The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote (original) (raw)
Abstract
The covalent modification of nucleosomal histones has emerged as a major determinant of chromatin structure and gene activity. To understand the interplay between various histone modifications, including acetylation and methylation, we performed a genome-wide chromatin structure analysis in a higher eukaryote. We found a binary pattern of histone modifications among euchromatic genes, with active genes being hyperacetylated for H3 and H4 and hypermethylated at Lys 4 and Lys 79 of H3, and inactive genes being hypomethylated and deacetylated at the same residues. Furthermore, the degree of modification correlates with the level of transcription, and modifications are largely restricted to transcribed regions, suggesting that their regulation is tightly linked to polymerase activity.
Keywords: Epigenetics, chromatin, histone, Drosophila, chromatin immunoprecipitation, microarray
Nucleosomal histones are the target of a variety of covalent modifications. It is now well established that posttranslational acetylation, methylation, phosphorylation, and ubiquitination of histones play an intrinsic role in transcriptional regulation and are potentially involved in the propagation of the transcriptional state through cell division (Turner 2002; Felsenfeld and Groudine 2003). On the basis of the large number of these covalent marks, it was suggested that these alterations form a histone code, which mediates specificity in transcriptional regulation (Strahl and Allis 2000).
However, although considerable progress has been made in determining the enzymes that alter histones, only limited information exists regarding the patterns of nucleosomal modifications at a given gene. Immunostaining of the nucleus with antibodies specific for a histone modification reveals whether a modification occurs in the euchromatic or the heterochromatic nuclear compartment. The heterochromatic compartment was first defined in higher eukaryotes by light microscopy as the part of the genome that stays highly compact throughout the cell cycle (Heitz 1928). It consists mainly of large blocks of pericentromeric repeats that lack genes. The euchromatic nuclear compartment contains the majority of genes interspersed by intergenic regions (Adams et al. 2000). Given that developmental and tissue-specific gene regulation occurs in this chromosomal environment, it is not surprising that the exact chromatin structure of the euchromatic nuclear compartment is far from uniform (Felsenfeld and Groudine 2003).
To determine the distribution of histone modification patterns in euchromatin, we analyzed the modification state of over 5000 genes in the Drosophila genome. We combined chromatin immunoprecipitation (ChIP) with subsequent microarray analysis to measure H3 and H4 acetylation, H3 Lys 4 dimethylation (H3-di-meK4) and trimethylation (H3-tri-meK4), and Lys 79 dimethylation (H3-di-meK79) at >40% of all genes in this organism. This analysis has resulted in a genome-wide distribution map of chromatin modifications in a higher eukaryote and allows us to determine whether modifications coincide at the same genes and whether their presence depends on the chromosomal position of a gene. Furthermore, by combining these data sets with our previous analysis of replication timing and transcription (Schübeler et al. 2002), we constructed an epigenomic matrix that allows us to analyze the interplay of each of these histone modifications with transcriptional activity and the timing of DNA replication.
Results
Chromatin profiling of the Drosophila genome
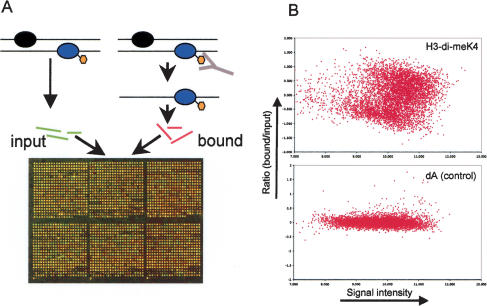
We used ChIP followed by hybridization to DNA microarrays to map the pattern of six different histone modifications in the Drosophila genome. Similar to our previous study of the genome-wide pattern of DNA replication, we used the karyotypically stable Drosophila Kc cell line (Schübeler et al. 2002). Chromatin was purified after formaldehyde cross-linking (= input) and immunoprecipitated either with antibodies that recognize a specific histone modification or without the addition of antisera as a control. DNA enriched for a specific modification (= bound) and DNA from the input material was isolated, labeled with different fluorescent dyes, and hybridized to a DNA microarray (Fig. 1A; Materials and Methods). Enrichment for a histone modification via immunoprecipitation results in a stronger fluorescence signal from the bound fraction, whereas absence of the modification results in a stronger signal from the input fraction. Figure 1B shows the signal and ratio of bound-over input fraction (Dudoit et al. 2002a) for the specific antibody against H3-di-meK4 and for an antibody-less control experiment. Because the observed enrichments are antibody specific, the ratio of the two dyes represents a quantitative measure of the studied modification.
Figure 1.
(A) Principle of genome-wide chromatin analysis. Chromatin is purified after cross-linking with formaldehyde and subsequently immunoprecipitated with an antibody specific for a histone modification. The enriched DNA fractions are purified, fluorescently labeled, and cohybridized to a spotted microarray with an input sample that has been labeled with a different dye. The resulting ratio of the signal from the bound-over input fraction is determined after the hybridization and used as a measure for enrichment. (B) Antibody-specific enrichment during ChIP is illustrated by comparing a ChIP with an antibody against a euchromatic histone modification (H3-di-meK4) with a control experiment (dA) in which no antibody has been added. The ratio of the bound-over input fraction versus the signal intensity is plotted (M/A plot, Dudoit et al. 2002a) for both experiments. Notice the increased spread of ratios in the H3-di-meK4 experiment compared with the control experiment, reflecting the specific enrichment during the immunoprecipitation.
