eIF4AIII enhances translation of nuclear cap-binding complex–bound mRNAs by promoting disruption of secondary structures in 5′UTR (original) (raw)
Significance
Two major cap-binding components can drive mammalian translation initiation: the nuclear cap-binding complex (CBC) and eukaryotic translation initiation factor 4E (eIF4E). Although eIF4E-dependent translation has been well characterized, the mechanism of CBC-dependent translation remains unclear. Here, we demonstrate the previously unappreciated role of eIF4AIII in CBC-dependent translation. eIF4AIII is traditionally considered a component of the exon junction complex loaded onto mRNAs after splicing. In addition, we found that eIF4AIII can be recruited to the 5′-end of CBC-associated mRNA and promotes the translation of CBC-associated mRNA by helping to unwind secondary structures in 5′UTR. Therefore, our data provide evidence that eIF4AIII is a translation initiation factor specifically required for translation of CBC-associated mRNAs.
Keywords: eIF4AIII, cap-binding complex, translation, CTIF, nonsense-mediated mRNA decay
Abstract
It has long been considered that intron-containing (spliced) mRNAs are translationally more active than intronless mRNAs (identical mRNA not produced by splicing). The splicing-dependent translational enhancement is mediated, in part, by the exon junction complex (EJC). Nonetheless, the molecular mechanism by which each EJC component contributes to the translational enhancement remains unclear. Here, we demonstrate the previously unappreciated role of eukaryotic translation initiation factor 4AIII (eIF4AIII), a component of EJC, in the translation of mRNAs bound by the nuclear cap-binding complex (CBC), a heterodimer of cap-binding protein 80 (CBP80) and CBP20. eIF4AIII is recruited to the 5′-end of mRNAs bound by the CBC by direct interaction with the CBC-dependent translation initiation factor (CTIF); this recruitment of eIF4AIII is independent of the presence of introns (deposited EJCs after splicing). Polysome fractionation, tethering experiments, and in vitro reconstitution experiments using recombinant proteins show that eIF4AIII promotes efficient unwinding of secondary structures in 5′UTR, and consequently enhances CBC-dependent translation in vivo and in vitro. Therefore, our data provide evidence that eIF4AIII is a specific translation initiation factor for CBC-dependent translation.
In mammalian cells, translation initiation is driven by two major cap-binding proteins (CBPs). One is the nuclear cap-binding complex (CBC), which is a heterodimer of nuclear CBP80 and CBP20 (CBP80/20) (1), and the other is the eukaryotic translation initiation factor (eIF) 4E, which is a major cytoplasmic CBP (2). The CBC binds the 5′-cap structure of a newly synthesized mRNA in the nucleus and then directly interacts with an eIF4G-like scaffold protein called CBP80/20-dependent translation initiation factor (CTIF) (3). Because CTIF is mostly present on the cytoplasmic side of the nuclear envelope and is found in the nucleus (3), the interaction between the CBC and CTIF may occur either in the nucleus or during mRNA export through the nuclear pore complex. In the cytoplasm, at the 5′-end of mRNA, the CBC–CTIF complex recruits the eIF3 complex (4) and then the 40S small subunit of the ribosome, thereby directing the so-called “first” (or pioneer) round of translation (1).
Translation of a CBC-bound mRNA precedes that of the eIF4E-bound mRNA, because the CBC-bound mRNA is a precursor form of the eIF4E-bound mRNA (5). The timing of the replacement of the CBC by eIF4E has not yet been elucidated; however, it is known that the replacement is mediated by the action of importin α/β in a translation-independent manner (6). The replaced eIF4E at the 5′-end of mRNA recruits a scaffold protein, eIF4GI or eIF4GII, which recruits eIF3 and eIF4AI/II and, eventually, the 40S ribosomal subunit. Then, the recruited 40S subunit scans the mRNA until it reaches the authentic translation initiation codon AUG. Efficient ribosome scanning requires either eIF4AI or eIF4AII helicase and the accessory factors eIF4B and eIF4H to disrupt possible RNA secondary structures in the 5′UTR of mRNA, as reviewed previously (7, 8). On the AUG codon, the large ribosomal subunit 60S joins the 40S subunit and forms the 80S ribosome complex. In general, translation of eIF4E-bound mRNA is responsible for the high expression of polypeptides in the cytoplasm.
It has long been thought that nonsense-mediated mRNA decay (NMD), which drives selective destruction of aberrant or physiological mRNAs containing a premature termination codon (9–11), occurs during the first round of translation driven by the CBC (1). The above view, however, was recently challenged by two reports showing that NMD occurs in the cytoplasm, regardless of whether the CBC or eIF4E drives the first round of translation (12, 13). In addition, several recent studies revealed that translation of CBC-bound mRNAs may occur multiple times (3, 14–16) and that, more intriguingly, the translation of replication-dependent histone mRNAs mainly occurs on CBC-bound mRNA (15). Therefore, for simplicity and clarity, we will hereafter refer to the two types of translation as CBC-dependent translation (CT), which takes place on CBC-bound mRNA, and eIF4E-dependent translation (ET), which occurs on eIF4E-bound mRNA, irrespective of the number of rounds of translation.
The critical difference between CBC-bound and eIF4E-bound mRNAs is that the former is imprinted by the exon junction complex (EJC), which is deposited ∼20–24 nt upstream of an exon–exon junction as a consequence of splicing (17). The EJC contains a heterotetrameric protein core that includes eIF4AIII, Magoh, Y14, metastatic lymph node 51 (MLN51; also known as Barentsz), and peripheral proteins. The EJC plays various roles in posttranscriptional regulation, including mRNA export from the nucleus, subcellular mRNA localization, translation, and NMD, as reported previously (18, 19).
EJCs are largely found in CBC-bound mRNA but not in eIF4E-bound mRNA (5), and the deposited EJCs are displaced by a translocating ribosome (20). Accordingly, it is believed that the EJC promotes efficiency of CT rather than ET. In line with this view, translational efficiency of intron-containing mRNA (spliced mRNA) is generally higher than that of intronless mRNA (identical mRNA not produced by splicing) (21–23). Such a difference is attributable, in part, to the EJC (deposited as a consequence of splicing) rather than to splicing per se. Furthermore, insulin treatment increases the enhanced translation of intron-containing mRNA compared with intronless mRNA through recruitment of activated ribosomal protein S6 kinase 1 (S6K1) to newly spliced mRNAs via the EJC component S6K1 Aly/REF-like substrate (SKAR) (24). Despite these advances in understanding of the role of EJCs in CT, molecular mechanisms of the possible communication between the CT complex and the EJC are poorly understood.
Here, we demonstrate that the core EJC component, eIF4AIII, is a constituent of the CT complex. eIF4AIII is preferentially recruited to the 5′-end of CBC-bound mRNAs via its direct interaction with CTIF. Interestingly, the recruitment of eIF4AIII occurs irrespective of the presence of introns. We also found that eIF4AIII promotes CT of a reporter mRNA by helping to unwind a stem-loop (SL) structure in the 5′UTR. Our data provide evidence of a previously unidentified role of eIF4AIII as a CT-specific translation initiation factor.
Results
NMD of mRNA Containing an SL in the 5′UTR Depends on eIF4AIII but Not on eIF4AI/II.
It is well known that eIF4AI and eIF4AII helicases are required for unwinding of secondary structures in the 5′UTR during ribosome scanning in ET (2, 7, 8). It is unknown whether CT requires the same factors for efficient ribosome scanning, and even whether CT depends on ribosome scanning. Therefore, our aim was to determine the possible role of the eIF4AI and eIF4AII helicases in CT. To this end, we used the NMD system, because it is known that NMD is mainly associated with the first round of translation (1, 12, 13). Because newly synthesized mRNAs are exported from the nucleus to the cytoplasm with the CBC bound to the 5′-end, the first round of translation, for the most part, is expected to be driven by the CBC rather than by eIF4E.
