Reconstitution of a minimal mtDNA replisome in vitro (original) (raw)
Abstract
We here reconstitute a minimal mammalian mitochondrial DNA (mtDNA) replisome in vitro. The mtDNA polymerase (POLγ) cannot use double-stranded DNA (dsDNA) as template for DNA synthesis. Similarly, the TWINKLE DNA helicase is unable to unwind longer stretches of dsDNA. In combination, POLγ and TWINKLE form a processive replication machinery, which can use dsDNA as template to synthesize single-stranded DNA (ssDNA) molecules of about 2 kb. The addition of the mitochondrial ssDNA-binding protein stimulates the reaction further, generating DNA products of about 16 kb, the size of the mammalian mtDNA molecule. The observed DNA synthesis rate is 180 base pairs (bp)/min, corresponding closely to the previously calculated value of 270 bp/min for in vivo DNA replication. Our findings provide the first biochemical evidence that TWINKLE is the helicase at the mitochondrial DNA replication fork. Furthermore, mutations in TWINKLE and POLγ cause autosomal dominant progressive external ophthalmoplegia (adPEO), a disorder associated with deletions in mitochondrial DNA. The functional interactions between TWINKLE and POLγ thus explain why mutations in these two proteins cause an identical syndrome.
Keywords: DNA polymerase γ, mitochondrion, mtSSB, TWINKLE
Introduction
Human mitochondria have a small double-stranded DNA (dsDNA) genome of 16.6 kb, which contains two origins of replication, OH and OL (Shadel and Clayton, 1997). According to the strand-asymmetric model for mitochondrial DNA (mtDNA) replication, DNA synthesis is continuous on both strands and takes place in a strand-asymmetric mode. The DNA synthesis from OH is unidirectional and proceeds to displace the parental heavy strand. After leading-strand synthesis has reached two-thirds of the genome, it activates OL and DNA synthesis then initiates in the opposite direction (Robberson et al, 1972; Tapper and Clayton, 1981; Kang et al, 1997). However, this strand-asymmetric model for mtDNA replication has recently been challenged by 2-D gel electrophoresis analyses, demonstrating the presence of conventional duplex mtDNA replication intermediates, indicative of coupled leading and lagging-strand DNA synthesis (Yang et al, 2002; Holt and Jacobs, 2003). Initiation of mtDNA replication is coupled to mitochondrial transcription. Transcription is initiated from two major mtDNA promoters, the light- and heavy-strand promoters (LSP and HSP) (Fernandez-Silva et al, 2003). Transcription from the two promoters produces near-genomic length transcripts that are released as individual mRNAs, tRNAs, and rRNAs after RNA processing. A separate transcription unit for the rRNA genes has also been reported in mammalian mitochondria (Montoya et al, 1982). The primer needed to initiate DNA replication at OH in mammalian cells is believed to be generated by the transcription from LSP and subsequent RNA processing (Chang and Clayton, 1985; Shadel and Clayton, 1997).
In vertebrates, POLγ is the only DNA polymerase devoted to mtDNA synthesis (Ropp and Copeland, 1996). The POLγ holoenzyme consists of a catalytic subunit with exonuclease activity (POLγA) and an accessory subunit (POLγB) that increases the processivity (Carrodeguas et al, 1999, 2001). The mtSSB also has a role in mtDNA replication, as the rate of DNA synthesis by Drosophila POLγ is increased nearly 40-fold after addition of mtSSB. Furthermore, flies with disruption of the mtSSB gene show a marked mtDNA depletion and defective mitochondrial respiration (Maier et al, 2001; Farr et al, 2004).
TWINKLE is a mitochondrial DNA helicase with 5′ to 3′ directionality and distinct substrate requirements. TWINKLE-dependent DNA unwinding of short stretches of dsDNA (20 base pairs (bp)) is specifically stimulated by mtSSB (Korhonen et al, 2003). Interestingly, TWINKLE displays sequence similarity to the C-terminal helicase part of the bacteriophage T7 gene 4 protein, which contains the DNA helicase and primase activities needed at the bacteriophage DNA replication fork (Spelbrink et al, 2001). It has been proposed that TWINKLE may be the helicase at the mitochondrial DNA replication fork (Spelbrink et al, 2001; Korhonen et al, 2003), but evidence for functional interactions between TWINKLE and POLγ has not previously been presented. We have now reconstituted a minimal mammalian mtDNA replisome with pure proteins and demonstrate here that POLγ, TWINKLE, and mtSSB act together at the DNA replication fork to form a macromolecular machinery resembling the bacteriophage T4 and T7 replisomes. The definition of the core components of the mammalian mtDNA minimal replisome will provide a new biochemical basis for investigating the mode of replication of mammalian mtDNA, an intensely debated topic.
Results
POL_γ_ is unable to use dsDNA as template for DNA synthesis
To construct a template for DNA replication, we annealed a 90-nt oligonucleotide to a 70-nt ssDNA mini-circle. The template formed contains a replication fork for loading the replication machinery, a 50-bp dsDNA region and a free 3′-hydroxyl terminus that can act as a primer for DNA synthesis (Figure 1A). Once initiated, leading-strand DNA synthesis coupled to continuous unwinding of the double-stranded template could in principle progress indefinitely. POLγ could on its own utilize the 3′-hydroxyl terminus on the mini-circle template and initiate DNA synthesis, but the enzyme failed to elongate through double-stranded regions and only formed a 110-nt product (Figure 1B and C). We could thus conclude that POLγ is unable to use dsDNA as template for DNA synthesis, consistent with previously reported biochemical properties of the T7 DNA polymerase (Engler et al, 1983).
Figure 1.
