Collateral Damage: Microbiota-derived Metabolites and Immune Function in the Antibiotic Era (original) (raw)
. Author manuscript; available in PMC: 2015 Aug 13.
Published in final edited form as: Cell Host Microbe. 2014 Aug 13;16(2):156–163. doi: 10.1016/j.chom.2014.07.009
SUMMARY
Our long-standing evolutionary association with gut-associated microbial communities has given rise to an intimate relationship, which affects many aspects of human health. Recent studies on the mechanisms that link these microbial communities to immune education, nutrition and protection against pathogens point to microbiota-derived metabolites as key players during these microbe-host interactions. A disruption of gut-associated microbial communities by antibiotic treatment can result in a depletion of microbiota-derived metabolites, thereby enhancing pathogen susceptibility, impairing immune homeostasis and contributing to the rise of certain chronic inflammatory diseases. Here, we highlight some of the recently elucidated mechanisms that showcase the impacts of microbiota-derived metabolites on human health.
Introduction
The large intestine is host to a diverse microbial community, the gut microbiota, which is composed predominantly of obligate anaerobic bacteria. Bifidobacterium species, which belong to the phylum Actinobacteria, dominate gut-associated microbial communities within the first year of life in breast fed infants. This preponderance of Bifidobacterium species is explained by their capability to break down milk oligosaccharides, constituents of milk that cannot be utilized by newborns for nutrition but instead serve to guide the composition of the developing infant gut microbiota (reviewed in (Garrido et al., 2013)). After weaning, bacteria belonging to the phyla Bacteroidetes and Firmicutes rise to dominance, while members of the phyla Actinobacteria, Proteobacteria are minor constituents commonly found within a balanced community (Eckburg et al., 2005; Koenig et al., 2011; Palmer et al., 2007). Occasionally representatives of Fusobacteria, Verrucomicrobia, Cyanobacteria or other phyla can be present within the community. The combined metabolic activities and host interactions of a balanced gut-associated microbial community confers benefit to the host by providing nutrition, protection against enteric pathogens and immune education. However, the mechanisms responsible for the beneficial properties of a balanced microbial community and some consequences of its interaction with the host immune system are just beginning to be worked out. Here we will review recent mechanistic insights into the benefits conferred by gut-associated microbial communities on the immune system and the adverse effects associated with their disruption by antibiotic treatment.
Delivering the goods: Microbiota-derived metabolites
Gut-associated microbial communities are highly diverse, vary between individuals on the species level and can change in composition over time (reviewed in (Lozupone et al., 2012)), which makes it unlikely that their health benefits can be reduced to the presence of individual bacterial species within the community alone. More likely, the beneficial properties of the gut microbiota in the large bowel are attributable to its combined metabolic activities and products, which are not immediately obvious from an analysis of its composition by sequencing 16S RNA genes. Thus, to understand how the gut microbiota affects health it is important to know which products and metabolites this community generates.
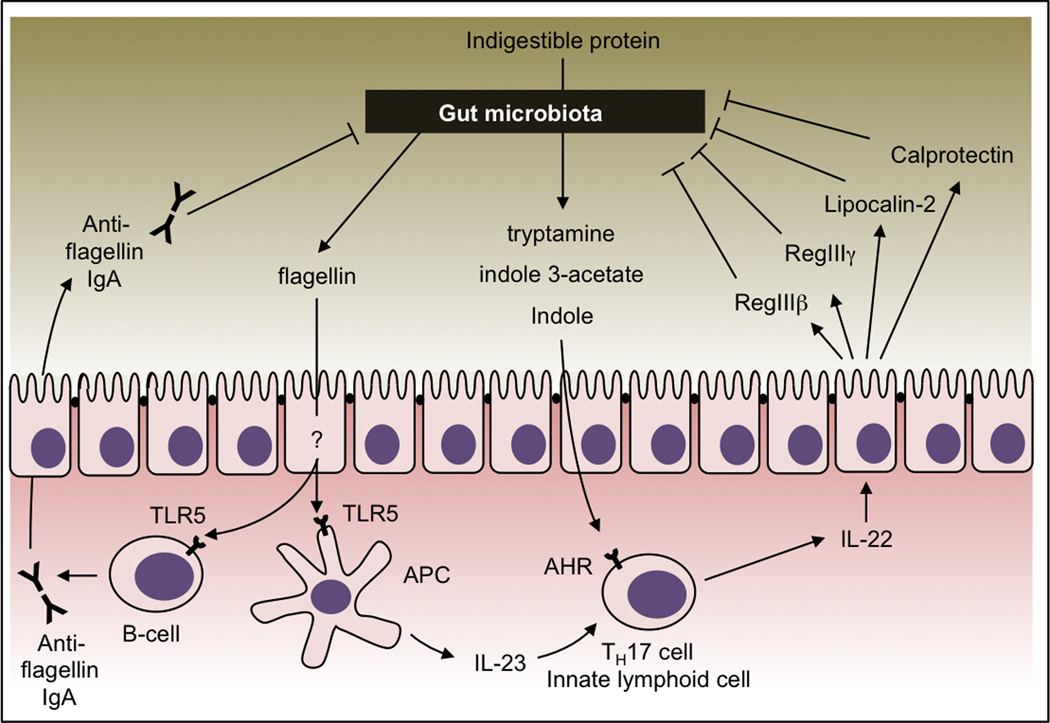
The gut microbiota can influence host responses by generating an abundance of microbe-specific molecules in the distal gut. Some of these molecules, termed pathogen-associated molecular patterns (PAMPs), are evolutionarily conserved and are used by the host’s innate immune system to distinguish microbes from self (Janeway, 1989). Although the name implies an association with pathogens, all microbes produce PAMPs, regardless of their pathogenic potential. We will thus refer to PAMPs as conserved microorganism-associated molecular patterns (MAMPs) throughout this review. As outlined below, flagella, produced by motile bacterial species (Figure 1), and certain polysaccharides covering the bacterial surface appear to be MAMPs that are particularly important for interactions between the host and its gut microbiota.
