Setup and use of a two-laser multiphoton microscope for multichannel intravital fluorescence imaging (original) (raw)
. Author manuscript; available in PMC: 2014 May 21.
Published in final edited form as: Nat Protoc. 2011 Sep 8;6(10):1500–1520. doi: 10.1038/nprot.2011.376
Abstract
Characterizing biological mechanisms dependent upon the interaction of many cell types in vivo requires both multiphoton microscope systems capable of expanding the number and types of fluorophores that can be imaged simultaneously while removing the wavelength and tunability restrictions of existing systems, and enhanced software for extracting critical cellular parameters from voluminous 4D data sets. We present a procedure for constructing a two-laser multiphoton microscope that extends the wavelength range of excitation light, expands the number of simultaneously usable fluorophores and markedly increases signal to noise via ‘over-clocking’ of detection. We also utilize a custom-written software plug-in that simplifies the quantitative tracking and analysis of 4D intravital image data. We begin by describing the optics, hardware, electronics and software required, and finally the use of the plug-in for analysis. We demonstrate the use of the setup and plug-in by presenting data collected via intravital imaging of a mouse model of breast cancer. The procedure may be completed in ~24 h.
INTRODUCTION
The ability of multiphoton microscopy to optically section deep into living tissue without damaging the sample has made it a primary tool in intravital imaging, thereby explaining the behavior of cells in their native microenvironments. The success of this tool in the discovery of new biological mechanisms involving interactions between different cell populations1–4 underscores the necessity of expanding multiphoton imaging to more simultaneous fluorescent channels to facilitate the study of interactions between multiple cell types.
Labeling of multiple targets has been hampered, though, by the limited range of probes that may be used simultaneously and still separated into individual channels.
Some attempts to expand the number of imaging channels have focused on artificially creating a new channel by the careful balancing of cross talk between two independent channels5 or on spectral unmixing of the signal acquired in multiple detectors6. Other attempts to meet this demand have focused on actually expanding the repertoire of probes (injectable or genetically encoded) that can be visualized simultaneously. These efforts have taken one of two approaches.
The first approach focuses on using a conventional multiphoton microscope and selecting specific compatible probes that may be excited with one femtosecond laser7,8. The second approach, however, aims to extend the wavelength range of excitation light beyond the capabilities of standard femtosecond laser systems (690–1,040 nm) and utilize the additional use of an optical parametric oscillator (OPO; 1,100–1,600 nm) to create a ‘broadband’ multiphoton platform. Use of this wavelength range enables the use of red-shifted fluorescent proteins9 including what we term Red (emission > 550 nm) fluorescent proteins (e.g., mCherry, TagRFP, mKate2 (refs. 10,11) and new far-red (emission > 650 nm) fluorescent proteins, such as TagRFP657 (ref. 12).
The development of custom multiphoton microscopes specifically designed to extend the capabilities beyond commercially available systems has a long history (Table 1), with many of the earlier efforts focused on modifying commercial point-scanning confocal microscopes. Recent efforts at building systems using OPOs have focused on modifying a commercially available multiphoton microscope using a single femtosecond pulsed laser as both an instrument for imaging and as a pump laser for an OPO13,14. At the time of writing this article, this system is commercially available (TriM Scope II, LaVision).
TABLE 1.
Overview of literature on custom-developed multiphoton systems.
| Year | Group | First application | Original system | Reference |
|---|---|---|---|---|
| Modified commercial systems | ||||
| 1990 | Webb, W. | Beads | BioRad MRC 500 | 24 |
| 1996 | Cannell, M.B. | Rat myocardial cells | Zeiss LSM410 | 25 |
| 1996 | Konig, K. | Rat macrophages in vitro | Zeiss LSM310 | 26 |
| 1999 | Ellisman, M.H. | Rat cardiac myocytes and E. coli in vitro | Nikon RCM8000 | 27 |
| 1999 | Loew, L.M. | Neuroblastoma in vitro | BioRad MRC 600 | 28 |
| 2000 | Yuste, R. | Neurons/Drosophila egg/leech muscle in vitro | Olympus FluoView | 29 |
| 2001 | Jain, R.K. | Mouse angiogenesis in vivo | BioRad MRC 600 | 30 |
| 2001 | Diaspro, A. | Cow artery ex vivo/yeast | Nikon PCM2000 | 31 |
| 2003 | White, J. | C. elegans eggs in vitro | BioRad MRC 1024 | 32 |
| 2004 | White, J. | Monkey kidney ex vivo | BioRad MRC 600 | 33 |
| 2004 | Miesenbock, G. | Fly olfactory neurons in vivo | Thermo-Noran Oz | 34 |
| 2007 | Diaspro, A. | Ultrathin fluorescent layers | Leica SP5 | 35 |
| 2007 | Low, P.S. | Tumor cells in blood flow in vivo | Olympus FluoView FV300 | 36 |
| 2008 | Brown, E.B. | Mouse breast tumor ex vivo | Olympus FluoView FV300 | 37 |
| 2009 | Friedl, P. | Cancer cells in human and mouse dermis in vivo and ex vivo | LaVision TriM Scope | 13 |
| Custom-built systems | ||||
| 1997 | Gratton, E. | Embryonic mouse fibroblasts in vitro | 38 | |
| 1999 | Neher, E. | CA-3 pyramidal cells (brain) ex vivo | 39 | |
| 1999 | Piston, D.W. | Pancreatic cells ex vivo | 40 | |
| 1999 | Svoboda, K. | Rat hippocampal brain slices ex vivo | 41 | |
| 2001 | Parker, I. | Cheese/pollen/neurons ex vivo | 42 | |
| 2002 | Robey, E. | Mouse thymocytes in vitro | 3 | |
| 2002 | Kleinfeld, D. | Rat brain in vivo/HeLa cells in vitro | 43 | |
| 2002 | Tromberg, B.J. | Human fibroblasts in vitro | 44 | |
| 2005 | Beaurepaire, E. | Drosophila embryos in vivo | 45 | |
| 2005 | Weissleder, R. | Mouse bladder, kidney, muscle, jejunum, liver in vivo | 46 | |
| 2006 | Toledo-Crow, R. | Skin and lymph node in mouse in vivo | 43 | |
| 2007 | Miller, M.J. | Leukocytes in mouse footpad in vivo | 47 | |
| 2009 | Condeelis, J.S. | Breast cancer in mouse in vivo | 22 |
However, as the OPO requires very specific pump wavelengths determined by the desired output wavelength range (see Table 2), and the great majority of the output power of the femtosecond laser, these systems are limited in wavelength tunability and range of available intensity, forcing the return, once again, to the limited situation of selecting specific compatible probes. In contrast, the system we have developed is a custom-built two-laser multiphoton microscope (TLMPM) capable of exciting fluorophores in the ranges of 750–1,040 and 1,100–1,600 nm, and detecting fluorescence in the range of 400–740 nm. Because this microscope uses two independent laser systems, the number and type of fluorophores is not limited by the source and we are able to excite fluorophores simultaneously and image rapid events (such as cell motility). By using four simultaneously acquiring physical detectors in combination with the color balancing technique previously developed in our lab5 (and described in this protocol), it is possible to simultaneously image five different fluorophores and separate their signals.
TABLE 2.
Summary of OPO pump wavelength requirements.
| Pump wavelength (nm) | Output wavelength range (nm) |
|---|---|
| 750 | 1,100–1,200 |
| 775 | 1,200–1,350 |
| 810 | 1,340–1,600 |
Once multiple cell types can be separately visualized, their interactions can only be understood via quantitative statistical analyses of cellular parameters extracted from the image data. However, until recently15,16, most available image processing tools were not optimized for high-resolution intravital or 3D imaging in tumors. They were commonly designed for 2D images or time-lapse sequences without the ability for measurement of axial movements or to follow movement of the same cell throughout a 4D (3D information over time) stack. The task is further complicated by difficulty in displaying 4D data and, as is often the case in intravital imaging, the inclusion of drifts in one or more of the x, y and z dimensions, as well as artifacts from irregular breathing.
As the number of groups using time-resolved imaging in 3D cultures or in vivo grows, solving these issues becomes increasingly important. Although some progress in quantitative extraction has been made, all techniques reported so far are based on automatic fluorescence intensity thresholding and segmentation algorithms, which are able to work well for cases in which there is clear physical separation among the cells being visualized17.
Unfortunately, the close 3D packing of cells in solid tissues and the decreased z axis resolution (when compared with the x and y axes) inherent in optical imaging generate images that cause these algorithms to break down, making this a task that cannot be fully automated.
To address this, we have developed a Java-based plug-in for NIH ImageJ (Rasband, W.S., ImageJ, US National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2009), which provides a user-friendly graphical interface that simplifies both the identification and tracking of objects in multidimensional image sets and the extraction of this quantitative data for further analysis in other software packages. The design of this plug-in makes it a useful tool not only for the analysis of cell motility in solid tumors, but also for any type of object tracking problem. Instructions for accessing the ROI_Tracker plug-in may be found on the Gruss Lipper Biophotonics Center website (http://www.einstein.yu.edu/biophotonics) under the ‘Innovation Laboratory’ link.
Thus, in this protocol, we describe the procedure for constructing a two-laser multiphoton microscope that extends the number of simultaneously usable fluorophores and for using a software plug-in that we have written for the free and widely used image analysis program ImageJ, which simplifies the quantitative tracking and analysis of 4D intravital image data available in these more complex multichannel imaging data sets.
Experimental design
Optical design
A block diagram of the optical layout of the microscope, detailed in Figure 1a, and laid out in a computer-aided design drawing in Figure 1b, shows the two laser sources. The first, a standard femtosecond pulsed Ti:Sapphire laser (Tsunami- Millennia), is used for excitation of fluorophores in the range of 750–950 nm (e.g., cyan fluorescent protein (CFP), green fluorescent protein (GFP), yellow fluorescent protein (YFP)). The second consists of an automatically tunable femtosecond pulsed Ti:Sapphire laser (Mai-Tai) and an OPO (Opal).
Figure 1.
Optical layout of custom-built TLMPM. (a) The TLMPM provides excitation at 750–1,040 nm and 1,100–1,600 nm and collection in four distinct channels. Wavelengths < 950 nm are excited by a Millennia-pumped Tsunami laser and those between 960 and 1,040 nm by a Mai Tai laser. For excitation between 1,100 and 1,600 nm, a pair of flip mirrors in the light path switch Mai Tai use from an illumination source to a pump beam for the Opal optical parametric oscillator. Both beams then pass through scanning galvos into the microscope and the generated fluorescence is collected, depending on the wavelength, into one of four PMT detectors. The letters A, B, C, D and E designate five separate functional groupings of optical components. The inset details the spectra of the blocking filters before the photodetectors. (b) Computer-aided design of the entire optical layout for the two-laser multiphoton microscope showing the two laser systems and the five functional units. Note that the body of the microscope is omitted. GVD, group velocity dispersion compensator; M, Millennia; MA, manual attenuator; MT, Mai Tai; OL, objective lens; OPO, optical parametric oscillator; PMT, photomultiplier tube; TS, Tsunami.