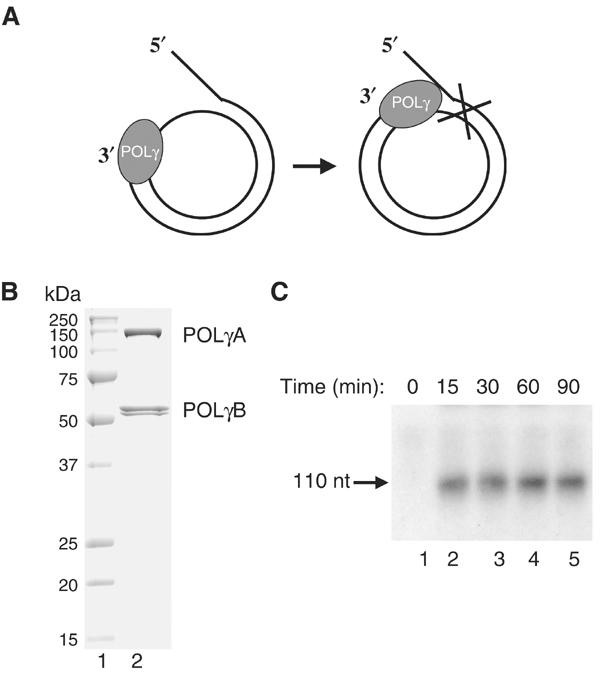
POLγ cannot use dsDNA as template for DNA synthesis. (A) The mini-circle template was prepared as described in Materials and methods. DNA synthesis is initiated at the 3′-hydroxy terminus and proceeds 20 nt before it encounters the dsDNA region of the template. (B) Recombinant POLγ (1 μg) purified over heparin sepharose was separated by SDS–PAGE (12.5%) and revealed with Coomassie brilliant blue staining. (C) POLγ (300 fmol) was incubated together with the mini-circle template at 37°C in a total reaction mixture of 75 μl as described in Materials and methods. At times indicated, 10 μl was removed and the reaction was terminated by addition of 10 μl of gel-loading buffer (98% formamide, 10 mM EDTA (pH 8.0), 0.025% xylene cyanol FF, and 0.025% bromophenol blue). The samples were analyzed on a 10% denaturing polyacrylamide gel.
TWINKLE is unable to unwind longer stretches of dsDNA
We annealed 32P-labeled oligonucleotides to the complementary regions of M13mp18 ssDNA to form helicase substrates with either a 20- or 55-bp double-stranded region and a 40-nt-long 5′ single-stranded tail. TWINKLE could efficiently unwind the 20-bp substrate (Figure 2A), but no unwinding was observed with the 55-bp substrate (Figure 2B). We have previously reported that mtSSB stimulates TWINKLE-dependent unwinding of short 20-bp dsDNA substrate (Korhonen et al, 2003), but we now failed to observe any effect of mtSSB on unwinding of the 55-bp dsDNA substrate. We used the replicative helicase–primase complex from herpes simplex virus type 1 (UL5/8/52) as a positive control. The UL5/8/52 complex is unable to unwind longer stretches of dsDNA, whereas it can unwind the 55-bp template in the presence of its cognate single-stranded DNA (ssDNA)-binding protein, ICP8 (Figure 2B) (Crute and Lehman, 1991). The inability of TWINKLE to unwind longer stretches of dsDNA, even in the presence of mtSSB, seemed to argue against a role for the protein at the mitochondrial replication fork.
Figure 2.
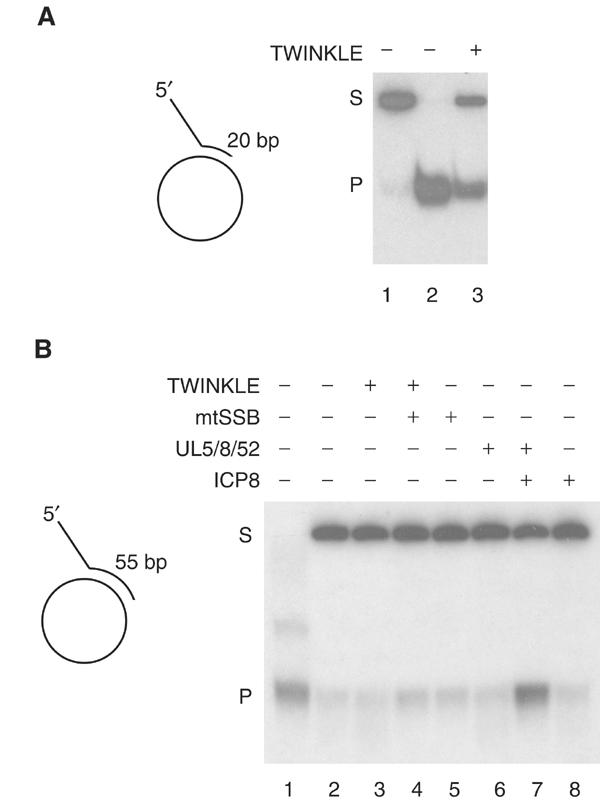
TWINKLE cannot unwind longer stretches of double-stranded DNA, even in the presence of mtSSB. (A) TWINKLE was added to a reaction mixture containing the short 20-bp double-stranded template as described in Materials and methods, and incubated for 30 min. Lane 1, untreated substrate; lane 2, substrate heated to 100°C before loading; lane 3, 550 fmol Twinkle; S, double-stranded substrate; P, single-stranded product. (B) TWINKLE is unable to unwind a 55-bp double-stranded template. Lane 3, 550 fmol Twinkle; lane 4, 550 fmol TWINKLE and 5 pmol mtSSB; lane 5, 5 pmol mtSSB; lane 6, 550 fmol UL5/52/8; lane 7, 550 fmol UL5/52/8 and 5 pmol ICP8; lane 8, 5 pmol ICP8.
