Assessing the clinical utility of cancer genomic and proteomic data across tumor types (original) (raw)
. Author manuscript; available in PMC: 2015 Jan 1.
Published in final edited form as: Nat Biotechnol. 2014 Jun 22;32(7):644–652. doi: 10.1038/nbt.2940
Abstract
Molecular profiling of tumors promises to advance the clinical management of cancer, but the benefits of integrating molecular data with traditional clinical variables have not been systematically studied. Here we retrospectively predict patient survival using diverse molecular data (somatic copy-number alteration, DNA methylation and mRNA, miRNA and protein expression) from 953 samples of four cancer types from The Cancer Genome Atlas project. We found that incorporating molecular data with clinical variables yielded statistically significantly improved predictions (FDR < 0.05) for three cancers but those quantitative gains were limited (2.2–23.9%). Additional analyses revealed little predictive power across tumor types except for one case. In clinically relevant genes, we identified 10,281 somatic alterations across 12 cancer types in 2,928 of 3,277 patients (89.4%), many of which would not be revealed in single-tumor analyses. Our study provides a starting point and resources, including an open-access model evaluation platform, for building reliable prognostic and therapeutic strategies that incorporate molecular data.
Introduction
The Cancer Genome Atlas (TCGA) project has yielded many biological insights through generating genomic, transcriptomic, epigenomic and proteomic data from a large number of patient samples in many cancer types1–6. However, the potential clinical utility of these data in aggregate remains largely unknown.
Large-scale molecular profiling data may be informative for multiple aspects of oncology practice. One key application for patients with primary disease is accurate prognosis, which helps stratify patients into different risk groups and choose both treatment and surveillance strategies. Traditionally, prognosis is based on clinical variables such as age and tumor stage. Recently, extensive efforts have been made to incorporate molecular information for better prognosis. For example, ER, PR and HER2 protein levels and HER2 genomic amplification are important biomarkers in breast cancer that have demonstrated high value in clinical use7. However, owing to the high cost of molecular profiling on a large scale, previous studies have either focused on a small number of selected genes or have employed only single-platform genomic data (e.g., microarrays). By convention, such studies have been limited to a single cancer lineage. Another important clinical application is to choose targeted therapies based on the alteration spectrum in an individual patient’s tumor. Multiple efforts have been initiated to apply high-throughput sequencing data in clinical strategies,8, 9 although alterations in clinically actionable genes have not been fully cataloged. Knowledge of this catalog may inform target selection for drug development as well as clinical trial design and identify patient populations that may benefit from emerging targeted therapeutics.
The overall goal of this study was to address how and to what extent TCGA molecular data could impact oncology practice. Thus, we evaluated two closely related but distinct aspects of clinical utility—prognostic utility (that is, predicting patient survival using various types of high-throughput molecular data across multiple tumor lineages) and therapeutic utility (that is, identifying the spectrum of somatic alterations in clinically actionable genes, which in the future may inform treatment selection). First, we examined the performance of molecular data (somatic copy number alteration (SCNA), DNA methylation and mRNA, microRNA and protein expression) alone or in combination with clinical variables in predicting censored or dichotomized patient survival data for four TCGA cancer types with high-quality overall survival data. Furthermore, to facilitate a broader community effort, we developed an open-access platform that allows researchers to build and evaluate survival prediction models on these datasets. Here we did not intend to generate prognostic models ready for clinical use, but rather we sought to provide insights into how to improve such models by incorporating informative molecular data. Second, we investigated the current spectrum of potentially clinically actionable alterations (somatic point mutations and small insertions/deletions) across 12 TCGA tumor types. By analyzing molecular data from multiple cancer types, we were able to evaluate prognostic models and identify alterations that would not have been obtained with single-tumor datasets.
Results
Assessment of prognostic power of diverse molecular data
We focused on four TCGA cancer types: kidney renal clear cell carcinoma (KIRC)6, glioblastoma multiforme (GBM)1, ovarian serous cystadenocarcinoma (OV)2 and lung squamous cell carcinoma (LUSC)4. These cancer types were chosen because their TCGA datasets included survival data with adequate follow-up time and sufficient samples characterized by multiple types of molecular data. The TCGA cohorts have overall survival patterns similar to those reported in previous publications10–13. For each cancer type, we compiled a core sample set in which each sample has information available for the overall survival time, clinical variables (e.g., gender, age, tumor stage and grade) and at least four out of the five types of molecular data related to gene expression (SCNA: Affymetrix Human SNP Array 6.0, ~100 arm or focal alterations; DNA methylation: Illumina DNA Methylation microarray, ~20,000 genes; mRNA expression: Agilent 244K microarray or Illumina mRNA-seq, ~20,000 genes; miRNA expression: Agilent Human miRNA-specific microarray or Illumina miRNA-seq, >500 microRNAs; protein expression: Reverse Phase Protein Array, ~170 proteins) (Table 1).
Table 1.
Overview of TCGA samples and high-throughput characterization platform information by cancer type
| Cancer | Overall Survival | SCNA | Methy | mRNA | miRNA | Protein | Core set |
|---|---|---|---|---|---|---|---|
| GBM | SNP_6 | 27k | AgilentG4502A | H-miRNA_8x15K | RPPA* | ||
| 565 | 563 | 287 | 492 | 491 | 214 | 210 | |
| KIRC | SNP_6 | 450k | HiseqV2 | GA+Hiseq | RPPA | ||
| 500 | 493 | 283 | 469 | 454 | 480 | 243 | |
| OV | SNP_6 | 27k | AgilentG4502A | H-miRNA_8x15K | RPPA | ||
| 563 | 559 | 600 | 558 | 586 | 412 | 379 | |
| LUSC | SNP_6 | 450k* | HiseqV2 | GA+Hiseq | RPPA | ||
| 305 | 343 | 225 | 220 | 351 | 195 | 121 |
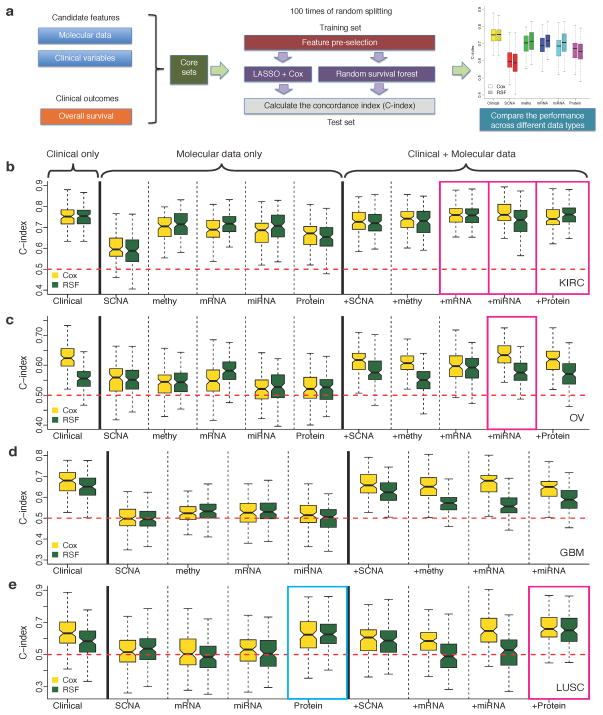
For each core sample set, we applied an approach of Monte Carlo cross-validation and assessed the predictive power of individual molecular data types or clinical variables using the concordance index (C-index)14. The C-index is a non-parametric measure to quantify the discriminatory power of a predictive model: C-index = 1 indicates perfect prediction accuracy and C-index = 0.5 is as good as a random guess (Online Methods). We compiled candidate features from molecular data or clinical data for each cancer type and randomly split the core set into training and test sets 100 times (Fig. 1a). We built the predictive models from the training set using two well-established, but highly complementary methods: (i) Cox, the multivariate Cox proportional hazards model with L1 penalized log partial likelihood (LASSO)15 for feature selection, and (ii) random survival forest (RSF)16.