Depending on the combination of fluorophores needed, the output from the Mai-Tai can be used directly, giving one source with a wavelength range of 950–1,040 nm, or it can be tuned to a pumping wavelength (Table 2) and used to pump the OPO where its energy is converted into femtosecond pulses in the range of 1,100–1,600 nm. A pair of flip mirrors allows selection between these two possible paths. Figure 2 details the spectra of combinations of fluorescent proteins commonly used in our lab along with laser wavelengths used to image them, and Table 3 summarizes the optical properties of commonly used fluorescent proteins gathered from the literature.
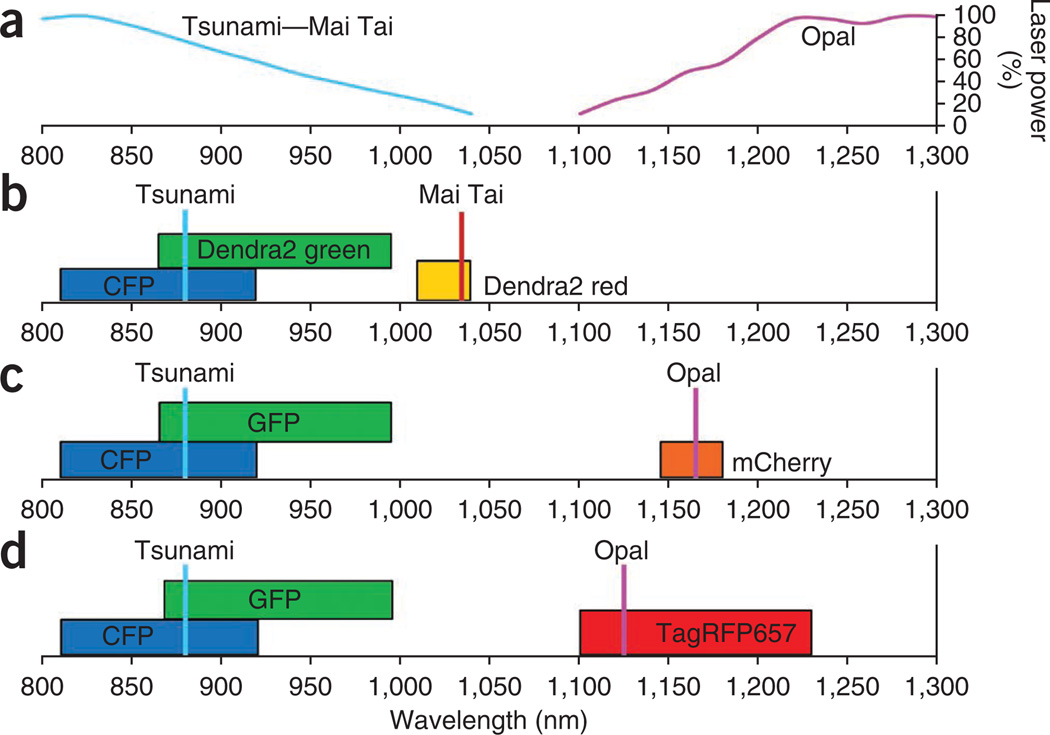
Figure 2.
Laser wavelengths and spectra of commonly used combinations of fluorescent proteins with the TLMPM. (a) Normalized output power for the two-laser systems in the TLMPM. The cyan and purple curves show the normalized power output of the Tsunami (or Mai Tai) laser and the Opal laser, respectively. (b) CFP (blue bar), Dendra2 green (green bar) and Dendra2 red (yellow bar) excitation bandwidths (full width at half-maximum; FWHM). Thin cyan and red vertical bars indicate the excitation wavelengths used for the Tsunami and Mai Tai, respectively. (c) CFP (blue bar), GFP (green bar) and mCherry (orange bar) excitation bandwidths (FWHM). Thin cyan and purple vertical bars indicate the excitation wavelengths used for the Tsunami and Opal, respectively. (d) CFP (blue bar), GFP (green bar) and TagRFP657 (red bar) excitation bandwidths (FWHM). Thin cyan, orange and purple vertical bars indicate the excitation wavelengths used for the Tsunami, Mai Tai and Opal, respectively.
TABLE 3.
Summary of 1p and 2p characteristics of commonly used fluorescent proteins.
| Protein | 2PE (GM) | 1P QE (%) | 1p EC (M−1 cm−1) | Peak (nm) | Bandwidth (nm) |
|---|---|---|---|---|---|
| ECFP | 47a | 40b | 32,500b | 870 | 120 |
| mKeima | 26a | 24c | 13,400c | 884a | 138 |
| Dendra2 Green | ND | 50d | 45,000d | 910e | ND |
| EGFP | 174a | 60b | 56,000b | 930a | 130 |
| YFP | 213f | 61b | 83,400b | 970f | 60 |
| Dendra2 Red | ND | 55d | 35,000d | 1,015e | ND |
| TagRFP | 36g | 48c | 100,000c | 1,030g | 110 |
| tdTomato | 60h | 69b | 69,000b | 1,053h | 125 |
| DsRed2 | 64h | 55b | 43,800b | 1,068h | 115 |
| mCherry | 5g | 22b | 72,000b | 1,133g | 95 |
| mKate2 | ND | 40i | 62,500c | 1,176e | ND |
| TagRFP657 | ND | 10j | 34,000j | 1,314e | ND |
With either choice of path, both beams are each passed through a shutter (SH-20-B), a manual attenuator (MA), a computer-controlled Pockels cell attenuator (Model 350–80 for 750–950 nm range and Model 360–40 for > 950 nm), and for the Tsunami beam, a custom-built group velocity dispersion (GVD) compensator (G; described in the next section).
A MA is used in addition to a computer-controlled Pockels cell to limit the maximum intensity available at the microscope objective to reasonable values that will not damage samples. This gives simple 0–100% control over the laser intensity at the computer interface. It is also important to note that the MA comprises a half-wave plate followed by a linear polarizer. This type of attenuator must be used instead of neutral density filters or reflective attenuators as these will be burned by the high peak pulse powers of the lasers.
One further important consideration for the Pockels cell is that of the linearity of its attenuation. On its own, a Pockels cell creates an attenuation response that is a sine-squared function of the control voltage. When the software is written to control this element, this functional dependence should be eliminated to produce a linear attenuation response.
Next, the two beams are combined using a dichroic mirror (Di, used at 45° incidence) and sent on to a galvanometer pair (VM500 +). A scan lens (SL) conjugate to the galvos focuses the beam at the back focal plane of the tube lens (TL), which is itself conjugated to the objective lens.
In many multiphoton systems, an additional set of lenses is typically included to expand the laser beam so as to properly fill the back aperture of the objective lens and ensure the full use of the numerical aperture (NA) of the objective. This expander can then be adjusted to accommodate the different objective lenses with different-sized back apertures and NAs. In our system, we make use of a single ×20, 0.95 NA objective lens for all our imaging. The combination of low magnification and high NA of this lens allows us to rapidly switch between low-magnification, histologictype imaging and high-magnification, high-resolution imaging by simply adjusting the range of the scanning galvanometers. Proper filling of our objective lens is accomplished by the telescope formed by our SL and TL. Doing so simplifies the optical alignment of the system and minimizes the amount of dispersive glass present.
The generated fluorescence light is separated from the excitation beam with a second dichroic (720DCXXR) and sent on to a light-tight detector box. The detector box is designed to hold three standard Olympus filter cubes (OMF) on magnetic mirror mounts. This design enables the rapid changing of filter sets, which can thus be tailored to each experiment, without compromising alignment. Two detector boxes may also be attached together, extending the number of filter cubes (FC) and detectors available. In our setup, four nondescanned (×3 H7422PA-40 and ×1 HC-125–02) detectors simultaneously collect the spectrally separated fluorescence light.
The optics surrounding the lasers and the actual microscope are divided into four functional units (Fig. 1, letters A through E) further detailed in Figures 3–7 and described in Table 4. See Supplementary Figure 1 for an interactive 3D .pdf file of the complete optical layout of the multiphoton microscope.
Figure 3.
Optical layout detail of multiphoton functional unit A—Tsunami beam path. (a) Computer-aided design detail of functional unit A shown in Figures 1 and 2. The black double-headed arrow indicates the motion of adjustment of prism P2, which allows fine-tuning of the dispersion compensation. (b) Optical block diagram of the area shown in a. I, iris; M, mirror; MA, manual attenuator; PC, Pockels cell; PS, periscope; S, shutter.
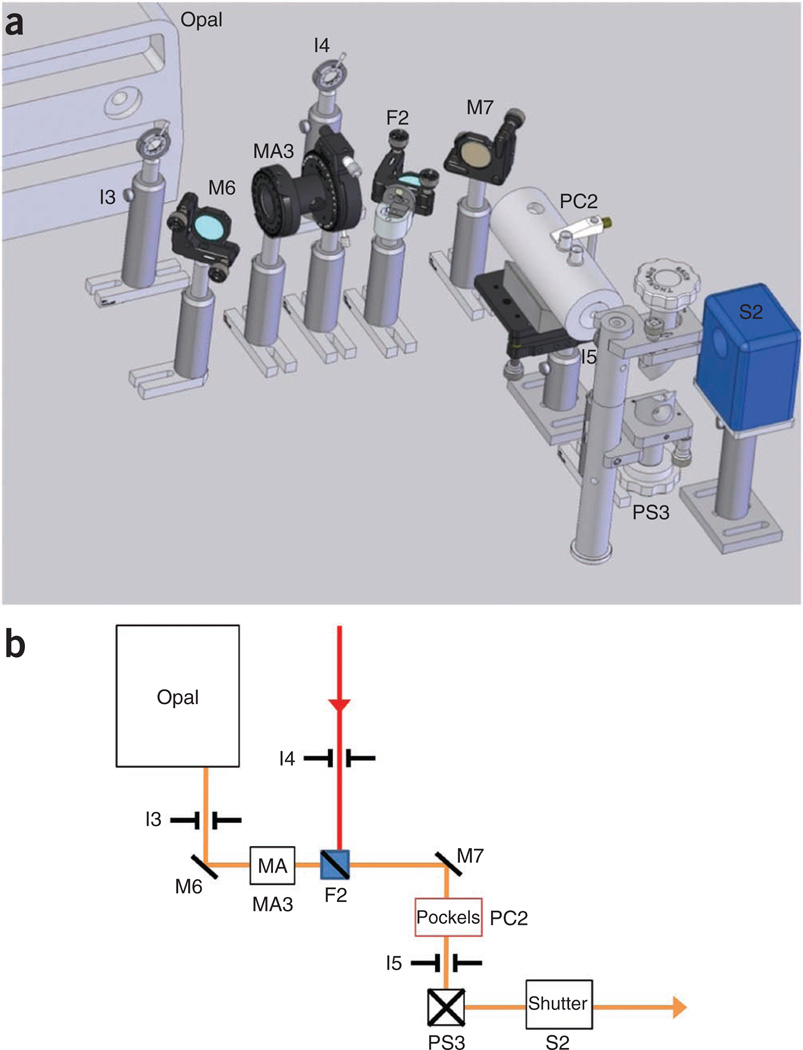
Figure 7.