TWINKLE and POL_γ_ can efficiently replicate the mini-circle template
We incubated the mini-circle template with POLγ at a molar ratio of 1:1 and added increasing amounts of TWINKLE to the reactions (Figures 3A and C; lanes 4–7). TWINKLE alone was unable to unwind the mini-circle template (Figure 3B), but caused a dramatic stimulation of POLγ-dependent DNA synthesis, allowing the polymerase to utilize dsDNA as template for rolling-circle synthesis of ssDNA molecules of about 2000 nt (Figure 3C). The stimulatory effect was first observed at a molar ratio of about 2:1 of the TWINKLE hexamer to POLγ (Figure 3C, lane 5). We thus conclude that although TWINKLE alone is unable to unwind longer stretches of dsDNA it can functionally interact with POLγ and support processive unwinding of dsDNA at a preformed replication fork. If TWINKLE indeed serves as the helicase at the mtDNA replication fork, it must support unwinding of the entire 16.6-kb mtDNA genome. We therefore investigated if mtSSB could stimulate the DNA synthesis further.
Figure 3.
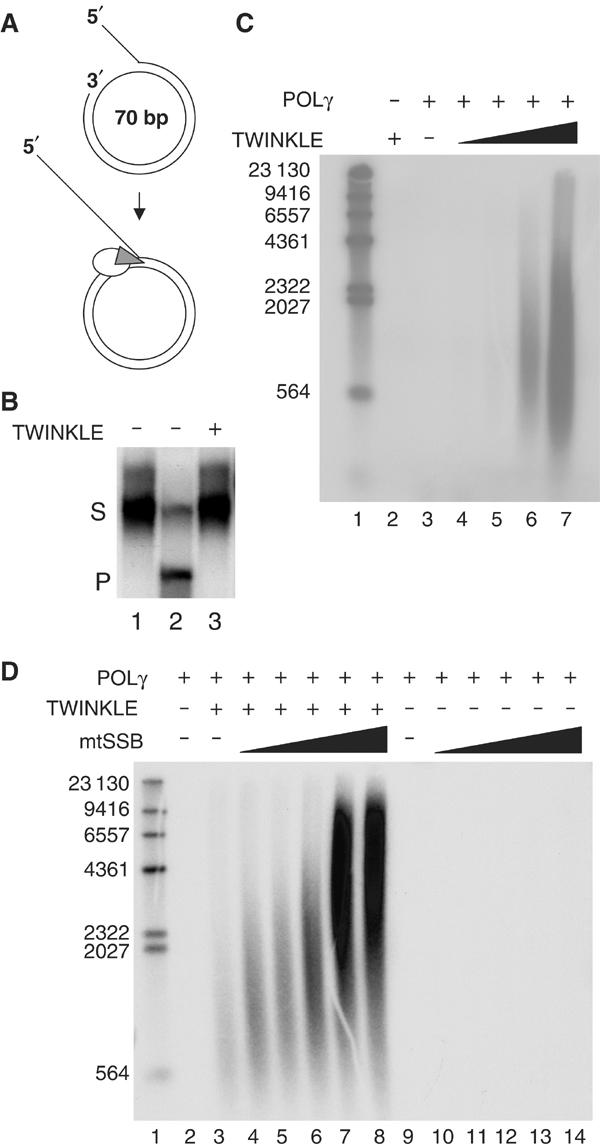
Rolling-circle DNA replication. (A) The mini-circle template was prepared as described in Materials and methods, and [α-32P]dCTP was used to preferentially label the leading strand. (B) TWINKLE alone cannot unwind the mini-circle template. TWINKLE was added to a reaction mixture containing the mini-circle template as described in Materials and methods, and incubated for 30 min. Lane 1, untreated substrate; lane 2, substrate heated to 100°C before loading; lane 3, 550 fmol TWINKLE; S, double-stranded substrate; P, single-stranded product. (C) Increasing amounts of TWINKLE were added to the mini-circle template together with 100 fmol POLγ as described under Materials and methods, and then analyzed on a 0.8% denaturating agarose gel. Lane 1, size marker; lane 2, 1.65 pmol TWINKLE (hexamer); lane 3, 0 fmol TWINKLE; lane 4, 55 fmol TWINKLE; lane 5, 180 fmol TWINKLE; lane 6, 550 fmol TWINKLE; lane 7, 1.65 pmol Twinkle. (D) A constant amount of TWINKLE (550 fmol, hexamer) and POLγ (100 fmol) was added when indicated, together with an increasing amount of mtSSB and analyzed as above. Lane 1, size marker; lane 3, 0 pmol mtSSB; lane 4, 0.1 pmol mtSSB; lane 5, 0.5 pmol mtSSB; lane 6, 1.0 pmol mtSSB; lane 7, 5 pmol mtSSB; lane 8, 10 pmol mtSSB; lane 9, 0 pmol mtSSB; lane 10, 0.1 pmol mtSSB; lane 11, 0.5 pmol mtSSB; lane 12, 1.0 pmol mtSSB; lane 13, 5 pmol mtSSB; lane 14, 10 pmol mtSSB.
We added increasing concentrations of mtSSB in the presence of constant amounts of both TWINKLE and POLγ to the mini-circle template (Figure 3D). The mtSSB had a strong stimulatory effect on the DNA synthesis reaction and allowed the formation of more than 15 000-nt-long stretches of ssDNA (Figure 3D; lanes 3–8) The observed stimulatory effect of mtSSB was dependent on the presence of TWINKLE, since we observed no additional DNA synthesis activity when mtSSB was added to POLγ on its own (Figure 3D; lanes 10–14). As expected for POLγ-dependent DNA synthesis, the rolling-circle DNA replication assay was inhibited by ddCTP, but unaffected by aphidicolin (Supplementary Figure S1).
