Analysis of optimized DNase-seq reveals intrinsic bias in transcription factor footprint identification (original) (raw)
. Author manuscript; available in PMC: 2014 Jul 1.
Published in final edited form as: Nat Methods. 2013 Dec 8;11(1):73–78. doi: 10.1038/nmeth.2762
Abstract
DNase-seq is a powerful technique for identifying cis-regulatory elements across the genome. We studied the key experimental parameters to optimize the performance of DNase-seq. We found that sequencing short 50-100bp fragments that accumulate in long inter-nucleosome linker regions is more efficient for identifying transcription factor binding sites than using longer fragments. We also assessed the potential of DNase-seq to predict transcription factor occupancy through the generation of nucleotide-resolution transcription factor footprints. In modeling the sequence-specific DNaseI cutting bias we found a surprisingly strong effect that varied over more than two orders of magnitude. This confounds DNaseI footprint analysis to the extent that the nucleotide resolution cleavage patterns at most transcription factor binding sites are derived from intrinsic DNaseI cleavage bias rather than from specific protein-DNA interactions. In contrast, quantitative comparison of DNaseI hypersensitivity between states can predict transcription factor occupancy associated with particular biological perturbations.
Keywords: DNaseI hypersensitivity, DNase-seq, DNaseI footprint, Chromatin dynamics, CTCF, Androgen receptor, Estrogen receptor, Transcription factor binding, Nucleosome
Introduction
DNase-seq adapts traditional DNaseI footprinting 1 and leverages modern DNA sequencing to identify regions of the genome where regulatory factors interact with DNA to modify chromatin structure and gene transcription1-5. DNase-seq has been applied to map regulatory regions in diverse cell and tissue types, revealing cell and lineage specific regulators, as well as regulatory regions that are present in a broad spectrum of cell types 5,6. These regulatory regions may also be used to help understand the biological role of noncoding genetic variants in physical traits and common diseases6,7. In addition, DNase-seq has been highly effective in comparing transcription factor binding profiles in treatment relative to control conditions 8-11.
To optimize DNase-seq and to characterize its biases, we studied the key parameters of DNaseI strength and selected fragment size. We sought to assess the ability of DNase-seq to detect transcription factor binding sites and to understand the nature of systematic biases that could influence the interpretation of DNase-seq data. We also addressed the use of DNase-seq footprinting to discover transcription factor binding sites at nucleotide resolution 5,12. We found that footprinting from DNase-seq, while informative for some transcription factors such as CTCF, was uninformative for many others such as the androgen receptor (AR). We show that intrinsic DNA cutting biases, a factor that has been largely ignored in footprinting studies, can be incorrectly interpreted as patterns induced by transcription factor binding.
Results
Fragment size and enzyme strength influence DNase-seq
One of the main goals of this study was to determine the optimal DNaseI digestion conditions to obtain high quality, reproducible, DNase-seq datasets for identifying genome-wide transcription factor (TF) binding sites. We conducted DNase-seq experiments using nominal DNaseI strength of 5U, 25U, 50U, 75U and 100U per reaction (500ml), selecting fragments in size ranges of 50-100bp, 100-200bp and 200-300bp for subsequent sequencing (Supplementary Fig. 1). In the online methods we provide a calibration approach to allow enzyme activity to be reproduced in other systems (Supplementary Fig. 1) and found high levels of concordance between replicates (Supplementary Fig. 2).
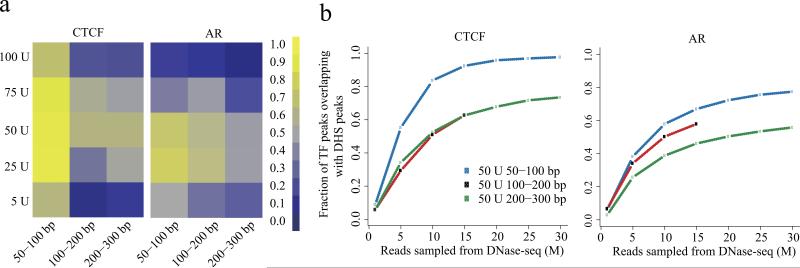
We compared the overlap of DNase-seq peaks identified by MACS 13 in the prostate cancer cell line, LNCaP, with high confidence CTCF, AR and FOXA1 ChIP-seq peaks in the same line. At a read depth down-sampled to 15M mapped reads, the short (50-100bp) fragments recovered a greater proportion of known sites than the intermediate (100-200bp) and long (200-300bp) fragments (Fig. 1a). A similar trend was observed for peaks of the enhancer and promoter associated histone modification H3K4me2 (Supplementary Fig. 3). DNaseI digestion with 25U and 50U performed better than other amounts for various fragment lengths (Fig. 1a), regardless of sequencing depth (Fig. 1b). Moreover, the use of longer (100-200bp or 200-300bp) fragments would require many fold greater sequencing depth to find the sites found using the 50-100bp fragments. Sites that were not detected tended to have lower ChIP-seq signal, and were less likely to be identified as strong ChIP-seq peaks (Supplementary Fig 4). Across ChIP-seq binding sites, as 5U 50-100bp DHS signal decreased so did the DHS signal in all other experimental conditions (Supplementary Fig. 5). Pooling samples with suboptimal conditions decreased performance (Supplementary Fig. 6). The results we observed in LNCaP could be applied to other cell lines. In MCF-7 the optimal conditions for DNase-seq to identify estrogen receptor (ER) and CTCF binding sites were similar to those found in LNCaP (Supplementary Fig. 7).
Figure 1.
Effect of digestion level and fragment size on recovering known transcription factor binding sites. (a) Proportion of ChIP-seq enriched regions discovered as DNaseI hypersensitive (DHS) sites for CTCF (left), androgen receptor (AR, center) and FOXA1 (right) in LNCaP cells. As the DNase-seq read depth strongly influences performance, for this comparison 15M reads were sampled from each experimental condition. In each heatmap, rows correspond to the DNaseI enzyme strength and columns represent fragment sizes. The colors represent the proportion of binding sites detected by DNase-seq. (b) Influence of read depth and fragment size on the overlap between TF binding sites and DHS sites. At the 50U strength the performance of the three size fractions are compared across a range of read depths. The results are consistent between different read depths, showing how shallow sampling is informative about the results obtained with deeper sequencing. Diminishing returns in performance with read depth, especially in the case of CTCF, shows that a vast increase in sequencing depth would be required before the 100-200bp and 200-300bp fragments could recover the proportion of CTCF binding sites that can be recovered by the 50-100bp fragments at a read depth of 30M.