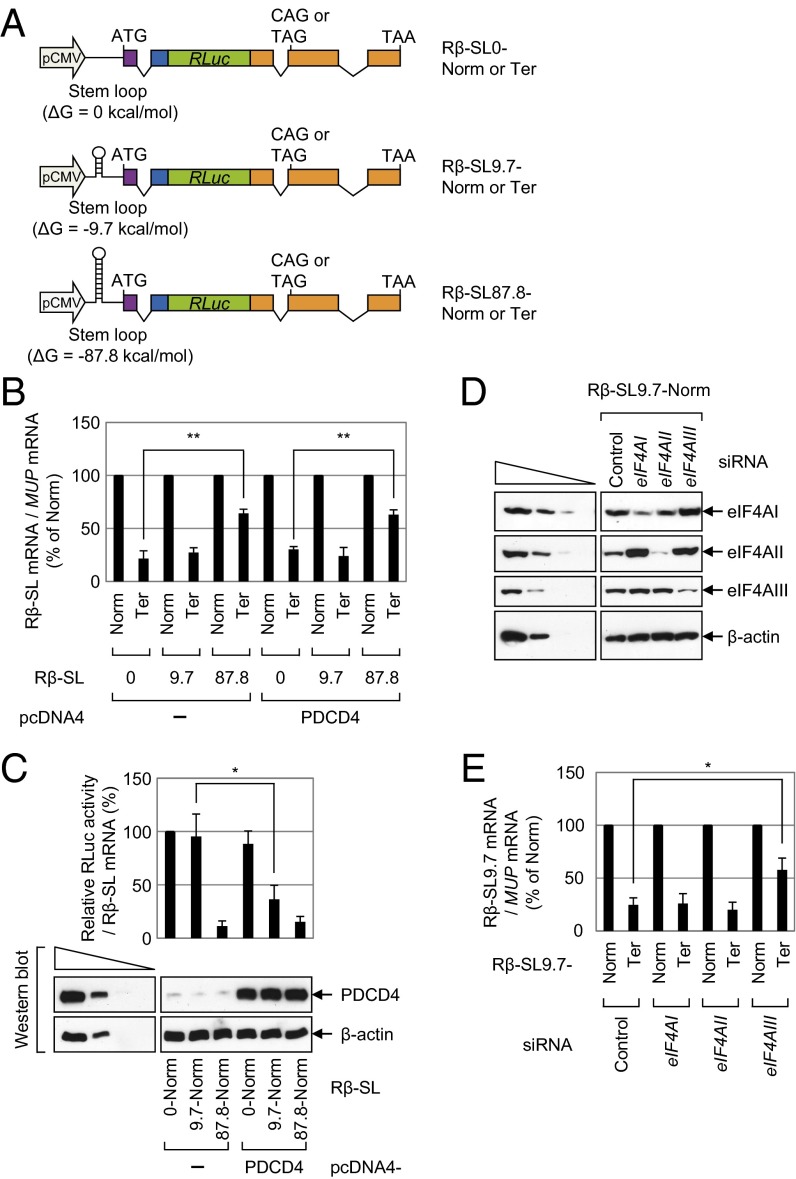
We constructed NMD reporter plasmids expressing intronless Renilla luciferase (RLuc) Rβ-SL0, Rβ-SL9.7, and Rβ-SL87.8 mRNAs, all of which contain RLuc cDNA, β-globin genomic DNA containing either a normal (Norm) or termination (Ter) codon at the 39th codon, and the SL structures with different stability (ΔG = 0, −9.7, and −87.8 kcal/mol, respectively) 80 nt downstream of the 5′-end and 21 nt upstream of the translation initiation codon (Fig. 1_A_). Other studies showed that the insertion of a very stable SL structure (ΔG = −75 kcal/mol) in the middle of the 5′UTR causes ribosome stalling and drastic inhibition of translation (25, 26). In contrast, translation is not significantly affected when a moderately stable SL structure (ΔG = −30 kcal/mol) is introduced into the 5′UTR, because eIF4AI/II can efficiently unwind this SL structure.
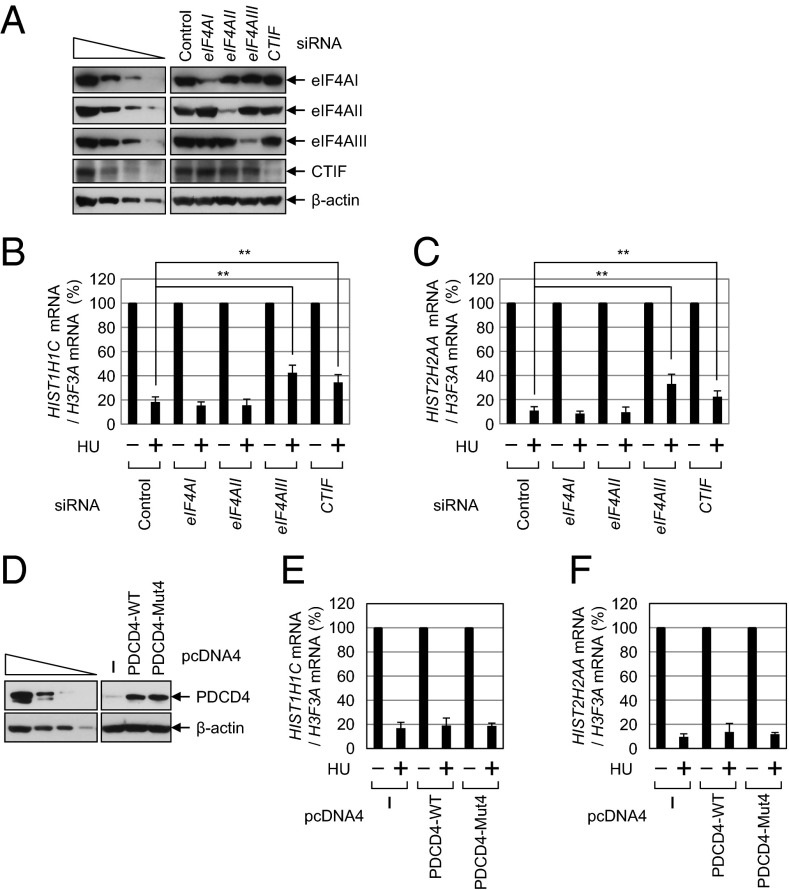
Fig. 1.
Translation preceding NMD requires helicase activity to unwind secondary structures in the 5′UTR in an eIF4AI/II-independent manner. (A) Schematic representation of NMD reporter plasmids, all of which contain (in the following order) triose phosphate isomerase (TPI) exon 6 (purple), TPI exon 7 (blue), RLuc cDNA (green), and β-globin genomic DNA (orange) containing either the normal codon CAG (Norm) or the premature termination codon TAG (Ter) in the 39th codon of the β-globin gene. An SL structure of various stability (ΔG = 0, −9.7, or −87.8 kcal/mol as calculated by the Mfold program) was inserted in the middle of the 5′UTR in each reporter. (B and C) HeLa cells were cotransfected with a reporter plasmid, the reference plasmid phCMV-MUP, and either pcDNA4 or pcDNA4-PDCD4. Two days after the transfection, total RNA and protein were isolated and analyzed using a qRT-PCR assay and a luciferase assay, respectively. (B) qRT-PCR assay of reporter mRNAs. The levels of the mRNAs tested were normalized to MUP mRNAs. Normalized levels of Norm mRNAs were arbitrarily set to 100%. (C) Translation efficiency of reporter mRNAs. (Upper) RLuc activities were measured using the extracts obtained from cells transfected with the Norm reporter plasmid. The RLuc activity was normalized to the level of RLuc mRNA. The normalized RLuc activity (translation efficiency) of Rβ-SL0-Norm in the absence of PDCD4 was arbitrarily set to 100%. (Lower) Expression levels of PDCD4 were determined by Western blotting. (D and E) HeLa cells were transfected with the indicated siRNA; 1 d later, they were retransfected with the reporter plasmid Rβ-SL9.7, either Norm or Ter, and the reference plasmid phCMV-MUP. Two days later, total RNA and protein were isolated. (D) Western blot analysis shows specific down-regulation by siRNA. (E) qRT-PCR assay of Rβ-SL9.7 mRNAs. The levels of Rβ-SL9.7 mRNA were normalized to MUP mRNAs. Normalized levels of Rβ-SL9.7-Norm mRNA were arbitrarily set to 100%. All data represent three independently performed transfections, qRT-PCR assays, and luciferase assays. Two-tailed, equal-sample variance Student t tests were used to calculate the P values. *P < 0.05; **P < 0.01.
HeLa cells were transfected with an NMD reporter construct and the reference plasmid phCMV-major urinary protein (MUP), which encodes the MUP. The MUP mRNA was used for adjustment of data for the variations observed during transfection and RNA purification. A quantitative real-time RT-PCR (qRT-PCR) analysis showed that the levels of Rβ-SL0 Ter mRNA and Rβ-SL9.7 Ter mRNA were reduced to 30% of the corresponding Norm mRNAs (Fig. 1_B_). In contrast, the level of Rβ-SL87.8 Ter mRNA was reduced to 60% of the corresponding Norm mRNA level (Fig. 1_B_), suggesting that the presence of a strong SL structure in the 5′UTR inhibits NMD.
We then measured the translation efficiency of the reporter mRNAs by calculating the ratio of RLuc activity to RLuc mRNA (Fig. 1_C_, Upper). As expected, translation of Rβ-SL87.8-Norm mRNA, but not Rβ-SL0-Norm mRNA or Rβ-SL9.7-Norm mRNA, was drastically inhibited (Fig. 1_C_, Upper), suggesting that the presence of a strong SL structure in the 5′UTR inhibits overall translation. Given that NMD is closely linked to translation, all these data, along with the results in Fig. 1_B_, indicate that the translation preceding NMD requires a ribosome scanning process.
We next used programmed cell death protein 4 (PDCD4), which binds to and inhibits the helicase activity of eIF4AI and eIF4AII (27, 28), to determine clearly whether the translation preceding NMD requires eIF4AI and eIF4AII (both of which are helicases known to be involved in the unwinding of secondary structures during ET). The NMD and translation levels of Rβ-SL0 mRNA, which lacked an SL structure in the 5′UTR, and consequently did not require eIF4AI/II helicase activity, were not significantly affected by the overexpression of PDCD4 (Fig. 1 B and C). In addition, the NMD and translation levels of Rβ-SL87.8 mRNA were not significantly affected by the overexpression of PDCD4 (Fig. 1 B and C); this result is logical because eIF4AI/II cannot unwind very strong SL structures (25, 26). In contrast, overexpression of PDCD4 reduced the efficiency of translation of Rβ-SL9.7 mRNA by 2.5-fold (Fig. 1_C_). These results indicate that the unwinding of a moderately stable SL structure in the middle of the 5′UTR of Rβ-SL9.7 mRNA requires eIF4AI/II helicase activity. Intriguingly, the levels of NMD of Rβ-SL9.7 mRNA were not significantly affected by the overexpression of PDCD4 (Fig. 1_B_) despite translational inhibition (Fig. 1_C_). The specific role of PDCD4 in the unwinding of secondary structures via eIF4AI/II was further demonstrated by overexpression of mutant PDCD4, PDCD4-Mut4 (Fig. S1), which contains four amino acid substitutions in two MA3 domains and fails to interact with eIF4AI/II (29). These results showed that both NMD and overall translation levels of Rβ-SL9.7 mRNA were not affected by overexpression of mutant PDCD4 (Fig. S1). All these observations suggest that the translation preceding NMD is largely independent of eIF4AI/II. In line with this notion, another report showed that down-regulation of eIF4AI or eIF4AII fails to block NMD of β-globin mRNA (30). We also found that NMD of Rβ-SL9.7 mRNA was significantly inhibited by down-regulation of eIF4AIII but not by individual or simultaneous down-regulation of eIF4AI and eIF4AII using siRNA (Fig. 1 D and E and Fig. S2). Therefore, all these results suggest that NMD of mRNA containing an SL in the 5′UTR requires eIF4AIII.
eIF4AIII Associates with the CT Complex via Its Direct Interaction with CTIF.
