Treg cells expressing the co-inhibitory molecule TIGIT selectively inhibit pro-inflammatory Th1 and Th17 cell responses (original) (raw)
. Author manuscript; available in PMC: 2015 Apr 17.
Summary
Foxp3+ T regulatory (Treg) cells regulate immune responses and maintain self-tolerance. Recent work shows that Treg cells are comprised of many subpopulations with specialized regulatory functions. Here we identified Foxp3+ T cells expressing the co-inhibitory molecule TIGIT as a distinct Treg cell subset that specifically suppresses pro-inflammatory T helper 1 (Th1) and Th17 cell, but not Th2 cell responses. Transcriptional profiling characterized TIGIT+ Treg cells as an activated Treg subset with high expression of Treg signature genes. Ligation of TIGIT on Treg cells induced expression of the effector molecule fibrinogen-like protein 2 (Fgl2), which promoted Treg cell-mediated suppression of T effector cell proliferation. In addition, Fgl2 was necessary to prevent suppression of Th2 cell cytokine production in a model of allergic airway inflammation. TIGIT expression therefore identifies a Treg cell subset that demonstrates selectivity for suppression of Th1 and Th17 cell but not Th2 cell responses.
Introduction
Regulatory T cells (Treg cells) are a subset of CD4+ T cells that are marked by expression of the transcription factor Foxp3 and which act as a central component in regulating immune responses to pathogens and in maintaining self-tolerance. Other regulatory populations also contribute to this balance, but Foxp3+ Treg cells are critical for maintaining immune homeostasis as demonstrated by the devastating multi-organ autoimmune disease caused by genetic deficiencies in Foxp3 (Brunkow et al., 2001; Wildin et al., 2001). A series of recent reports has led to the emerging concept that Foxp3+ Treg cells are not all identical, but comprised of multiple, functionally diverse subtypes with distinct phenotypes and specialized functions. Foxp3+ Treg cells have been shown to specialize to selectively regulate specific effector T cell responses and control inflammation at defined anatomical tissue sites (Chaudhry et al., 2009; Cipolletta et al., 2012; Koch et al., 2009; Zheng et al., 2009). Although the transcription factors that differentially induce specialized suppressor functions in Treg cells have been identified, the molecules that mediate these selective effector functions remain largely unknown. Identification of cytokines and cell surface molecules that mediate specialization of Treg cell function would allow the development of therapeutic approaches that target Treg cells to selectively regulate specific types of T cell responses.
In conventional T cells, cytokines and co-stimulatory molecules act in concert to control differentiation and acquisition of effector functions. For example, OX40 (CD134) augments Th2 responses by increasing IL-4 secretion to favor the induction of Th9 cells (Flynn et al., 1998; Xiao et al., 2012). Similarly, inducible costimulator (ICOS) regulates T follicular helper (Tfh) cell expansion and critically contributes to Th17 function by regulating IL-23 receptor expression in an IL-21 and c-Maf-dependent manner (Bauquet et al., 2009). In Treg cells, co-inhibitory molecules, such as programmed cell death 1 (PD-1) and cytotoxic T-lymphocyte antigen 4 (CTLA-4) promote suppressive function. PD-1 plays an important role in iTreg cell stability and suppressive function (Francisco et al., 2009). CTLA-4 is essential for Treg cell function (Wing et al., 2008) and can mediate suppression by enabling Treg cells to compete with effector T cells for co-stimulatory signals on APCs and by inducing the production of indoleamine 2,3-dioxygenase (IDO) in APCs, thereby limiting T cell proliferation (Fallarino et al., 2003). While costimulatory molecules have been shown to promote effector functions of defined T helper lineages, there are no reports that implicate co-inhibitory molecules in the specialized function of Treg cell subsets, despite their important role in promoting the suppressive function of Treg cells in general.
Recently, the co-inhibitory molecule TIGIT has gained attention as an inhibitor of autoimmune responses (Joller et al., 2011; Levin et al., 2011). TIGIT can inhibit T cell responses by binding the ligand CD155 on DCs and thereby inhibiting IL-12 while inducing IL-10 production (Yu et al., 2009). In addition, TIGIT engagement also directly inhibits T cell activation and proliferation (Joller et al., 2011; Levin et al., 2011; Lozano et al., 2012). Like other co-inhibitory molecules, TIGIT is highly expressed on Treg cells (Levin et al., 2011; Yu et al., 2009); however, whether it plays a functional role in these cells has not been explored.
In this study we examined the role of TIGIT on Treg cells. Our results show that TIGIT expression defines a functionally distinct Treg cell subset with an activated phenotype. TIGIT not only acts as a marker for this Treg cell subset but contributes to the selective Treg cell-mediated suppression of pro-inflammatory Th1 and Th17 cells but not Th2 responses by inducing the secretion of the soluble effector molecule fibrinogen-like protein 2 (Fgl2).
Results
TIGIT expression on Treg cells defines a functionally distinct Treg cell subset
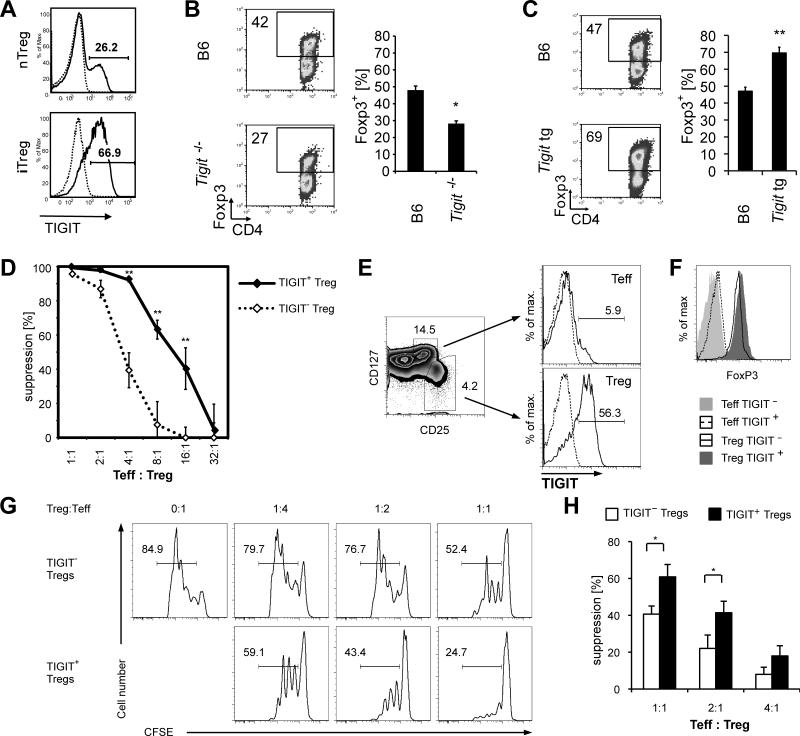
Previous reports have shown that TIGIT is expressed on Treg cells (Levin et al., 2011; Yu et al., 2009). We first tested whether TIGIT was expressed in natural as well as in vitro differentiated induced Treg cells (nTreg and iTreg cells, respectively) and detected expression of TIGIT on both Treg cell populations (Figure 1A). To address whether TIGIT functionally contributes to Treg cell differentiation we tested the ability of TIGIT-deficient T cells to differentiate into Foxp3+ iTreg cells in vitro. Indeed, TIGIT-deficient T cells generated a lower frequency of Foxp3+ T cells after 4 days of TGF-β-mediated differentiation (Figure 1B). Conversely, transgenic overexpression of TIGIT led to the generation of a greater frequency of iTreg cells, confirming that TIGIT promotes iTreg cell differentiation (Figure 1C). As iTreg cells expressed high amounts of TIGIT, we asked whether TIGIT+ Treg cells present in vivo might also be generated peripherally. However, TIGIT+ Treg cells were primarily Neuropilin-1+ and expressed high amounts of Helios, indicating that the majority of TIGIT+ Treg cells are nTreg cells (Figure S1A and B). In addition, TIGIT+ Treg cells do not appear to be a terminally differentiated lineage as both TIGIT+ and TIGIT− Treg cells can convert into the other subset, as evidenced by the loss of TIGIT from TIGIT+ Treg cells upon adoptive transfer and conversely gain of TIGIT expression in transferred TIGIT− Treg cells. However, the vast majority of TIGIT+ Treg cells maintain their TIGIT+ phenotype (Figure S1C).
