A G-Quadruplex-Containing RNA Activates Fluorescence in a GFP-Like Fluorophore (original) (raw)
. Author manuscript; available in PMC: 2015 Feb 1.
Published in final edited form as: Nat Chem Biol. 2014 Jun 22;10(8):686–691. doi: 10.1038/nchembio.1561
Abstract
Spinach is an in vitro selected RNA aptamer that binds a GFP-like ligand and activates its green fluorescence.Spinach is thus an RNA analog of GFP, and has potentially widespread applications for in vivo labeling and imaging. We used antibody-assisted crystallography to determine the structures of Spinach both with and without bound fluorophore at 2.2 and 2.4 Å resolution, respectively. Spinach RNA has an elongated structure containing two helical domains separated by an internal bulge that folds into a G-quadruplex motif of unusual topology. The G-quadruplex motif and adjacent nucleotides comprise a partially pre-formed binding site for the fluorophore.The fluorophore binds in a planar conformation and makes extensive aromatic stacking and hydrogen bond interactions with the RNA. Our findings provide a foundation for structure-based engineering of new fluorophore-binding RNA aptamers.
Introduction
The discovery, characterization and development of green fluorescent protein (GFP) have revolutionized biomedical research. By virtue of the hydroxybenzylideneimidazolinone (HBI) fluorophore that forms auto-catalytically from residues in the β-barrel cage of the nascent protein1, GFP and its derivatives have become indispensable biological agents for in vivo labeling and imaging2. Inspired by the structure and mechanism of GFP, engineering and grafting have produced a family of colored fluorescent proteins that span a broad spectrum of emission wavelengths from cyan to infrared3,4.
The demand for analogous techniques for investigation of RNA biology sparked the recent development of fluorescent RNA modules. In vitro selections of RNA aptamers that bind a range of synthetic GFP-like HBI fluorophores have generated a novel family of RNA-fluorophore complexes lighting up with diverse colors5,6. One of these aptamers, named Spinach, and its more stable variant, Spinach26, mimics the fluorescent properties of enhanced GFP (EGFP). Spinach binds the phenolate form of an HBI derivative, 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI) and selectively activates its fluorescence. This fluorophore is cell permeable and undergoes minimal photobleaching when bound to Spinach, making it an excellent modality for imagingand labeling5–7. Recently, Spinach has been adapted for use as a genetically encoded RNA sensor for metabolite imaging8,9 as well as a tool for synthetic biology applications10.
We crystallized the minimal form of Spinach RNA (aptamer 24-2-min5, referred to simply as “Spinach” throughout this manuscript) using the antibody-assisted RNA crystallography approach developed in our laboratory11 and obtained the structure of the DFHBI-bound and unbound states at 2.2 and 2.4 Å resolution, respectively. (Supplementary Results, Supplementary Table 1). We show that Spinach adopts an elongated conformation, with two helical segments flanking a unique G-quadruplex motif that serves as a platform for fluorophorebinding. Our findings provide a foundation for structure-based engineering of new fluorophore-binding RNA aptamers.
Results
Antibody-assisted crystallography
We replaced the wild-type stem-loop (UUCG) of Spinach helix P2 with a pentaloop hairpin graft from the class I ligase ribozyme to create a binding site for the crystallization chaperone Fab BL3-612 (Fig. 1a, nucleotides 37–43). The Fab-RNA complex formed with high affinity (KD = 25 ± 6 nM; Supplementary Fig. 1a), comparable to that previously reported for Fab BL3-6 binding to either the class I ligase ribozyme or the stem-loop in isolation12. Neither the hairpin graft nor the bound Fab affected the fluorescence spectrum of the Spinach-DFHBI complex relative to that of the original aptamer (Supplementary Fig. 1b).
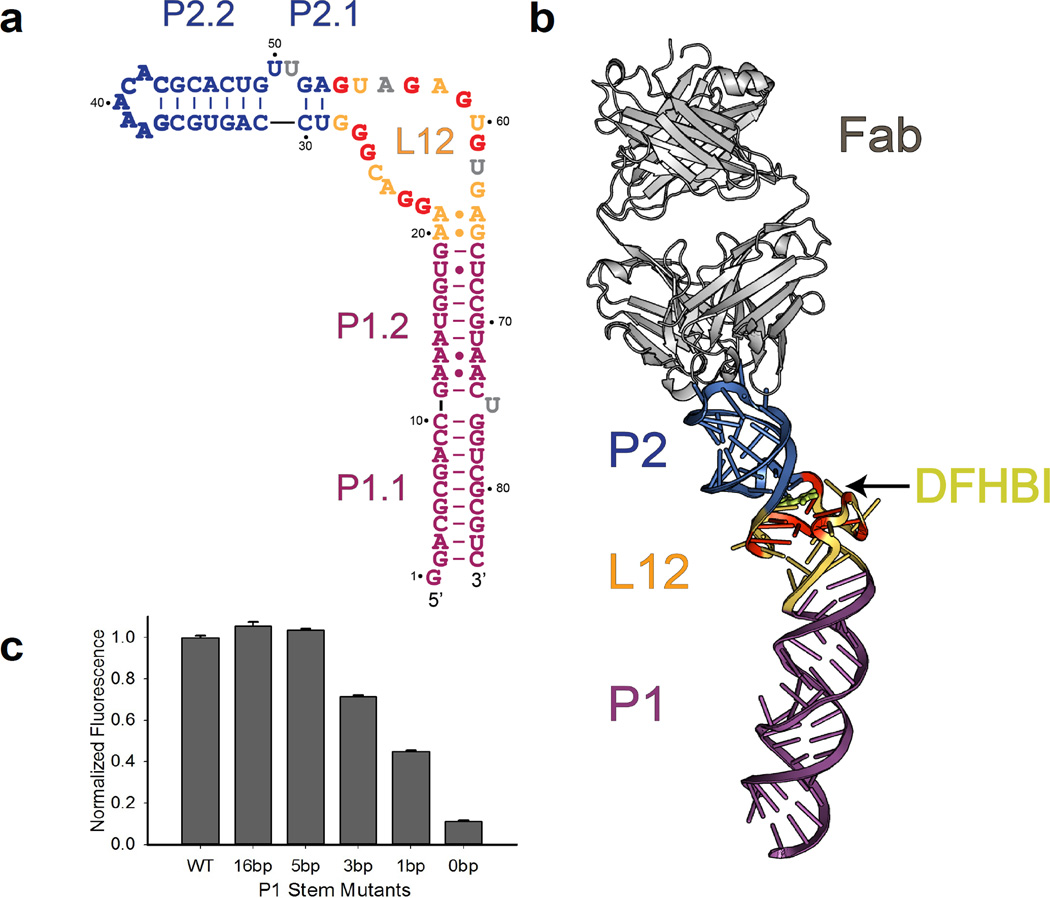
Figure 1. Global structure of the Spinach RNA-Fab complex.
(a). Observed secondary structure of Spinach construct containing G37AAACAC43 antigenic tag (bold blue letters). The L12 region (brown-yellow) contains a G-quadruplex motif, with participating Gs in bold red letters. Flipped-out nucleotides with partial electron densities are in grey. (b). Overview of the Spinach RNA structure in complex with the BL3-6 Fab (grey). The RNA forms a long, slightly bent helical domain that docks into the Fab heavy chain CDRs via binding interactions with the GAAACAC tag (blue). The core G-quadruplex region in L12, colored yellow and red, forms a platform for stacking of the DHFBI ligand (lemon). (c). Fluorescence activation by P1 stem truncation mutants.Data represent mean values ± s.d. from three measurements. The entire P1 stem (P1.1 and P1.2) is replaced with a designated number of Watson-Crick base pairs in each truncate as shown in Supplementary Fig. 10. A Spinach construct containing a five base-pair P1 stem retains WT levels of fluorescence activation. Sequences of them and other mutants are all included in Supplementary Table 3.
