Whole exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer (original) (raw)
. Author manuscript; available in PMC: 2014 Nov 1.
Published in final edited form as: Nat Biotechnol. 2014 Apr 20;32(5):479–484. doi: 10.1038/nbt.2892
Abstract
Comprehensive analyses of cancer genomes promise to inform prognoses and precise cancer treatments. A major barrier, however, is inaccessibility of metastatic tissue. A potential solution is to characterize circulating tumor cells (CTCs), but this requires overcoming the challenges of isolating rare cells and sequencing low-input material. Here we report an integrated process to isolate, qualify and sequence whole exomes of CTCs with high fidelity, using a census-based sequencing strategy. Power calculations suggest that mapping of >99.995% of the standard exome is possible in CTCs. We validated our process in two prostate cancer patients including one for whom we sequenced CTCs, a lymph node metastasis and nine cores of the primary tumor. Fifty-one of 73 CTC mutations (70%) were observed in matched tissue. Moreover, we identified 10 early-trunk and 56 metastatic-trunk mutations in the non-CTC tumor samples and found 90% and 73% of these, respectively, in CTC exomes. This study establishes a foundation for CTC genomics in the clinic.
Enabling precision medicine for each cancer patient depends on the ability to access samples that accurately represent the genomic features of their tumor1. Two critical bottlenecks, however, are that metastatic tissue is often inaccessible and the purity and yield of biopsy samples are low. To date, genomic characterization of cancer has emphasized large-scale sequencing of primary tumors and in few cases, metastatic lesions2. Both circulating tumor DNA (ctDNA)3 and circulating tumor cells (CTCs)4 represent alternative sources that may overcome these sampling challenges. Comprehensive sequencing and confident determination of genomic variants in CTCs could provide routine monitoring of transiting cells with potential for metastatic colonization to complement the static sampling of resected or biopsied lesions5.
Technologies for enriching and enumerating CTCs have provided prognostic value4,6,7, and characterizing specific regions, genes or patterns of gene expression in CTCs is both possible and useful. PCR-based methods, array CGH and high-throughput sequencing have revealed somatic single nucleotide variants (SSNVs) and copy number alterations8–10, and RNA sequencing has shown pathways implicated in metastasis11. For example, exome sequencing of lung cancer CTCs can uncover mutations shared with metastases10. However, without comprehensive power statistics, it remains difficult to assess the fraction of CTC exomes that are being robustly and accurately sequenced and whether such approaches apply to other cancers like prostate cancer.
Robust and accurate detection of SSNVs from CTCs is challenging. CTCs in a vial of blood are sparse12, and whole genome amplification (WGA) is necessary to construct sequencing libraries. Yields of amplified DNA vary among CTCs13, and WGA introduces amplification bias and polymerase errors14,15. “Census-based sequencing” of multiple libraries from the same sample (requiring a variant to be present in more than one), therefore, has helped to distinguish private mutations from polymerase errors with some fidelity14,15. Despite technical capabilities demonstrated for sequencing CTCs, no generalizable framework exists for confidently calling SSNVs, and design optimization of the experimental processes could provide a critical foundation for future comprehensive surveys of the genomics of CTCs across large numbers of samples.
Based on these considerations, we developed a modular set of experimental and analytical protocols for census-based whole exome sequencing (WES) and confident calling of SSNVs from prostate CTCs. We show these techniques can provide a window into the genetics of metastatic prostate cancer in a manner that is potentially useful clinically.
We first created a standardized process to generate and qualify multiple independent libraries for WES from CTCs recovered from one vial of blood. The process involves cell enrichment and isolation, genomic amplification, library qualification and census-based sequencing (Fig. 1a, Supplementary Fig. 1). We used the Illumina MagSweeper to enrich EpCAM-expressing CTCs16. The recovered cells, enriched with CTCs, were deposited into dense arrays of subnanoliter wells and imaged by automated epifluorescence imaging (Fig. 1b). Individual EpCAM (+) CD45 (−) CTCs were recovered by robotic micromanipulation for WGA using multiple displacement amplification (MDA). In all, this combined process reliably isolated single CTCs in a highly automated fashion (Supplementary Fig. 2).
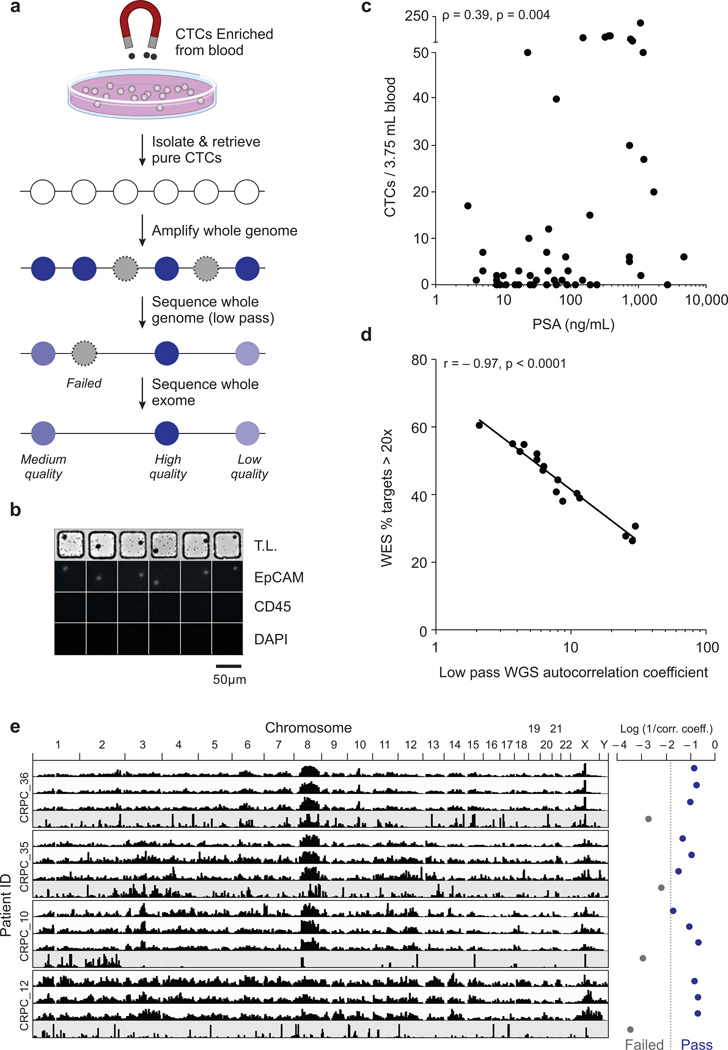
Figure 1.