Figure 1.
Microbial products induce mucosal barrier functions. Concomitant activation of TLR5 during presentation of flagellin by antigen presenting cells (APC) renders flagellin a dominant B cell antigen. Flagellin-specific immunoglobulin A (IgA) helps control motile bacteria in the gut lumen. Indole derivatives (tryptamine, indole 3-acetate and indole) are produced by the gut microbiota during the break down of indigestible proteins. These metabolites induce the release of IL-22 by activating the aryl hydrocarbon receptor (AHR) on host cells. IL-22 stimulates epithelial cells to release antimicrobial proteins (such as RegIIIβ, RegIIIγ, lipocalin-2, and calprotectin) that help control luminal bacteria.
In addition to producing MAMPs, the fermentative growth of microbes in the lower gastrointestinal tract generates a multitude of metabolic end products. Complex carbohydrates and proteins that cannot be degraded by the host enter the large bowel where they support growth of the gut microbiota. Obligate anaerobic bacteria belonging to the classes Bacteroidia (phylum Bacteroidetes) and Clostridia (phylum Firmicutes) break down complex carbohydrates from fiber or mucus, which results in the release of metabolic end products, including hydrogen, organic acids, such as lactate and succinate, alcohols, such as 1,2 propanediol and short-chain fatty acids (SCFAs), such as formate, acetate, propionate and butyrate (Figure 2) (reviewed in (Fischbach and Sonnenburg, 2011)). Hydrogen is consumed by obligate anaerobic sulfate reducing bacteria of the family Desulfovibrionaceae (phylum Proteobacteria), a process yielding the metabolite hydrogen sulfide (H2S) (Deplancke et al., 2000; Fite et al., 2004; Zinkevich and Beech, 2000). Members of the phyla Bacteroidetes, Firmicutes and Fusobacteria degrade indigestible proteins, such as gluten, into amino acids to support their growth by fermentation. Fermentation of the amino acid tryptophan by facultative anaerobic bacteria of the class Bacilli (phylum Firmicutes) yields a number of metabolites, such as indole, indole 3-acetate and tryptamine, which accumulate in the intestine (Figure 1) (Jin et al., 2014; Zelante et al., 2013). Furthermore, dietary phospholipids are broken down by the microbiota anaerobically to yield the metabolites ethanolamine and trimethyl amine (TMA) (de la Huerga and Popper, 1951; Oresic et al., 2009; Tang et al., 2013). These examples illustrate that the combined metabolic activities of gut-associated microbial communities lead to the accumulation of a variety of microbiota-derived metabolites in the lumen of the distal gut. We will discuss below how many of these metabolites influence host physiology.
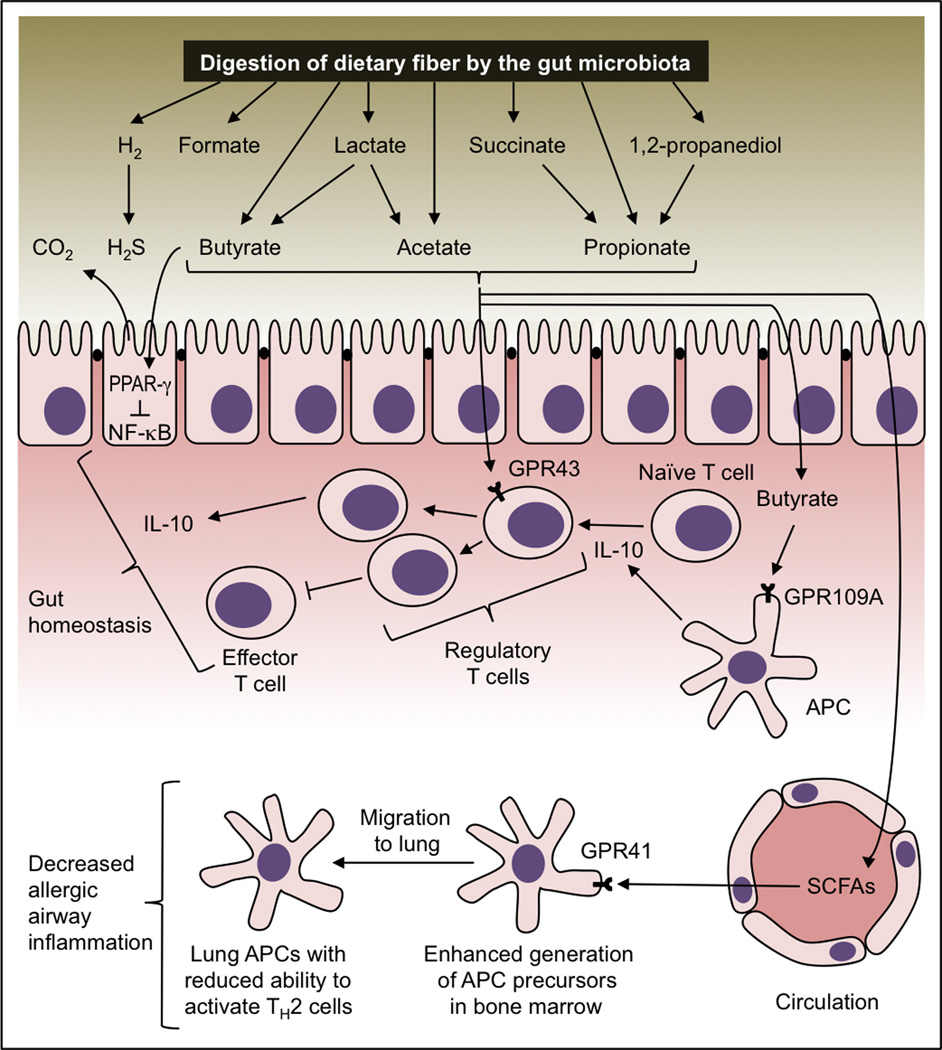
Figure 2.