Inter-nucleosome spacing filters DNase-seq fragments
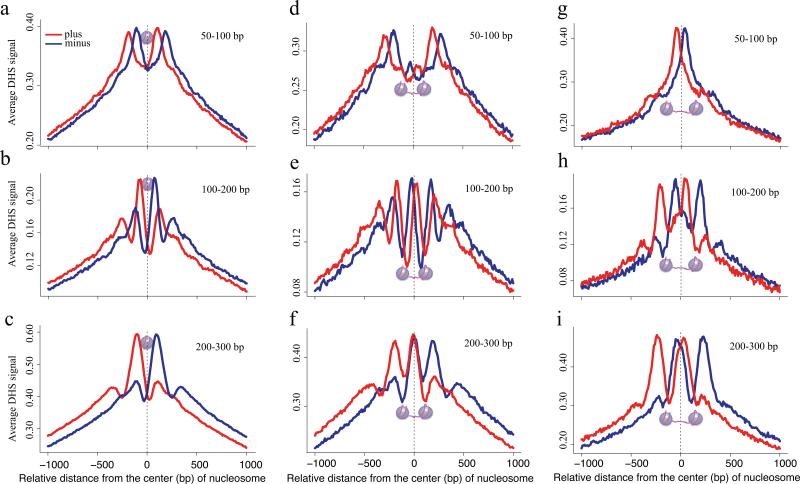
To probe chromatin structural effects that underlie fragment size differences we examined strand specific DNase-seq read distributions as a function of the distance from experimentally identified H3K4me2 marked nucleosome centers (Fig. 2) 14. We found striking differences in the patterns produced by the different fragment sizes, although the nucleosome occupied region was depleted of cuts in all cases. The 50-100bp fragments, being shorter than the 147bp of DNA associated with a nucleosome, cannot span a nucleosome, therefore are constrained to lie within the linker regions between two nucleosomes (Fig. 2a,b). Of the intermediate size 100-200bp fragments a proportion between 147 and 200bp can span a nucleosome, consistent with the observed pattern, therefore the 100-200bp fragments are likely to have cuts in adjacent linker regions flanking a nucleosome (Fig. 2c). The long 200-300bp fragments also showed cut sites in adjacent linkers where the linkers might span slightly broader nucleosome depleted regions (Fig. 2d). This suggests that inter-nucleosomal linker length influences the fragment size distribution at a locus. To test this, we selected pairs of positioned nucleosomes, identified by H3K4me2 MNase-ChIP-seq 14, separated from each other by an inter-nucleosomal linker length of 20-50bp, too short to accommodate entire 50-100bp DNase digestion fragments (Fig. 2e). Consistently, we noted a depletion of cuts from this fragment range in these linker regions (Fig. 2f). Longer fragments can span the nucleosomes and accordingly we saw cuts from the 100-200bp and 200-300bp fragment ranges inside these short linker regions (Fig. 2g,h). Extending the linker lengths to 100-130bp both ends of 50-100bp fragments could, in principle, be accommodated in the linker (Fig. 2i). Indeed, we saw that the cut sites from the short fragments were enriched in these longer linker regions (Fig. 2j) along with the ends of the longer fragments (Fig. 2k, l). DNase-seq tag density is therefore not simply a measure of DNA accessibility but is a function of the relative probability of cleaving both DNA strands at two loci separated by a narrow genomic interval. Genomic loci that are compatible with nucleosome spanning fragments are more common than long linker regions needed to accommodate 50-100bp fragments. The scarcity of these longer linker regions and their overlap with transcription factor binding sites accounts for the efficiency of the 50-100bp fragments.
Figure 2. Nucleosome positioning effects on DNase-seq results.
(a) Schematic figure shows fragments less than 147bp in length, cannot span a nucleosome. (b, c, d) Distribution of DNaseseq tags relative to the center of nucleosomes identified by MNase digestion and H3K4me2 immunoprecipation for (b) 50-100bp, (c) 100-200bp, and (d) 200-300bp fragments in LNCaP. Tags from both the plus and minus strands for the 50-100bp fragments fall in the regions that flank the nucleosome. Plus strand and minus strand mapped ends of (c) 100-200bp and (d) 200-300bp fragments accumulate on opposite sites of the nucleosome. (e) Illustration of the 50-100bp fragments being too long to be contained entirely in the short linker (20-50bp) and too short to span the nucleosomes. (f-h) Distribution of DNase-seq tags from (f) 50-100bp, (g) 100-200bp and, (h) 200-300bp fragments relative to pairs of nucleosomes selected to have short, 20-50bp, inter-nucleosomal linker distances. The 50-100bp fragments (f) show no peak in the linker but rather show peaks on either side of the paired nucleosomes. The longer (g) 100-200bp and (h) 100-300bp fragments show peaks that are consistent with tags spanning each nucleosome in the nucleosome pair. (i-l) Nucleosome pairs with longer, 100-130bp, linkers can accommodate the (j) Short fragments entirely. (k) A minor proportion of the 100-200bp fragments can be accommodated in the linker while the majority span the nucleosome. (l) The 200-300bp fragments cannot be accommodated in the linker but they can still span the nucleosomes.
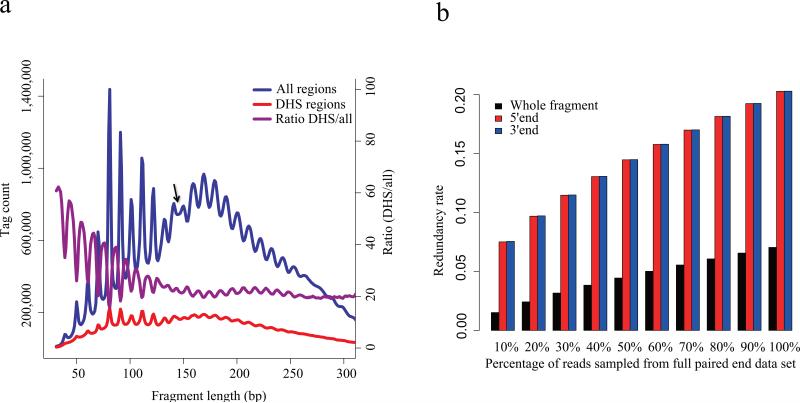
To examine fragment size effects at a higher resolution we conducted paired-end sequencing of DNaseI digested chromatin (at 50U). The fragment size distribution was dominated by a periodicity of approximately 10.4 base pairs (Fig. 3a), consistent with one complete turn of the double helix, and may be attributed to the accessibility of the minor groove of nucleosome associated DNA15. Interestingly we observed the periodic pattern in fragments longer than the nucleosome size, 147bp, and a phase shift between the sub-nucleosomal and super-nucleosomal patterns (Fig. 3a). This pattern may represent a dominant signal coming from heterochromatic regions with a relatively fixed linker length16. Fragments that overlap with DNase-seq peaks showed a weaker periodic pattern (Fig. 3a), suggesting that the periodic pattern arises primarily from fragments distributed widely across the genome and not at clustered hypersensitive regions. As a function of fragment length the proportion of reads that fall into the DHS regions is strongly biased towards the shorter ones (Fig. 3a). Paired-end or full length sequencing of DNase-seq fragments allows the fragment size distribution to be precisely characterized and allows amplification artifacts to be filtered while retaining multiple hits at the same nucleotide (Fig. 3b).