In addition to H3-di-meK4, similar chromatin profiling experiments were performed for four other euchromatic histone modifications (H3-tri-meK4, H3-Ac, H4-Ac, and H3-di-K79; see following). To verify enrichments detected by the microarray hybridizations, we performed semiquantitative PCR controls for a subset of genes, which confirmed the microarray results (Fig. 2; data not shown). As an additional control, we performed chromatin profiling with an antibody to a histone modification that we hypothesized to be distributed more evenly throughout the genome. Phosphorylation of Ser 10 of histone H3 (H3-S10) is involved in chromosome condensation and present homogenously on mitotic chromosomes (Wei et al. 1999). Although this modification can be detected at a subset of Drosophila promoters in inter-phase (Labrador and Corces 2003), it is highly up-regulated in the G2 phase of the cell cycle (Hsu et al. 2000). The Kc cells used in our analysis also display a strong enrichment of this modification in G2 (data not shown), a cell cycle phase that is prominent in logarithmically growing Kc cultures (see Fig. 1 in Schübeler et al. 2002). Using Kc cell chromatin immunoprecipitated with an H3-S10 antibody as probe, we observed that this modification is enriched to a similar extent at the vast majority of genes (Fig. 3; see following), an observation that is in agreement with the uniform and high abundance of H3-S10 on mitotic chromosomes.
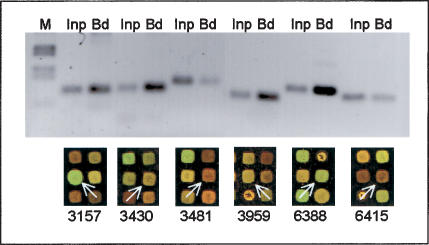
Figure 2.
Control of microarray results by gene-specific PCR. We amplified sequences from input and from immunoprecipitated (= bound) chromatin and compared it with the detection on the microarray. Enrichment by ChIP is indicated by a stronger signal in the PCR from the bound fraction (Bd) and in a stronger microarray signal from the fluorescent dye used to label the bound fraction (here Cy3 = green fluorescence). Lack of enrichment is indicated by a weak PCR signal in the bound fraction compared with input (Inp) and a stronger microarray signal from the input material (here Cy5 = red fluorescence). Shown are six sequences after ChIP against H3-di-meK4 that are analyzed by PCR and microarray. Microarray sections are taken from the same slide with identical color settings. Array spots corresponding to the PCR products are labeled with white arrows. In each case, the PCR confirms the microarray analysis and similar results were obtained for other modifications (data not shown).
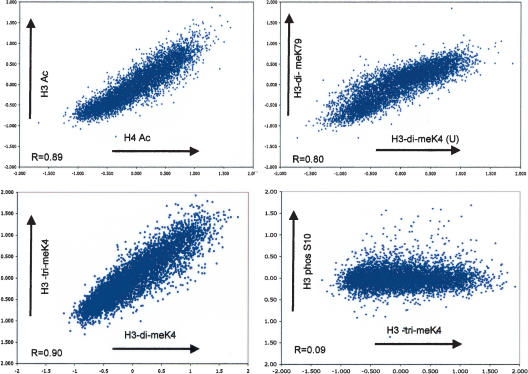
Figure 3.
Pairwise comparisons of different euchromatic histone modifications. Shown are four scatterplots comparing the log2 ratios of bound-over input material for H3-Ac versus H4-Ac, H3-di-meK4 versus H3-di-meK79, H3-tri-meK4 versus H3-di-meK4, and H3phos-S10 versus H3-tri-meK4. A high degree of positive correlation is observed between the euchromatic modifications, indicating that these marks are shared by the same genes. This correlation, however, is not observed when a euchromatic modification is compared with the evenly distributed H3-S10 phosphorylation (see also Table 1).
Each immunoprecipitation was repeated three times independently, starting with cells from different passages. These experiments proved to be highly reproducible, as indicated by a low variation (average covariance of 13%). The resulting set of chromatin profiles for six different histone modifications contained 5375 single-copy genes, representing >40% of all predicted Drosophila genes.
Histone acetylation and Lys 4 and Lys 79 methylation are enriched at the same genes
The actual level of histone acetylation is dynamic and depends on the regulated interplay between histone acetylases (HATs) and histone deacetylases (HDACs; Turner 2002). In vitro studies suggest that HATs and HDACs can vary widely in their histone preference and furthermore in their preference for a certain lysine (Kuo and Allis 1998; Johnson et al. 2002; Robyr et al. 2002). On the other hand, most histone methylases seem to modify only a defined arginine or lysine residue (Zhang and Reinberg 2001; Kouzarides 2002). Compared with acetylation, overall histone methylation has a long half-life (Waterborg 1993), possibly due to the absence of specific histone demethylases, which have not yet been conclusively identified (Bannister et al. 2002).