It has long been thought that eIF4AIII is necessary for proper loading or formation of the EJC on a spliced mRNA (18, 19, 31). As expected, disruption of EJCs by the down-regulation of eIF4AIII inhibited NMD of Rβ-SL9.7 mRNA (Fig. 1 D and E and Fig. S2). However, considering that (i) immunoprecipitation (IP) studies in our previous report revealed that eIF4AIII is enriched in the CT complex (3) and (ii) eIF4AIII is an RNA helicase homologous to eIF4AI/II (32), an alternative interpretation of our results is possible: eIF4AIII may help to unwind secondary structures during CT, and consequently increase the efficiency of NMD. Therefore, we first tested whether eIF4AIII associates with the CT complex.
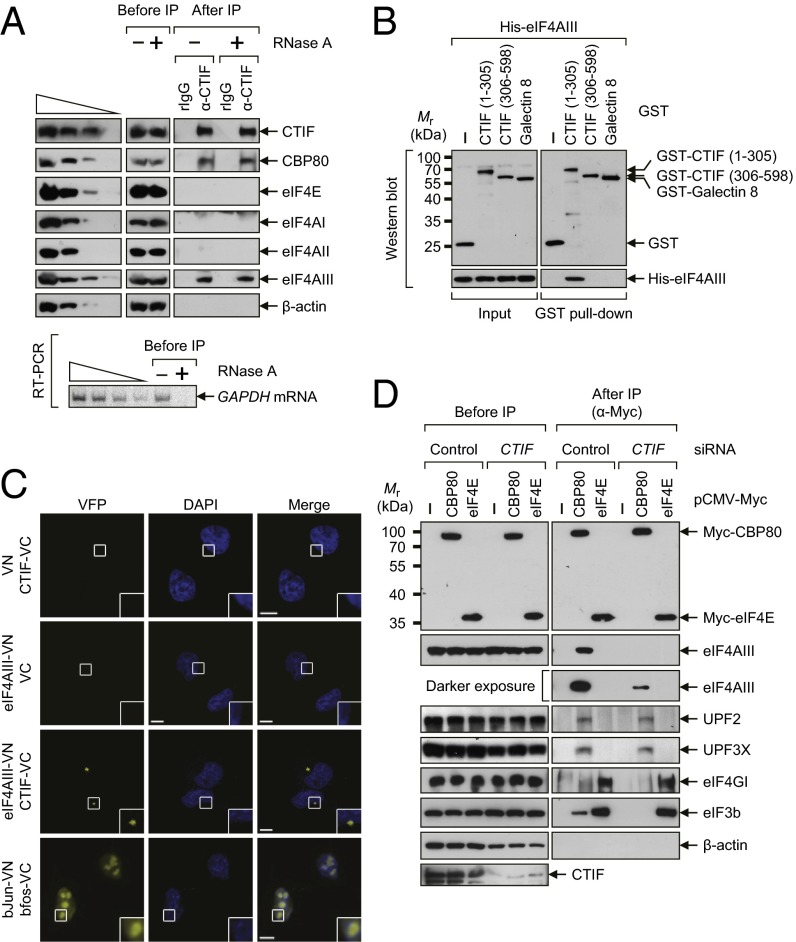
First, the IP results showed that endogenous eIF4AIII and CBP80, but not eIF4AI/II and eIF4E, coimmunopurified with endogenous CTIF in an RNase A-resistant manner (Fig. 2_A_). Second, endogenous eIF4AI/II and eIF4GI were mostly present in the IP of Myc-eIF4E, but endogenous eIF4AIII and CTIF were detected mostly in the IP of Myc-CBP80 in an RNase A-resistant manner (Fig. S3_A_). Third, endogenous CBP80 and CTIF (but not eIF4E, eIF4GI, or β-actin) coimmunopurified with FLAG-eIF4AIII in an RNase A-resistant manner (Fig. S3_B_). Fourth, endogenous CBP80 and eIF4AIII coimmunopurified with FLAG-CTIF WT and FLAG-CTIF (1–305), but not with FLAG-CTIF (306–598), in an RNase A-resistant manner (Fig. S3_C_). Fifth, the GST pull-down assays showed that recombinant His-eIF4AIII copurified with GST-CTIF (1–305) but not GST-CTIF (306–598) (Fig. 2_B_). Sixth, the results of bimolecular fluorescence complementation (BiFC) assays (33), in which fluorescent signals are emitted when two tested proteins interact within a cell, showed that coexpression of the two fusion proteins, an eIF4AIII-fused N-terminal fragment of yellow Venus-enhanced fluorescent protein (eIF4AIII-VN) and a CTIF-fused C-terminal fragment of yellow Venus-enhanced fluorescent protein (CTIF-VC), or vice versa, resulted in the emission of moderate fluorescent signals in the cytoplasm (Fig. 2_C_ and Fig. S3_D_). All these data indicate that eIF4AIII directly interacts with the N-terminal half of CTIF in vivo and in vitro.
Fig. 2.
eIF4AIII interacts with CTIF in vivo and in vitro. (A) IP of endogenous CTIF. Extracts of HEK293T cells were either treated or not treated with RNase A. IPs were performed using either α-CTIF antibody or a nonspecific rabbit IgG (rIgG). (Upper) Cell extracts before IP and immunopurified proteins after IP were analyzed by Western blotting using the indicated antibodies. (Lower) RT-PCR analysis using α-[32P]-dATP and specific oligonucleotides for amplification of cellular GAPDH mRNA demonstrated efficient digestion of cellular transcripts by RNase A. For quantitative analysis, threefold serial dilutions of cell extracts (Upper) and twofold serial dilutions of RNA samples (Lower) obtained before IP were loaded in the four leftmost lanes. (B) In vitro GST pull-down assays. The E. coli extracts expressing GST, GST-CTIF (1–305), GST-CTIF (306–598), or GST-Galectin 8, which served as a negative control, were mixed with purified recombinant His-eIF4AIII. After the GST pull-down, purified proteins were subjected to Western blotting using α-GST antibody (Upper) or α-His antibody (Lower). (C) In vivo BiFC analysis of the interaction between CTIF and eIF4AIII. HeLa cells were transiently transfected with the indicated plasmids, and the cells were fixed and examined under a confocal microscope 6 h later. Nuclei were stained with DAPI. The combination of a bJun–N-terminal fragment of yellow Venus-enhanced fluorescent protein (VN) and bfos–C-terminal fragment of yellow Venus-enhanced fluorescent protein (VC) was used as a positive control. Enlarged images of the boxed areas are shown in the lower right corner of each image. (Scale bars: 10 μm.) (D) IP of Myc-CBP80 and Myc-eIF4E after CTIF down-regulation. HEK293T cells were transiently transfected with either CTIF siRNA or nonspecific control siRNA. Two days later, the cells were retransfected with pCMV-Myc, pCMV-Myc-CBP80, or pCMV-Myc-eIF4E. One day later, the total-cell extracts were subjected to IP using α-Myc antibody. The cellular protein β-actin served as a negative control. All data represent at least two independently performed transfections, IPs, and confocal microscopy observations.
Next, we tested whether the association of eIF4AIII with the CT complex is attributable to the direct interaction of CTIF and eIF4AIII. The results of IPs using the extracts of cells depleted of CTIF revealed that eIF4AIII was enriched in the IP of Myc-CBP80, but not in the IP of Myc-eIF4E, in a CTIF-dependent manner (Fig. 2_D_). In contrast, other EJC components, UPF2 and UPF3X, were enriched in the IP of Myc-CBP80 in a CTIF-independent manner. Consistent with other studies (3, 4, 15), endogenous eIF4GI was largely detected in the IP of Myc-eIF4E, whereas endogenous eIF3 was detected in both IPs. It should be noted that eIF3 is recruited to CT and ET complexes via its interaction with CTIF and eIF4GI/II, respectively (4). Because of direct interaction between eIF4AIII and CTIF (Fig. 2_B_), these findings suggest that eIF4AIII binds to the CT complex via its direct interaction with CTIF.
eIF4AIII Aassociates with Intronless mRNAs as Well as Intron-Containing (Spliced) mRNAs.
