenous | Bossi et al., 2006; Vaughan et al., 2012a |
| R175H, R248H | BE-13 | stable/transient | Hsiao et al., 1994 |
| R175H, R273H, D281G | H1299 | stable/transient | Liu et al., 2011; Scian et al., 2004b) |
| C176F, P223L, R273H, R282Q | PC-3 | stable/transient | Shi et al., 2002 |
| M246I | H23 | endogenous | Vaughan et al., 2012b |
| R248W, D281G | 10(3) | stable/transient | Loging and Reisman, 1999; Scian et al., 2004a |
| R249S | KNS-62 | endogenous | Vaughan et al., 2012a |
| R267P | H1437 | endogenous | Vaughan et al., 2012a; Vaughan et al., 2012b |
| R273C | H1048 | endogenous | Vaughan et al., 2012b |
| R273H | HT-29, MDA MB468, H2405 | endogenous | Bossi et al., 2006; Gurtner et al., 2010; Vaughan et al., 2012a; Wang et al., 2013a |
| R273H/ P309S | SW480 | endogenous | Bossi et al., 2006; Yan et al., 2008 |
| R273H/ R248W | Mia-Paca-2 | endogenous | Yan et al., 2008 |
| R280T | SWO-38 | endogenous | Lin et al., 2012 |
| Drug Resistance/Avoidance of Cell Death | |||
| A135V, R248W, R273H | M1/2 cells, LN-308 | stable/transient | Li et al., 1998; Matas et al., 2001; Pohl et al., 1999; Trepel et al., 1998 |
| R175H | MEC, 10(3), HEC-50 | stable/transient | Dong et al., 2012; Murphy et al., 2000; Pugacheva et al., 2002 |
| R175H | SK-BR3 | endogenous | Bossi et al., 2006; Di Agostino et al., 2006; Vaughan et al., 2012b |
| R175H, P223L + V274F | Pc-3 | stable/transient | Gurova et al., 2003; Zalcenstein et al., 2003 |
| R175H, R245S, R273H, D281G | Saos-2 | stable/transient | Atema and Chène, 2002; El-Hizawi et al., 2002; Kawamata et al., 2007; Tsang et al., 2005; Wong et al., 2007 |
| R175H, R248W, R273H | SKOV-3 | stable/transient | Buganim et al., 2006; Liu et al., 2011; Pugacheva et al., 2002 |
| R175H, R248W, R273H | H1299 | stable/ transient | Blandino et al., 1999; Di Como et al., 1999; Pugacheva et al., 2002; Zalcenstein et al., 2006 |
| Y220S | fibroblasts | stable/transient | Capponcelli et al., 2005 |
| M237? | T98G | endogenous | Wang et al., 2013b |
| R248Q | HEC-1 | endogenous | Dong et al., 2012 |
| G266E | MDA MB435 | endogenous | Vaughan et al., 2012b |
| R273? | U138 | endogenous | Wang et al., 2013b |
| R273C | C33A, H1048 | endogenous | Liu et al., 2011; Vaughan et al., 2012b |
| R273H | C33A | endogenous | Liu et al., 2011 |
| R273H | HT-29, MDA MB468 | endogenous | Bossi et al., 2006; Vaughan et al., 2012b |
| R273H/ P309S | SW480 | endogenous | Bossi et al., 2006; Di Agostino et al., 2006 |
| R273H/ R248W | Mia-Paca-2 | endogenous | Do et al., 2012 |
| V143A, R175H, R248W, R273H | Hep3B | stable/transient | Schilling et al., 2010 |
| Anchorage-Independent Growth/Anoikis | |||
| Y126C, R175H, H214R, G245S, R273C, R273H, V273F, R280T, R282Q | SAOS-2 | stable/transient | Dittmer et al., 1993; Shi et al., 2002; Sun et al., 1993 |
| P151S | TU-138 | endogenous | Xie et al., 2013 |
| Increased Colony Formation | |||
| V143A | BEAS-2B | stable/transient | Gerwin et al., 1992 |
| V143A, R175H, R248W, R273H | H1299 | stable/transient | Kalo et al., 2012; Liu et al., 2011; Weisz et al., 2004 |
| V143A, Y163C, R175H, L194R, R273H, D281G, R282W | 10(3) | stable/transient | Scian et al., 2004a |
| G144P, R158H, Y163N, H168Y, V173L, Y234C, R248W | REFb | stable/transient | Smith et al., 1999 |
| C174Y | Saos-2 | stable/transient | Preuss et al., 2000 |
| R172H (human R175H) | MEF | endogenous | Lang et al., 2004 |
| R175H | SK-BR3 | endogenous | Bossi et al., 2006 |
| C194T | T47D | endogenous | Nguyen et al., 2013; Vikhanskaya et al., 2007 |
| A220G | Huh-7 | endogenous | Vikhanskaya et al., 2007 |
| R270C | IP3 | stable/transient | Halevy et al., 1990 |
| R273H | HT-29, MDA MB 468, U373, SNB19 | endogenous | Bossi et al., 2006, 2008; Huang et al., 2013; Wang et al., 2013a |
| R273H | MCF10Ab | stable/transient | Nguyen et al., 2013 |
| R273H/ P309S | SW480 | endogenous | Bossi et al., 2006; Yan and Chen, 2009, 2010; Yan et al., 2008 |
| R273H/ R248W | Mia-Paca-2 | endogenous | Yan and Chen, 2009; Yan et al., 2008 |
| Genomic Instability | |||
| R172H (human R175H) | primary mouse oral tumor | endogenous | Acin et al., 2011 |
| R175H | MEC | stable/transient | Murphy et al., 2000 |
| R175H, R248W, R273H | MEF | stable/transient | Agapova et al., 1996 |
| N236S (human N239S) | MEF | endogenous | Jia et al., 2012 |
| R248W | primary mouse cells | endogenous | Song et al., 2007 |
| R248W, R273H | K562 KMV | stable/transient | Restle et al., 2008 |
| Spheroid Disorganization/Mammary Architecture Disruption | |||
| R273H, R280K | MDA MB 468, MDA MB231 | endogenous | Freed-Pastor et al., 2012 |
| R175H, G245S, R248W, R273H | MCF10Ab | stable/transient | Zhang et al., 2011 |
| Stem Cell Dedifferentiation/Propagation | |||
| V143A, R175H, R273H | 10(3) | stable/transient | Yi et al., 2012 |
| R172H (human R175H) | MEF | endogenous | Sarig et al., 2010 |
| Xenograft Growth (Cell Line Injected Subcutaneously or in the Mammary Fat Pad) | |||
| V143A, R175H, R248W, R273H, R281D, D281G | (10) 3 | stable/transient | Dittmer et al., 1993; Lányi et al., 1998 |
| R172H (human R175H) | primary mouse oral tumor | endogenous | Acin et al., 2011 |
| R175H, R273H, | H1299 | stable/transient | Liu et al., 2011 |
| N236S (human N239S) | MEF | endogenous | Jia et al., 2012 |
| R267P | H1437 | endogenous | Vaughan et al., 2012a |
| R273C | H1048 | endogenous | Vaughan et al., 2012b |
| R273H | HT29, MDA MB 468 | endogenous | Bossi et al., 2008; Wang et al., 2013a |
| P278S | ABC1 | endogenous | Vaughan et al., 2012a |
| R280K | MDA MB 231 | endogenous | Adorno et al., 2009 |
| R280T | SAOS-2 | stable/transient | Sun et al., 1993 |
| Intravenous Injection (Formation of Lung Metastasis) | |||
| R175H, R248G, R213G | BE-13c | stable/transient | Hsiao et al., 1994 |
| C236F | D3S2 | endogenous | Adorno et al., 2009 |
| R280K | MDA MB231 | endogenous | Adorno et al., 2009 |
| Elongated Cell Morphology/EMT | |||
| C135Y, R175H, R273H | HEC-50 | stable/transient | Dong et al., 2012 |
| V143A | HCT116−/− | stable/transient | Roger et al., 2010 |
| R175H | H1299 | stable/transient | Adorno et al., 2009 |
| R175H, R273H | 10(3) | stable/transient | Gloushankova et al., 1997 |
| R248Q | HEC-1 | endogenous | Dong et al., 2012 |
| R273H | SW620 | endogenous | Roger et al., 2010 |
| R175H, G245S, R248W, R273H | MCF10Ab | stable/transient | Zhang et al., 2011 |
| Polyploidy | |||
| V143A | NHF3 cellsb | stable/transient | Gualberto et al., 1998 |
| R248W, R249S, R175H | H1299 | stable/transient | Noll et al., 2012 |
| Angiogenesis | |||
| Δ126 | T24 | endogenous | Zhu et al., 2013 |
| R175Hd | H1299 | stable/transient | Fontemaggi et al., 2009 |
| Y220S | fibroblasts | stable/transient | Capponcelli et al., 2005 |
| Cell Survival | |||
| V157F | Hs578T | endogenous | Braicu et al., 2013 |
| C194T | T47D | endogenous | Lim et al., 2009 |
| P223L/V274F | DU-145 | endogenous | Zhu et al., 2011 |
| R273H | MDA MB468, U373, SNB19 | endogenous | Huang et al., 2013; Lim et al., 2009 |
| R273H | H1299 | stable/transient | Kalo et al., 2012 |
| R280K | MDA MB231 | endogenous | Ali et al., 2013; Hui et al., 2006 |
| R280T | 5637 | endogenous | Zhu et al., 2013 |
| Mammosphere Formation | |||
| R175H | MESC, HEC-50 | endogenous | Lu et al., 2013; Dong et al., 2012 |
| R248Q | HEC-1 | endogenous | Dong et al., 2012 |
Mutant p53 inhibition of TAp63 can be modeled by deletion of TAp63, which results in an aggressive tumor profile and metastases similar to that seen in mice expressing mutant p53 (Su et al., 2010). However, a direct comparison of mutant p53 expression with loss of TAp63 in a mouse model of pancreatic ductal adenocarcinoma (PDAC) showed that loss of TAp63 is less potent in inducing metastases, suggesting that mutant p53 does more than inhibiting TAp63 (Tan et al., 2013). This is not surprising, because mutant p53 interacts with a wide variety of other proteins, resulting in interference in a multitude of cellular pathways, some of which are likely to contribute to metastasis (Freed-Pastor and Prives, 2012; Muller and Vousden, 2013; Walerych et al., 2012). Besides inhibiting p63, mutant p53 inhibits and interacts with other proteins including the MRE11-Rad51-NSB complex, p73, and SP-1 to induce genomic instability, chemoresistance, or proliferation (Chicas et al., 2000; Gaiddon et al., 2001; Song et al., 2007). Furthermore, mutant p53 can also promote the function of proteins including SREBP, NF-Y, VDR, ETS2, or NRF2, resulting in increased proliferation, cholesterol synthesis, accumulation of reactive oxygen species, and enhanced cell survival (Do et al., 2012; Freed-Pastor et al., 2012; Kalo et al., 2012; Liu et al., 2011; Stambolsky et al., 2010). All of these proteins and pathways affected by mutant p53 are thoroughly described in three recent reviews (Freed-Pastor and Prives, 2012; Muller and Vousden, 2013; Walerych et al., 2012).