Figure 3. Pair-end sequencing of DHS.
(a) Fragment size distribution of DNase-seq data produced through paired end sequencing. The overall distribution (blue) exhibits an approximately 10.4bp periodicity that is consistent with one complete turn of the double helix. This phenomenon is likely to arise from nucleosomal DNA where DNase cleavage is possible only at exposed sites on the nucleosome. The arrow marks the point at which there is a shift in this periodic pattern. This periodicity is weaker in the distribution of fragment lengths in DHS regions (red). The ratio of fragments in the DHS regions relative to the entire fragment populations (purple) shows that the short fragments are enriched in the DHS regions. The periodicity in this ratio reflects a depletion of nucleosome associated fragments in the DHS regions. (b) Redundancy rate calculated from sampling pair-end DNase-seq data. Whole fragments as determined by the pair-end sequencing of both ends of DNA fragments are far less redundant than the 5’ and 3’ ends taken in isolation from each other.
Sequence bias confounds DNase-seq footprint analysis
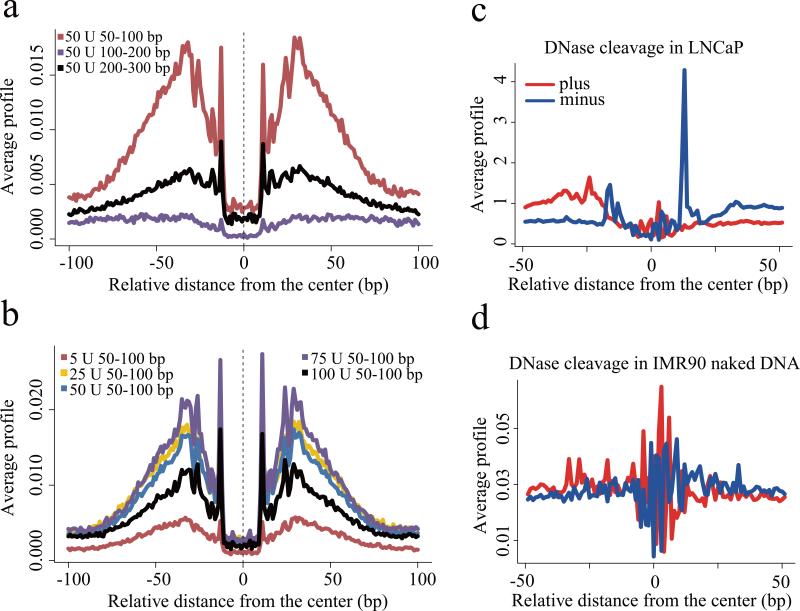
The binding of a TF to DNA can modify the pattern of DNaseI sensitivity at and around the site of binding, typically producing a “footprint” pattern of low sensitivity in the region of contact and high sensitivity at positions immediately flanking the binding site. This pattern of sensitivity has been exploited to discover DNaseI footprints in DNase-seq data to reveal the precise binding sites associated with a broad array of DNA sequence motifs 5. To assess the influence of digestion conditions on footprinting we investigated the pattern of cuts around the CTCF sequence motif in CTCF loci identified by ChIP-seq. Consistent with previous results we found the strongest footprint signal in the short fragment at 50U conditions (Fig. 4a and 4b, Supplementary Fig. 8). Interestingly, orienting DNase-seq reads relative to the CTCF motif revealed a strong directionality in the cut pattern (Fig. 4c). Contrasting this pattern with the pattern of DNaseI cleavage across CTCF motifs in DNase-seq of naked DNA derived from the IMR90 cell line17 we saw a clearly different pattern of cut sites (Fig. 4d), indicating that the footprints we observed for CTCF were related to CTCF binding and were not an artifact of the intrinsic DNaseI cleavage bias at the CTCF motif.
Figure 4. CTCF footprint.
(a) Nucleotide resolution DNase cleavage frequencies across CTCF recognition sequences at CTCF ChIP-seq peaks in LNCaP. DNase-seq signals were normalized to 1M reads in a non-strand specific manner. Short 50-100bp fragments produce clearer cleavage signals than 100-200bp or 200-300bp fragments. (b) DNaseI enzyme strength is most effective for detecting CTCF cleavage patterns in the 25U-75U range. (c) The positional distribution of oriented tags relative to the CTCF motif at CTCF ChIP-seq peaks in LNCaP reveals a strong directionality in the DNaseI cleavage pattern. Heatmaps show cleavage patterns at each locus for plus (red) and minus (blue) strands independently. The heatmap rows are ranked by the total DNase-seq tag count in each 100bp region. (d) The pattern of cleavage across the CTCF recognition sequence in naked DNA derived from the IMR90 cell line is very different from that observed in LNCaP chromatin at CTCF binding sites.
Encouraged by this result, we applied the same footprint characterization procedure to the AR motif in AR ChIP-seq peaks. The pattern we found (Fig. 5a), unlike the trough-like pattern observed for CTCF, was nevertheless consistent with the gapped pattern of AR interaction with specific nucleotides and exhibited a pattern of evolutionary conservation (Supplementary Fig. 8 and 9). In addition, the observed pattern was the same as that reported independently for AR using a different DNaseI protocol12. Curiously, when we focused on sites of high DNaseI sensitivity with an AR DNA sequence motif but no ChIP-seq evidence of AR binding we found a remarkably similar pattern (Supplementary Fig. 10). Constructing DHS footprint profiles associated with the AR DNA motif we found the same pattern in MCF-7 and in independent DNase-seq data from several other cell lines (Supplementary Fig. 10). Examining the cleavage pattern from IMR90 naked DNA that was not bound by AR or any other protein (Fig. 5b) it was clear that the DNaseI cleavage pattern across the AR motif closely resembled the AR “footprint” observed in LNCaP chromatin.
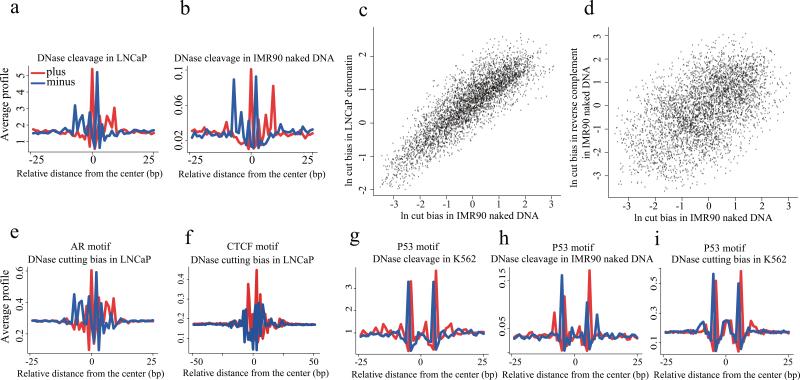
Figure 5. DNaseI cleavage bias as revealed by AR and P53 binding.