The list of enzymes that potentially acetylate, deacetylate, or methylate nucleosomal histones has grown substantially in recent years, and it is likely that each modification is catalyzed by one or several enzymes. Thus, each modification could have a unique genomic distribution reflecting its function and regulation. To address whether such diversity in histone-modifying enzymes yields diversity in histone modification patterns, we generated scatterplots and calculated the Pearson correlation coefficient (R) for all potential pairs between the different modifications and the control (Fig. 3; Table 1). We found that all studied euchromatic modifications display a high degree of positive correlation (R from 0.7 to 0.9) with the exception of both control experiments (H3-phos-S10 and no antibody, R from 0.02 to 0.16). Thus a gene that is dimethylated on Lys 4 has a high likelihood of also being trimethylated on Lys 4, dimethylated on Lys 79, and acetylated at histone H3 and H4 (and vice versa). This surprisingly high coincidence of various euchromatic histone modifications is independent of the short (acetylation) or long (methylation) half-life time of the modification and might reflect a concerted regulation.
Table 1.
The relationship between the studied histone modifications
| R | H4-Ac | H3-Ac | H3-di-meK4(U) | H3-di-meK4(T) | H3-tri-meK4 | H3-di-meK79 | H3-phos-S10 | dA (ctr) |
|---|---|---|---|---|---|---|---|---|
| H4-Ac | X | |||||||
| H3-Ac | 0.89 | X | ||||||
| H3-di-meK4(U) | 0.85 | 9.83 | X | |||||
| H3-di-meK4(T) | 0.83 | 0.88 | 0.87 | X | ||||
| H3-tri-meK4 | 0.77 | 0.81 | 0.79 | 0.90 | X | |||
| H3-di-meK79 | 0.71 | 0.75 | 0.80 | 0.80 | 0.72 | X | ||
| H3-phos-S10 | 0.03 | 0.07 | 0.02 | 0.07 | 0.09 | 0.02 | X | |
| dA (ctr) | 0.11 | 0.09 | 0.09 | 0.14 | 0.16 | 0.04 | 0.53 | X |
The transcriptional state reflects the pattern of histone modifications
Recent studies have revealed that histone acetylation and H3-K4 or H3-K79 methylation are rarely present on heterochromatin and are not directly involved in transcriptional repression (Turner 2002). Consequently, these modifications should be present on genes that are not repressed and thus are either transcribed or in an activatable state. To study the relationships of the analyzed histone modifications with the transcriptional state of a given gene, we compared the histone modification data sets with our previously described microarray expression data obtained with the identical microarray and Kc cell line (Schübeler et al. 2002). Genes were scored as transcriptionally active or inactive as described (Schübeler et al. 2002). A comparison between the transcriptional status and the enrichment for each of the analyzed euchromatic histone modifications reveals a strong correlation between enrichment and the likelihood of transcription (Fig. 4; Supplementary Fig. 1). This correlation is unique to the euchromatic modifications because it was not observed in the control experiments, including the H3-phos-S10 data set (Supplementary Fig. 1). Thus, we conclude that histone H3 and H4 acetylation, as well as Lys 4 and Lys 79 methylation, occur mostly at genes that are actively transcribed.
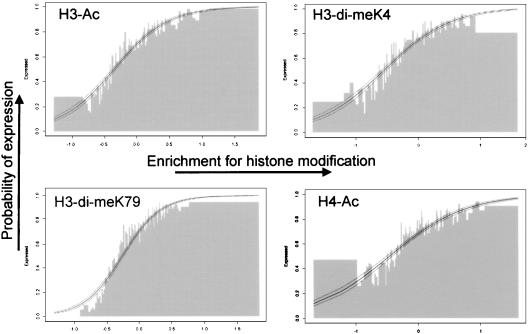
Figure 4.
Relationship between transcriptional status and chromatin structure. To determine whether enrichment for the tested euchromatic histone modifications depends on the expression status, we compared the enrichment for a histone modification with the transcriptional status for all single-copy genes present on the array. Genes were ranked according to their enrichment for a particular modification and divided into groups of 50 genes. The percentage of active genes (that is, the probability of expression) in each group was calculated by using an existing expression analysis (Schübeler et al. 2002). The percentage of active genes (_Y_-axis) is plotted versus the enrichment for a histone modification (_X_-axis). This presentation shows that genes that are enriched in euchromatic modifications are almost all transcriptionally active, whereas genes that are not enriched are mostly inactive. To validate this observation with the appropriate statistical analysis, we performed logistic regression for all studied modifications. Logistic regression is used if there are only two potential outcomes (binominal) for one of the two variables, which in this case is the transcriptional status (on/off). The resulting logistic regression curve (thick line) and the 95% confidence interval (outer lines) are plotted and in each case show a very strong correlation between enrichment for euchromatic histone modifications and transcriptional activity (a complete set of plots is shown in the Supplemental Material).
Clearly, our finding does not rule out that these coinciding modifications are distributed unevenly along the transcribed regions or that other histone modifications display a more variable genomic distribution. For example, methylation of Lys 20 of histone H4 has been shown to be enriched on inactive genes in Drosophila polytene chromosomes (Fang et al. 2002; Nishioka et al. 2002). Using our experimental conditions, however, we were not successful in immunoprecipitating chromatin with various antisera against H4-di-meK20 (data not shown).
Nevertheless, the strong positive correlation that we report for five different euchromatic modifications suggests there is a general binary chromatin state of euchromatic genes, in which the transcriptional “on” and “off” configurations are each marked by a common histone modification pattern, suggesting a coordinated regulation of different histone-modifying enzymes.