Optical layout detail of multiphoton functional unit E—microscope and detector box. (a) Computer-aided design detail of functional unit E shown in Figures 1 and 2. (b) Optical block diagram of the area shown in a. The optical elements required within the microscope (body not shown) and the detector box. PD1 is the blue PMT module and PD2–4 are the green, red and far-red PMT modules. The arrangement of the filters within the filter cubes is as follows: FC1, blue dichroic and blue blocking filter (Semrock, FF02-447/60-25); FC2, green dichroic and green blocking filter; FC3, red dichroic, red blocking filter (top) and far-red blocking filter (left). Part numbers for the PMTs and filters are listed in the general equipment section. CL, collection lens; D, dichroic mirror; FC, filter cube; M, mirror; L, lens; OL, objective lens; PD, photodetector; TL, tube lens.
TABLE 4.
Descriptions of microscope functional units.
| Unit | Title | Description |
|---|---|---|
| A | Tsunami beam path | Conditioning optics for Tsunami beam |
| B | Mai Tai–Mai Tai pump beam path | Optics selecting use of Mai Tai as laser source or pump beam |
| C | Mai Tai–Opal beam path | Conditioning optics for both Opal and Mai Tai beams |
| D | Combining optics and scanning optics | Galvanometers, scan and tube lenses |
| E | Microscope and detector box | Fluorescence collection optics |
GVD compensator
Although care has been taken in the optical design to minimize the number of dispersive optical elements between the laser and the sample, significant pulse broadening is to be expected even with this minimum number of elements. For the OPO beam, this dispersion can be somewhat compensated for by using GVD compensation prisms built into the oscillator cavity. However, for the Tsunami beam, the range of GVD compensation available is limited by the necessity of maintaining mode lock within the cavity.
As such, we have built an external GVD compensator18 using a double-pass arrangement. The optical layout is shown in Figure 3. Here the laser beam enters the GVD compensator by way of a pair of mirrors forming a periscope (PS1) and is sent on to mirror M1, which redirects the light to the apex of the first prism (P1). The prism angle is adjusted so as to produce a minimum beam deflection. Negative dispersion of the pulses then accrues as the beam then traverses the distance from P1 to a fold mirror (M2) and on to a second prism (P2) mounted on a linear translation stage. The angle of this second prism is also adjusted so as to produce a minimum beam deflection, and a mirror (M3) is placed so as to retroreflect the beam back along its original path. The incoming and outgoing beams are then separated by slightly tilting M3 vertically. This causes the return beam to pass over M1 and to be reflected by the final mirror (M4).
By properly adjusting the distance between P1, M2 and P2, the amount of negative dispersion added to the beam can be tailored to cancel out the amount of positive dispersion created by the two prisms and all of the other optical elements in the microscope (i.e., Pockels cell, objective lens and so on). Fine tuning of the amount of dispersion can then be accomplished by adjusting the micrometer screw for P2 (double-headed arrow), and adding or removing an additional thickness of glass. It should be noted that adjustment of these prisms will induce small deviations into the beam path and result in a slight misalignment of the microscope. These deviations may be quickly compensated for, however, by adjusting the exiting mirrors of the GVD compensator (M4 from Fig. 3 and M9 from Fig. 6) and using the beam alignment irises (I6 and I7 from Fig. 6) placed before the microscope to ensure that the beam returns to its original path.
Figure 6.
Optical layout detail of multiphoton functional unit D—combining optics and scanning optics. (a) Computer-aided design detail of functional unit D shown in Figures 1 and 2. (b) Optical block diagram of the area shown in a. The optical elements required for combining the Tsunami and Mai Tai or Opal OPO beams, the scanning optics, as well as the scan and tube lenses. Irises I6 and I7 comprise the beam alignment irises depicted in Figures 1 and 2. D, dichroic mirror; I, iris; G, galvanometers; GVDC, group velocity dispersion compensator; S, shutter; SL, scan lens; TL, tube lens.
Using an autocorrelator (Carpe) we measured the pulse widths of the beam at various locations. Without any GVD compensation, dispersion of our optical elements increases the 95-fs pulses coming out of the laser to 255 fs after the objective lens. With GVD compensation, these pulses are again narrowed down to 85 fs at the sample (see Fig. 8). To compare the effect that GVD compensation has upon the collected fluorescence signal, we imaged an excised PyMT-generated tumor expressing CFP both with and without GVD compensation. All other parameters (e.g., laser power, photomultiplier tube (PMT) gain) were maintained constant. For the same field of view, we observe a 300% increase in fluorescence signal with GVD compensation (Fig. 8.)
Figure 8.
Effect of GVD compensator. (a) By adjusting the prism (P2 in Fig. 4) position via its linear translation stage, more or less glass (positive dispersion) can be added to the optical path. When the positive and negative dispersions are balanced, a minimum of pulse width is observed. We have observed the pulse width reduced to 85 fs at the sample as compared with 255 fs without GVD compensation. (b) Left, a CFP-labeled tumor generated in a PyMT transgenic mouse without GVD compensation. Right, the same sample with GVD compensation. The presence of GVD compensation increases the fluorescence signal by 300%. All animals were used according to protocols that have been reviewed and approved by Einstein’s Institutional Animal Care and Use Committee.
Electronics design
The timing and control signals generating the galvanometer raster scan are created with a high-speed analog output board (PXI-6713). This board also generates the control signals for the Pockels cells and the photodetector gain signals.
Blanking of the laser during fly-back is accomplished by using the H-Sync signal to turn off the Pockels cell control voltage via a custom-designed electronic circuit (Fig. 9) based on a wideband video switch (DG541). The photodetector signals are captured and digitized with two data acquisition (DAQ) boards (PXI-6115).
Figure 9.
Electronic schematic of blanking circuit. Blanking of the beam during the fly-back portion of the raster scan is accomplished by zeroing the Pockels cell control voltage. The horizontal synchronization signal is used as the TTL input to a wide-band video switch. Although connections for only one channel are shown, each integrated circuit chip contains four equal circuits.
An overview of the microscope control electronics is given in Figure 10. This block diagram lays out all the connections between the computer, the control/DAQ boards and each of the external devices. In this figure, the internal, software-configured connections between the two input/output boards and the analog output board are represented by the red real-time system integration lines. These connections are further detailed in Figure 11 showing how the onboard clock is used to generate a pixel clock (PCLK) and a clock for data acquisition (DAQ clock). The DAQ clock drives the rate at which DAQ progresses, and the PCLK is used to generate the rest of the raster generation signals.
Figure 10.
Electronic schematic of two-laser multiphoton microscope. The computer controls the XY stage and z motor for the microscope directly via RS-232 connections. The three control boards are connected directly to the computer via a dedicated MXI-4 line within a PXI chassis. All the rest of the electronics are connected directly to these three boards with the connection channel indicated in the diagram. Internal connections between boards are shown by the real-time system integration (RTSI) connections in red; H-sync, horizontal synchronization; IO, input/output; V-sync, vertical synchronization.
Figure 11.
Internal connections for generating timing signals and clocking acquisition. An onboard oscillator creates a master clock that is used both for the generation of timing signals and for clocking the acquisition boards. This design ensures that all signals can remain synchronized and gives the flexibility of allowing the over-clocked acquisition described in the text. DAQ, data acquisition; H-sync, horizontal synchronization; PCLK, pixel clock; V-sync, vertical synchronization.
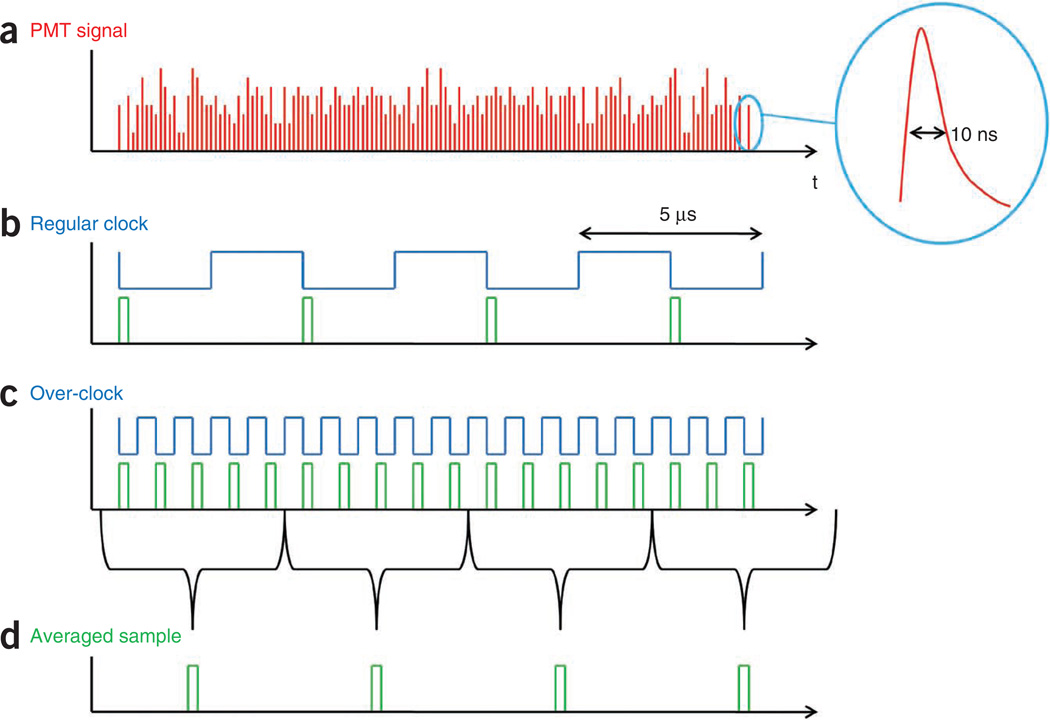
Although the fastest scan rate of the microscope system is limited by the inertial response of the galvanometers to ~1 frame per second, the higher bandwidth capabilities of the photodetectors and acquisition electronics gives an opportunity for signal-to-noise improvement without sacrifice of acquisition time. A typical multiphoton PMT photosignal (shown in Fig. 12a) comprises many electronic pulses (inset), each in the order of 10 ns in duration. With the conventional DAQ schemes, one data point is acquired for each scan position in an image and is triggered by a PCLK (Fig. 12b). The finite and typically fast sampling time (~100 ns) of DAQ board means that the majority of the available PMT pulses go undetected. To capture these lost pulses, we use an ‘over-clocking’ technique in which the acquisition clock is run a multiple faster than the PCLK rate (Fig. 12c). This allows the acquisition of several data points for each pixel in the scan, which are then averaged together (Fig. 12d).