Maintenance of immune homeostasis by microbiota-derived metabolites. Digestion of dietary fiber leads to an accumulation in the gut lumen of microbiota-derived metabolites that have anti-inflammatory properties. Activation of the SCFA receptor PPAR-γ inhibits pro-inflammatory responses by inhibiting NF-κB activation. SCFAs cause an expansion of regulatory T cells through mechanisms that depend on SCFA receptors GPR43 and GPR109A. In turn, regulatory T cells maintain gut homeostasis by resolving inflammation through inhibition of effector T cell function and increasing production of IL-10. SCFAs can enter the circulation and enhance the generation of APC precursors in the bone marrow through a GPR41-dependent mechanism. After seeding the lung, APCs generated through this process exhibit a reduced ability to promote T helper type 2 (TH2) cell effector function, thereby conferring protection from allergic airway inflammation.
Nursing the host: Microbiota-derived butyrate energizes colonocytes
One of the beneficial properties of a balanced gut microbiota is to provide nutrition for the host. A good example is the mitochondrial oxidation of microbiota-derived butyrate into carbon dioxide, which represents the primary energy source for colonocytes that comprise the colonic epithelium (Figure 2) (Donohoe et al., 2012). Exposure to SCFAs, such as butyrate, triggers profound changes in epithelial gene expression in vitro (Basson et al., 2000; Sanderson, 2004), which are mediated at least in part through the SCFA sensor PPAR-γ (peroxisome proliferator activated receptor-γ) (Alex et al., 2013). The absence of SCFAs in the large bowel of germ-free mice leads to major changes in the energy metabolism of colonocytes, which switch from oxidizing butyrate to fermenting glucose into lactate (Donohoe et al., 2012). Thus, microbiota-derived metabolites can have a marked influence on the energy metabolism of host cells. An increased concentration of fecal SCFAs in obese individuals compared to subjects with normal weight suggests that concentrations of these metabolites might be relevant for the development of metabolic syndrome (Schwiertz et al., 2010).
Fortifying the barrier: Microbial products help control the community
Some products of the gut microbiota elicit immune responses that are aimed at maintaining mucosal barrier integrity by controlling luminal microbes. For instance, flagellin, the structural protein subunit of the flagellar filament, is a MAMP recognized by Toll-like receptor 5 (TLR5) (Hayashi et al., 2001). Its TLR5 agonist activity makes flagellin a dominant antigen for CD4 T cells and B cells (Atif et al., 2014; Cullender et al., 2013). Although it remains unclear how flagellin crosses the epithelial barrier, flagellin-specific immunoglobulin reduces bacterial motility in the gut lumen, thereby strengthening epithelial barrier function (Figure 1) (Cullender et al., 2013).
Probiotic Lactobacillus species produce tryptophan metabolites, such as indole, indole 3-acetate and tryptamine (Zelante et al., 2013), which are ligands of the aryl hydrocarbon receptor (AHR) expressed on T helper type 17 (TH17) cells, innate lymphoid cells and dendritic cells (Heath-Pagliuso et al., 1998; Miller, 1997). TH17 responses are important for the control of luminal pathogens (Zheng et al., 2008). AHR activation leads to the production of interleukin 22 (IL-22), a TH17 cytokine that acts on epithelial cells to induce the luminal release of antimicrobial proteins, such as lipocalin-2, calprotectin, RegIII-β (regenerating islet-derived protein 3 beta) and RegIII-γ (Figure 1) (Fukumoto et al., 2014; Monteleone et al., 2011; Zelante et al., 2013). Production of IL-22 in the intestine can also be elicited by bacterial flagellin through a TLR5-dependent mechanism (Kinnebrew et al., 2010). The epithelial release of antimicrobial proteins can alter the microbiota composition by favoring growth of bacteria that express the corresponding resistance mechanisms (Behnsen et al., 2014; Raffatellu et al., 2009). Furthermore, growth of bacteria that are susceptible to these antimicrobial responses is reduced. Through this mechanism, the IL-22-induced epithelial release of antimicrobial proteins protects the mucosal surface from the attaching and effacing pathogen Citrobacter rodentium (phylum Proteobacteria) (Zheng et al., 2008), reduces colonization of opportunistic pathogens, such as Candida albicans (a fungus belonging to the phylum Ascomycota) (Zelante et al., 2013) or vancomycin resistant Enterococcus species (phylum Firmicutes, class Bacilli) (Kinnebrew et al., 2010), and helps control the growth of pathobionts, such as segmented filamentous bacteria (SFB) (phylum Firmicutes, class Clostridia) (Qiu et al., 2013).
SFBs are non-culturable obligate anaerobic bacteria that live in tight association with epithelial cells in the murine intestine. Colonization with SFBs induces a robust TH17 response, which clearly distinguishes these pathobionts from other, more benign members within the community (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009; Salzman et al., 2010). Although SFBs are not part of the human gut microbiota, we may engage in a similar relationship with Helicobacter pylori (phylum Proteobacteria), a pathogen entering the stomach early in life followed by stable colonization that continues throughout adulthood. H. pylori colonization influences the development of our immune system, as illustrated by its protective role in esophageal adenocarcinoma (Blaser, 2008).
Maintaining balance: Metabolites preserve gut immune homeostasis
While the presence of a balanced gut microbiota can help to fortify the epithelial barrier, arguably the most important characteristic of a balanced microbial community is its ability to maintain immune homeostasis. One mechanism by which the gut microbiota maintains immune homeostasis was gleaned from studies on Bacteroides fragilis. B. fragilis produces surface polysaccharide A, a MAMP that stimulates TLR2 expressed on regulatory T cells (Round et al., 2011), thereby inducing their activation and subsequent suppression of inflammatory responses in the intestine (Mazmanian et al., 2008). However, polysaccharide A production is not a trait that is widely distributed among members of the class Bacteroidia, which predicts the existence of additional mechanisms that are more broadly conserved evolutionarily.