Figure 1. Comparison of the survival predictive power of clinical variables, molecular data and their combinations.
(a) An overview of the computational approach. (b)–(e) C-indexes by models trained from clinical variables, individual molecular data alone or in combination with clinical variables in (b) KIRC (Ntotal = 243), (c) OV(Ntotal = 379), (d) GBM (Ntotal = 210) and (e) LUSC (Ntotal = 121). For each cancer type, during each of the 100 times of random splitting, 80% of the total samples were used to train the model and the remaining 20% as the test set for C-index calculations. The blue box highlights the model built from individual molecular data that shows comparable performance to that based on clinical variables (two-sided Wilcoxon signed rank test, P > 0.05); and the magenta boxes highlight the models integrating molecular data and clinical variables that show better performance than that based on only clinical variables (two-sided Wilcoxon signed rank test, FDR < 0.01).
For each cancer type, the clinical-variable-only models showed substantial predictive power, with C-indexes significantly higher than 0.5 (range: 0.624–0.754; P = 0) (Fig. 1b–e and Supplementary Figure 1). In 9 out of 18 cases, the models built from individual molecular data sets alone showed statistically significant predictive power (Supplementary Figure 1), but in only one case, the model built from LUSC protein expression data had predictive power similar to that of the corresponding clinical-variable-only model (Fig. 1e, C-index 0.632 vs. 0.626, P = 0.40, Wilcoxon signed rank test). The relative predictive power of individual molecular datasets strongly depended on the cancer type: for example, the prognostic power was much higher for KIRC than for the other three cancer types. In general, the trends observed with the Cox models were similar to those observed using the RSF models.
To examine whether genomic and proteomic data can provide additional prognostic power when used with clinical variables, we built predictive models by integrating clinical variables with each type of molecular data (both gene-level features and molecular subtype features) (Online Methods). Notably, the integrated models resulted in statistically significantly improved predictive power compared to those clinical-variable-only models in three cancer types, including mRNA, microRNA and protein expression in KIRC, miRNA expression in OV and protein expression in LUSC (one-sided Wilcoxon signed rank test, KIRC clinical + mRNA: P < 3.3×10−3, FDR < 0.035; clinical + miRNA: P < 1.2×10−4, FDR < 2.1×10−3; clinical + protein: P < 8.4×10−5, FDR < 2.1×10−3; OV clinical + miRNA: P < 7.0×10−5, FDR < 2.1×10−3; LUSC clinical + protein: P < 7.9×10−4, FDR < 0.011) (Fig. 1b–e). However, in terms of quantitative gain (i.e., the median value of Somers’ D14 across the 100 splits, a measurement for C-index change), the increase was limited (KIRC clinical + mRNA: 4.0%, clinical + miRNA: 7.4%, clinical + protein: 2.2%; OV clinical + miRNA: 13.7%; LUSC clinical + protein: 23.9%). In addition, we examined the effects of machine learning algorithms, feature selection and sample size of the training set on model performance (Supplementary text and Supplementary Figs 2–4).
To facilitate a broader community effort for such modeling, we developed an open-access platform that allows researchers to evaluate and submit survival prediction models in a “collaborative competition” research framework (Supplementary Fig. 5). The homepage of the TCGA Pan-Cancer Survival Prediction challenge can be accessed in Synapse (doi:10.7303/syn1710282). The site contains all models used in this study, including provenance records and transparent source code that allows each model to be inspected, rerun, or improved upon. Each model is linked to a standard set of meta-data annotations that provide online querying capability (e.g., corresponding to the cancer type or learning algorithm), allowing for the comparison of models based on user-defined criteria. The C-index scores for each model, as reported here, are displayed in the form of a real-time leaderboard.
Biological insights from top-performing prognostic models
For the top prognostic models highlighted in Figure 1, we further examined important molecular features included in each model to gain some mechanistic insights. The LUSC protein expression model is the only case where the molecular data alone showed a performance similar to that of the clinical-variable-only model. Features in the model with high predictive ability16 were dominated by proteins involved in DNA repair and microsatellite instability (e.g., MSH2) and metabolism (e.g., ACC1) (Supplementary Table 1).
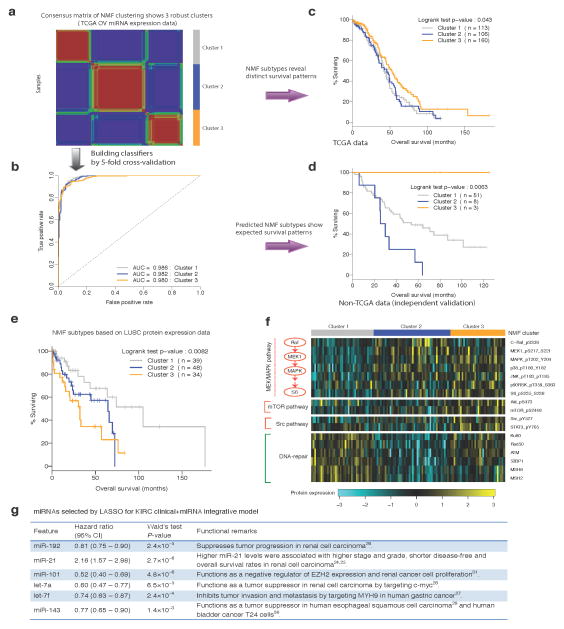
As shown in Figure 1, molecular data in five integrative models conferred additional prognostic power given clinical variables. Notably, in four of these models, the only contributing molecular feature was the molecular subtype derived from the corresponding expression data (through consensus nonnegative matrix factorization (NMF)17). Molecular subtypes can be regarded as higher-level assembles of individual gene features and therefore may act as a more robust predictor than an individual marker or small marker sets. Indeed, the NMF subtypes (derived from OV miRNA expression, LUSC protein expression and KIRC mRNA and protein expression data, respectively) showed distinct survival patterns in the respective cancer types(log-rank test, Fig. 2c, P < 0.043; Fig. 2e, P < 8.2×10−3; Supplementary Fig. 6a, P < 9.8×10−5; Supplementary Fig. 7a, P < 1.1×10−4).
Figure 2. Biological insights from the top prognostic models.