Crystallization of the Fab-RNA-DFHBI complex is described in Online Methods. We obtained initial phases by molecular replacement using Fab BL3-6 (Protein Data Bank accession code: 3IVK) as a search model (Supplementary Table 1). After model building and refinement at 2.2Å resolution, the final values of Rfree and Rwork were 0.211 and 0.179, respectively. The interactions between the Fab and RNA agree with those observed previously in the ligase ribozyme-Fab complex involving four of the six CDRs12 (Supplementary Fig. 2a and 3). The Fab provided most of the intermolecular contacts that form the crystal lattice (Supplementary Fig. 2b and 4): Fab-RNA contacts buried 1,689 Å2 of otherwise solvent-accessible surface area (per complex), and Fab-Fab contacts buried 896 Å2, mostly between Fab light chains from symmetry-related molecules (651 Å2; Supplementary Fig. 4c). In contrast, intermolecular RNA-RNA contacts contributed only one bidentate hydrogen bond (37 Å2; Supplementary Fig. 5).Including the Fab-RNA binding interface within the asymmetric unit (821 Å2), Fab-mediated contacts accounted for about 99% (3406 Å2 out of 3443 Å2) of buried surface area in the lattice, confirming that BL3-6 can serve as an effective crystallization chaperone11,12.
Overall structure of Spinach
Spinach adopts an elongated fold containing two sets of coaxially stacked helical stems (P1.1/P1.2 and P2.1/P2.2) that flank the ligand-binding L12 region (Fig. 1b). We found no long-range tertiary interactions. Strikingly, the L12 region of Spinach folds into a G-quadruplex motif, which forms by the intertwining of the two strands of L12, with the flanking helices (P1 and P2) apparently localizing the strands and providing stacking continuity. The G-quadruplex motif, together with flanking nucleotides in the L12 region, serves as a platform for DFHBI binding and fluorescence activation. As observed in DNA quadruplexes previously13, two purine:purine base pairs occur at the P1 duplex - quadruplex junction (here A20·G65 and A21·A64; Supplementary Fig. 6a).
The Spinach secondary structure derived from our crystallographic analysis (Fig. 1a) differs significantly from the preliminary mFold analysis described previously, probably due to difficulties in predicting G-quadruplex motifs. The predicted secondary structure contains one terminal stem and three stem-loops connected by single-stranded bulges5,14 (Supplementary Fig. 7). Our structure contains only two of these (P1.1 and P2.2; Fig. 1a). This discrepancy and the extensive protein-RNA interactions observed in the crystal lattice led us to examine the formal possibility that chaperone-mediated crystal-packing interactions might have altered the structure of Spinach despite the sparsity of contacts in the ligand-binding region. We therefore performed several assays to assess the correlation of the solution and crystal structures. First, we assessed solvent accessibility of the RNA backbone in the presence and absence of the Fab using hydroxyl radical footprinting. Consistent with high solvent accessibility throughout the RNA calculated from our structure, we found no protected regions indicative of long-range tertiary interactions other than the antigenic P2.2 loop that interacts with the Fab CDRs (Supplementary Fig. 8). Second, we performed small-angle X-ray scattering (SAXS) to interrogate the relationship between the crystal structure and the predominant solution conformation. The observed scattering profiles and the resulting molecular envelopes resembled those calculated from our structure (Online Methods; Supplementary Fig. 9). The data presented above do not rule out the presence of an alternative, transient active conformation.However, the crystals of the Fab-Spinach complex fluoresced green in the presence of DFHBI, indicating that the crystallized conformation binds DFHBI and activates its fluorescence.
The crystal structure, but not the mFold structure, predicts that most of the P1 stem is dispensable. To test this idea, we constructed truncates of the Spinach aptamer in which the P1 stem was replaced with Watson:Crick-paired duplexes of varying length (16, 5, 3, 1, or 0 base pairs; Supplementary Fig. 10).We found that construct containing only five Watson-Crick base pairs in P1 retained WT levels of fluorescence (Fig. 1c), even though that construct lacks entirely the largest helical stem predicted by mFold (Supplementary Fig. 7). These observations supported the P1 secondary structure assignment inferred from the crystal structure and showed that the U75 bulge and the non-canonical pairs in P1.2 (A12·A73, A13·A72 and U18·U67) have little functional significance (Fig. 1a;Supplementary Fig. 6b, c). The data described here and above agree with the overall RNA structure observed in our crystal.
L12 adopts a novel G-quadruplex fold
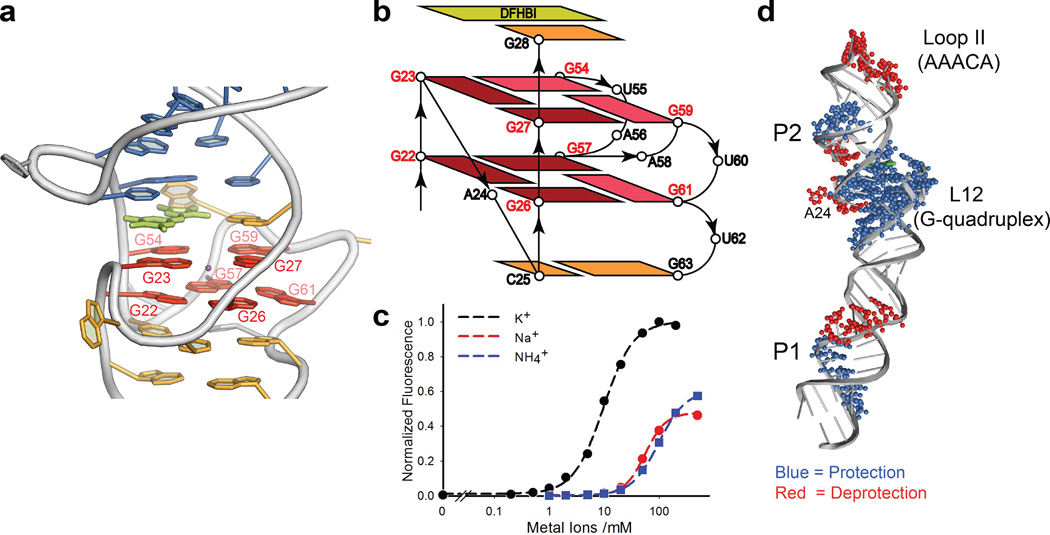
The L12 region folds into a novel RNA quadruplex motif that contains only two layers of G-tetrads (Fig. 2a). Each strand of L12 contributes four Gs to the quadruplex, forming two parallel segments via double chain reversal loops15,16 (Fig. 2b). Within the 5’ side of L12, there are two pairs of consecutive residues with _anti_-glycosidic conformations (G22, G23 and G26, G27) each of which forms a corner of the quadruplex connected through the double-chain reversal loop A24. Within the 3’-side of L12, pairs of noncontiguous guanosine residues (G54, G57 and G59, G61) form the other two corners of the quadruplex, connected through the double-chain reversal loop A58 and looped out residues U55, A56, U60, and U62. Within this configuration, G54, G59, and G61 adopt the _syn_-glycosidic conformation and G57 adopts the _anti_-conformation. The guanine bases show the same hydrogen bonding pattern in both layers. The intrinsic complexity of the topology, which has not been observed previously in DNA or RNA, along with the presence of only two tetrad layers17, likely makes any prediction of this type of G-quadruplex from the primary sequence extremely difficult.
Figure 2. The Two-Layer G-quadruplex in L12 of Spinach.
(a). Highlight of the G-quadruplex (red); The G-quadruplex forms a platform for stacking of the DFHBI fluorophore (lemon). A potassium ion (purple) sits in the center of the quadruplex between two layers. The color codes of other nucleotides match Fig. 1a. (b). Topological diagram of the two-layer G-quadruplex region. The _anti_-glycosidic guanines are highlighted with dark red. (c). Monovalent cationdependence of fluorescence activation. A fit of the data to the Hill equation gave an apparent midpoint (K1/2 = 9.6 ± 0.4 mM) and Hill slope (n) of 1.6 ± 0.1 for potassium. (d). SHAPE analysis outcome of Spinach.The SHAPE footprints of the standard-folded WT Spinach [Mg+/K+/DFH+] (Supplementary Fig. 12 for the original gel image) are superimposed onto the crystal structure. A more detailed interpretation of this data is in Online Methods.