Figure 12.
Over-clocked acquisition of PMT signals. Improvements in signal to noise may be achieved by acquiring PMT photosignals at a rate faster than the scan rate. (a) Typical photosignals from PMT comprise many pulses due to the high PMT bandwith. (b) Traditional acquisition clocking. In a traditional data acquisition scheme, one sample is taken for each pixel in the final image and is clocked by a pixel clock. The fast acquisition time of data acquisition boards leaves many of the pulses undetected. (c) Over-clocked acquisition. Running the acquisition clock faster than the scan rate allows acquisition of these lost pulses. (d) Averaging of over-clocked acquisition. Averaging over-clocked pulses leads to improved signal-to-noise ratio without decreasing the acquisition rate.
Software design
The control software is an area that contains the most flexibility in its design. At a minimum, the control software must generate the raster scan signals, capture the incoming data and save them to disk. Additional features can be included to facilitate ease of use (e.g., real-time, on-display merging of imaging channels), or add additional imaging capabilities (e.g., z-stack imaging, time-lapse imaging).
In our system, custom software (written in LabVIEW) centralizes control of the entire system within one interface and allows several types of imaging modalities including time series, z-stacks, mosaics and combinations thereof.
Future design considerations
Future expansion of the system is simplified as all electronics and software have been designed from the start to accommodate eight simultaneous acquisition channels. These extra imaging channels could allow simultaneous detection of up to four different cell types, two second harmonic signals and two reflectance confocal channels in the same living animal.
In addition, the gap between the Ti:Sapphire and OPO light sources, which spans 1,050–1,100 nm, can be eliminated through the use of the second harmonic of the OPOs additional ‘idler beam’ output.
Support software for multiphoton 4D imaging
The interface to our software, called ROI_Tracker, is shown in Figure 13. To fully leverage existing capabilities and minimize development time, our software was developed as a plug-in for the open-source image-processing package, ImageJ (version 1.41o or later). The plug-in is designed to access the pixel information contained in 3D, 4D or 5D (x, y, z, t and channel) image data, which has been formatted as ImageJ hyperstacks. A hyperstack is a method of logically organizing sets of multidimensional image data into depth slices, time sequences and channels, which are then navigated using sliders. For ease of development, the code was written within the open-source integrated development environment Eclipse (version Ganymede) and used the Visual Editor package (version 1.4.0) for graphical user interface development.
Figure 13.
The ROI tracker user interface. The ROI_Tracker software allows the rapid identification, marking and grouping of series of selections into Tracks. For each selection, the region parameters are calculated and displayed in tabular format. Export of region parameters and individual selection points to tab delimited ASCII files is possible, enabling further numerical analyses in other software packages.
The plug-in is thus designed to allow users to easily outline and track the location of cellular features with region of interest (ROI) indicators as the features progress throughout time and change in depth within the hyperstack. Marking these features can be done with a variety of types of selection tools, including point, polygon or freehand selections. Once a particular cellular feature has been followed and marked throughout time and depth of the image set, the ROIs are linked together to form a Track. Multiple features may be followed in a single set of images by creating multiple Tracks. In addition, if reference points can be identified within the 4D data, their locations may serve as the basis of a reference Track, which may be used to compensate for overall xyz drift within the image set. From these Tracks, region parameters (e.g., area, centroid location, velocity and so on) are calculated and displayed.
Capabilities have been added to allow saving and loading of Tracks (enabling analysis over multiple sessions) and exporting of calculated data and individual ROI coordinates to tab delimited ASCII files for further analysis in other software packages. For presentation purposes, all of the ROIs for all time points can be displayed in one image, or movies of the selected ROIs, along with their paths of travel and centroid locations, can be created on either a black background or overlaid on the image data.
Note concerning equipment
For many of the parts listed in MATERIALS, there are several vendors offering alternative products of equivalent quality. In the cases in which these alternative products may be substituted, we have marked the item with ‘or equivalent’. When not marked as such, we have found the indicated item to be superior to possible alternatives. To account for the multitude of optomechanical parts required, we have recommended purchasing kits rather than listing them individually. More than one kit of each type may be required. For the few parts that have been custom designed, drawings are available upon request.
MATERIALS
EQUIPMENT
General equipment
- Femtosecond laser (2; Spectra Physics, Tsunami or Mai Tai HP Ti:Sapphire laser—or equivalent)
- Optical parametric oscillator (1; Spectra Physics, Opal—or equivalent)
- Optical table with air damped legs (1; Newport, ST-UT Series Upgradable Smart Table, 5 ft × 8 ft—or equivalent)
- Aluminum breadboard, 24 inches × 24 inches × 1/2 inch (1; Thorlabs, cat. no. MB2424)
- Vertical bracket for breadboards (2; Thorlabs, cat. no. VB01)
- Optical power meter and detector head (1; Coherent, FieldMax II and PM10—or equivalent)
- Autocorrelator (1; APE, Carpe)
- Spectrometer (1; Ocean Optics, cat. no. USB2000 +—or equivalent)
- Oscilloscope (Tektronix, cat. no. TDS2024—or equivalent)
- Screw Kit (1/4 inch −20, Thorlabs, cat. no. HW-KIT2—or equivalent)
- Screw Kit (8–32; Thorlabs, cat. no. HW-KIT1—or equivalent)
- Post holders and bases (Thorlabs, cat. no. ESK01—or equivalent)
- Posts and accessories (Thorlabs, cat. no. ESK03—or equivalent)
- Irises and mirror mounts (Thorlabs, cat. no. ESK05—or equivalent)
- Mirrors (Newport, cat. no. 10D20ER.2)
- Infrared (IR) viewer (FJW Optical Systems, FIND-R-SCOPE Model 84499A—or equivalent)
- Flip mounts (2; Thorlabs, cat. no. FM90)
- Periscope assembly (2; Thorlabs, cat. no. RS99)
- Laboratory power supply (1; GW Instek, cat. no. GPS-3303)
- Threaded frosted alignment disk (1; Thorlabs, cat. no. DG10-1500-H1-MD)
Software
- Programming language (National Instruments, LabVIEW Developer Suite—or equivalent)
- Visual Editor package (version 1.4.0)
- ImageJ (NIH, ImageJ v.1.44e or later)
- Dual processor computer (Dell, Precision 690—or equivalent)
- ROI_Tracker plug-in (can be downloaded from the Gruss Lipper Biophotonics Center website: http://www.einstein.yu.edu/biophotonics under the Innovation Laboratory link. Note that the software is available free of charge to academic and/or nonprofit researchers for their noncommercial research, but that commercial entities acquiring the software need to obtain a license).
Microscope
- Inverted fluorescence microscope (1; Olympus, IX71 with Laser Port B option and Laser Port C option and epifluorescence filters for 4,6-diamidino-2-phenylindole (DAPI), FITC, TRITC and Cy5)
- Silver mirror (1; Chroma, 21010)
- Objective lens (1; Olympus, ×20 0.95NA water)
- Motorized xy stage (1; ASI, MS-2000 XY System for Olympus IX71—or equivalent)
- Motorized z axis drive and controller (1; ASI, MFC-2000—or equivalent)
- Filter cubes (4; Olympus, U-MF2)
- Excitation-emission separating dichroic mirror (1; Chroma, 720DCXXR)
GVD compensator equipment
- Aluminum breadboard (1; Thorlabs, cat. no. MB2424—or equivalent)
- Glass-dispersing prism (2; Newport, cat. no. 10FS10)
- Translation stage (1; Thorlabs, cat. no. PT1—or equivalent)
Scanning optics equipment
- Galvanometer pair and driver (1; GSI Lumonics, cat. no. VM500 +, LNISD Driver)
- Galvanometer power supply (1; International Power, cat. no. IHCC24-2.4 with over voltage protection module)
Attenuators and shutters
- Pockels cell and controller (2; ConOptics, cat. no. 350-80 for 750–1040 nm range and cat. no. 360-40 for 950–1,600 nm range)
- Platform mount (2; Thorlabs, cat. no. KM200B)
- Linear polarizer (3; Nova Phase, cat. no. ZZQ-POLARIZER)
- Half-wave plate (3; Nova Phase, cat. no. ACH 12.7-950H-M for the 750–1,040 range and ACH-12.7-1430H-M for the 100–1,600 nm range)
- Linear polarizer mount (3; Thorlabs, cat. no. PRM1GL10)
- Half-wave plate mount (3; Thorlabs, cat. no. RSP1)
- Shutter and driver (2; EOPC, cat. nos. SH-20-B and DSH-20-110)
Filters and lenses
- Beam combining dichroic mirror (1; Semrock, cat. no. LP02-1064RU-25)
- Blue dichroic (1; Semrock, cat. no. FF495-Di02)
- Blue blocking filter (1; Semrock, cat. no. FF02-447/60-25)
- Green dichroic (1; Semrock, cat. no. FF560-Di01)
- Green blocking filter (1; Semrock, cat. no. FF01-520/35-25)
- Red blocking filter (1; Semrock, cat. no. FF01-580-/60-25)
- Red dichroic (1; Semrock, cat. no. FF650-Di01)
- Far-red blocking filter (1; cat. no. FF01-697/75-25-D)
- IR-coated scan lens (1; Thorlabs, cat. no. AC254-045-C)
- IR-coated tube lens (1; Olympus, cat. no. IX2-RFACB2-TL)
- Fluorescence collection lens (1; Thorlabs, cat. no. LA1979-A)
- PMT focusing lenses (4; Thorlabs, cat. no. LA1422-A)
Acquisition/control electronics
- Electronics chassis (1; National Instruments, cat. no. PXI-1033)
- Multifunction input/output board (2; National Instruments, cat. no. PXI-6115)
- Digital-to-analog converter board (1; National Instruments, cat. no. PXI-6713)
Blanking circuit electronics
- Wide-band video switch integrate circuit chip (1; Vishay, cat. no. DG541)
- Prototyping board (1; Vector Electronics, cat. no. 8015-1—or equivalent)
- BNC connectors (9; Amphenol, cat. no. 031-10-RFXG1—or equivalent)
- Instrument enclosure box (1; Hammond, cat. no. 1598BBK)
- Ohm resistors, 10K (8; Panasonic, cat. no. ECG, ERD-S2TJ103V—or equivalent)
Photodetectors
- Green, red, far-red PMT (3; Hamamatsu, cat. no. H7422PA-40)
- Green, red, far-red PMT controller (3; Hamamatsu, cat. no. M9012)
- Green, red, far-red PMT trans-impedance Amplifier (3; Hamamatsu, cat. no. HC130-03)
- Blue PMT module (1; Hamamatsu, cat. no. HC-125-02)
PROCEDURE
Setting up the table, lasers and microscope ● TIMING 6 h
- 1|
Have the vendor set up the optical table. - 2|
Have the vendor set up the lasers as shown in Figure 1.
