Recurrent Hemizygous Deletions in Cancers May Optimize Proliferative Potential (original) (raw)
. Author manuscript; available in PMC: 2014 May 20.
Published in final edited form as: Science. 2012 May 24;337(6090):104–109. doi: 10.1126/science.1219580
Abstract
Tumors exhibit numerous recurrent hemizygous focal deletions that contain no known tumor suppressors and are poorly understood. To investigate whether these regions contribute to tumorigenesis, we searched genetically for genes with cancer-relevant properties within these hemizygous deletions. We identified STOP and GO genes, which negatively and positively regulate proliferation, respectively. STOP genes include many known tumor suppressors, whereas GO genes are enriched for essential genes. Analysis of their chromosomal distribution revealed that recurring deletions preferentially over represent STOP genes and under represent GO genes. We propose a hypothesis called the cancer gene island model whereby gene islands encompassing high densities of STOP genes and low densities of GO genes are hemizygously deleted to maximize proliferative fitness through cumulative haploinsufficiencies. Because hundreds to thousands of genes are hemizygously deleted per tumor, this mechanism may help drive tumorigenesis across many cancer types.
Cancer progression is directed by alterations in oncogenes and tumor suppressor genes (TSGs) that provide a competitive advantage to increase proliferation, survival, and metastasis (1-3). The cancer genome is riddled with amplifications, deletions, rearrangements, point mutations, loss of heterozygosity (LOH), and epigenetic changes that collectively result in tumorigenesis (4-7). How these changes contribute to the disease is a central question in cancer biology. In his “two-hit hypothesis” Knudson proposed that two mutations in the same gene are required for tumorigenesis, indicating a recessive disease (8). In addition, there are now several examples of haploinsufficient TSGs (9-11). Current models do not explain the recent observation that hemizygous recurrent deletions are found in most tumors (12, 13). Whether multiple genes within such regions contribute to the tumorigenic phenotype remains to be elucidated.
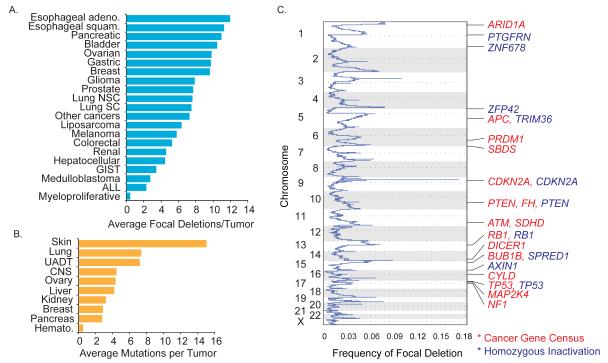
Recent analysis of 3131 tumors revealed 82 regions of recurrent focal deletion (13), averaging six deletions per tumor and 24 genes per deletion (Figure 1C, fig. S1A, table S1) (14). Breast, gastric, bladder, pancreatic and ovarian cancers average ≥10 deletions/tumor (Fig. 1A). Several possible explanations exist for the roles of these deletions in tumorigenesis. First, they may contain a recessive TSG where mutation or epigenetic silencing of the second allele is necessary for tumorigenesis. Second, they may recur because they mark unstable genomic regions, such as fragile sites (12). Finally, it is possible that single-copy loss may provide a selective advantage irrespective of changes in the remaining allele.
Fig. 1. Most recurrent cancer deletions do not contain known or putative recessive TSGs.
A) Average recurrent focal deletions per tumor for cancer subtypes (adeno: adenocarcinoma; squam: squamous; NSC: non-small cell; SC: small cell; GIST, gastrointestinal stromal tumor; ALL: acute lymphoblastic leukemia)
B) Loss-of-function mutations per tumor from COSMIC, averaged for various cancers (UADT: upper aero-digestive tract, CNS: central nervous system, Hemato: hematopoietic)
C) A subset of recurrent cancer deletions contains known or putative tumor suppressors. The frequency of focal deletion was plotted by chromosome location. Red gene names denote the presence of a known TSG from the Cancer Gene Census; blue gene names denote a homozygously inactivated gene from COSMIC whole-genome sequencing.