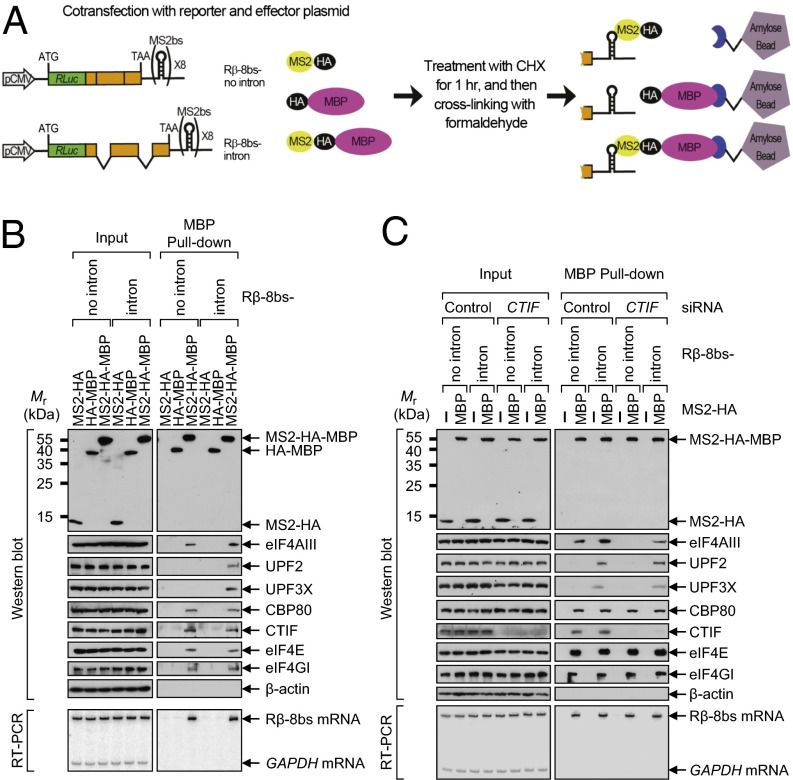
The finding that eIF4AIII directly associates with the CT complex (Fig. 2) led us to hypothesize that eIF4AIII could associate with the 5′-end of mRNA as a CT component, as well as with exon–exon junctions of a spliced mRNA as an EJC component. To test this hypothesis, we used pull-down assays with fusion proteins containing the bacteriophage MS2 coat protein (MS2), which binds to a specific RNA-hairpin structure (MS2-binding site; MS2bs), and maltose-binding protein (MBP). This assay takes advantage of the strong and specific binding of MS2 to MS2bs and allows for recovery of the MS2bs-containing messenger ribonucleoproteins (mRNPs) on amylose beads via the MBP (34). Two reporter constructs, Rβ-8bs-no intron and Rβ-8bs-intron, were generated, which contained (in the following order) RLuc cDNA lacking a translation termination codon, β-globin (either genomic or cDNA sequence) with a translation termination codon (TAA), and MS2bs (Fig. 3_A_).
Fig. 3.
eIF4AIII associates with both intronless mRNAs and intron-containing (spliced) mRNAs. (A) Diagram of the experimental procedure of the MBP pull-down assay. The reporter plasmid contains (in the following order) RLuc cDNA without a translation termination codon, the β-globin gene containing either no intron or two introns, the translation termination codon (TAA), and eight tandem MS2bs repeats (8bs) within the 3′UTR. HEK293T cells were cotransfected with a reporter plasmid (either Rβ-8bs-no intron or Rβ-8bs-intron) and an effector plasmid expressing MS2-HA, HA-MBP, or MS2-HA-MBP. Two days after the transfection, the cells were treated with 100 μg/mL cycloheximide for 1 h and then cross-linked with formaldehyde. Total-cell extracts were prepared and then precipitated with amylose beads. (B) MBP pull-down assay using the reporter mRNAs Rβ-8bs-no intron and Rβ-8bs-intron. (Upper) Proteins purified in the MBP pull-down assay were analyzed by Western blotting using the indicated proteins. (Lower) RT-PCR assays of Rβ-8bs mRNAs and GAPDH mRNAs using specific oligonucleotides and α-[32P]-dATP demonstrated that the reporter mRNAs were selectively purified only in the presence of MS2-HA-MBP. (C) Experiments as described in B, except that the cells were transfected with CTIF siRNA or nonspecific control siRNA and retransfected with a plasmid expressing either MS2-HA or MS2-HA-MBP.
Transiently expressed HA-MBP and MS2-HA-MBP, but not MS2-HA, were enriched by the MBP pull-down (Fig. 3_B_, Upper), and Rβ-8bs mRNAs were present only in the pull-down of MS2-HA-MBP (Fig. 3_B_, Lower). These data demonstrated the efficacy of this system under our experimental conditions. The copurified proteins were analyzed by Western blotting (Fig. 3_B_, Upper). The CT components, CBP80 and CTIF, and the ET components, eIF4E and eIF4GI, copurified with Rβ-8bs-no intron mRNAs and Rβ-8bs-intron mRNAs without a significant difference. In contrast, the EJC components, UPF2 and UPF3X, copurified only with Rβ-8bs-intron mRNAs; this result is consistent with the role of these proteins in EJCs. Intriguingly, eIF4AIII was detected in the Rβ-8bs-no intron mRNP. Furthermore, greater amounts of eIF4AIII were detected in the Rβ-8bs-intron mRNP. These data indicate that eIF4AIII associates with intronless mRNAs as a CT component and with intron-containing (spliced) mRNAs as both an EJC and CT component.
The finding that the interaction between CTIF and eIF4AIII is necessary for recruitment of eIF4AIII to the CT complex (Fig. 2_D_) was further confirmed by repeating the same MBP pull-down assay with extracts of cells depleted of CTIF (Fig. 3_C_). Down-regulation of CTIF to ∼30% of the normal level significantly reduced the levels of copurified eIF4AIII in the Rβ-8bs-no intron mRNP and Rβ-8bs-intron mRNP, without affecting the levels of other copurified proteins tested (Fig. 3_C_). These results demonstrate that eIF4AIII associates with intronless and intron-containing (spliced) mRNAs via its direct interaction with CTIF.
eIF4AIII Selectively Increases CT Efficiency in Vivo.
Given the specific interaction between eIF4AIII and the CT complex and the sequence similarity between eIF4AIII and ET-specific RNA helicases (eIF4AI and eIF4AII), we next wanted to see whether eIF4AIII is selectively involved in unwinding of secondary structures in a 5′UTR during CT. To exclude the effects of eIF4AIII as an EJC component and to test the role of eIF4AIII in CT of mRNA with secondary structures in the 5′UTR, we first constructed a reporter plasmid that expresses intronless RLuc (R) mRNAs with or without a SL structure (R-SL9.7 mRNA and R-SL0 mRNA, respectively) that has an ΔG of −9.7 kcal/mol (SL9.7; Fig. 4_A_). Of note, the introduction of SL9.7 into the 5′UTR of the NMD reporter mRNA rendered the mRNA more vulnerable to translational repression mediated by PDCD4, although NMD of the reporter mRNA was not abrogated (Fig. 1 and Fig. S1).
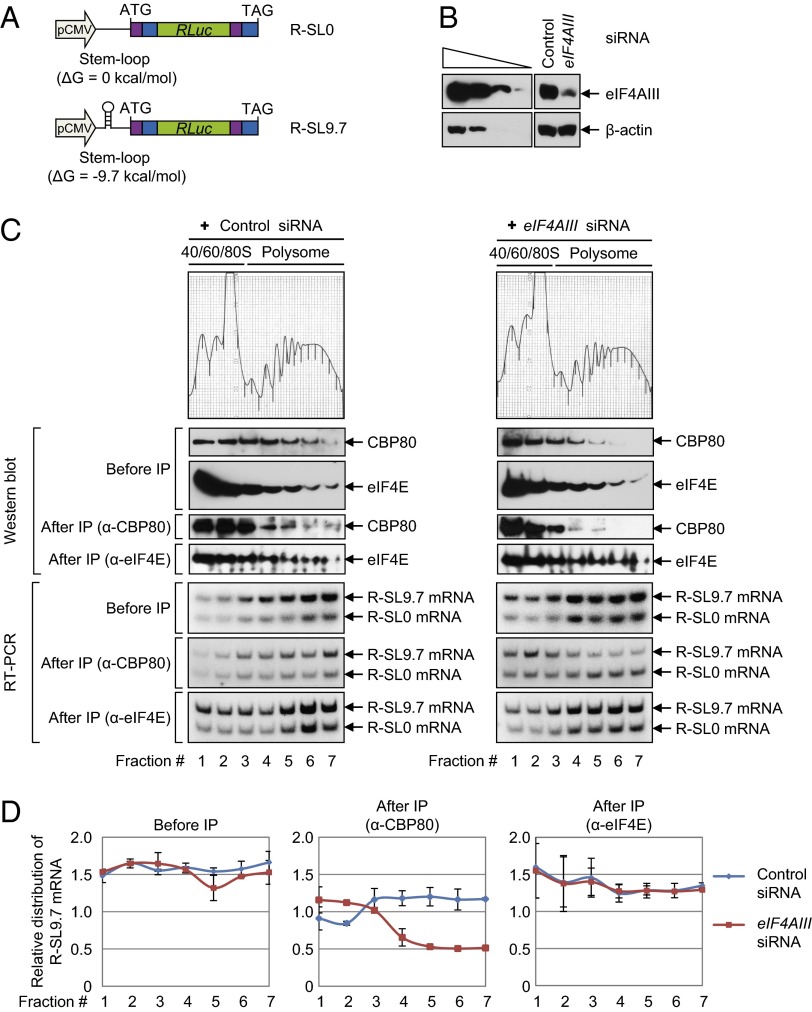
Fig. 4.
eIF4AIII increases CT efficiency in vivo. (A) Schematic representation of reporter plasmids R-SL0 and R-SL9.7, which contain (in the following order) TPI exon 6 (purple), TPI exon 7 (blue), RLuc cDNA (green), TPI exon 6 (purple), and TPI exon 7 (blue). The plasmid R-SL9.7 contains an SL structure in the middle of the 5′UTR. ATG, translation initiation codon; TAG, translation termination codon. (B_–_D) HEK293FT cells were transiently transfected with either eIF4AIII siRNA or nonspecific control siRNA. One day later, the cells were retransfected with reporter plasmids R-SL0 and R-SL9.7. Two days later, the cells were harvested and the cytoplasmic extracts were prepared and subjected to polysome fractionation using the 10–50% (wt/vol) sucrose gradient. Then, the CT complex and ET complex were separated by subjecting each fraction to IP using either the α-CBP80 or α-eIF4E antibody. (B) Western blot analysis demonstrates efficient down-regulation of eIF4AIII. (C) Analyses of polysome-fractionated samples. The polysome-fractionated samples before and after IP were analyzed by Western blotting using the indicated antibodies (Upper) and RT-PCR assays using specific oligonucleotides and α-[32P]-dATP (Lower). (D) Quantitative representation of the relative levels of R-SL9.7 mRNAs. The levels of R-SL9.7 mRNA and R-SL0 mRNA in each fraction (C, Lower) were quantitated. The levels of R-SL9.7 mRNA were normalized to R-SL0 mRNA.