We observed non-nucleoside electron density in the center of the quadruplex between the two layers (Online Methods) and ascribed it to K+, based on its octadentate coordination geometry, appropriate K+ - O distances18, and the following assays showing the significance of potassium ions for fluorescence activation. In the presence of 5 mM Mg2+ and DFHBI (20 µM), fluorescence increased as the K+ concentration increased. A fit of the data to the Hill equation gave K1/2 = 9.6 ± 0.4 mM with a Hill coefficient of 1.6 ± 0.1 (Fig. 2c). The positive cooperativity parallels the cooperative folding of RNA G-quadruplexes containing tracts of two G’s17. Other cations, NH4+ and Na+, also activated fluorescence but with K1/2 values about one order of magnitude higher and fluorescence “maxima” about two fold lower than observed for K+. Thus at the final crystallization drop concentrations [K+ (50mM), Na+ (30mM), and NH4+ (16mM)], K+ could account for 95% of the fluorescence, whereas Na+ and NH4+ would at most account for 10% and 3%, respectively (Fig. 2c). These data, along with similar data in the absence of Mg2+ (Supplementary Fig. 11) indicate that K+ is more efficient than Na+ or NH4+ in activating Spinach fluorescence, presumably by stabilizing the G-quadruplex structure.
The ability of K+, Na+, and NH4+ to enhance fluorescence also conforms to the known ability of these cations to support quadruplex formation19, and the preference for K+ parallels that observed for short-loop sized DNA quadruplexes20. The divalent ion, Mg2+, enhanced fluorescence activation in the presence of monovalent cations5 but could not support fluorescence activation on its own (Fig. 2c; Supplementary Fig. 11). Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) revealed that the L12 region of Spinach became strongly resistant to acylation in the presence of K+ (Fig. 2d; Supplementary Fig. 12), consistent with K+-induced formation of the G-quadruplex. Taken together, these structural and functional data show that L12 forms a G-quadruplex motif that plays a role in DFHBI binding and fluorescence activation.
Fluorophore configuration
DFHBI may exist as two geometric isomers, (Z) and (E), with respect to the double bond between the two rings. The steric influence of the keto oxygen is expected to render the (Z)-configuration more favorable. 1H NMR analysis showed the vinylic hydrogen as a single peak (Online Methods), suggesting that the synthesis yielded one predominant configuration. After recrystallization from ethanol, X-ray structure determination revealed DFHBI inthe (Z)-configuration (Supplementary Fig. 13; Supplementary Table 2). Nevertheless, recent studies have estimated that upon excitation of the Spinach-DFHBI complex, up to 25% of the Spinach-bound DFHBI photoisomerizes to the (E)-configuration and returns to (Z) via thermal isomerization in the dark21,22. This raises the possibility that a fraction of the complexes in our crystal contain DFHBI in the (E)-configuration.
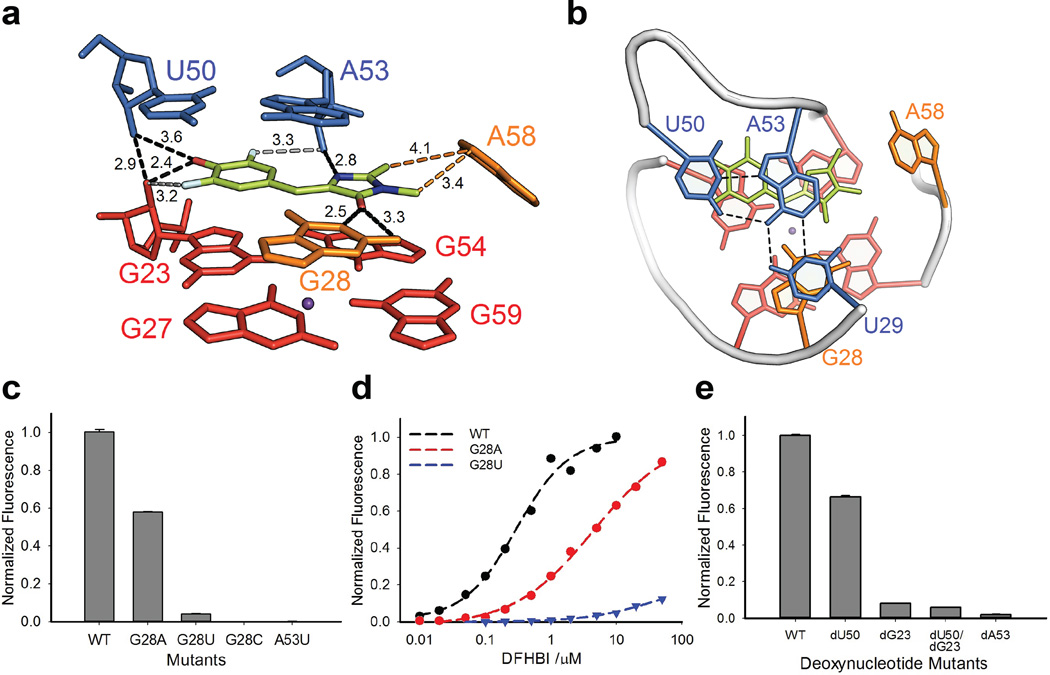
While the refined structure factors clearly showed electron density corresponding to two rings in a planar configuration, on the basis of this data alone we could not ascertain definitively the mode of ligand binding. Moreover, the density does not encompass the ligand fully, possibly reflecting multiple ligand binding modes, the presence of both geometric isomers, or the high B-factors associated with this region of the structure. With careful consideration of the steric, hydrophobic, and electrostatic features of the binding site (Online Methods), we modeled in DFHBI as the (Z)-isomer based on its higher stability and ability to make favorable interactions with the binding site. The resulting orientation directs the hydrophobic methyl groups away from the polar hydroxyl groups of G23 and U50 towards the aromatic face of A58 (Fig. 3a and Supplementary Fig. 14).
Figure 3. The fluorophore binding site in Spinach.
(a). Side view of the DFHBI binding site. U29 is omitted for clarity. Black dashes (with the distances numbered in Å) represent inferred hydrogen bonds from DFHBI to the G28 nucleobase and several 2’-hydroxyl groups. Potential hydrogen bonds from fluorine are in grey dashes with distances. The imidazolinone methyl groups of DFHBI may engage in hydrophobic interactions (orange dashes) with A58 (orange). Ligand atoms: C = lemon, N = blue, F = cyan, O = red. DFHBI geometry, positioning, and orientation were deduced as described in Online Methods and Supplementary Figs. 13, 14, and 15, respectively. (b). Top-down view showing DFHBI (lemon) and adjacent nucleotide G28 stacking between a U50-A53-U29 Hoogsteen base triple (blue) above and a layer of G-quadruplex (red) below. (c). Mutations at G28 or A53 diminish fluorescence activation. Data represent mean values ± s.d. from three measurements. DFHBI concentration was 5 µM. (d). Dependence of fluorescence on DFHBI concentration. Fits of the data to the Hill equation give KD = 300 ± 68 nM and 4.4 ± 0.8 µM for WT and G28A, respectively. Weak fluorescence activation by the G28U mutant precluded KD determination. (e). Effect of deoxynucleotide mutants on corresponding positions (G23, U50 and A53) in Spinach. Data represent mean values ± s.d. from three measurements.
To investigate the ligand orientation further, we performed crystallization trials with other fluorophore analogs. Spinach complexes bound to BrBI and DFHBI-1T23(Supplementary Fig. 15), containing a bromophenyl group in place of the difluorophenolate group and an N-trifluoroethyl group in place of the N-methyl group, respectively, diffracted to lower resolution (2.5 Å and 3.1 Å, respectively; Supplementary Table 1) than did the DFHBI complex but clearly revealed the locations of the –Br and –CF3 groups. These additional data sets agreed with the inferred mode of ligand binding (Supplementary Fig. 15; Online Methods). Although these studies implicate a preferred mode of ligand binding, in light of the photophysical studies21,22 we do not exclude the possibility that a fraction of the complexes in the crystal contain a distinct configuration and orientation of the ligand.
The fluorophore binding site
The structure revealed the DFHBI fluorophore bound in a planar configuration (Fig. 3a), fitting into a binding site formed by the G-quadruplex platform and other adjacent nucleotides (G28 and A58 of L12; U29, U50, and A53 of P2.1). The G28 nucleobase stacks on quadruplex-G27 and forms hydrogen bonds with the DFHBI keto oxygen (Fig. 3a); together they complete a stacking layer above the G-quadruplex platform. A Hoogsteen base triple involving U50-A53-U29 forms the base of the P2 stem and stacks against the DFHBI:G28 layer to seal the fluorophore binding site from the top (Fig. 3b, blue). At the side of the contiguous stack, the fluorophore’simidazolinone N-methyl group points towards C-2 of A58 located 3.4 Å away, possibly forming a hydrophobic interaction. The stacking configuration also positions the 2’-OH groups of G23, U50, and A53 within hydrogen bonding distance of DFHBI’s F3, phenolate oxygen, F5, and imidazolinone-N3 (Fig. 3a).