▲ CRITICAL STEP Ensure that enough space is left between the Mai Tai and the Opal for the inclusion of an iris and a flip mirror. About 11 inches between the side of the Mai Tai and the input end of the Opal should be sufficient.
! CAUTION The Periscope (PS2) and lens holders L1 and L2 shown in Figure 4 are set up by the vendors, and care should be taken not to disturb them; otherwise, the operation of the Opal will be affected. It is not necessary, but is highly recommended, that training be received on how to align the Opal system. - 3|
Have the vendor set up the microscope, install the Chroma silver mirror in the Laser Port B FC (this side port will be the input port for the lasers) and install the Chroma excitation-emission separating dichroic mirror (Di) in the Laser Port C FC (this port will be positioned where the fluorescence light is collected). Remove the TL and its holder from the Laser Port B turret.
Figure 4.
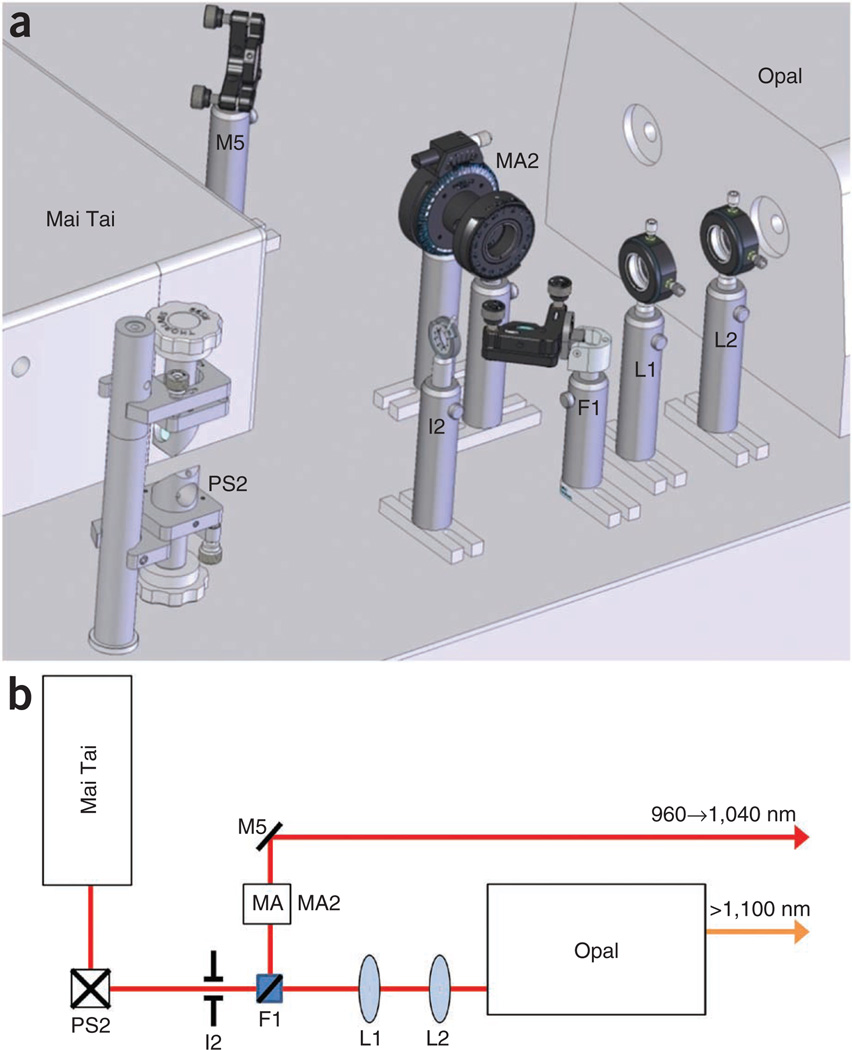
Optical layout detail of multiphoton functional unit B—Mai Tai– Mai Tai pump beam path. (a) Computer-aided design detail of functional unit B shown in Figures 1 and 2. (b) Optical block diagram of the area shown in a. The optical elements required for mating the Mai Tai to the Opal OPO and for selecting the use of Mai Tai as a laser source or a pump beam. F, flip mirror; I, iris; M, mirror; MA, manual attenuator; PS, periscope.
Setting up the components in Mai Tai–Mai Tai pump beam path ● TIMING 30 min
- 4|
Turn on and tune the laser to a wavelength easily visible to the eye (e.g., 690 nm) (see Fig. 4). - 5|
Install the iris (I2) so that the beam coming from the top periscope (PS2) mirror passes centered through its aperture.
▲ CRITICAL STEP This element, as with all of the irises in the system, may be used in the future as references to regain beam positioning if an upstream element is accidently misaligned. - 6|
Install the flip mirror (F1) so as to deflect the beam 90° to the left when the mirror is in place.
? TROUBLESHOOTING - 7|
Install the linear polarizer portion of the MA (MA2) in the beam path and rotate it to produce a minimum in the intensity of transmitted light. - 8|
Install the half-wave plate portion of the MA in the beam path just before the linear polarizer and rotate it to allow only about 50 mW of transmitted light.
! CAUTION Back reflections from either the linear polarizer or the half-wave plate that enter the laser will disrupt the mode-locking ability and/or the stability of the laser output. It is thus critical to rotate each of these elements slightly so that their back reflections miss entering the laser aperture. - 9|
Install mirror M5 to deflect the beam down the length of the Opal system.
Setting up the optical components in the Mai Tai-Opal beam path ● TIMING 2 h
- 10|
Tune the Mai Tai to 690 nm. No signal beam will be emitted from the OPO at this pump wavelength (see Fig. 5). - 11|
Move the flip mirror that was installed in the previous section out of the beam path so that the beam enters the Opal. - 12|
Remove the blocking filter from the output window of the Opal. Only the residual pump beam should be visible.
! CAUTION When properly tuned, the light output from the Opal is a combination of the signal output (the desired wavelength), a residual amount of the pump wavelength, and the sum frequency between the pump and the idler beams. These beams are not collinear and only the signal beam should be used for alignment. The filter attached to the output Brewster window eliminates all beams except for the signal.
? TROUBLESHOOTING - 13|
Install an iris (I3) close to the output of the Mai Tai and center it so that the beam passes evenly through its aperture. This iris may now be used to attempt to recover the Opal alignment if the input pump beam is accidentally moved. - 14|
Tune the Mai Tai to 750 nm and tune the Opal so that it generates 1,200-nm light at its output.
? TROUBLESHOOTING - 15|
Replace the blocking filter at the output window of the Opal. - 16|
Using the IR viewer, install the mirror M6 as shown in Figure 5, so that the beam is centered on the mirror and the reflected beam travels 90° to the original path and parallel to the table. - 17|
Install the linear polarizer of the MA (MA3) into the beam path and rotate it to produce a maximum of transmission.
▲ CRITICAL STEP This is opposite of what was required for the Step 7. Although both the Opal and the Mai Tai have horizontal polarization for their output beams, the Mai Tai beam traverses a periscope with a right angle turn that changes its polarization to vertical. Setting this element to produce a maximum of transmission returns both beams to the same vertical polarization. - 18|
Install the half-wave plate portion of the MA (MA3) in the beam path just before the linear polarizer and rotate it to produce less than 50 mW of transmitted power. - 19|
Flip the mirror (F1) from Figure 4 into the beam path. - 20|
Install the flip mirror (F2) so that the Mai Tai beam strikes the center of the mirror and the beam is reflected 90° to the left and parallel to the table. - 21|
Flip the mirror (F1) from Figure 4 back out of the beam path. - 22|
Flip the mirror (F2) out of the beam path. - 23|
Install mirror (M7) so as to redirect the beam 90° to the right. - 24|
Install the periscope (PS3) and adjust its bottom mirror so that the Opal beam strikes its center.
▲ CRITICAL STEP Ensure that enough space is retained for the Pockels cell (PC2). - 25|
Install the Pockels cell according to the vendor’s instructions.
▲ CRITICAL STEP When properly aligned, the Pockels cell should transmit > 90% of the input light when set for full retardation and < 0.01% when set for zero retardation. - 26|
Tune the Mai Tai to 1,040 nm and flip both F1 and F2 into the beam path. - 27|
By using mirrors M5 and F2, adjust the Mai Tai beam so that it passes through the Pockels cell (PC2) and has > 90% transmission when set for full retardation and < 0.01% when set for zero retardation.
? TROUBLESHOOTING - 28|
Install the shutter (S2) to block the beam reflected off of the top mirror of the periscope (PS3). These elements will be further aligned as described in the following section.
Figure 5.
Optical layout detail of multiphoton functional unit C—Mai Tai– Opal beam path. (a) Computer-aided design detail of functional unit C shown in Figures 1 and 2. (b) Optical block diagram of the area shown in a. The optical elements required for conditioning both the Opal OPO beam and the Mai Tai beam. F, flip mirror; I, iris; M, mirror; MA, manual attenuator; PC, Pockels cell; PS, periscope; S, shutter.
Installing the microscope and scanning optics ● TIMING 2 h
- 29|
Position the microscope on the optical table ~30 inches from the right side of the table and bolt it to the table (Fig. 6). - 30|
Place a sample containing small (approximately 5–10 µm) absorbing particles on the stage. Dried highlighter on a cover slip often works well. - 31|
View the sample using transillumination and adjust the microscope to attain Koehler illumination. - 32|
Center a small particle in the ocular field of view. - 33|
Turn the filter turret to the position containing the Laser Port B mirror and increase the transillumination lamp to its maximum intensity. Light will exit from the Laser Port B aperture. - 34|
Install the TL into the lens mount and position it so that a sharp image of the particle is transmitted straight out from the side of the microscope and travels parallel to the table.
▲ CRITICAL STEP The flat side of the lens should face the microscope. - 35|
Install the SL 225 mm away from the TL, ensuring that the transmitted light travels parallel to the table.
▲ CRITICAL STEP The flat side of the lens should face away from the microscope. - 36|
Install the galvanometer assembly next to the SL and using the three linear stages, position it so that all of the light strikes the top mirror and that the focus comes to a focus in between the two galvos. - 37|
Connect the power supply for the galvanometers and connect the laboratory power supply to the control voltage inputs. - 38|
Carefully adjust the control voltages, reversing polarity if necessary, to make the reflected image travel 90° from the SL path and parallel to the table. Record the voltages required for later use. - 39|
Install and position the two irises (I6 and I7) so that the image of the particle is centered through their apertures. These two irises now define the scanner’s incoming optical axis. - 40|
Install mirror M8 so that the image of the particle strikes the mirror center, and rotate it so that the beam is reflected by 90° to the left and travels parallel to the table.