Figure 1. TIGIT is expressed on highly suppressive Treg cells and promotes iTreg cell differentiation.
(A) CD4+ T cells were purified from Foxp3-GFP.KI mice and the Foxp3+ and Foxp3− cells were sorted. Foxp3+ nTreg cells were stained directly for TIGIT (solid line) or with an isotype control (dotted line) and analyzed by flow cytometry. Foxp3+ induced Treg (iTreg) cells were analyzed after 4 days of stimulation with TGF-β. (B, C) Naïve CD4+CD62L+ T cells were sorted from WT, _Tigit_−/− (B) or Tigit tg (C) mice and differentiated as in (A). Foxp3 expression was analyzed by flow cytometry. (D) CD4+Foxp3+TIGIT+ (◆) or CD4+Foxp3+TIGIT− (◇) Treg cells were sorted from Foxp3-GFP.KI mice and titrated onto Foxp3−GFP− effector T cells stimulated with anti-CD3 and APCs. Proliferation was measured after 72 h by 3H-thymidine incorporation. (Mean ± s.d.; * P<0.01; representative experiment of >10 independent experiments). (E) Sorting strategy of ex vivo FACS sorted human effector T cells (CD4+CD25+CD127+) and Treg cells (CD4+CD25highCD127−) sorted into TIGIT+ and TIGIT−. (F) Treg cells sorted as outlined in (E) showed >96% purity in both subsets measured by Foxp3 staining after isolation. (G) Representative suppression assay with human CD4+CD25highCD127-TIGIT+ and TIGIT− Treg cells co-cultured with CFSE-labeled CD25-depleted CD4+ effector T cells for 4 days. (H) Statistical summary of (G) of six healthy donors (mean ± SEM; * P<0.05).
We then tested whether TIGIT+ and TIGIT− Treg cells also display functional differences by comparing their ability to suppress CD4+ Foxp3− effector T cells in vitro. TIGIT+ nTreg cells showed an increased ability to suppress TCR-stimulated proliferation of conventional T cells (Figure 1D). TIGIT therefore marks a functionally distinct subset of nTreg cells with superior suppressive capacity in vitro.
Next, we determined whether TIGIT+ Treg cells are also detected in humans and whether they might represent a similarly potent Treg cell subset as in mice. We first analyzed TIGIT expression in human CD4+ T cells and found that a large proportion of human Treg cells are TIGIT+ (Figure 1E). We then performed in vitro suppression assays to test whether human TIGIT+ and TIGIT− Treg cells also differ in their suppressive capacity. Indeed, we detected increased suppression by TIGIT+ Treg cells compared to TIGIT− Treg cells (Figures 1F-H), indicating that TIGIT+ Treg cells are highly suppressive and may represent a functionally distinct Treg cell subset in humans.
TIGIT+ Treg cells display an activated phenotype
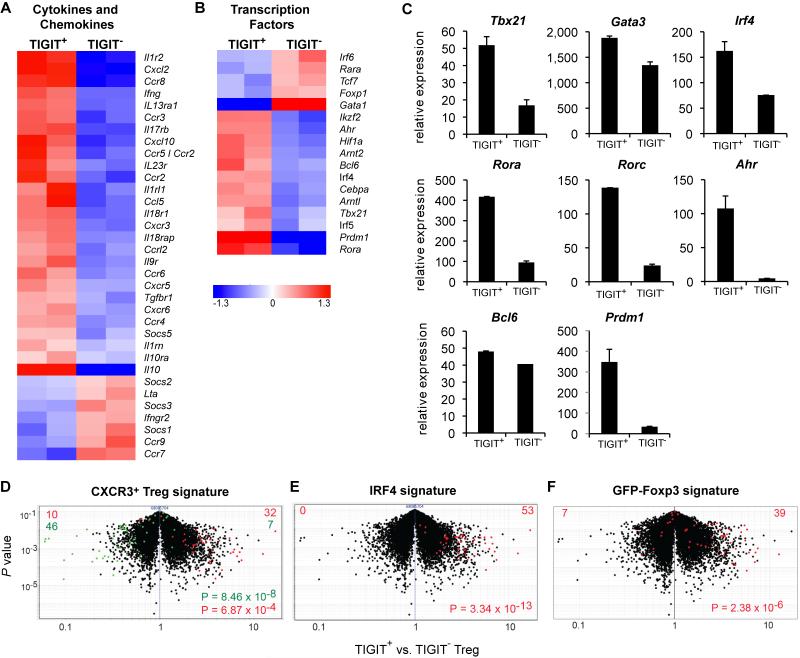
To better understand the differences between TIGIT+ and TIGIT− Treg cells, we analyzed their gene expression patterns by microarray profiling. Overall, a total of 472 and 184 genes were over- or under-expressed in TIGIT+ Treg cells relative to their TIGIT− counterparts (with an arbitrary cut-off of at fold change > 2 and t-test p<0.05; Figure S1D, Table S1). These belonged to several functional families including chemokines/cytokines or their receptors, transcription factors, and costimulatory and other surface receptors, as well as molecules typical of Treg cells. Overall, TIGIT+ Treg cells seemed to display a more activated phenotype than their TIGIT− counterparts (Figure S1E and F). Furthermore, TIGIT+ Treg cells expressed 3-fold higher levels of Ki67 and incorporated four times higher amounts of BrdU than their TIGIT− counterparts when we labeled proliferating cells in vivo (Figure S1G-H). The activated phenotype of TIGIT+ Treg cells observed by transcriptional profiling therefore translates into a higher rate of proliferation in vivo.
TIGIT+ Treg cells share features with pro-inflammatory T cell lineages
Treg cells share a number of features with the effector population they suppress, including the expression of chemokine receptors as well as the transcription factors that induce the development of those effector T cells (Chaudhry et al., 2009; Chung et al., 2011; Koch et al., 2009; Linterman et al., 2011; Zheng et al., 2009). The pattern of chemokine receptors expressed by TIGIT+ Treg cells does not overlap with that of any particular Th effector subset (Figure 2A), but includes receptors that are expressed by several lineages, mainly by the pro-inflammatory Th1 and Th17 subsets (Ccr2, Ccr5, Ccr6, Cxcr3, and Cxcr6), but to a lesser degree also those expressed by Th2 (Ccr3) or Tfh cells (Cxcr5). This might indicate that TIGIT+ Treg cells are equipped to target a broad spectrum of effector cells and tissues, specifically under pro-inflammatory conditions.
Figure 2. Expression profiling of TIGIT+ Treg cells.
Heat map of chemokine (receptor) and cytokine (receptor) (A) or transcription factor (B) genes that are differentially expressed (>1.5-fold) in CD4+Foxp3+TIGIT+ and CD4+Foxp3+TIGIT− Treg cells (duplicate samples are shown). (C) Differential expression of a selection of genes from (B) was determined by quantification of mRNA in CD4+Foxp3+TIGIT+ and CD4+Foxp3+TIGIT− Treg cells by RT-PCR. Mean ± s.d. of at least 3 independent experiments is shown. (D-F) Volcano plots comparing the P value versus fold-change for probes from TIGIT+ versus TIGIT− Treg cells. Treg cell signatures generated from (D) CXCR3+ versus CXCR3− Treg cells, (E) WT versus IRF4 KO Treg cells and (F) Treg cells from GFP-Foxp3 fusion protein reporter mice versus Foxp3-IRES-GFP mice are highlighted in red (overexpressed) and green (underrepresented). P values form a chi-squared test. Genes and Probe IDs included in the signatures are listed in Table S2.
Similarly, the transcription factors that are more highly expressed in TIGIT+ Treg cells do not specifically fall within the fingerprint of a particular effector lineage (Figure 2B). Transcription factors that are expressed in higher amounts by TIGIT+ Treg cells include those that are specific for Th1 (Tbx21) and Th17 cells (Rora, Rorc, Irf4, Ahr), while only minor or no differences could be observed in the expression of the Th2 lineage factor Gata3 and the Tfh-lineage specific transcription factor Bcl6 (Figure 2C). Prdm1, a transcription factor that transactivates IL-10 expression (Cretney et al., 2011), was expressed in higher amounts in TIGIT+ Treg cells, consistent with increased production of IL-10 by these cells (Figure 2A, C and S2A).