We also tested key features of the structural model by assaying the ability of mutated Spinach variants to activate DFHBI fluorescence in the presence of 100 mM K+ and 5 mM Mg2+. As expected, mutating either layer of Gs into pyrimidines completely abolished fluorescence activation, likely reflecting disruption of the entire quadruplex motif, as did mutating three Gs (G23, G28, and G54) close to the DFHBI binding site (Supplementary Fig. 16). We performed more extensive analysis on G28, which is coplanar with and hydrogen bonded to DFHBI. Mutation to any other nucleotide decreased fluorescence, with changes to pyrimidines being most drastic (G28C > G28U > G28A; Fig. 3c). SHAPE analysis of these mutations revealed that G28C completely disrupted the quadruplex fold, probably by forming a Watson:Crick pair with one of the quadruplex G’s, whereas G28U and G28A retained the quadruplex motif (Supplementary Fig. 12). Analysis of fluorescence activation versus DFHBI concentration showed that decreased fluorescence of G28A and G28U reflected weaker affinity for DFHBI (Fig.3d). Indeed, at sufficiently high DFHBI concentrations (> 10 µM) G28A activated fluorescence with near WT efficiency. These results clearly demonstrate an important role for nucleotide G28 in fluorescence activation by Spinach, consistent with the observed structure.
We next tested the role of the 2’-hydroxyl groups of U50, G23, and A53, which interact directly with the bound DFHBI, by replacing those residues with deoxynucleotides. To facilitate site-specific incorporation of 2’-deoxynucleotides into Spinach, we used the Spinach truncate (having 5 base pairs in P1; Fig. 1c). Using enzymatic ligation of synthetic oligonucleotides, we constructed four deoxynucleotide variants of this minimal form of Spinach (dU50, dG23, dU50-dG23 and dA53). The fluorescence signal from the latter three variants was almost completely lost, whereas the signal from dA53 was 35% lower than that of the unmodified minimal Spinach (5 µM in the presence of 20 µM DFHBI; Fig. 3e). The dU50 variant exhibited modestly weakened DFHBI affinity (KD = 1.1 ± 0.3 µM) and at higher ligand concentrations activated fluorescence as efficiently as did the unmodified Spinach (Supplementary Fig. 17). In contrast, higher DFHBI concentrations failed to activate the dG23 and dA53 variants. These effects, which could reflect structural disruption of the binding site or weakened interactions with the ligand, support the functional significance of the hydroxyl groups observed to line the DFHBI binding site in the crystal structure.
Lastly, we assessed the apparent binding affinity of Spinach for a variety of fluorophore analogs (Supplementary Fig. 18). For those analogues that form a phenolate anion at neutral pH, we monitored fluorescence activation directly (DFHBI, MFHBI, and HBI). For the remaining analogues we used a competition assay, monitoring the decrease in DFHBI fluorescence resulting from the presence of the analogue. We observed that modifications to the phenol ring had modest effects on binding (< 4-fold), including removal of the phenolic oxygen, removal of one or both fluorine atoms, or protonation/methylation of the phenolate oxygen. Considering these results, the strongly deleterious effect of removing the 2’-OH at G23 may result from structural disruption of the binding site rather than from loss of a hydrogen bond to the phenolic oxygen. Alternatively, the 2’-OH interaction may serve a critical role in accommodating the negative charge of the phenolate anion in the context of the polyanionic RNA but have little significance for binding neutral ligands. In contrast to the modest effects from phenol ring modifications, modification of the imidazolinone ring dramatically affected binding. Whereas FBI bound with Ki = 780 nM, FBO, which contained an oxygen atom in place of the N-CH3 group, failed to compete with DFHBI fluorescence at any of the concentrations tested. Overall, the strong effects from modification of the imidazolinone ring and its associated interaction partners (G28 and 2’-OH of A53) support the proposed orientation of DFHBI in the ligand binding site.
Similarities between Spinach and EGFP
At neutral pH, free DFHBI (pKa = 5.5) populates predominantly the deprotonated, phenolate form (Supplementary Fig. 13; Supplementary Table 2), and its binding interactions with Spinach bear much similarity to fluorophore interactions observed within EGFP2,24 (Supplementary Fig. 19). Consistent with the extensive aromatic stacking of the fluorophore in our structure, binding to Spinach shifts the absorbance maximum of the fluorophore by 60 nm relative to the unbound form5. Analogously in yellow fluorescent protein (YFP), mutation of threonine 203 to tyrosine creates a stacking interaction with the fluorophore that red-shifts the absorbance maximum by 20 nm25. Quadruplex stacking also features prominently in recognition of DNA G-quadruplexes by synthetic ligands, which frequently contain extended aromatic systems26,27.
DFHBI binding induces local structural changes in Spinach
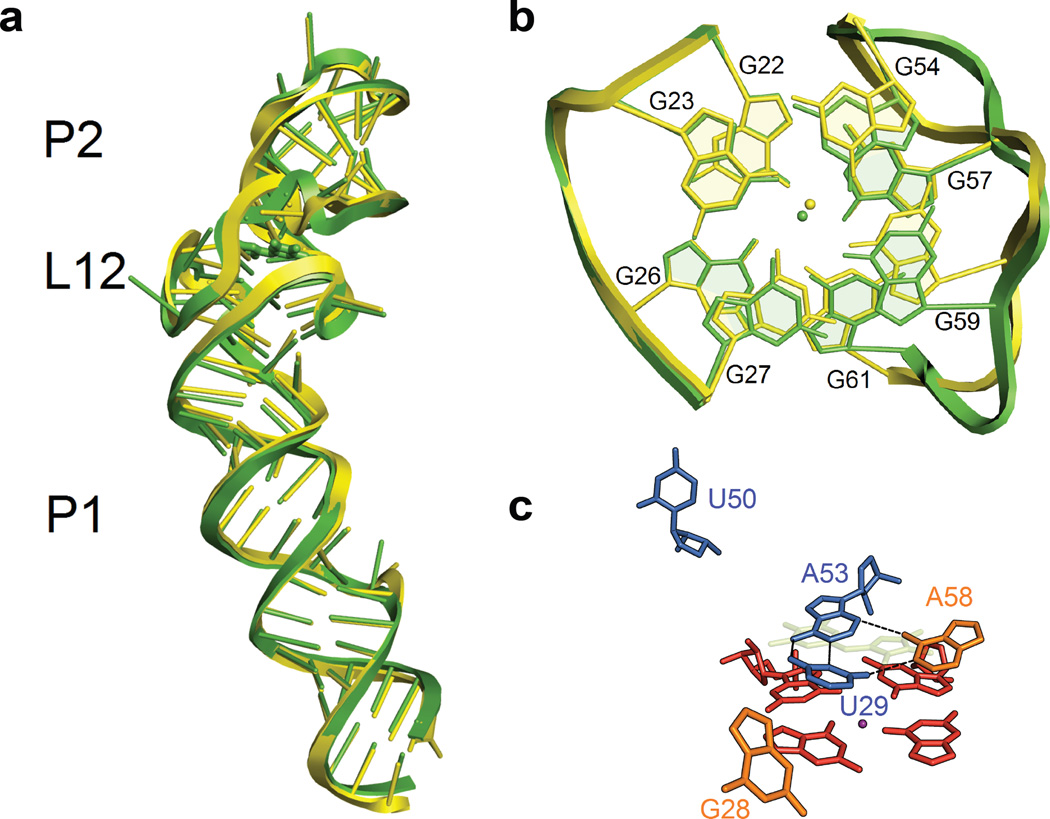
We also determined the crystal structure of the same Spinach construct in the absence of bound DFHBI at 2.4-Å resolution (Supplementary Table 1). The global architecture, including the two-layer G-quadruplex, still forms in the absence of DFHBI (Fig. 4a,b), consistent with SHAPE and hydroxyl-radical footprinting (Supplementary Fig. 8, 12). Nevertheless, DFHBI binding does cause significant local changes in the RNA structure, possibly accounting for the relatively slow rate of ligand association (kon= 6× 104 or 8× 104 M−1 s−1)21,22. In the absence of DFHBI, both the A53-U29 Watson-Crick pair from the Hoogsteen base triple and A58, which formed the hydrophobic interaction with the fluorophore, collapse on the quadruplex platform into a new base triple (Fig. 4c). This collapse extrudes two other nucleotides involved in the DHFBI stack: G28, which contacted DFHBI in the ligand plane, and U50, which formed the Hoogsteen base triple with the A53-U29 pair (Fig. 4c).