(a) Consensus nonnegative matrix factorization (NMF) clustering of the TCGA OV miRNA expression data reveals three molecular subtypes (clusters). (b) The ROC curves of the multiclass classifier against NMF subtypes trained from the TCGA OV miRNA expression data through five-fold cross-validation. (c) The Kaplan-Meier plot of the patients from the TCGA OV core set stratified by OV miRNA-expression NMF subtypes. (d) The Kaplan-Meier plot of the patients from the independent OV cohort stratified by predicted miRNA NMF subtypes using the classifier in (b). (e) The Kaplan-Meier plot of the patients from the LUSC core set stratified by LUSC protein-expression NMF subtypes. (f) The top differentially expressed protein markers among LUSC protein-expression NMF subtypes grouped by pathways/functions. (g) The miRNAs selected by LASSO for the KIRC clinical + miRNA integrative model.
Given the limited availability of suitable independent data in the public domain, we evaluated the performance of the OV clinical + miRNA model. Using the multiclass classifier built from TCGA OV miRNA expression data (Fig. 2a, Online Methods, the area under the receiver operating characteristic curve [AUC] = 0.98, Fig. 2b), we recovered the survival pattern of the NMF subtypes observed for the TCGA core set (Fig. 2c, log-rank test P < 0.043) in an independent cohort18 (Fig. 2d, log-rank test P < 6.3×10−3): the patients in cluster 3 have better survival than those in clusters 1 and 2 (prognostic miRNAs in each cluster are shown in Supplementary Table 2). Further, applying this trained model to the independent cohort yielded the expected improvement for including the miRNA NMF subtypes.
For the molecular subtypes defined by LUSC protein expression, pMEK1 and pMAPK and the downstream target pS6 were among the top markers expressed at higher levels in patients with shorter survival times (clusters 2 and 3, Fig. 2e,f). Clinical and preclinical data suggest that MEK inhibitors are active in specific subsets of non-small cell lung cancer, such as KRAS-mutated lung adenocarcinomas19, 20. Our results suggest that patients with high-risk forms of LUSC have relatively greater activation of the RAS/MEK/MAPK pathway and that MEK targeting warrants further exploration in this population as well. In addition, the mTOR and Src pathways may also be more active in cluster 3, whereas DNA-repair protein levels were low in both clusters 2 and 3 (Fig. 2f). Gene signatures associated with KIRC mRNA expression subtypes were aligned with their reported biological roles and survival patterns: many proteins involved in acute-phase response signaling and several pro-metastatic matrix metalloproteases were highly expressed in the groups with worse survival outcomes (clusters 2 and 3)21, 22, whereas death receptor signaling proteins were down-regulated in these groups23 (Supplementary Fig. 6a,b). The survival pattern of the NMF subtypes by KIRC protein expression also matches the survival correlations of individual protein markers (Supplementary Fig. 7a,b).
Finally, the KIRC clinical + miRNA model was the only integrative model for which individual gene features, instead of a molecular subtype derived from the complete expression dataset, provided additional prognostic power. Each of the six miRNAs comprising the signature was significantly correlated with survival, and their hazard ratio matched with previously reported roles in cancer progression. Although up-regulation of miR-21, which has growth promoting activity, is associated with a worse prognosis24, 25, the remaining miRNAs (miR-192, miR-101, let-7a, let-7f and miR-143) suppress tumor growth, with higher expression being associated with a better prognosis26–31 (Fig. 2g).
Patient survival prediction using cross-tumor models
To test whether molecular data could identify commonalities across different tumor types, we assessed whether a model trained using molecular data in one cancer type could predict survival in other cancer types that share the same type of molecular data generated by the same platform (Online Methods). In the vast majority of cases, the C-index was around 0.5, suggesting little predictive power across tumor types (Supplementary Fig. 8).
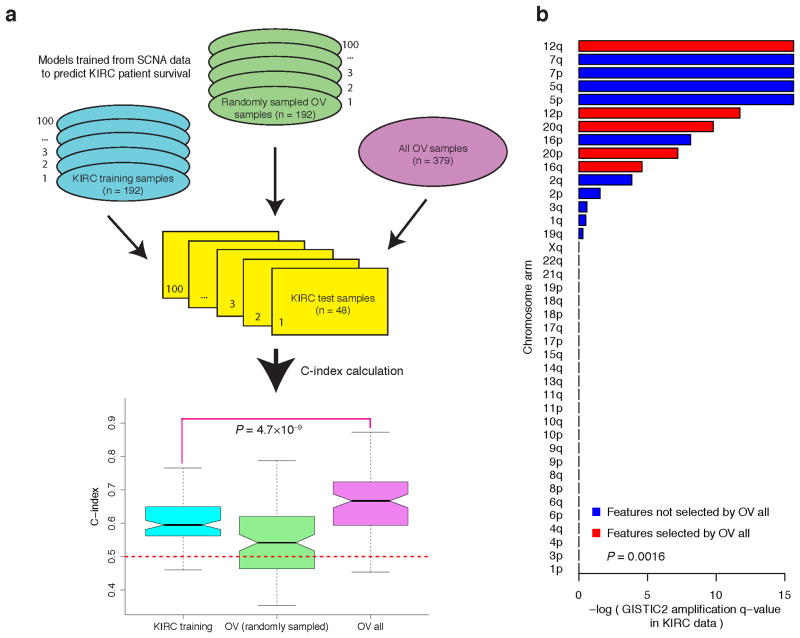
However, a model trained from OV SCNA data was predictive of survival for patients with KIRC, with a median C-index of 0.67 (Fig. 3a, Supplementary Fig. 9). Furthermore, given the same KIRC test sets, the OV model showed higher predictive power than a model trained from SCNA data of the KIRC training samples (Fig. 3a, median C-index, 0.67 vs. 0.59, P = 4.7×10−9, Wilcoxon signed rank test). When we randomly sampled the same number of OV samples as the KIRC training sets to build the predictive model, the C-index dropped from 0.67 to 0.54, suggesting that the higher predictive power was largely due to the larger sample size of the OV core set. We further confirmed this pattern using an independent approach and an independent sample partition (Supplementary Fig. 10, Online Methods). Closer examination revealed one common feature (“12q”) among the features selected from SCNA of the whole OV core set and KIRC core set, which may be crucial for the cross-tumor predictive power. In addition to 12q, there were another four arm-level SNCA features included in the model trained from OV SCNA data: 12p, 16q, 20p and 20q, all of which showed significant amplification of q-values in KIRC according to GISTIC232 (Fig. 3b). Indeed, the KIRC q-values of the features selected from OV SCNA data were lower than those not selected (Fig. 3b, P = 1.6×10−3, Wilcoxon rank sum test). The shared biological features identified above provide key insights into mechanistic connections between the two cancer types.
Figure 3. Models trained from OV SCNA data can predict survival of individuals with KIRC.
(a) From left to right: C-index for the models trained from KIRC SCNA data (Ntraining= 192), C-index for the models trained from SCNA of OV sample sets with the same size as the KIRC training sets (Ntraining = 192), and C-index for the model trained from SCNA of the whole OV core set (Ntraining = 379). The OV model trained from the whole OV core set showed higher predictive power than the model trained from SCNA data of independent KIRC training samples (two-sided Wilcoxon signed rank test, P = 4.7 × 10−9). (b) The bar plot of amplification Q-value of arm-level SCNA features from GISTIC2. The features included in the model trained from OV SCNA data are shown in red. The KIRC q-values of the features selected from OV SCNA data were lower than those not selected (two-sided Wilcoxon rank sum test, P = 1.6 × 10−3).