We analyzed the expression profile of TIGIT+ Treg cells in relation to signatures of previously described Treg cell subsets. TIGIT+ Treg cells were enriched for a gene set that distinguishes CXCR3+ Treg cells; cells that express T-bet and were shown to be specialized in suppression of Th1 responses (Koch et al., 2012; Koch et al., 2009) (Figure 2D). IRF4 expression in Treg cells is important for control of Th2 responses as demonstrated by the dysregulated Th2 responses observed in mice that lack IRF4 in Foxp3+ Treg cells (Zheng et al., 2009). Many of the IRF4-dependent genes are upregulated in TIGIT+ Treg cells (Figure 2E), which is in line with the increased expression of IRF4 by TIGIT+ Treg cells (Figure 2C). Finally, we found that TIGIT+ Treg cells share features with Treg cells from mice in which Foxp3 is modified by an N-terminal fusion with GFP, leading to modified interaction with Foxp3 cofactors (Bettini et al., 2012; Fontenot et al., 2005) (Figure 2F). In these mice the GFP-fusion altered the molecular characteristics of Foxp3, reducing its interaction with HIF-1α and promoting interaction with IRF4, modifying the Treg cell transcriptome and resulting in enhanced suppression of Th2 and Th17 cell responses but weaker suppression of Th1 responses (Bettini et al., 2012; Darce et al., 2012). Overall these data suggest that, rather than representing a subset specialized to suppress a specific T effector lineage, TIGIT+ Treg cells express features of multiple effector and regulatory T cell subsets.
TIGIT+ Treg cells express a gene profile indicative of a highly suppressive Treg cell subset
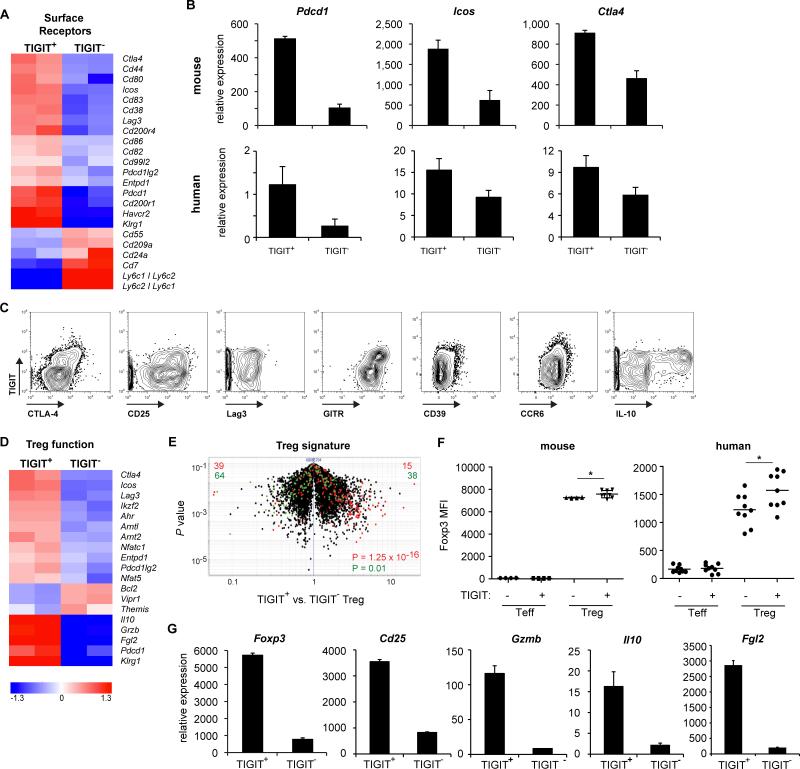
Effector as well as regulatory T cell function is shaped by the cytokine environment as well as engagement of co-stimulatory ligands. We therefore examined the transcriptional profile of TIGIT+ and TIGIT− Treg cells for differential expression of membrane receptors and detected a distinct pattern of co-stimulatory molecule expression in TIGIT+ Treg cells. TIGIT+ Treg cells expressed higher amounts of the co-stimulatory molecule ICOS, and had increased expression of a number of co-inhibitory molecules, such as CTLA-4, PD-1 (Pdcd1), Lymphocyte activation gene 3 (Lag3), and T cell Immunoglobulin and Mucin 3 (Tim3, Havcr2) (Figure 3A-C, Figure S2). Co-inhibitory molecules such as CTLA-4 and PD-1 not only serve as markers for T cell activation but also contribute to Treg cell stability and function (Francisco et al., 2009; Wing et al., 2008), indicating TIGIT+ Treg cells might be better equipped for mediating suppression. Indeed, TIGIT+ Treg cells expressed higher amounts of CTLA-4, CD25, and GITR but showed no or only slight differences in the expression of Lag3 (Figure 3C-G and S2). In mice, CD39 and chemokine receptor 6 (CCR6) expression in TIGIT+ Treg cells is comparable to TIGIT− Treg cells while human TIGIT+ Treg cells upregulated expression of these markers (Figure 3C and S2B). When comparing TIGIT+ to TIGIT− Treg cells, the immunosuppressive cytokine IL-10 was primarily produced by TIGIT+ Treg cells (Figure 3C). qPCR and flow cytometry analysis showed TIGIT+ Treg cells to express higher amounts of Foxp3 than TIGIT− Treg cells (Figure 3F and G). In addition, CD25 and Treg cell effector molecules, such as Granzyme B, IL-10 and Fgl2, were also expressed in higher amounts in TIGIT+ Treg cells (Figure 3C, D and G and Figure S2A). TIGIT+ Treg cells therefore display a transcriptional profile that suggests an activated, highly suppressive Treg cell subset.
Figure 3. TIGIT+ Treg cells express a gene profile indicative of a highly suppressive Treg cell subset.
(A) Heat map of surface receptor genes that are differentially expressed (>1.5-fold, duplicate samples) in CD4+Foxp3+TIGIT+ and CD4+Foxp3+TIGIT− Treg cells. Quantitative RT-PCR (B) and flow cytometric (C) confirmation for a selection of genes from (A) and (D). (D) Heat map of differentially expressed genes involved in Treg cell differentiation and function. (E) Volcano plot comparing the P value versus fold-change for probes from TIGIT+ versus TIGIT− Treg cells. The canonical Treg cell signature is highlighted in red (transcripts upregulated in Treg cells) and green (transcripts downregulated in Treg cells). (F) Foxp3 protein expression was quantified by flow cytometry in mouse Foxp3− (Teff) or Foxp3+ (Treg) and human memory T cells (CD4+CD127+CD25med; Teff) and Treg cells (CD4+CD127lowCD25high) (n=9; *p<0.05). (G) Relative expression of the indicated genes in CD4+Foxp3+TIGIT+ and CD4+Foxp3+TIGIT− Treg cells was determined by quantitative PCR.
TIGIT ligation induces the Treg cell effector molecule Fgl2
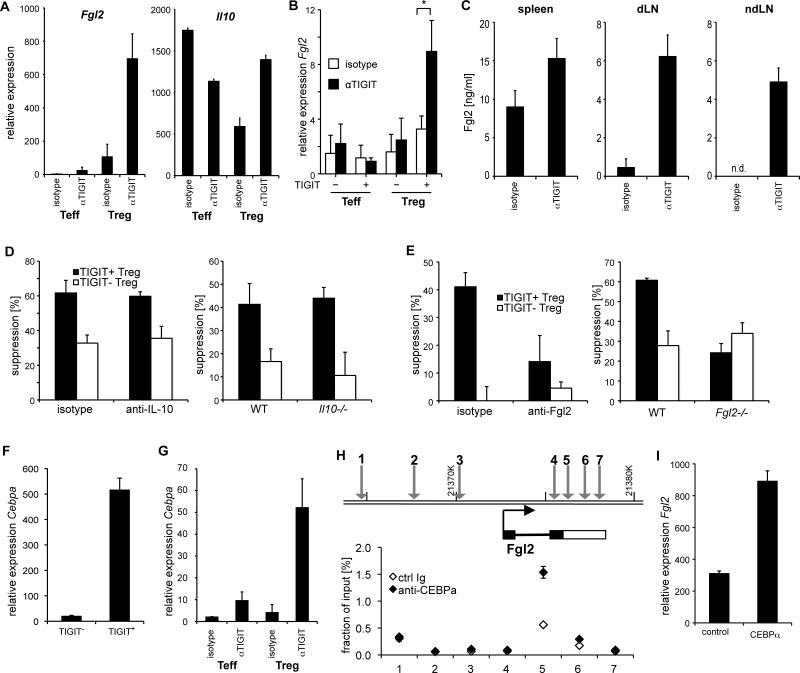
We next tested whether TIGIT ligation could directly induce Treg cell effector molecules. Among these, IL-10 and Fgl2 stood out as particularly interesting molecules as both were highly expressed in TIGIT+ Treg cells (Figure 3G) and are able to suppress pro-inflammatory responses (Chan et al., 2003; Kuhn et al., 1993). To test whether expression of Fgl2 and IL-10 could be induced through TIGIT, we isolated regulatory T cells and stimulated them in vitro in the presence of an agonistic anti-TIGIT antibody (Ab). TIGIT ligation triggered a 2-fold increase in Il10 gene expression by Treg cells in vitro. Similarly, Fgl2 expression in Treg cells was increased in the presence of agonistic anti-TIGIT Ab demonstrating that TIGIT signaling induces Fgl2 transcription in TIGIT+ Treg cells (Figure 4A and B). We went on to determine whether TIGIT was also able to induce Fgl2 and IL-10 in vivo. As our anti-TIGIT Ab also acts agonistically in vivo and does not affect Treg cell frequencies or composition (Figure S3A-B), we tested whether treatment of immunized mice with the Ab would result in induction of either effector molecule. Cells were isolated 10 days after immunization, restimulated for 2 days in vitro, and culture supernatants were analyzed for Fgl2 and IL-10. IL-10 could not be detected in these cultures (not shown). However, Fgl2 was increased in cell supernatants from anti-TIGIT treated mice Treg cell (Figure 4C). TIGIT therefore induces Fgl2 in vitro and in vivo.