Figure 4. The structure of Spinach RNA in the absence of DFHBI.
(a). Overlay of Spinach structures obtained in the presence (green) and absence (yellow) of bound DFHBI fluorophore. DFHBI binding has minimal influence on global architecture (RMS = 0.92Å on RNA). (b). Overlay of the G-quadruplex motifs in the presence (green) and absence (yellow) of bound DFHBI fluorophore (RMS = 0.72 Å). (c). The DFHBI binding site collapses in the absence of DFHBI ligand. The transparent, lemon structure indicates the position of DFHBI in the bound structure. The color codes of other adjacent nucleotides and molecular orientation of the RNA match Figure 3a.
Discussion
Structural features of the DFHBI binding site provide insight into how Spinach can activate the fluorescence pathway of a chromophore. First, the two-layer G-quadruplex serves as a unique floor to support the fluorophore on a hydrophobic stacking platform. With the quadruplex motif as a buttress, neighboring nucleotides including a base triple above, form a binding site that likely stabilizes the planar conformation of the DFHBI fluorophore and restricts its motion. As seen in the GFP-family fluorophores, the planar conformation predominantly favors energy emission via the fluorescence pathway2. Upon ligand binding Spinach undergoes only local structural perturbations rather than a global conformational change. In this respect, Spinach resembles the lysine and SAM-I riboswitches, for which crystal structures and solution probing data of ligand-free and ligand-bound forms reveal relatively small, local perturbations28.
The architecture of the Spinach-DFHBI complex revealed here offers new opportunities to understand, manipulate, and fine-tune the spectroscopic properties of the aptamer for diverse applications. Additionally, the structure will guide the design of constructs for in vivo studies that fuse Spinach with RNAs of interest8,9. Current widespread efforts to target G-quadruplexes with small molecule ligands may also benefit from the structural principles of quadruplex-ligand recognition presented here27.
G-quadruplex motifs readily form in vitro within DNA and RNA sequences containing consecutive runs of G-tracts17,29,30. These motifs have emerged frequently in DNA aptamers and deoxyribozymes selected from random libraries31,32. In contrast, the motif has been relatively rare in RNA aptamers. Biochemical studies and NMR analysis have revealed a quadruplex structure present in small RNA aptamers selected against the bovine prion proteinand an RGG peptide-binding RNA33–35. In the latter case, the RGG peptide binds the RNA aptamer at the quadruplex-duplex junction analogous to DFHBI binding to Spinach RNA. Notably, the other reported fluorophore-binding RNA aptamers have G-rich sequences and therefore may contain a G-quadruplex motif for fluorophore recognition and activation5. The unusual features (sequence, topology, and structural context) of the G-quadruplex in Spinach raise questions about whether transcriptomes will form such structures in vivo36,37. On the other hand, natural RNAs known to adopt complex tertiary architectures (tRNAs, riboswitches, ribosomes, self-splicing introns, and endonucleolytic ribozymes) exhibit a conspicuous absence of the G-quadruplex motif.
Online Methods
DFHBI synthesis
(Z)-2,6-Difluoro-4-((2-methyl-5-oxooxazol-4(5H)-ylidene)methyl)phenyl acetate
The published method5 was used to obtain the desired compound in 80% yield. 1H NMR (CDCl3/TMS) δ 7.77 (d, 2H, J = 8.0 Hz), 6.96 (s, 1H), 2.43 (s, 3H), 2.40 (s, 3H).
(Z)-4-(3,5-Difluoro-4-hydroxybenzylidene)-1,2-dimethyl-1H-imidazol-5(4H)-one (DFHBI)
A mixture of (Z)-2,6-difluoro-4-((2-methyl-5-oxooxazol-4(5_H_)-ylidene)methyl)phenyl acetate (955 mg, 3.40 mmol), ethanol (15 mL), 40% aqueous methylamine (1.0 mL), and potassium carbonate (700 mg) was heated at reflux for 3 h. After cooling the reaction mixture in an ice bath, the precipitate containing the product was collected by filtration and further purified by silica gel chromatography, eluting with ethyl acetate to give 0.569 g of product (66% yield). 1H NMR (CD3OD/TMS) 7.78 (dd, 2H, J = 8.0, 1.5 Hz), 6.89 (s, 1H), 3.18 (s, 3H), 2.40 (s, 3H). The two-step overall yield was 53%.
Construct preparation
The BL3-6 antibody Fab used as the chaperone for crystallography was expressed from a pFab plasmid and purified as described previously12.
The gene fragment containing Spinach RNA sequences was produced by gBlocks (Integrated DNA Technologies), then subcloned into the EcoRI and HindIII sites of pUC19 (New England Biolabs) under a T7 promoter. The relevant sequence of the insert in pSpinach plasmid is: taatacgactcactataGGACGCGACCGAAATGGTGAAGGACGGGTCCAGTGCTTCGGCACTGTTGAGTAGAGTGTGAGCTCCGTAACTGGTCGCGTC
The relevant sequence for the design with the BL3-6 Fab recognition tag in pSpinach_tag is: taatacgactcactataGGACGCGACCGAAATGGTGAAGGACGGGTCCAGTGCGAAACACGCACTGTTGAGTAGAGTGTGAGCTCCGTAACTGGTCGCGTC
The lowercase letters indicate the T7 promoter.
All other Spinach mutants (RNA sequences) used in this work are summarized in Supplementary Table 3.
RNA synthesis and purification
Templates for transcription reactions were prepared by PCR amplification of the plasmids pSpinach and pSpinach_tag. The primer sequences were: 5'-ATC GAA TTC CGT AAT ACG ACT CAC TAT AG-3' and 5'-mGmAC GCG ACC AGT TAC GG-3' for the forward and the reverse primer, respectively. The first two nucleotides of the reverse primer (those preceded by a lowercase “m”) contained 2'-OMe modifications to reduce transcriptional heterogeneity at the 3'-end38.
RNA was prepared by in vitro transcription for 2 h at 37 °C in buffer containing 40 mMTris-HCl pH 7.9, 2 mMspermidine, 10 mMNaCl, 25 mM MgCl2, 10 mM DTT, 30 U/mL RNase Inhibitor (NEB), 2.5 U/mL TIPPase (NEB), 5 mM each of NTPs, 30 pmol/mL DNA template and 40 µg/mL T7 RNA polymerase. Transcription reactions were quenched by addition of 5 U/mL DNase I (Promega) and incubation at 37 °C for 30 min.
The RNA purification strategy was designed based on the outlined scheme39. RNA was phenol-chloroform extracted (pH 4.3) three times and loaded into a NAP-10 column pre-equilibrated with gel filtration (GF) buffer (10 mMTris pH 7.5, 100 mMKCl, 5 mM MgCl2, referring to the RNA selection conditions5). RNA was eluted with 1.5 mL of GF buffer and loaded onto a 120 mL HiLoad 16/60 Superdex 200pg gel filtration column (GE Healthcare). All gel filtration runs were carried out in GF buffer at 4 °C at a rate of 1 mL/min. Elution peaks were collected and concentrated using an Amicon Ultra-15 column (10 kDa cut-off). The concentrated RNA was incubated for 30 minutes at 37 °C, in the presence or absence of 1 equivalent of DFHBI fluorophore, before being aliquoted and frozen into stocks stored at −80 °C.
Fluorescence determination
The fluorescence emission of each Spinach design or mutant was measured at 5 µM, 20 °C, using a Fluorolog-3 spectrofluorometer equipped with a thermo controller (Horiba Inc.) at the excitation wavelength of 468 nm; results were the average of three measurements and were normalized using molar concentrations of Spinach (WT).