To address the possibility that recurrent deletions are enriched for recessive TSGs, we analyzed these regions for the presence of known recessive or putative TSGs. For this purpose we used a list from the Cancer Gene Census (15) and a list of putative TSGs we identified with homozygous loss-of-function (termination codon or frameshift) mutations from whole-genome sequencing of 526 tumors in COSMIC (Fig. 1B and tables S2 and S3) (16). Only 14 of 82 recurrent deletions contained a known TSG, and only 10 had a mutant or putative TSG, 6 of which were in a region with a known TSG (Figure 1C and fig. S1). Thus, only 18 of 82 deletions can be explained by known or putative recessive TSGs. This number may increase if gene silencing is as prevalent as point mutation for gene inactivation, but this remains to be determined across all cancers. These data suggest that in addition to the two-hit mechanism, an alternative mechanism may function to provide a selective advantage to these deletions.
Of the many altered processes promoting tumorigenesis, proliferation is likely to encompass the most genes as it is integrated into all developmental decisions. Cancer evolution relies on alterations that provide incremental increases in cell number- a function of cell duplication frequency coupled with cell survival efficiency. The average fitness increase of a single alteration in tumors is estimated to be 0.4% (17). Because subtle changes in proliferation rates can have profound effects on tumor fitness and clonal selection, we examined whether recurrent deletions affect regulators of cell proliferation. We define proliferation regulators as falling into two categories: suppressors of tumorigenesis and/or proliferation (STOP genes) that restrain proliferation, and growth enhancers and oncogenes (GO genes) that promote proliferation. By definition, STOP genes contain prominent TSGs that restrain proliferation (e.g. Cdk inhibitors, Rb, and p53), whereas GO genes include essential genes and some that simply enhance proliferation rates. The interplay between STOP and GO genes controls proliferation.
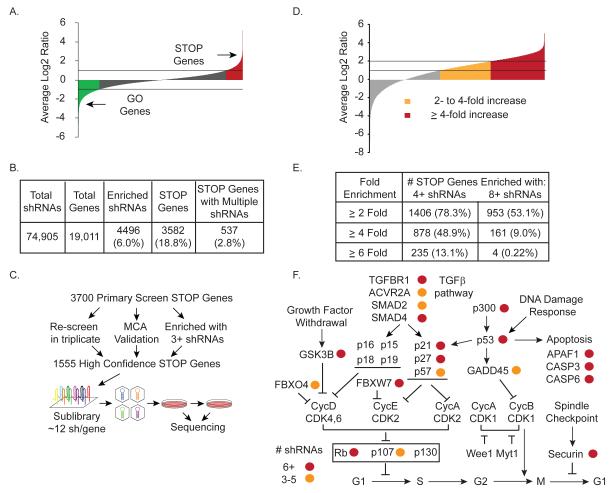
To identify candidate STOP genes, we performed a proliferation screen with a library containing 74,905 short hairpin RNAs (shRNAs) targeting 19,011 genes (18-20) in telomerase-immortalized human mammary epithelial cells (HMECs) (fig. S2A). We chose HMECs because they have intact TSG pathways and should hypothetically be a model for proliferation effects in early tumorigenesis where the neoplasms are less abnormal. By comparing the ratio of each shRNA’s abundance (end vs. initial sample) after eight population doublings, we identified enriched shRNAs (Fig. 2A, red). Screen data were analyzed as described (20) using significance analysis of microarray (SAM) to identify shRNAs consistently enriched by a factor of 1.8 or more across triplicates [false discovery rate (FDR) = 5%], representing ≥ 7.5% increase in cell number per generation. This identified 4496 (6.0%) enriched shRNAs targeting 3582 (18.8%) candidate STOP genes (Fig. 2B and table S4). Of the shRNAs tested, 51% recapitulated in a 5-day multi-color competition assay (MCA) (21) (figs. S2, B, and C).
Fig. 2. Identification of STOP gene candidates that restrain cell proliferation.