Experimental process for sequencing of CTCs. (a) Schematic of workflow for the enrichment, isolation and sequencing of CTCs. (b) Sample micrographs of CTCs isolated in nanowells are shown with matched transmitted light (T.L.) and immunophenotyping for EpCAM, CD45, and DAPI by epifluorescence. Scale bar denotes 50 µm. (c) Scatter plot of the number of CTCs enumerated versus levels of PSA from 51 blood samples from 36 prostate cancer patients (Supplementary Table 1) screened using the MagSweeper for enrichment. CTC numbers in blood correlated with PSA levels (p = 0.004; Spearman, two-tailed). (d) Scatter plot of the percentage of target bases covered > 20× from whole exome sequencing (WES) versus the autocorrelation coefficient (Online Methods) calculated from low pass whole genome sequencing (WGS) over chromosome 1 for patient CRPC_36 (p < 0.0001; Pearson, two-tailed). WES yielded 124 ± 12× mean target coverage (Supplementary Table 2). WGS yielded mean coverage over chromosome 1 between 0.0003× and 0.03×, with a median of 0.017×. (e) Genome-wide read densities (1 Mb bins) from low pass WGS of CTC libraries from four different patients (CRPC_10, CRPC_12, CRPC_35, CRPC_36). Examples of three quality libraries and one poor library are shown per patient. The log of the inverse correlation coefficient was used to select high-quality libraries, with a cut off of −1.8 used here.
We next validated our method for isolating CTCs. The yield of tumor cells spiked into whole blood was ≥85% (Supplementary Fig. 3) and concurred with an independent method for enrichment (Veridex CellSearch) (Supplementary Fig. 4). We also performed low-coverage single-cell RNA-sequencing on cells recovered through our process from prostate cancer patients to confirm our isolated EpCAM (+) cells expressed prostate-specific antigen (PSA), confirming their prostate origins (Supplementary Fig. 5). We then enriched and enumerated CTCs from 51 blood samples of 36 patients with metastatic castration-resistant prostate cancer (CRPC) (Supplementary Table 1). The automated process was used for 45 of the 51 samples, and manual picking was performed for the remaining 6 samples. These samples yielded 0 to 200 CTCs per 3.75 mL of blood (median of 7 for samples with ≥1 CTCs, with 27% having no detectable CTCs); 45% of samples had ≥5, consistent with volume-adjusted counts previously reported in metastatic prostate cancer12. The number of CTCs also correlated with serum levels of PSA (p = 0.004; Spearman, two-tailed) (Fig. 1c).
We reasoned that establishing methods to assess the quality and uniformity of genome-wide coverage of CTC-derived sequencing libraries prior to in-depth WES or whole-genome sequencing (WGS) would help make census-based genomic sequencing of CTCs cost-efficient and facilitate subsequent analysis of SSNVs. To address this challenge, we first performed WGA on all single CTCs isolated from five patients with 20 or more CTCs and in which matched tumor tissue was available in tumor banks for comparison. As expected, the rates of success in amplification of single prostate CTCs varied widely (11–100%) (Supplementary Fig. 1), consistent with variability reported for amplifying lung cancer CTCs13. To assess the level of amplification bias in the recovered products, we developed a rapid and cost-effective method using low-pass WGS (< 0.05×) and autocorrelation analysis of single-base coverage to qualify libraries prior to WES (Online Methods). This metric accurately predicted the fraction of well-covered targets in subsequent WES (p < 0.0001; Pearson, two-tailed) (Fig. 1d). It also corresponded well to low-resolution views (1 Mb) of genome-wide read densities, highlighting prominent hallmarks of copy-number variants in prostate cancer, including chromosome 8q amplification, 8p deletion and amplification of chromosome X (Fig. 1e)17. When a library generated insufficient coverage (~ 0.0001×) to calculate an autocorrelation coefficient accurately, visual inspection of genome-wide read density could provide a qualitative means for qualification (Supplementary Fig. 6). Together, these examples demonstrate that our integrated experimental approach generates independent, high-quality libraries from single CTCs for WES and that low pass WGS can reliably predict which single CTC sequencing libraries are likely to yield high-quality data.
We then implemented a sensitive method to detect SSNVs from CTC libraries. We selected WES to generate high-coverage sequencing data of maximally informative genomic regions to enable cost-effective discovery of SSNVs. We sequenced 19 single CTC libraries of patient CRPC_36 to 124 ± 12× mean target coverage (Supplementary Table 2). As expected, individual libraries exhibited non-uniform coverage with only a fraction of the exome present (Supplementary Fig. 7a)15, and bimodal allelic distortion at sites of germline heterozygous SNPs, compared to normal distributions from bulk sequencing (Supplementary Fig. 7b). This effect caused improper genotyping at 38.3 ± 9.8% of such covered sites (Supplementary Fig. 8a). Coverage of either the alternate or both alleles ranged from 33.5 to 79.6% for individual CTCs and strongly correlated with the autocorrelation metric used for quality control (p < 0.0001; Spearman, two-tailed) (Fig. 2a).
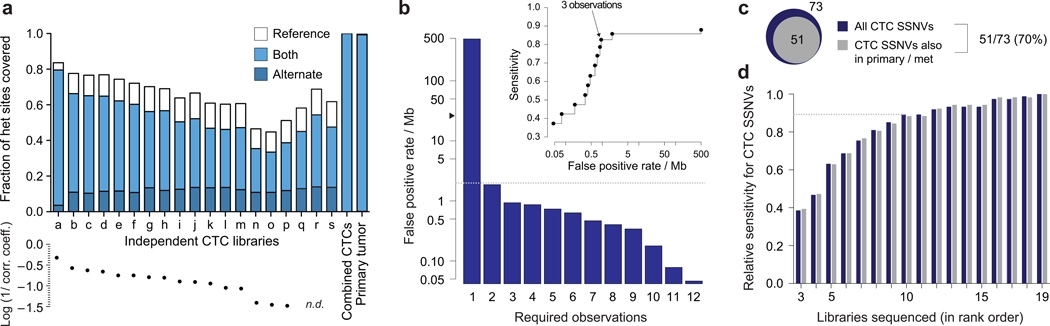
Figure 2.