In the metal-ion dependent fluorescence assay and ion titration, the Spinach RNA was first denatured and isolated in its pellet form from ethanol precipitation, followed by a standard refolding protocol:
- Redissolve in distilled, deionized water
- Heat treatment for 1 min at 90 °C
- Refolding in the corresponding buffer (supplied as 10x) for 15min at 50 °C
- Incubation in the presence of DFHBI for 30 min at 37 °C
The fluorescence emission was then measured at room temperature, as described above. Data were normalized and plotted using SigmaPlot.
RNA-ligand affinity measurements
Dissociation constants (KD) for the RNA-fluorophore complexes were determined by measuring the increase in fluorescence as a function of increasing fluorophore concentration in the presence of 30 nM RNA aptamer. For each concentration of fluorophore measured, a background signal for fluorophore alone in buffer was also measured and subtracted from the signal measured for RNA and fluorophore together. Curves were fitted to the general Hill equation: C = C0 + (Cmax*[DFH]n) / ([DFH]n + KDn)), where KD is the binding constant; C0 and Cmax are the minimum and maximum fluorescence counts after normalization; and n is the Hill coefficient.
Competitive inhibition constants (Ki) for the non-fluorescent RNA-ligand complexes were determined by measuring the decrease in fluorescence as a function of increasing ligand concentration in the presence of 300 nM 1:1 RNA-DFHBI complexes. A background signal for competitive ligand alone in buffer was also measured and subtracted from the measured signal. IC50 was derived by fitting to the Hill equation, and Ki was calculated with Cheng-Prusoff equation: Ki = IC50 / (1 + [Complex] / KD) = IC50 / 2, in the simplified circumstance where both RNA and ligand at 300 nM around KD. All data were normalized and plotted using SigmaPlot.
SHAPE analysis
Cassette construction, primer design and SHAPE analysis of Spinach and its mutants were carried out as described in a previous protocol40. WT Spinach was folded under different folding conditions as indicated in Supplementary Fig. 12, while G28 mutants were all folded under the standard condition (10 mMTris-Cl pH 7.5, 100 mMKCl, 5 mM MgCl2). Sequencing lanes of G or U are also included, by incorporating 10 mMddCTP or ddATP into reverse transcription, respectively. The quenched reactions were fractionated by 8% denaturing PAGE. Gels were exposed to Phosphorimager screens, and scanned on a Typhoon Trio imager (GE Healthcare), followed by ImageQuant processing.
Data interpretation: The presence of K+ caused protection from acylation in the L12 regions of the RNA (Supplementary Fig. 12; compare lane #3 with lanes #2, #4 or #5), consistent with K+-induced formation of the G-quadruplex implied by our structures. DFHBI had no effect on the SHAPE profile (compare lane #4 with lane #5), while Mg2+ alone only induced the formation of Loop II and exposed L12 (compare lane #3 with lane #1). Mutants G28A and G28U showed the same SHAPE profile as WT under standard folding conditions (compare lanes #6 and #7 with lane #5). In contrast, G28C rendered L12 susceptible to NMIA acylation (compare lane #8 with lane #3 and #5), indicating that the mutation has disrupted the G-quadruplex motif.
Fab-RNA Complex formation and crystallization
An aliquot of RNA was rapidly thawed, incubated with 1.1 equivalents of BL3-6 in its binding buffer at RT for 30 min, and concentrated to 6 mg/mL using an Amicon Ultra-15 column (10 kDa cut-off). The formation of Fab-RNA complex was confirmed using a HiLoad 16/60 Superdex 200pg gel filtration column (GE) under native conditions as described above (Supplementary Fig. 1c). To decrease the number of nucleation events41, RNA was then passed over Millipore centrifugal filter units (0.2 µm cutoff). A Mosquito liquid handling robot (TTP Labtech) was used to set up RT high-throughput hanging-drop vapor-diffusion crystallization screens using commercially premade screening kits (Hampton Research). The best-diffracting crystals of the Fab-Spinach complex were obtained in a condition from the PEG/Ion Screen: 8% Tacsimate pH 7.0, 20% PEG 3,350. Similarly, for the complex without the DFHBI fluorophore, the optimal conditions were 8% Tacsimate pH 7.0, 20% PEG 3,350, 0.1 M HEPES pH 7.2; for the complex with the BrBI ligand, the conditions were 8% Tacsimate pH 7.0, 16% PEG 3,350, 0.1M cacodylate pH 7.1, seeded with ligand-free crystals; for the complex with DFHBI-1T ligand, the conditions were 8% Tacsimate pH 7.0, 20% PEG 3,350. Crystals appeared and grew to full size within 2–3 days, or within 1 week when the trial was repeated in larger 1 µL + 1 µL hanging drops on siliconized glass slides. Crystals of the complex in the presence of DFHBI showed green fluorescence. For cryoprotection, drops containing suitable crystals were brought to 21% sucrose (for crystals containing DFHBI) or 24% glycerol (for other crystals), keeping all other compositions isotonic. Crystals were immediately flash-frozen in liquid nitrogen after being fished out from the drops.
Data collection and processing
All datasets were collected at the Advanced Photon Source (APS) NE-CAT section and GM/CA sectionbeamlines at Argonne National Lab, with 0.97950 Å (native) or 0.91840 Å (bromine anomalous data) wavelength at 77 K, then integrated and scaled using its on-site RAPD automated programs (https://rapd.nec.aps.anl.gov/rapd). Initial phases were obtained from molecular replacement (MR) using Phaser42, with BL3-6 Fab-only coordinates (PDB code: 3IVK) as the searching model, and then BL3-6 Fab plus the Spinach RNA as the model for other data sets. The coordinates were refined for each data set, with crystallographic statistics available (Supplementary Table 1). Model building was completed in COOT43 with the aid of RCrane44. Refinement was carried out with the Phenix and ERRASER pipeline45,46. Ligand fitting was done in LigandFit47. Protein backbones were optimized by using a realistic backbone move set48. Metal ions were assigned on the basis of coordination distance and temperature factors18,49. Composite omit maps were calculated by simulated annealing using SFCHECK50. Simulated annealing maps omitting the ligand and feature-enhanced maps were calculated using Phenix46. Solvent-accessible surface areas were calculated using PDBePISA51. Sugar pucker configurations were identified using AMIGOS II52. All figures were made in Pymol (DeLano Scientific LLC).
Temperature factors and model construction
Spinach RNA in the structure has a high average temperature factor of 70.2Å2 (43.9 Å2 of all atoms in the structure). This elevated temperature factor reflects high solvent content (68%) in the lattice and lack of crystal contacts to the RNA, especially around the G-quadruplex region. The observed electron density of this region was lower than that of the Fab, with density for the flipped-out nucleotides being partial or absent for several residues (U51, A56, U62, U75). These nucleotides were still retained in the structure to maintain sequence integrity and were indicated in grey color (Fig. 1a). All other nucleotides were built and checked according to the 2|Fobs|-|Fcal| maps contoured at 1.0σ (Supplementary Fig. 20). In particular, the G-quadruplex guanines provided sufficient electron density to serve as beacons upon which to construct the scaffold.
The location of the fluorophore and its stacking interactions with the G-tetrad are clearly supported by the |Fobs|-|Fcal| simulated annealing omit map shown in Supplementary Fig. 14a (contoured at 3.5σ). The best fit of the fluorophore into the electron density was initially inferred from steric, hydrophobic and electrostatic complementarity: the imidazolinone ring with its two methyl groups was positioned at the hydrophobic side of the binding site close to A58, and the keto oxygen was directed toward the W-C face of G28. We also modeled in the(E)-isomer into the Spinach binding site, but no orientation fit as well as did the (Z)-isomer (Fig. 3a against Supplementary Fig. 21). Testing refinements with a series of configuration combinations ranging from Z(100%)/E(0%) to Z(0%)/E(100%) resulted in negligible differences in the overall Rfree values (0.2108 to 0.2111). Overall, while the native data revealed the location of the ligand, from that alone we could not unambiguously establish the ligand geometry or its binding orientation.