One such mechanism is mediated by SCFAs, metabolites produced by many of the phylogenetic groupings represented within gut-associated microbial communities. The host can detect and respond to the presence of SCFAs using the intracellular receptor PPAR-γ, the surface located G-protein-coupled receptor 43 (GPR43, also known as free fatty acid receptor 2 [FFAR2]), GPR41 (FFAR3) and the butyrate receptor GPR109A (also known as hydroxycarboxylic acid receptor 1 [HAC2] or HM74A) (Alex et al., 2013; Brown et al., 2003; Le Poul et al., 2003; Taggart et al., 2005). Furthermore, SCFAs inhibit the activities of histone deacetylases (HDACs) in host cells (Siavoshian et al., 2000). The anti-inflammatory properties of SCFAs, most importantly acetate, propionate and butyrate, have been long recognized (reviewed in (Al-Lahham et al., 2010; Tan et al., 2014)). For example, after surgical interventions that require a diversion of the fecal stream, concentrations of SCFAs drop in segments of the colorectum, resulting in diversion colitis, which can be brought into remission by irrigation with SCFA solution (Harig et al., 1989).
SCFAs can suppress inflammation through several distinct mechanisms. SCFAs can suppress inflammation through a GPR41-dependent pathway, although the underlying mechanism has not been fully resolved (Trompette et al., 2014). Butyrate down-regulates lipopolysaccharide-induced responses in intestinal macrophages by inhibiting HDACs in vitro, however this mechanism does not influence the severity of chemically induced colitis in mice (Chang et al., 2014). Activation of PPAR-γ by SCFAs can reduce host inflammatory responses in vitro by promoting nuclear export of the NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) subunit RelA (Figure 2) (Kelly et al., 2004). SCFA can promote the resolution of inflammation in a mouse model of chemically induced colitis by activating GPR43 (Maslowski et al., 2009). Here, the underlying mechanism is a stimulation of GPR43 on regulatory T cells, which causes this cell population to expand in the colon, thereby limiting the proliferation of effector CD4+ T cells and increasing the expression of the anti-inflammatory cytokine IL-10 (Arpaia et al., 2013; Atarashi et al., 2011; Furusawa et al., 2013; Smith et al., 2013). Butyrate can also induce IL-10 production by antigen presenting cells (APCs) (Millard et al., 2002; Wang et al., 2008). The mechanism behind this activity of butyrate is an activation of GPR109A expressed on APCs in the colon, which induces the release of IL-10, a process that promotes the maturation of naïve T cells into regulatory T cells (Singh et al., 2014). Mice deficient for Niacr1, the gene encoding GPR109A, exhibit increased susceptibility to chemically induced colitis (Singh et al., 2014). Tryptophan metabolites might exhibit properties that are similar to those of SCFAs, because activation of AHR also reduces the severity of inflammation in a mouse model of hapten-induced colitis by inducing an expansion of regulatory T cells (Benson and Shepherd, 2011).
The picture emerging from above studies is that microbiota-derived metabolites display prominent anti-inflammatory properties that are important for maintaining intestinal immune homeostasis. Conditions that lead to a drop in the concentration of microbiota-derived metabolites would thus be predicted to disrupt immune homeostasis and predispose to inflammatory diseases.
Side effects: Antibiotics deplete microbiota-derived metabolites
Antibiotic treatment results in a severe disruption of the gut microbiota of humans, which is characterized by a decrease in both the taxonomic richness and diversity of the community (Dethlefsen et al., 2008). Within four weeks after withdrawal of antibiotics the microbiota returns to an overall composition that resembles the pretreatment state, although some changes in the community can persist for longer periods of time (Dethlefsen et al., 2008; Dethlefsen and Relman, 2011; Jernberg et al., 2007). It is perhaps not surprising that the rapid loss in diversity of the gut microbiota observed after antibiotic treatment has profound effects on its combined metabolic activities and products.
Treatment of mice with a combination of metronidazole, neomycin and vancomycin lowers intestinal expression of the antimicrobial peptide RegIII-γ, suggesting that the ability of the gut-associated microbial community to fortify barrier functions can be impaired by antibiotic treatment (Brandl et al., 2008). There is direct evidence to suggest that changes in the composition of the gut microbiota that are associated with antibiotic treatment can lead to a drop in the concentrations of some microbiota-derived metabolites. For example, dietary phosphatidylcholine is metabolized by the gut microbiota to generate TMA, which is absorbed and then oxidized in host tissue to form trimethyl amine oxide (TMAO). Measurements of TMAO concentrations in human plasma and urine reveal a marked decline after the administration of antibiotics and a succeeding increase after withdrawal of antibiotics (Tang et al., 2013). Antibiotics also lower the concentrations of metabolites derived from complex carbohydrate fermentation, because treatment of mice with streptomycin, metronidazole or vancomycin reduces SCFA concentrations in the cecum (Garner et al., 2009; Meynell, 1963; Smith et al., 2013). Similarly, volunteers treated orally with metronidazole, vancomycin or bacitracin excrete significantly reduced levels of fecal SCFAs (Hoverstad et al., 1986; Lewis et al., 2005). Oral doxycycline, co-trimoxazole, nalidixic acid, or ofloxacin treatment does not produce this effect in volunteers, which illustrates that not all antibiotics reduce the metabolic activities of gut-associated microbial communities to the same extent, perhaps due to differences in their pharmacokinetics or to factors that remain to be worked out. Nonetheless, it is clear from the above data that at least some antibiotics cause a disruption of the gut microbiota that is severe enough to significantly reduce the concentrations of microbiota-derived metabolites. We will discuss below how this side effect of antibiotic treatment can have several adverse consequences for the host.
Undesired consequences: Metabolite depletion disrupts gut homeostasis
Disruption of the gut microbiota by vancomycin treatment lowers intestinal SCFA concentrations, thereby reducing stimulation of GPR43 on regulatory T cells, which leads to a contraction of this cell population in the colonic mucosa, but not in other organs (Atarashi et al., 2011; Smith et al., 2013). However, the numbers of regulatory T cells in the colon remain unchanged when mice are treated with a combination of vancomycin and SCFAs (Smith et al., 2013). These data suggest that antibiotic treatment can lower the numbers of regulatory colonic T cells by reducing the concentration of SCFAs in the distal gut.