Factors affecting prediction of dichotomized survival data
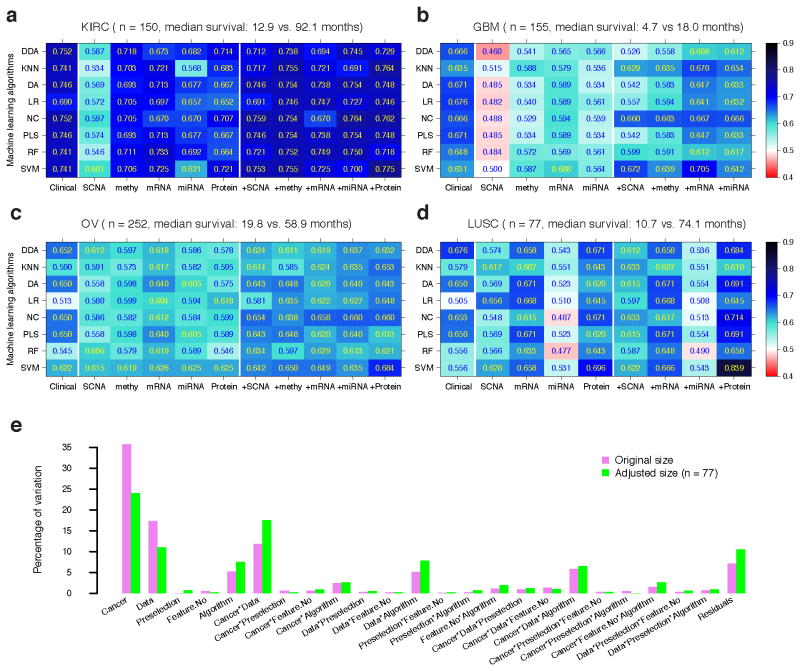
In addition to analysis on censored survival data, we examined the power of molecular data in predicting dichotomized overall survival data. Unlike censored survival data, there are many machine-learning algorithms for classifying binary clinical outcomes. Although the process of dichotomization will lose some information, this practice enables us to systematically survey many modeling scenarios and assess the effect of different factors on predicting survival data. For each cancer type, we dichotomized the censored continuous survival data through a designated cutoff time (survival milestone) (Online Methods), and then constructed a series of models from individual molecular data alone or with clinical variables by (i) using eight common classification algorithms, (ii) applying two feature pre-selection strategies and (iii) including different numbers of final features in the model. In total, we assessed the performance of >5,000 models through 10-fold cross-validation based on the threshold-independent AUC score.
Figure 4a–d and Supplementary Table 3 show the AUC score for each algorithm, with the optimal setting for each dataset and each cancer. Overall, as observed for continuous survival data, the predictive power of molecular data strongly depended on the cancer type. Clinical variables showed better performance than individual molecular data except for LUSC, where the protein expression data showed better performance than the clinical variables given the best-performing algorithms (Fig. 4d). Moreover, the integration of molecular data with clinical variables improved the predictive power, especially for the following datasets: DNA methylation and protein expression in KIRC using most algorithms (Fig. 4a), mRNA expression in GBM using K-nearest neighbor (KNN), nearest centroid (NC) and support vector machine (SVM) (Fig. 4b), and protein expression in OV and LUSC using most algorithms (Fig. 4c,d). To quantify the effects of specific factors on survival prediction, we performed an analysis of variance (ANOVA) on the AUC scores, and found that cancer type, data type and their interactions were the three dominant sources of variability, respectively explaining 35.7%, 17.4% and 11.8% of the variability of the prediction performance (Fig. 4e, violet bars). In contrast, the effect of machine-learning algorithms was moderate (5.2%). We obtained similar results when the sample size was kept consistent across all cancer types (Fig. 4e, green bars, Online Methods). These results were consistent with the recent microarray quality control (MAQC)-II study33.
Figure 4. Predictive performance of clinical variables, molecular data and their combination on dichotomized survival data.
The best AUC achieved by each classification algorithm for each clinical/molecular/combination dataset in (a) KIRC (Ntotal = 150), (b) GBM (Ntotal = 155), (c) OV (Ntotal = 252) and (d) LUSC (Ntotal = 77). (e) Variation explained by modeling factors and their interactions. Abbreviations for classification algorithms: diagonal discriminant analysis (DDA), K-nearest neighbor (KNN), discriminant analysis (DA), logistic regression (LR), nearest centroid (NC), partial least square (PLS), random forest (RF) and support vector machine (SVM). AUCs were calculated based on 10-fold cross-validation.
Somatic alterations in clinically relevant genes
Finally, we assessed the therapeutic utility of TCGA data by analyzing somatic mutations and small insertion/deletions (indels) in 3,277 patients across 12 tumor types. We applied a heuristic algorithm34, 35 to score the clinical importance of each alteration in 121 clinically relevant genes. Clinically relevant genes were defined as those that, when somatically altered, may predict resistance or response to a therapy and/or have diagnostic or prognostic relevance for a particular tumor type36 (Online Methods). It is important to emphasize that not all aberrations in clinically relevant genes will act as “drivers” and portend response to therapeutic targeting, and that a majority of the alterations in these clinically relevant genes remain variants of uncertain clinical significance and require further experimental and clinical evaluation.