Next, we performed polysome fractionation experiments followed by IPs using extracts of cells that transiently expressed both R-SL0 mRNA and R-SL9.7 mRNA and were either depleted or not depleted of eIF4AIII using siRNA. Western blotting results showed that the siRNA transfection reduced expression of the endogenous eIF4AIII protein to 15% of the normal level (Fig. 4_B_). Upon polysome fractionation, CBP80 and eIF4E were detected in most fractions before IP in undepleted cells (Fig. 4_C_, Left; before IP), consistent with other reports (1, 3). Distribution of eIF4E was not significantly changed by the eIF4AIII down-regulation (Fig. 4_C_, Right; before IP). On the other hand, distribution of CBP80 was marginally shifted from polysome to subpolysome fractions after the eIF4AIII down-regulation (Fig. 4_C_, compare Left and Right; before IP).
Distribution of the reporter mRNAs was analyzed by means of RT-PCR assays with specific oligonucleotides and α-[32P]-dATP. The reporter mRNAs R-SL0 and R-SL9.7 were present in all fractions before IP (Fig. 4_C_, before IP). Quantitative analysis showed that the relative distribution of R-SL9.7 mRNA, which was normalized to R-SL0 mRNA, was not significantly affected by the eIF4AIII down-regulation (Fig. 4_D_, before IP).
To distinguish the CT and ET complexes, IPs were performed using an α-CBP80 antibody or α-eIF4E antibody (Fig. 4_C_, after IP). Immunopurified CBP80 and eIF4E in each fraction were analyzed by means of Western blotting (Fig. 4_C_, after IP). The coimmunopurified mRNAs were analyzed using RT-PCR (Fig. 4_C_, after IP). The normalized levels of R-SL9.7 mRNA in the IP of CBP80, but not in the IP of eIF4E, significantly decreased in polysome fractions and moderately increased in subpolysome fractions after the eIF4AIII down-regulation (Fig. 4_D_, after IP). It should be noted that the polysome distribution of R-SL0 mRNA was marginally affected by the eIF4AIII down-regulation in the IP of CBP80 and eIF4E (Fig. 4_C_, after IP). All these results provided strong evidence that eIF4AIII preferentially enhances CT of mRNAs containing an SL structure. In agreement with the role of eIF4AIII in CT, CTIF that was artificially tethered to an intercistronic region of intronless dicistronic mRNA increased the translation of a downstream cistron fourfold. The translational enhancement was inhibited twofold by down-regulation of eIF4AIII but not eIF4AI, eIF4AII, UPF2, or UPF3X (Fig. S4).
ATPase/Helicase Activity of eIF4AIII Is Important for Promotion of CT Efficiency of mRNAs Containing an SL Structure.
The selective role of eIF4AIII in CT of mRNAs containing an SL structure was tested in experiments similar to those experiments in Fig. 4, using the extracts of cells depleted of eIF4AIII or codepleted of eIF4AI and eIF4AII (Fig. S5A). After polysome fractionation using a sucrose gradient, the subpolysome fractions (40S, 60S, and 80S) and polysome fractions were pooled separately. The pooled fractions were subjected to IPs using either an α-CBP80 antibody or an α-eIF4E antibody (Fig. S5_B_). Then, the relative polysome/subpolysome distributions of R-SL9.7 mRNA were determined using qRT-PCR assays. The results showed that eIF4AIII down-regulation reduced the relative polysome/subpolysome ratio of R-SL9.7 mRNA in the IP of CBP80 but not in the IP of eIF4E (Fig. S5_C_). On the other hand, simultaneous down-regulation of eIF4AI and eIF4AII reduced the relative polysome/subpolysome ratio of R-SL9.7 mRNA in the IP of eIF4E but not in the IP of CBP80 (Fig. S5_C_). Intriguingly, the reduction in the relative polysome/subpolysome ratio of R-SL9.7 mRNA in the IP of CBP80 was completely reversed via expression of siRNA-resistant eIF4AIII-WT but not of an ATPase/helicase mutant, eIF4AIII-E188Q (Fig. S6). It is known that the introduction of the E188Q substitution into the Walker boxes of eIF4AIII abolishes ATPase/helicase activity without affecting the function of eIF4AIII in EJC assembly and NMD of a reporter mRNA lacking a clearly defined SL structure (35, 36). These results suggest that (i) eIF4AIII and eIF4AI/II are specifically involved in CT and ET, respectively, of mRNAs containing an SL structure and (ii) the ATPase/helicase activity of eIF4AIII is necessary for promotion of CT of mRNAs containing an SL structure.
eIF4AIII Selectively Increases CT Efficiency in an in Vitro Translation System.
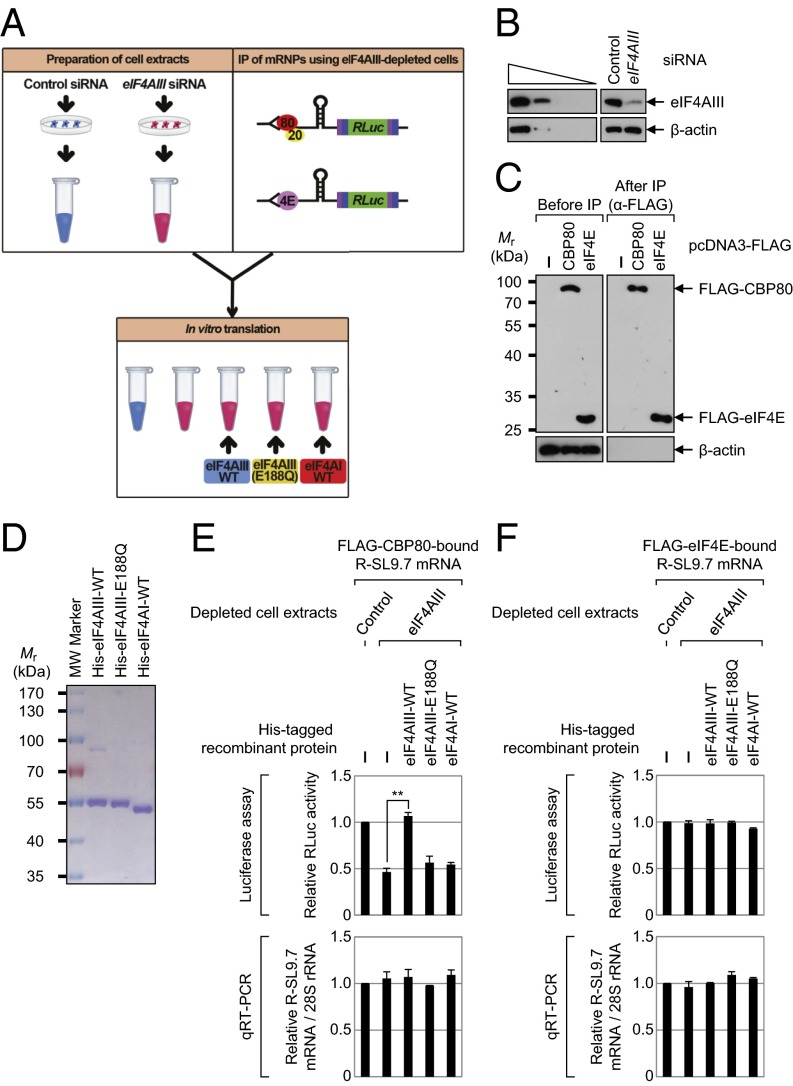
The selective increase by eIF4AIII of the CT of mRNAs containing an SL structure was recapitulated in an in vitro translation system. As shown in Fig. 5_A_, the cytoplasmic extracts for in vitro translation were obtained from HeLa cells either depleted or undepleted of eIF4AIII. For preparation of mRNAs for in vitro translation, we used either immunopurified CBP80-bound or eIF4E-bound intronless mRNA to eliminate the effects of EJCs on translation. We also needed to rule out the possible effects of preexisting eIF4AIII, which had already associated with immunopurified mRNAs. To this end, IPs were performed using α-FLAG antibody-conjugated agarose beads and the extracts of eIF4AIII-depleted HEK293T cells transiently expressing R-SL9.7 mRNA and either FLAG-CBP80 or FLAG-eIF4E. After that procedure, FLAG-CBP80–bound or FLAG-eIF4E–bound mRNPs were eluted from the resin using FLAG peptides. In vitro translation reactions were performed by mixing the cytoplasmic extracts, the eluted mRNPs, and one of recombinant proteins (eIF4AIII-WT, eIF4AIII-E188Q, eIF4AI-WT, or BSA as a control). To minimize possible exchange of the CBC for eIF4E during the reaction, the reaction mixtures were incubated for only 15 min.
Fig. 5.
Down-regulation of endogenous eIF4AIII and add-back of purified recombinant eIF4AIII inhibits and restores, respectively, the CT of intronless mRNAs with SL structures. (A) Diagram of the experimental procedure of the in vitro-reconstituted translation system. Cytoplasmic cell extracts and either FLAG-CBP80–bound or FLAG-eIF4E–bound R-SL9.7 mRNPs for in vitro translation were obtained from HeLa cells and HEK293T cells, respectively. The details are described in Results. (B) Western blot analysis shows specific down-regulation of eIF4AIII in HeLa cells. Comparable down-regulation of eIF4AIII was observed in HEK293T cells (Fig. S8_A_). (C) Western blot analysis demonstrates specific IPs. β-Actin served as a negative control. (D) Coomassie Blue staining of purified recombinant His-eIF4AIII-WT, His-eIF4AIII-E188Q, and His-eIF4AI-WT. (E) In vitro translation of FLAG-CBP80–bound R-SL9.7 mRNAs. The levels of in vitro-translated RLuc protein were analyzed using luciferase assays. (Upper) Value of RLuc activity in the control siRNA-transfected cell extracts was set to 1.0. **P < 0.01. (Lower) After the in vitro translation reaction, total RNA was purified and analyzed by qRT-PCR assay. The levels of RLuc mRNAs were normalized to endogenous 28S ribosomal RNAs (rRNAs). The normalized level of RLuc mRNAs in control siRNA-transfected cell extracts was set to 1.0. (F) In vitro translation of FLAG-eIF4E–bound R-SL9.7 mRNAs. Experiments were conducted as described in E, except that the FLAG-eIF4E–bound R-SL9.7 mRNAs were analyzed using in vitro translation. The columns and bars represent the mean and SD of three independent transfections and in vitro translation reactions.