An antibody-mediated depletion of regulatory T cells does not trigger overt intestinal inflammation in mice, but it increases the severity of chemically induced colitis (Boehm et al., 2012). An antibiotic-induced lowering of intestinal SCFA concentrations and consequent reduction in the numbers or regulatory T cells is therefore predicted to render the host more susceptible to colitis. In line with this prediction, antibiotic treatment enhances the severity of chemically induced colitis in mice (Rakoff-Nahoum et al., 2004; Singh et al., 2014; Spees et al., 2013b). Similarly, the severity of colitis caused by enteric pathogens, such as Citrobacter rodentium or Salmonella enterica serovar Typhimurium (family Enterobacteriaceae, phylum Proteobacteria), is enhanced in mice treated with streptomycin, vancomycin or metronidazole (Barthel et al., 2003; Spees et al., 2013b; Wlodarska et al., 2011). Consistent with a mechanism that depends on a disruption of the community of SFCA producers, the enhanced severity of _S. enterica_-induced colitis after antibiotic treatment is independent of pathogen colonization, but correlates with differences in the composition of the gut microbiota (Ferreira et al., 2011) and reduced concentrations of SCFAs in the large bowel (Garner et al., 2009).
Antibiotic treatment also lowers ‘colonization resistance’ against enteric pathogens belonging to the family Enterobacteriaceae. The concept that the gut microbiota confers ‘colonization resistance’ against Enterobacteriaceae was first established in the 1950’s and 60’s by demonstrating that treatment with streptomycin greatly enhances the ability of S enterica or Escherichia coli to colonize the large bowel of mice (Bohnhoff et al., 1954; Saito, 1961a, b). Early work on the underlying mechanism revealed that streptomycin treatment lowers the concentration of SCFAs and increases the redox potential in the cecum to conditions that approximate an aerobic broth culture (Meynell, 1963). More recent results show that treatment with antibiotics leads to a subtle elevation of some inflammatory markers in the intestinal mucosa (Spees et al., 2013b; Wlodarska et al., 2011), which might be a consequence of lowering the concentrations of microbota-derived metabolites that possess anti-inflammatory properties, such as SCFAs (Garner et al., 2009; Hoverstad et al., 1986; Lewis et al., 2005; Meynell, 1963; Smith et al., 2013). For instance, streptomycin treatment of mice markedly increases cecal expression of inducible nitric oxide synthase (iNOS) encoded by the NOS2 gene (Spees et al., 2013b), the expression of which is suppressed in human colonocytes by the SCFA sensor PPAR-γ (Marion-Letellier et al., 2008). Furthermore, mild inflammatory infiltrates of neutrophils, inflammatory monocytes and natural killer cells are observed in the intestinal mucosa of mice treated with streptomycin or metronidazole (Spees et al., 2013b; Wlodarska et al., 2011). In humans, repeated courses of antibiotics can lead to the development of irritable bowel syndrome (IBS), a condition characterized by low-level intestinal inflammation, diarrhea, lower concentrations of fecal SCFAs (Kopecny and Simunek, 2002; Treem et al., 1996) and an increased abundance of Enterobacteriaceae in the fecal microbial population (Carroll et al., 2012; Matto et al., 2005).
The observation that antibiotic treatment can induce iNOS expression (Spees et al., 2013b) might help explain the increased redox potential in the cecum of streptomycin-treated mice first observed in 1963 (Meynell, 1963). Nitric oxide generated by iNOS can react with superoxide produced by epithelial NADPH oxidases to give rise to peroxynitrite, which can be further converted to nitrate (Szabo et al., 2007). Nitrate is an excellent respiratory electron acceptor, because the redox potential of the nitrate/nitrite redox couple (E° = 433 mV) is second only to that of the oxygen/water redox couple (E° = 818 mV) (Thauer et al., 1977). Nitrate respiration enhances growth of E. coli in the large bowel of streptomycin-treated mice (Jones et al., 2011) and in mice with chemically induced colitis (Winter et al., 2013a), but not in untreated mice (Spees et al., 2013b). Furthermore, the fitness advantage conferred upon E. coli by nitrate respiration is abrogated in streptomycin-treated _NOS2_-deficient mice, suggesting that nitrate in the gut lumen after antibiotic treatment is host-derived (Spees et al., 2013b).
The evidence reviewed above raises the possibility that antibiotics can disrupt immune homeostasis by disrupting the community of SCFA producers, which in turn increasing the inflammatory tone of the intestinal mucosa. This low-degree of inflammation might lead to an enhanced susceptibility to colitis and is at least in part responsible for reducing ‘colonization resistance’ against Enterobacteriaceae (Spees et al., 2013a). Enterobacteriaceae benefit from low-level intestinal inflammation because by-products of the inflammatory host response, such as nitrate, can selectively boost the growth of these facultative anaerobic bacteria in the large bowel (Lopez et al., 2012; Spees et al., 2013b; Winter et al., 2013b). For example, an analysis of 2,476 genomes representing all phylogenetic groupings found within the gut microbiota suggests that sequences predicted to encode nitrate reductase activity are present at the highest frequency within members of the Enterobacteriaceae (Winter and Baumler, 2014). Thus, the generation of host-derived nitrate after antibiotic treatment might selectively boost the growth of Enterobacteriaceae simply because its members are more likely to encode the enzymes to utilize this electron acceptor.