A) A genome-wide proliferation screen identifies STOP and GO genes. Average log2 ratios of end vs. initial samples for > 74,000 shRNAs across triplicates were plotted. Enriched shRNAs are denoted as STOP genes (red); lethal shRNAs are denoted as GO genes (green).
B) Numbers and percentages of STOP genes identified with single and multiple shRNAs, enriched shRNAs scoring in the primary screen, and total shRNAs and genes screened.
C) Generation and screening of a validation sublibrary containing multiple shRNAs per gene. A sublibrary targeting 1555 high-confidence STOP genes was designed containing 12+ additional shRNAs per gene, shRNAs that enriched by a factor of 2 in the primary screens, and control shRNAs. The sublibrary was synthesized using parallel microarray synthesis, cloned, and screened for the ability to increase cell proliferation using Illumina sequencing for pool deconvolution.
D) Average log2 ratios of end vs. initial samples across triplicates for 21,768 shRNAs from the validation screen were normalized for sequencing reads per sample and to the mean of 50 negative control shRNAs targeting FF. shRNAs that increased proliferation (relative to FF controls) by a factor of 2 to 4 are in orange; those that increased proliferation by a factor of 4 or more are in red.
E) Validation of STOP genes as assessed by multiple shRNAs. Numbers and percentages of STOP genes with multiple shRNAs are shown, according to increased proliferation by a factor of ≥ 2, ≥ 4, ≥ 6.
F) Known pathways with multiple shRNAs in the validation screen. Genes that validated in the secondary screen by a factor ≥ 4 with multiple shRNAs are denoted with circles corresponding to three to five (orange) and six or more shRNAs (red).
To validate more genes and eliminate off-target effects, we used a large-scale sublibrary validation (Fig. 2C). From 3700 candidate STOP genes from multiple screens (Fig. 2, A and B, fig. S3, A and B, and tables S4 and S5), we chose 1555 genes for validation studies by including only those genes that either: i) increased proliferation upon depletion in an independent triplicate re-screen, ii) validated by MCA, or iii) were enriched by a factor >2 with three or more independent shRNAs. We synthesized a sublibrary against this higher-confidence list with 12 shRNAs per gene and 50 negative control shRNAs targeting firefly luciferase (FF). We performed a secondary validation screen in triplicate and deconvolved samples by Illumina sequencing. Data were normalized for the number of sequencing reads per sample and the mean of 50 FF shRNAs (table S6) and analyzed using SAM with a FDR = 5%. Sixty percent of the shRNAs increased cell proliferation by a factor of 2 or more (Fig. 2D and table S7). Many STOP candidates validated with 4 or more shRNAs enriched by a factor of ≥ 2 (1406 genes), ≥ 4 (878 genes), or ≥ 6 (235 genes) (Fig. 2E). Furthermore, we observed a much larger fraction of shRNAs strongly enriching by a factor of ≥ 4 (30.2%) or ≥ 6 (13.3%) relative to our primary screen. Examination of shRNAs against the known proliferation regulators p53 and Rb revealed that 9 of 13 p53 shRNAs and 9 of 12 Rb shRNAs increased cell proliferation by a factor of 2 or more (fig. S4A). These data indicate that the validation screen can distinguish between authentic regulators of cell proliferation and false-positives.