Figure 4. TIGIT ligation induces Fgl2 expression.
(A) Foxp3− (Teff) and Foxp3+ (Treg) cells were sorted from Foxp3-GFP.KI mice, stimulated with anti-CD3 and anti-CD28 in the presence of agonistic anti-TIGIT Ab. After 3 days RNA was extracted and Fgl2 and Il10 mRNA was quantified by quantitative RT-PCR. (B) Ex vivo human memory T cells (CD4+CD127+CD25med) and Treg cells (CD4+CD127lowCD25high) were sorted gating into TIGIT+ and TIGIT−. After isolation, cells were cultured in the presence of agonistic anti-TIGIT or isotype control for 4 days. FGL2 expression was quantified by RT-PCR (n=6; * P<0.05). (C) Mice were immunized s.c. with MOG35–55 peptide in CFA and treated with anti-TIGIT or isotype control antibody. On day 10 cells were re-stimulated with MOG35-55 peptide for 48h. Fgl2 concentrations in the supernatants were determined by ELISA. (D, E) CD4+CD25+TIGIT+ (closed bars) or CD4+CD25+TIGIT− (open bars) Treg cells were sorted from WT, IL-10-deficient (D) or Fgl2-deficent (E) mice and co-cultured with CD25− effector T cells stimulated with anti-CD3 and APCs at a ratio of 1:8. Where indicated neutralizing anti-IL-10 (D) or anti-Fgl2 (E) Ab or the respective isotype control Ab was added to the culture. Proliferation was measured after 72 h by 3H-thymidine incorporation. (F) CD4+Foxp3+TIGIT+ and CD4+Foxp3+TIGIT− Treg cells were sorted from Foxp3-GFP.KI mice and mRNA for Cebpa was measured by RT-PCR. (G) Cells were isolated and stimulated as in (A) and on day 3 Cebpa mRNA was examined by quantitative RT-PCR. (H) ChIP assays were performed on P815 cells expressing TIGIT using anti-CEBPα antibody or an isotype control. The precipitated chromatin was analyzed by quantitative PCR with primers specific for 3 promoter and 4 intragenic regions of the Fgl2 gene with predicted CEBPα binding sites. Signals are displayed as % of the total input chromatin. (I) CD4+Foxp3+ Treg cell cells were sorted from Foxp3-GFP.KI mice and transfected with a CEBPα over-expression construct (CEBPα) or the empty vector as control (control) and stimulated with anti-CD3/CD28 Dynabeads. Relative expression of Fgl2 mRNA was determined by RT-PCR 4 days later. (all panels represent mean ± s.d)
We next addressed whether neutralizing IL-10 and Fgl2 would abolish the difference in in vitro suppressive capacity of TIGIT+ and TIGIT− Treg cells. In line with previous reports, blocking or deletion of IL-10 had no effect on suppression by TIGIT+ or TIGIT− Treg cells in vitro (Thornton and Shevach, 1998) (Figure 4D). Similarly, neutralizing or deleting Fgl2 had no effect on the suppression by TIGIT− Treg cells, which express only minimal amounts of Fgl2 (Figure 4A and 4E). In contrast, after neutralization or deletion of Fgl2 the suppressive activity of TIGIT+ Treg cells was similar to that observed for TIGIT− Treg cells (Figure 4E), indicating that Fgl2 drives increased suppression by TIGIT+ Treg cells in vitro. In contrast, Fgl2 deficiency had no effect on the TIGIT-mediated induction of Foxp3 or IL-10 (Figure S3C and D). Therefore, TIGIT ligation triggers secretion of Fgl2 by Treg cells, which enables them to act as potent suppressors.
To better understand how TIGIT induced Fgl2 expression, we searched the genomic region of Fgl2 for binding sites of transcription factors that showed differential expression in the microarray analysis of TIGIT+ vs. TIGIT− Treg cells (Figure 2B). Fgl2 was found to contain binding sites for the transcription factor CEBPα, which was differentially expressed in TIGIT+ Treg cells. Quantitative PCR confirmed that CEBPα is highly expressed in TIGIT+ but not TIGIT− Treg cells (Figure 4F) and that TIGIT engagement is able to upregulate Cebpa transcription in Treg cells (Figure 4G). Chromatin Immunoprecipitation (ChIP)-PCR using an anti-CEBPα Ab together with primer pairs specific for the Fgl2 genomic region confirmed that CEBPα binds to the Fgl2 gene as predicted (Figure 4H). To further analyze whether CEBPα can promote transcription of Fgl2, we overexpressed CEBPα in nTreg cells and observed an increase in Fgl2 expression (Figure 4I). TIGIT might therefore promote Treg cell suppression by inducing CEBPα, and thereby Fgl2 expression.
TIGIT+Treg cells inhibit Th1 and Th17 cell but not Th2 cell differentiation
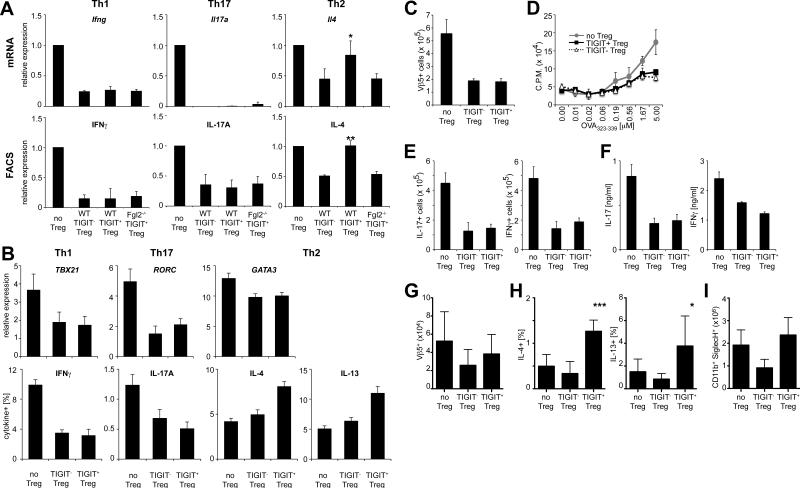
Fgl2 not only suppresses effector T cell proliferation, it also shifts the cytokine profile towards a Th2 response as it inhibits Th1 responses while promoting Th2 polarization and induction of IL-10 and IL-4 (Chan et al., 2003; Shalev et al., 2008). Furthermore, Fgl2 is important for Treg cell function in vivo as Fgl2-deficient Treg cells show impaired control of effector T cell expansion in lymphopenic hosts (Figure S4A). As TIGIT+ Treg cells produce high amounts of Fgl2, we reasoned that they might differentially suppress distinct T helper cell lineages. To test this, we co-cultured naïve T cells and TIGIT− and TIGIT+ Treg cells under differentiation conditions for Th1, Th2, and Th17 cells and assessed expression of lineage-specific cytokines. TIGIT+ Treg cells showed no difference in suppression of Th1 cell differentiation when compared to TIGIT− Treg cells as indicated by reduced expression of IFNγ (Figure 5A and S4B). Similarly, both subsets potently suppressed expression of IL-17 by Th17 cells. However, in contrast to TIGIT− Treg cells, TIGIT+ Treg cells did not suppress differentiation of Th2 cells, as indicated by comparable IL-4 production as in unsuppressed controls (Figure 5A and S4B). This effect was dependent on Fgl2, as loss of Fgl2 in TIGIT+ Treg cells restored the ability to suppress Th2 differentiation, indicating that Fgl2 produced by TIGIT+ Treg cells interferes with suppression of Th2 responses (Figure 5A and S4B). Although Fgl2 contributes to suppression of effector T cell proliferation under Th1, Th2 and Th17 conditions (Figure S4C), it does not contribute to suppression of Th1 and Th17 cell cytokine production. It is, however, needed to prevent suppression of the Th2 cytokine IL-4. In line with these results, analysis of human effector T cells cultured in the presence of TIGIT+ or TIGIT− Treg cells were also inhibited in Th1 and Th17 cell but not Th2 cell cytokine production (Figure 5B).