Census-based variant calling from whole exome sequencing of CTCs from patient CRPC_36. (a) Characterization of allelic coverage in each CTC sequencing library from the same patient, compared to those libraries combined and primary tumor, as determined by 22,054 germline heterozygous SNP sites; an allele was scored as covered if there were ≥ 3 total reads of the particular allele(s). For reference, the autocorrelation coefficient is plotted below all CTC libraries except for three CTC libraries (n.d., not determined) that had insufficient low pass WGS coverage but passed quality control prior to exome sequencing based on visual inspection of genome-wide read densities (Supplementary Fig. 6). Coverage of the alternate allele (either alternate alone or both alleles) at germline heterozygous SNPs was correlated with the autocorrelation metric for individual CTC libraries (p < 0.0001; Spearman, two-tailed). When the individual CTC libraries were combined (“combined CTCs”), 99.995% of sites were covered by both alleles, similar to bulk sequencing of the primary tumor. (b) Estimation of false positive rate / Mb among 19 independent CTC libraries after requiring the variant to be observed in at least N independent CTC libraries (Supplementary Fig. 10). Grey dashed line indicates the reported mutation rate in bulk tumor sequencing of treated prostate cancer (~2 / Mb)19; black arrow head indicates the false positive rate / Mb observed for a single CTC library. Inset shows sensitivity versus false positive rate / Mb as a function of the required number N of independent observations of the variant. (c) The number of SSNVs called in total among 19 CTC libraries (73) and those that were validated as being present in matched tumor tissue (51) are shown. (d) Relative sensitivity to call CTC SSNVs (fraction of the total number called using 19 CTC libraries) as a function of the number of libraries sequenced, ranked in order by the autocorrelation coefficient (blue bars). A sustained improvement in sensitivity was observed. Additionally, considering only the 51 CTC SSNVs also observed in bulk whole exome sequencing of matched tumor tissue, we observed a very similar increase in sensitivity for each additional library sequenced (grey bars).
We hypothesized that combining data from independent CTC libraries would improve sensitivity. Indeed, the total coverage of both alleles (99.995%) at 22,054 SNP sites among 19 independent CTC libraries compared well to a representative bulk library from the primary tumor—only 0.005% of sites were improperly genotyped (Fig. 2a, Supplementary Fig. 8a). Analysis of whole exomes of CTCs from a second patient (CRPC_10) revealed similar extents of amplification bias that were also overcome by sequencing multiple independent libraries (Supplementary Fig. 7c,d, 8b,c). (We found that amplifying a single pool of CTCs was sensitive to the same allelic distortion as any other individual MDA-derived library (Supplementary Fig. 9a).) Together, these observations are consistent with stochastic loss of DNA from single cells, random preferential amplification of alleles and the lack of systematic coverage biases in MDA products14; they also confirm that sequencing multiple independent libraries of CTCs for a patient can enable robust, highly sensitive determination of variants.
We next sought to assess the specificity of this approach. We estimated an upper bound for the rate of false positives by assuming all variants, identified using MuTect18, not present in bulk tumor samples from the same patient were false positives (Online Methods). This assumption is conservative because contemporary CTCs may have diverged biologically from previously resected samples. Although amplifying and sequencing a pool of CTCs exhibited a false positive rate (~10/Mb) less than that for a single CTC library (~25/Mb), this rate was still insufficient for accurate calling of mutations on its own (Supplementary Fig. 9b). When combining multiple single CTC libraries, however, the false positive rate of called SSNVs dropped substantially from ~500 per Mb (N = 1 library) to a rate below the expected mutational rate in treated prostate cancer (~2 per Mb) when observed across two or more CTCs (Fig. 2b)14,15,19. When increasing the number of multiple observations required (Supplementary Fig. 10), the false positive rate further diminished to 0.9 per Mb (N = 3) with an estimated sensitivity of 82.6% (Fig. 2b, Inset). Analysis of six CTCs from CRPC_10 supported our statistical predictions from CRPC_36 that census-based sequencing also improves specificity (Supplementary Fig. 11a).
Applying this analytical method to the 19 CTCs from CRPC_36, we detected 73 SSNVs (N = 3; Supplementary Table 3). We found that 51 of these SSNVs (70%) were also present among nine cores from matched primary tumor and a lymph node metastasis, confirming that these EpCAM (+) cells were genetically related to the primary prostate cancer in this patient (Fig. 2c). Similarly applying this analytical method to the six CTCs from CRPC_10 sequenced to 89 ± 8 mean target coverage (Supplementary Table 2), 12 of 22 CTC SSNVs called (55%) (Supplementary Table 4) were also present among 12 cores from the primary tumor (Supplementary Fig. 11b). Overall, the sensitivity of this technique increases with the number of CTC libraries included in the analysis, and reached a relative sensitivity of 88% using 10 of 19 libraries from CRPC_36 (Fig. 2d).
The results above suggest that comprehensive sequencing of prostate CTCs and accurate assessments of SSNVs are possible with our approach. We then hypothesized that CTC sequencing could have clinical utility, perhaps providing a reasonable proxy for metastatic sampling in disseminated cancer. Clinical sequencing in metastatic prostate cancer is challenging because metastatic tissue is not routinely sampled20 and computed topographic (CT)–guided biopsy has a poor success rate with low purity of biopsied lesions21. For patient CRPC_36, there was widespread metastatic disease (Fig. 3a). Although the vast majority of the metastases (>10) were not available for sequencing, one neck lymph node had been resected six months prior to CTC collection. Of the 93 SSNVs detected in this metastasis, 47 (51%) were detected in CTCs (Fig. 3b). Owing to the timing of the sample acquisition, the CTCs sequenced could not have derived from this particular metastasis, so non-overlapping mutations could reflect divergent evolution at different sites, as previously demonstrated in prostate cancer22,23.