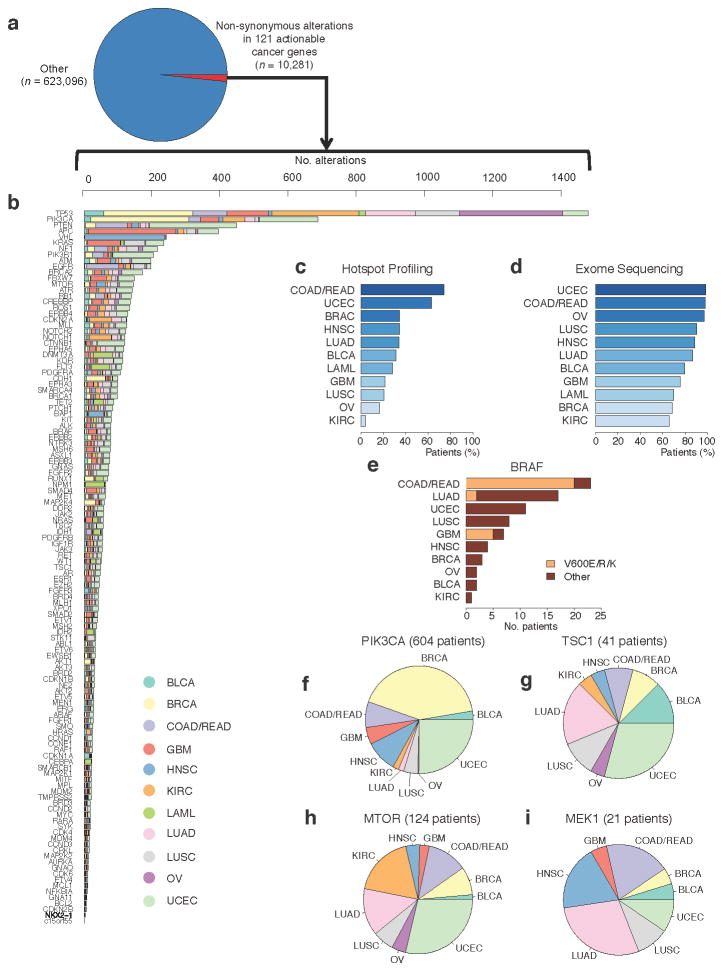
In 89.4% (2,928/3,277) of the TCGA patient samples, 10,281 somatic non-synonymous alterations (1.62% of all alterations, synonymous or non-synonymous, in this combined cohort) in 121 clinically relevant cancer genes were observed (Fig. 5a,b). Of these, 1,287 alterations in 31.4% (1,028/3,277) of patients were observed in genomic hotspots that were tested for in representative prospective clinical settings using a panel that probes events in 41 genes9. As expected, by extending genomic profiling to cover all exons of the same gene set, we observed a large increase in the number of observed alterations in clinically relevant genes in all 12 tumor types (Fig. 5c,d). This result reflects the gap in the understanding of the clinical relevance of a majority of alterations in genes potentially linked to clinical actions. To exclude the effect of hypermutated tumors in this cohort (e.g., colorectal, endometrial)3, 5, we repeated the analysis for tumors with mutation rates of ≤ 10 mutations/Mb (n = 2,892) as suggested in previous TCGA studies. We observed 3.5% (6,153/177,977) somatic alterations in the 121 clinically relevant genes (Supplementary Figure 11).
Figure 5. Alterations in clinically relevant genes across 12 tumor types.
(a)–(b) Examination of mutations and indels in 3,277 patients representing 12 tumor types reveals a long tail of the frequency distribution of alterations in clinically relevant genes that warrant further exploration across 12 tumor types. Expanding tumor profiling beyond hotspot profiling technologies (c) to whole exome sequencing (d) increases the percentage of patients in all tumor types that may harbor clinically relevant alterations. (e) Hotspot alterations in known cancer genes occur at low frequencies in unexpected tumor types. (f)–(i) Alterations in emerging genes with potential clinical relevance are observed across tumor types. For a key to the tumor types, see Supplementary Table 4.
These results highlight multiple themes pertaining to the use of genomics in clinical oncology. First, well-characterized clinically relevant alterations can rarely be observed in unexpected tumor types. For instance, one patient with cervical cancer harbored a somatic _BRAF_K601E mutation (Fig. 5e). While not previously reported in this tumor type, such a patient may warrant consideration for therapies that target these alterations37. This descriptive example highlights the potential benefit for comprehensive profiling in clinical settings, although prospective implementation of this approach is needed to determine the general applicability to clinical oncology.
Next, by combining mutation data from 12 tumor types, we observed a “tail” of low-frequency alterations in clinically relevant cancer genes that warrant clinical investigation but would not be apparent with smaller, single-tumor cohorts. This is demonstrated by a series of observations involving genes in the phosphatidylinositide 3-kinase/mTOR signaling pathway. In PIK3CA, both hotspot alterations and those requiring preclinical and clinical evaluation were observed in 604 patients (Fig. 5f). Somatic mutations in TSC1, which have been implicated in everolimus sensitivity in urothelial carcinomas38, 39, were noted in 41 patients with a diverse array of tumor types (Fig. 5g). Somatic alterations in MTOR itself, which may predict response and/or resistance to rapamycin analogs or mTOR catalytic domain inhibitors40, were observed in 124 patients (Fig. 5h).
Finally, comprehensive profiling may also be useful for identifying patients who may be intrinsically resistant to certain therapies. For instance, 21 therapy-naïve patients harbored MEK1 somatic mutations (Fig. 5i); a subset of these mutations may predict resistance to RAF and/or MEK inhibitors in specific clinical contexts41, 42. Critically, RAF and MEK inhibitors are currently being studied in numerous cancer types, so prospective knowledge of MEK1 status may impact treatment selection for patients with _MEK_1 mutations. Broadly, these results demonstrate how global surveys of mutational patterns in clinically relevant genes may impact clinical trial design and treatment selection.
Discussion
In contrast to previous studies driven by a single cancer type or data type, we systematically evaluated patient survival prediction from different molecular data types and described the potential prognostic and/or therapeutic relevance revealed across multiple cancer types, raising several important issues related to the potential clinical utility of large-scale molecular data.
Currently, only a few gene expression–based molecular prognostic markers have been established in clinical practice. For the cancers surveyed here, none of the previously reported gene expression signatures are routinely used in current clinical practice in lung and kidney cancer. For GBM, although the status of a few molecular markers (e.g., MGMT promoter methylation) is frequently ordered for patients, that finding exerts limited influence on clinical decision making43. For OV, CA125 is the only marker accepted for clinical use44. Our systematic assessment helped address one key issue related to the lack of prognostic markers with clinical utility: statistical significance vs. magnitude difference. Across the four TCGA patient cohorts, clinical variables appeared to be the most informative resources (C-index: 0.624–0.754); and molecular data alone often (9 out of 18 cases) had statistically significant predictive power above a random guess (C-index: 0.544–0.718). Given the clinical-variable-only models, incorporating molecular data statistically boosts the model performance (5 out of 18 cases) in three cancer types, including mRNA, miRNA and protein expression, especially their molecular subtype information. However, the absolute magnitude gains were very limited (Somers’ D, 2.2~22.9%; a 2.2% gain in Somers’ D corresponds to a 2.2% increase of rank correlation coefficient between the predicted risk score and the actual survival of the patients), suggesting that the information content of clinical variables and molecular data are largely redundant in terms of patient survival stratification. This echoes the observation that the number of cancer prognostic molecular markers in clinical use is pitifully small, despite decades of protracted and tremendous efforts45, 46. Currently, many investigators conclude the utility of their markers of interest by heavily relying on _P_-value rather than the size of the difference in patient outcomes47. Our study calls attention to the criteria of magnitude difference that should be emphasized in future publications of tumor prognostic markers.
Another important related issue is reliability and reproducibility. The literature of tumor biomarkers is plagued by publication bias and selective/incomplete reporting46, which especially poses an acute challenge in the post-genomic era. In this regard, we have developed an open-access model assessment platform for TCGA pan-cancer survival prediction, which will (i) reduce the barrier to analyzing TCGA data by providing access to well curated, computable datasets used as inputs to all models in our study; (ii) increase the transparency and reproducibility of prognostic models by providing our models as re-runnable source code and providing this capability to other researchers; and (iii) improve the objectivity and rigor of future model assessments by providing a baseline set of model scores based on pre-defined criteria and evaluation scores posted on a real-time, publically available “leaderboard”. Such an effort not only helps improve prognostic models though a community-based challenge48, 49, but also ensures transparency and reproducibility for tumor biomarker identification. We expect to seed such a community effort to release the whole dataset, prognostic models and evaluation criteria for future studies of clinically usable prognostic models.