(a) The pattern of DNase cleavage across AR ChIP-seq enriched AR recognition sequences in the LNCaP cell line. (b) The DNaseI cleavage pattern produced from IMR90 naked DNA using the same AR sites as in (a). (c) The cleavage ratio represents, for each possible DNA hexamer, the number of observed cleavage sites between the 3rd and 4th bases of that hexamer relative to the number of such hexamers in the mappable genome. Cleavage ratios in IMR90 naked DNA are highly correlated with the ratios in LNCaP chromatin, showing consistency in bias across samples. (d) The log of the cleavage ratios for hexamers in DNaseI digested naked DNA and their reverse complements are plotted, showing a broad range of ratios. (e) The DNaseI cleavage pattern predicted from DNA sequence at the AR sites in (a), using the hexamer model of intrinsic DNaseI cleavage bias. (f) The pattern of cleavage predicted from a hexamer model of DNaseI cutting bias at CTCF binding sites in LNCaP. This pattern is similar to that seen in IMR90 naked DNA but different from the DNaseI cleavage pattern in chromatin at CTCF binding sites. (g) The observed DNaseI cleavage pattern in K562 chromatin at imputed p53 binding sites. (h) The DNaseI cleavage pattern produced from IMR90 naked DNA using the same p53 sites as in (g). (i) The DNaseI cleavage pattern predicted from DNA sequence using the hexamer model of intrinsic DNaseI cleavage bias at the p53 sites used in (g). Heatmaps in (a,b,e-i) show cleavage patterns at each locus for plus (red) and minus (blue) strands independently. The heatmap rows are ranked by the total DNase-seq tag count in each 50bp region.
To understand the intrinsic sequence bias of DNaseI cleavage we analyzed the ratio of observed to potential cleavage sites for 2-,4-,6- and 8-mers. As the 6-mer bias model captured more variation than the 2-mer and 4-mer models, and the 8-mer did not improve substantially on the 6-mer (Supplementary Figs. 11 and 12), we selected the 6-mer for subsequent analyses (Supplementary Table 1). There is a strong correlation between the 6-mer cut biases in IMR90 naked DNA and LNCaP open chromatin (Fig. 5c), as well as other DNase-seq datasets (Supplementary Fig. 13). This correlation between different samples is higher than the correlation between forward strand cuts on the 6-mers, and forward strand cuts on their reverse complements in the same naked DNA sample (Fig. 5d), suggesting the strand oriented nature of the bias, due to the single stranded nicking action of DNaseI 18. This phenomenon is not limited to DNase-seq but is also evident in Benzonase-seq and Cyanase-seq, although the precise nature of the sequence bias varies between DNaseI and the other nucleases (Supplementary Fig. 14) 19. We applied the predicted DNase cleavage bias to AR motifs in regions that were determined to be AR bound by ChIP-seq in LNCaP cells, obtaining cut profiles (Fig. 5e) that closely resemble the pattern for AR in chromatin (Fig. 5a) and the pattern observed at the same sites in naked DNA (Fig. 5b). Similar analysis on the CTCF motif shows that the predicted cleavage bias (Fig. 5f) is distinctly different from the pattern for CTCF in chromatin (Fig. 4c) but resembles the pattern observed in naked DNA (Fig. 4d). While the contribution of sequence bias to AR cleavage patterns was substantial, the contribution of sequence bias to CTCF footprints was minor and modeling this bias would have minimal impact on CTCF footprint discovery (Supplementary Fig. 15).
We investigated the DNase-seq footprint of p53, finding a DNase-seq cut pattern (Fig. 5g) that closely resembled the one reported 5. This pattern was also very similar to cleavage patterns derived from both naked DNA (Fig. 5h) and the 6-mer model (Fig. 5i). This suggests that the pattern observed for p53, like AR, is more likely to be a product of intrinsic sequence bias rather than protein binding effects.
Footprinting quality is factor dependent
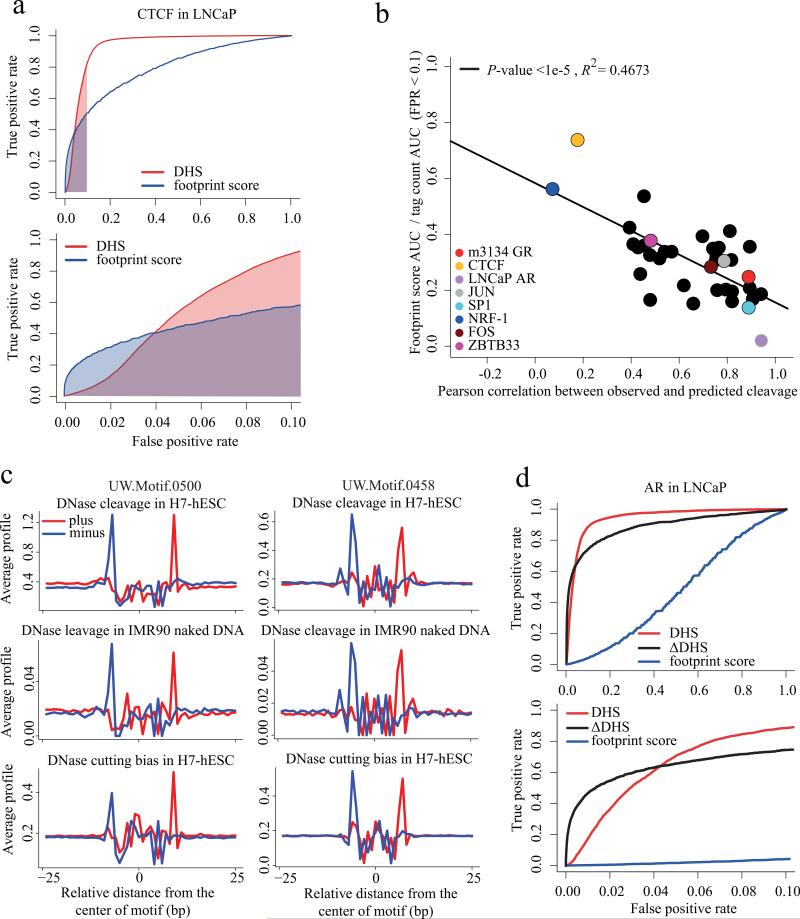
We next asked whether most transcription factors are like CTCF, in displaying a strong DNaseseq footprint, or like AR and p53, virtually indistinguishable from the naked DNA background. We systematically assessed the DNase-seq footprints for 34 transcription factors with ChIP-seq data and well-defined binding motifs 20 in the K562 cell line 21, along with AR in LNCaP and the glucocorticoid receptor (GR) in mouse mammary 3134 cells. For each of the 36 transcription factors we used two methods to predict whether a factor's sequence motif occurrence in the genome would be enriched in the ChIP-seq data. The first prediction was based simply on the number of DNase-seq tags (DHS) falling in a 200bp window centered on the TF recognition sequence. The second was based on the DNaseI footprint score based on the ratio of reads in the regions flanking the TF motif over the motif center regions (online methods). For each of the 36 factors we plotted a receiver-operating characteristic (ROC) curve as illustrated for CTCF (Fig. 6a). In this figure we see the footprint score outperforms tag count at low false positive rates (FPR) and underperforms at higher FPR. To summarize the performance of the footprint score relative to the tag count at low FPR, where it performs best, we calculated the ratio of the area under the curve (AUC) for the footprint score to the AUC for the DNase-seq tag count for FPR ranging from 0 to 0.1.