Figure 5. TIGIT+ Treg cells suppress Th1 and Th17 cell but not Th2 cell responses.
(A) Naïve effector T cells, WT Foxp3+TIGIT−, WT Foxp3+TIGIT+ Treg cells, and _Fgl2_−/− Foxp3+TIGIT+ Treg cells were sorted and co-cultured at a ratio of 1:10 under Th1, Th2, or Th17 polarizing conditions. After 3 days mRNA was measured by quantitative RTPCR. On day 5 intracellular cytokines in CD45.1+ T effector cells were determined by flow cytometry (values normalized to unsuppressed controls, mean ± SEM; * P<0.05, ** P<0.001, paired student's t-test). (B) Human TIGIT+ and TIGIT− Treg cells (CD4+CD25highCD127neg) were sorted and co-cultured with CFSE-labeled CD25-depleted CD4+ T effector cells. Gene expression (qRT-PCR) and intracellular cytokines (flow cytometry) were measured on day 4 (mean ± SEM; n=6). (C-F) CD25− effector OT-II cells and CD25high OT-II Treg cells (TIGIT−, TIGIT+ or no Treg cell control) were transferred i.v. into WT recipients and mice were immunized with OVA in CFA. (C) Expansion of Vβ5+ OT-II T cells and (D) proliferation in response to OVA323-339 were determined 10 days later. (E) Intracellular cytokine levels were determined by flow cytometry and (F) cytokine concentration in the culture supernatants was determined by cytometric bead array. (G-I) CD25− effector OT-II cells and CD25high OT-II Treg cells (TIGIT−, TIGIT+ or no Treg cell control) were transferred i.v. into WT recipients. Mice were then sensitized with OVA (i.p.) on days 0 and 7 and challenged with aerosolized OVA on days 14–17 to induce allergic airway inflammation. (G) Total numbers of Vβ5+ OT-II cells in lungs, (H) intracellular cytokine levels from lung-infiltrating CD4+ T cells, and (I) total eosinophil numbers in bronchio-alveolar lavage fluid were determined by flow cytometry. Pooled data from two experiments are shown (mean ± SEM; n=8).
TIGIT+Treg cells inhibit Th1 and Th17 cell but not Th2 cell responses in vivo
To test, whether our in vitro results also translated into selective suppression of Th1 and Th17 cell vs. Th2 cell responses in vivo, we transferred TIGIT+ and TIGIT− OVA-specific OT-II Treg cells together with OT-II effector cells into WT recipients and analyzed the ability of the different Treg cell subsets to suppress Th1, Th17, and Th2 cell responses upon immunization. Under conditions that induce a Th1 and Th17 cell dominated response (OVA in CFA), TIGIT+ and TIGIT− Treg cells were equally capable of suppressing effector T cell expansion as determined by the number of Vβ5+ OT-II cells. In addition, in vivo differentiation of Th1 and Th17 cells was suppressed equally well by TIGIT+ vs. TIGIT− Treg cells as judged by IFNγ and IL-17 production (Figure 5C-F). When mice were immunized for induction of Th2-mediated allergic airway inflammation, TIGIT− Treg cells were able to suppress the disease. In contrast, TIGIT+ Treg cells failed to inhibit recruitment of antigen-specific Vβ5+ OT-II cells to the lung and production of Th2 cytokines (IL-4 and IL-13) was significantly higher than in mice that had received TIGIT− Treg cells (Figure 5G, H). Consistent with an increase in Th2 cells in the presence of TIGIT+ Treg cells, we observed high numbers of eosinophils in the bronchio-alveolar lavage of these mice (Figure 5I). Taken together these data indicate that TIGIT+ Treg cells selectively suppress pro-inflammatory Th1 and Th17 cell, but not Th2 cell, responses.
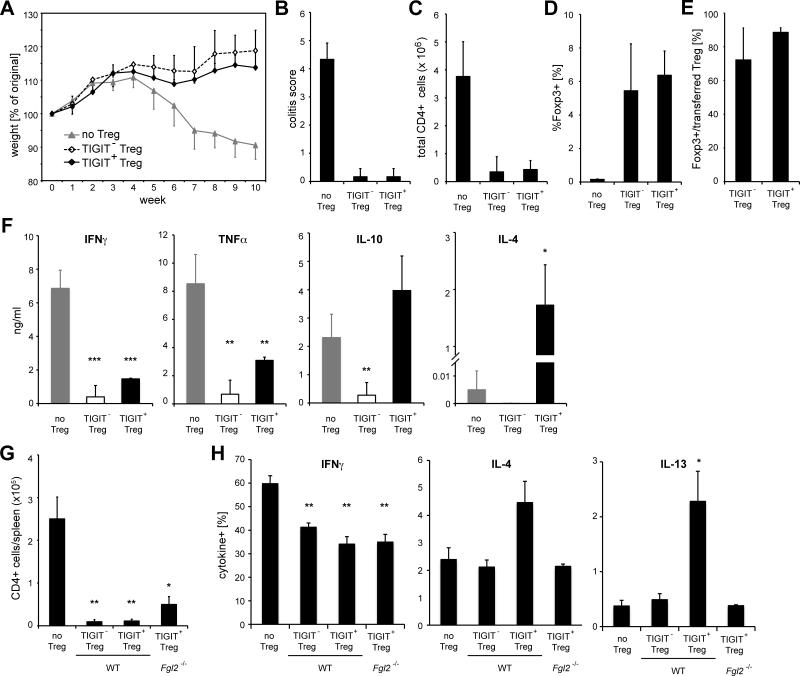
Based on these results we wanted to test whether TIGIT+ Treg cells might shift effector T cell responses towards a Th2 phenotype, if the system is not biased by immunization with adjuvant and thus took advantage of the Rag-transfer model of colitis (Izcue et al., 2008). To induce disease, congenically marked effector T cells were transferred into _Rag1_−/− recipients; either alone or together with TIGIT+Foxp3+ or TIGITFoxp3+ Treg cells. Mice that received the effector T cells alone lost weight over time, while co-transfer of either TIGIT+ or TIGIT− Treg cells promoted disease suppression (Figure 6A). Despite the difference in suppressive capacity that we observed in vitro, both Treg cell populations were able to suppress the disease equally well. Co-transfer of either Treg cell population prevented tissue inflammation, as indicated by the histopathological colitis score (Figure 6B), and suppressed the expansion of effector T cells in vivo (Figure 6C). In addition, TIGIT+ and TIGIT− Treg cells displayed comparable ability to expand and persist in vivo (Figure 6D and E). TIGIT+ as well as TIGIT− Treg cells were able to suppress pro-inflammatory cytokines as T cells from the mesenteric lymph nodes (LNs) produced significantly lower amounts of IFN-γ and TNF-γ than those from mice that did not receive Treg cells (Figure 6F and S5). In contrast, TIGIT+ Treg cells did not suppress, or may have even increased the expression of IL-10 and the Th2 cytokine IL-4 when compared to the control group (no Treg cells) (Figure 6F). Intracellular cytokine staining revealed that, while IL-10 was produced by both effector T cells and Treg cells, IL-4 was entirely produced by effector T cells (Figure S5). Next, we tested whether the sparing of Th2 cells was also dependent on Fgl2 in vivo. To this end, effector T cells and either WT or Fgl2-deficient Treg cells were co-transferred into Rag1-deficient mice, which were then immunized and boosted with OVA in Alum on days 0 and 7. In line with our in vitro results, Fgl2 contributed to suppression of T cell proliferation but was not required for inhibition of IFNγ-secretion by effector cells (Figure 6G and H). In contrast, the inability of TIGIT+ Treg cells to suppress Th2 responses was again dependent on Fgl2 (Figure 6H). TIGIT+ Treg cells therefore suppress pro-inflammatory responses in vivo, while sparing or promoting Th2-like responses in an Fgl2-dependent manner.