By exploring the spectrum of clinically actionable somatic alterations among 12 tumor types, we identified multiple instances where alterations in clinically relevant genes were observed in enough patients to rationalize clinical trial development across tumor types. This was true even if these alterations were rarely observed in any single tumor type, leading to so-called “bucket” trials. We also revealed how the potential applications of precision oncology can be expanded to numerous additional patients with more extensive forms of profiling. As the number of identified clinically actionable cancer genes and alterations continues to rise8, prospective genomic profiling may inform individualized treatment plans for patients with metastatic or localized disease, as these patients are guided toward genomically-driven clinical trials. Importantly, many of the alterations observed in clinically actionable cancer genes have either no known functional effect that may be consistent with a clinical action or even have the converse effect. Through our cross-tumor analysis, we expect that many of these alterations will emerge in preclinical and clinical studies, thereby informing their relative impact in specific clinical settings from a predictive/prognostic standpoint when linked to relevant clinical outcomes.
Although our study provides important insights into the translation of biological data into clinical utility, it has some limitations. First, we employed purely data-mining approaches to prognostic modeling. Such a practice comes with a cost: we may miss some informative individual features that could be identified by a candidate gene approach driven by prior knowledge. Second, we did not analyze somatic mutations for prognostic utility because the mutation data are binary and sparse across the patient cohorts. New methods should be developed for assessing the prognostic power of large-scale mutation data. Third, effectively combining multiple types of molecular data remains a technical challenge owing to the over-fitting issue and widespread co-linearity of large-scale biological data. Therefore, one important future direction is to build prognostic models that incorporate clinical variables and multiple types of molecular data, which may provide crucial complementary information. In that regard, more effective feature selection strategies should be developed. Finally, because TCGA patient samples were collected from multiple source sites for the purpose of comprehensive molecular profiling and were characterized at different centers, this practice may introduce both heterogeneity and bias. In addition, the resulting clinical annotation of patient samples may not be as rigorous and complete as those obtained from clinical trials. Therefore, further efforts, especially independent validations with clinical trial-grade follow-up, are crucial for assessing our findings from TCGA data.
Online Methods
Core dataset compilation
We downloaded overall survival data and SCNA data from Firehose (https://confluence.broadinstitute.org/display/GDAC/Home). We obtained the clinical variables from TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/) and the molecular data (including DNA methylation, mRNA, miRNA and protein expression) from the Pan-cancer project (syn300013) on Synapse (http://www.synapse.org). Specifically, molecular data from the following platforms were used in our study. For SCNA, the platform is Affymetrix Genome-Wide Human SNP Array 6.0 (arm-level and focal-level copy number calls were derived from Firehose: https://confluence.broadinstitute.org/display/GDAC/Home). For DNA methylation, the platform is Illumina Infinium Human DNA Methylation 27K (for GBM and OV) or 450K (for KIRC and LUSC, we retained only the probes that most negatively correlated with gene expression according to Firehose). For mRNA expression, the platform was either Agilent 244K Custom Gene Expression G4502A (for GBM and OV) or Illumina HiSeq 2000 RNA Sequencing V2 (for KIRC and LUSC). For miRNA expression, the platform was either Agilent 8×15K Human miRNA-specific microarray platform (for GBM and OV) or Illumina Genome Analyzer/HiSeq 2000 miRNA sequencing platform (for KIRC and LUSC). For protein expression, the platform was the MD Anderson Reverse Phase Protein Array (RPPA) Core platform and both total protein and phosphorylated protein were included in this study as distinct molecular features. For each cancer type, we defined the sample intersection across all the platforms as the core sample set. We included neither DNA methylation data in the LUSC core, nor protein expression data in the GBM core in order to preserve a statistically sufficient sample size. For each molecular data type, in additional to gene-level features, we included the NMF subtypes from Firehose analyses reported on January 16, 2013. All core clinical and genomic/proteomic data used to construct survival models, as well as the training and test dataset splits, are available at the Synapse homepage of our project (accession number syn1710282, doi:10.7303/syn1710282). The full Pan-Cancer dataset is available at the Synapse Pan-Cancer home page (accession number syn300013, doi:10.7303/syn300013).
Model training and performance comparison
For each core set, we randomly split the samples into two groups: 80% as the training set and 20% as the test set. On the training set, we first performed a pre-selection step to keep the top significant features correlated with overall survival (univariate Cox model, likelihood ratio test P < 0.05). To obtain better convergence of the training model, we required that the retained feature number did not exceed the number of events (deaths) in the training set. We used two computational methods to train the models: (i) Cox: the Cox proportional hazards model with LASSO for feature selection, and (ii) RSF: random survival forest. The univariate and multivariate Cox models were built with the R package “survival”; the LASSO was performed using the R package “glmnet” and the penalty parameter λ was chosen based on the 5-fold cross-validation within the training set; and the RSF models were built using the R package “RandomSurvivalForest” with the recommended default parameters. We then applied the models thereby obtained to the test set for prediction, and calculated the C-index using the R package “survcomp”. For each core set, the above procedure was repeated 100 times to generate 100 C-indexes. To compare the performance across different data types, we first chose the better performing method (Cox or RSF) and then used its results based on the Wilcoxon signed rank test to calculate the P value (using 0.05 as the significance cutoff).
To assess the predictive power of integrating molecular data with clinical variables, we slightly modified the Cox method to include both clinical and molecular features. We used the clinical features (such as patient age and gender, tumor stage and grade, and Karnofsky performance score, upon availability) that were significantly correlated with survival (likelihood ratio test P < 0.05 in both the univariate Cox model and the full model with all clinical variables) as the baseline to build the clinical Cox model. We then combined the gene-level features that better fit the existing model (through performing a feature selection step against the residuals) or the subtype features with the clinical variables to build a new multivariate Cox model. We performed the RSF method as before.
To evaluate the effect of feature selection, for the data exhibiting striking discrepancies (i.e., clinical + molecular data for GBM and LUSC), we applied different feature selection methods before RSF: i) the same LASSO approach as for Cox; ii) minimal depth variable selection; and iii) variable hunting. We calculated the C-indexes when applying these new models to the same test sets. To evaluate the effect of sample size, for the molecular models with substantial predictive power (median C-index > 0.6), we conducted a serial sampling of various portions (ranging from 0.2–1, with a increment of 0.1) of the original training samples as the new training set, from which we built the models using the same approach and calculated the C-index when applying these models to the same test set.
Building multiclass classifier from known NMF subtypes
Using the TCGA OV miRNA expression data as the explanatory variables and the three-class NMF subtypes derived from these expression data by Firehose as the response variable, we built the multiclass classifier (multinomial logistic regression model) using a scheme adapted from Yuan et al50. We first cleaned out the expression data by retaining the common features between TCGA data and the independent dataset and performed sample-wise centering. We then built the multiclass classifier from the TCGA data and evaluated its performance through 5-fold cross validation. During each of the five iterations, feature selection by LASSO (class = “multinomial”) and tuning of the penalty parameter λ were based on 80% of the data, and the prediction (using the R package “glmnet”) was made to the remaining 20% of the data, where class labels was assigned according to the class with the largest probability. The predictions from the five iterations were combined and the AUCs were calculated by the R package “ROCR”. Finally, we trained the classifier using the whole TCGA data and applied it to the independent OV dataset for the final prediction.