Hidden costs: Beyond the hygiene hypothesis
While it may be unsurprising that a severe disruption of gut-associated microbial communities by antibiotics influences immune homeostasis and pathogen susceptibility in the gut, it is less obvious that it also impacts immune responses in distal organs, such as the airways. An increase in allergies observed since the second half of the 20th century has been attributed by the 'hygiene hypothesis' to decreased exposure to sporadic infections in early childhood (Strachan, 1989). The progressive disappearance of H. pylori in high-income countries over the course of the 20th century is inversely correlated to childhood asthma (Chen and Blaser, 2008; Reibman et al., 2008), which led to the advancement of the 'disappearing microbiota hypothesis’ for explaining the rise in allergies by a lack of continued exposure to H. pylori during childhood (Blaser, 2008). These hypotheses suggest that sporadic exposure ('hygiene hypothesis') or continued exposure ('disappearing microbiota hypothesis’) to pathogens in early childhood has a protective role against allergic diseases by influencing the development of the neonate's immune system. However, recent evidence suggests that in addition to pathogen exposure, the continued exposure to metabolites produced by the commensal gut microbiota plays a protective role in preventing allergic airways disease (Bjorksten, 2009; Isolauri et al., 2009; Shreiner et al., 2008).
One reason for such distal effects is that some microbiota-derived metabolites enter the circulation and influence cells that are located within peripheral tissues. For example, fermentation of complex carbohydrates by the gut microbiota increases systemic levels of SCFAs, which in turn influence dendritic cell hematopoiesis and functionality by a mechanism that depends on GPR41, a SCFA receptor expressed on the surface of host cells (Trompette et al., 2014). Stimulation of GPR41 with propionate does not lead to an accumulation of regulatory T cells, but protects against allergic inflammation in the lung through a mechanism that involves decreased activation of lung dendritic cells (Trompette et al., 2014) (Figure 2). The relevance of this observation is highlighted by clinical studies documenting an increased risk for developing allergic asthma after exposure to antibiotics early in life (Marra et al., 2009; Martel et al., 2009; Murk et al., 2011). Furthermore, low fecal SCFA levels in children are associated with allergy (Sandin et al., 2009). A vancomycin-mediated disruption of the community of SCFA producers in neonatal mice, but not in adult mice, enhances subsequent susceptibility to allergic asthma (Russell et al., 2012), suggesting that an antibiotic-mediated disruption of the gut microbiota might be most consequential during the developmental period.
While the 'hygiene hypothesis' and the 'disappearing microbiota hypothesis’ propose exposure to pathogens as a mechanism protecting from allergies, the data reviewed above provide evidence for a second mechanism that involves exposure to metabolites produced by commensal, non-pathogenic gut microbes early in life. The terms 'microbial deprivation hypothesis' (Bjorksten, 2009) and 'microflora hypothesis’ (Shreiner et al., 2008) have been advanced to describe the idea that continued exposure to a balanced gut microbiota during childhood, more than sporadic infections, are important factors for the prevention of allergy. Antibiotics can disrupt a continued exposure to microbiota-derived metabolites, thereby increasing susceptibility to allergies.
In conclusion, microbes inhabiting our gastrointestinal tract during the first years of life shape our physiology and may have lifelong influences on our immune system. A combination of an increasing childhood exposure to antibiotics starting in the second half of the 20th century and a progressive disappearance of H. pylori throughout the last century impairs immune education, thereby contributing to the concomitant rise in allergies observed in high-income countries.
Conclusions
Our lifelong association with a large microbial community inhabiting our intestine provides local as well as systemic benefits. The combined metabolic activities of the gut microbiota promote host nutrition, mucosal barrier function, gut immune homeostasis, ‘colonization resistance’ and immune education. Recent research is beginning to illuminate the consequences of perturbing this homeostasis, for example by depleting microbiota-derived metabolites during antibiotic therapy. A better understanding of these microbe-host interactions will aid in the development of strategies to prevent undesired side effects of antibiotic therapy and improve human health.
ACKNOWLEDGEMENTS
Work in AJB’s laboratory is supported by Public Health Service Grants AI044170, AI096528 and AI107393. CAL is supported by Public Health Services Grant AI112241. DDK is supported by Public Health Services Grant OD11147. EMV is supported by Public Health Services Grant OD010931.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE
- Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta. 2010;1801:1175–1183. doi: 10.1016/j.bbalip.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Alex S, Lange K, Amolo T, Grinstead JS, Haakonsson AK, Szalowska E, Koppen A, Mudde K, Haenen D, Al-Lahham S, Roelofsen H, Houtman R, van der Burg B, Mandrup S, Bonvin AM, Kalkhoven E, Muller M, Hooiveld GJ, Kersten S. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 2013;33:1303–1316. doi: 10.1128/MCB.00858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atif SM, Uematsu S, Akira S, McSorley SJ. CD103-CD11b+ dendritic cells regulate the sensitivity of CD4 T-cell responses to bacterial flagellin. Mucosal Immunol. 2014;7:68–77. doi: 10.1038/mi.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson MD, Liu YW, Hanly AM, Emenaker NJ, Shenoy SG, Gould Rothberg BE. Identification and comparative analysis of human colonocyte short-chain fatty acid response genes. J Gastrointest Surg. 2000;4:501–512. doi: 10.1016/s1091-255x(00)80093-1. [DOI] [PubMed] [Google Scholar]
- Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, Raffatellu M. The Cytokine IL-22 Promotes Pathogen Colonization by Suppressing Related Commensal Bacteria. Immunity. 2014;40:262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson JM, Shepherd DM. Aryl hydrocarbon receptor activation by TCDD reduces inflammation associated with Crohn's disease. Toxicol Sci. 2011;120:68–78. doi: 10.1093/toxsci/kfq360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorksten B. The hygiene hypothesis: do we still believe in it? Nestle Nutr Workshop Ser Pediatr Program. 2009;64:11–18. doi: 10.1159/000235780. discussion 18–22, 251–257. [DOI] [PubMed] [Google Scholar]