The degree of euchromatic histone modifications is coupled to the amount of RNA produced
As our experiments indicate that the transcriptional status of a gene is tightly correlated with a common histone modification pattern, we wished to address whether the level of modification is correlated with the level of transcription. Previously, we scored a gene as “on” if the RNA signal obtained from the microarray hybridization was above the background signal (Schübeler et al. 2002). This measurement reflects the presence of mRNA but is not a robust indicator for the RNA quantity, because it does not account for sequence-specific differences that can affect the spot intensity during the array hybridization. To account for spot characteristics in signal intensity, we normalized the expression data by dividing it by the average signal obtained from genomic DNA. The resulting value was used as a measure of the amount of transcript present. This procedure leads to a better approximation of the actual level of cytoplasmic RNA, but it should be noted that any measure of steady-state RNA cannot account for differences in transcript half-life.
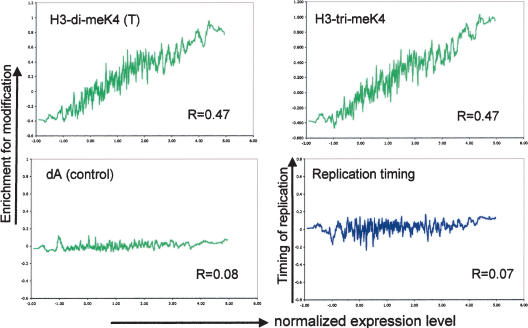
A comparison of the normalized transcript value with the chromatin profiles (Fig. 5; Supplementary Fig. 2) reveals a significant correlation between the level of transcript detected and the degree of euchromatic histone modification (R between 0.44 and 0.51). This correlation is rather uniform for all studied modifications; thus, histone H3 and H4 acetylation and Lys 4 and Lys 79 methylation are equally linked to transcript abundance, which is indicative of transcriptional activity.
Figure 5.
Genome-wide relationship between transcription rate, euchromatic histone modifications, and timing of DNA replication. The expression level of all active genes was compared with the enrichment for each histone modification and with the timing of DNA replication. The moving average (n = 30) of the normalized expression value is plotted against the enrichment for H3-di-meK4, H3-tri-meK4, the control IP, and the timing of DNA replication. In addition, the Pearson correlation coefficient (R) was calculated for each data set (without averaging). For active genes, we find that the degree of each euchromatic histone modification depends on the level of transcript (additional graphs are in the Supplemental Material), whereas such dependency is observed neither for the control experiment nor for the timing of DNA replication. This indicates that although both histone modifications and replication timing correlate with the transcriptional status of a gene, only euchromatic histone modifications correlate with the level of transcription.
Di- and trimethylation of H3 Lys 4 are present on the same genes
Surprisingly, our analysis of 5375 Drosophila genes does not reveal a difference between H3-di-meK4 and H3-trimeK4 and their relation to transcriptional activity. This observation contrasts with studies in budding yeast, which suggested that Lys 4 dimethylation marks active and activatable genes, whereas Lys 4 trimethylation is an exclusive mark for highly expressed genes (Santos-Rosa et al. 2002; Ng et al. 2003b). However, a recent analysis in chicken erythrocytes did not reveal such difference in the presence of di- and trimethylation of this residue; instead, di- and trimethylation seemed to peak at the same sequences (Schneider et al. 2004). Together with our observed genome-wide pattern in Drosophila, this might indicate a difference in the regulation of K4 methylation between budding yeast and multicellular organisms.
Genome-wide distribution of H3-di-meK79 methylation
Methylation of Lys 79 of histone H3 was first described in Saccharomyces cerevisiae as a histone modification that occurs at the majority of nucleosomes and that can affect gene silencing even though it is absent from silenced loci (Ng et al. 2002; van Leeuwen et al. 2002). Furthermore, studies in mammalian cell lines showed that H3-di-meK79 is hypermethylated in euchromatin (Ng et al. 2003a). Here we find that the presence of this modification in the Drosophila euchromatic compartment is restricted to actively transcribed genes. H3-K79 methylation is less abundant in flies than in budding yeast (McKittrick et al. 2004). This disparity could reflect the difference in gene density between these organisms. In S. cerevisiae, two-thirds of the genome consists of annotated genes (Harrison et al. 2002), and intergenic regions are short in length. As 70%-80% of all genes are active in this organism under standard growth conditions (E. Oakeley, pers. comm.), the majority of nucleosomes reside in, or are in close proximity to, actively transcribed regions. Consequently, the high abundance of H3-di-meK79 methylation in yeast is consistent with the hypothesis that this modification is preferentially associated with transcriptionally active genes, as our data suggest for Drosophila.
Timing of DNA replication is not an indicator of the level of transcription
We have shown previously that genes that replicate early in S-phase are more likely to be active than late replicating genes (Schübeler et al. 2002). However, in contrast to the histone modifications, we find that the level of transcript from active genes is independent of their replication timing (Fig. 5). In other words, although more early replicating genes are active compared with late replicating genes, the expression level of late replicating active genes is similar to that of early replicating active genes. Thus, in combination, these results indicate that the studied euchromatic histone modifications correlate not only with the transcriptional status (“on” or “off”), but also with the level of transcription, whereas replication timing is an indicator only of transcriptional status.