Analysis of important biological features in the top prognostic models
The Kaplan-Meier survival curves were drawn according to the samples’ original NMF subtypes or predicted classes, and the log-rank test _P_-values were calculated using the R package “survival”. The association of individual features with survival (better or worse) was decided based on the hazard ratio (HR) from the univariate Cox model, where an HR greater than 1 represented a worse prognosis. Wald’s test _P-_values were used to assess the significance. For expression values obtained through sequencing (i.e., KIRC miRNA expression), the raw values were log2 transformed before the univariate Cox analysis. The enriched gene pathways were identified through IPA (Ingenuity Systems®, www.ingenuity.com).
Cross–tumor-type survival prediction
For the cancers that shared the same platforms for the same type of genomic data (e.g., microarray for GBM and OV mRNA expression, RNA-seq for KIRC and LUSC mRNA expression), we first obtained the common features shared by any two cancers in a pairwise manner. Then we trained the Cox model from the shared molecular features of one cancer and applied the model trained to the same 100 test sample sets utilized in the global analysis of the other cancer. C-indexes were calculated accordingly. To test the effect of sample size, for the OV-KIRC case, we randomly sampled the same number of OV samples as the KIRC training size and repeated the whole analysis. We used the Wilcoxon signed rank test to compare the performance of the model trained with data from different cancers. We further confirmed this result using an independent approach and an independent sample partition (Supplementary Fig. 11). The sample partition was done using standard 5-fold cross-validation on the KIRC data. For each fold, we trained non-parametric unregularized Cox proportional hazards models using the glmnet package in R. We repeated the cross-validation process 30 times, randomly choosing a 5-fold split each time. Finally, the KIRC patients were stratified using the SCNA signatures derived from OV, and the Kaplan-Meier curves were drawn based on the median risk score.
Dichotomization of survival data
For each cancer type, we dichotomized the censored continuous survival data by assigning a cutoff time (survival milestone) of 1 year for individuals with GBM, 2 years for LUSC, 3 years for OV and 4 years for KIRC. The individuals who lived beyond the cutoff time were labeled as 1; the deceased were labeled as 0. The individuals with survival times that were censored before the cutoff were excluded. The different cutoffs were chosen in order to reach a balance between the event ratio and the sample size. The dichotomized survival data were deposited in Synapse (syn1748545).
Classification algorithms for dichotomized survival data
For each dichotomized survival dataset, we used two pre-selection strategies (ANOVA and shrinking centroids) and 8 classification algorithms: diagonal discriminant analysis (DDA), K-nearest neighbor (KNN), discriminant analysis (DA), logistic regression (LR), nearest centroid (NC), partial least square (PLS), random forest (RF) and support vector machine (SVM). The performance was assessed in 10-fold cross-validation, and AUCs were calculated as the measurement. For KNN, we varied the number of candidate neighbors from 1 to 9 (odd numbers only) and used the Euclidean distance as the measure of distance. For NC, we assigned equal prior probabilities. For DDA, DA, and PLS, we chose the linear discriminant function with equal prior probabilities. For SVM, we chose the radial basis kernel. The parameter C (the cost) ranged from 1 to 1001, in increments of 100, and gamma ranged from 10−10 to 10−2, moving one decimal place per time. For RF, 1000 trees were used. The other parameters were chosen by default. The number of features after pre-selection ranged from 10 to 50, in increments of 10.
Variability analysis for modeling factors
There are five factors that can potentially affect the performance of binary classification: cancer type, data type (clinical or individual molecular features), pre-selection strategies, the number of features after pre-selection and classification algorithms. We used ANOVA to assess the variability contributed by these factors and their interactions, and the Akaike information criterion in stepwise model selection for significant factors and interactions. The estimated variance components were then divided by their total in order to compare the proportion of variability explained by each modeling factor. To remove the effect of sample size, we performed a size-adjusted analysis, in which we kept the sample size consistent across all cancer types by randomly sampling 77 samples according to the smallest sample (which was LUSC) from KIRC, GBM and OV, and repeated the same procedure for the original dichotomized sets.
Identification of somatic alterations in clinically relevant genes
Somatic mutations and indels called from exome sequencing of matched tumor and normal genome pairs from 12 TCGA projects were aggregated using mutation annotation format (MAF) files from Synapse (syn1710680). Each alteration was ranked for clinical relevance using a heuristic algorithm34, 35. Clinical actionability was defined at the gene level: any gene that, when somatically altered in cancer, predicted response or resistance to a specific therapy, had diagnostic potential, or had prognostic significance, was considered a clinically actionable gene. These genes were derived by a review of the primary literature, consultation with experts, and manual curation. A complete list is available at www.broadinstitute.org/cancer/cga/target. For the purpose of understanding the distribution of hotspot alterations in BRAF and PIK3CA, the following definitions were assigned: hotspot alterations in BRAF were defined as those leading to V600E, V600K, and V600R protein changes. Similarly, hotspot alterations in PIK3CA were restricted to those that resulted in E545K and H1047R protein changes. All code for this effort was generated using the R statistical package.
Supplementary Material
1
Acknowledgments
We gratefully acknowledge contributions from the TCGA Research Network and the TCGA Pan-Cancer Analysis Working Group (contributing consortium members are listed in the Supplementary Note). The TCGA Pan-Cancer Analysis Working Group is coordinated by J. M. Stuart, C. Sander and I. Shmulevich. This study was supported by the National Institutes of Health (CA143883 to G.B.M. and J.N.W., CA175486 to H.L., and CCSG grant CA016672); UTMDACC – G.S. Hogan Gastrointestinal Research Fund, NCI-MDACC Uterine SPORE career development award and the Lorraine Dell Program in Bioinformatics for Personalization of Cancer Medicine to J.N.W.; Dana-Farber Leadership Council to E.M.V.; Conquer Cancer Foundation to E.M.V.
Footnotes
Disclosures: Consultant/Equity holders in Foundation Medicine (N.W., L.A.G.), and Consultant for Kew (G.G.).
Author Contributions
L.A.G., A.A.M., G.G. and H.L. conceived and designed the study; Y.Y., E.M.V., L.O., N.W., A. A.-M., A.S., L.A.B., Y.X., K.R.H., L.D., L.H., X.H., M.S.L., J.N.W., J.M.S., G.B.M., L.A.G., A.A.M., G.G. and H.L. performed data analysis; Y.Y., E.M.V., A.A.M. and H.L. wrote the manuscript with inputs from other authors; and H.L. supervised the whole project.
The authors declare no competing interests.