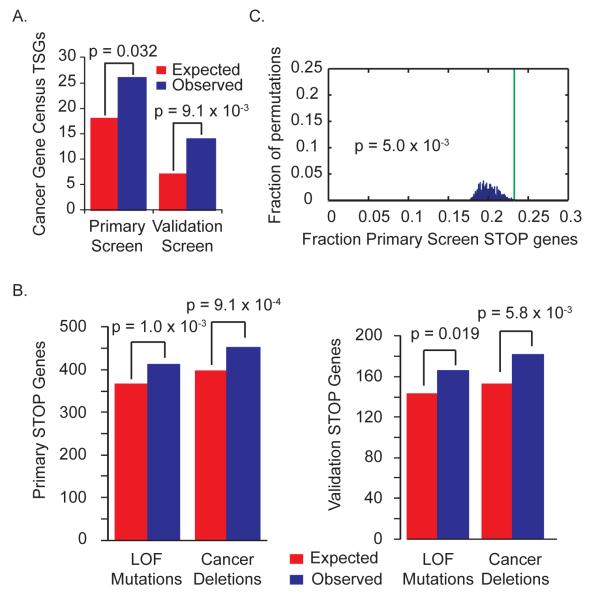
Using a stringent cutoff, analysis of the 878 STOP genes for which 4 or more shRNAs each resulted in a factor of ≥ 4 increase in cell proliferation revealed many genes involved in cell cycle regulation, apoptosis and autophagy (Fig. 2F and table S7) and numerous TSGs (fig. S4B). To establish statistical significance for TSG enrichment, we compared our primary and validation gene sets to the list of known TSGs defined by the Cancer Gene Census (Fig. 3A and table S2) (15). This comparison revealed significant enrichment with 44.4% more TSGs than expected in the primary STOP gene set (P = 0.032) and 100% more TSGs than expected in the validation screen (P = 9.1 × 10−3). We also compared the STOP candidates to the list of loss-of-function mutations in the 526 tumors in the COSMIC database (Fig. 3B and table S8) (16) and found significant enrichment of primary and validation screen STOP genes, with 12.9% more primary STOP genes (P = 1.0 × 10−3) and 16.1% more validation STOP genes (P = 0.019) exhibiting loss-of-function mutations in cancers. These data indicate that our STOP lists are likely to be enriched for novel TSGs and that genes with loss-of-function mutations found in tumors are enriched for negative regulators of proliferation, arguing that cell proliferation in this HMEC system is relevant to in vivo tumorigenesis.
Fig. 3. STOP genes are enriched for known TSGs and genes mutant or deleted in cancer.
A) STOP genes are significantly enriched for known TSGs. Candidates from the primary or validation screens were compared to Cancer Gene Census TSGs using Fisher’s exact test.
B) STOP gene candidates are significantly enriched for genes exhibiting loss-of-function (LOF) mutations and deletions in cancer. Genes containing loss-of-function mutations were determined using COSMIC whole-genome sequencing data. STOP genes from the primary or validation screens were mapped to genomic locations. Comparing STOP genes from the primary or validation screens to these lists of mutant or focally deleted genes revealed significant enrichment by Fisher’s exact test.
C) Multiple STOP genes cluster in cancer deletions. The primary STOP gene set was mapped to genomic location, and cancer deletion peak regions were overlaid. The percent of STOP genes found in cancer deletion peaks (green line) was compared to the distribution observed after 1000 permutations of the deletion peak regions across the genome (14).
To examine deletions, we mapped the chromosomal locations of STOP genes relative to recurrent deletions from 3131 tumors (13) (Fig. 3B and table S9). We observed a significant enrichment (P = 9.1 × 10−4) of genes located in recurring deletions in the primary STOP gene set, with 13.6% more genes than expected. The enrichment improved with the validation set to 19.1% more genes than expected (P = 5.8 × 10−3). This enrichment indicates that a significant proportion of the STOP genes identified are likely to functionally restrain tumorigenesis. Additionally, of the 451 observed overlapping genes, to our knowledge only 6 have been previously implicated as bona fide TSGs (SMAD4, RB1, ATM, APC, PTEN, and TP53), suggesting the existence of many previously undescribed TSGs on this list. The observation that hemizygous recurrent focal deletions contain more STOP genes than expected suggests that multiple genes in each region contribute to tumorigenesis, possibly through haploinsufficiency.
The recurrent deletions contain more STOP genes than predicted if only one gene per deletion were contributing to the phenotype. If these deletions preferentially select regions with the highest densities of STOP genes, then one might expect that the STOP gene distribution would be significantly different in the recurrent deletions than in other regions of the genome. Thus, we examined the density of STOP gene location within the 82 recurrent deletion peaks relative to the rest of the genome (13). To determine the likelihood of observing the same degree of STOP gene clustering as seen in actual recurrent deletions, we performed a Monte Carlo permutation analysis in which we compared the genes in the original 82 deletion peaks to those generated by random permutation of regions (containing the same number of genes) across a circularized genome. During permutation, the distance between the deletions was fixed to avoid deletion overlaps, and the original deletions were masked from the genome to prevent resampling. We performed 1000 permutations to determine how frequently the same or greater density of STOP genes was observed in randomized deletions, and found that existing cancer deletions specifically encompass regions with high STOP gene density (Fig. 3C and fig. S5A). Significant clustering of STOP genes within recurrent cancer deletions was observed with gene sets from our primary (P = 5.0 × 10−3) and validation screens (P = 7.0 × 10−3). Thus, STOP gene densities equal to that found in recurring deletions were identified only 5 to 7 out of 1000 permutations.