H3 Lys 4 dimethylation is restricted to transcribed regions
The correlation between euchromatic histone modifications and transcriptional status and level argues for a process of chromatin modification that is intrinsically transcription coupled. As such, these observations are compatible with a model in which histone-modifying enzymes interact directly with the polymerase complex. A prediction of such a scenario is that these modifications should largely be restricted to the transcribed region. Although the use of a microarray that consists of cDNAs permits a survey of the histone modifications of a large number of genes, the absence of intergenic sequence probes limits conclusions regarding the chromosomal extent of the analyzed modification. Thus, we used a novel Drosophila genomic DNA array consisting of all nonrepetitive sequences from chromosome 2L (D.M. MacAlpine and S.P. Bell, in prep.) to map the local extent of histone H3 Lys 4 dimethylation for a large contiguous region. To ensure comparability, we hybridized the same ChIP samples to the chromosomal array that were used for the cDNA hybridizations.
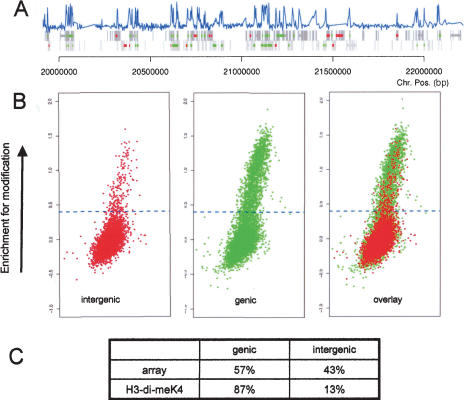
A comparison of chromosomal fragments that are enriched for Lys 4 methylation of histone H3 reveals that ∼87% of them are in genic regions (Fig. 6A,B). Only 13% of enriched sequences are intergenic, even though they represent half of the spotted sequences (Fig. 6). Given our observed link between Lys 4 methylation and transcription, it will be interesting to determine if enriched intergenic sequences reside in genes that have not been annotated (Hild et al. 2003) or if they reflect intergenic transcription, which has been described for a number of multigene loci (Gribnau et al. 2000; Bae et al. 2002; Rank et al. 2002). Clearly, however, Lys 4 dimethylation is largely absent from intergenic regions and mostly limited to genic regions. A similar restriction to transcribed regions has been observed in S. cerevisiae for H3-dimeK4 (Bernstein et al. 2002). Furthermore, the only S. cerevisiae enzyme that catalyzes H3-K4 methylation (Set1) has been shown to interact directly with the elongating polymerase (Ng et al. 2003b). Our results predict a similar mechanism for H3-K4 methylation in higher eukaryotes.
Figure 6.
Chromosome-wide distribution of H3-K4 methylation in Drosophila. Sequences enriched for H3-di-meK4 were hybridized together with an input control to a DNA microarray representing all nonrepetitive sequences of the Drosophila chromosome 2L, and the ratio of bound-over input material was calculated as a measure of enrichment. (A) The enrichment for H3-dimeK4 of a 2-Mb region is shown. The blue bar represents the enrichment for Lys 4 methylation of histone H3 for each sequence that is present on the microarray. The gray boxes indicate genic regions, with each strand shown separately. For a subset of these genic regions, the transcriptional status has been determined on the cDNA array. These genes are labeled green if active and red if inactive. In the majority of cases, the active genes coincide with a peak in H3-di-meK4 methylation that is largely restricted to the transcribed sequence. (B) Shown are three plots (M/A, see Fig. 1) of fluorescent signal versus enrichment for H3-di-meK4 for the complete chromosomal arm. The enrichment for H3-dimeK4 (_Y_-axis) is plotted against the signal intensity (_X_-axis). Sequences that are enriched in the modification are above the dotted line. The left plot shows the enrichments for intergenic regions, of which only a few are enriched. The plot in the middle shows genic regions (independent of their actual transcriptional activity). The comparison of both plots reveals that almost all H3-di-meK4-positive sequences are in genic regions. The right plot shows all spots present on the array. The ratio of genic to intergenic sequences is 1:1 for the complete microarray and 7:1 for the H3-di-meK4 experiment, revealing the restriction of this modification to transcribed regions.
Discussion
Cotranscriptional chromatin modification?
Using a genome-wide analysis of chromatin structure, we report a strong interplay between transcription and a set of euchromatic histone modifications. Our principal findings include the following: (1) There is a binary pattern of histone modifications for euchromatic genes, with active genes consistently marked by all of the euchromatic histone modifications analyzed and the absence of any of these modifications on nontranscribed genes; (2) the level of transcript abundance is positively correlated with the degree of euchromatic histone modifications; and (3) the chromosomal extent of the modification coincides with, and is limited to, the transcribed region. Our surprising observation of an “all-or-none” pattern of histone modification for euchromatic genes suggests a concerted mechanism for the placing of these marks. For example, the euchromatic modifications could be restricted to nucleosomes containing a certain histone H3 variant. The replication-independent deposition of the H3 variant 3.3 (Ahmad and Henikoff 2002a,b) raises the possibility that in Metazoa the majority of euchromatic histone H3 modifications may occur on H3.3. Indeed, histone H3.3 has recently been reported to be enriched in acetylated lysines and in methylated Lys 4 and Lys 79 (McKittrick et al. 2004).