To investigate the ligand binding mode further, we co-crystallized Spinach with the additional ligands BrBI and DFHBI-1T23 (Supplementary Fig. 15; Supplementary Table 1).Crystals of theBrBI and DFHBI-1T Spinach complexes diffracted to lower resolution than the crystals of DFHBI-Spinach complex (2.5 Å and 3.1 Å, respectively) but both yielded new information about ligand positioning. The bromine atom in BrBI appeared as a single 6.0σ peak in an anomalous difference map, which clearly established the location of the phenyl ring. The data for DFHBI-1T revealed the presence of additional density (4.0σ peak) near the opposite ring in the |FDFHBI-1T(obs)|-|Fcal| map. Using the ring density and both Br and –CF3 signals, we modeled in a dummy ligand that contained both groups. Among 8 possible ring orientations (four each for the (Z) and (E) isomers, Supplementary Fig. 15), only one allowed simultaneous positioning of the bromine atom, the –CF3 group, and the rings into their respective densities. This orientation corresponded well to the one inferred from the steric and electronic features of the binding site (Fig. 3a).
The potassium ion was suggested by positive density that appeared in both the |Fobs|-|Fcal| and 2|Fobs|-|Fcal| maps during refinement, and later verified by omit maps in which the K+ omitted before refinement, where it appeared with a peak height of 4.8 sigma. The B-factor of the K+ refined to 100.0Å2(with the occupancy set to 1.0), which is slightly higher than the average B for the 8 oxygen atoms that coordinate it.
Determination of unbound DFHBI configuration by crystallography
Powdered DFHBI was dissolved into boiling ethanol, and the solution was cooled to 4 °C overnight. The resulting crystals were collected by removing the solvent and rinsed with a small amount of cold ethanol. An irregular broken fragment (0.08 × 0.20 × 0.28 mm) was selected using a stereo-microscope; the crystals were kept immersed in Fluorolube oil during microscopy to prevent reaction with air. The crystal was removed from the oil using a tapered glass fiber that also served to hold the crystal for data collection. A data set was obtained approximately 100% of reciprocal space to a resolution of 0.90 Å. Data collection was performed at 100 °K.
Integration of intensities and refinement of cell parameters were done using SAINT. Absorption corrections were applied using SADABS based on redundant diffraction. The space group was determined to be P21/c. Direct methods were used to locate all atoms from the E-map. Repeated difference Fourier maps allowed assignment of F, O, N and C atoms. Following anisotropic refinement of all non-H atoms, ideal H atom positions were calculated. Final refinement was anisotropic for non-H atoms and isotropic-riding for H atoms.The structural output was checked using the CheckCIF routine from the International Union of Crystallography and is reported in Supplementary Dataset 1.
Filter-binding affinity assay
Approximately 50,000 cpm of 5’-32P-radiolabeled Spinach RNA was diluted to a total volume of 900 µL with binding buffer (described above, plus 0.2 mM EDTA), supplemented with 0.5 mg/mL heparin and 0.1 U/µL RNase inhibitor (Amersham). The final RNA concentration was < 1 nM. RNA was refolded at 37°C for 30 min in the presence of 5 µM DFHBI fluorophore, mixed with Fabs (2 nM to 1 µM final concentrations) and incubated for 30 min at room temperature. A nitrocellulose membrane (Whatman) and HyBond n+ filter (Amersham) were placed in a 96-well Dot-Blot apparatus (BioRad), and the wells were pre-equilibrated with 100 µL binding buffer. The Fab-RNA mixture was applied, and the filter was washed with another 100 µL of binding buffer. The membrane and filter were air-dried, exposed to Phosphorimager screens, and scanned with a Typhoon Trio imager (GE Healthcare). The amount of radiolabeled RNA bound to each filter was quantitated with ImageQuant software (Molecular Dynamics). Binding constants were obtained by fitting the fraction of nitrocellulose-bound RNA to the following Hill equation: F = F0 + (Fmax*[Fab]n) / ([Fab]n + KDn)), where KD is the dissociation constant; F0 and Fmax are the minimum and maximum fractions of RNA bound; and n is the Hill coefficient.
Hydroxyl radical (Fe-EDTA) footprinting
Footprinting reactions contained 1 µL (~100,000 cpm) of 5-labeled RNA folded under indicated conditions (with or without 5 µM DFHBI), 9 µL each. 1 µL of 10x Fab BL3-6 (5 µM) was added, and the RNA-Fab mixture allowed to incubate at room temperature for 30 minutes for Fab binding. A 10x footprinting reagent was prepared separately and contained 1 mM Fe(NH4)2(SO4)2, 1.25 mM EDTA pH 8.0, and 60 mM sodium ascorbate. To initiate each footprinting reaction, 1 µL of the 10x footprinting reagent was added to each well to make a final 11 µL reaction containing 100,000 cpm 5’-labeled RNA. Footprinting reactions proceeded for 30 minutes at room temperature, and were quenched by the addition of 5 µL of a thiourea stop solution (9 M urea, 300 mMthiourea, 0.04% each bromophenol blue and xylene cyanol).
Each gel included an RNase T1 size standard lane, an input lane (RNA in water), as well as an “input + Fab” control. To prepare the T1 ladder, 7 µL 9 M urea was mixed with 1 µL 0.2 M citrate pH 5.0, 1 µL 5’-labeled ligase product (~100,000 cpm), and 1 µL 1 U/µL RNase T1 (Roche). The reaction was incubated at 50 °C for 10 min and quenched with thiourea stop solution. The quenched footprinting and T1 reactions were fractionated by 15% denaturing PAGE (15% acrylamide, 0.5x TBE, 7 M urea). Gels were exposed to Phosphorimager screens, and scanned on a Typhoon Trio imager (GE Healthcare). SAFA was used to analyze footprinting data and make color plots53.
Enzymatic ligation
Donor and acceptor oligonucleotides were purchased from Dharmacon (Supplementary Table 3), deprotected by following the suppliers deprotection protocol and purified by 15% DPAGE. DNA as splint oligonucleotide was purchased from Integrated DNA Technologies (IDT) and used without further purification.
For the ligation reaction, the DNA splint oligo was mixed with acceptor and donor RNA oligos at a 1:4:2 ratio of acceptor : splint : donor, the final concentration of acceptor RNA oligo 62.5 µM and were annealed (90 °C for 3 min followed by 80 cycles of 2 min with 1 °C decreased each cycle) in the presence of 10 mMTris-HCl and 120 mMKCl. After annealing T4 RNA ligase buffer (50 mMTris-HCl, 2 mM MgCl2, 1 mM DTT, 400 µM ATP, pH 7.5) and T4 RNA ligase 2 (NEB, 20 units) were added and resulting reaction mixture was incubated at 37 °C for 6 h. Ligation reactions were diluted with TE buffer (10 mMTris-HCl, pH 8, 1mM EDTA), PCI extracted followed by ethanol precipitation and purified by 10% dPAGE. Concentrations for pure oligos were determined using Nanodrop (Thermo Scientific).
Small-angle X-ray scattering (SAXS)
SAXS experiments were conducted on the SIBYLS beamline at the Advanced Light Source synchrotron as previously described54. The samples were purified and prepared as described above. For each experiment, concentrated samples (1.0 and 2.0 mg/mL in 25 µL) were placed in a 96-well plate. SAXS data were collected continuously from Q = 0.01 to 0.32 Å−1, with a frame duration of 1.0 s. The buffer (10 mMTris pH 7.5, 100 mMKCl, 5 mM MgCl2) scattering was then subtracted from the signal. Scattered intensity curves were calculated from the atomic coordinates of the crystallographic structure (PDB code: 4KZD) using CRYSOL with 50 harmonics and 256 data points55. This program was also used to fit the calculated curve to the experimental one derived from the 1.0 mg/mL sample. SAXS data collection, scattering derived parameters, and programs used for analysis are presented in Supplementary Table 4. Data analysis is presented in Supplementary Figure 9.
Databases and Accession codes
Protein data bank (PDB): The coordinates and structure factors have been deposited to RCSB Protein Data Bank under PDB ID codes 4KZD (in the presence of DFHBI), 4KZE (in the absence of DFHBI), 4Q9Q (in the presence of BrBI), and 4Q9R (in the presence of DFHBI-1T).
Cambridge Structural Database (CSD): The DFHBI fluorophore coordinates and structure factors have been deposited under code CCDC-956054.