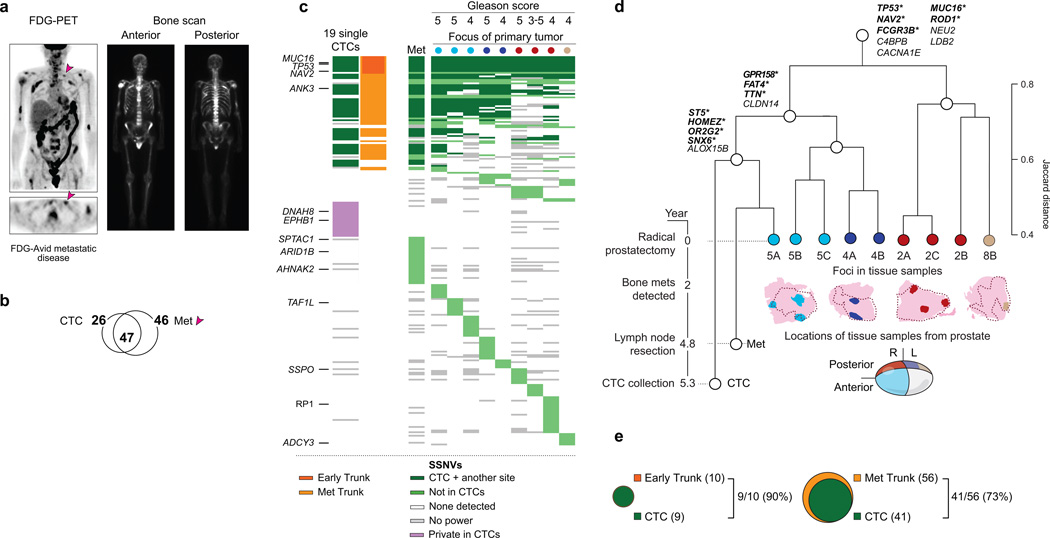
Figure 3.
Comparison of mutation pattern across CTCs, primary cores and metastasized tumor from patient CRPC_36. (a) FDG-PET and bone scans show widespread metastatic disease. FDG-PET Maximum Intensity Projection (MIP) image (top left) and axial FDG-PET slice (bottom left) demonstrate multifocal FDG-avid skeletal metastases throughout the axial and appendicular skeleton as well as bilateral cervical, left supraclavicular (arrow), retroperitoneal and bilateral common iliac metastatic lymphadenopathy. Bone scan demonstrates widespread bone metastasis. (b) Venn diagram representing mutations called in the CTCs and metastasis. Of note, 51% of mutations in the metastasis were called in CTCs. (c) Hierarchical clustering using the Jaccard index for mutations called across the nine primary cores, metastasized tumor and CTCs (when observed in ≥3 out of 19 single CTCs). Only sites in the exomes that were considered to be powered for mutation calling, as described in Online Methods, were included in this analysis. Shading of green represents presence in CTCs and at least one other sample (dark green) or not present in CTCs (light green). Genes highlighted indicate non-synonymous mutations present in >2 patients from a previous sequencing study in prostate cancer31. Of note, one of the cores included regions of both Gleason 3 and Gleason 5 cancer. (d) Dendrogram representing hierarchical clustering by the Jaccard index, and timeline of sample acquisition. SSNVs detected in all individual cores of tissue (early trunk), or in all cores that belong to only one of the two branchpoints of the clustering dendrogram are listed. Non-synonymous mutations are highlighted in bold with “*”. The areas shaded in pink represent the pathology blocks from which cores of tissue were obtained (drawn to scale). The dotted lines represent the area with histological presence of tumor within each block. The sites from which the individual cores of tissue were obtained for sequencing are displayed in colors corresponding to the cluster dendrogram and Fig. 3c. The regions of the prostate from which the pathology blocks were retrieved are schematized. (e) The number of mutations found in the metastasis and at least one core of the primary tumor (metastatic trunk), the early trunk mutations, and the overlap of these with CTC mutations are shown, excluding sites that were consistently underpowered in greater than half of the samples. The one early trunk mutation not detected in CTCs (N ≥ 3) was observed in 2 CTCs.
We next asked whether sequencing CTCs could uncover mutations present early in tumor evolution (trunk mutations) or in the inferred metastatic precursor (metastatic trunk mutations)24. Such founder mutations in other cancer types (e.g. BRAF in malignant melanoma or KIT in GIST) represent excellent therapeutic targets25,26. Detecting such mutations in metastatic patients from other tumor types via simple blood draws might therefore have considerable clinical utility.
We compared the landscape of mutations in CTCs and the metastatic sample to multiple samples from the patient’s primary prostate tumor resected 5.3 years earlier. Because prostate cancer is often histologically multifocal27, we sequenced nine spatially distinct foci of the primary tumor in regions of uniform Gleason grade (Gleason 4 or 5) with one exception noted (Fig. 3c). To assess relationships between these foci, the CTCs and the metastasis sample, we performed hierarchical clustering, excluding sites consistently underpowered in more than half of the samples (owing to lack of coverage) (Fig. 3c). Indeed, the primary tumor foci exhibited marked heterogeneity, but as expected, ones from similar physical regions of the tumor were more closely related to each other than those from other locations (Fig. 3c,d). Notably, we also identified one particular focus that most closely resembled the CTCs and the metastasis, suggesting that this focus may share a more recent common ancestor with the CTCs than other foci. Alhough this focus was Gleason 5, the score itself did not predict the likely metastatic precursor since other Gleason 5 regions were not in this evolutionary branch.
We found ten SSNVs, including a mutation in TP53, ubiquitous among all primary foci and metastasis, suggesting the cancer arose from a single ancestor with divergent evolution thereafter (Fig. 3d). The CTCs had nine of ten (90%) of these early trunk mutations (Fig. 3e). Notably, despite allelic distortion, these were present in a greater fraction of CTCs (corrected for power) than the non-trunk mutations on average (p = 0.012; Wilcoxon rank sum test). Fifty-six mutations were present in both the metastasis and primary tumor (any foci), and the CTCs had 41 (73%) of these metastatic trunk mutations (Fig. 3e). For patient CRPC_10, we found the CTCs had all three early trunk mutations for which they were powered (out of nine) (Supplementary Fig. 11c). Together, these proof-of-concept data support the notion that CTC sequencing can reveal early mutations in tumor evolution and those that could be shared among metastatic sites. As such trunk mutations are likely to be present in the majority of sites in advanced cancer patients, these results suggest clinical utility for systematic CTC genomics.