Although it is currently unclear whether these euchromatic modifications can be set prior to nucleosome assembly and deposition, there is ample evidence for post-deposition modification of histones. For example, a link between the elongating polymerase complex and several histone-modifying enzymes (Hampsey and Reinberg 2003), including Set1 (an H3-K4 methylase; Ng et al. 2003b), Set2 (an H3-K36 methylase; Krogan et al. 2003b; Xiao et al. 2003), and Sas3 (a HAT; John et al. 2000), has been demonstrated in S. cerevisiae. Furthermore, genetic evidence from S. cerevisiae suggests that Dot1, the H3-K79 methylase, may also be recruited to chromatin by the elongating polymerase complex (Krogan et al. 2003a). These findings in budding yeast indicate a coupling of histone modifications and transcription. Our genome-wide analysis in Drosophila cells strongly supports these findings and further argues that such interactions may be an integral component of transcriptional elongation in metazoans.
More than 25 years ago, it was observed that chromatin of active genes is more sensitive to DNaseI digestion than that of inactive genes (Weintraub and Groudine 1976). Although, to date, the nature of this sensitivity has been elusive, we propose that it reflects the presence of euchromatic tail modifications. Why does such a “switch” between two chromatin configurations involve a large set of histone modifications? Each modification may participate in creating a chromatin structure that facilitates transcription, either by changing nucleosomal interactions or by serving as a binding substrate for other proteins. The use of multiple modifications would make such system more robust. Regardless, our results reveal a tight coupling between transcription and euchromatic histone modifications. On recruitment, these modifications may serve to facilitate polymerase elongation and reinitiation and to propagate the transcriptional state through cell division.
Materials and methods
Tissue culture and chromatin cross-linking
Drosophila melanogaster Kc cells were cultured as described (van Steensel et al. 2001). Cells (5 × 108) were cross-linked in insect media with formaldehyde as described (Schübeler et al. 2000a,b) with minor modifications. Sonication was performed five times for 30 sec in 40 mM Tris-HCl (pH 8.0), 1% Triton X-100, 4 mM EDTA, 300 mM NaCl.
Antibodies and ChIP
After centrifugation, the supernatant was used for the immunoprecipitation, which was performed as described (Schübeler et al. 2000a) except that immunoprecipitation of H3-phos-S10 was performed in the presence of phosphatase inhibitors (50 mM NaF and 0.2 mM Na3VO4). Part of the supernatant was set aside and served as the input fraction. Polyclonal antibodies against acetylated H3 and H4 and for H3-phos-S10 were purchased from Upstate Biotechnology. Two antisera against H3-di-meK4 were used. One was purchased from Upstate Biotechnology [H3-di-meK4(U)] and the second [H3-di-meK4(T)] was raised by immunizing rabbits with the peptide ARTme2KQTARKSC, where m2K is dimethyl lysine, using the procedures described previously (White et al. 1999). Both antibodies yielded similar enrichments. The antiserum against H3-tri-meK4 was raised by immunization with a peptide that was the same except for trimethyl lysine at position 4. Specificity of the antibodies for dior trimethylated Lys 4 was verified by peptide competition assay (L.P. O'Neill and B.M. Turner, unpubl.). The antiserum against H3-di-meK79 was raised against the peptide IAQDFme2KTDLRF (F. van Leeuwen and D.E. Gottschling, unpubl.).
PCR amplification and fluorescent labeling
DNA from the antibody-bound and input fraction was isolated after reversal of the cross-link and amplified as described (Schübeler et al. 2002) using a primer that was labeled with either Cy3 or Cy5 (Qiagen). Size distribution and fluorescence of the amplified product were confirmed by agarose gel electrophoresis followed by fluorescent scanning (Molecular Dynamics). We carried out four PCR reactions for each input and bound sample and pooled them before the hybridization. We did three independent repeats starting with different passages of Kc cells and performed one hybridization for each repeat. To account for potential influences of the fluorescent dyes, we reversed the dye combination for the input and bound sample in one of the three hybridizations.
Control PCR
Primers were designed to amplify products of 80-120 bp to control for enrichments for histone modifications as detected by microarray hybridization. Five-nanogram template (genomic DNA or DNA-enriched for a histone modification) was used in each reaction using standard conditions and 27 rounds of amplification. PCR products were separated by gel electrophoresis and visualized by ethidium bromide staining of the gel. Detailed conditions and primer sequences are available on request.
Array analysis
The fluorescent scans were analyzed essentially as described using the GenePix software package (Axon) combined with improved background correction (Kooperberg et al. 2002). The ratio of the two fluorescent dyes was log2 transformed and normalized using intensity-dependent normalization (Yang et al. 2001). We then used the average value from the three independent repeats for further analysis. The high reproducibility of these experiments is indicated by a low mean covariance of 13%. The resulting data set is available on our Web site (http://www.fmi.ch/members/dirk.schubeler/supplemental.htm).
The chromosomal arrays were analyzed with Spot software (CSIRO) using the sma package for R (Dudoit et al. 2002b).