- Blaser MJ. Disappearing microbiota: Helicobacter pylori protection against esophageal adenocarcinoma. Cancer Prev Res (Phila) 2008;1:308–311. doi: 10.1158/1940-6207.CAPR-08-0170. [DOI] [PubMed] [Google Scholar]
- Boehm F, Martin M, Kesselring R, Schiechl G, Geissler EK, Schlitt HJ, Fichtner-Feigl S. Deletion of Foxp3+ regulatory T cells in genetically targeted mice supports development of intestinal inflammation. BMC Gastroenterol. 12:97. doi: 10.1186/1471-230X-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnhoff M, Drake BL, Miller CP. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med. 1954;86:132–137. doi: 10.3181/00379727-86-21030. [DOI] [PubMed] [Google Scholar]
- Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, Dowell SJ. The Orphan G protein- coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2012;24:521–530. doi: 10.1111/j.1365-2982.2012.01891.x. e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Blaser MJ. Helicobacter pylori colonization is inversely associated with childhood asthma. J Infect Dis. 2008;198:553–560. doi: 10.1086/590158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, Walter J, Vijay-Kumar M, Gewirtz AT, Ley RE. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Huerga J, Popper H. Urinary excretion of choline metabolites following choline administration in normals and patients with hepatobiliary diseases. J Clin Invest. 1951;30:463–470. doi: 10.1172/JCI102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Hristova KR, Oakley HA, McCracken VJ, Aminov R, Mackie RI, Gaskins HR. Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl Environ Microbiol. 2000;66:2166–2174. doi: 10.1128/aem.66.5.2166-2174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe DR, Wali A, Brylawski BP, Bultman SJ. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS One. 2012;7:e46589. doi: 10.1371/journal.pone.0046589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, Croxen MA, Finlay BB. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One. 2011;6:e20338. doi: 10.1371/journal.pone.0020338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fite A, Macfarlane GT, Cummings JH, Hopkins MJ, Kong SC, Furrie E, Macfarlane S. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut. 2004;53:523–529. doi: 10.1136/gut.2003.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S, Toshimitsu T, Matsuoka S, Maruyama A, Oh-Oka K, Takamura T, Nakamura Y, Ishimaru K, Fujii-Kuriyama Y, Ikegami S, Itou H, Nakao A. Identification of a probiotic bacteria-derived activator of the aryl hydrocarbon receptor that inhibits colitis. Immunol Cell Biol. 2014 doi: 10.1038/icb.2014.2. [DOI] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Garner CD, Antonopoulos DA, Wagner B, Duhamel GE, Keresztes I, Ross DA, Young VB, Altier C. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar typhimurium murine model of infection. Infect Immun. 2009;77:2691–2702. doi: 10.1128/IAI.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Dallas DC, Mills DA. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology. 2013;159:649–664. doi: 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harig JM, Soergel KH, Komorowski RA, Wood CM. Treatment of diversion colitis with short-chain-fatty acid irrigation. N Engl J Med. 1989;320:23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- Heath-Pagliuso S, Rogers WJ, Tullis K, Seidel SD, Cenijn PH, Brouwer A, Denison MS. Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry. 1998;37:11508–11515. doi: 10.1021/bi980087p. [DOI] [PubMed] [Google Scholar]
- Hoverstad T, Carlstedt-Duke B, Lingaas E, Norin E, Saxerholt H, Steinbakk M, Midtvedt T. Influence of oral intake of seven different antibiotics on faecal short-chain fatty acid excretion in healthy subjects. Scand J Gastroenterol. 1986;21:997–1003. doi: 10.3109/00365528608996411. [DOI] [PubMed] [Google Scholar]
- Isolauri E, Kalliomaki M, Rautava S, Salminen S, Laitinen K. Obesity -extending the hygiene hypothesis. Nestle Nutr Workshop Ser Pediatr Program. 2009;64:75–85. doi: 10.1159/000235784. discussion 85–79, 251–257. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. Isme J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- Jin UH, Lee SO, Sridharan G, Lee K, Davidson LA, Jayaraman A, Chapkin R, Alaniz R, Safe SH. Microbiome-derived Tryptophan Metabolites and Their Aryl Hydrocarbon Receptor-Dependent Agonist and Antagonist Activities. Mol Pharmacol. 2014 doi: 10.1124/mol.113.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Gibson T, Maltby RC, Chowdhury FZ, Stewart V, Cohen PS, Conway T. Anaerobic respiration of Escherichia coli in the mouse intestine. Infection and immunity. 2011;79:4218–4226. doi: 10.1128/IAI.05395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, Pettersson S, Conway S. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecny J, Simunek J. Cellulolytic bacteria in human gut and irritable bowel syndrome. Acta Veterinaria Brno. 2002;71:421–427. [Google Scholar]
- Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, Brezillon S, Dupriez V, Vassart G, Van Damme J, Parmentier M, Detheux M. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- Lewis S, Brazier J, Beard D, Nazem N, Proctor D. Effects of metronidazole and oligofructose on faecal concentrations of sulphate-reducing bacteria and their activity in human volunteers. Scand J Gastroenterol. 2005;40:1296–1303. doi: 10.1080/00365520510023585. [DOI] [PubMed] [Google Scholar]
- Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Baumler AJ. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio. 2012;3:e00143–00112. doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion-Letellier R, Butler M, Dechelotte P, Playford RJ, Ghosh S. Comparison of cytokine modulation by natural peroxisome proliferator-activated receptor gamma ligands with synthetic ligands in intestinal-like Caco-2 cells and human dendritic cells--potential for dietary modulation of peroxisome proliferator-activated receptor gamma in intestinal inflammation. Am J Clin Nutr. 2008;87:939–948. doi: 10.1093/ajcn/87.4.939. [DOI] [PubMed] [Google Scholar]