More recent studies are identifying further GOF activities of mutant p53, such as a role in cell reprogramming and expansion or in the maintenance and interaction with tumor stroma. Wild-type p53 was characterized as a suppressor of somatic stem cell reprogramming, the process in which differentiated somatic cells can be reprogrammed into a pluripotent stem cell to allow for unlimited expansion (Kawamura et al., 2009; Marión et al., 2009). Loss of p53 promoted the dedifferentiation of somatic cells and some, but not all, mutant p53s could potentiate the reprogramming (Sarig et al., 2010; Yi et al., 2012). An expansion of hematopoietic and mesenchymal stem cell progenitors is also seen in mutant p53 R248Q transgenic mice (Hanel et al., 2013). Consistently, in breast tissue with a Wnt transgene, loss of wild-type p53 generally promoted the formation of one distinct tumor, whereas mutant p53 R175H expression promoted the initiation of multiple different tumors that could be expanded in mammosphere assays (Lu et al., 2013). Together, these data suggest that mutant p53 can initiate tumor formation by promoting the generation and expansion of pluripotent stem cells.
The role of stroma tissue, including extracellular matrix, proteases, cytokines, immune cells, epithelial cells, and cancer-associated fibroblasts (CAFs), in tumorigenesis has become very evident (Pietras and Ostman, 2010). CAFs, the most abundant cell type in the stroma, secrete cytokines, hormones, and growth factors including hepatocyte growth factor and TGF-β (Bhowmick et al., 2004; Ostman and Augsten, 2009), both of which have been shown to mediate mutant p53-dependent invasion and metastasis (Adorno et al., 2009; Muller et al., 2012). In addition, a recent report highlights an important function for mutant p53 in promoting the inflammatory environment of colorectal tumors by prolonging NF-κB activation and cell survival (Cooks et al., 2013). It seems clear, therefore, that the presence of a mutant p53 in tumor cells will have an influence on how the tumor and stromal cells interact. In co-culture experiments, H1299 cells (regardless of p53 status) upregulated interferon-β (IFN-β) secretion in CAFs. This would normally cause inhibition of cell migration, but mutant p53-expressing tumor cells counteracted this response by enhancing STAT phosphorylation to promote invasion (Madar et al., 2013). Although interesting, these experiments are difficult to interpret, because the IFN-β secreted by the fibroblasts also reduced mutant p53 expression (Madar et al., 2013). Alternatively, it is possible that TP53 mutations occur in the stroma surrounding tumors to promote tumor growth (Narendran et al., 2003; Patocs et al., 2007). Mutant p53-expressing fibroblasts were shown to promote tumor growth better than p53 null fibroblasts, suggesting that mutant p53 has a pro-oncogenic GOF role not only in tumor cells, but also in stromal cells (Addadi et al., 2010). However, whether stromal cells that have sustained mutations in p53 are prevalent, and how they are affected by (or affect) tumor cells remains unclear.
Are All Mutant p53s the Same?
Although most experimental studies have focused on the activity of a few most commonly detected p53 mutations that are clustered at codons 175, 245, 248, 249, 273, and 282, almost every codon within the DNA binding domain of p53 has been found to be mutated in cancer. Mutations have also been found in other domains, but their contribution to carcinogenesis is largely unknown (Leroy et al., 2013). Different tumor types show different spectra of TP53 mutations—in some cases, reflecting the mutagenic event was thought to contribute to that type of cancer (e.g., aflatoxin and liver, UV light, and skin) or geographic variation in other cases. The frequency of missense mutations also differs in different subclasses of tumors of the same organ. For example, luminal breast cancers almost all carry point mutations in TP53, while alterations resulting in p53 truncations were more frequently detected in basal breast tumors (Dumay et al., 2013).
Whereas p53 mutants are often considered to be equivalent, evidence is accumulating to indicate that different mutants show a distinct profile with respect to loss of wild-type p53 activity, the ability to inhibit wild-type p53, and the acquisition of gain of function (Table 1; Halevy et al., 1990; Petitjean et al., 2007). The large number of p53 mutations complicates such analyses, as does the realization that different mutants may function differently in different tissues, potentially reflecting differences in the expression of targets of mutant p53 such as TAp63. To date, mutant p53s have been considered in two different categories: the first affecting amino acids that contact DNA and so preventing wild-type transcriptional activity without dramatically affecting the conformation of the p53 protein (known as contact mutants), and the second comprising mutations that clearly disrupt the three-dimensional structure of the protein (termed conformational mutants). Data from cell lines suggest that conformational and contact mutants can cooperate via different mechanisms with the H-Ras signaling pathway, leading to similar gene expression profiles and tumorigenesis (Solomon et al., 2012). However, this classification of mutants is clearly an oversimplification, because different mutations can lead to subtly different alterations in the structure and conformational stability of the p53 protein (Joerger and Fersht, 2007). Various mouse models have shown that both conformational and contact mutants can promote metastasis compared to p53 null mice. These differences appear to be dependent on the nature of the substitution, but caution should be taken when interpreting data from mouse models using different strain backgrounds that are being studied in different laboratories, and in some cases mutate the mouse gene and in others examine humanized TP53 sequences in the mouse. Models of R172H or R270H (prototype examples of a conformation and a contact hotspot mutation, equivalent to R175H and R273H in humans) both showed GOF activity (Lang et al., 2004; Olive et al., 2004), whereas no GOF was seen in R246S (the mouse equivalent of human R249S) and the humanized G245S mutant p53 mouse models, although the R246S could dominant-negatively inhibit wild-type p53 to promote cell survival after radiation exposure (Hanel et al., 2013; Lee et al., 2012). R248Q (humanized) p53 knock-in mice showed an earlier onset of tumor formation with a significantly reduced lifespan compared to p53 null mice (Hanel et al., 2013), although this reduction in overall survival was not evident in any of the other mutant p53 models. Consistently, Li-Fraumeni patients carrying an R248Q mutation display an earlier onset of cancer compared to inherited null mutations or the G245S mutation (Hanel et al., 2013). These findings suggest that the R248Q p53 functions in a different manner than other p53 mutants that have been studied so far. Remarkably, not only the position of the mutation, but also the nature of the substitution may influence the activity of the resulting mutant protein. For example, both R248Q and R248W are structural mutants, but the humanized R248W p53 knock-in mouse does not display reduced lifespan or earlier disease onset (Song et al., 2007). Understanding the consequences of each p53 mutation in relationship to disease progression and response to therapy therefore promises to be an extremely complex undertaking.
Consequences of Mutant p53 Expression to Tumor Therapy
The realization that loss of p53 and expression of mutant p53 may not be analogous has also raised the question of whether the presence of a mutant p53 protein may affect the response to therapy. Whereas there is evidence that the presence of mutant p53 may dampen the response to restoration of wild-type p53 (Wang et al., 2011), reflecting a dominant negative activity of mutant p53, more recent studies have indicated that the retention of wild-type p53 can be detrimental to the therapeutic response in breast cancer. This effect is seen in tumors that express both mutant and wild-type p53 alleles (Jackson et al., 2012). Such studies highlight the possibility that in some tumor types wild-type p53 can be dominant over mutant, and that studies of patient response based on p53 status must take into account heterozygosity at the TP53 locus, as well as the presence of mutant or wild-type p53 (Jackson and Lozano, 2013).