Western blotting results demonstrated specific down-regulation of eIF4AIII (Fig. 5_B_) and selectivity of IP (Fig. 5_C_). The purity and integrity of the recombinant proteins were demonstrated by Coomassie Blue staining (Fig. 5_D_). In vitro translation reactions and add-back experiments revealed that translation of CBP80-bound R-SL9.7 mRNA decreased twofold in the eIF4AIII-depleted cell extracts and was restored by the addition of recombinant eIF4AIII-WT but not eIF4AIII-E188Q or eIF4AI-WT (Fig. 5_E_, Upper). In contrast, translation of eIF4E-bound R-SL9.7 mRNAs was not significantly changed in the eIF4AIII-depleted cell extracts relative to the undepleted cell extracts and was not affected by the addition of a recombinant protein (Fig. 5_F_, Upper). In addition, there was no significant change in the levels of R-SL9.7 mRNAs after in vitro translation (Fig. 5 E and F, Lower). Of note, the relative translation efficiency of CT (RLuc activity divided by the R-SL9.7 mRNA level) in the control cell extracts was slightly lower compared with ET, without statistical significance (Fig. S7). This finding suggests that CT is as efficient as ET under our conditions. All these results, along with the data from the polysome fractionation (Fig. 4) and artificial tethering experiments (Fig. S4), indicate that eIF4AIII preferentially promotes CT of mRNAs containing an SL structure in vitro and in vivo. In line with this notion, the results of in vitro translation reactions and add-back experiments using FLAG-CBP80–bound or FLAG-eIF4E–bound R-SL0 mRNPs lacking an SL structure in the 5′UTR showed that CT and ET of R-SL0 mRNA were not dependent on eIF4AIII (Fig. S8).
eIF4AIII–mRNA Interactions Are Enriched at the 5′-End and in the Translation Initiation Codon of Intronless as Well as Intron-Containing (Spliced) mRNAs.
Recently, a transcriptome-wide interaction map of eIF4AIII was generated using high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation [HITS-CLIP; also named cross-linking immunoprecipitation sequencing (CLIP-Seq)], elucidating that eIF4AIII-binding sites are largely located in the canonical EJC deposition region and in the noncanonical region within the exons (37). Given our finding that eIF4AIII is recruited to the 5′-end of mRNA via its direct interaction with CTIF, we expected to see in the eIF4AIII CLIP data that eIF4AIII–mRNA interactions would be enriched in 5′UTRs of mRNAs. For this purpose, we retrieved the previously reported eIF4AIII CLIP data (two replicates performed in HeLa cells) and further filtered reproducible binding regions (clusters) that contained overlapping CLIP tags between two replicates [biological complexity (BC) = 2] and peak height (PH > 10)] (Fig. S9_A_). By analyzing locations of such robust eIF4AIII CLIP clusters in mRNAs, we found that eIF4AIII–mRNA interactions were enriched in 5′-end regions, mostly with peaks in the translation initiation codon (Fig. S9_B_). Such enrichment, however, was not observed either in the stop codon or at the 3′-end. The eIF4AIII showed slight enrichment upstream of the translation termination codon, but the result was not statistically significant relative to control clusters for mRNA length (Fig. S9_C_, Left). We also found that the binding of eIF4AIII downstream of the termination codon or near the 3′-end was significantly less (Fig. S9_C_, Right). Furthermore, we observed the same distribution pattern in intronless mRNAs, although there was sharply higher enrichment at the 5′-end (Fig. S9_D_). Taken together, these data suggest that eIF4AIII interacts with the 5′-end and translation initiation codon of mRNAs, consistent with its biochemical interaction with CTIF and its role in CT observed in this study.
Down-Regulation of eIF4AIII Inhibits Rapid Degradation of Replication-Dependent Histone mRNAs Induced by Inhibition of DNA Replication.
To demonstrate further the role of eIF4AIII in CT, we used replication-dependent histone mRNAs, which lack introns and a poly(A) tail. The stability of replication-dependent histone mRNAs is well known to be closely linked to translation, as reviewed elsewhere (38). In particular, the majority of replication-dependent histone mRNAs that are translated in the cytoplasm bind to CBP80/20 rather than to eIF4E (15, 34). Furthermore, rapid degradation of replication-dependent histone mRNAs, which is induced by treatment with hydroxyurea (HU; a potent inhibitor of DNA replication), largely occurs during CT (15, 34). Therefore, our present finding that eIF4AIII is preferentially involved in CT prompted us to test whether eIF4AIII is involved in the stability of replication-dependent histone mRNAs (Fig. 6 and Fig. S10). For this purpose, we depleted HeLa cells of eIF4AI, eIF4AII, eIF4AIII, or CTIF or codepleted those cells of eIF4AI and eIF4AII. Then, the levels of cellular HIST1H1C and HIST2H2AA mRNAs, which are replication-dependent histone mRNAs, were measured using qRT-PCR assays. We also measured the levels of cellular H3F3A mRNA, which is a replication-independent histone mRNA containing a poly(A) tail, and used them as a control for variations in this experiment.
Fig. 6.
Rapid degradation of replication-dependent histone mRNAs upon inhibition of DNA replication is dependent on eIF4AIII. (A_–_C) HeLa cells were transfected with the indicated siRNAs. Three days later, the cells were treated with HU for 40 min before cell harvesting, and total-cell protein and RNA were then purified. (A) Western blot analysis shows specific down-regulation by siRNAs. The qRT-PCR assays of HIST1H1C mRNAs (B) and HIST2H2AA mRNAs (C) are shown. The levels of HIST1H1C and HIST2H2AA mRNAs were normalized to H3F3A mRNA. Normalized levels of HIST1H1C mRNAs (B) and HIST2H2AA mRNAs (C) in the absence of HU treatment were set to 100%. **P < 0.01. The columns and bars represent the mean and SD of four independent transfections and qRT-PCR assays. (D_–_F) HeLa cells were transfected with a plasmid expressing PDCD4. Two days later, the cells were treated with HU for 40 min before cell harvesting, and total-cell protein and RNA were then purified. (D) Western blot analysis shows overexpression of PDCD4-WT or Mut4. The qRT-PCR analyses of HIST1H1C mRNAs (E) and HIST2H2AA mRNAs (F) are shown. The columns and bars represent the mean and SD of four (A_–_C) or three (D_–_F) independent transfections and qRT-PCR assays.
The specific down-regulation of cellular proteins by siRNA treatment was confirmed by Western blotting (Fig. 6_A_). The qRT-PCR results showed that HU treatment triggered rapid degradation of HIST1H1C and HIST2H2AA mRNAs (Fig. 6 B and C). In agreement with our previous report (15), down-regulation of CTIF attenuated the rapid degradation of replication-dependent histone mRNAs induced by HU treatment.
The rapid degradation of replication-dependent histone mRNAs was attenuated by down-regulation of eIF4AIII but not by individual or simultaneous down-regulation of eIF4AI and eIF4AII (Fig. 6 B and C and Fig. S10 A_–_C). This inhibition was reversed by expression of eIF4AIII-WT but not eIF4AIII-E188Q (Fig. S10 D_–_F). These data suggest that CT of intronless replication-dependent histone mRNAs requires eIF4AIII but not eIF4AI/II. In line with this result, degradation of replication-dependent histone mRNAs was not significantly affected by overexpression of either PDCD4-WT or -Mut4 (Fig. 6 D_–_F).
Discussion
In this study, we demonstrate a previously unidentified role of eIF4AIII in translation driven by the CBC. We report that eIF4AIII is recruited to the CT complex via direct interaction with CTIF, and that it triggers CT of mRNAs with secondary structures in the 5′UTR. Based on our in vivo and in vitro results, we propose a model in which eIF4AIII preferentially promotes CT efficiency (Fig. 7).
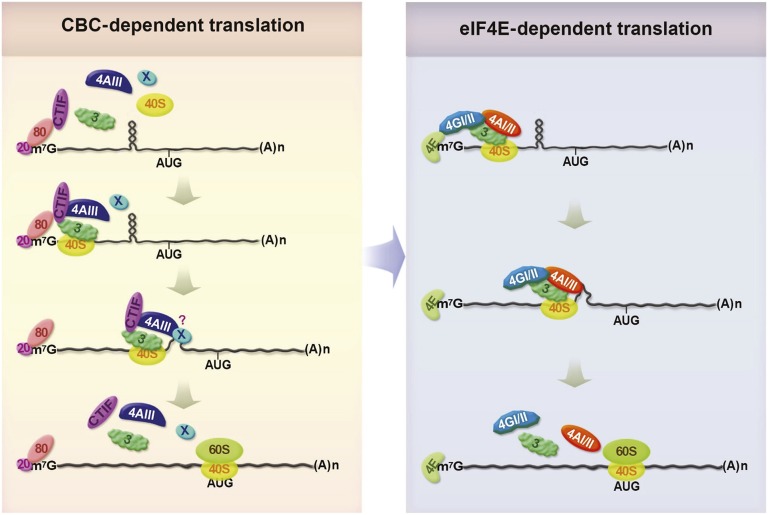
Fig. 7.