Figure 6. Predicting transcription factor binding from DHS.
(a) Receiver-operator curve comparing the performance of the DNase-seq footprint with the absolute DNase-seq tag count (DHS, red). From amongst all CTCF recognition sequences genome wide we predicted the ones that are CTCF ChIP-seq enriched using the DNase-seq footprint score (blue) and the number of DNase-seq tags in a 200bp window centered in the CTCF site (red). Only at low false positive rates (FPR) does the footprint score perform better than the tag count. The footprint score area under the curve (AUC) for FPRs less than 0.1 is shaded blue. Similarly the red shaded region is the AUC for the absolute tag count for FPR < 0.1. (b) For 36 transcription factors with known DNA binding motifs and ChIP-seq we constructed ROC curves like (a). The y-axis represents the footprint score relative to tag count performance as the ratio of the footprint score AUC to the tag count AUC for FPRs < 0.1. For CTCF this is the ratio of blue to red shaded areas in (a). The x-axis represents the Pearson correlation between the observed DNase cleavage pattern and that predicted from the hexamer intrinsic bias model. This shows how the footprint score performance deteriorates as the correlation between observed and predicted cleavage patterns increases. (c)
Comparison of observed, predicted and naked DNA cleavage bias in de novo motifs UW.Motif.0500 and UW.Motif.0458. (d) Receiver-operator curve for AR in LNCaP, comparing the performance of the DNase-seq footprint (blue) with the absolute tag count (DHS, red) and the ΔDHS score (black). While the footprint score is uninformative, the ΔDHS score, which compares DNase-seq between hormone stimulated and unstimulated conditions, performs better than the tag count at low FPRs.
We next examined the ratio of footprint score AUC to DNase-seq tag count AUC at low FPR for the 36 transcription factors versus the correlation between the observed DNaseI cleavage pattern and the 6-mer predicted background cleavage pattern (Fig. 6b, Supplementary Table 2). We found that the strength of the footprint (p < 10-4) (Supplementary Fig. 16a) as well as footprint performance (p < 10-5) (Fig. 6b) were inversely related to the correlation between the observed DNaseI cleavage and intrinsic bias. With the exception of CTCF at FPR < 0.04, the footprint score performed worse than the DNase-seq tag count at all points on the ROC curve for all of the factors analyzed.
To test whether there is residual footprint signal after adjusting for DNaseI cutting bias, we normalized the cleavage signal, taking the 6-mer bias into account, and compared this result with a similar uniform normalization that assumes all 6-mers are cut with equal likelihood (Supplementary Fig. 17). Modeling 6-mer bias dampened most of the cleavage signal across AR and SP1 binding sites (motif and ChIP-seq peak) (Supplementary Figures 17a and 17b). The different normalizations across CTCF sites had little effect on the cleavage pattern (Supplementary Fig. 17c). For JUN (Supplementary Fig. 17d) and ZBTB33 (Supplementary Fig. 17e) the normalization reduced the bias induced signal, revealing a trough-like footprint. A sequence bias normalization of the footprint score however did not improve the TF binding site prediction performance (Supp. Fig. 16c). While improved analysis techniques may improve the performance for factors that have footprints different from the underlying nuclease cleavage bias, our analysis found that there are a substantial number of transcription factors like AR, that leave negligible footprints in DNase-seq data obtained by current methods (Fig. 6d). Not surprisingly, although we did not observe DNaseI footprints at AR motifs in AR ChIP-seq peaks, we could find many instances of DNaseI footprints at CTCF and NRF1 motifs within AR ChIP-seq peak regions (Supplementary Fig. 18).
Recently DNase-seq has been used to identify 289 novel regulatory factor DNA binding motifs that are not represented in the major motif databases TRANSFAC, JASPAR and UniPROBE 5. Many of these novel motifs were reported to display nearly identical DNaseI footprint patterns in human cell lines and in mouse liver 5. On this basis it was claimed that these novel motifs correspond to transcriptional regulatory factors that are evolutionarily conserved between the two species. The cleavage patterns in the embryonic stem (ES) cell line H7 for the de novo motifs, UW.Motif.0500 and UW.Motif.0458, which were reported to be ES cell type specific, were nearly identical to the 6-mer prediction, as well as the naked DNA pattern (Figure 6c). The same was seen in K562 and mouse liver (Supplementary Fig. 19). Whereas the known motifs with informative footprints, such as CTCF, tended to be weakly correlated with predicted bias, all 15 of the de novo motifs showed strong correlation coefficients in the H7 ES and K562 cell lines (Supplementary Fig. 20). The similarity of footprint patterns between human and mouse that was reported as a sign of conserved transcription factor activity 5 may be the result of DNaseI cleavage bias instead (Supplementary Figs. 14g and 19). While this analysis shows that DNaseseq derived footprinting does not support the identity of these novel motifs, it does not rule out the possibility that novel cell type specific motifs may be present in DHS regions. In contrast, differential DHS (ΔDHS) 11 between conditions can predict differential transcription factor occupancy for factors such as AR (Figure 6d) and GR (Supplementary Fig. 21).
Discussion
In this study we highlight the role that nucleosome linker lengths play in shaping patterns of DNaseI cleavage and producing distinct fragment size distributions of digested chromatin. Fragments longer than 147bp in length are compatible with short inter-nucleosomal linker lengths, therefore can be produced over a broader span of the genome than shorter fragments. Transcription factor binding is often associated with nucleosome displacement and, consequently, long inter-nucleosomal linker lengths. Short 50-100bp fragments are therefore more highly enriched in TF binding sites than longer fragments, thus more efficient in identifying transcription factor binding sites. To optimize digestion conditions for effective identification of transcription factor binding sites, we provide quantitative guidelines to help experimentalists determine effective enzyme strength using constitutive CTCF binding sites and housekeeping gene promoters. Comparing DNase-seq read counts between treatment and control conditions appears to be the most informative strategy for identifying differentially used enhancers..
Footprinting analysis of DNaseI cutting patterns at nucleotide resolution has been proposed to identify the precise locations of TF-DNA interaction. While the DNaseI cutting pattern shows short regions of high DNaseI sensitivity flanking a region of low DNaseI sensitivity on CTCF motifs in CTCF binding sites, that associated with the AR motif in AR binding sites did not match this pattern but rather a pattern of interleaved high and low sensitivity. Comparison of DNaseI cleavage preferences in DNase-seq derived from chromatin and naked DNA show a strong correlation. This suggests that sequence bias, not protein-DNA interaction, is the likely cause of the interleaved DNaseI cleavage patterns for many factors including AR and p53. Previous work investigating the effect of intrinsic sequence bias on footprint detection found it to be minimal5. Sequence bias becomes abundantly clear in the analysis of cleavage patterns in aggregate, with DNA lined up by specific sequence motifs. Using this approach, we found a very high correlation between the novel motif “footprints” reported in ES cells5 and the DNaseI cleavage bias, suggesting that these footprints may be artifacts rather than evidence of novel motifs of transcription factor binding.