Here we have demonstrated the feasibility of sequencing whole exomes of prostate CTCs and confidently calling SSNVs to provide a minimally invasive window into the mutational landscape of metastatic prostate cancer. We implemented a systematic process to obtain, qualify and sequence whole exomes of CTCs and call SSNVs. Applying this process to two individual patients showed that sequencing of multiple independent CTC libraries can achieve full coverage of the exome territory accessible in bulk sequencing and a false positive rate below the expected mutational rate in prostate cancer.
As implemented, the current process works well for patients from whom five or more single CTCs are recovered from 3.75mL of blood and high-quality libraries generated for sequencing. The numbers of CTCs can vary substantially among different types of cancer12, so sequencing of CTCs may not directly apply to all cancer patients. Nonetheless, advancing the individual technologies used to recover and amplify as many CTCs as possible from patients would increase the numbers of cancers for which this approach could benefit. For instance, enriching CTCs based on physical separation or microfluidics rather than expression of EpCAM6, processing greater volumes of blood or using other means of WGA that improve uniformity in genome-wide coverage of the amplified DNA could all increase the numbers of CTC libraries available15. The designed modularity of our approach can accommodate new emerging operations for the enrichment and isolation of CTCs, WGA30 and sequencing platforms. Furthermore, as the costs of library preparation, hybrid selection and sequencing decline, we anticipate census-based sequencing will become more cost-effective for monitoring more patients.
The approach here does not emphasize private mutations that may be held by individual CTCs. Such analysis of heterogeneity is extremely challenging because variants in individual cells can either be absent for technical (allelic distortion, false positive) or biological (subclonality in the population) reasons. Although we identified many mutations in single libraries, our census-based approach deprioritizes them because we could not distinguish them from false positives. Exome sequencing of ctDNA has also demonstrated concordance of variants with tumor biopsies29. Although such DNA is fragmented and has similarly low abundance, these materials may provide a complementary source for reducing false positive calls or revealing other mutations not sampled among sparse CTCs. Nonetheless, our results suggest that CTC sequencing could augment both large-scale efforts to map the genetics of cancer and clinical sequencing from individual cancer patients where a focus on evolutionarily early and shared metastatic events such as those identified by the proof-of-concept case herein are critical for precision medicine. The integrated process may also enable longitudinal monitoring of the genetic state of disseminated cancer, revealing important insights in tumor evolution, metastatic dissemination and the resistance to therapeutics.
ONLINE METHODS
Patient recruitment
Eligible patients were metastatic castration-resistant prostate cancer (CRPC) patients who had 1) progression on a phase II study of abiraterone in combination with dutasteride (DFCI Protocol # 10–448, IRB expiration date 2/7/2014) or 2) PSA >20 ng/ml and progressive disease based on rising PSA and scan progression to enrich for patients likely to have detectable circulating tumor cells. There is no PSA cutoff for the phase II study itself. The Prostate Clinical Research Information System (CRIS) database at Dana-Farber Cancer Institute was used to identify metastatic CRPC patients. The CRIS system comprises data-entry software, a central data repository, collection of patient data including comprehensive follow-up of all patients, and tightly integrated security measures, as previously described33. All patients provided written informed consent to allow the collection of tissue and blood and the analysis of clinical and genetic data for research purposes (DFCI Protocol # 01–045, IRB expiration date 3/20/2014). After initial screening of patients with metastatic CRPC, chart review was performed by a physician to identify those who had progressive disease described above. Blood specimens were prospectively collected from eligible patients. Refer to Supplementary Table 1 for patient information. Blood was drawn in EDTA tubes and transported at room temperature to the Broad Institute within 3 hours.
Patient characteristics
The clinical course of the patient described here was as follows: At the age of 54, the patient was diagnosed with a T3, Gleason 9, PSA 10 prostate cancer. He was treated on a research trial of neoadjuvant docetaxel and bevacizumab. At prostatectomy he had a pT3b, N1 tumor. He next received adjuvant radiation and androgen deprivation therapy (ADT). Upon rising PSA continuous ADT was reinitiated and he was treated with sequential bicalutamide and nilutamide. Seventeen months after initial diagnosis he developed metastases and was treated with the following therapies until his death 6 years after his presentation: docetaxel, phase 1 trial of PI3-kinase/MEK kinase inhibitor, sipuleucel-T, XL-184, abiraterone, enzalutamide, cabazetaxel, palliative radiation to bone and combined abiraterone/enzalutamide. At the time of CTC isolation, he received abiraterone/enzalutamide, and a lymph node biopsy was performed while he received enzalutamide. A basic summary of the timeline of events is illustrated in Fig. 3d.
Enrichment of CTCs from blood
For each 3.75 mL of blood, red blood cell (RBC) lysis was first performed using 1× Pharm Lyse solution (BD Biosciences). The RBC-depleted sample was then incubated with 4 tests of FITC-anti-CD45 (eBiosciences; clone HI30) for 30 min at 4 °C, followed by incubation with anti- EpCAM magnetic beads (Illumina)16 for 30 min at 4 °C. PE-anti-EpCAM (BD Biosciences; clone EBA-1) was then added for 30 min at 4 °C prior to immunomagnetic isolation using the Illumina MagSweeper16.
Isolation of CTCs
Isolation of CTCs from the enriched samples from the MagSweeper was performed using either the nanowell-based method with automated imaging and robotic retrieval of single cells, or a 6-well dish with manual imaging and pipetting. For the nanowell-based approach, enriched samples were loaded into the wells of a 1 × 3 inch polydimethylsiloxane nanowell device containing a 24 × 72 array of 7 × 7 wells, each of the dimensions 50 × 50 × 50 µm (Supplementary Fig. 2)34. Automated epifluorescence imaging of the array was performed (Zeiss) and images were processed using a custom software program. Following manual review of candidate cells via a custom CTC analysis software (EVA), candidate CTCs (DAPI− CD45− EpCAM+) were retrieved from individual wells of the device using an automated robotic micromanipulator (CellCelector, ALS) and deposited within 3 µL droplets of Superblock/PBS (Thermo Scientific) into empty wells of a 96 well PCR plate. For the manual approach, candidate CTCs (DAPI− CD45− EpCAM+) were recovered from Superblock/PBS by pipetting 3 µL into a 96 well PCR plate. PCR plates was frozen down at −80 °C until ready for further processing.