Model illustrates the role of eIF4AIII in CT (details are provided in Discussion). AUG, translation initiation codon; m7G, Cap; (A)n, poly(A) tail; 20, CBP20; 80, CBP80; 40S, 40S ribosomal subunit; 60S, 60S ribosomal subunit; 3, eIF3; 4AIII, eIF4AIII; 4E, eIF4E; 4GI/II, eIF4GI/II; 4AI/II, eIF4AI/II; X, unidentified factor. It remains unknown whether eIF4AIII directly melts the structures or induces remodeling of the complex to promote unwinding of the structures.
The newly synthesized intron-containing or intronless mRNAs are exported from the nucleus to the cytoplasm with the CBC bound to the 5′-end (Fig. 7, Left). During the export, the perinuclear protein CTIF joins the 5′-end of the mRNAs via direct interaction between CTIF and CBP80 (3). In the cytoplasm, the CBC–CTIF complex recruits eIF4AIII and eIF3, both of which are known to interact directly with CTIF (as shown in Fig. 2 and ref. 4). Alternatively, formation of the complex may occur before mRNA export because a significant fraction of CTIF and eIF4AIII is found in the nucleus (3, 30, 39–41). The resulting CT complex can recruit the 40S ribosomal subunit to the 5′-end of the mRNAs via eIF3, triggering the ribosome scanning process. When the scanning ribosome encounters secondary structures, eIF4AIII may melt the structures or induce remodeling of the complex to promote unwinding of these structures. As a result, the ribosome can continue scanning the mRNA until the ribosome reaches an authentic translation initiation codon. It is likely that eIF4AIII comigrates with the scanning ribosome until the ribosome reaches the authentic translation initiation codon because reanalysis of the previously reported eIF4AIII CLIP-seq data (37) shows significant enrichment of eIF4AIII CLIP clusters in translation initiation codons of both total mRNAs and intronless mRNAs (Fig. S9). These observations provide additional evidence of the existence of noncanonical eIF4AIII (or EJC)-binding sites that were reported previously (37, 42). Although we do not know the exact timing of the replacement of the CBC by eIF4E, this event is triggered by importin α/β (6). After that, the eIF4E directs efficient ET (Fig. 7, Right).
The ATP-dependent helicase activity of recombinant or purified eIF4AI is weak; however, the RNA helicase activity of eIF4AI is stimulated by cofactors, such as eIF4B and eIF4H (43–45). Like eIF4AI, purified eIF4AIII has relatively weak helicase activity (32, 45, 46). It was reported, however, that RNA unwinding by eIF4AIII is stimulated by MLN51, which is another EJC component (46), but not by eIF4B or eIF4H (45). Furthermore, it was reported recently that MLN51 enhances translation of intron-containing mRNAs and, to a lesser extent, intronless mRNAs via direct interaction with eIF3 (47). These observations, together with our present results, show that MLN51 may promote eIF4AIII-mediated unwinding of secondary structures or eIF4AIII-mediated remodeling of the CT complex by enhancing the helicase activity of eIF4AIII. In particular, the MLN51-mediated enhancement of CT via eIF4AIII may be further coordinated by an interaction between MLN51 and eIF3 (47–49).
In general, the translational efficiency of intron-containing (spliced) mRNA is higher than that of intronless mRNA, in part, because of EJCs deposited as a consequence of splicing (21–23). Such an EJC-mediated increase in the translation of intron-containing (spliced) mRNAs over intronless mRNAs could be explained, in part, by our present observations. An intron-containing (spliced) mRNA may be subjected to a microenvironment with a high local concentration of eIF4AIII because translocating ribosomes displace EJCs from an mRNA. Therefore, free eIF4AIII and the eIF4AIII displaced from EJCs may associate with CTIF at the 5′-end of mRNAs more frequently, and thereby promote CT efficiency. Further studies should elucidate molecular details of the above possibility.
Materials and Methods
Construction of Plasmids.
These details are described in SI Materials and Methods.
Cell Culture and Transfection.
HeLa, HEK293T, and HEK293FT cells were grown in DMEM (HyClone) containing 10% (vol/vol) FBS (HyClone) and 1% penicillin/streptomycin (HyClone). The cells were transiently transfected with the indicated plasmids and 100 nM in vitro-synthesized siRNA (Invitrogen) using Lipofectamine 2000 (Invitrogen) and Oligofectamine (Invitrogen), respectively, as described previously (3, 50, 51). To achieve simultaneous down-regulation of eIF4AI and eIF4AII, we transfected the cells with either 50 nM (Fig. S5) or 100 nM (Figs. S2 and S10 A_–_C) siRNA. The total amount of siRNA used in this experiment was adjusted to either 100 or 200 nM using control siRNA. The following siRNA sequences were used in this study: 5′-r(GCCCAAUCUGGGACUGGGA)d(TT)-3′ for eIF4AI and 5′-r(AGGAGUCUCUCCUUCGUGG)d(TT)-3′ for eIF4AII. The siRNA sequences for eIF4AIII (40), CTIF (3), UPF2 (52), UPF3X (52), and the nonspecific control (52) were described previously. At 2 or 3 d after the transfection, the cells were harvested, and total protein and RNA were purified as described previously (3, 52).
When indicated, the cells were incubated with 5 mM HU (Sigma–Aldrich) for 40 min before cell harvesting.
Immunoprecipitation.
Immunoprecipitation (IP) was performed as described previously (3, 50, 53). When indicated, the cells were treated with a final concentration of 1% formaldehyde (Sigma–Aldrich) in PBS for 10 min before cell harvesting.
Western Blotting.
Antibodies against the following proteins were used: FLAG (Sigma), Myc (Calbiochem), HA (Roche), GFP (Santa Cruz Biotechnology), eIF4E [an antibody raised in rabbits (Cell Signaling Technology) or mice (BD Bioscience)], eIF3b (Santa Cruz Biotechnology), eIF4GI [a gift from S. K. Jang (Pohang University of Science and Technology, Pohang, Korea)], CTIF (3), CBP80 (4), PDCD4 (Bethyl), eIF4AI (Abcam), eIF4AII (Abcam), eIF4AIII (53), UPF2 (53), UPF3X [a gift from L. E. Maquat (University of Rochester, Rochester, NY)], β-actin (Sigma), His (GE Healthcare), and GST (Amersham).
RT-PCR Analysis Using Specific Oligonucleotides and α-[32P]-dATP and qRT-PCR Analyses.
The details of the RT-PCR assays are as described previously (3, 50, 51). The qRT-PCR analyses were performed using a LightCycler system (Roche Diagnostics).
The mRNAs for the Rβ-SL reporters (Rβ-SL0-Norm or Rβ-SL0-Ter, Rβ-SL9.7-Norm or Rβ-SL9.7-Ter, and Rβ-SL87.8-Norm or Rβ-SL87.8-Ter), Rβ-8bs (either intron or no intron), R-SL0, and R-SL9.7 were analyzed by means of qRT-PCR using two specific oligonucleotides: 5′-ATGGTGCACCTGACTCCTGA-3′ (sense) and 5′-GGGTTTAGTGGTACTTGTGAGC-3′ (antisense) for the Rβ-SL and Rβ-8bs mRNAs, and 5′-AATACGACTCACTATAGGGA-3′ (sense) and 5′-GGAGCTTCTCGTGTACTTCC-3′ (antisense) for the R-SL0 and R-SL9.7 mRNAs. Oligonucleotides used for amplification of mRNAs of MUP, RLuc, β-actin, and GAPDH have been described elsewhere (3, 15, 50, 51). Oligonucleotides used for amplification of cellular HIST1H1C and HIST2H2AA mRNAs, which are replication-dependent histone mRNAs (54, 55), and cellular H3F3A mRNA, which is a replication-independent histone mRNA containing a poly(A) tail (54, 55), have been described elsewhere (15).
Polysome Fractionation.
Polysome fractionations using HEK293FT cells (two 150-mm culture dishes) were performed as described previously (50).
BiFC Assays.
BiFC assays were performed as described previously (15, 50).
Protein Expression, Purification, and GST Pull-Down Assay.
GST, GST-CTIF (1–305), GST-CTIF (306–598), and GST-Galectin 8, which served as a negative control, were overexpressed in Escherichia coli BL21(DE3)pLysS by adding 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when the OD at 600 nm was 0.5; the cultures were then incubated at 37 °C for 3 h.
The His-eIF4AIII-WT, His-eIF4AIII-E188Q, and His-eIF4AI-WT proteins were also overexpressed in E. coli BL21(DE3)pLysS by adding 1 mM IPTG when the OD at 600 nm reached 0.5, and the cells were subsequently incubated either for 20 h at 18 °C (His-eIF4AIII-WT) or for 16 h at 16 °C (His-eIF4AIII-E188Q and His-eIF4AI-WT). The cells were then harvested, resuspended in lysis buffer [20 mM Hepes-NaOH (pH 7.5), 100 mM NaCl, 10% (vol/vol) glycerol, and 1 mM PMSF], and sonicated. Total-cell extracts were loaded onto a HisTrap HP column (GE Healthcare), and the column was washed with the binding buffer [20 mM Hepes-NaOH (pH 7.5), 100 mM NaCl, and 10% (vol/vol) glycerol]. The bound proteins were eluted at ∼250 mM imidazole in the binding buffer. The purified proteins were dialyzed against the buffer [10 mM Hepes (pH 7.4), 120 mM potassium acetate, 1.5 mM magnesium acetate, 4 mM DTT, and 10% (vol/vol) glycerol].
In the in vitro GST pull-down assays, the purified recombinant His-eIF4AIII-WT and the extracts of E. coli expressing the GST-tagged proteins were mixed and incubated in 500 μL of binding buffer [10 mM Tris⋅HCl (pH 8.0), 1% (vol/vol) glycerol, 150 mM NaCl, 0.1% Triton X-100, 2 mM benzamidine, 1 mM PMSF, 0.001% BSA, and protease inhibitor mixture tablet] at 4 °C for 1 h. Glutathione Sepharose 4B resin (Amersham–Pharmacia Biotech) was added, and the mixture was incubated for 1 h and washed five times with the binding buffer. The resin-bound proteins were separated using SDS/PAGE and subjected to Western blotting.