Our analysis of DNase-seq and ChIP-seq for 36 TFs shows that the efficiency of DNaseI footprints in recovering TF binding sites is associated with the degree to which the observed cleavage pattern differs from the intrinsic cleavage bias. The strong influence of DNA sequence on DNaseI cleavage efficiency has been associated with the width of the minor groove 17. The interactions between TFs and DNA in permitting or hindering DNaseI cleavage may account for the variety of footprinting effects. Alternatively TF-DNA interaction times in vivo might vary greatly between TFs8. Current DNase-seq methods do not allow these alternatives to be distinguished, which calls for new methods to probe transcription factor occupancy in vivo at high spatial and temporal resolution.
Online methods
Cell line and culture conditions
The prostate cancer cell line LNCaP and breast cancer cell line MCF7 were obtained from the American Type Culture Collection. LNCaP cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2mM glutamine, 100 U/mL penicillin and 100 mg/mL streptomycin. MCF-7 cells were maintained in DMEM medium supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/mL penicillin, 100 mg/mL streptomycin. LNCaP and MCF-7 cells were starved in phenol-red-free medium supplemented with 10% charcoal stripped FBS for 3 days before hormone stimulation.
DNaseI hypersensitivity mapping
DNaseI hypersensitivity mapping was performed as previously described with brief modifications 9-11. LNCaP cells were starved for 3 days in phenol-red-free medium supplemented with 10% charcoal stripped FBS and then treated with ethanol or active androgen 5α-dihydrotestosterone (DHT) at a final concentration of 10 nM for 4 hours. MCF-7 cells were starved the same way and then treated with ethanol or 17β-estrodial (E2) at a final concentration of 10 nM for 45 min. The cells were trypsinized and pelleted prior to washing and resuspension in buffer A (15 mM Tris-Cl (pH 8.0), 15 mM NaCl, 60 mM KCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0), 0.5 mM spermidine and 0.15 mM spermine). Nuclei were extracted by adding Buffer A containing NP-40. The nuclei were washed with buffer A and resuspended in prewarmed lysis buffer at a concentration of 5 M/mL and then digested with different amounts of DNase I for 5 min at 37 °C. The reactions were terminated by the addition of an equal volume of stop buffer and incubated at 55 °C. After 15 min, Proteinase K (final concentration of 20 μg/mL) was added to each digestion reaction and incubated for 2 hours at 55 °C. DNA was extracted by careful phenol-chloroform purification using phase lock gel. qPCR on the 3 CTCF sites and 3 housekeeping gene promoters were performed to determine the ideal digestion level. DNA fragments of 50-100, 100-200, 200-300 bp long or 50-300 bp (for pair-end sequencing) were selected using low melting agarose gel. The sequencing libraries were prepared following the Rubicon Genomics ThruPLEX-FD library preparation protocol. DNase-seq libraries were sequenced at the Center for Cancer Computational Biology (CCCB) at the Dana-Farber Cancer Institute.
DNaseI strength calibration
As the strength of the DNaseI enzyme may vary by manufacturer and batch, tends to decay over time, and may be inhibited in cellular extracts, we use a calibration approach to allow the enzyme strength optimized in this experiment to be reproduced in other systems. We assess the degree to which DNA is digested by DNaseI using loci selected to be stable across cell lines and conditions. CTCF ChIP-seq and DNase-seq experiments across multiple cell lines have shown the transcription factor CTCF to bind broadly across diverse cell types in DNaseI hypersensitive locations 21. We selected a set of 25 constitutive CTCF binding sites with low variability in DHS tags across 77 DNase-seq data sets in 43 cell types for enzyme calibration. After initial testing, primer pairs spanning 3 of these sites together with 3 house keeping gene promoters, provided in Supplementary Table 3, were used to measure the proportion of uncleaved loci within the cell population over a range of DNaseI strength. In three different cell lines tested (Supplementary Fig. 1c), a sharp change in the proportion occurs for all loci in the range between 5 U and 100 U, enabling accurate calibration of DNaseI enzyme activity in this range.
Performance evaluation of DNase conditions
DNase-seq reads were mapped to the human reference genome hg19 using the bowtie software. Mapped reads were randomly sampled without replacement from the set of all mapped reads. At each sampling level peaks were identified using MACS2 with options --keep-dup=1 with the default p-value cutoff of 10-5. ChIP-seq peaks were compared with DNase-seq peaks by trimming ChIP-seq peaks to 600 bp around the peak summit. Peaks were considered to be overlapping if they had at least 1bp overlap. We selected the most significant 10,000 ChIP-seq peaks from each ChIP-seq data set.
Differential DNase-seq
We used the methodology that we have described previously11 to compute a ΔDHS score representing the change in DNase sensitivity for each DHS site, ΔDHS = (nT/mean(nT))1/2 – (nC/mean(nC))1/2, where nT and nC represent the tag counts within 100bp from the motif center in treatment and control conditions respectively. ΔDHS was calculated using 50U DNaseI data in LNCaP DHT and vehicle conditions. Androgen receptor, almost completely absent in the control condition, was induced to bind in the treatment condition. To test if changes in DHS are associated with AR binding we ranked loci by ΔDHS score and grouped these ranked loci into bins of 500. We then assessed the proportion of sites in each bin that overlapped with a ChIP-seq identified AR site.
Step-to-step DNase-seq protocol
1. Nuclei isolation
- Dissociate tissue or cultured cells to single cells (~10M cells) with trypsin.
- Centrifuge at 900 rpm for 3mins, remove supernatant.
- Wash once with buffer A+ (on ice), spin down at 900rpm for 3 minute.
- Remove supernatant, and resuspend pellet in 6 ml buffer A+.
- Add 2 ml 0.2% NP40 (in buffer A), dropwise, to reach the final concentration of 0.05%, invert gently to mix, digest on ice for 5-10 minutes. (You can count nuclei at the end of this step, and centrifuge in the meantime.)
- Centrifuge at 2500 rpm for 3 minutes at 40C, remove supernatant, transfer pellet to a 2ml microcentrifuge tube, wash once with 1ml buffer A+.
2. DNaseI digestion
- Resuspend into 5M/ml in buffer A and aliquot 500ul to 1.5ml tubes, spin down and resuspend in 500ul pre-warmed 370C 1x digestion buffer for 4x1.5ml tubes, snap free the remaining (if any).
- Add 0U, 25U, 50U and 75U of DNaseI (Roche) respectively. Invert to mix. Incubate at 37 0C for 5mins.
- Add 500ul of stop buffer (with spermine, spermidine, and 2ul of RNase (Roche)) to each reaction, and mix by inverting. Incubate at 55 0C for 15 mins.
- Add 2ul 20mg/ml PK, digest at 55 0C for at least 2h (no more than 16h).
- Extract DNA using 1X VOL phenol/chloroform, shake vigorously, centrifuge at top speed for 15 min. (Recommend using phase lock gel to reduce protein contamination)
- Take top aqueous layer and add 2-3X VOL 100% EtOH, add 2ul glycogen.
- Precipitate at 800C for at least 30 min or overnight. Centrifuge at top speed for 15 mins. Carefully remove EtOH, then add 1ml 70% EtOH to wash. Invert gently, centrifuge for 3-5 min at top speed. Remove EtOH.
- Air dry the pellet, and resuspend in 50ul TE or nuclease-free H2O.
- Remove residual RNA by adding 1ul RNase (Roche) and incubate at 37 0C for 30mins, go to step 11 or bring sample vol to 400 ul then purify DNA using 1X VOL phenol/chloroform (add sodium acetate for precipitation).
- Measure DNA concentration using Nano-drop, digest 2.5ug of undigested control DNA (0U) with 0.05-0.2U DNaseI (Roche) at 37 0C for 5mins (25ul reactions system), stop reaction by adding 5ul 25mM EDTA, incubate at 65 0C for 10mins, mix well. Chose the one with size range between 50 and 300bp. The digested DNA will be used as Input for DNase-seq (optional)
25ul rxn sys: 2.5 ul 10X buffer, 2.5 ug DNA, 0.05-0.2U DNaseI (use 1ul after dilution), add ddH2O to 25 ul total, incubate for 5mins, add 2.5ul EDTA to stop rxn
3. Post DNaseI digestion
- Perform qPCR using undigested control DNA (0U) and DNaseI digested DNA as templates. Digestion level is measured by comparing the qPCR signal of digested DNA with undigested control DNA. We have selected three constitutive CTCF sites and three housekeeping gene promoters to determine the digestion level (see online methods section “DNase strength calibration”).
- Appropriate digestion level is 0.6-0.8 for the three CTCF sites and 0.05-0.2 for the three housekeeping genes (see primers at the end of this protocol). Digestion level lower than 0.6 and 0.05 at CTCF sites and housekeeping genes respectively is considered as over digestion. The digestion level higher than 0.8 and 0.2 at CTCF sites and housekeeping genes respectively is considered as under digestion (see Supplementary Fig.1c).
- Run DNA on a 2% agarose gel (low melting temperature). See Supplementary Figure 1c for examples of smears for under, appropriately and over digested samples.
- Cut the gel to select desired fragments. Purify size selected DNA using Qiagen Gel Purification Kit or QIAEX II Gel Extraction kit. We suggest selecting fragments of 50- 100 bp to most efficiently identify transcription factor binding sties.
4. Sequencing library preparation
- Measure DNA concentration using Qubit (Invitrogen). For 2.5M cells, the total amount of DNA varies from 0.1ng to 30ng depends on the digestion level and size you select.
- Samples with optimal digestion levels can be pooled.
- Prepare sequencing library following Rubicon Genomics ThruPLEX-FD (R40012) library preparation protocol. This protocol allows the use of as low as 50pg DNA to construct library, but increasing the amount of starting material can increase the library complexity and decrease redundancy rate.
5. Reagents used in this protocol
| Buffer A (store at 4°C): | ||
|---|---|---|
| Final Concentration | Stock concentration | Amount used from stock |
| 15 mM Tris-Cl, pH 8.0 | 1 M Tris-Cl, ph 8.0 | 15 ml |
| 15 mM NaCl | 5 M NaCl | 3 ml |
| 60 mM KCl | 1 M KCl | 60 ml |
| 1 mM EDTA, pH 8.0 | 0.5 M EDTA, pH 8.0 | 2 ml |
| 0.5 mM EGTA, pH 8.0 | 500 mM EGTA, pH 8.0 1 ml |
| Spermine | 0.1M | 0.15mM | 15ul |
|---|---|---|---|
| Spermidine | 0.1M | 0.5mM | 50ul |
| PIC | 50 × | 1 × | 200ul |
| PMSF | 1M | 1mM | 10ul |
| DTT | 1M | 0.5mM | 5ul |
| 10X DnaseI Digestion Buffer (Roche) | ||
|---|---|---|
| For 10ml (pH7.9) | ||
| Final concentration | Stock concentration | Amount used from stock |
| 60 mM MgCl2 | 1 M MgCl2 | 600 ul |
| 100 mM NaCl | 5 M NaCl | 200ul |
| 10 mM CaCl2 | 1 M CaCl2 | 100 ul |
| 400mM Tris-HCl | 1 M Tris-HCl | 4ml |
| Stop Buffer (per Liter) | ||
|---|---|---|
| Final concentration | Stock concentration | Amount used from stock |
| 50 mM Tris-Cl, pH 8.0 | 1 M Tris-Cl, pH 8.0 | 50 ml |
| 100 mM NaCl | 5 M NaCl | 20 ml |
| 0.10 % SDS | 20% SDS | 5 ml |
| 100 mM EDTA, pH 8.0 | 0.5 M EDTA, pH 8.0 | 200 ml |
| Make fresh, for 10ml stop buffer, add | |||
|---|---|---|---|
| Spermine | 0.1M | 0.3mM | 30ul |
| Spernidine | 0.1M | 0.1mM | 100ul |
6. Primers to determine digestion level
| Forward | Reverse | |
|---|---|---|
| CTCF4 | CCCCAGAGAGTAGGGAACAG | GGCACGCAAAGACATACTGA |
| CTCF10 | AGAGCACCCCCTACTGGCTAA | TAAGAAGCTGTGCGCGATGAC |
| CTCF15 | CTTAGGGGACCTTTTCTACAGGA | GAGCACTTGTAAACTCGTCTGCT |
| GAPDH_pro | AAAAGCGGGGAGAAAGTAGG | GCTGCGGGCTCAATTTATAG |
| B-ACT_pro | TCGAGCCATAAAAGGCAACT | TCTCCCTCCTCCTCTTCCTC |
| RPS28_pro | CGGCAGCTGACACGTAAGTC | CAATGCAGAGCGACACTCAC |
Analysis of nucleosome position effects on DNase-seq reads
Positions of nucleosomes marked by H3K4me2 were computed in the same way as previously described14 (GSE33216) using the NPS software 22. Profiles of tag densities relative to the centers of these nucleosomes were derived based on the 5’ end of the DNase-seq tags.