The TWINKLE helicase activity requires NTP hydrolysis (Korhonen et al, 2003). We therefore monitored DNA synthesis in the absence and presence of different nucleoside 5′-triphosphates (NTPs) (Figure 4). In the absence of NTPs, no DNA synthesis reaction was observed. When added, both ATP and GTP could efficiently support rolling-circle replication, whereas UTP and CDP were poor coeffectors. We conclude that the TWINKLE helicase activity indeed unwinds dsDNA at the mini-circle DNA replication fork, because the in vitro DNA synthesis reaction is dependent on ATP or GTP.
Figure 4.
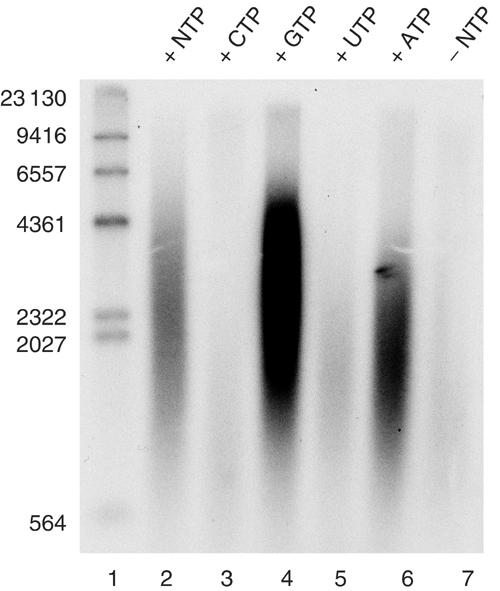
The mitochondrial replication machinery can utilize both ATP and GTP as energy sources. TWINKLE (0.55 pmol), DNA POLγ (100 fmol), and mtSSB (5 pmol) were added to the reaction, together with 4 mM of the indicated NTP. In lane 2, we added 1 mM of each NTP. The reactions were incubated at 37°C for 60 min, and treated as described in Materials and methods. The reactions were analyzed on a 0.8% denaturating agarose gel.
The functional interaction between POL_γ_ and TWINKLE is specific
TWINKLE-dependent stimulation of POLγ appears specific, since TWINKLE failed to stimulate the heterologous T7 DNA polymerase (Figure 5B) although using the same conditions the T7 DNA polymerase was highly active on a single-stranded template (Figure 6D). TWINKLE also failed to stimulate the T4 bacteriophage DNA polymerase even in the presence of the T4 ssDNA-binding protein, gp32 (Figure 5B). The observed specificity could reflect transient protein–protein interactions formed only at the migrating mitochondrial DNA replication fork. In support of this notion, TWINKLE and POLγ do not form a stable complex during gel filtration (data not shown). We also substituted TWINKLE with the heterologous UL5/8/52 helicase (Figure 5A), which has previously been shown to support T7 DNA polymerase-dependent DNA synthesis on a double-stranded template (Falkenberg et al, 1998). There was no stimulation of DNA synthesis activity by the UL5/8/52 helicase under conditions where TWINKLE produced a strong stimulation of POLγ activity. In the presence of its cognate ssDNA-binding protein (ICP8), the herpes UL5/8/52 helicase is highly processive and can unwind long stretches of dsDNA (Falkenberg et al, 1998). The ssDNA formed in this way can be used as template for POLγ-dependent DNA synthesis (data not shown).
Figure 5.
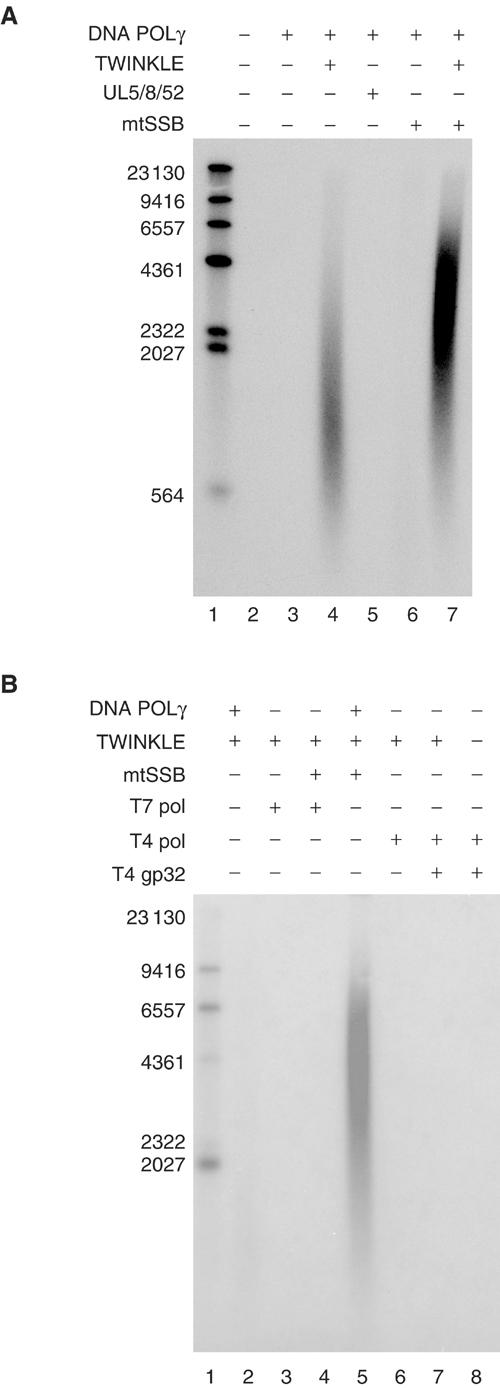
(A) The UL5/8/52 DNA helicase cannot replace TWINKLE at the mtDNA replication fork. We added POLγ (100 fmol), TWINKLE (0.9 pmol), mtSSB (5 pmol), and UL5/52/8 (0.9 pmol). The reactions were incubated for 60 min at 37°C and treated as described under Materials and methods. (B) The T4 and T7 DNA polymerases cannot replace POLγ at the mtDNA replication fork. The replication reactions were as described in (A), with T4 pol (1 U), T7 (1 U), and T4 gp32 (5 pmol). Reactions were performed with 4 mM ATP, but identical results were obtained with GTP as cofactor (data not shown).