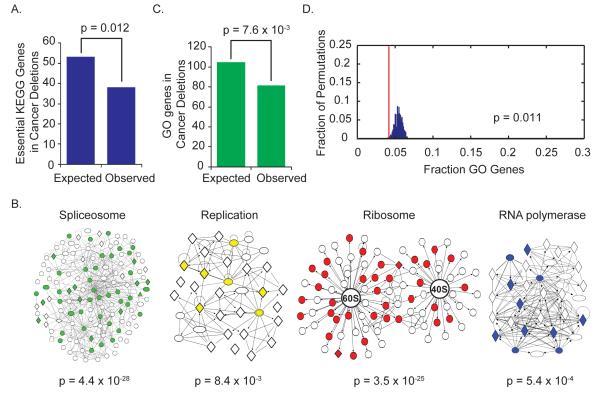
Loss of multiple STOP genes per deletion suggests that cancer cells optimize their proliferative fitness. Increased frequencies of deletions with clusters of STOP genes could occur because the cell now has multiple options for losing the second allele of a recessive TSG or because of combined haploinsufficiencies. If the latter were a primary driving force by which hemizygous deletions fuel cancer, one would expect that deletions would avoid loss of one copy of essential genes that would limit fitness. To test this, we assembled an in silico list of high probability essential GO genes involved in critical cellular processes as annotated by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis including DNA, RNA, protein, and fatty acid synthesis (table S10) encompassing 473 genes. This set demonstrated a significant depletion (P = 0.012) from recurrent deletion regions with 28.3% fewer genes present in deletion regions than expected (Fig. 4A). This in silico analysis suggests that the loss of a single copy of GO genes has a negative impact on cellular fitness.
Fig. 4. Hemizygous focal deletions avoid essential GO genes.
A) Essential KEGG genes are depleted from cancer deletion peak regions. An essential KEGG gene set representing essential GO genes was assembled using all genes from the following processes: basal transcription and RNA polymerase, the spliceosome, the ribosome, DNA replication, fatty acid biosynthesis, amino-acyl tRNA synthesis, and mRNA transport. Genes were mapped to chromosomal locations and compared to genes found in recurrent cancer deletions. Significant depletion from recurrent deletions was observed using Fisher’s exact test.
B) GO genes are enriched for genes involved in transcription, splicing, translation, and DNA replication. All genes included in the ribosome, spliceosome, RNA polymerase, and DNA replication KEGG pathways were assembled into interaction modules using Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA). GO genes are colored red, green, blue, and yellow, respectively, within each module to demonstrate enrichment for these pathways within the GO gene set using Fisher’s exact test.
C) GO gene density is lower than expected in cancer deletions. GO genes were mapped to genomic locations and compared to genes found in deletions. Significant depletion was observed between GO genes and genes found in cancer deletion peaks using Fisher’s exact test.
D) GO genes are significantly depleted from cancer deletions. The GO gene set was mapped to genomic location, and cancer deletion peak regions were overlaid. The percentage of GO genes found in cancer deletions (red line) was compared to the distribution observed after 1000 permutations of the deletion peak regions across the genome.