Lysis and whole genome amplification (WGA) of CTCs
Each PCR plate containing frozen CTCs was thawed on ice and the volume of individual wells was diluted to 5 µL using UltraPure water (Invitrogen). 5 µL of lysis buffer, containing 0.4 M KOH (Sigma Aldrich) and 80 mM DTT (Qiagen), was added to each well and the plate was sealed, gently shaken to mix, spun down at 300 rcf for 1 min, and incubated for 10 min at 50 °C using a thermal cycler (Eppendorf). Following lysis, the plate was spun down at 300 rcf for 1 min, and 5 µL of 0.4 M HCl (Fluka), was added to each well, and the plate was kept on ice. Master mix for whole genome amplification by multiple displacement amplification (MDA) was prepared by adding, for each reaction, 26.25 µL of sterile water, 5 µL of 10× reaction buffer from the RepliPHI kit (Epicentre), 0.5 µL of 10 mg/mL BSA (NEB), 0.2 µL of 1 M DTT (Qiagen), 0.8 µL of 25 mM dNTPs from the RepliPHI kit (Epicentre), 1.25 µL of 10 mM random hexamers (NNNN*N*N) from IDT, and 1 µL of RepliPHI enzyme from the RepliPHI kit (EpiCentre). 35 µL of this master mix was added to each well of the PCR plate, containing lysed genomic DNA, and incubated for 2 h at 30 °C on a thermal cycler (Eppendorf). Following the MDA reaction, clean up was performed using AmpureXP beads (Beckman Coulter). Briefly, 100 µL of AmpureXP beads was added to each sample and incubated for 5 min at room temperature. The samples were then placed on a 96-well plate magnet (Invitrogen) and incubated for 5 min. Supernatant was removed from each sample and 100 µL of fresh 70% ethanol (Koptec) was added and removed twice to wash the beads. Following complete removal of the ethanol and drying for 10 min at room temperature, beads were resuspended in 60 µL of Tris-EDTA buffer, pH 8 (Teknova), incubated for 5 min at room temperature, placed back on the magnet for 5 min, then cleaned-up products were removed to a new PCR plate. These MDA products were quantified using the Quant-IT PicoGreen dsDNA assay kit (Invitrogen), and products with concentrations greater than the negative control were selected for low-pass whole-genome sequencing (WGS).
Library preparation and low-pass whole genome sequencing
Whole genome sequencing libraries were prepared using the Nextera DNA Sample Prep Kit (Illumina), quantified using the Library Quantification Kit for Illumina (Kapa Biosystems), and pooled and loaded at 12 pM onto the Illumina MiSeq sequencer using the MiSeq Reagent Kit v2 (Illumina). Up to 96 libraries can be multiplexed in the same run. The MiSeq Reporter (Illumina) was used to align reads and generate BAM files, and IGV Tools (Broad Institute) was used to bin the genome for coverage at 1 Mb intervals and generate TDF files. The TDF files were viewed in the Integrative Genomics Viewer (IGV) to visually inspect genome-wide uniformity in coverage of each MDA product.
Calculation of autocorrelation coefficient and selection of CTC libraries
Using the data from low-pass whole genome sequencing, we computed the degree of correlation in single-base coverage over various distances, normalized by the mean target coverage, for each library. The Depth Of Coverage module from GATK was used to compute single-base coverage (http://www.broadinstitute.org/gatk/) using a minimum mapping quality of 5, and the autocorrelation coefficient represents the magnitude (not the length-scale or genomics distance of correlation) of the correlation in single base coverage at 1 kb distances normalized by the mean sequencing depth. 1 kb represents a length scale well above the average fragment length, yet short enough to capture local biases in coverage due to preferential overamplification in WGA. In our study, the autocorrelation analysis was computed over chromosome 1 because it is the largest chromosome, does not have visible copy-number alterations from both WGS and WES read coverage, and provides the analysis the most statistical power. The analysis could have been performed on other chromosomes too. Chromothripsis, although rare, would only affect the correlation near translocation junctions, which would be a small fraction of the chromosome and would have negligible effects on the analysis.
Libraries were ranked based on autocorrelation coefficient. In this study, we selected libraries for whole exome sequencing that had the logarithm of (1 / autocorrelation coefficient) greater than −1.8. In the rare event of insufficient coverage (~0.0001×) for computing of the autocorrelation coefficient, visual inspection of genome-wide read densities may be used to include samples with seemingly uniform genomic coverage, as demonstrated in Supplementary Fig. 6.
Isolation of genomic DNA from blood and tumor tissue
Genomic DNA was isolated from blood to control for germline variants using the DNeasy Blood and Tissue Kit (Qiagen). 100 µL of anticoagulated blood was added to 20 µL proteinase K and adjusted to 220 µL volume with PBS. 200 µL of Buffer AL was added, mixed, and incubated at 56 °C for 10 minutes. All subsequent steps were performed per the manufacturer’s recommendations.
Genomic DNA and RNA were isolated from primary tumor tissue using the AllPrep DNA/RNA Mini Kit (Qiagen), and from metastatic tumor tissue using AllPrep DNA/RNA Micro Kit (Qiagen) following the manufacturer’s recommendations. Primary tumor tissue consisted of blocks of fresh frozen tissue acquired at the time of radical prostatectomy, frozen in OCT medium, and stored in liquid nitrogen at the Gelb Center for Translational Research. Accompanying slides were reviewed by a pathologist and tumor boundaries were marked at the time of storage. Slides were re-reviewed at the time of retrieval for presence of tumor, and regions of Gleason 3, 4, and 5 within the tumor were identified and areas were marked. Nine representative tumor foci were chosen to maximize distance between the cores, favoring higher Gleason grade regions. Each block was removed from the cassette, placed on a petri dish on dry ice to keep cold, and aligned with the accompanying marked slide to identify the selected foci. Blocks were cored using a Miltex 2mm Disposable Biopsy Punch with Plunger, placed into a DNA LoBind Eppendorf tube, and stored at −80 °C until time of nucleic acid extraction.