Figure 6. TIGIT+ Treg cells suppress pro-inflammatory responses in vivo.
To induce colitis CD45RBhi effector T cells (CD45.1) were transferred into Rag1−/− mice together with TIGIT+ or TIGIT− Treg cells (CD45.2) or no Treg cells as controls (Teff:Treg cell ratio was 4.4:1 for TIGIT+ Treg cells and 3.6:1 for TIGIT− Treg cells). (A) Mice were monitored for weight loss over 10 weeks and (B) total colitis scores were determined by histopathology. (C-E) At 10 weeks after transfer mesenteric LNs were harvested and (C) total number of infiltrating CD4+ T cells, (D) proportion of Foxp3+ Treg cells among CD4+ T cells, and (E) Foxp3 expression among the transferred Treg cell population (CD45.2+) were determined by flow cytometry. (F) Mesenteric LN cells were re-stimulated in vitro with 0.5μg/ml anti-CD3 for 3 days and cytokine secretion was determined by cytometric bead array in supernatants. (G, H) CD45RBhi effector T cells (CD45.1) and WT or _Fgl2_−/− TIGIT+ or TIGIT− Treg cells (CD45.2) were co-transferred into _Rag1_−/− mice on day -1, which were then immunized with OVA in alum on day 0 and 7. On day 14 (G) total number of CD4+CD45.1+ effector T cells and (H) frequencies of cytokine producing cells upon re-stimulated with PMA/Ionomycin were determined by flow cytometry. (P<0.05 (*), P<0.01 (**), P<0.005 (***)).
Discussion
In this study, we identified TIGIT+Foxp3+ T cells as a distinct Treg cell subset that specifically suppresses pro-inflammatory Th1 and Th17 cell but not Th2 cell responses through the secretion of Fgl2, which was induced by engagement of TIGIT. TIGIT+ Treg cells therefore shift the balance towards Th2 responses by suppressing pro-inflammatory effector Th1 and Th17 cell responses, but not Th2 cell responses. This is one of the first examples of how a co-inhibitory molecule can mediate selective inhibition of certain effector responses while leaving others intact.
TIGIT was first described as an inhibitory molecule that suppresses immune responses indirectly by regulating DC function. By interacting with its ligand CD155 on DCs, TIGIT was shown to induce IL-10 and suppress IL-12 production in DCs and thereby inhibit Th1 responses (Yu et al., 2009). In our previous studies we showed that TIGIT also has T cell intrinsic inhibitory effects (Joller et al., 2011). As Treg cells are the primary cell type that constitutively expresses TIGIT, we suspect that many of the DC effects that have been observed might be mediated by TIGIT+ Treg cells. In addition to TIGIT-induced IL-10 produced by the DCs themselves, we propose that increased amounts of IL-10 and Fgl2 produced by TIGIT+ Treg cells may also contribute to the generation of tolerogenic DCs and thereby inhibit the generation of effector T cell responses. Although TIGIT-induced IL-10 was shown to suppress both IL-12p35 and IL-12p40 (Yu et al., 2009), the effect of Fgl2 in suppressing these key differentiating cytokines has not been evaluated. We propose that IL-10 and Fgl2 secreted by TIGIT+ Treg cells may act in concert to suppress both IL-12 and IL-23 production from activated DCs and thereby inhibit development of both Th1 and Th17 cell responses.
It has become clear that Treg cells represent a very heterogeneous population that encompasses many specialized subpopulations. While Foxp3 is necessary to equip T cells with basic Treg cell functions (Fontenot et al., 2005), additional factors are required for efficient suppression of effector T cell responses in vivo and for maintaining immune tolerance. Several transcription factors have been identified that drive additional programs in Treg cells to efficiently control certain classes of effector T cells and immune responses in defined target tissues. For instance, Treg cells that are specialized in controlling specific effector T cell lineages co-express lineage-specific transcription factors from T helper cells, such as T-bet, IRF4, Stat3, or Bcl6 to fulfill their subset-specific inhibitory functions (Chaudhry et al., 2009; Chung et al., 2011; Koch et al., 2009; Linterman et al., 2011; Zheng et al., 2009). TIGIT+ Treg cells share features with several different Treg cell subsets and express more T-bet and IRF4, as well as Th17-specific transcription factors such as RORα and RORγ, than TIGIT− Treg cells. The fact that TIGIT+ Treg cells express elevated amounts of IRF4 would suggest that they are well equipped for suppression of Th2 responses as IRF4-deficieny in Foxp3+ T cells results in spontaneous Th2 pathology (Zheng et al., 2009). However, our data suggest that TIGIT+ Treg cells do not suppress Th2 responses but effectively inhibit pro-inflammatory Th1 and Th17 cell responses. It should be noted that IRF4 is not only expressed in Th2 cells but is also required for Th17 cell differentiation. While conditional deletion of IRF4 in Foxp3+ Treg cells most prominently affects control of Th2 cell responses, these mice also have slightly elevated amounts of IL-17 (Zheng et al., 2009) and in settings in which the immune response is dominated by Th17 effector cells, such as arthritis, diminished function of IRF4 in Treg cells results in impaired control of Th17 cell responses (Darce et al., 2012). While TIGIT+ Treg cells seem to share the inability to suppress Th2 cell responses with IRF4-deficient Treg cells, the fact that they potently suppress Th17 cell responses distinguishes them from IRF4-deficient Treg cells.
Our findings suggest that in addition to the lineage- and tissue-specific transcription factors, co-inhibitory molecules like TIGIT also contribute to the functional specialization of Treg cells by inducing a distinct set of suppressive mediators that can selectively suppress certain classes of effector T cell responses. While we found that the specialized TIGIT+ and TIGIT− Treg cell subsets display a certain degree of plasticity, both populations are sufficiently stable to mediate differential TIGIT-dependent biological effects in the in vivo models of asthma and colitis, which span a 3 or 10 week time period, respectively. In the case of TIGIT+ Treg cells, expression of Fgl2 allows them to selectively spare Th2 responses, while maintaining efficient suppression of Th1 and Th17 responses. Co-inhibitory receptors might therefore tailor the suppressive function of Foxp3+ Treg cells to what is required in a specific inflammatory environment. The expression pattern of these receptors and/or engagement through their ligands in a particular tissue environment could thereby alter the molecular signature of Treg cells and equip them with specialized suppressive mechanisms that are tailored for a specific tissue or type of inflammation.
Besides transcription factors, we now show that cell surface molecules like TIGIT expressed on Foxp3+ Treg cells can differentially suppress effector T cell responses, providing a target by which defined subsets of Treg cells can be manipulated to regulate immune and autoimmune responses.
Experimental Procedures
Animals
C57BL/6 (B6), B6.SJL-PtprcaPepcb/BoyJ (CD45.1), B6.129P2-Il10_tm1Cgn_/J (_Il10_−/−) and B6.129S7-Rag1tm1Mom/J (_Rag1_−/−) mice were purchased from the Jackson Laboratories. Foxp3-GFP.KI reporter mice (Bettelli et al., 2006), _Tigit_−/− mice (Levin et al., 2011), TIGIT transgenic mice (Levin et al., 2011), and _Fgl2_−/− mice (Shalev et al., 2008) have been previously described. Animals were maintained in a conventional, pathogen-free facility at the Harvard Institutes of Medicine (Boston, MA) and all experiments were carried out in accordance with guidelines prescribed by the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School.
Human samples
Peripheral venous blood was obtained from healthy control volunteers in compliance with Institutional Review Board protocols at Yale University School of Medicine. Total CD4+ T cells were isolated by negative selection (CD4+ T cell isolation kit II, Miltenyi Biotec, Auburn, CA) and then sorted by flow cytometry.