Figure 6.
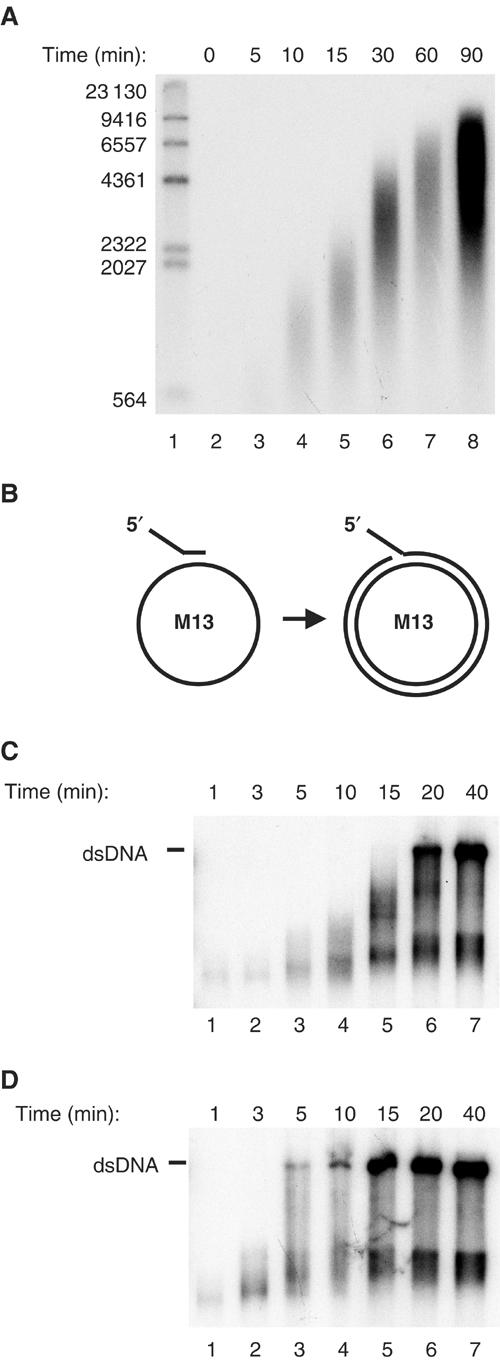
The mitochondrial DNA replication machinery can synthesize ssDNA at a rate of 180 bp/min on the dsDNA mini-circle template. (A) We incubated TWINKLE (2.7 pmol), DNA POLγ (500 fmol, mtSSB (25 pmol), and the mini-circle template (175 fmol) at 37°C in a total reaction volume of 125 μl as described in Materials and methods. At times indicated, we removed 12.5 μl and the reaction was stopped as described in Materials and methods. The reactions were analyzed on a 0.8% denaturating agarose gel. (B) A schematic representation of the single-stranded M13 mp18 DNA template used to measure DNA synthesis rate. (C) The DNA synthesis rate of POLγ on single-stranded M13 mp18 DNA in the presence of mtSSB was about 350 nt/min. DNA POLγ (500 fmol) and 25 pmol mtSSB were incubated together with a primed M13 ssDNA at 37°C in a total reaction mixture of 125 μl as described in Materials and methods. At times indicated, 12.5 μl was removed and the reaction was terminated by addition of 3-μl of stop solution. The reactions were analyzed by nondenaturing agarose gel electrophoresis. (D) The rate of T7 DNA polymerase together with mtSSB on ssDNA was 1500 nt/min. The reactions were as described in (C), but T7 DNA polymerase (5 U) was used instead of POLγ.
The minimal replisome DNA synthesis rate
The mini-circle template was incubated with TWINKLE, POLγ, and mtSSB and the reaction was followed in a time-course experiment. We observed a linear increase in the size of the ssDNA products and calculated an estimated rate constant of 180 nt/min (Figure 6A). For comparison, we measured the DNA synthesis rate of both POLγ and T7 DNA polymerase on single-stranded M13 mp18 DNA (Figure 6B) in the presence of mtSSB and obtained values of about 350 nt/min for POLγ (Figure 6C) and 1500 nt/min for T7 DNA polymerase (Figure 6D). We could thus conclude that, in the presence of TWINKLE, the DNA synthesis rate of POLγ on dsDNA is only marginally lower than the rate observed on an ssDNA template. The DNA synthesis rate of 180 bp/min corresponds closely to the calculated value of 270 bp/min for in vivo mtDNA replication.
Discussion
We have combined TWINKLE, POLγ, and mtSSB and thus reconstituted the human mitochondrial DNA minimal replisome in vitro. The reconstituted system can efficiently utilize dsDNA as template and synthesize ssDNA molecules of more than 15 000 nt in length. The estimated rate of the system was 180 nt/min, which corresponds well to the estimated in vivo replication rate of 270 nt/min (Clayton, 1982). The functional interactions observed between the mitochondrial DNA replication proteins appear specific, since the TWINKLE helicase cannot support T4 and T7 DNA polymerase-dependent rolling-circle DNA replication, even in the presence of the T4 ssDNA-binding protein.