To independently test this hypothesis, we turned to the other arm of our screen that identified candidate GO genes whose depletion limits proliferation and survival. Because both normal and cancer cells are dependent on these essential GO genes, we analyzed data from proliferation screens on HMECs, one normal prostate epithelial cell line, and seven breast or prostate cancer cell lines for shRNAs that reduced cell proliferation and viability by a factor of ≥ 1.5 in five of the nine cell lines (table S11). This GO gene set is enriched for essential core cellular machinery such as the ribosome, spliceosome, RNA polymerase, and DNA replication required for proliferation (Fig. 4B). We observed a significant depletion (P = 7.6 × 10−3) of GO genes located in recurrent deletions, with 22.1% fewer GO genes than expected (Fig. 4C). When we examined the location of GO genes within recurrent deletions, we found that more than half (58.5%) of deletion regions contained zero GO genes. In contrast to STOP genes, Monte Carlo permutation analysis confirmed that recurrent deletions exist in regions with unusually low GO gene density (P = 0.011) (Fig. 4D).
A potential caveat to the interpretation that GO gene depletion reflects haploinsufficiency is that a substantial fraction of recurrent deletions might actually be homozygous. However, our examination of 611 cell lines used in the recurrent deletion analysis (13) revealed only 5.4% of all genes were ever homozygously deleted, similar to the 11% reported previously (12). Fewer than 1% of genes within deletion regions were homozygous, which suggests that the majority of focal deletions are hemizygous. This low level of homozygous deletion cannot account for the 22 to 28% depletion of GO genes observed, indicating that the absence is more likely due to haploinsufficiency of hemizygous deletions. If such frequent haploinsufficiency occurs among GO genes, by analogy, it is likely that other genes such as the STOP genes also display a similar frequency of haploinsufficiency; if so, this would imply that haploinsufficiency of both STOP and GO genes in sporadic tumors drives tumorigenesis. One possible explanation for this higher than expected frequency of haploinsufficiency is monoallelic expression, a phenomenon in which there is an imbalance in expression levels from the two alleles of a given gene. This imbalance may occur in up to 10% of genes (22). Deletion of the higher expressing allele could produce a more penetrant haploinsufficient phenotype.
Our analysis found that only 22% of recurrent deletion regions could potentially be explained by known or putative recessive TSG (Fig. 1C and fig. S1). If most recurrent deletions primarily represent passenger alterations caused by location in a deletion-prone region such as a fragile site, the genes in these regions should possess no special properties. However, we find the opposite to be true, namely that STOP and GO genes exhibit significantly skewed distributions in regions frequently deleted across cancers. Thus, an additional mechanism of cancer evolution may exist that involves selection of hemizygous somatic deletions encompassing high densities of STOP genes and low densities of GO genes. This strategy promotes net proliferation and survival due to the cumulative reduction in dosage of genes with tumor suppressive properties while avoiding deleterious effects due to reduced dosage of genes that promote proliferation.
Our analysis suggests that ~20% of human genes might display haploinsufficiency, which could have important implications for human health and development given the wide copy number variation seen in humans. Supporting this hypothesis of widespread haploinsufficiency, a number of genes thought to be classical two-hit tumor suppressors also display haploinsufficiency (9, 23, 24). To provide a simple way to discuss this hypothetical cumulative mechanism, we refer to it as the “cancer gene island model”. This model is consistent with the theory of clonal evolution because these deletions provide a selective value to the cell by allowing them to clonally expand, unlike a truly recessive TSG mutation.
Our study provides experimental and statistical evidence that large hemizygous deletions containing islands of clustered proliferation inhibitory genes are preferentially selected during tumorigenesis, indicating that cancers may exhibit properties of a contiguous gene syndrome. Partial gene dosage due to deletion of multiple adjacent genes in a single deletion region is known to cause several classical contiguous gene syndromes, such as 22q11.2 deletion syndrome. Although we have analyzed proliferation and survival genes, cancer-relevant haploinsufficient genes affecting other aspects of tumorigenesis may also exist in these deletions regions.
If a halving of gene dosage can cause a phenotype, then subtle increases in gene dosage may also. In addition to deletions, recurrent amplifications are also found in cancers (13). If the observations from the cancer gene island model can be extended to gain-of-function mutations, then amplification regions may show enrichment for GO genes whose overproduction enhances proliferation. Recent functional analyses of gene amplifications in hepatocellular carcinoma (HCC) revealed that adjacent genes in the 11q13.3 amplicon (CCND1 and FGF19) and the 11q22 amplicon (BIRC2 and YAP1) are cancer-driving oncogenes in HCC (25, 26). Thus, some amplifications in cancer may also represent contiguous gene syndromes.