Treg cell differentiation and suppression assays
Cells were cultured in DMEM supplemented with 10% (vol/vol) FCS, 50 mM mercaptoethanol, 1 mM sodium pyruvate, nonessential amino acids, L-glutamine, and 100 U/ml penicillin and 100 g/ml streptomycin. CD4+ T cells from splenocytes and lymph node cells were isolated using anti-CD4 beads (Miltenyi). For in vitro Treg cell differentiation, naïve CD4+CD62L+CD44− cells were sorted by flow cytometry and stimulated with plate bound anti-CD3 (145-2C11, 0.3 μg/ml) and anti-CD28 (PV-1, 2 μg/ml) in the presence of 2.5ng/ml TGF-μ (R&D). Foxp3 expression was assessed by flow cytometry 4 days later. For suppression assays, CD4+Foxp3− responder cells and CD4+Foxp3+ Treg cells were flow sorted from Foxp3-GFP.KI reporter mice based on GFP expression. CD4+Foxp3− (2 × 104/well) and CD4+Foxp3+ cells were cultured in triplicate in the presence of soluble anti-CD3 (1 μg/ml) and irradiated splenic APCs (1.2 × 105/well). After 48 h cells were pulsed with 1 μCi [3H]thymidine for an additional 18h, harvested and [3H]thymidine incorporation was analyzed to assess proliferation. Percentage of suppression = 100 − C.P.M. of well with the indicated ratio of effector : Treg cells / mean C.P.M. of wells with CD4+Foxp3− effectors alone. Where indicated, anti-Fgl2 Ab (clone 6D9, 30 μg/ml, Abnova), anti-IL-10 Ab (clone JES5-16E3, Biolegend) or an isotype control was added to the cultures. For human Treg cell suppression assays, CD25-depleted T cells were CFSE– labeled and co-cultured with FACS-sorted Treg cells (TIGIT+ or TIGIT−) at indicated ratios. Cells were stimulated with Treg cell Inspector Beads (Miltenyi) at manufacturer's recommended concentration. At day 4, cells were stained with LIVE/DEAD Fixable Dead Cell Stain Kit (Molecular Probes) to allow gating on viable cells and proliferation was measured by CFSE dilution. Samples were analyzed by flow cytometry.
Microarray
CD4+ T cells were pre-purified from splenocytes and lymph node cells of naïve Foxp3-GFP.KI reporter mice using Dynal beads (Invitrogen) and CD4+Foxp3+TIGIT+ and CD4+Foxp3+TIGIT− cells were sorted by flow cytometry. CD4+Foxp3+CXCR3+ and CD4+Foxp3+CXCR3+ were similarly sorted from spleens of Foxp3-GFP.KI mice. All cells were double-sorted for purity, the final sort being directly into TRIzol (Invitrogen). RNA was extracted and used to prepare probes for microarray analysis on the Affymetrix Mouse Gene1.0ST platform, using ImmGen protocols (Heng and Painter, 2008). Microarray data was analyzed using the GeneSpring 11 (Agilent, Santa Clara, CA; quantile normalization) or GenePattern (RMA normalization) software. Genes of interest (fold change >1.5) were handpicked and two-way hierarchical clustering using Euclidean distance metric was performed to generate heat-maps. Analysis of signature genes within the TIGIT+/TIGIT− comparison used previously determined genesets: a T cell activation/proliferation signature from in vivo activated T cells (Hill et al., 2007); the canonical Treg/Tconv signature (Hill et al., 2007); a geneset that distinguishes Treg cells which express the chimeric GFP-Foxp3 fusion protein (Darce et al., 2012), and the IRF4-dependent signature in Treg cells (Zheng et al., 2009). P values form a chi-squared test. The genes and Probe IDs included in these signatures are listed in Table S2.
Flow cytometry
Surface staining was performed for 20 minutes at 4°C in PBS containing 0.1% sodium azide and 0.5% BSA. For intracellular cytokine stainings, cells were re-stimulated with phorbol 12-myristate 13-acetate (PMA, 50 ng/ml, Sigma), ionomycin (1 μg/ml, Sigma), and GolgiStop (1 μl/1ml, BD Bioscience) at 37°C in 10% CO2 for 4 h before staining was performed using the Cytofix/Cytoperm kit (BD Biosciences). Intracellular staining for Foxp3 was performed using the Foxp3 Staining Buffer Set (eBioscience). Antibodies were from BioLegend except for anti-Foxp3 (eBioscience), and anti-Ki67 (BD Biosciences). 7AAD was purchased from BD Biosciences. Samples were acquired on a FACSCalibur or LSRII flow cytometer (BD Biosciences) and analyzed using the FlowJo software (Tree Star).
Quantitative RT-PCR
RNA was extracted with RNAeasy mini Kits (Qiagen) and cDNA was prepared using the iScript cDNA synthesis kit (BioRad). Real-time PCR (RTPCR) was performed using Taqman probes and the 7500 Fast Real-Time PCR system (Applied Biosystems). All samples were normalized to b-actin internal control.
In vitro antibody treatment
CD4+Foxp3− effector T cells and CD4+Foxp3+ Treg cellss were sorted from Foxp3-GFP.KI reporter mice and stimulated at a density of 1 × 106/ml with plate bound anti-CD3 (145-2C11, 1 μg/ml), anti-CD28 (PV-1, 2 μg/ml), and anti-TIGIT (4D4, 100 μg/ml) or isotype control antibody. RNA was was isolated on day 3. Antibodies to human TIGIT were kindly provided by ZymoGenetics, Inc. (a wholly-owned subsidiary of Bristol-Myers Squibb). Cells were stimulated with anti-CD3 (UCHT1, 1 μg/ml), anti-CD28 (28.2, 1 μg/ml) and IL-2 (10 U/ml) in the presence of agonistic anti-TIGIT at 20 μg/ml or IgG isotype control. Gene expression was assessed on day 4.
In vivo antibody treatment
Mice were immunized s.c. with 200 μl of an emulsion containing 100 μg of MOG35–55 peptide (MEVGWYRSPFSRVVHLYRNGK) in adjuvant oil (CFA) on day 0 and treated i.p. with 100 μg of anti–TIGIT (clone 4D4) or isotype control Ab (armenian hamster IgG) on days 0, 2, 4, 10 and 17. For antigen-specific proliferation assays spleens and lymph nodes were collected on day 10 and 2.5 × 106 cells / ml were re-stimulated with 50 μg/ml MOG35-55 peptide. After 48h Fgl2 concentrations in culture supernatants were determined by ELISA (Biolegend).
ChIP-PCR and over-expression
ChIP assays were performed on P815 cells expressing TIGIT using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling) according to the manufacturers instructions. Lysates were immunoprecipitated using anti-C/EBPα antibody (8μg; Santa Cruz Biotech, sc-61) or rabbit IgG isotype control. Quantitative PCR reactions were performed with SYBR-Green on ChIP-bound and input DNA. % input = 2% × 2(CT 2% input sample – CT sample). For CEBPα over-expression CD4+Foxp3+ Treg cells were flow sorted from Foxp3-GFP.KI reporter mice. 5×105 Treg cells / ml were stimulated with Mouse T-Activator CD3/CD28 Dynabeads (Invitrogen) and transfected with 10μg/ml of Cebpa cDNA in pCMV6-Kan/Neo or the empty vector, which had been pre-incubated with FuGene 6 (Roche Diagnostics). RNA was extracted on day 4 with RNAeasy mini Kits (Qiagen), samples were treated with DNAse (RNAse-free DNAse set, Qiagen) and cDNA was prepared using the iScript cDNA synthesis kit (BioRad). Cebpa over-expression was verified by Taqman PCR.
Suppression of Th differentiation
For in vitro experiments CD4+CD62L+ naive T cell from CD45.1 mice and CD4+Foxp3+TIGIT+ or TIGIT− Treg cells from Foxp3-GFP.KI mice (CD45.2) were sorted and cultured at 105 Teff and 104 Treg cells/well. Cells were stimulated with Mouse T-Activator CD3/CD28 Dynabeads (Invitrogen, 0.6μl/well) in the presence of polarizing cytokines (Th1: 4ng/ml IL-12; Th2: 4ng/ml IL-4; Th17: 10ng/ml IL-6, 2ng/ml TGF-β; all cytokines from R&D). RNA was extracted after 3 days and flow cytometric analysis was performed on day 5.