The human mitochondrial and bacteriophage T7 replication machineries are structurally and functionally related. TWINKLE shares primary sequence homology with the gp4 helicase–primase (Spelbrink et al, 2001). Both TWINKLE and gp4 catalyze ATP-dependent unwinding of a DNA duplex with a distinct polarity (5′ → 3′) (Ahnert and Patel, 1997; Korhonen et al, 2003). The proteins also have very similar helicase substrate requirements with a single-stranded 5′-DNA-loading site and a short 3′-tail to initiate unwinding. Sequence similarities also exist between the T7 DNA polymerase and POLγ, both of which are classified as family A DNA polymerases (Ito and Braithwaite, 1990; Ye et al, 1996). The catalytic subunit POLγ A associates with the processivity factor POLγ B, which allows for efficient utilization of RNA primers. POLγ B is structurally homologous to tRNA synthetases and appears to have been added ad hoc to the replication machinery during evolution (Carrodeguas et al, 2001). The T7 DNA polymerase forms a heterodimer with the bacterially encoded thioredoxin, which functions as a processivity factor for the bacteriophage polymerase. The stimulatory activity is distinct from the role of thioredoxin in redox regulation and rather reflects a structural role for the protein at the DNA replication fork. The X-ray structure of the T7 DNA polymerase including thioredoxin has been determined at 2.2 Å resolution, and suggests two possible mechanisms for increased processivity. The thioredoxin could swing across the DNA-binding groove to encircle the DNA or it could simply extend the DNA-binding site of the T7 DNA polymerase (Huber et al, 1987; Doublie et al, 1998).
The replication mechanism differs between the two bacteriophages T4 and T7. The T4 DNA replication machinery includes a dimeric DNA polymerase. The polymerase subunit on the lagging-strand recycles in an ATP-dependent manner and requires a sliding clamp and a clamp loader. In contrast, recycling of the processive T7 DNA polymerase is independent of ATP and it does not form a homodimer. It has instead been suggested that physical interactions between the T7 ssDNA-binding protein and the leading-strand polymerase coordinate leading- and lagging-strand synthesis (Benkovic et al, 2001). POLγ is also monomeric and no sliding clamp or clamp loader have been identified in mitochondria. It has recently been suggested that mtDNA synthesis is not strand-asymmetric and that leading- and lagging-strand mtDNA synthesis are coordinated at the replication fork (Holt et al, 2000; Yang et al, 2002; Holt and Jacobs, 2003). In this respect, it is interesting to note that the monomeric T7 ssDNA-binding protein, which coordinates leading- and lagging-strand synthesis in the bacteriophage, is structurally distinct from the mtSSB. The mtSSB is instead similar to the tetrameric Escherichia coli ssDNA-binding protein, SSB (Lohman and Ferrari, 1994), which is not essential for strand-coordinated DNA synthesis at the bacterial replication fork (Benkovic et al, 2001). If coordinated leading- and lagging-strand DNA synthesis takes place at the mitochondrial replication fork, the coordination must be achieved by mechanisms, which are functionally distinct from what has previously been described in and E. coli, bacteriophage T4, and T7.
UTP is an efficient cofactor for the TWINKLE helicase activity, but only supports low levels of rolling-circle DNA replication. It is thus possible that interactions with POLγ at the DNA replication fork induce a structural change in TWINKLE, which increases processivity and at the same time changes the nucleotide triphosphate specificity. In agreement with this notion, GTP constantly generates higher levels of rolling-circle DNA replication than ATP, although both GTP and ATP are potent activators of the TWINKLE helicase activity (Korhonen et al, 2003).
We believe that the data presented here represent an important step toward a biochemical understanding of mammalian mitochondrial DNA replication, but also have certain limitations. The mtDNA is negatively supercoiled in vivo, which may assist unwinding of the DNA during replication and further increase the rate of DNA synthesis. In addition, the mtDNA replicated by the mitochondrial replication machinery in vivo will not be naked, but in complex with a variety of proteins. The mammalian TFAM protein binds nonspecifically and may in fact fully coat mtDNA in mammalian cells. How TFAM binding to mtDNA affects the minimal replisome is an open question. In the future, we will need to address these and other related questions, in order to reach a complete understanding of the mitochondrial replication machinery. We will in future studies also utilize the reconstituted minimal replisome to investigate naturally occurring mutations in POLγ and TWINKLE, which have been shown to cause adPEO, a mitochondrial disorder associated with multiple mtDNA deletions. These mutations may help us to better understand how the mitochondrial DNA polymerase and TWINKLE work together at the replication fork. We will also characterize how the replication machinery responds to damage DNA templates and how it interacts with the repair systems present in the mammalian mitochondria.
Materials and methods
Recombinant proteins
We used human cDNA as a template and amplified a DNA fragment encoding the B subunit of POLγ by PCR. A cDNA plasmid encoding the A subunit of POLγ was a gift from Dr William C Copeland, National Institute of Environmental Health Sciences, NC, USA. We cloned the fragments encoding the two subunits of POLγ into the vector pBacPAK9 (Clontech), creating pBac-POLγA and pBac-POLγB. We used the plasmid constructs to prepare Autographa californica nuclear polyhedrosis virus recombinant for the proteins as described in the BacPAK manual (Clontech). The generation of recombinant baculoviruses expressing mtSSB and TWINKLE has been described earlier (Korhonen et al, 2003). The TWINKLE and mtSSB proteins were isolated from Spodoptera frugiperda (Sf9) cells as described. For purification of POLγ, we co-infected Sf9 cells with recombinant baculoviruses encoding His6-tagged versions of POLγ subunits A and B. Whole-cell extract was generated and recombinant POLγ was purified on Ni2+-agarose, as previously described for the human mitochondrial RNA polymerase (Falkenberg et al, 2002). The peak of eluted protein was dialyzed against buffer B (20 mM Tris–HCl (pH 8.0), 0.5 mM EDTA (pH 8.0), 1% glycerol, and 1 mM DTT) containing 0.1 M NaCl. The proteins were further purified on a 1-ml HiTrap Heparin column (Amersham Biosciences) equilibrated in buffer B (0.1 M NaCl). After washing the column with three column volumes of buffer B (0.1 M NaCl), we used a linear gradient (10 ml) of buffer B (0.1–1.0 M NaCl) to elute the column. The POLγA was eluted as a complex with POLγB at about 0.8 M NaCl. The yield from 400 ml of culture was 4 mg. We estimated the purity of the proteins to be at least 95% by SDS–PAGE with Coomassie blue staining. In the text, the holoenzyme containing both POLγA and POLγB will be referred to POLγ.