The enrichment for genes localized to deletions suggests that we have identified dozens of new TSGs in recurrent deletions. We have also likely identified more TSGs outside of these regions because the STOP gene set is (i) enriched for known TSGs, many of which are not found in recurrent deletions, and (ii) enriched for genes that undergo somatic loss-of-function mutation. Finally, this work also suggests that cells possess a substantial number of genes that restrain proliferation in vitro, which could be inactivated to promote clonal expansion during tumorigenesis in addition to the traditional driver genes currently known.
Given the prevalence of multiple, large, recurring hemizygous deletions encompassing skewed distributions of growth control genes in tumors, we propose that the elimination of cancer gene islands that optimize fitness through cumulative haploinsufficiencies may play an important role in driving tumorigenesis with implications on the way in which we think about cancer evolution.
Supplementary Material
Supplemental Online Data
Supplementary Table
Acknowledgements
We thank S. Forbes, A. Futreal, and M. Stratton for generously providing whole genome sequencing data from the COSMIC database, and C. Shaw, D. MacPherson, M. Emanuele, C. Thoma, and T. Westbrook, and members of the Elledge lab for helpful discussions and critical reading of this manuscript. Supported by grants from the National Human Genome Research Institute-funded Cancer Genome Atlas project (M.M); NIH grant U54CA143798 (R.B.); NIH, Stand Up to Cancer and the U.S. Department of Defese (S.J.E.); Susan G. Komen for the Cure Foundation postdoctoral fellowship KG080087 (N.L.S); American Cancer Society postdoctoral fellowship 116410-PF-09-078-01-MGO (Q.X.); and National Institute of General Medical Sciences Medical Sciences Training Program award T32GM07753 (C.H.M). S.J.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplementary Materials: Materials and Methods Supplementary Text Figs. S1 to S6 Tables S1 to S12 References: (27-32)
References and Notes
- 1.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009 Mar 6;136:823. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144:646. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004 Aug;10:789. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 4.Bass AJ, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009 Nov;41:1238. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005 Jul 7;436:117. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 6.Nanjundan M, et al. Amplification of MDS1/EVI1 and EVI1, located in the 3q26.2 amplicon, is associated with favorable patient prognosis in ovarian cancer. Cancer Res. 2007 Apr 1;67:3074. doi: 10.1158/0008-5472.CAN-06-2366. [DOI] [PubMed] [Google Scholar]
- 7.Scott KL, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009 Jun 25;459:1085. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knudson AG., Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971 Apr;68:820. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santarosa M, Ashworth A. Haploinsufficiency for tumour suppressor genes: when you don’t need to go all the way. Biochim Biophys Acta. 2004 Jun 7;1654:105. doi: 10.1016/j.bbcan.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Sherr CJ. Principles of tumor suppression. Cell. 2004 Jan 23;116:235. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 11.Kumar MS, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009 Dec 1;23:2700. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010 Feb 18;463:893. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010 Feb 18;463:899. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worster DT, et al. Akt and ERK control the proliferative response of mammary epithelial cells to the growth factors IGF-1 and EGF through the cell cycle inhibitor p57Kip2. Science signaling. 2012 Mar 6;5:ra19. doi: 10.1126/scisignal.2001986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004 Mar;4:177. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011 Jan;39:D945. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bozic I, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci U S A. 2010 Oct 26;107:18545. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paddison PJ, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 19.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 20.Schlabach MR, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008 Feb 1;319:620. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007 Apr 20;129:289. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zwemer LM, et al. Autosomal monoallelic expression in the mouse. Genome Biol. Feb 20;13:R10. doi: 10.1186/gb-2012-13-2-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011 Aug 11;476:163. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solimini NL, et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012 Jul 6;337:104. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawey ET, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell. 2011 Mar 8;19:347. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006 Jun 30;125:1253. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Online Data
Supplementary Table