Therapeutic Strategies to Restore Wild-Type Activity to Mutant p53
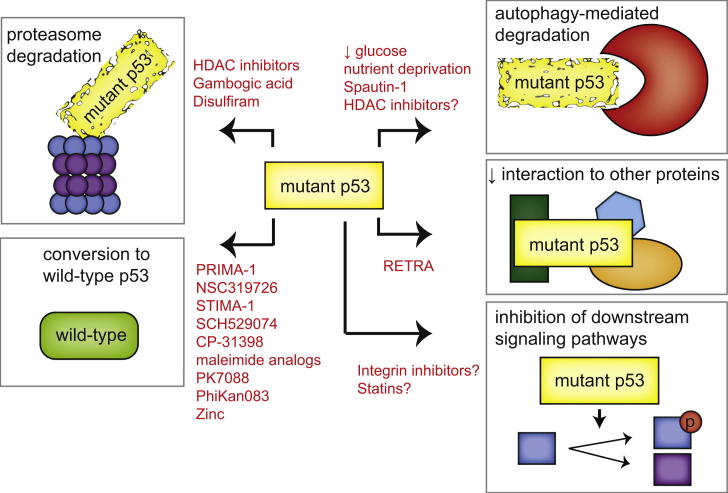
With so many different mutations and phenotypes it is not surprising that a variety of strategies are being explored to target tumors expressing mutant p53s (summarized in Figure 1). Wild-type p53 is a potent inducer of apoptosis and senescence when expressed in tumor cells, making the reactivation of some level of wild-type function in mutant p53 (which is generally expressed at high levels in cancer cells) an attractive therapeutic avenue. Interestingly, loss of wild-type function introduced by some destabilizing tumor-derived mutations can be rescued by additional point mutations that serve to stabilize the conformation of p53 protein, showing that the loss of structure is intrinsically reversible (Joerger and Fersht, 2008). In addition, a variety of compounds that might restore wild-type p53 function have been characterized and are reviewed in several recent publications (Lehmann and Pietenpol, 2012; Maslon and Hupp, 2010; Wiman, 2010). Small molecules that bind to a site in p53 formed in the Y220C mutant (PhiKan083 and PK7088) function by stabilizing the structure of this mutant p53, and so increasing the level of p53 with a wild-type conformation and activity (Boeckler et al., 2008; Liu et al., 2013). Other compounds bind to multiple mutant p53 proteins (e.g., PRIMA-1, or the soluble derivative PRIMA-met/APR-246, CP-31398, and SCH29074; Bykov et al., 2002; Demma et al., 2010; Foster et al., 1999), interacting with the DNA binding domain, thereby promoting proper folding of the mutant protein and restoration of p53 function. However, the precise mechanistic function of these compounds and others, such as maleimide analogs and STIMA-1, remain to be elucidated (Bykov et al., 2005; Zache et al., 2008).
Figure 1.
Strategies that Are Currently Being Explored to Target Mutant p53
Depicted in red are schematics of the strategies that are currently being explored to target p53 mutant-expressing cancers. These strategies include promotion of mutant p53 degradation through the proteasome and autophagy pathways, restoration of wild-type p53 activity, interference with the interaction between mutant p53 and other proteins, and interference in signaling pathways downstream of mutant p53.
Whereas wild-type p53 requires binding to the metal ion Zn(2+) to fold correctly (Loh, 2010; Verhaegh et al., 1998), the R175H p53 mutant was found to be impaired in zinc binding (Butler and Loh, 2003). Loss of metallothioneins that chelate and store intracellular zinc promotes a wild-type conformation of misfolded p53 (Puca et al., 2009) and addition of zinc to the conformational mutants G245C and G245D p53 partially restored the wild-type conformation (Pintus et al., 2013). The potential use of zinc to recover wild-type folding has therefore been explored and this approach has been shown to restore chemosensitivity to anticancer drugs in cells expressing endogenous mutant p53 (Puca et al., 2011). In addition, the thiosemicarbazone metal ion chelator NSC31926 was found to restore wild-type function in a variety of different mutant p53-expressing cell lines, possibly through increasing the bioavailability of zinc to (mutant) p53 (Yu et al., 2012).
Of all the compounds that restore wild-type activity, the most progress has been made with PRIMA-1 analogs, with the demonstration of safety in a phase I clinical study (Lehmann et al., 2012). PRIMA-1 is rapidly converted to other compounds, including MQ, which can bind to both mutant p53 and wild-type p53 (Lambert et al., 2009), although the precise mechanisms underlying the p53 reactivation are currently unknown. Under some circumstances, p53 can adopt an unfolded conformation and behave like a mutant p53 protein to promote invasion (Trinidad et al., 2013). Unfolded wild-type p53 seen in tumor cells grown under hypoxia (Gogna et al., 2012) could be restored by PRIMA-1 treatment (Rieber and Strasberg-Rieber, 2012). It will therefore be interesting to explore whether both wild-type and mutant p53 tumors might benefit from PRIMA-1 treatment.
Therapeutic Strategies to Promote Mutant p53 Degradation
An alternative approach to targeting mutant p53 is to remove the proteins by enhancing turnover (Figure 1). Both wild-type and mutant p53 can be targeted for proteasomal degradation in otherwise normal cells by the ubiquitin ligase MDM2. Inhibition of MDM2 in response to stress underlies the activation of wild-type p53, but is also thought to lead to the overexpression of mutant p53 seen in cancer cells. Indeed, stress induced stabilization of mutant p53 seems to be a prerequisite for its GOF (Suh et al., 2011). In addition to MDM2, another chaperone-associated E3 ubiquitin ligase, CHIP, was shown to be important for mutant p53 degradation (Esser et al., 2005; Lukashchuk and Vousden, 2007). To be stabilized, mutant p53 interacts with the Hsp70 and Hsp90 chaperone complex that requires an interaction with HDAC6 for proper functioning (Li et al., 2011b). Abrogation of HDAC6 binding results in the dissociation of the heat shock proteins from mutant p53 and allows for mutant p53 degradation by MDM2 and CHIP (Li et al., 2011b). HDAC inhibitors such as SAHA show promise in destabilizing mutant p53 by preventing HDAC6 from interacting with Hsp90 (Li et al., 2011a). However, SAHA and the pan-HDAC inhibitor NaB were recently shown to not only regulate mutant p53 stability, but also its transcription via the p53 activator HoxA5 (Yan et al., 2013). This activity was not confined to mutant p53 and also extended to decreasing wild-type p53 expression (Yan et al., 2013), indicating that care should be taken to determine the p53 status of tumors when HDAC inhibitors are used as therapeutic agents. Small molecule activators of SIRT1 have also been shown to lead to the deacetylation of p53 and reduction of overall mutant p53 levels (Yi et al., 2013). In other studies, Stathmin—a transcriptional target of wild-type p53 and mutant p53 (through the regulation of miR-223)—promoted mutant p53 activity by regulating phosphorylation and stability in ovarian cancers (Sonego et al., 2013).
Autophagy also plays a role in mutant p53 degradation. Macro-autophagy is the process by which intracellular contents such as proteins or organelles are engulfed and degraded through lysosomes. This can provide a means to recycling intracellular content, providing an alternative energy source to allow cells to survive transient starvation, and also functioning to remove damaged or excess organelles (Mizushima et al., 2008). The role of autophagy in cancer is complex and can both promote and inhibit tumor development, depending on the targets of the autophagic process and the timing during tumor evolution (Liu and Ryan, 2012). Macro-autophagy induced by glucose restriction selectively promoted mutant p53 degradation, whereas wild-type p53 was stabilized under similar conditions (Rodriguez et al., 2012). The degradation of mutant p53 was promoted by proteasomal inhibition and depended on functional autophagy machinery (Choudhury et al., 2013; Rodriguez et al., 2012). Glucose starvation combined with confluent growth conditions could promote mutant p53 degradation by a specialized form of autophagy known as chaperone-mediated autophagy (Vakifahmetoglu-Norberg et al., 2013). In contrast to the findings of Rodriguez et al. (2012), degradation of mutant p53 via this specialized autophagy pathway was enhanced by inhibition of macro-autophagy (Vakifahmetoglu-Norberg et al., 2013), suggesting conditional aspects to glucose deprived mutant p53 degradation. Furthermore, both mutant and wild-type p53 can inhibit autophagy when localized in the cytoplasm (Morselli et al., 2008; Tasdemir et al., 2008), indicating that the relationship between autophagy and mutant p53 is complex.
Therefore, while targeting mutant p53 for degradation seems feasible, there remains a concern as to how effective simple removal of mutant p53 (without replacement by degradation-resistant wild-type p53) might be in driving a therapeutic response. Some comfort has been provided by many studies showing reduction of mutant p53 levels (either by siRNA or spautin treatment) results in increased apoptosis, indicating that these cells may have become dependent on mutant p53 for their survival (Table 1; Ali et al., 2013; Braicu et al., 2013; Huang et al., 2013; Hui et al., 2006; Lim et al., 2009; Vakifahmetoglu-Norberg et al., 2013; Xie et al., 2013; Zhu et al., 2011, 2013). However, whether decreasing mutant p53 levels is sufficient as a means of therapy in vivo and in the long term requires confirmation.
Targeting Mutant p53 Regulated Pathways
Instead of targeting mutant p53 directly, another approach is to identify commonalities in the mechanisms through which mutant p53 proteins function and to target and exploit these downstream pathways (Figure 1). Despite the clear differences between mutant p53s, a large number of them interact and inhibit p63 and p73. A small molecule named RETRA, identified by serendipity in a screen to identify drugs to stabilize wild-type p53, has been suggested to destabilize the p73 mutant p53 interaction (Kravchenko et al., 2008). RETRA-induced release of p73 resulted in the activation of p73 target genes and a concomitant decreased tumor cell survival and suppression of xenograft tumor growth (Kravchenko et al., 2008). Whether RETRA impairs the interaction of mutant p53s with other target proteins has not been reported, but this could be a more general approach to block the oncogenic effect of mutant p53s that share binding partners.
Downstream pathways activated by mutant p53 may also be targets for therapeutic intervention. An attractive possibility here is the cholesterol synthesis pathway through which mutant p53 disrupts the morphology of mammary tumors (Freed-Pastor et al., 2012). Inhibition of cholesterol synthesis restored the morphology and decreased survival of mutant p53 cells (Freed-Pastor et al., 2012). This is of particular interest because statins (cholesterol inhibitors) are among the most commonly prescribed drugs worldwide to prevent cardiovascular diseases and have shown promise as preventive anticancer agents (Singh and Singh, 2013). It will therefore be interesting and relatively straightforward to determine the utility of statins as a therapeutic strategy for mutant p53 tumors.
Finally, several studies have described a role for mutant p53 in enhancing receptor tyrosine kinase (RTK) signaling (Adorno et al., 2009; Muller et al., 2009; Sauer et al., 2010; Wang et al., 2013a). A multitude of inhibitors of the kinase activity of RTKs or their downstream mediators have been described, including EGFR inhibitors, MET inhibitors and MAPK inhibitors. Selective efficacy of these compounds in the treatment of mutant p53 expressing cancers remains to be explored. The specific role of RTK and integrin recycling may also provide an additional attractive target, since various integrin antibodies and drugs that inhibit integrin recycling are currently on the market and have shown some promise as anticancer agents (Desgrosellier and Cheresh, 2010).
Future Directions
A number of hurdles still need to be overcome before the studies of mutant p53 can be translated into clinical practice. While there is clear evidence that mutant p53 promotes various oncogenic responses, the relative importance of survival, motility, invasion, and metabolic changes, or the critical pathways through which these responses are mediated remain unclear. How different mutations affect p53 function also remains underexplored, as does the comparative importance of loss of wild-type, dominant-negative, and GOF phenotypes. The fact that most mutant p53s are expressed at very high levels in cancer cells (leading to the immunohistochemical detection of p53 being used as a proxy for the presence of mutant p53) makes these proteins tremendously attractive therapeutic targets, and the efficacy of inhibiting the activity of these mutant p53s or even re-establishing some wild-type function, as described above, holds great promise. Such approaches depend, however, on designing efficient mechanisms through which to target mutant p53, an understanding of the activities and function of the many different mutants, and the capacity to identify which mutation a tumor carries (the latter likely to be the most easily attainable goal).
Maybe a more effective approach will be to explore the possibility of synthetic lethality as a therapeutic strategy. Recently, a computational approach using gene expression from the NCI-60 panel, the GBM (glioblastoma multiforme) project and the TCGA (the cancer genome) project revealed a number of genes and pathways that may result in synthetic lethality when targeted in mutant p53-expressing tumors (Wang and Simon, 2013). The majority of these genes were involved in the cell cycle, perhaps reflecting the loss of wild-type p53 function, and an interesting candidate identified in several of the data sets is polo-like kinase 1 (PLK1), which is involved in the regulation of mitosis. PLK1 was found to be upregulated in breast cancers with mutant p53 expression; the presence of both coincided with a worse prognosis than cancers with either PLK1 upregulation or mutant p53 expression alone (King et al., 2012). Because PLK1 can be inhibited by a variety of compounds (Strebhardt, 2010), it will be interesting to follow up this lead.
Conclusions
Recent data reveal that mutant p53 is not just one protein, but a multitude of proteins that can contribute to a wide range of oncogenic processes. Designing drug strategies to target mutant p53 tumors is therefore highly challenging and will require a deeper understanding of the degradation pathways, interaction partners, and downstream signaling pathways in mutant p53 cells. However, we are optimistic that our ever-expanding knowledge of mutant p53 function will translate into some useful therapeutic strategies in the future.
Acknowledgments
We thank Cancer Research UK, AICR, and the Royal Society/Wellcome trust for funding support.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Patricia A.J. Muller, Email: pm292@le.ac.uk.
Karen H. Vousden, Email: k.vousden@beatson.gla.ac.uk.
References
- Acin S., Li Z., Mejia O., Roop D.R., El-Naggar A.K., Caulin C. Gain-of-function mutant p53 but not p53 deletion promotes head and neck cancer progression in response to oncogenic K-ras. J. Pathol. 2011;225:479–489. doi: 10.1002/path.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addadi Y., Moskovits N., Granot D., Lozano G., Carmi Y., Apte R.N., Neeman M., Oren M. p53 status in stromal fibroblasts modulates tumor growth in an SDF1-dependent manner. Cancer Res. 2010;70:9650–9658. doi: 10.1158/0008-5472.CAN-10-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adorno M., Cordenonsi M., Montagner M., Dupont S., Wong C., Hann B., Solari A., Bobisse S., Rondina M.B., Guzzardo V. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Agapova L.S., Ilyinskaya G.V., Turovets N.A., Ivanov A.V., Chumakov P.M., Kopnin B.P. Chromosome changes caused by alterations of p53 expression. Mutat. Res. 1996;354:129–138. doi: 10.1016/0027-5107(96)00062-0. [DOI] [PubMed] [Google Scholar]
- Ali A., Wang Z., Fu J., Ji L., Liu J., Li L., Wang H., Chen J., Caulin C., Myers J.N. Differential regulation of the REGgamma-proteasome pathway by p53/TGF-beta signalling and mutant p53 in cancer cells. Nature communications. 2013;4:2667. doi: 10.1038/ncomms3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atema A., Chène P. The gain of function of the p53 mutant Asp281Gly is dependent on its ability to form tetramers. Cancer Lett. 2002;185:103–109. doi: 10.1016/s0304-3835(02)00318-x. [DOI] [PubMed] [Google Scholar]
- Bhowmick N.A., Neilson E.G., Moses H.L. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandino G., Levine A.J., Oren M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene. 1999;18:477–485. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- Boeckler F.M., Joerger A.C., Jaggi G., Rutherford T.J., Veprintsev D.B., Fersht A.R. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc. Natl. Acad. Sci. USA. 2008;105:10360–10365. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi G., Lapi E., Strano S., Rinaldo C., Blandino G., Sacchi A. Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene. 2006;25:304–309. doi: 10.1038/sj.onc.1209026. [DOI] [PubMed] [Google Scholar]
- Bossi G., Marampon F., Maor-Aloni R., Zani B., Rotter V., Oren M., Strano S., Blandino G., Sacchi A. Conditional RNA interference in vivo to study mutant p53 oncogenic gain of function on tumor malignancy. Cell Cycle. 2008;7:1870–1879. doi: 10.4161/cc.7.12.6161. [DOI] [PubMed] [Google Scholar]
- Bougeard G., Sesboüé R., Baert-Desurmont S., Vasseur S., Martin C., Tinat J., Brugières L., Chompret A., de Paillerets B.B., Stoppa-Lyonnet D., French LFS working group Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families. J. Med. Genet. 2008;45:535–538. doi: 10.1136/jmg.2008.057570. [DOI] [PubMed] [Google Scholar]
- Braicu C., Pileczki V., Irimie A., Berindan-Neagoe I. p53siRNA therapy reduces cell proliferation, migration and induces apoptosis in triple negative breast cancer cells. Mol. Cell. Biochem. 2013;381:61–68. doi: 10.1007/s11010-013-1688-5. [DOI] [PubMed] [Google Scholar]
- Brázdová M., Navrátilová L., Tichý V., Nĕmcová K., Lexa M., Hrstka R., Pečinka P., Adámik M., Vojtesek B., Paleček E. Preferential binding of hot spot mutant p53 proteins to supercoiled DNA in vitro and in cells. PLoS ONE. 2013;8:e59567. doi: 10.1371/journal.pone.0059567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Kalo E., Brosh R., Besserglick H., Nachmany I., Rais Y., Stambolsky P., Tang X., Milyavsky M., Shats I. Mutant p53 protects cells from 12-O-tetradecanoylphorbol-13-acetate-induced death by attenuating activating transcription factor 3 induction. Cancer Res. 2006;66:10750–10759. doi: 10.1158/0008-5472.CAN-06-0916. [DOI] [PubMed] [Google Scholar]
- Butler J.S., Loh S.N. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 2003;42:2396–2403. doi: 10.1021/bi026635n. [DOI] [PubMed] [Google Scholar]
- Bykov V.J., Issaeva N., Shilov A., Hultcrantz M., Pugacheva E., Chumakov P., Bergman J., Wiman K.G., Selivanova G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- Bykov V.J., Issaeva N., Zache N., Shilov A., Hultcrantz M., Bergman J., Selivanova G., Wiman K.G. Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J. Biol. Chem. 2005;280:30384–30391. doi: 10.1074/jbc.M501664200. [DOI] [PubMed] [Google Scholar]
- Capponcelli S., Pedrini E., Cerone M.A., Corti V., Fontanesi S., Alessio M., Bachi A., Soddu S., Ribatti D., Picci P. Evaluation of the molecular mechanisms involved in the gain of function of a Li-Fraumeni TP53 mutation. Hum. Mutat. 2005;26:94–103. doi: 10.1002/humu.20192. [DOI] [PubMed] [Google Scholar]
- Chee J.L., Saidin S., Lane D.P., Leong S.M., Noll J.E., Neilsen P.M., Phua Y.T., Gabra H., Lim T.M. Wild-type and mutant p53 mediate cisplatin resistance through interaction and inhibition of active caspase-9. Cell Cycle. 2013;12:278–288. doi: 10.4161/cc.23054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A., Molina P., Bargonetti J. Mutant p53 forms a complex with Sp1 on HIV-LTR DNA. Biochem. Biophys. Res. Commun. 2000;279:383–390. doi: 10.1006/bbrc.2000.3965. [DOI] [PubMed] [Google Scholar]
- Choudhury S., Kolukula V.K., Preet A., Albanese C., Avantaggiati M.L. Dissecting the pathways that destabilize mutant p53: the proteasome or autophagy? Cell Cycle. 2013;12:1022–1029. doi: 10.4161/cc.24128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffill C.R., Muller P.A., Oh H.K., Neo S.P., Hogue K.A., Cheok C.F., Vousden K.H., Lane D.P., Blackstock W.P., Gunaratne J. Mutant p53 interactome identifies nardilysin as a p53R273H-specific binding partner that promotes invasion. EMBO Rep. 2012;13:638–644. doi: 10.1038/embor.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooks T., Pateras I.S., Tarcic O., Solomon H., Schetter A.J., Wilder S., Lozano G., Pikarsky E., Forshew T., Rosenfeld N. Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell. 2013;23:634–646. doi: 10.1016/j.ccr.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demma M., Maxwell E., Ramos R., Liang L., Li C., Hesk D., Rossman R., Mallams A., Doll R., Liu M. SCH529074, a small molecule activator of mutant p53, which binds p53 DNA binding domain (DBD), restores growth-suppressive function to mutant p53 and interrupts HDM2-mediated ubiquitination of wild type p53. J. Biol. Chem. 2010;285:10198–10212. doi: 10.1074/jbc.M109.083469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrosellier J.S., Cheresh D.A. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Agostino S., Strano S., Emiliozzi V., Zerbini V., Mottolese M., Sacchi A., Blandino G., Piaggio G. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Di Como C.J., Gaiddon C., Prives C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol. 1999;19:1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer D., Pati S., Zambetti G., Chu S., Teresky A.K., Moore M., Finlay C., Levine A.J. Gain of function mutations in p53. Nat. Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- Do P.M., Varanasi L., Fan S., Li C., Kubacka I., Newman V., Chauhan K., Daniels S.R., Boccetta M., Garrett M.R. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 2012;26:830–845. doi: 10.1101/gad.181685.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P., Xu Z., Jia N., Li D., Feng Y. Elevated expression of p53 gain-of-function mutation R175H in endometrial cancer cells can increase the invasive phenotypes by activation of the EGFR/PI3K/AKT pathway. Mol. Cancer. 2009;8:103. doi: 10.1186/1476-4598-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P., Karaayvaz M., Jia N., Kaneuchi M., Hamada J., Watari H., Sudo S., Ju J., Sakuragi N. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene. 2012 doi: 10.1038/onc.2012.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle B., Morton J.P., Delaney D.W., Ridgway R.A., Wilkins J.A., Sansom O.J. p53 mutation and loss have different effects on tumourigenesis in a novel mouse model of pleomorphic rhabdomyosarcoma. J. Pathol. 2010;222:129–137. doi: 10.1002/path.2748. [DOI] [PubMed] [Google Scholar]
- Dumay A., Feugeas J.P., Wittmer E., Lehmann-Che J., Bertheau P., Espie M., Plassa L.F., Cottu P., Marty M., Andre F. Distinct tumor protein p53 mutants in breast cancer subgroups. International journal of cancer Journal international du cancer. 2013;132:1227–1231. doi: 10.1002/ijc.27767. [DOI] [PubMed] [Google Scholar]
- El-Hizawi S., Lagowski J.P., Kulesz-Martin M., Albor A. Induction of gene amplification as a gain-of-function phenotype of mutant p53 proteins. Cancer Res. 2002;62:3264–3270. [PubMed] [Google Scholar]
- Esser C., Scheffner M., Höhfeld J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J. Biol. Chem. 2005;280:27443–27448. doi: 10.1074/jbc.M501574200. [DOI] [PubMed] [Google Scholar]
- Fontemaggi G., Dell’Orso S., Trisciuoglio D., Shay T., Melucci E., Fazi F., Terrenato I., Mottolese M., Muti P., Domany E. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat. Struct. Mol. Biol. 2009;16:1086–1093. doi: 10.1038/nsmb.1669. [DOI] [PubMed] [Google Scholar]
- Foster B.A., Coffey H.A., Morin M.J., Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286:2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- Frank A.K., Pietsch E.C., Dumont P., Tao J., Murphy M.E. Wild-type and mutant p53 proteins interact with mitochondrial caspase-3. Cancer Biol. Ther. 2011;11:740–745. doi: 10.4161/cbt.11.8.14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor W.A., Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor W.A., Mizuno H., Zhao X., Langerød A., Moon S.H., Rodriguez-Barrueco R., Barsotti A., Chicas A., Li W., Polotskaia A. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiddon C., Lokshin M., Ahn J., Zhang T., Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwin B.I., Spillare E., Forrester K., Lehman T.A., Kispert J., Welsh J.A., Pfeifer A.M., Lechner J.F., Baker S.J., Vogelstein B. Mutant p53 can induce tumorigenic conversion of human bronchial epithelial cells and reduce their responsiveness to a negative growth factor, transforming growth factor beta 1. Proc. Natl. Acad. Sci. USA. 1992;89:2759–2763. doi: 10.1073/pnas.89.7.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardini J.E., Napoli M., Piazza S., Rustighi A., Marotta C., Radaelli E., Capaci V., Jordan L., Quinlan P., Thompson A. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 2011;20:79–91. doi: 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Gloushankova N., Ossovskaya V., Vasiliev J., Chumakov P., Kopnin B. Changes in p53 expression can modify cell shape of ras-transformed fibroblasts and epitheliocytes. Oncogene. 1997;15:2985–2989. doi: 10.1038/sj.onc.1201483. [DOI] [PubMed] [Google Scholar]
- Gogna R., Madan E., Kuppusamy P., Pati U. Chaperoning of mutant p53 protein by wild-type p53 protein causes hypoxic tumor regression. J. Biol. Chem. 2012;287:2907–2914. doi: 10.1074/jbc.M111.317354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhler T., Jäger S., Warnecke G., Yasuda H., Kim E., Deppert W. Mutant p53 proteins bind DNA in a DNA structure-selective mode. Nucleic Acids Res. 2005;33:1087–1100. doi: 10.1093/nar/gki252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grugan K.D., Vega M.E., Wong G.S., Diehl J.A., Bass A.J., Wong K.K., Nakagawa H., Rustgi A.K. A common p53 mutation (R175H) activates c-Met receptor tyrosine kinase to enhance tumor cell invasion. Cancer Biol. Ther. 2013;14:14. doi: 10.4161/cbt.25406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto A., Aldape K., Kozakiewicz K., Tlsty T.D. An oncogenic form of p53 confers a dominant, gain-of-function phenotype that disrupts spindle checkpoint control. Proc. Natl. Acad. Sci. USA. 1998;95:5166–5171. doi: 10.1073/pnas.95.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurova K.V., Rokhlin O.W., Budanov A.V., Burdelya L.G., Chumakov P.M., Cohen M.B., Gudkov A.V. Cooperation of two mutant p53 alleles contributes to Fas resistance of prostate carcinoma cells. Cancer Res. 2003;63:2905–2912. [PubMed] [Google Scholar]
- Gurtner A., Starace G., Norelli G., Piaggio G., Sacchi A., Bossi G. Mutant p53-induced up-regulation of mitogen-activated protein kinase kinase 3 contributes to gain of function. J. Biol. Chem. 2010;285:14160–14169. doi: 10.1074/jbc.M109.094813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy O., Michalovitz D., Oren M. Different tumor-derived p53 mutants exhibit distinct biological activities. Science. 1990;250:113–116. doi: 10.1126/science.2218501. [DOI] [PubMed] [Google Scholar]
- Hanel W., Marchenko N., Xu S., Yu S.X., Weng W., Moll U. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20:898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao M., Low J., Dorn E., Ku D., Pattengale P., Yeargin J., Haas M. Gain-of-function mutations of the p53 gene induce lymphohematopoietic metastatic potential and tissue invasiveness. Am. J. Pathol. 1994;145:702–714. [PMC free article] [PubMed] [Google Scholar]
- Huang X., Zhang Y., Tang Y., Butler N., Kim J., Guessous F., Schiff D., Mandell J., Abounader R. A novel PTEN/mutant p53/c-Myc/Bcl-XL axis mediates context-dependent oncogenic effects of PTEN with implications for cancer prognosis and therapy. Neoplasia. 2013;15:952–965. doi: 10.1593/neo.13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L., Zheng Y., Yan Y., Bargonetti J., Foster D.A. Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D. Oncogene. 2006;25:7305–7310. doi: 10.1038/sj.onc.1209735. [DOI] [PubMed] [Google Scholar]
- Jackson J.G., Lozano G. The mutant p53 mouse as a pre-clinical model. Oncogene. 2013;32:4325–4330. doi: 10.1038/onc.2012.610. [DOI] [PubMed] [Google Scholar]
- Jackson E.L., Olive K.P., Tuveson D.A., Bronson R., Crowley D., Brown M., Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Jackson J.G., Pant V., Li Q., Chang L.L., Quintás-Cardama A., Garza D., Tavana O., Yang P., Manshouri T., Li Y. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell. 2012;21:793–806. doi: 10.1016/j.ccr.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S., Zhao L., Tang W., Luo Y. The gain of function of p53 mutant p53S in promoting tumorigenesis by cross-talking with H-RasV12. Int. J. Biol. Sci. 2012;8:596–605. doi: 10.7150/ijbs.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger A.C., Fersht A.R. Structure-function-rescue: the diverse nature of common p53 cancer mutants. Oncogene. 2007;26:2226–2242. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- Joerger A.C., Fersht A.R. Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- Kalo E., Kogan-Sakin I., Solomon H., Bar-Nathan E., Shay M., Shetzer Y., Dekel E., Goldfinger N., Buganim Y., Stambolsky P. Mutant p53R273H attenuates the expression of phase 2 detoxifying enzymes and promotes the survival of cells with high levels of reactive oxygen species. J. Cell Sci. 2012;125:5578–5586. doi: 10.1242/jcs.106815. [DOI] [PubMed] [Google Scholar]
- Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata H., Omotehara F., Nakashiro K., Uchida D., Shinagawa Y., Tachibana M., Imai Y., Fujimori T. Oncogenic mutation of the p53 gene derived from head and neck cancer prevents cells from undergoing apoptosis after DNA damage. Int. J. Oncol. 2007;30:1089–1097. [PubMed] [Google Scholar]
- Kawamura T., Suzuki J., Wang Y.V., Menendez S., Morera L.B., Raya A., Wahl G.M., Izpisúa Belmonte J.C. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S.I., Purdie C.A., Bray S.E., Quinlan P.R., Jordan L.B., Thompson A.M., Meek D.W. Immunohistochemical detection of Polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcom. Breast Cancer Res. 2012;14:R40. doi: 10.1186/bcr3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko J.E., Ilyinskaya G.V., Komarov P.G., Agapova L.S., Kochetkov D.V., Strom E., Frolova E.I., Kovriga I., Gudkov A.V., Feinstein E., Chumakov P.M. Small-molecule RETRA suppresses mutant p53-bearing cancer cells through a p73-dependent salvage pathway. Proc. Natl. Acad. Sci. USA. 2008;105:6302–6307. doi: 10.1073/pnas.0802091105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.M., Gorzov P., Veprintsev D.B., Söderqvist M., Segerbäck D., Bergman J., Fersht A.R., Hainaut P., Wiman K.G., Bykov V.J. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15:376–388. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Lang G.A., Iwakuma T., Suh Y.A., Liu G., Rao V.A., Parant J.M., Valentin-Vega Y.A., Terzian T., Caldwell L.C., Strong L.C. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Lányi A., Deb D., Seymour R.C., Ludes-Meyers J.H., Subler M.A., Deb S. ‘Gain of function’ phenotype of tumor-derived mutant p53 requires the oligomerization/nonsequence-specific nucleic acid-binding domain. Oncogene. 1998;16:3169–3176. doi: 10.1038/sj.onc.1201857. [DOI] [PubMed] [Google Scholar]
- Lee M.K., Teoh W.W., Phang B.H., Tong W.M., Wang Z.Q., Sabapathy K. Cell-type, dose, and mutation-type specificity dictate mutant p53 functions in vivo. Cancer Cell. 2012;22:751–764. doi: 10.1016/j.ccr.2012.10.022. [DOI] [PubMed] [Google Scholar]
- Lehmann B.D., Pietenpol J.A. Targeting mutant p53 in human tumors. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30:3648–3650. doi: 10.1200/JCO.2012.44.0412. [DOI] [PubMed] [Google Scholar]
- Lehmann S., Bykov V.J., Ali D., Andren O., Cherif H., Tidefelt U., Uggla B., Yachnin J., Juliusson G., Moshfegh A. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol. 2012;30:3633–3639. doi: 10.1200/JCO.2011.40.7783. [DOI] [PubMed] [Google Scholar]
- Leroy B., Fournier J.L., Ishioka C., Monti P., Inga A., Fronza G., Soussi T. The TP53 website: an integrative resource centre for the TP53 mutation database and TP53 mutant analysis. Nucleic Acids Res. 2013;41(Database issue):D962–D969. doi: 10.1093/nar/gks1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Sutphin P.D., Schwartz D., Matas D., Almog N., Wolkowicz R., Goldfinger N., Pei H., Prokocimer M., Rotter V. Mutant p53 protein expression interferes with p53-independent apoptotic pathways. Oncogene. 1998;16:3269–3277. doi: 10.1038/sj.onc.1201867. [DOI] [PubMed] [Google Scholar]
- Li D., Marchenko N.D., Moll U.M. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011;18:1904–1913. doi: 10.1038/cdd.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Marchenko N.D., Schulz R., Fischer V., Velasco-Hernandez T., Talos F., Moll U.M. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol. Cancer Res. 2011;9:577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.Y., Vidnovic N., Ellisen L.W., Leong C.O. Mutant p53 mediates survival of breast cancer cells. Br. J. Cancer. 2009;101:1606–1612. doi: 10.1038/sj.bjc.6605335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Liang Y., Zhu H., Zhang J., Zhong X. R280T mutation of p53 gene promotes proliferation of human glioma cells through GSK-3β/PTEN pathway. Neurosci. Lett. 2012;529:60–65. doi: 10.1016/j.neulet.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Liu E.Y., Ryan K.M. Autophagy and cancer—issues we need to digest. J. Cell Sci. 2012;125:2349–2358. doi: 10.1242/jcs.093708. [DOI] [PubMed] [Google Scholar]
- Liu K., Ling S., Lin W.C. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol. Cell. Biol. 2011;31:4464–4481. doi: 10.1128/MCB.05574-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wilcken R., Joerger A.C., Chuckowree I.S., Amin J., Spencer J., Fersht A.R. Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res. 2013;41:6034–6044. doi: 10.1093/nar/gkt305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loging W.T., Reisman D. Elevated expression of ribosomal protein genes L37, RPP-1, and S2 in the presence of mutant p53. Cancer Epidemiol Biomarkers Prev. 1999;8:1011–1016. [PubMed] [Google Scholar]
- Loh S.N. The missing zinc: p53 misfolding and cancer. Metallomics: integrated biometal science. 2010;2:442–449. doi: 10.1039/c003915b. [DOI] [PubMed] [Google Scholar]
- Lu X., Liu D.P., Xu Y. The gain of function of p53 cancer mutant in promoting mammary tumorigenesis. Oncogene. 2013;32:2900–2906. doi: 10.1038/onc.2012.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashchuk N., Vousden K.H. Ubiquitination and degradation of mutant p53. Mol. Cell. Biol. 2007;27:8284–8295. doi: 10.1128/MCB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madar S., Harel E., Goldstein I., Stein Y., Kogan-Sakin I., Kamer I., Solomon H., Dekel E., Tal P., Goldfinger N. Mutant p53 attenuates the anti-tumorigenic activity of fibroblasts-secreted interferon beta. PLoS ONE. 2013;8:e61353. doi: 10.1371/journal.pone.0061353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marión R.M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslon M.M., Hupp T.R. Drug discovery and mutant p53. Trends Cell Biol. 2010;20:542–555. doi: 10.1016/j.tcb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Matas D., Sigal A., Stambolsky P., Milyavsky M., Weisz L., Schwartz D., Goldfinger N., Rotter V. Integrity of the N-terminal transcription domain of p53 is required for mutant p53 interference with drug-induced apoptosis. EMBO J. 2001;20:4163–4172. doi: 10.1093/emboj/20.15.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E., Tasdemir E., Maiuri M.C., Galluzzi L., Kepp O., Criollo A., Vicencio J.M., Soussi T., Kroemer G. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–3061. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- Morton J.P., Timpson P., Karim S.A., Ridgway R.A., Athineos D., Doyle B., Jamieson N.B., Oien K.A., Lowy A.M., Brunton V.G. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc. Natl. Acad. Sci. USA. 2010;107:246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P.A., Vousden K.H. p53 mutations in cancer. Nat. Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- Muller P.A., Caswell P.T., Doyle B., Iwanicki M.P., Tan E.H., Karim S., Lukashchuk N., Gillespie D.A., Ludwig R.L., Gosselin P. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- Muller P.A., Trinidad A.G., Timpson P., Morton J.P., Zanivan S., van den Berghe P.V., Nixon C., Karim S.A., Caswell P.T., Noll J.E. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene. 2012;32:1252–1265. doi: 10.1038/onc.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.L., Dennis A.P., Rosen J.M. A gain of function p53 mutant promotes both genomic instability and cell survival in a novel p53-null mammary epithelial cell model. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2000;14:2291–2302. doi: 10.1096/fj.00-0128com. [DOI] [PubMed] [Google Scholar]
- Narendran A., Ganjavi H., Morson N., Connor A., Barlow J.W., Keystone E., Malkin D., Freedman M.H. Mutant p53 in bone marrow stromal cells increases VEGF expression and supports leukemia cell growth. Exp. Hematol. 2003;31:693–701. doi: 10.1016/s0301-472x(03)00159-0. [DOI] [PubMed] [Google Scholar]
- Neilsen P.M., Noll J.E., Mattiske S., Bracken C.P., Gregory P.A., Schulz R.B., Lim S.P., Kumar R., Suetani R.J., Goodall G.J., Callen D.F. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene. 2012;32:2992–3000. doi: 10.1038/onc.2012.305. [DOI] [PubMed] [Google Scholar]