References
- 1.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weigel MT, Dowsett M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocrine-related cancer. 2010;17:R245–262. doi: 10.1677/ERC-10-0136. [DOI] [PubMed] [Google Scholar]
- 8.Garraway L. Genomics-driven oncology: framework for an emerging paradigm. Journal of Clinical Oncology. 2013;31:1806–1814. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 9.MacConaill L, et al. Profiling critical cancer gene mutations in clinical tumor samples. PloS One. 2009;4 doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berchuck A, et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clinical Cancer Research. 2005;11:3686–3696. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
- 11.Douillard JY, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol. 2006;7:719–727. doi: 10.1016/S1470-2045(06)70804-X. [DOI] [PubMed] [Google Scholar]
- 12.Heng DY, et al. External validation and comparison with other models of the International Metastatic Renal-Cell Carcinoma Database Consortium prognostic model: a population-based study. Lancet Oncol. 2013;14:141–148. doi: 10.1016/S1470-2045(12)70559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson DR, O’Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. Journal of Neuro-oncology. 2012;107:359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 14.Harrell F, Lee K, Mark D. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statistics in Medicine. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 15.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B. 1996;58:267–288. [Google Scholar]
- 16.Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. The Annals of Applied Statistics. 2008:841–860. [Google Scholar]
- 17.Brunet JP, Tamayo P, Golub T, Mesirov J. Metagenes and molecular pattern discovery using matrix factorization. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shih K, et al. A microRNA survival signature (MiSS) for advanced ovarian cancer. Gynecologic oncology. 2011;121:444–450. doi: 10.1016/j.ygyno.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Janne PA, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14:38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- 20.Ohashi K, et al. Characteristics of lung cancers harboring NRAS mutations. Clinical Cancer Research. 2013;19:2584–2591. doi: 10.1158/1078-0432.CCR-12-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falconer JS, et al. Acute-phase protein response and survival duration of patients with pancreatic cancer. Cancer. 1995;75:2077–2082. doi: 10.1002/1097-0142(19950415)75:8<2077::aid-cncr2820750808>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Kallakury BV, et al. Increased expression of matrix metalloproteinases 2 and 9 and tissue inhibitors of metalloproteinases 1 and 2 correlate with poor prognostic variables in renal cell carcinoma. Clinical Cancer Research. 2001;7:3113–3119. [PubMed] [Google Scholar]
- 23.Antoon JW, et al. Altered death receptor signaling promotes epithelial-to-mesenchymal transition and acquired chemoresistance. Scientific reports. 2012;2:539. doi: 10.1038/srep00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faragalla H, et al. The Clinical Utility of miR-21 as a Diagnostic and Prognostic Marker for Renal Cell Carcinoma. J Mol Diagn. 2012;14:385–392. doi: 10.1016/j.jmoldx.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Zaman MS, et al. Up-regulation of microRNA-21 correlates with lower kidney cancer survival. PLoS One. 2012;7:e31060. doi: 10.1371/journal.pone.0031060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khella H, et al. miR-192, miR-194 and miR-215: a convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis. 2013 doi: 10.1093/carcin/bgt184. [DOI] [PubMed] [Google Scholar]
- 27.Liang S, et al. MicroRNA let-7f inhibits tumor invasion and metastasis by targeting MYH9 in human gastric cancer. PLoS One. 2011;6:e18409. doi: 10.1371/journal.pone.0018409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Yin B, Zhang C, Zhou L, Fan J. Hsa-let-7a functions as a tumor suppressor in renal cell carcinoma cell lines by targeting c-myc. Biochemical and Biophysical Research Communications. 2012;417:371–375. doi: 10.1016/j.bbrc.2011.11.119. [DOI] [PubMed] [Google Scholar]
- 29.Ni Y, et al. MicroRNA-143 functions as a tumor suppressor in human esophageal squamous cell carcinoma. Gene. 2012 doi: 10.1016/j.gene.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 30.Noguchi S, et al. MicroRNA-143 functions as a tumor suppressor in human bladder cancer T24 cells. Cancer Letters. 2011;307:211–220. doi: 10.1016/j.canlet.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Sakurai T, et al. The enhancer of zeste homolog 2 (EZH2), a potential therapeutic target, is regulated by miR-101 in renal cancer cells. Biochemical and Biophysical Research Communications. 2012;422:607–614. doi: 10.1016/j.bbrc.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 32.Mermel C, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biology. 2011;12 doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi L, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nature biotechnology. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Allen EM, Wagle N, Levy MA. Clinical analysis and interpretation of cancer genome data. Journal of Clinical Oncology. 2013;31:1825–1833. doi: 10.1200/JCO.2013.48.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Allen EM, et al. Whole-exome sequencing and clinical interpretation of FFPE tumor samples to guide precision cancer medicine. Nature Medicine. 2014 doi: 10.1038/nm.3559. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagle N, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discovery. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim KB, et al. Phase II Study of the MEK1/MEK2 Inhibitor Trametinib in Patients With Metastatic BRAF-Mutant Cutaneous Melanoma Previously Treated With or Without a BRAF Inhibitor. Journal of Clinical Oncology. 2013;31:482–489. doi: 10.1200/JCO.2012.43.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyer G, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krueger DA, et al. Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis. N Engl J Med. 2010;363:1801–1811. doi: 10.1056/NEJMoa1001671. [DOI] [PubMed] [Google Scholar]
- 40.Wagle N, et al. Activating mTOR Mutations in a Patient with an Extraordinary Response on a Phase I Trial of Everolimus and Pazopanib. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-13-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Allen EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagle N, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holdhoff M, et al. Use of personalized molecular biomarkers in the clinical care of adults with glioblastomas. Journal of Neuro-oncology. 2012;110:279–285. doi: 10.1007/s11060-012-0968-3. [DOI] [PubMed] [Google Scholar]
- 44.Sturgeon C, et al. National Academy of Clinical Biochemistry laboratory medicine practice guidelines for use of tumor markers in testicular, prostate, colorectal, breast, and ovarian cancers. Clinical Chemistry. 2008;54:79. doi: 10.1373/clinchem.2008.105601. [DOI] [PubMed] [Google Scholar]
- 45.McShane LM, Altman DG, Sauerbrei W. Identification of clinically useful cancer prognostic factors: What are we missing? Journal of the National Cancer Institute. 2005;97:1023–1025. doi: 10.1093/jnci/dji193. [DOI] [PubMed] [Google Scholar]
- 46.McShane LM, Hayes DF. Publication of Tumor Marker Research Results: The Necessity for Complete and Transparent Reporting. Journal of Clinical Oncology. 2012;30:4223–4232. doi: 10.1200/JCO.2012.42.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henry NL, Hayes DF. Uses and abuses of tumor markers in the diagnosis, monitoring, and treatment of primary and metastatic breast cancer. Oncologist. 2006;11:541–552. doi: 10.1634/theoncologist.11-6-541. [DOI] [PubMed] [Google Scholar]
- 48.Bilal E, et al. Improving breast cancer survival analysis through competition-based multidimensional modeling. PLoS Computational Biology. 2013;9 doi: 10.1371/journal.pcbi.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Margolin A, et al. Systematic analysis of challenge-driven improvements in molecular prognostic models for breast cancer. Science Translational Medicine. 2013;5 doi: 10.1126/scitranslmed.3006112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan Y, Xu Y, Xu J, Ball R, Liang H. Predicting the lethal phenotype of the knockout mouse by integrating comprehensive genomic data. Bioinformatics. 2012;28:1246–1252. doi: 10.1093/bioinformatics/bts120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1