Metastatic tumor tissue consisted of blocks of fresh frozen tissue acquired from excision of left supraclavicular lymph node and frozen in OCT medium. Tumor shavings were obtained at the time of sectioning the OCT block for immunohistochemistry. About 100–300 micron shavings were obtained using a cryostat, placed into a DNA LoBind Eppendorf tube, and stored at −80 °C until time of nucleic acid extraction.
Selection of libraries and whole exome sequencing
Whole exome sequencing was performed as previously described35. Briefly, 100 ng of DNA from each sample was used for library preparation, which included shearing and ligation of sequencing adaptors. Exome capture was performed using the Agilent v2 Human Exon bait kit. Captured DNA was sequenced using the Illumina HiSeq platform, and paired-end sequencing reads were generated for each sample. Initial alignment and quality control were performed using the Picard and Firehose pipelines at the Broad Institute. Picard generates a single BAM file for each sample that includes reads, calibrated quantities, and alignments to the genome. Firehose represents a set of tools for analyzing sequencing data from tumor and matched normal DNA. The pipeline uses GenePattern as its execution engine, and performs quality control, local realignment, mutation calling, and coverage calculations, among other analyses. Complete details of this pipeline can be found in Stransky et al 201136 or at the following website: www.broadinstitute.org/cancer/cga/. Sequencing was performed to an average target coverage of > 120×.
Calling of SSNVs from whole exome sequencing data
Reads were aligned to the reference human genome build hg19 through implementation of the Burrows-Wheeler Aligner37 and processed through Picard38. The Firehose pipeline (www.broadinstitute.org/cancer/cga) was used to manage input and output files and submit analyses for execution. MuTect was used to identify somatic single-nucleotide variants (SSNVs) in targeted exons by Bayesian statistical analysis of bases and their qualities in the tumor and normal BAMs at each given genomic locus; the MuTect publication describes the specificity and sensitivity of the method18. Reads from all SSNV candidates were then realigned more stringently by disregarding read pair information to reduce alignment based artifacts. All SSNVs were subjected to filtering against a large panel of normal samples, in order to remove common artifacts that escaped the original calling algorithms39. Further, only sites within chromosomes one through 22 and X were considered. Sites in the exomes of primary samples and metastasis were considered to be powered for mutation calling if the number of reads at a site allowed for a 0.9 probability of observing 3 supporting reads of the alternate allele, considering purity of the sample and assuming a minimum cancer cell fraction of 0.1. Sites were considered powered in CTCs if 5 CTCs had coverage of > 3 reads (this achieves ~98% power to detect a clonal mutation, based on a maximum loss of coverage of 14.4% of the alternate allele only across the CTCs as determined in Fig. 2a).
Calculation of False Positive Rate
To calculate the effective false positive rate of the method, we computed the total number of potential false positive events, and the territory at risk for these events. To calculate an upper bound for the number of events, we assumed every event was a false positive after removing events seen independently in at least one of the primary tumor cores or metastasis. To calculate the denominator of the false positive rate, the number of bases that were at risk for a mistake being made, we considered the effects of bi-allelic dropout as this leads to regions of the genome with no coverage, and thus no possibility of false positive occurring. Bi-allelic drop out was calculated from germline heterozygous single-nucleotide polymorphisms (SNPs) (see Fig. 2b). We then used a binomial model to calculate the probability of having k or more observations in n CTCs using 1 minus the median value of the estimated fraction of sites without bi-allelic dropout (0.33) as the probability of each CTC having coverage and multiplied by the approximate size of the original targeted exome territory (~32 Mb) to arrive at the total number of bases at risk.
RNA sequencing of single CTCs
Single CTCs were recovered, using the nanowell-based isolation platform, into individual wells of a 96 well plate containing 10 µL of buffer TCL (Qiagen) supplemented with 1% 2-mercaptoethanol (Sigma), spun down, snap frozen on dry ice, and stored at −80 °C until further processing. Next, RNA from each single CTC was isolated, reverse transcribed, and amplified using the SMARTer Ultra-low RNA kit (Clontech) as previously described32. Afterwards, cDNA libraries were prepared using Nextera XT DNA Sample preparation reagents (Illumina) as per the manufacturer’s recommendations, with minor modifications. Specifically, reactions were run at ¼ the recommended volume, the tagmentation step was extended to 10 minutes, and the extension time during the PCR step was increased from 30s to 60s. After the PCR step, all 96 samples were pooled without library normalization, cleaned twice with 0.9× AMPure XP SPRI beads (Beckman Coulter), and eluted in Tris-EDTA buffer, pH 8 (Teknova). The pooled libraries were quantified using Quant-IT DNA High-Sensitivity Assay Kit (Invitrogen) and examined using a high sensitivity DNA chip (Agilent). Finally, samples were sequenced using a MiSeq sequencer (Illumina).
Analysis of RNA sequencing data
Raw sequencing data were processed as described previously32, except that there was no need to trim SMARTer short and long adapter sequences due to the Nextera library preparation40. Short sequencing reads were aligned to the UCSC hg19 transcriptome. These alignments were used to estimate transcriptomic alignment rates, and were also used as input in RSEM v 1.1241 to quantify gene expression levels (transcripts per million; TPM) for all UCSC hg19 genes in all samples. Genomic mappings were performed with Tophat v. 1.4142, and the resulting alignments were used to calculate genomic mapping rates, rRNA contamination, and 3’ and 5’ positional bias38.
Cell spike-in experiments
LNCaP prostate cancer cells (ATCC) were cultured in RPMI-1640 medium (Corning, Cellgro) supplemented with 10% fetal bovine serum (FBS, Sigma). To determine surface expression of EpCAM, LNCaP cells were stained with anti PE-anti-EpCAM (BD Biosciences; clone EBA-1) and the level of expression was determined by flow cytometry on a LSRII (BD Biosciences), compared to unstained control. For technical validation of the MagSweeper enrichment procedure, LNCaP cells were labeled with carboxyfluorescein diacetate succinimidyl ester (CFDA, Invitrogen) and spiked into normal blood (obtained from Research Blood Components, LLC) at the indicated concentrations prior to MagSweeper enrichment. Successful isolation of LNCaP cells was determined by determining the number of CFDA-labeled cells by microscopy.