Supplementary Material
1
Acknowledgements
We are grateful to I.M. Steele for assistance in determining the structure of unbound DFHBI. We thank L. Zhang for the advice on structure and model building. We thank J.R. Fuller for refinement software support. We also thank F.C. Chou and R. Das for their aid with ERRASER software, K.N. Dyer, T.R. Sosnick and J.R. Hinshaw for the help with SAXS experiments. We thank members of the Piccirilli group, J.P. Staley, and D.M.J. Lilley for helpful discussions and comments on the manuscript. We also thank the reviewers for valuable insights. The work is supported by NIH grants R01-AI081987 and R01-GM102489 (to J.A.P.), NIH training grant T32GM007183 (to N.B.S.), and US NIGMS Medical Scientist NRSA no. 5 T32GM07281 (to Y.K.). This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamline 24-ID-C&E, GM/CA beamline 23-ID-D, and Advanced Light Source beamline 12.3.1 SIBYLS, all supported by U.S.A. Department of Energy (DOE).
Footnotes
Author contributions
H.H. and J.A.P. designed project; H.H. conducted most biochemical and biophysical assays and crystallography; N.B.S. and P.A.R. made essential contributions to crystallography; N.S.L. synthesized DFHBI and analogues; S.A.S and M.E.E. constructed and characterized truncation mutants; Y.K. developed the Fab BL3-6 chaperone; J.A.P. provided overall project supervision. The manuscript was prepared by H.H.,P.A.R. and J.A.P.
Competing financial interests
The authors declare no competing financial interests.
References and Notes
- 1.Yang F, Moss LG, Phillips GNJ. The molecular structure of green fluorescent protein. Nat. Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 2.Tsien RY. The green fluorescence protein. Ann. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 3.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 4.Shu X, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paige JS, Wu KY, Jaffrey SR. RNA Mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat. Methods. 2013;10:1219–1224. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strack RL, Jaffrey SR. New approaches for sensing metabolites and proteins in live cells using RNA. Curr. Opin. Chem. Biol. 2013;17:651–655. doi: 10.1016/j.cbpa.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335:1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kellenberger CA, Wilson SC, Sales-Lee J, Hammond MC. RNA-Based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc. 2013;135:4906–4909. doi: 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pothoulakis G, Ceroni F, Reeve B, Ellis T. The Spinach RNA aptamer as a characterization tool for synthetic biology. ACS Synth. Biol. 2013;3:182–187. doi: 10.1021/sb400089c. [DOI] [PubMed] [Google Scholar]
- 11.Ye JD, et al. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc. Natl. Acad. Sci. 2008;105:82–87. doi: 10.1073/pnas.0709082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koldobskaya Y, et al. A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat. Struct. Mol. Biol. 2010;18:100–106. doi: 10.1038/nsmb.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim KW, Phan AT. Structural basis of DNA quadruplex-duplex junction formation. Angew. Chem. Int. Ed. 2013;52:8566–8569. doi: 10.1002/anie.201302995. [DOI] [PubMed] [Google Scholar]
- 14.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adrian M, Heddi B, Phan AT. NMR spectroscopy of G-quadruplexes. Methods. 2012;57:11–24. doi: 10.1016/j.ymeth.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullen MA, Assmann SM, Bevilacqua PC. Toward a digital gene response: RNA G-quadruplexes with fewer quartets fold with higher cooperativity. J. Am. Chem. Soc. 2012;134:812–815. doi: 10.1021/ja2096255. [DOI] [PubMed] [Google Scholar]
- 18.Harding MM. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr. D Biol. Crystallogr. 2006;62:678–682. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- 19.Hud NV, Schultze P, Sklenar V, Feigon J. Binding sites and dynamics of ammonium ions in a telomere repeat DNA quadruplex. J. Mol. Biol. 1999;285:233–243. doi: 10.1006/jmbi.1998.2327. [DOI] [PubMed] [Google Scholar]
- 20.Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang P, et al. Photochemical properties of Spinach and its use in selective imaging. Chem. Sci. 2013;4:2865–2873. [Google Scholar]
- 22.Han KY, Leslie BJ, Fei J, Zhang J, Ha T. Understanding the photophysics of the Spinach-DFHBI RNA aptamer-fluorogen complex to improve live-cell RNA imaging. J. Am. Chem. Soc. 2013;135:19033–19038. doi: 10.1021/ja411060p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song W, Strack RL, Svensen N, Jaffrey SR. Plug-and-play fluorophores extend the spectral properties of Spinach. J. Am. Chem. Soc. 2014;136:1198–1201. doi: 10.1021/ja410819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Royant A, Noirclerc-Savoye M. Stabilizing role of glutamic acid 222 in the structure of enhanced green fluorescent protein. J. Struct. Biol. 2011;174:385–390. doi: 10.1016/j.jsb.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Wachter RM, Elsliger MA, Kallio K, Hanson GT, Remington SJ. Structural basis of spectral shifts in the yellow-emission variants of green fluorescent protein. Structure. 1998;6:1267–1277. doi: 10.1016/s0969-2126(98)00127-0. [DOI] [PubMed] [Google Scholar]
- 26.Yang P, De Cian A, Teulade-Fichou MP, Mergny JL, Monchaud D. Engineering bisquinolinium/thiazole orange conjugates for fluorescent sensing of G-quadruplex DNA. Angew. Chem. Int. Ed. 2009;48:2188–2191. doi: 10.1002/anie.200805613. [DOI] [PubMed] [Google Scholar]
- 27.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 28.Liberman JA, Wedekind JE. Riboswitch structure in the ligand-free state. Wiley Int. Rev. RNA. 2012;3:369–384. doi: 10.1002/wrna.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 31.Macaya RF, Schultze P, Smith FW, Roet JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang XB, Kong RM, Lu Y. Metal ion sensors based on DNAzymes and related DNA molecules. Annu. Rev. Anal. Chem. 2011;4:105–128. doi: 10.1146/annurev.anchem.111808.073617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss S, et al. RNA aptamers specifically interact with the prion protein PrP. J. Virol. 1997;71:8790–8797. doi: 10.1128/jvi.71.11.8790-8797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mashima T, Matsugami A, Nishikawa F, Nishikawa S, Katahira M. Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein. Nucleic Acids Res. 2009;37:6249–6258. doi: 10.1093/nar/gkp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phan AT, et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kao C, Rüdisser S, Zheng M. A simple and efficient method to transcribe RNAs with reduced 3′ heterogeneity. Methods. 2001;23:201–205. doi: 10.1006/meth.2000.1131. [DOI] [PubMed] [Google Scholar]
- 39.Kim I, McKenna SA, Viani Puglisi E, Puglisi JD. Rapid purification of RNAs using fast performance liquid chromatography (FPLC) RNA. 2006;13:289–294. doi: 10.1261/rna.342607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 41.Chayen R. Rigorous filtration for protein crystallography. J. Appl. Crystallogr. 2009;42:743–744. [Google Scholar]
- 42.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Keating KS, Pyle AM. Semiautomated model building for RNA crystallography using a directed rotameric approach. Proc. Natl. Acad. Sci. 2010;107:8177–8182. doi: 10.1073/pnas.0911888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chou FC, Sripakdeevong P, Dibrov SM, Hermann T, Das R. Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat. Methods. 2012;10:74–76. doi: 10.1038/nmeth.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Afonine PV, Grosse-Kunstleve RW, Adams PD. The Phenix refinement framework. CCP4 Newslett. 2005;42:8. [Google Scholar]
- 47.Terwilliger TC, Adams PD, Moriarty NW, Cohn JD. Ligand identification using electron-density map correlations. Acta Crystallogr. D Biol. Crystallogr. 2006;63:101–107. doi: 10.1107/S0907444906046233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haddadian Esmael J, et al. Automated real-space refinement of protein structures using a realistic backbone move set. Biophys. J. 2011;101:899–909. doi: 10.1016/j.bpj.2011.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stahley MR, Adams PL, Wang J, Strobel SA. Structural metals in the Group I Intron: A ribozyme with a multiple metal ion core. J. Mol. Biol. 2007;372:89–102. doi: 10.1016/j.jmb.2007.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedure for evaluating the quality of macromolecular structure-factor data and their agreement with atomic model. Acta Crystallogr. D Biol. Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 51.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 52.Wadley LM, Pyle AM. The identification of novel RNA structural motifs using COMPADRES: an automated approach to structural discovery. Nucl. Acids Res. 2004;32:6650–6659. doi: 10.1093/nar/gkh1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Das R. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat. Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svergun DI, Barberato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1