- Nguyen C.H., Lang B.J., Chai R.C., Vieusseux J.L., Kouspou M.M., Price J.T. Heat-shock factor 1 both positively and negatively affects cellular clonogenic growth depending on p53 status. Biochem. J. 2013;452:321–329. doi: 10.1042/BJ20130098. [DOI] [PubMed] [Google Scholar]
- Noll J.E., Jeffery J., Al-Ejeh F., Kumar R., Khanna K.K., Callen D.F., Neilsen P.M. Mutant p53 drives multinucleation and invasion through a process that is suppressed by ANKRD11. Oncogene. 2012;31:2836–2848. doi: 10.1038/onc.2011.456. [DOI] [PubMed] [Google Scholar]
- Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Ostman A., Augsten M. Cancer-associated fibroblasts and tumor growth—bystanders turning into key players. Curr. Opin. Genet. Dev. 2009;19:67–73. doi: 10.1016/j.gde.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Patocs A., Zhang L., Xu Y., Weber F., Caldes T., Mutter G.L., Platzer P., Eng C. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N. Engl. J. Med. 2007;357:2543–2551. doi: 10.1056/NEJMoa071825. [DOI] [PubMed] [Google Scholar]
- Petitjean A., Achatz M.I., Borresen-Dale A.L., Hainaut P., Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- Pietras K., Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp. Cell Res. 2010;316:1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- Pintus S.S., Ivanisenko N.V., Demenkov P.S., Ivanisenko T.V., Ramachandran S., Kolchanov N.A., Ivanisenko V.A. The substitutions G245C and G245D in the Zn(2+)-binding pocket of the p53 protein result in differences of conformational flexibility of the DNA-binding domain. J. Biomol. Struct. Dyn. 2013;31:78–86. doi: 10.1080/07391102.2012.691364. [DOI] [PubMed] [Google Scholar]
- Pohl U., Wagenknecht B., Naumann U., Weller M. p53 enhances BAK and CD95 expression in human malignant glioma cells but does not enhance CD95L-induced apoptosis. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 1999;9:29–37. doi: 10.1159/000016300. [DOI] [PubMed] [Google Scholar]
- Preuss U., Kreutzfeld R., Scheidtmann K.H. Tumor-derived p53 mutant C174Y is a gain-of-function mutant which activates the fos promoter and enhances colony formation. Int. J. Cancer. 2000;88:162–171. doi: 10.1002/1097-0215(20001015)88:2<162::aid-ijc3>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Puca R., Nardinocchi L., Bossi G., Sacchi A., Rechavi G., Givol D., D’Orazi G. Restoring wtp53 activity in HIPK2 depleted MCF7 cells by modulating metallothionein and zinc. Exp. Cell Res. 2009;315:67–75. doi: 10.1016/j.yexcr.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Puca R., Nardinocchi L., Porru M., Simon A.J., Rechavi G., Leonetti C., Givol D., D’Orazi G. Restoring p53 active conformation by zinc increases the response of mutant p53 tumor cells to anticancer drugs. Cell Cycle. 2011;10:1679–1689. doi: 10.4161/cc.10.10.15642. [DOI] [PubMed] [Google Scholar]
- Pugacheva E.N., Ivanov A.V., Kravchenko J.E., Kopnin B.P., Levine A.J., Chumakov P.M. Novel gain of function activity of p53 mutants: activation of the dUTPase gene expression leading to resistance to 5-fluorouracil. Oncogene. 2002;21:4595–4600. doi: 10.1038/sj.onc.1205704. [DOI] [PubMed] [Google Scholar]
- Quante T., Otto B., Brázdová M., Kejnovská I., Deppert W., Tolstonog G.V. Mutant p53 is a transcriptional co-factor that binds to G-rich regulatory regions of active genes and generates transcriptional plasticity. Cell Cycle. 2012;11:3290–3303. doi: 10.4161/cc.21646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restle A., Färber M., Baumann C., Böhringer M., Scheidtmann K.H., Müller-Tidow C., Wiesmüller L. Dissecting the role of p53 phosphorylation in homologous recombination provides new clues for gain-of-function mutants. Nucleic Acids Res. 2008;36:5362–5375. doi: 10.1093/nar/gkn503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieber M., Strasberg-Rieber M. Hypoxia, Mn-SOD and H(2)O(2) regulate p53 reactivation and PRIMA-1 toxicity irrespective of p53 status in human breast cancer cells. Biochem. Pharmacol. 2012;84:1563–1570. doi: 10.1016/j.bcp.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Rodriguez O.C., Choudhury S., Kolukula V., Vietsch E.E., Catania J., Preet A., Reynoso K., Bargonetti J., Wellstein A., Albanese C., Avantaggiati M.L. Dietary downregulation of mutant p53 levels via glucose restriction: mechanisms and implications for tumor therapy. Cell Cycle. 2012;11:4436–4446. doi: 10.4161/cc.22778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger L., Jullien L., Gire V., Roux P. Gain of oncogenic function of p53 mutants regulates E-cadherin expression uncoupled from cell invasion in colon cancer cells. J. Cell Sci. 2010;123:1295–1305. doi: 10.1242/jcs.061002. [DOI] [PubMed] [Google Scholar]
- Sarig R., Rivlin N., Brosh R., Bornstein C., Kamer I., Ezra O., Molchadsky A., Goldfinger N., Brenner O., Rotter V. Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. J. Exp. Med. 2010;207:2127–2140. doi: 10.1084/jem.20100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer L., Gitenay D., Vo C., Baron V.T. Mutant p53 initiates a feedback loop that involves Egr-1/EGF receptor/ERK in prostate cancer cells. Oncogene. 2010;29:2628–2637. doi: 10.1038/onc.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling T., Kairat A., Melino G., Krammer P.H., Stremmel W., Oren M., Müller M. Interference with the p53 family network contributes to the gain of oncogenic function of mutant p53 in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2010;394:817–823. doi: 10.1016/j.bbrc.2010.03.082. [DOI] [PubMed] [Google Scholar]
- Scian M.J., Stagliano K.E., Deb D., Ellis M.A., Carchman E.H., Das A., Valerie K., Deb S.P., Deb S. Tumor-derived p53 mutants induce oncogenesis by transactivating growth-promoting genes. Oncogene. 2004;23:4430–4443. doi: 10.1038/sj.onc.1207553. [DOI] [PubMed] [Google Scholar]
- Scian M.J., Stagliano K.E., Ellis M.A., Hassan S., Bowman M., Miles M.F., Deb S.P., Deb S. Modulation of gene expression by tumor-derived p53 mutants. Cancer Res. 2004;64:7447–7454. doi: 10.1158/0008-5472.CAN-04-1568. [DOI] [PubMed] [Google Scholar]
- Shi X.B., Nesslinger N.J., Deitch A.D., Gumerlock P.H., deVere White R.W. Complex functions of mutant p53 alleles from human prostate cancer. Prostate. 2002;51:59–72. doi: 10.1002/pros.10072. [DOI] [PubMed] [Google Scholar]
- Singh S., Singh P.P. Statin a day keeps cancer at bay. World journal of clinical oncology. 2013;4:43–46. doi: 10.5306/wjco.v4.i2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.D., Crossland S., Parker G., Osin P., Brooks L., Waller J., Philp E., Crompton M.R., Gusterson B.A., Allday M.J., Crook T. Novel p53 mutants selected in BRCA-associated tumours which dissociate transformation suppression from other wild-type p53 functions. Oncogene. 1999;18:2451–2459. doi: 10.1038/sj.onc.1202565. [DOI] [PubMed] [Google Scholar]
- Solomon H., Buganim Y., Kogan-Sakin I., Pomeraniec L., Assia Y., Madar S., Goldstein I., Brosh R., Kalo E., Beatus T. Various p53 mutant proteins differently regulate the Ras circuit to induce a cancer-related gene signature. J. Cell Sci. 2012;125:3144–3152. doi: 10.1242/jcs.099663. [DOI] [PubMed] [Google Scholar]
- Sonego M., Schiappacassi M., Lovisa S., Dall’Acqua A., Bagnoli M., Lovat F., Libra M., D’Andrea S., Canzonieri V., Militello L. Stathmin regulates mutant p53 stability and transcriptional activity in ovarian cancer. EMBO molecular medicine. 2013;5:707–722. doi: 10.1002/emmm.201201504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Hollstein M., Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat. Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- Stambolsky P., Tabach Y., Fontemaggi G., Weisz L., Maor-Aloni R., Siegfried Z., Shiff I., Kogan I., Shay M., Kalo E. Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell. 2010;17:273–285. doi: 10.1016/j.ccr.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S., Fontemaggi G., Costanzo A., Rizzo M.G., Monti O., Baccarini A., Del Sal G., Levrero M., Sacchi A., Oren M., Blandino G. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J. Biol. Chem. 2002;277:18817–18826. doi: 10.1074/jbc.M201405200. [DOI] [PubMed] [Google Scholar]
- Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat. Rev. Drug Discov. 2010;9:643–660. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- Su X., Chakravarti D., Cho M.S., Liu L., Gi Y.J., Lin Y.L., Leung M.L., El-Naggar A.K., Creighton C.J., Suraokar M.B. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y.A., Post S.M., Elizondo-Fraire A.C., Maccio D.R., Jackson J.G., El-Naggar A.K., Van Pelt C., Terzian T., Lozano G. Multiple stress signals activate mutant p53 in vivo. Cancer Res. 2011;71:7168–7175. doi: 10.1158/0008-5472.CAN-11-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nakamura K., Wendel E., Colburn N. Progression toward tumor cell phenotype is enhanced by overexpression of a mutant p53 tumor-suppressor gene isolated from nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. USA. 1993;90:2827–2831. doi: 10.1073/pnas.90.7.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E.H., Morton J.P., Timpson P., Tucci P., Melino G., Flores E.R., Sansom O.J., Vousden K.H., Muller P.A. Functions of TAp63 and p53 in restraining the development of metastatic cancer. Oncogene. 2013 doi: 10.1038/onc.2013.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E., Maiuri M.C., Galluzzi L., Vitale I., Djavaheri-Mergny M., D’Amelio M., Criollo A., Morselli E., Zhu C., Harper F. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepel M., Groscurth P., Malipiero U., Gulbins E., Dichgans J., Weller M. Chemosensitivity of human malignant glioma: modulation by p53 gene transfer. J. Neurooncol. 1998;39:19–32. doi: 10.1023/a:1005910323338. [DOI] [PubMed] [Google Scholar]