Installing the detector box ● TIMING 3 h
- 41|
Assemble the detector box without any FCs or photodetectors (Fig. 7). - 42|
Change the position of the excitation-emission separation dichroic so that it is in the imaging path. Light will be emitted out of Laser Port C. - 43|
Install the threaded, frosted alignment disk into the port in the detector box opposite the collection lens. - 44|
Install the detector box onto the aperture of Laser Port C and adjust its height and position so that the image of the particle is centered on the alignment disk. - 45|
Install the FCs on the magnetic mounts into the detector box. - 46|
Move the alignment disk from the back wall to the first PMT position. - 47|
Adjust the actuator screws on the first FC to center the image of the particle on the alignment disk.
? TROUBLESHOOTING - 48|
Repeat Steps 46 and 47 for each subsequent FC and PMT position. - 49|
Install the PMTs, their lenses and their controller electronics.
▲ CRITICAL STEP The flat side of the lenses should face the PMTs. - 50|
Slide the excitation-emission separating dichroic out of the beam path.
Installing the components in the Tsunami beam path ● TIMING 2 h
- 51|
Install the optical breadboard of the GVD compensator on the table next to the Tsunami laser. - 52|
Turn on the Tsunami and adjust it such that the output is set to a wavelength close to the expected operating wavelength and is less than 50 mW in power. - 53|
Install the first mirror of the periscope (PS1) shown in Figure 3, such that the beam is centered on the mirror and is reflected directly up to the ceiling. - 54|
Install the second mirror of the periscope (PS1) so that the beam is centered on the mirror and is reflected 90° to the right and parallel to the table and to the GVD breadboard. - 55|
Install the square mirror (M1) so that the beam is centered horizontally on the mirror, but strikes the mirror about two beam diameters from its top. Adjust its angle so that the beam is reflected 90° toward the ceiling and parallel to the table. - 56|
Install the shutter so that the beam passes through the center of its aperture and open the shutter. - 57|
Install the linear polarizer portion of the MA (MA1) into the beam path. - 58|
Install the half-wave plate portion of the MA into the beam path. - 59|
Increase the power on the Tsunami laser to its normal operating power and rotate it such that there is <50 mW of transmitted power. - 60|
Install the Pockels cell according to the vendor’s instructions and connect its electronics. - 61|
Adjust the Pockels cell offset bias so that the transmitted beam power is < 50 mW. Note the power transmitted by the Pockels cell. - 62|
Install the stationary prism assembly (P1) so that the beam strikes the prism about two beam diameters from the bottom of the prism and two beam diameters from the apex of the prism. Note: the apex should be pointed toward the ceiling. - 63|
Rotate the prism in its post holder and observe the deflected beam. The beam will move in the same direction that the prism is being turned until a point at which the beam stops and reverses direction. Adjust the prism so that the beam is reflected to this maximum deflection point. - 64|
Install the fold mirror (M2) so that the beam strikes about one beam diameter below its center and is reflected by ~10°. - 65|
Install the moving prism assembly so that the beam passes about two beam diameters below the center of the prism and about two beam diameters from the apex of the prism. - 66|
Rotate the prism assembly in its post holder and observe the deflected beam. Adjust the prism so that the beam is maximally deflected. - 67|
Install mirror (M3) so that the deflected beam strikes its center and is reflected directly back down the path. - 68|
Adjust the vertical actuator on mirror (M3) to deflect the retroreflected beam so that it passes over the top of mirror M1 by about two beam diameters. - 69|
Install mirror (M4) such that the beam strikes the mirror in the center, is reflected by 90° to the left and travels parallel to the optical table. - 70|
Measure the power of the beam after mirror M4. The power loss should be < 20% when compared with that measured in Step 59.
? TROUBLESHOOTING - 71|
Shutter the Tsunami.
Mating the beam paths ● TIMING 2 h
- 72|
Install mirror (M8) such that the image of the particle is centered on the mirror, is reflected by 90° to the right and travels parallel to the table (Fig. 6). Position the mirror on the table such that the reflected beam strikes in the same plane as the top periscope mirror (PS3 from Fig. 5). - 73|
Adjust the height of the top periscope mirror (PS3 from Fig. 5), such that the image of the particle is centered on the mirror. - 74|
Install the beam-combining Di such that the image of the particle is centered on the mirror, is reflected by 90° to the left and travels parallel to the table. - 75|
Install mirror (M9) such that the image of the particle is centered on the mirror, is reflected by 90° to the left and travels parallel to the table. - 76|
Position the mirror (M9) such that the reflected image lies in plane with the last mirror (M4 in Fig. 5) of the GVD compensator. - 77|
Adjust the height of the mirror (M4 in Fig. 3) such that the image strikes the mirror center. - 78|
Open the Tsunami shutter. - 79|
Using the actuators on M9 and on the Di, guide the beam so that it passes through the center of both I6 and I7. - 80|
Close the Tsunami shutter. - 81|
Open the Opal shutter. - 82|
Using the actuators on PS3 and M8, guide the beam so that it passes through the center of both I6 and I7. - 83|
Flip the two mirrors F1 and F2 out of the beam path. - 84|
Adjust the Mai Tai to 750 nm and verify that the Opal is generating light at 1,200 nm. - 85|
By using the actuators on mirrors M6 and M7, guide the beam so that it passes though the center of both I6 an I7.
Fine tuning the GVD compensator ● TIMING 2 h
- 86|
Close the Opal shutter. - 87|
Open the Tsunami shutter, tune the Tsunami to 880 nm and ensure that it is pulsing. - 88|
Install the autocorrelator into the beam path according to the manufacturer’s instructions and place the detector head on the microscope stage. - 89|
Adjust the movable prism stage P2 (shown in Fig. 3) and measure the pulse width of the beam as a function of prism position (e.g., Fig. 8). - 90|
If the pulse width does not attain a minimum, adjust the spacing between P1 and P2, realign the alignment Tsunami beam and repeat Step 89 until a minimum is attained within the range of motion of the movable stage. This completes the optical alignment of the microscope.
Setting up the electronics ● TIMING 4 h
- 91|
Install the electronic boards in the computer. - 92|
Solder together the components for the blanking circuit according to the diagram shown in Figure 9. - 93|
Connect all of the electronics as shown in the diagram in Figure 10.
Color balancing and separation of channels ● TIMING 10 min
- 94|
While recording images, adjust laser powers and PMT gains so that the signal level recorded from a fluor that bleeds into two channels is balanced. This is most easily done when viewing a color-merged image. For example, when blue and green signals are balanced, they will appear cyan in color.
! CAUTION Care must be taken to avoid using laser powers high enough to cause photodamage or photobleaching to the sample. - 95|
Save the four channels of data in either 8-bit or 16-bit images. - 96|
Subtract the first channel from the second to create one separated channel. For example, when using CFP and GFP together, subtracting the blue data from the green data will leave an image containing only the GFP signal. Any signal that existed in both channels (as CFP will be) will result in a zero value in the resultant image, and any signal that existed solely in the CFP (such as a second harmonic signal) channel will result in a zero value in the resultant image because negative pixel values are set to zero in 8-bit or 16-bit images. - 97|
Subtract the second channel from the first to create another separated channel. For example, subtracting the green from the blue channels will result in an image residing solely in the blue channel for the same reasons stated in Step 96. - 98|
Steps 96 and 97 may be repeated for other overlapping channels (e.g., Texas Red and TagRFP657). Proper use of the technique can be verified by using a mixture of cells, in vitro, each separately labeled by a single fluorophore. This is demonstrated in Figure 14 using CellTracker Red CMPTX dye, a dye with spectral characteristics very similar to Texas Red.
Figure 14.
An in vitro demonstration of color subtraction technique. In vitro culture of MTLn3 tumor cells labeled with either CFP, GFP or TagRFP657, along with macrophages labeled by 2-h incubation in CellTracker Red CMTPX dye, all coinjected into a rat tail collagen I matrix gel. (a) The unprocessed signals acquired from each photodetector. (b) Unprocessed, merged image of four-channel data as acquired by multiphoton microscopy. (c) Spectral bleed-through of CFP into the GFP channel, as well as of Texas Red Dextran into the TagRFP657 channel, can be eliminated by balancing the signals within the respective detectors and then subtracting the blue from the green and the red from the farred channels. Cyan, SHG from collagen I fibers; blue, CFP-labeled MTLn3 tumor cells; green, GFP-labeled MTLn3 tumor cells; red, CellTracker Red CMTPX dye–labeled macrophages; white, TagRFP657-labeled MTLn3 tumor cells. (d) Merged image of the five separated channels obtained using the subtraction method described in this protocol. All animals were used according to protocols that have been reviewed and approved by Einstein’s Institutional Animal Care and Use Committee.
Set up plug-in for ImageJ ● TIMING 10 min
- 99|
Download and install a version of Java higher than 1.6. - 100|
Download and install a version of ImageJ higher than 1.41o. - 101|
Copy the ROI_Tracker plug-in jar file to the plug-ins folder under the ImageJ installation directory.
Using the ROI_Tracker plug-in ● TIMING 30 min
- 102|
Start ImageJ and import your 4D data set into ImageJ by using the ‘File/Import image sequence’ option. - 103|
Convert the data set to a hyperstack, specifying the number of channels, sections and time points present within. (Instructions for this conversion for various types of data sets are included in the documentation distributed with ROI_Tracker.) - 104|
Use ‘Image →Properties’ option to set voxel size and units, as well as the interval between time points (‘frame interval’). - 105|
Choose ROI_Tracker from the ‘Plug-ins’ menu. - 106|
Draw a selection around an object of interest (e.g., cell) using one of the ImageJ selection tools (freehand, point, elliptical, rectangular or line). - 107|
Click on ‘Add ROI’ to add the ROI to the Track. The program will calculate region parameters and display them within a table on the screen (Fig. 13). All of the region parameters (e.g., velocity, segment length) will be calculated in three dimensions.
? TROUBLESHOOTING - 108|
Advance to the next time point/_z_-section and repeat Steps 106 and 107 for this same object for each image in the data set. As more regions are added to a Track, markers for the centroid and a path connecting the centroids are drawn on the screen to facilitate visualization of the object progress. - 109|
When all time points have been completed, click on the ‘Add Track’ button to begin analyzing the movement of another object of interest. The parameters for the various Tracks may be viewed by selecting the appropriate Track using the Track dropdown field. - 110|
To access the ROI-associated data for further analysis and statistics, click on ‘Export’ and an ASCII file will be saved. To access the individual points of each ROI, click on the ‘Export ROI’ and an ASCII file will be saved. Ensure that the background option is checked and click on ‘Animate’ to generate a movie of the image sequence with animated ROIs overlaid.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 5.