For in vivo experiments 1-2 × 105 CD4+CD62L+CD25− sorted naive effector T cell and 2.5-5 × 104 CD4+CD25+TIGIT+ or TIGIT− Treg cells (Teff: Treg cell 4:1) from OT-II mice were transferred i.v. into WT recipients one day before immunization. To elicit a Th1 and Th17 cell response, mice were immunized with 10μg OVA (Sigma) emulsified in CFA and spleens and draining LN were analyzed 10 days later. Allergic airway inflammation was induced as described previously (Haworth et al., 2008; Rogerio et al., 2012). Briefly, mice were sensitized with 10μg OVA in alum i.p. on days 0 and 7 and challenged with 6% (wt/vol) OVA aerosol for 25 min on days 14, 15, 16 and 17. Cells from lung and bronchioalveolar lavage were analyzed directly following challenge on day 17.
Alternatively, 7.5 × 105 CD4+CD62L+CD25− sorted naive effector T cell from CD45.1 mice and 1.5 × 105 CD4+CD25+TIGIT+ or TIGIT− Treg cells from WT or _Fgl2_−/− mice (CD45.2) transferred i.v. into _Rag1_−/− recipients one day before immunization. Mice were immunized with 10μg OVA and 45μg anti-IFNγ (clone AN-18, Biolegend) in alum i.p. on days 0 and 7 and analyzed on day 14.
Colitis and histopathology
CD4+CD45RBhigh naive T cell from CD45.1 mice and CD4+Foxp3+ Treg cells from Foxp3-GFP.KI mice were purified by cell sorting after enrichment for CD4+ cells using anti-CD4 MACS beads. 8 × 105 CD4+CD45RBhigh cells were transferred i.v. into RAG1−/− mice, either alone or with Treg cells (4:1 effector T cell:Treg cell ratio) and mice were weighed weekly. At the time of sacrifice small and large intestine samples were fixed in neutral buffered formalin.
Routinely processed, paraffin-embedded tissue samples were stained with hematoxylin and eosin (H&E). The presence and severity of colitis was evaluated in a blinded manner and graded semi-quantitatively from 0 to 3 for the three following criteria: epithelial hyperplasia; leukocyte infiltration; and the presence of crypt abscesses. Scores for each criterion were added to give an overall inflammation score for each sample of 0–9.
Statistical analysis
Statistical significance was assessed either by 2-tailed Student's T-test (two groups) or ANOVA for multiple groups with a post hoc Tukey's test; P values < 0.05 were considered statistically significant. Statistical significance values indicated as follows: p < 0.05 (*), p < 0.01 (**) and p < 0.005 (***).
Supplementary Material
01
02
03
Highlights.
- TIGIT defines a distinct Foxp3+ Treg cell subset
- TIGIT induces transcription and secretion of the effector molecule Fgl2 in Tregs.
- TIGIT+ Treg cells suppress pro-inflammatory Th1 and Th17 cells but not Th2 responses.
- Selective suppression by TIGIT+ Treg cells is Fgl2-dependent.
Acknowledgements
We would like to thank Gary Levi for the Fgl2 reagents and KO mice, Deneen Kozoriz for cell sorting, Jenna Sullivan for technical assistance and members of the Kuchroo group, especially Anneli Peters, for discussions. This work was supported by grants P01AI056299 (to V.K.K., A.H.S. and C.B.), P01AI039671 (to V.K.K., D.A.H. and A.H.S.), RO1 NS 30843, R01NS035685 (to V.K.K.), 5T32HL007633 (to P.R.B.), K01DK090105 (to S.X.), AI075285 and AI093903 (to F.J.Q.) from NIH and RG4111A1 (to F.J.Q) and RG2751 (to V.K.K) from the National Multiple Sclerosis Society. N.J. is supported by the Swiss Multiple Sclerosis Society and the Arthritis National Research Foundation. E. L. is recipient of a Postdoctoral Scholarship in the Beatriu de Pinós Programme (2011-2013) (AGAUR-Government of Catalonia).
Abbreviations
Treg
regulatory T cell
TIGIT
T cell immunoglobin and ITIM domain
MOG
myelin oligodendrocyte glycoprotein
EAE
experimental autoimmune encephalomyelitis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, Kuchroo VK. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- Bettini ML, Pan F, Bettini M, Finkelstein D, Rehg JE, Floess S, Bell BD, Ziegler SF, Huehn J, Pardoll DM, Vignali DA. Loss of epigenetic modification driven by the Foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity. 2012;36:717–730. doi: 10.1016/j.immuni.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- Chan CW, Kay LS, Khadaroo RG, Chan MW, Lakatoo S, Young KJ, Zhang L, Gorczynski RM, Cattral M, Rotstein O, Levy GA. Soluble fibrinogen-like protein 2/fibroleukin exhibits immunosuppressive properties: suppressing T cell proliferation and inhibiting maturation of bone marrow-derived dendritic cells. J Immunol. 2003;170:4036–4044. doi: 10.4049/jimmunol.170.8.4036. [DOI] [PubMed] [Google Scholar]
- Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, Kallies A. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- Darce J, Rudra D, Li L, Nishio J, Cipolletta D, Rudensky AY, Mathis D, Benoist C. An N-terminal mutation of the Foxp3 transcription factor alleviates arthritis but exacerbates diabetes. Immunity. 2012;36:731–741. doi: 10.1016/j.immuni.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallarino F, Grohmann U, Hwang KW, Orabona C, Vacca C, Bianchi R, Belladonna ML, Fioretti MC, Alegre ML, Puccetti P. Modulation of tryptophan catabolism by regulatory T cells. Nat Immunol. 2003;4:1206–1212. doi: 10.1038/ni1003. [DOI] [PubMed] [Google Scholar]
- Flynn S, Toellner KM, Raykundalia C, Goodall M, Lane P. CD4 T cell cytokine differentiation: the B cell activation molecule, OX40 ligand, instructs CD4 T cells to express interleukin 4 and upregulates expression of the chemokine receptor, Blr-1. J Exp Med. 1998;188:297–304. doi: 10.1084/jem.188.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth O, Cernadas M, Yang R, Serhan CN, Levy BD. Resolvin E1 regulates interleukin 23, interferon-gamma and lipoxin A4 to promote the resolution of allergic airway inflammation. Nat Immunol. 2008;9:873–879. doi: 10.1038/ni.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Izcue A, Hue S, Buonocore S, Arancibia-Carcamo CV, Ahern PP, Iwakura Y, Maloy KJ, Powrie F. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity. 2008;28:559–570. doi: 10.1016/j.immuni.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joller N, Hafler JP, Brynedal B, Kassam N, Spoerl S, Levin SD, Sharpe AH, Kuchroo VK. Cutting edge: TIGIT has T cell-intrinsic inhibitory functions. J Immunol. 2011;186:1338–1342. doi: 10.4049/jimmunol.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Thomas KR, Perdue NR, Smigiel KS, Srivastava S, Campbell DJ. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor beta2. Immunity. 2012;37:501–510. doi: 10.1016/j.immuni.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- Levin SD, Taft DW, Brandt CS, Bucher C, Howard ED, Chadwick EM, Johnston J, Hammond A, Bontadelli K, Ardourel D, et al. Vstm3 is a member of the CD28 family and an important modulator of T-cell function. Eur J Immunol. 2011;41:902–915. doi: 10.1002/eji.201041136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, et al. Foxp3(+) follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano E, Dominguez-Villar M, Kuchroo V, Hafler DA. The TIGIT/CD226 Axis Regulates Human T Cell Function. J Immunol. 2012 doi: 10.4049/jimmunol.1103627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogerio AP, Haworth O, Croze R, Oh SF, Uddin M, Carlo T, Pfeffer MA, Priluck R, Serhan CN, Levy BD. Resolvin D1 and aspirin-triggered resolvin D1 promote resolution of allergic airways responses. J Immunol. 2012;189:1983–1991. doi: 10.4049/jimmunol.1101665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev I, Liu H, Koscik C, Bartczak A, Javadi M, Wong KM, Maknojia A, He W, Liu MF, Diao J, et al. Targeted deletion of fgl2 leads to impaired regulatory T cell activity and development of autoimmune glomerulonephritis. J Immunol. 2008;180:249–260. doi: 10.4049/jimmunol.180.1.249. [DOI] [PubMed] [Google Scholar]
- Thornton AM, Shevach EM. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J Exp Med. 1998;188:287–296. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- Xiao X, Balasubramanian S, Liu W, Chu X, Wang H, Taparowsky EJ, Fu YX, Choi Y, Walsh MC, Li XC. OX40 signaling favors the induction of T(H)9 cells and airway inflammation. Nat Immunol. 2012;13:981–990. doi: 10.1038/ni.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, Tom I, Ivelja S, Refino CJ, Clark H, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03