- Trinidad A.G., Muller P.A., Cuellar J., Klejnot M., Nobis M., Valpuesta J.M., Vousden K.H. Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol. Cell. 2013;50:805–817. doi: 10.1016/j.molcel.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang W.P., Ho F.Y., Fung K.P., Kong S.K., Kwok T.T. p53-R175H mutant gains new function in regulation of doxorubicin-induced apoptosis. International journal of cancer Journal international du cancer. 2005;114:331–336. doi: 10.1002/ijc.20818. [DOI] [PubMed] [Google Scholar]
- Tucci P., Agostini M., Grespi F., Markert E.K., Terrinoni A., Vousden K.H., Muller P.A.J., Dötsch V., Kehrloesser S., Sayan B.S. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc. Natl. Acad. Sci. USA. 2012;109:15312–15317. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakifahmetoglu-Norberg H., Kim M., Xia H.G., Iwanicki M.P., Ofengeim D., Coloff J.L., Pan L., Ince T.A., Kroemer G., Brugge J.S., Yuan J. Chaperone-mediated autophagy degrades mutant p53. Genes Dev. 2013;27:1718–1730. doi: 10.1101/gad.220897.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan C.A., Frum R., Pearsall I., Singh S., Windle B., Yeudall A., Deb S.P., Deb S. Allele specific gain-of-function activity of p53 mutants in lung cancer cells. Biochem. Biophys. Res. Commun. 2012;428:6–10. doi: 10.1016/j.bbrc.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan C.A., Singh S., Windle B., Sankala H.M., Graves P.R., Andrew Yeudall W., Deb S.P., Deb S. p53 mutants induce transcription of NF-κB2 in H1299 cells through CBP and STAT binding on the NF-κB2 promoter and gain of function activity. Arch. Biochem. Biophys. 2012;518:79–88. doi: 10.1016/j.abb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaegh G.W., Parat M.O., Richard M.J., Hainaut P. Modulation of p53 protein conformation and DNA-binding activity by intracellular chelation of zinc. Mol. Carcinog. 1998;21:205–214. doi: 10.1002/(sici)1098-2744(199803)21:3<205::aid-mc8>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Vikhanskaya F., Lee M.K., Mazzoletti M., Broggini M., Sabapathy K. Cancer-derived p53 mutants suppress p53-target gene expression—potential mechanism for gain of function of mutant p53. Nucleic Acids Res. 2007;35:2093–2104. doi: 10.1093/nar/gkm099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walerych D., Napoli M., Collavin L., Del Sal G. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 2012;33:2007–2017. doi: 10.1093/carcin/bgs232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Simon R. Identification of potential synthetic lethal genes to p53 using a computational biology approach. BMC Med. Genomics. 2013;6:30. doi: 10.1186/1755-8794-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Suh Y.A., Fuller M.Y., Jackson J.G., Xiong S., Terzian T., Quintás-Cardama A., Bankson J.A., El-Naggar A.K., Lozano G. Restoring expression of wild-type p53 suppresses tumor growth but does not cause tumor regression in mice with a p53 missense mutation. J. Clin. Invest. 2011;121:893–904. doi: 10.1172/JCI44504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Cheng B., Miao L., Mei Y., Wu M. Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression. Cell Death Dis. 2013;4:e574. doi: 10.1038/cddis.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen J.X., Liu J.P., You C., Liu Y.H., Mao Q. Gain of function of mutant TP53 in glioblastoma: prognosis and response to temozolomide. Ann. Surg. Oncol. 2013 doi: 10.1245/s10434-013-3380-0. Published online November 19, 2013. [DOI] [PubMed] [Google Scholar]
- Weisz L., Zalcenstein A., Stambolsky P., Cohen Y., Goldfinger N., Oren M., Rotter V. Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res. 2004;64:8318–8327. doi: 10.1158/0008-5472.CAN-04-1145. [DOI] [PubMed] [Google Scholar]
- Weisz L., Oren M., Rotter V. Transcription regulation by mutant p53. Oncogene. 2007;26:2202–2211. doi: 10.1038/sj.onc.1210294. [DOI] [PubMed] [Google Scholar]
- Wiman K.G. Pharmacological reactivation of mutant p53: from protein structure to the cancer patient. Oncogene. 2010;29:4245–4252. doi: 10.1038/onc.2010.188. [DOI] [PubMed] [Google Scholar]
- Wong R.P., Tsang W.P., Chau P.Y., Co N.N., Tsang T.Y., Kwok T.T. p53-R273H gains new function in induction of drug resistance through down-regulation of procaspase-3. Mol. Cancer Ther. 2007;6:1054–1061. doi: 10.1158/1535-7163.MCT-06-0336. [DOI] [PubMed] [Google Scholar]
- Xie T.X., Zhou G., Zhao M., Sano D., Jasser S.A., Brennan R.G., Myers J.N. Serine substitution of proline at codon 151 of TP53 confers gain of function activity leading to anoikis resistance and tumor progression of head and neck cancer cells. Laryngoscope. 2013;123:1416–1423. doi: 10.1002/lary.23846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Chen X. Identification of GRO1 as a critical determinant for mutant p53 gain of function. J. Biol. Chem. 2009;284:12178–12187. doi: 10.1074/jbc.M900994200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Chen X. Characterization of functional domains necessary for mutant p53 gain of function. J. Biol. Chem. 2010;285:14229–14238. doi: 10.1074/jbc.M109.097253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Liu G., Scoumanne A., Chen X. Suppression of inhibitor of differentiation 2, a target of mutant p53, is required for gain-of-function mutations. Cancer Res. 2008;68:6789–6796. doi: 10.1158/0008-5472.CAN-08-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Liu S., Xu E., Zhang J., Zhang Y., Chen X., Chen X. Histone deacetylase inhibitors suppress mutant p53 transcription via histone deacetylase 8. Oncogene. 2013;32:599–609. doi: 10.1038/onc.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeudall W.A., Vaughan C.A., Miyazaki H., Ramamoorthy M., Choi M.Y., Chapman C.G., Wang H., Black E., Bulysheva A.A., Deb S.P. Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration. Carcinogenesis. 2012;33:442–451. doi: 10.1093/carcin/bgr270. [DOI] [PubMed] [Google Scholar]
- Yi L., Lu C., Hu W., Sun Y., Levine A.J. Multiple roles of p53-related pathways in somatic cell reprogramming and stem cell differentiation. Cancer Res. 2012;72:5635–5645. doi: 10.1158/0008-5472.CAN-12-1451. [DOI] [PubMed] [Google Scholar]
- Yi Y.W., Kang H.J., Kim H.J., Kong Y., Brown M.L., Bae I. Targeting mutant p53 by a SIRT1 activator YK-3-237 inhibits the proliferation of triple-negative breast cancer cells. Oncotarget. 2013;4:984–994. doi: 10.18632/oncotarget.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K., Hamada J., Tada M., Kameyama T., Nakagawa K., Suzuki Y., Ikawa M., Hassan N.M., Kitagawa Y., Moriuchi T. Mutant p53 R248Q but not R248W enhances in vitro invasiveness of human lung cancer NCI-H1299 cells. Biomed. Res. 2010;31:401–411. doi: 10.2220/biomedres.31.401. [DOI] [PubMed] [Google Scholar]
- Yu X., Vazquez A., Levine A.J., Carpizo D.R. Allele-specific p53 mutant reactivation. Cancer Cell. 2012;21:614–625. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zache N., Lambert J.M., Rökaeus N., Shen J., Hainaut P., Bergman J., Wiman K.G., Bykov V.J. Mutant p53 targeting by the low molecular weight compound STIMA-1. Mol. Oncol. 2008;2:70–80. doi: 10.1016/j.molonc.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalcenstein A., Stambolsky P., Weisz L., Müller M., Wallach D., Goncharov T.M., Krammer P.H., Rotter V., Oren M. Mutant p53 gain of function: repression of CD95(Fas/APO-1) gene expression by tumor-associated p53 mutants. Oncogene. 2003;22:5667–5676. doi: 10.1038/sj.onc.1206724. [DOI] [PubMed] [Google Scholar]
- Zalcenstein A., Weisz L., Stambolsky P., Bar J., Rotter V., Oren M. Repression of the MSP/MST-1 gene contributes to the antiapoptotic gain of function of mutant p53. Oncogene. 2006;25:359–369. doi: 10.1038/sj.onc.1209061. [DOI] [PubMed] [Google Scholar]
- Zerdoumi Y., Aury-Landas J., Bonaïti-Pellié C., Derambure C., Sesboüé R., Renaux-Petel M., Frebourg T., Bougeard G., Flaman J.M. Drastic effect of germline TP53 missense mutations in Li-Fraumeni patients. Hum. Mutat. 2013;34:453–461. doi: 10.1002/humu.22254. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yan W., Chen X. Mutant p53 disrupts MCF-10A cell polarity in three-dimensional culture via epithelial-to-mesenchymal transitions. J. Biol. Chem. 2011;286:16218–16228. doi: 10.1074/jbc.M110.214585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Mao Q., Lin Y., Yang K., Xie L. RNA interference targeting mutant p53 inhibits growth and induces apoptosis in DU145 human prostate cancer cells. Med. Oncol. 2011;28(Suppl 1):S381–S387. doi: 10.1007/s12032-010-9679-9. [DOI] [PubMed] [Google Scholar]
- Zhu H.B., Yang K., Xie Y.Q., Lin Y.W., Mao Q.Q., Xie L.P. Silencing of mutant p53 by siRNA induces cell cycle arrest and apoptosis in human bladder cancer cells. World J. Surg. Oncol. 2013;11:22. doi: 10.1186/1477-7819-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]