The UL5/8/52 helicase–primase and the ICP8 single-stranded protein was purified as described (Falkenberg et al, 2000), and was a kind gift from Dr Per Elias, Göteborg University, Sweden.
DNA substrates
To generate the helicase substrates, a 60-nt oligonucleotide (5′-ACATGATAAGATACATGGATGAGTTTGGACAA ACCACAACGTAAAACGACGGCCAGTGCC-3′) or a 95-nt oligonucleotide (5′-ACATGATAAGATACATGGATGAGTTTGGACAA ACCACAACGTAAAACGACGGCCAGTGCCAAGCTTG CATGCCTGCAGGTCGACTCTAGAGGATC-3′) was labeled with 32P at its 5′-terminus with T4 polynucleotide kinase (Stratagene) and annealed to M13mp18 ssDNA (Amersham Biosciences) to generate a 20-bp (short substrate) or 55-bp double-stranded (long substrate) region with a 40-nucleotide 5′ tail. Unannealed oligonucleotides were removed by filtration through an ultra-filter with a 100-kDa molecular weight cutoff (Centricon 100 from Amicon), in a buffer containing 20 mM Tris–HCl (pH 7.6), 100 mM NaCl, and 0.1 mM EDTA.
The mini-circle template for rolling-circle DNA replication was generated as previously described (Falkenberg et al, 2000). We also prepared a mini-circle substrate for helicase assays following the same protocol, but with the 90-mer labeled with 32P at its 5′-terminus before annealing.
Helicase assay
The reaction mixture (25 μl) contained 15 fmol of DNA substrate (short, long, or mini-circle substrate), 25 mM Tris–HCl (pH 7.6), 10 mM magnesium chloride, 1 mM DTT, 100 μg/ml BSA, 4 mM ATP, 10% glycerol, 250 μM dATP, 250 μM dTTP, 250 μM dGTP, and 250 μM dCTP. Proteins (TWINKLE, the UL5/UL52/UL8 helicase–primase, mtSSB, and ICP8) were added as indicated in the figure legends. The reactions were incubated at 32°C for 50 min and stopped by the addition of 3 μl of stop solution (90 mM EDTA (pH 8.0), 6% SDS, 30% glycerol, 0.25% bromophenol, and 0.25% xylene cyanol). We did not observe any significant levels of spontaneous reannealing of unwound DNA under the assay conditions used. The products were separated by electrophoresis through a 10% nondenaturing polyacrylamide gel, which was dried onto DE81 (Whatman) and autoradiographed overnight at −80°C with an intensifying screen.
DNA synthesis on the mini-circle template
The mini-circle template (final concentration 1.4 nM) was added to a reaction mixture containing 25 mM Tris–HCl (pH 7.6), 10 mM magnesium chloride, 1 mM DTT, 100 μg/ml BSA, 4 mM ATP, 10% glycerol, 250 μM dATP, 250 μM dTTP, 250 μM dGTP, 10 μM dCTP, 2 μCi [α-32P]dCTP, and the different amounts of replication factors indicated in the figure legends. The hexameric concentration is used for TWINKLE. The reaction was incubated at 37°C and terminated after 90 min if nothing else is indicated. For detection of short DNA products, an equal amount of gel-loading buffer (98% formamide, 10 mM EDTA (pH 8.0), and 0.025% xylene cyanol FF, 0.025% bromophenol blue) was added to the reactions. The samples were heated at 95°C for 5 min, and analyzed on a 10% denaturing polyacrylamide gel in 1 × TBE buffer, dried onto DE81 (Whatman), and autoradiographed overnight at −80°C with an intensifying screen.
For detection of longer, rolling-circle DNA products, we stopped the reactions at the times indicated by adding 200 μl of stop buffer (10 mM Tris–HCl (pH 8.0), 0.2 M NaCl, 1 mM EDTA, and 0.1 mg/ml glycogen). We treated the samples with 0.5% SDS and 100 μg/ml proteinase K for 45 min at 42°C, and precipitated them by adding 0.6 ml of ice-cold 95% ethanol. We dissolved the pellets in 20 μl water. Denaturing agarose gel electrophoresis was performed by adding 4 μl of alkaline loading buffer containing 18% (w/v) ficoll, 300 mM NaOH, 60 mM EDTA (pH 8.0), 0.15% (w/v) bromocresol green, and 0.25% (w/v) xylene cyanol FF to the reaction mixture. The samples were subjected to electrophoresis through a 0.8% alkaline agarose gel in 50 mM NaOH and 1 mM EDTA at 1.5 V/cm for 20 h, and then dried onto DE81 (Whatman) and autoradiographed for 2 h at −80°C with an intensifying screen.
Measurement of DNA polymerase activity
The reaction mixture (125 μl) contained 25 mM Tris–HCl (pH 8.0), 1 mM dithiothreitol, 10 mM MgCl2, 4 mM ATP, 250 μM dATP, dGTP, dTTP, 10 μM dCTP, 2 μCi of [α-32P]dCTP (3000 Ci/mmol), and 20 fmol of tailed M13 mp18 ssDNA (the short helicase substrate). The reaction mixture also contained 50 pmol mtSSB and either 500 fmol of POLγ or 5 U of T7 DNA polymerase. Incubation was at 37°C and at the times indicated 12.5 μl samples were removed and the reaction was stopped by addition of 3 μl of stop solution (90 mM EDTA (pH 8.0), 6% SDS, 30% glycerol, 0.25% bromophenol blue, and 0.25% xylene cyanol). The samples were analyzed by gel electrophoresis in 0.8% agarose gels in Tris–borate–EDTA (TBE) buffer. The gels were run at 150 V for 3 h and after drying they were autoradiographed with an intensifying screen.