Expression normalization
In order to normalize our previously published expression analysis (Schübeler et al. 2002) for spot intensity, we divided the normalized RNA fluorescence by the normalized genomic signal of the control experiment (dA).
Microarray preparation
Two different spotted microarrays were used in this study: a previously published cDNA array containing 5543 expressed sequence tags from D. melanogaster (Schübeler et al. 2002) and a newly developed chromosomal array representing all nonrepetitive sequences from the Drosophila chromosome 2L. The chromosomal array contains 11,816 unique 1.5-kb PCR products that tile the entire sequenced region of the left arm of chromosome 2 (D.M. MacAlpine and S.P. Bell, in prep.). Sample preparation and hybridization were essentially as described (van Steensel et al. 2001).
Acknowledgments
We thank Matthew Lorincz, Fang-Lin Sun, and Muhammad Tariq for critical reading of the manuscript; Bas van Steensel, Steve Henikoff, Ed Oakeley, and members of the Groudine and Schübeler labs for suggestions; and the staff of the Genomics Resource unit at the FHCRC for microarray printing and processing. M.G. is supported by NIH grants DK44746 and HL57620, D.E.G. by GM43893, and C.K. by CA 74841. D.S. and C.W. are supported by the Novartis Research Foundation. Work by S.P.B. and D.M.M. is funded by the Howard Hughes Medical Institute. D.M.M. is a fellow of the Damon Runyon Cancer Research Foundation. Grant support for B.M.T. is from BBSRC and Cancer Research UK. L.P.O'N. is a Royal Society Research Fellow. F.v.L. is a Special Fellow of The Leukemia and Lymphoma Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1198204.
Supplemental material is available at http://www.genesdev.org.
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287**:** 2185-2195. [DOI] [PubMed] [Google Scholar]
- Ahmad, K. and Henikoff, S. 2002a. Epigenetic consequences of nucleosome dynamics. Cell 111**:** 281-284. [DOI] [PubMed] [Google Scholar]
- ____. 2002b. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9**:** 1191-1200. [DOI] [PubMed] [Google Scholar]
- Bae, E., Calhoun, V.C., Levine, M., Lewis, E.B., and Drewell, R.A.. 2002. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 99**:** 16847-16852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister, A.J., Schneider, R., and Kouzarides, T. 2002. Histone methylation: Dynamic or static? Cell 109**:** 801-806. [DOI] [PubMed] [Google Scholar]
- Bernstein, B.E., Humphrey, E.L., Erlich, R.L., Schneider, R., Bouman, P., Liu, J.S., Kouzarides, T., and Schreiber, S.L. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. 99**:** 8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudoit, S., Yang, Y.H., Callow, M.J., and Speed, T.P. 2002a. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statistica Sinica 12**:** 111-139. [Google Scholar]
- Dudoit, S., Yang, Y.H., and Bolstad, B. 2002b. Using R for the analysis of DNA microarray data. R News 2**:** 24-32. [Google Scholar]
- Fang, J., Feng, Q., Ketel, C.S., Wang, H., Cao, R., Xia, L., Erdjument-Bromage, H., Tempst, P., Simon, J.A., and Zhang, Y. 2002. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 12**:** 1086-1099. [DOI] [PubMed] [Google Scholar]
- Felsenfeld, G. and Groudine, M. 2003. Controlling the double helix. Nature 421**:** 448-453. [DOI] [PubMed] [Google Scholar]
- Gribnau, J., Diderich, K., Pruzina, S., Calzolari, R., and Fraser, P. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell 5**:** 377-386. [DOI] [PubMed] [Google Scholar]
- Hampsey, M. and Reinberg, D. 2003. Tails of intrigue: Phosphorylation of RNA polymerase II mediates histone methylation. Cell 113**:** 429-432. [DOI] [PubMed] [Google Scholar]
- Harrison, P.M., Kumar, A., Lang, N., Snyder, M., and Gerstein, M. 2002. A question of size: The eukaryotic proteome and the problems in defining it. Nucleic Acids Res. 30**:** 1083-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz, E. 1928. Das Heterochromatin der Moose. Jahrb Wiss Botanik 69**:** 762-818. [Google Scholar]
- Hild, M., Beckmann, B., Haas, S.A., Koch, B., Solovyev, V., Bu-sold, C., Fellenberg, K., Boutros, M., Vingron, M., Sauer, F., et al. 2003. An integrated gene annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome. Genome Biol. 5**:** R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, J.Y., Sun, Z.W., Li, X., Reuben, M., Tatchell, K., Bishop, D.K., Grushcow, J.M., Brame, C.J., Caldwell, J.A., Hunt, D.F., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102**:** 279-291. [DOI] [PubMed] [Google Scholar]
- John, S., Howe, L., Tafrov, S.T, Grant, P.A., Sternglanz, R., and Workman, J.L. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes & Dev. 14**:** 1196-1208. [PMC free article] [PubMed] [Google Scholar]
- Johnson, C.A., White, D.A., Lavender, J.S., O'Neill, L.P., and Turner, B.M. 2002. Human class I histone deacetylase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. J. Biol. Chem. 277**:** 9590-9597. [DOI] [PubMed] [Google Scholar]
- Kooperberg, C., Fazzio, T.G., Delrow, J., and Tsukiyama, T. 2002. Improved background correction for spotted DNA microarrays. J. Comput. Biol. 9**:** 55-66. [DOI] [PubMed] [Google Scholar]
- Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12**:** 198-209. [DOI] [PubMed] [Google Scholar]