Supplementary Material
1
2
3
4
ACKNOWLEDGEMENTS
J.G.L. was supported by a Conquer Cancer Foundation Young Investigator Award, and NIH grant 5P50CA100707-10 (DF/HCC SPORE), and the Wong Family Award. V.A.A. was supported in part by a graduate fellowship from the National Science Foundation. A.D.C is supported by the Prostate Cancer Foundation Young Investigator Award and the DoD Physician Scientist Training Award. J.C.L. is a Camille Dreyfus Teacher-Scholar. The authors acknowledge the Arthur and Linda Gelb Center for Translational Research for the acquisition and annotation of clinical samples, and Amanda Abbott and Annick Van Den Abbeele from the DFCI Department of Imaging for PET images. The authors also acknowledge Priscilla K. Brastianos and Ian Dunn for contributing samples for CTC analysis, David Peck for help with technology development, Olga Voznesensky and Steven Balk for purification of DNA from the metastatic tumor for sequencing, Charlie Whittaker and Stuart S Levine for advice on sequencing and analysis, and the Broad Genomics Platform for the development of new sequencing approaches used here. This work was also supported in part by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute, and we thank the Koch Institute Swanson Biotechnology Center for technical support, specifically the BioMicroCenter. This work was also supported in part by Janssen Pharmaceuticals, Inc. The authors dedicate this paper to the memory of Officer Sean Collier, for his caring service to the MIT community and for his sacrifice.
Footnotes
AUTHOR CONTRIBUTIONS
J.G.L., V.A.A. designed and performed experiments, analyzed data and wrote the manuscript. K.C., M.R developed computational methods, analyzed data and wrote the manuscript. A.D.C. provided clinical samples, patient data, and analyzed clinical data. P.C.G., N.T., S.K. performed experiments for isolating CTCs. J.F. developed single-cell sequencing methods and designed experiments. C.Z.Z. analyzed data and applied the autocorrelation methods. A.K.S., R.S., J.T.T., D.L. performed single-cell RNA sequencing and data analysis. N.T. developed code for determining CTCs to recover from nanowells. B.S.B. performed early technology development. C.S., D.A. performed sample and data management and gave conceptual advice. A.L., A.L. performed experiments comparing process to the Veridex CellSearch System. E.M.V. analyzed sequencing data. M.N., G.M.L., T.L., M.S.C. coordinated and acquired clinical samples. R.T.L. reviewed pathology slides and guided selection of clinical samples. T.E.C. provided samples and validated methods for isolating CTCs. M.T., M.L., A.R., M.M., W.C.H., P.W.K. supervised experiments, sample and data collection and edited manuscript. T.R.G., G.G., J.S.B. and J.C.L. designed the experimental strategy, supervised the analysis, and wrote the manuscript. All authors discussed the results and implications and reviewed the manuscript.
COMPETING FINANCIAL INTERESTS
J.C.L. is a founder and shareholder of Enumeral Biomedical Corp., holding a license for a patent on the specific design of the nanowells used in this study. All other authors declare no conflict of interest.
ACCESSIONS
phs000717.v1.p1
REFERENCES
- 1.Garraway LA. Genomics-driven oncology: framework for an emerging paradigm. J. Clin. Oncol. 2013;31:1806–1814. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 2.International Cancer Genome Consortium et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson S-J, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 4.Cristofanilli M, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 2013;5:180ra48. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu M, et al. A. Circulating tumor cells: approaches to isolation and characterization. J. Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen SJ, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 8.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heitzer E, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 10.Ni X, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc. Natl. Acad. Sci. 2013 doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu M, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allard WJ, et al. Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas but not in Healthy Subjects or Patients With Nonmalignant Diseases. Clin. Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 13.Swennenhuis JF, et al. Efficiency of whole genome amplification of Single Circulating Tumor Cells enriched by CellSearch and sorted by FACS. Genome Med. 2013;5:106. doi: 10.1186/gm510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 15.Zong C, et al. Genome-Wide Detection of Single-Nucleotide and Copy-Number Variations of a Single Human Cell. Science. 2012;338:1622. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cann GM, et al. mRNA-Seq of single prostate cancer circulating tumor cells reveals recapitulation of gene expression and pathways found in prostate cancer. PLoS One. 2012;7:e49144. doi: 10.1371/journal.pone.0049144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Gammal AT, et al. Chromosome 8p deletions and 8q gains are associated with tumor progression and poor prognosis in prostate cancer. Clin. Cancer Res. 2010;16:56–64. doi: 10.1158/1078-0432.CCR-09-1423. [DOI] [PubMed] [Google Scholar]
- 18.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beltran H, et al. New strategies in prostate cancer: translating genomics into the clinic. Clin. Cancer Res. 2013;19:517–523. doi: 10.1158/1078-0432.CCR-12-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross RW, et al. Predictors of prostate cancer tissue acquisition by an undirected core bone marrow biopsy in metastatic castration-resistant prostate cancer--a Cancer and Leukemia Group B study. Clin. Cancer Res. 2005;11:8109–8113. doi: 10.1158/1078-0432.CCR-05-1250. [DOI] [PubMed] [Google Scholar]
- 22.Robbins CM, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nickerson ML, et al. Somatic alterations contributing to metastasis of a castration-resistant prostate cancer. Hum. Mutat. 2013;34:1231–1241. doi: 10.1002/humu.22346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinrich MC, et al. Kinase Mutations and Imatinib Response in Patients With Metastatic Gastrointestinal Stromal Tumor. J. Clin. Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 27.Lindberg J, et al. Exome sequencing of prostate cancer supports the hypothesis of independent tumour origins. Eur. Urol. 2013;63:347–353. doi: 10.1016/j.eururo.2012.03.050. [DOI] [PubMed] [Google Scholar]
- 28.Keats JJ, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120:1067–1076. doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murtaza M, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 30.Gole J, et al. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat. Biotechnol. 2013 doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh WK, et al. Development of an integrated prostate cancer research information system. Clin. Genitourin. Cancer. 2006;5:61–66. doi: 10.3816/CGC.2006.n.019. [DOI] [PubMed] [Google Scholar]
- 34.Love JC, et al. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat. Biotechnol. 2006;24:703–707. doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 35.Fisher S, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramsköld D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B, Dewey C. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4