TABLE 5.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 6 | Reflected beam position not repeatable when flip mirror is moved out of and returned to beam path | Mounting screws not tight enough | Tighten all mounting screws |
| 12 | No residual pump observed | Beam is blocked | Ensure beam is entering OPO properly |
| Beam entering OPO is misaligned | Realign/have vendor realign OPO system | ||
| 14 | No output light is generated | OPO is misaligned | Realign/have vendor realign OPO system |
| 27 | Pockels cell transmission is too low when set for full retardation or too high when set for zero retardation | Bias voltage not set correctly | Adjust bias voltage |
| Pockels cell misaligned | Realign according to manufacturer’s instructions | ||
| 47 | Actuators run out of range of motion and image is still not centered | Magnetic mounts are installed at an angle | Loosen and adjust magnetic mounts to decrease angle |
| 70 | Power loss is more than 20% | The polarization of the beam is incorrectly set | Repeat Steps 57 to 61 to ensure that the polarization of the transmitted beam is correct |
| 107 | Velocities are displayed as infinity symbols | The frame interval is not set | Save the Tracks, click on Image→Properties from the dropdown menus and enter the proper frame interval and then reload the Tracks |
● TIMING
- Steps 1–3, Setting up the table, lasers and microscope: 6 h
- Steps 4–9, Setting up the components in Mai Tai–Mai Tai pump beam path: 30 min
- Steps 10–28, Setting up the optical components in the Mai Tai-Opal beam path: 2 h
- Steps 29–40, Installing the microscope and scanning optics: 2 h
- Steps 41–50, Installing the detector box: 3 h
- Steps 51–71, Installing the components in the Tsunami beam path: 2 h
- Steps 72–85, Mating the beam paths: 2 h
- Steps 86–90, Fine tuning the GVD compensator: 2 h
- Steps 91–101, Setting up the electronics: 4 h
- Steps 102–110, Using the ROI_Tracker plug-in: 30 min.
ANTICIPATED RESULTS
The two-laser multiphoton microscope described in this protocol enables the simultaneous acquisition of fluorescence signal in four separate physical detectors covering the entire visible spectrum (blue, green, red and far-red) (Fig. 15). A typical image of the unprocessed data collected by the system is shown in Figure 15b, where signal from CFP-, GFP- and TagRFP657- labeled MTLn3 tumor cells, Texas Red Dextran–labeled macrophages and second harmonic generation from collagen I fibers are collected from a living mouse with the four simultaneously acquiring detectors (Fig. 15a). By using the signal balancing techniques described in this protocol, the five signals (Fig. 15d) detected by the four detectors may be completely separated from each other (Fig. 15c).
Figure 15.
In vivo images of exogenously grown tumor in immunodeficient mouse. With four simultaneously acquiring detectors and the color-balancing technique described in this protocol, five separate fluorescence signals coming from multiple cell types and stromal features may be observed. (a) The unprocessed signals acquired from each photodetector. Blue, SHG from collagen I fibers; cyan, CFP-labeled MTLn3 tumor cells; green, GFP-labeled MTLn3 tumor cells; red, Texas Red Dextran in the bloodstream and taken up by macrophages; white, TagRFP657-expressing MTLn3 tumor cells. (b) Unprocessed, merged image of four-channel data as acquired by multiphoton microscopy. (c) Spectral bleed-through of CFP into the GFP channel, as well as of Texas Red Dextran into the TagRFP657 channel, can be eliminated by balancing the signals within the respective detectors and then subtracting the blue from the green and the red from the far-red channels. Cyan, SHG from collagen I fibers; blue, CFP-labeled MTLn3 tumor cells; green, GFP-labeled MTLn3 tumor cells; red, Texas Red Dextran in the bloodstream and taken up by macrophages; white, TagRFP657-expressing MTLn3 tumor cells. (d) Merged image of the five separated channels. All animals were used according to protocols that have been reviewed and approved by Einstein’s Institutional Animal Care and Use Committee.
Further, the use of the over-clocking technique described in this protocol results in a marked improvement in signal-to-noise and image clarity that is readily observable in Figure 16. It is important to reiterate that this improvement is gained without sacrifice of scan rate.
Figure 16.
Comparison between traditional and over-clocked acquisition. Two images of a GFP-expressing PyMT-generated mammary tumor taken in vivo, demonstrating the difference in signal-to-noise and image quality between traditional image acquisition and over-clocked acquisition. Green, GFP-expressing tumor cells; red, macrophages labeled with Texas Red Dextran; blue, second harmonic generation from collagen I fibers. (a,b) The two images were acquired with (a) traditional acquisition scheme, and (b) over-clocking using 10 acquisition clocks per pixel. All other imaging parameters were kept constant. All animals were used according to protocols that have been reviewed and approved by Einstein’s Institutional Animal Care and Use Committee.
The unique capabilities of this microscope are particularly well suited to photoconversion microscopy. In this type of microscopy, cells of interest are labeled with a fluorescent protein, the excitation and emission spectra of which can be altered from one state to another. An example of this is the photoconvertable protein Dendra2 (ref. 19) which, in its unconverted state, has excitation and emission spectra that closely resemble that of GFP. However, on exposure to 405-nm light, both spectra shift to the red (Fig. 2). This red fluorescence then persists for a period of up to 2 weeks, during which time, the cells that have been selectively exposed to the 405-nm light can be easily distinguished from the rest of the green, nonconverted cells, and their progression and fate can be tracked. Our group has used this technique in several different studies20–23.
In particular, we have made a transgenic mouse that spontaneously generates mammary tumors that express Dendra2 (MMTV-iCre × CAGCAT-Dendra2 × MMTV-PyMT)1,22. Use of this transgenic mouse enables the establishment of a zero time point in the study of living tumors after which the stages of tumor progression may be examined.
To study the dissemination of tumor cells to sites distant from the primary tumor mass, we have utilized a 405-nm diode array to photoconvert the entire tumor transdermally and establish a zero time point. Before photoconversion, the entire tumor mass fluoresces green with very little red signal (Fig. 17a). After photoconversion, a large increase in red signal is observed (Fig. 17b).
Figure 17.
Use of the two-laser multiphoton microscope to study dissemination. By using a 405-nm diode array, a Dendra2-expressing PyMT-generated mammary tumor was photoconverted transdermally. (a) Before photoconversion, the tumor expresses the Dendra2 protein in its green form (left) and very little red signal is observed (right). (b) After photoconversion, a portion of the green protein (left) is photoconverted to red and the red signal is easily dectable using the 1,035-nm light from the Mai Tai laser (right). (c) After 5 d, tumor cells can be observed within the lungs (left and right). The green cells are those that have disseminated prior to photoconversion, and the red cells are those that disseminated after photoconversion. All animals were used according to protocols that have been reviewed and approved by Einstein’s Institutional Animal Care and Use Committee.
Examination of the lungs after 5 d reveals fluorescent tumor cells that have disseminated from the primary tumor mass. The tumor cells fluorescing green are those cells that have disseminated from the lung before photoconversion, and those fluorescing in red are the cells that had been photoconverted at time zero (Fig. 17c).
To study the migration and intravasation of tumor cells from the primary tumor mass, we have photoconverted a field of Dendra2-expressing mammary tumor cells next to a blood vessel (Fig. 18, dashed lines). Figure 18a shows time 0, immediately after conversion. After 8 min, one of the cells at the periphery invades out of the tumor mass and intravasates into the blood vessel (Fig. 18b). Although the large amount of scattering from erythrocytes greatly diminishes the brightness of the single cell, analysis and presentation of the data are greatly facilitated by use of the ROI_Tracker plug-in (Fig. 18c).
Figure 18.
ROI _Tracker analysis. Tumor migration and intravasation is imaged within a Dendra2-expressing PyMT-generated mammary tumor. (a) The circular region in the center of the field of view was photoconverted using the epi lamp with a 4,6-diamidino-2-phenylindole (DAPI) filter and with the field diaphragm stopped down, establishing a zero time point. (b) After 8 min, a tumor cell that has migrated from the photoconverted tumor mass can be observed in the blood vessel (arrow). (c) A series of stills taken every 2 min show the migration toward the blood vessel (0 and 2 min) and subsequent intravasation and passive transport of the tumor cell in the blood vessel (4–8 min). The average velocity of the migrating tumor cell before intravasation is 3.4 µm min−1, and is 10.0 µm min−1 after entering the blood vessel. Analysis within the ROI_Tracker software provides cell outlining and enables extraction of cellular parameters such as directionality and cell speed. The blood vessel is indicated by the yellow dashed lines. All animals were used according to protocols that have been reviewed and approved by Einstein’s Institutional Animal Care and Use Committee.
The ROI_Tracker plug-in enables the quantitative analysis of these cells. Figure 19 shows the type of analyses that can be done with the data extracted using ROI_Tracker. Depicted is a screenshot of the data extracted from ROI_Tracker and displayed in a spreadsheet. The fields colored in cyan and pink are the data directly exported from ROI_Tracker. The fields colored in green are values calculated from the data generating the Net Path Length, the Directionality and the Average Turning Frequency of the cells. ROI_Tracker also simplifies the quantification of parameters for objects moving along Tracks that cross imaging planes. An example of this type of movement is depicted in Figure 20. This figure shows a single tumor cell migrating from a tumor mass over a period of 8 min (2 min per frame). As the cell progresses, it disappears from the first imaging plane and appears in a deeper imaging plane. Analyzing this type of 3D movement using ROI_Tracker is simple, as all parameters are calculated on the basis of the full 3D coordinates at each time point.
Figure 19.
ROI _Tracker analysis. Screenshot of typical analysis of ROI_Tracker data in Excel. Fields in cyan and pink are unaltered data exported from ROI_Tracker. Fields in green are calculated values for Average Turning Frequency, Net Path, Directionality and so on. The inserted chart demonstrates the ability to visualize both the cellular paths and outlines in a quantitative manner.
Figure 20.
Tracking ability in ROI_Tracker Software. ROI_Tracker enables the tracking of 3D movement of tumor cells. A mosaic of time series data shows a cell moving between two imaging planes within solid tumor tissue. In the top row, 65 µm beneath the surface of CFP-labeled PyMT-generated tumor, a tumor cell (cyan) can be seen migrating along collagen I fibers (magenta). Partway into the sequence, the tumor cell disappears from the 65-µm plane and reappears deeper in the tissue at the 70-µm plane. Frames interval is 2 min. Cyan, PyMT tumor cells; magenta, second harmonic generation from collagen I fibers; blue, macrophages; red, Texas Red Dextran. All animals were used according to protocols that have been reviewed and approved by Einstein’s Institutional Animal Care and Use Committee.
Supplementary Material
Supp Material
Acknowledgments
This work was supported by grants to J.C. from the US National Institutes of Health (NCI100324), the National Cancer Institute’s Tumor Microenvironment Network, the Gruss Lipper Biophotonics Center and Mouse Models of Human Cancers Consortium; and grants to V.V.V. from the US National Institutes of Health (GM073913). B.G. was supported by a Charles H. Revson fellowship. We thank M. Metz, member of the Gruss Lipper Biophotonics Center, for his help with design and development. We also thank the members of the V.V.V. lab for useful discussions and M. Roh-Johnson for preparing the in vitro cell cultures.