- Krogan, N.J., Dover, J., Wood, A., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Ryan, O.W., Golshani, A., Johnston, M., et al. 2003a. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol. Cell 11**:** 721-729. [DOI] [PubMed] [Google Scholar]
- Krogan, N.J., Kim, M., Tong, A., Golshani, A., Cagney, G., Canadien, V., Richards, D.P., Beattie, B.K., Emili, A., Boone, C., et al. 2003b. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23**:** 4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, M.H. and Allis, C.D. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20**:** 615-626. [DOI] [PubMed] [Google Scholar]
- Labrador, M. and Corces, V.C. 2003. Phosphorylation of histone H3 during transcriptional activation depends on promoter structure. Genes & Dev. 17**:** 43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick, E., Gafken, P.R., Ahmad, K., and Henikoff, S. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. 101**:** 1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H.H., Feng, Q., Wang, H., Erdjument-Bromage, H., Tempst, P., Zhang, Y., and Struhl, K. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes & Dev. 16**:** 1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H.H., Ciccone, D.N., Morshead, K.B., Oettinger, M.A., and Struhl, K. 2003a. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. 100**:** 1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, H.H., Robert, F., Young, R.A., and Struhl, K. 2003b. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11**:** 709-719. [DOI] [PubMed] [Google Scholar]
- Nishioka, K., Rice, J.C., Sarma, K., Erdjument-Bromage, H., Werner, J., Wang, Y., Chuikov, S., Valenzuela, P., Tempst, P., Steward, R., et al. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 9**:** 1201-1213. [DOI] [PubMed] [Google Scholar]
- Rank, G., Prestel, M., and Paro, R. 2002. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 22**:** 8026-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr, D., Suka, Y., Xenarios, I., Kurdistani, S.K., Wang, A., Suka, N., and Grunstein, M. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109**:** 437-446. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa, H., Schneider, R., Bannister, A.J., Sherriff, J., Bernstein, B.E., Emre, N.C., Schreiber, S.L., Mellor, J., and Kouzarides, T. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419**:** 407-411. [DOI] [PubMed] [Google Scholar]
- Schneider, R., Bannister, A.J., Myers, F.A., Thorne, A.W., Crane-Robinson, C., and Kouzarides, T. 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6**:** 73-77. [DOI] [PubMed] [Google Scholar]
- Schübeler, D., Francastel, C., Cimbora, D.M., Reik, A., Martin, D.I., and Groudine, M. 2000a. Nuclear localization and histone acetylation: A pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes & Dev. 14**:** 940-950. [PMC free article] [PubMed] [Google Scholar]
- Schübeler, D., Lorincz, M.C., Cimbora, D.M., Telling, A., Feng, Y.Q., Bouhassira, E.E., and Groudine, M. 2000b. Genomic targeting of methylated DNA: Influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol. Cell. Biol. 20**:** 9103-9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler, D., Scalzo, D., Kooperberg, C.K., Van Steensel, B., Delrow, J., and Groudine, M. 2002. Genome-wide DNA replication profile for Drosophila melanogaster: A link between transcription and replication timing. Nat. Genet. 32**:** 438-442. [DOI] [PubMed] [Google Scholar]
- Strahl, B.D. and Allis, C.D. 2000. The language of covalent histone modifications. Nature 403**:** 41-45. [DOI] [PubMed] [Google Scholar]
- Turner, B.M. 2002. Cellular memory and the histone code. Cell 111**:** 285-291. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, F., Gafken, P.R., and Gottschling, D.E. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109**:** 745-756. [DOI] [PubMed] [Google Scholar]
- van Steensel, B., Delrow, J., and Henikoff, S. 2001. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 27**:** 304-308. [DOI] [PubMed] [Google Scholar]
- Waterborg, J.H. 1993. Dynamic methylation of alfalfa histone H3. J. Biol. Chem. 268**:** 4918-4921. [PubMed] [Google Scholar]
- Wei, Y., Yu, L., Bowen, J., Gorovsky, M.A., and Allis, C.D. 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97**:** 99-109. [DOI] [PubMed] [Google Scholar]
- Weintraub, H. and Groudine, M. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193**:** 848-856. [DOI] [PubMed] [Google Scholar]
- White, D.A., Belyaev, N.D., and Turner, B.M. 1999. Preparation of site-specific antibodies to acetylated histones. Methods 19**:** 417-424. [DOI] [PubMed] [Google Scholar]
- Xiao, T., Hall, H., Kizer, K.O., Shibata, Y., Hall, M.C., Borchers, C.H., and Strahl, B.D. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes & Dev. 17**:** 654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.H., Dudoit, S., Luu, P., and Speed, T.P. 2001. Normalization for cDNA microarrays data. In Microarrays: Optical technologies and informatics (eds. M.L. Bittner et al.), pp. 141-152. International Society for Optical Engineering, San Jose, CA.
- Zhang, Y. and Reinberg, D. 2001. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 15**:** 2343-2360. [DOI] [PubMed] [Google Scholar]