- Marra F, Marra CA, Richardson K, Lynd LD, Kozyrskyj A, Patrick DM, Bowie WR, Fitzgerald JM. Antibiotic use in children is associated with increased risk of asthma. Pediatrics. 2009;123:1003–1010. doi: 10.1542/peds.2008-1146. [DOI] [PubMed] [Google Scholar]
- Martel MJ, Rey E, Malo JL, Perreault S, Beauchesne MF, Forget A, Blais L. Determinants of the incidence of childhood asthma: a two-stage case-control study. Am J Epidemiol. 2009;169:195–205. doi: 10.1093/aje/kwn309. [DOI] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matto J, Maunuksela L, Kajander K, Palva A, Korpela R, Kassinen A, Saarela M. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome--a longitudinal study in IBS and control subjects. FEMS immunology and medical microbiology. 2005;43:213–222. doi: 10.1016/j.femsim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Meynell GG. Antibacterial mechanisms of the mouse gut. II. The role of Eh and volatile fatty acids in the normal gut. Br J Exp Pathol. 1963;44:209–219. [PMC free article] [PubMed] [Google Scholar]
- Millard AL, Mertes PM, Ittelet D, Villard F, Jeannesson P, Bernard J. Butyrate affects differentiation, maturation and function of human monocyte-derived dendritic cells and macrophages. Clin Exp Immunol. 2002;130:245–255. doi: 10.1046/j.0009-9104.2002.01977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA., 3rd Expression of the human aryl hydrocarbon receptor complex in yeast. Activation of transcription by indole compounds. J Biol Chem. 1997;272:32824–32829. doi: 10.1074/jbc.272.52.32824. [DOI] [PubMed] [Google Scholar]
- Monteleone I, Rizzo A, Sarra M, Sica G, Sileri P, Biancone L, MacDonald TT, Pallone F, Monteleone G. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141:237–248. doi: 10.1053/j.gastro.2011.04.007. 248 e231. [DOI] [PubMed] [Google Scholar]
- Murk W, Risnes KR, Bracken MB. Prenatal or early-life exposure to antibiotics and risk of childhood asthma: a systematic review. Pediatrics. 2011;127:1125–1138. doi: 10.1542/peds.2010-2092. [DOI] [PubMed] [Google Scholar]
- Oresic M, Seppanen-Laakso T, Yetukuri L, Backhed F, Hanninen V. Gut microbiota affects lens and retinal lipid composition. Exp Eye Res. 2009;89:604–607. doi: 10.1016/j.exer.2009.06.018. [DOI] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Bäumler AJ. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Reibman J, Marmor M, Filner J, Fernandez-Beros ME, Rogers L, Perez-Perez GI, Blaser MJ. Asthma is inversely associated with Helicobacter pylori status in an urban population. PLoS One. 2008;3:e4060. doi: 10.1371/journal.pone.0004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, Finlay BB. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO reports. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K. [Studies on the habitation of pathogenic Escherichia coli in the intestinal tract of mice. I. Comparative experiments on the habitation of each type of resistant pathogenic Escherichia coli under an administration of streptomycin] Paediatria Japonica. 1961a;65:385–393. [PubMed] [Google Scholar]
- Saito K. [Studies on the habitation of pathogenic Escherichia coli in the intestinal tract of mice. II. Experimental inoculation of type 055 Escherichia coli after long-term administration of streptomycin] Paediatria Japonica. 1961b;65:394–399. [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson IR. Short chain fatty acid regulation of signaling genes expressed by the intestinal epithelium. J Nutr. 2004;134:2450S–2454S. doi: 10.1093/jn/134.9.2450S. [DOI] [PubMed] [Google Scholar]
- Sandin A, Braback L, Norin E, Bjorksten B. Faecal short chain fatty acid pattern and allergy in early childhood. Acta Paediatr. 2009;98:823–827. doi: 10.1111/j.1651-2227.2008.01215.x. [DOI] [PubMed] [Google Scholar]
- Schwiertz A, Taras D, Schafer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- Shreiner A, Huffnagle GB, Noverr MC. The "Microflora Hypothesis" of allergic disease. Adv Exp Med Biol. 2008;635:113–134. doi: 10.1007/978-0-387-09550-9_10. [DOI] [PubMed] [Google Scholar]
- Siavoshian S, Segain JP, Kornprobst M, Bonnet C, Cherbut C, Galmiche JP, Blottiere HM. Butyrate and trichostatin A effects on the proliferation/differentiation of human intestinal epithelial cells: induction of cyclin D3 and p21 expression. Gut. 2000;46:507–514. doi: 10.1136/gut.46.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spees AM, Lopez CA, Kingsbury DD, Winter SE, Baumler AJ. Colonization resistance: battle of the bugs or Menage a Trois with the host? PLoS Pathog. 2013a;9:e1003730. doi: 10.1371/journal.ppat.1003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, Tsolis RM, Baumler AJ. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio. 2013b;4 doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- Taggart AK, Kero J, Gan X, Cai TQ, Cheng K, Ippolito M, Ren N, Kaplan R, Wu K, Wu TJ, Jin L, Liaw C, Chen R, Richman J, Connolly D, Offermanns S, Wright SD, Waters MG. (D)-beta-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J Biol Chem. 2005;280:26649–26652. doi: 10.1074/jbc.C500213200. [DOI] [PubMed] [Google Scholar]
- Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treem WR, Ahsan N, Kastoff G, Hyams JS. Fecal short-chain fatty acids in patients with diarrhea-predominant irritable bowel syndrome: in vitro studies of carbohydrate fermentation. Journal of pediatric gastroenterology and nutrition. 1996;23:280–286. doi: 10.1097/00005176-199610000-00013. [DOI] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- Wang B, Morinobu A, Horiuchi M, Liu J, Kumagai S. Butyrate inhibits functional differentiation of human monocyte-derived dendritic cells. Cell Immunol. 2008;253:54–58. doi: 10.1016/j.cellimm.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Winter SE, Baumler AJ. Dysbiosis in the inflamed intestine: Chance favors the prepared microbe. Gut Microbes. 2014;5 doi: 10.4161/gmic.27129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Lopez CA, Baumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO reports. 2013a;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Baumler AJ. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013b;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infection and immunity. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- Zinkevich VV, Beech IB. Screening of sulfate-reducing bacteria in colonoscopy samples from healthy and colitic human gut mucosa. FEMS Microbiol Ecol. 2000;34:147–155. doi: 10.1111/j.1574-6941.2000.tb00764.x. [DOI] [PubMed] [Google Scholar]