Preparation of Cytoplasmic Extracts.
HeLa cells were transfected with either eIF4AIII siRNA or nonspecific control siRNA, and the cells were harvested and resuspended in hypotonic buffer [10 mM Hepes (pH 7.4), 10 mM potassium acetate, 1.5 mM magnesium acetate, and 2.5 mM DTT] 3 d later. After incubation for 30 min on ice, the cells were ruptured by passing 10 times through a 27-gauge needle attached to a 3-mL syringe. The cell homogenate was centrifuged at 13,000 × g for 15 min at 4 °C. The supernatant was collected and used for in vitro translation.
In Vitro Translation of Coimmunopurified mRNAs.
In vitro translation reactions were performed as described previously (3). Briefly, HEK293T cells were transfected with eIF4AIII siRNA; 1 d later, the cells were retransfected with the reporter plasmid expressing either R-SL9.7 mRNA (Fig. 5) or R-SL0 mRNA (Fig. S8) and a plasmid expressing a FLAG-tag only, FLAG-CBP80, or FLAG-eIF4E. Two days after the transfection, IP was performed using α-FLAG antibody-conjugated agarose beads, and resin-bound ribonuclease (RNP) complexes were eluted using 3×FLAG peptides (Sigma–Aldrich).
In vitro translation was performed for 15 min at 37 °C in 20-μL reaction mixtures containing 4 μL of immunoprecipitated CBP80-bound RNP or eIF4E-bound RNP and 1 μg of either purified BSA or a recombinant protein. The activity of in vitro-translated luciferase was measured as described previously (51, 56).
Analysis of the eIF4AIII CLIP Data.
For bioinformatic analysis of eIF4AIII CLIP tags, we retrieved relevant data from the Gene Expression Omnibus database (accession no. GSE40778) and used a previously described analytical method (57). In brief, CLIP data from duplicate experiments were downloaded in bedGraph format, further processed, and combined as reproducible cluster regions (regions with overlapping tags; BC = 2, PH > 10; PH was defined as the maximal number of overlapping tags in each cluster region). Based on the position of clusters in the genome and annotation in the Reference Sequence database, relative positions of the clusters were calculated and plotted. To adjust the data for differences in the length of transcripts, we followed the normalization method described elsewhere (57).
Supplementary Material
Supplementary File
Acknowledgments
We thank C. D. Hu for the BiFC plasmids, K. H. Klempnauer for the plasmids encoding PDCD4 and its variant, N. Sonenberg for pcDNA3-HA-eIF4AIII, S. K. Jang for pET28a-eIF4AI-WT and the α-eIF4GI antibody, L. E. Maquat for the α-UPF3X antibody, and M. J. Moore for the plasmids no intron and 5′-intron. This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by Ministry of Education, Science, and Technology Grant 2012R1A2A1A01002469. S.W.C. was supported, in part, by Korean Health Technology Research and Development Project, Ministry of Health and Welfare Grant A111989. I.R. was supported, in part, by a NRF Grant 2013R1A1A2063273.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106(5):607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 2.Pestova TV, Lorsch JR, Hellen CU. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- 3.Kim KM, et al. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes Dev. 2009;23(17):2033–2045. doi: 10.1101/gad.1823409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choe J, et al. Translation initiation on mRNAs bound by nuclear cap-binding protein complex CBP80/20 requires interaction between CBP80/20-dependent translation initiation factor and eukaryotic translation initiation factor 3g. J Biol Chem. 2012;287(22):18500–18509. doi: 10.1074/jbc.M111.327528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 2002;21(13):3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato H, Maquat LE. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin beta. Genes Dev. 2009;23(21):2537–2550. doi: 10.1101/gad.1817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marintchev A. Roles of helicases in translation initiation: A mechanistic view. Biochim Biophys Acta. 2013;1829(8):799–809. doi: 10.1016/j.bbagrm.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsyan A, et al. mRNA helicases: The tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12(4):235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 9.Karam R, Wengrod J, Gardner LB, Wilkinson MF. Regulation of nonsense-mediated mRNA decay: Implications for physiology and disease. Biochim Biophys Acta. 2013;1829(6-7):624–633. doi: 10.1016/j.bbagrm.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13(4):246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay—Mmechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013;1829(6-7):612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Rufener SC, Mühlemann O. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2013;20(6):710–717. doi: 10.1038/nsmb.2576. [DOI] [PubMed] [Google Scholar]
- 13.Durand S, Lykke-Andersen J. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat Struct Mol Biol. 2013;20(6):702–709. doi: 10.1038/nsmb.2575. [DOI] [PubMed] [Google Scholar]
- 14.Apcher S, et al. Major source of antigenic peptides for the MHC class I pathway is produced during the pioneer round of mRNA translation. Proc Natl Acad Sci USA. 2011;108(28):11572–11577. doi: 10.1073/pnas.1104104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choe J, et al. Rapid degradation of replication-dependent histone mRNAs largely occurs on mRNAs bound by nuclear cap-binding proteins 80 and 20. Nucleic Acids Res. 2013;41(2):1307–1318. doi: 10.1093/nar/gks1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K. Thriving under stress: Selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012;8(3):e1002612. doi: 10.1371/journal.ppat.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19(24):6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Hir H, Séraphin B. EJCs at the heart of translational control. Cell. 2008;133(2):213–216. doi: 10.1016/j.cell.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16(3):279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137(3):536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 21.Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci USA. 2003;100(20):11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18(2):210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gudikote JP, Imam JS, Garcia RF, Wilkinson MF. RNA splicing promotes translation and RNA surveillance. Nat Struct Mol Biol. 2005;12(9):801–809. doi: 10.1038/nsmb980. [DOI] [PubMed] [Google Scholar]
- 24.Ma XM, Yoon SO, Richardson CJ, Jülich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133(2):303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 25.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1986;83(9):2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vassilenko KS, Alekhina OM, Dmitriev SE, Shatsky IN, Spirin AS. Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res. 2011;39(13):5555–5567. doi: 10.1093/nar/gkr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang HS, et al. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol Cell Biol. 2004;24(9):3894–3906. doi: 10.1128/MCB.24.9.3894-3906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang HS, et al. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003;23(1):26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wedeken L, Singh P, Klempnauer KH. Tumor suppressor protein Pdcd4 inhibits translation of p53 mRNA. J Biol Chem. 2011;286(50):42855–42862. doi: 10.1074/jbc.M111.269456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferraiuolo MA, et al. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci USA. 2004;101(12):4118–4123. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14(9):1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, et al. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol Cell Biol. 1999;19(11):7336–7346. doi: 10.1128/mcb.19.11.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques. 2006;40(1):61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- 34.Choe J, Ahn SH, Kim YK. The mRNP remodeling mediated by UPF1 promotes rapid degradation of replication-dependent histone mRNA. Nucleic Acids Res. 2014;42(14):9334–9349. doi: 10.1093/nar/gku610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibuya T, Tange TO, Stroupe ME, Moore MJ. Mutational analysis of human eIF4AIII identifies regions necessary for exon junction complex formation and nonsense-mediated mRNA decay. RNA. 2006;12(3):360–374. doi: 10.1261/rna.2190706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Krainer AR. Splicing remodels messenger ribonucleoprotein architecture via eIF4A3-dependent and -independent recruitment of exon junction complex components. Proc Natl Acad Sci USA. 2007;104(28):11574–11579. doi: 10.1073/pnas.0704946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saulière J, et al. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19(11):1124–1131. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- 38.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet. 2008;9(11):843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan CC, et al. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10(2):200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427(6976):753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 41.Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11(4):346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 42.Singh G, et al. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151(4):750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grifo JA, Abramson RD, Satler CA, Merrick WC. RNA-stimulated ATPase activity of eukaryotic initiation factors. J Biol Chem. 1984;259(13):8648–8654. [PubMed] [Google Scholar]
- 44.Rogers GW, Jr, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem. 2001;276(33):30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 45.Rozovsky N, Butterworth AC, Moore MJ. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA. 2008;14(10):2136–2148. doi: 10.1261/rna.1049608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noble CG, Song H. MLN51 stimulates the RNA-helicase activity of eIF4AIII. PLoS ONE. 2007;2(3):e303. doi: 10.1371/journal.pone.0000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chazal PE, et al. EJC core component MLN51 interacts with eIF3 and activates translation. Proc Natl Acad Sci USA. 2013;110(15):5903–5908. doi: 10.1073/pnas.1218732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ballut L, et al. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12(10):861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 49.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126(4):713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell. 2009;33(1):75–86. doi: 10.1016/j.molcel.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 51.Choe J, Cho H, Lee HC, Kim YK. microRNA/Argonaute 2 regulates nonsense-mediated messenger RNA decay. EMBO Rep. 2010;11(5):380–386. doi: 10.1038/embor.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120(2):195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 53.Cho H, et al. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell. 2012;46(4):495–506. doi: 10.1016/j.molcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Brush D, Dodgson JB, Choi OR, Stevens PW, Engel JD. Replacement variant histone genes contain intervening sequences. Mol Cell Biol. 1985;5(6):1307–1317. doi: 10.1128/mcb.5.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wells D, Kedes L. Structure of a human histone cDNA: Evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc Natl Acad Sci USA. 1985;82(9):2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choe J, Cho H, Chi SG, Kim YK. Ago2/miRISC-mediated inhibition of CBP80/20-dependent translation and thereby abrogation of nonsense-mediated mRNA decay require the cap-associating activity of Ago2. FEBS Lett. 2011;585(17):2682–2687. doi: 10.1016/j.febslet.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 57.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File