Footnotes
AUTHOR CONTRIBUTIONS D.E., J.W. and J.C. designed the microscope and plug-in. D.E. built the microscope and wrote the plug-in. B.G. transfected proteins into cells and grew the mouse tumors. E.T.R. provided the ROI_Tracker analysis data. V.V.V. developed the TagRFP657 protein and its stably expressing MTLn3 tumor cell line. J.W.P., J.W. and J.C. developed the transgenic Dendra2 mouse model. D.E., J.W., B.G. and J.C. wrote the paper. J.C. defined the microscope performance characteristics required to address the biological application, and overall design was done by D.E. and J.C.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Wyckoff J, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 2.Hickman HD, Bennink JR, Yewdell JW. Caught in the act: intravital multiphoton microscopy of host-pathogen interactions. Cell Host Microbe. 2009;5:13–21. doi: 10.1016/j.chom.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bousso P, Bhakta NR, Lewis RS, Robey E. Dynamics of thymocyte-stromal cell interactions visualized by two-photon microscopy. Science. 2002;296:1876–1880. doi: 10.1126/science.1070945. [DOI] [PubMed] [Google Scholar]
- 4.Stoll S, Delon J, Brotz TM, Germain RN. Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science. 2002;296:1873–1876. doi: 10.1126/science.1071065. [DOI] [PubMed] [Google Scholar]
- 5.Sahai E, et al. Simultaneous imaging of, GFP, CFP and collagen in tumors in vivo using multiphoton microscopy. BMC Biotechnol. 2005;5:14. doi: 10.1186/1472-6750-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ducros M, et al. Spectral unmixing: analysis of performance in the olfactory bulb in vivo. PLoS One. 2009;4:e4418. doi: 10.1371/journal.pone.0004418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawano H, Kogure T, Abe Y, Mizuno H, Miyawaki A. Two-photon dual-color imaging using fluorescent proteins. Nat. Methods. 2008;5:373–374. doi: 10.1038/nmeth0508-373. [DOI] [PubMed] [Google Scholar]
- 8.Tillo SE, Hughes TE, Makarov NS, Rebane A, Drobizhev M. A new approach to dual-color two-photon microscopy with fluorescent proteins. BMC Biotechnol. 2010;10:6. doi: 10.1186/1472-6750-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piatkevich KD, Verkhusha VV. Advances in engineering of fluorescent proteins and photoactivatable proteins with red emission. Curr. Opin. Chem. Biol. 2010;14:23–29. doi: 10.1016/j.cbpa.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drobizhev M, Tillo S, Makarov NS, Hughes TE, Rebane A. Absolute two-photon absorption spectra and two-photon brightness of orange and red fluorescent proteins. J. Phys. Chem. B. 2009;113:855–859. doi: 10.1021/jp8087379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shcherbo D, et al. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 12.Morozova KS, et al. Far-red fluorescent protein excitable with red lasers for flow cytometry and superresolution STED nanoscopy. Biophys. J. 2010;99:L13–L15. doi: 10.1016/j.bpj.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andresen V, et al. Infrared multiphoton microscopy: subcellular-resolved deep tissue imaging. Curr. Opin. Biotechnol. 2009;20:54–62. doi: 10.1016/j.copbio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Herz J, et al. Expanding two-photon intravital microscopy to the infrared by means of optical parametric oscillator. Biophys. J. 2010;98:715–723. doi: 10.1016/j.bpj.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klauschen F, et al. Quantifying cellular interaction dynamics in 3D fluorescence microscopy data. Nat. Protoc. 2009;4:1305–1311. doi: 10.1038/nprot.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klauschen F, Qi H, Egen JG, Germain RN, Meier-Schellersheim M. Computational reconstruction of cell and tissue surfaces for modeling and data analysis. Nat. Protoc. 2009;4:1006–1012. doi: 10.1038/nprot.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296:1869–1873. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 18.Fork RL, Martinez OE, Gordon JP. Negative dispersion using pairs of prisms. Opt. Lett. 1984;9:150–152. doi: 10.1364/ol.9.000150. [DOI] [PubMed] [Google Scholar]
- 19.Gurskaya NG, et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat. Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 20.Kedrin D, et al. Intravital imaging of metastatic behavior through a mammary imaging window. Nat. Methods. 2008;5:1019–1021. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dovas A, et al. Visualisation of actin polymerization in invasive structures of macrophages and carcinoma cells using photoconvertible Beta-actin—Dendra2 fusion proteins. PloS One. 2011;6:e16485. doi: 10.1371/journal.pone.0016485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyckoff J, Gligorijevic B, Entenberg D, Segall J, Condeelis J. In: Live Cell Imaging: A Laboratory Manual. 2nd edn. Swedlow J, Goldman R, Spector D, editors. Cold Spring Harbor Press; 2009. pp. 409–422. [Google Scholar]
- 23.Gligorijevic B, Condeelis J. Stretching the timescale of intravital imaging in tumors. Cell Adh. Migr. 2009;3:313–315. doi: 10.4161/cam.3.4.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 25.Soeller C, Cannell MB. Construction of a two-photon microscope and optimisation of illumination pulse duration. Pflugers Arch. 1996;432:555–561. doi: 10.1007/s004240050169. [DOI] [PubMed] [Google Scholar]
- 26.Konig K, Simon U, Halbhuber KJ. 3D resolved two-photon fluorescence microscopy of living cells using a modified confocal laser scanning microscope. Cell Mol. Biol. (Noisy-le-grand) 1996;42:1181–1194. [PubMed] [Google Scholar]
- 27.Fan GY, et al. Video-rate scanning two-photon excitation fluorescence microscopy and ratio imaging with cameleons. Biophys. J. 1999;76:2412–2420. doi: 10.1016/S0006-3495(99)77396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campagnola PJ, Wei MD, Lewis A, Loew LM. High-resolution nonlinear optical imaging of live cells by second harmonic generation. Biophys. J. 1999;77:3341–3349. doi: 10.1016/S0006-3495(99)77165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majewska A, Yiu G, Yuste R. A custom-made two-photon microscope and deconvolution system. Pflugers. Arch. 2000;441:398–408. doi: 10.1007/s004240000435. [DOI] [PubMed] [Google Scholar]
- 30.Brown EB, et al. In vivo measurement of gene expression, angiogenesis and physiological function in tumors using multiphoton laser scanning microscopy. Nat. Med. 2001;7:864–868. doi: 10.1038/89997. [DOI] [PubMed] [Google Scholar]
- 31.Diaspro A, et al. Two-photon microscopy and spectroscopy based on a compact confocal scanning head. J. Biomed. Opt. 2001;6:300–310. doi: 10.1117/1.1382809. [DOI] [PubMed] [Google Scholar]
- 32.Wokosin DL, Squirrell JM, Eliceiri KW, White JG. Optical workstation with concurrent, independent multiphoton imaging and experimental laser microbeam capabilities. Rev. Sci. Instrum. 2003;74:193–201. doi: 10.1063/1.1524716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bird DK, Eliceiri KW, Fan CH, White JG. Simultaneous two-photon spectral and lifetime fluorescence microscopy. Appl. Opt. 2004;43:5173–5182. doi: 10.1364/ao.43.005173. [DOI] [PubMed] [Google Scholar]
- 34.Roorda RD, Hohl TM, Toledo-Crow R, Miesenbock G. Video-rate nonlinear microscopy of neuronal membrane dynamics with genetically encoded probes. J. Neurophysiol. 2004;92:609–621. doi: 10.1152/jn.00087.2004. [DOI] [PubMed] [Google Scholar]
- 35.Vicidomini G, et al. Characterization of uniform ultrathin layer for z-response measurements in three-dimensional section fluorescence microscopy. J. Microsc. 2007;225:88–95. doi: 10.1111/j.1365-2818.2007.01718.x. [DOI] [PubMed] [Google Scholar]
- 36.He W, Wang H, Hartmann LC, Cheng JX, Low PS. In vivo quantitation of rare circulating tumor cells by multiphoton intravital flow cytometry. Proc. Natl. Acad. Sci. USA. 2007;104:11760–11765. doi: 10.1073/pnas.0703875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han X, Burke RM, Zettel ML, Tang P, Brown EB. Second harmonic properties of tumor collagen: determining the structural relationship between reactive stroma and healthy stroma. Opt. Express. 2008;16:1846–1859. doi: 10.1364/oe.16.001846. [DOI] [PubMed] [Google Scholar]
- 38.Masters BR, So PT, Gratton E. Multiphoton excitation fluorescence microscopy and spectroscopy of in vivo human skin. Biophys. J. 1997;72:2405–2412. doi: 10.1016/S0006-3495(97)78886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan YP, Llano I, Hopt A, Wurriehausen F, Neher E. Fast scanning and efficient photodetection in a simple two-photon microscope. J. Neurosci. Methods. 1999;92:123–135. doi: 10.1016/s0165-0270(99)00103-x. [DOI] [PubMed] [Google Scholar]
- 40.Piston DW, Knobel SM. Quantitative imaging of metabolism by twophoton excitation microscopy. Methods Enzymol. 1999;307:351–368. doi: 10.1016/s0076-6879(99)07023-8. [DOI] [PubMed] [Google Scholar]
- 41.Mainen ZF, et al. Two-photon imaging in living brain slices. Methods. 1999;18:231–239. 181. doi: 10.1006/meth.1999.0776. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen QT, Callamaras N, Hsieh C, Parker I. Construction of a two-photon microscope for video-rate Ca(2+) imaging. Cell Calcium. 2001;30:383–393. doi: 10.1054/ceca.2001.0246. [DOI] [PubMed] [Google Scholar]
- 43.Tsai PS, et al. In: In Vivo Optical Imaging of Brain Function. Frostig RD, editor. CRC Press; 2002. pp. 113–171. [PubMed] [Google Scholar]
- 44.Zoumi A, Yeh A, Tromberg BJ. Imaging cells and extracellular matrix in vivo by using second-harmonic generation and two-photon excited fluorescence. Proc. Natl. Acad. Sci. USA. 2002;99:11014–11019. doi: 10.1073/pnas.172368799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Supatto W, et al. In vivo modulation of morphogenetic movements in Drosophila embryos with femtosecond laser pulses. Proc. Natl. Acad. Sci. USA. 2005;102:1047–1052. doi: 10.1073/pnas.0405316102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alencar H, Mahmood U, Kawano Y, Hirata T, Weissleder R. Novel multiwavelength microscopic scanner for mouse imaging. Neoplasia. 2005;7:977–983. doi: 10.1593/neo.05376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zinselmeyer BH, Lynch JN, Zhang X, Aoshi T, Miller MJ. Video-rate two-photon imaging of mouse footpad—a promising model for studying leukocyte recruitment dynamics during inflammation. Inflamm. Res. 2008;57:93–96. doi: 10.1007/s00011-007-7195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 49.Shcherbo D, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Material