Estimation of intrinsic DNA induced nuclease cut bias
Intrinsic cut bias in chromatin and naked DNA was estimated from ratios of observed to background cleavage sites. For each n-mer we counted the number of DNase-seq tags mapped to the reference genome in such a way that the tag mapped to the + strand with 5’ nucleotide aligned with the (n/2+1)th base of the n-mer spanning positions [_i-n/2, i+n/2-1_]. This count was compared with the number of all occurrences of that n-mer in the background set of genomic intervals. In the case of chromatin derived DNase-seq this background included 400bp from each MACS determined peak region. Background in naked DNA samples included all regions with a mappability index greater than 0.95. We use ai+ to denote the intrinsic sequence bias at genomic position i based on the n-mer ratio associated with the n-mer spanning positions [_i-n/2, i+n/2-1_]. ai− is calculated in a consistent way based on nucleotides spanning genomic positions [_i-n/2-1, i+n/2_]. In scatter plots representing these bias ratios the bias ratios are scaled by a constant so that in each case the bias ratio is 1.0
DNaseI footprint analysis contrast against intrinsic bias
To generate aggregate plots and heatmaps we identified the motif matches that coincided with regions with an mappability index greater than 0.95 (genome.ucsc.edu table wgEncodeCrgMapabilityAlign36mer), DNase-seq and ChIP-seq peaks (determined by MACS) in the same cell line. The positions of the 5’ end of sequence tags were recorded separately for tags mapping to the plus and minus strands. We calculated the aggregate plot using the trimmed mean of tag counts at every position, filtering out the highest and lowest 1% of tag counts. Any correlation coefficient of aggregate value is based on these trimmed mean summaries. In this analysis of DNase-seq peaks in LNCaP, the peaks were determined using MACS combining all DNase-seq data from 5U to 75U, including all fragment lengths.
Inferrence of p53 binding sites in K562
p53 binding sites in K562 where inferred as the intersection as regions matching the p53 DNA sequence motif, K562 DNase-seq ChIP-seq peaks and p53 ChIP-seq peaks in Saos-2.
Comparison of observed cleavage and sequence bias prediction
We use the Pearson correlation coefficient to compare observed cleavage with simulated sequence bias within the 50bp region centered on every motif center. In the sequence bias prediction, the strand (s ∈ {+, −} oriented 5’ end DNase-seq tag count is predicted at genomic position (i) by distributing the total number (Nis=∑k=i−25i+24nks) of observed strand s 5’ tag ends within a 50bp window centered on nucleotide i in proportion to their sequence bias contribution yis=ais∕∑k=i−25i+24aks.ai+, for example, is the intrinsic sequence estimated mer for nucleotides spanning positions [_i-3,i+2_]. The predicted count is n^is=Nisyis
Analysis of footprint performance
ROC curves were generated using the absolute DNase-seq tag count (DHS), the footprint score and differential tag count (ΔDHS) on every motif site to predict the binding of transcription factor represented by MACS peak calling. The absolute tag count refers to the number of tags located within 100bp of the motif match center. The footprint score was calculated using the formula f =-((nC+1)/(nR+1)+(nC+1)/(nL+1)), where nC, nR, nL represent respectively the tag count in the motif region, and the flanking regions to the right and left of the motif. The lengths of the flanks are both the same as that of the motif. We calculated the Pearson correlation coefficient of the DNaseI cleavage to cutting bias based on 25bp upstream and downstream from the motif center. The performance of the footprint score relative to the tag count is represented by the ratio of the areas of the footprint score ROC curve to the tag count ROC curve for the false positive rate range of [0,0.1]. Ordinary least squares regression was used to show the correlation between the similarity and the prediction power.
Uniform and sequence bias normalizations
In the uniform normalization we calculate the log ratio of the observed 5’ tag counts relative to uniformly distributed tag counts. Specifically, at each position (i) the observed strand (s) specific tag count nis is compared with the average per base strand specific tag count in the 50bp region centered at that position,n‒is=∑k=i−25i+24nks∕50. The uniform normalized DNaseI sensitivity is uis=log(nis+1)−log(n‒is+1). In the sequence bias normalization DNaseI cleavage is normalized by the predicted count, n^is=Nisyis. The sequence bias normalization is zis=log(nis+1)−log(n^is+1). The similarity of these two normalization approaches, when applied to the same data, allows for a comparison on the same scale.
Supplementary Material
1
Acknowledgements
This work was supported by grants from the National Institute of Health (1R01 GM099409 to X.S.L.; 1U41 HG007000 to X.S.L. and C.A.M; 2P50 CA090381-06 to C.A.M. and M.B.; 2R01 DK074967-06 to M.B. and X.S.L, 1K99CA172948-01 to H.H.H); the Mazzone Award (to X.S.L.), the Department of Defense (W81XWH-10-1-0557 to H.H.H) and the Prostate Cancer Foundation (to M.B.).
Footnotes
Author contributions. H.H.H., C.A.M., H.W.L. X.S.L. and M.B. designed the experiments and wrote the manuscript. M.W.C, H.H.H. performed the experiments with the help from Y.L., P.R. and T.F. C.A.M., S. H and H.H.H. conducted the data analysis with the help from C.Z. and H.X.
Accession codes NCBI Short Read Archive: LNCaP and MCF-7 DNase-seq raw fastq and processed bed files have been deposited under accession code GSEXXXX. ChIP-seq data in K562 and LNCaP CTCF from ENCODE: GSE32465, GSE31477, GSE30263 and GSM822289; AR in LNCaP ChIP-seq: GSM353644; TP53 ChIP-seq from Saos-2 cell: GSE15780; DNase in K562 from ENCODE: GSM646567; DNase in IMR90 naked DNA: SRA068503; mouse DNase/Benzonase/Cyanase: GSE39982. ER and CTCF in MCF7 are from http://cistrome.dfci.harvard.edu/NR_Cistrome.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song L, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle AP, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–464. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voss TC, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ling G, Sugathan A, Mazor T, Fraenkel E, Waxman DJ. Unbiased, genome-wide in vivo mapping of transcriptional regulatory elements reveals sex differences in chromatin structure associated with sex-specific liver gene expression. Mol Cell Biol. 2010;30:5531–5544. doi: 10.1128/MCB.00601-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He HH, et al. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res. 2012;22:1015–1025. doi: 10.1101/gr.133280.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tewari AK, et al. Chromatin accessibility reveals insights into androgen receptor activation and transcriptional specificity. Genome Biol. 2012;13:R88. doi: 10.1186/gb-2012-13-10-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaffney DJ, et al. Controls of nucleosome positioning in the human genome. PLoS Genet. 2012;8:e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarovici A, et al. Probing DNA shape and methylation state on a genomic scale with DNase I. Proc Natl Acad Sci U S A. 2013;110:6376–6381. doi: 10.1073/pnas.1216822110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell VW, Jackson DA. The effect of divalent cations on the mode of action of DNase I. The initial reaction products produced from covalently closed circular DNA. J Biol Chem. 1980;255:3726–3735. [PubMed] [Google Scholar]
- 19.Grontved L, et al. Rapid genome-scale mapping of chromatin accessibility in tissue. Epigenetics Chromatin. 2012;5:10. doi: 10.1186/1756-8935-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Shin H, Song JS, Lei Y, Liu XS. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics. 2008;9:537. doi: 10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1