Supplementary Material
Supplementary Figure
Acknowledgments
Work in the MF laboratory was supported by grants from the Swedish Research Council, Åke Wiberg Foundation, Magn. Bergwalls Foundation, Emil and Wera Cornell's foundation, Jeansson's foundations, and the Swedish National Board for Laboratory Animals.
References
- Ahnert P, Patel SS (1997) Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J Biol Chem 272: 32267–32273 [DOI] [PubMed] [Google Scholar]
- Benkovic SJ, Valentine AM, Salinas F (2001) Replisome-mediated DNA replication. Annu Rev Biochem 70: 181–208 [DOI] [PubMed] [Google Scholar]
- Carrodeguas JA, Kobayashi R, Lim SE, Copeland WC, Bogenhagen DF (1999) The accessory subunit of Xenopus laevis mitochondrial DNA polymerase gamma increases processivity of the catalytic subunit of human DNA polymerase gamma and is related to class II aminoacyl-tRNA synthetases. Mol Cell Biol 19: 4039–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrodeguas JA, Theis K, Bogenhagen DF, Kisker C (2001) Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer. Mol Cell 7: 43–54 [DOI] [PubMed] [Google Scholar]
- Chang DD, Clayton DA (1985) Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci USA 82: 351–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DA (1982) Replication of animal mitochondrial DNA. Cell 28: 693–705 [DOI] [PubMed] [Google Scholar]
- Crute JJ, Lehman IR (1991) Herpes simplex virus-1 helicase-primase. Physical and catalytic properties. J Biol Chem 266: 4484–4488 [PubMed] [Google Scholar]
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature 391: 251–258 [DOI] [PubMed] [Google Scholar]
- Engler MJ, Lechner RL, Richardson CC (1983) Two forms of the DNA polymerase of bacteriophage T7. J Biol Chem 258: 11165–11173 [PubMed] [Google Scholar]
- Falkenberg M, Elias P, Lehman IR (1998) The herpes simplex virus type 1 helicase-primase. Analysis of helicase activity. J Biol Chem 273: 32154–32157 [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM (2002) Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet 31: 289–294 [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Lehman IR, Elias P (2000) Leading and lagging strand DNA synthesis in vitro by a reconstituted herpes simplex virus type 1 replisome. Proc Natl Acad Sci USA 97: 3896–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr CL, Matsushima Y, Lagina AT III, Luo N, Kaguni LS (2004) Physiological and biochemical defects in functional interactions of mitochondrial DNA polymerase and DNA-binding mutants of single-stranded DNA-binding protein. J Biol Chem 279: 17047–17053 [DOI] [PubMed] [Google Scholar]
- Fernandez-Silva P, Enriquez JA, Montoya J (2003) Replication and transcription of mammalian mitochondrial DNA. Exp Physiol 88: 41–56 [DOI] [PubMed] [Google Scholar]
- Holt IJ, Jacobs HT (2003) Response: the mitochondrial DNA replication bubble has not burst. Trends Biochem Sci 28: 355–356 [DOI] [PubMed] [Google Scholar]
- Holt IJ, Lorimer HE, Jacobs HT (2000) Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 100: 515–524 [DOI] [PubMed] [Google Scholar]
- Huber HE, Tabor S, Richardson CC (1987) Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J Biol Chem 262: 16224–16232 [PubMed] [Google Scholar]
- Ito J, Braithwaite DK (1990) Yeast mitochondrial DNA polymerase is related to the family A DNA polymerases. Nucleic Acids Res 18: 6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Miyako K, Kai Y, Irie T, Takeshige K (1997) In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J Biol Chem 272: 15275–15279 [DOI] [PubMed] [Google Scholar]
- Korhonen JA, Gaspari M, Falkenberg M (2003) TWINKLE has 5′ → 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem 278: 48627–48632 [DOI] [PubMed] [Google Scholar]
- Lohman TM, Ferrari ME (1994) Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem 63: 527–570 [DOI] [PubMed] [Google Scholar]
- Maier D, Farr CL, Poeck B, Alahari A, Vogel M, Fischer S, Kaguni LS, Schneuwly S (2001) Mitochondrial single-stranded DNA-binding protein is required for mitochondrial DNA replication and development in Drosophila melanogaster. Mol Biol Cell 12: 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G (1982) Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc Natl Acad Sci USA 79: 7195–7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberson DL, Kasamatsu H, Vinograd J (1972) Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc Natl Acad Sci USA 69: 737–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropp PA, Copeland WC (1996) Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics 36: 449–458 [DOI] [PubMed] [Google Scholar]
- Shadel GS, Clayton DA (1997) Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem 66: 409–435 [DOI] [PubMed] [Google Scholar]
- Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, Larsson C (2001) Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet 28: 223–231 [DOI] [PubMed] [Google Scholar]
- Tapper DP, Clayton DA (1981) Mechanism of replication of human mitochondrial DNA. Localization of the 5′ ends of nascent daughter strands. J Biol Chem 256: 5109–5115 [PubMed] [Google Scholar]
- Yang MY, Bowmaker M, Reyes A, Vergani L, Angeli P, Gringeri E, Jacobs HT, Holt IJ (2002) Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell 111: 495–505 [DOI] [PubMed] [Google Scholar]
- Ye F, Carrodeguas JA, Bogenhagen DF (1996) The gamma subfamily of DNA polymerases: cloning of a developmentally regulated cDNA encoding Xenopus laevis mitochondrial DNA polymerase gamma. Nucleic Acids Res 24: 1481–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure