A neurodegeneration-specific gene expression signature and immune profile of acutely isolated microglia from an ALS mouse model (original) (raw)
. Author manuscript; available in PMC: 2014 Dec 20.
Abstract
Microglia are resident immune cells of the CNS that are activated by infection, neuronal injury and inflammation. Here we utilize flow cytometry and deep RNA sequencing of acutely isolated spinal cord microglia to define their activation in vivo. Analysis of resting microglia identified 29 genes that distinguish microglia from other CNS cells and peripheral macrophages/monocytes. We then analyzed molecular changes in microglia during neurodegenerative disease activation using the SOD1G93A mouse model of ALS. We find that SOD1G93A microglia are not derived from infiltrating monocytes, and that both potentially neuroprotective and toxic factors are concurrently up-regulated, including Alzheimer’s disease genes. Mutant microglia differed from SOD1WT, LPS activated microglia, and M1/M2 macrophages, that define an ALS-specific phenotype. Concurrent mRNA/FACS analysis revealed post-transcriptional regulation of microglia surface receptors, and T cell-associated changes in the transcriptome. These results provide insights into microglia biology and establish a resource for future studies of neuroinflammation.
Keywords: Microglia, transcriptome, FACS, ALS, neuroimmunology, neurodegeneration
Introduction
Microglia are motile cells that dynamically survey the CNS parenchyma for invading pathogens and cell death (Nimmerjahn et al., 2005). In addition, microglia actively participate in synaptic pruning during development (Schafer et al., 2012). In response to tissue injury, microglia undergo a rapid transition from resting, ramified forms to amoeboid morphologies (Davalos et al., 2005). At present, little is known about molecular changes that occur in microglia during their activation, and whether these changes are beneficial or harmful to neuron survival. Specific transcriptional responses of microglia may vary depending on the neuro-pathological condition and type of molecular stimuli encountered (Ransohoff and Perry, 2009).
A common feature of neurodegenerative diseases is the presence of activated microglia in areas of neuronal death (Frank-Cannon et al., 2009). ALS is a neurodegenerative disease characterized by progressive loss of motor neurons, leading to paralysis and death. Microglia activation occurs robustly in ALS patient tissue and in spinal cords of mutant SOD1 transgenic mice (Hall et al., 1998; Mcgeer and Mcgeer, 2002). Analysis of chimeric mice have shown that myeloid cells expressing mutant SOD1 is harmful to neurons (Beers et al., 2006; Boillee et al., 2006), consistent with a toxic role of microglia or peripheral macrophages, which infiltrate motor axons during disease progression (Chiu et al., 2009; Dibaj et al., 2011; Graber et al., 2010). The depletion of proliferating microglia does not affect survival in ALS mice (Gowing et al., 2008). Additional studies have shown that T cells enter the ALS spinal cord and slow disease progression by directing microglia to express neurotrophic factors (Beers et al., 2008; Chiu et al., 2008). Thus, the complex functional role of microglia ALS remains to be determined.
Myeloid immune cells induce specific transcriptional programs in response to various inflammatory stimuli (Gordon, 2003). In macrophages, classical activation by TH1 cytokine IFN-γ or Lipopolysaccharides (LPS) activates an “M1” phenotype, while the TH2 cytokine IL-4 induces “M2” alternative macrophages (Gordon, 2003). Although microglia are ontologically distinct from peripheral macrophages, they have nevertheless been classified as M1 or M2-like in ALS and Alzheimer’s disease (Beers et al., 2011; Hickman et al., 2008; Michelucci et al., 2009). ALS microglia have been reported to be pro-inflammatory and neurotoxic in vitro (Sargsyan et al., 2011; Xiao et al., 2007). However, the situation in the diseased spinal cord, where molecular changes are likely influenced by a myriad of surrounding cell-types, is likely to be more complex. Thus, the analysis of acutely isolated microglia from the diseased spinal cord may accurately reflect the complex micro-environment of microglia in diseased spinal cord,.
In this study, we perform molecular and immunological profiling of microglia acutely isolated from ALS transgenic and LPS injected mice. Correlative analysis of spinal cord T cells, surface protein expression, and transcriptome changes reveals intricate phenotypic changes occurring in microglia during activation in vivo.
Results
Deep RNA sequencing of the spinal cord microglia transcriptome
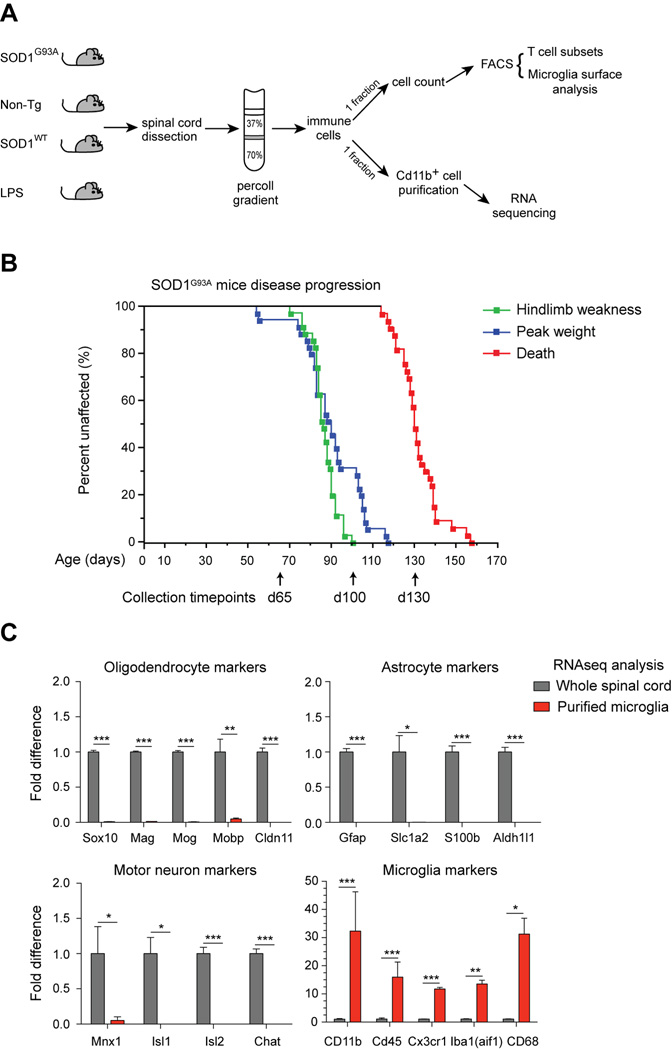
To analyze the transcriptomes of acutely isolated microglia during activation, spinal cords were dissociated and centrifuged over percoll gradients to isolate leukocytes (Sedgwick et al., 1991). One fraction of leukocytes was analyzed by flow cytometry to collect data on lymphocyte and microglia surface phenotypes (Fig. 1A), and a separate fraction was used for microglia purification using CD11b magnetic separation, as described previously (Cardona et al., 2006a). CD11b+CD45lo microglia were obtained with >96% purity from all samples at different disease timepoints (Supplementary Fig. S1). Total RNA was extracted and deep sequenced with paired ends, resulting in transcriptome data that could be directly correlated with FACS analysis of the spinal cords (Fig. 1A).
Fig. 1.
Transcriptional profiling of purified spinal cord microglia and parallel flow cytometry analysis. (A) Percoll gradient isolated leukocytes from SOD1G93A, non-Tg littermate, SOD1WT, and LPS injected mice were subjected to FACS analysis (T cells, microglia) and CD11b+ microglia purification for RNAseq. (B) Kaplan Meier curves of disease progression in SOD1G93A mice (n=35), showing onset of hindlimb weakness, peak weight, and death. Spinal cords were dissected at day 65, day 100, and day 130/end stage for microglia analysis. (C) Plots of relative transcript levels of purified microglia compared to whole spinal cord, examining oligodendrocyte (Sox10, Mag, Mog, Mobp, Cldn11), astrocyte (Gfap, Slc1a2, S100b, Aldh1l1), motor neuron (Mnx1, Isl1, Isl2, Chat), and microglia (Cd11b, Cd45, Cx3cr1, Iba1, Cd68) markers. Values are fold-change ± s.e.m. (*, p<0.05; **, p<0.01; ***, p<0.001 by t-test). See also Figure S1.
We used SOD1G93A mice to study microglia activation. These mice display progressive motor neuron degeneration and death (Gurney et al., 1994) (Fig. 1B). Non-Tg and SOD1WT microglia were analyzed as controls. Three timepoints were chosen based on disease progression in our SOD1G93A colony (n=35; 17 females, 18 males, Fig. 1B). Motor weakness onset was defined by observation of hind limb tremors (mean±sem; 86.4 ± 1.05 days of age) and measurement of peak weight (90.23±2.44 days). Disease end-stage occurred at 131.6 ±1.72 days of age. Microglia were isolated at day 65 (pre-symptomatic), day 100 (early symptomatic), and day 130/end-stage (Fig. 1B, arrows and Fig. S1A–B). We also analyzed microglia activated by a single, moderate dose of intra-peritoneal LPS injection, and SOD1G93A microglia from both B6/SJLF1 and B6 strain backgrounds (Fig. S1A).
Purity of microglia was verified by comparison of acutely isolated microglia and whole spinal cord RNAseq data. Oligodendrocyte (Sox10, Mag, Mog, Mobp, Cldn11), astrocyte (Gfap, Slc1a2, S100b, Aldh1l1), and motor neuron (Mnx1, Isl1, Isl2, Chat) markers were not detectable, while known microglia markers (Cd11b, Cd45, Cx3Cr1, Aif1, Cd68) were highly enriched following purification (Fig. 1C). 42 microglia samples were sequenced, 1.251 billion reads mapped to the genome, with an average of 29.79 million reads/sample (Supplemental Table S1). Normalized gene expression levels (reads per kilobase million [RPKM]) showed minimal variation between biological replicates, whereas significant differences were found between disparate sample groups (i.e. mutant or wild-type microglia) (Fig. S1C).
Identification of microglia-specific marker genes
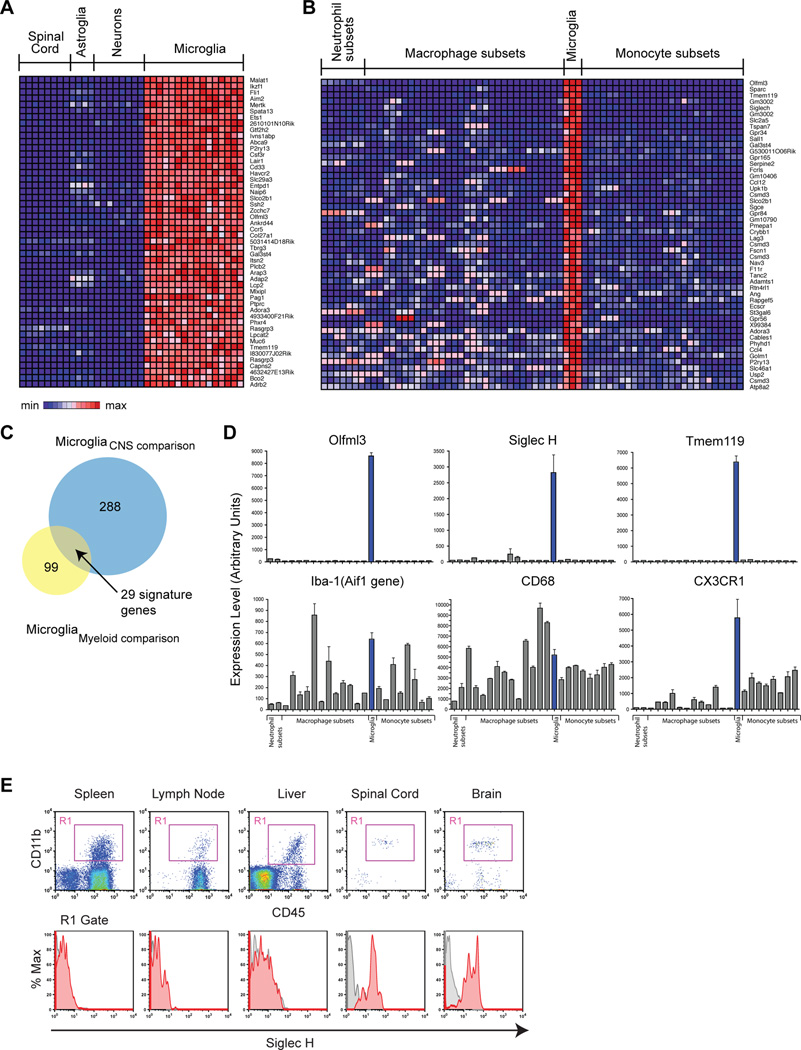
Currently, few specific markers are known that distinguish microglia from peripheral monocytes and macrophages (Frank-Cannon et al., 2009; Ransohoff and Perry, 2009). We used our whole transcriptome analysis to define specific markers for microglia. First, we compared microglia RNAseq data with RNAseq data obtained from astroglia, motor neurons, and whole spinal cord. This comparison yielded 288 genes enriched in microglia by 5-fold compared to these other CNS cell-types (q<0.05; heat-map, Fig. 2A). We next compared microglia microarray data with 22 other myeloid cell-types collected by the Immunological Genome Project (ImmGen), including subsets of neutrophils, peripheral macrophages, and monocytes (Gautier et al., 2012). This comparison yielded 99 genes enriched in microglia 5-fold relative to other myeloid immune cells (q<0.05; heat-map, Fig. 2B). The overlap between these 2 distinct dataset comparisons (Venn diagram, Fig. 2C) yielded 29 highly specific markers for microglia relative to both CNS cell-types and peripheral myeloid immune cells (Table S3). The most enriched microglia genes were Olfml3, Tmem119, and Siglec-H, which were expressed at very high levels in microglia but barely detected in macrophages and monocytes (Fig. 2D, top panels). By contrast, Iba1 (Aif1) and CD68, two widely used microglia markers, showed expression in macrophages, neutrophils, and monocytes (Fig. 2D, bottom panels). Cx3cr1 is extensively used to characterize microglia dynamics in vivo (Cardona et al., 2006b; Nimmerjahn et al., 2005; Schafer et al., 2012). However, Cx3cr1 ranks 24th on our list of 29 microglia enriched genes in comparison to other myeloid cells, and is not as specific as Olfml3, Tmem119, or Siglec-H (Fig. 2D and Table S3). We note that Cx3cr1 remains a good marker for microglia, as it is not expressed on neuroepithelial cells (Cardona et al., 2006a) and is not characteristic of CNS-infiltrating mononuclear myeloid cells (Mizutani et al., 2012).
Fig. 2.
Microglia marker identification by transcriptome comparisons with other CNS and myeloid immune cell-types. (A) Heat map of top 50 genes enriched in microglia compared to CNS cell-types by RNAseq, ranked by fold-change (q<0.05, >5-fold enrichment). (B) Heat map of top 50 genes enriched in microglia compared to neutrophil, macrophage, and monocyte subsets by microarray, ranked by fold-change (q<0.05, >5-fold enrichment). (C) Venn diagrams show that 29 genes distinguish microglia from other CNS and peripheral myeloid cells. (D) Plots of transcript levels across neutrophil, macrophage, monocyte subtypes, and microglia (n=3–5 arrays/group; mean±sem). Olfml3, SiglecH, and Tmem119 are highly enriched markers for microglia. By contrast, Iba1 (Aif1), Cd68, and Cx3cr1 are expressed by other myeloid cell-types. (E) FACS of Siglec H expression on CD11b+CD45+ myeloid cells isolated from spleen, lymph node, liver, spinal cord, and brain of the same animals. Siglec H expression was restricted to CNS microglia (red histograms; grey, unstained cells). Error bars, mean±s.e.m. See also Figure S2.
We confirmed microglia specific expression of Siglec-H and Olfml3 by flow cytometry and immunostaining (Fig. 2E, Supplemental Fig. S2, and Fig. 3C). By FACS, spleen, liver, and lymph node macrophages did not express Siglec-H, while it was expressed on spinal cord and brain microglia (Fig. 2E). We note that although these markers distinguish microglia from peripheral macrophages, they maybe expressed by other immune cells. For example, Siglec-H is a known marker for plasmacytoid dendritic cells (DC), a lymphoid cell-type and source of type 1 interferons (Loschko et al., 2011). We confirmed that splenic and lymph node plasmacytoid DC (CD11b+CD11c+B220+) expressed Siglec-H, but not myeloid DC (CD11b+CD11c+) or macrophages (CD11b+CD11c−) (Fig. S2).
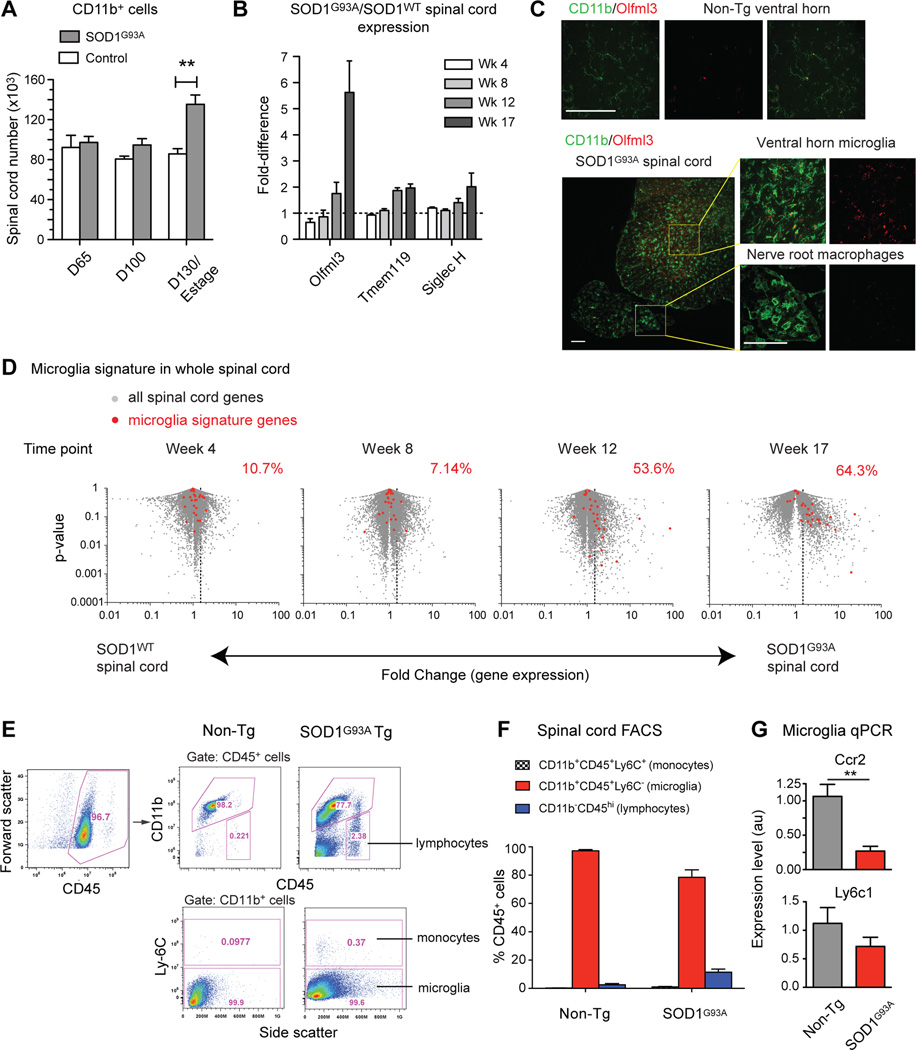
Fig. 3.
SOD1G93A spinal cord immune activation is characterized by resident microglia (CD11b+Olfml3+) and not peripheral monocytes (CD11b+Ly6c+). (A) FACS analysis and cell quantification show increases in CD11b+ myeloid cells in SOD1G93A spinal cords over time. (**, p<0.01). (B) Transcript level comparisons of whole spinal cord RNA show increases in the microglia markers Olfml3, Tmem119, and Siglec H in SOD1G93A vs. SOD1WT mice. (C) Immunostaining shows Olfml3 expression in resting microglia (CD11b+) in non-Tg spinal cord. At end-stage, activated SOD1G93A microglia in ventral horns express Olfml3, but nerve root activated macrophages do not. Scale bars, 100 µm. (D) Microglia signature genes (red) show progressive increases on volcano plots of SOD1G93A vs.SOD1WT spinal cord RNAseq data (fold-change vs. p-value). Percentages denote proportion of microglia genes expressed >1.5-fold (dotted line) in SOD1G93A spinal cord. (E) CD45+ spinal cord leukocytes were analyzed for lymphocyte, monocyte, and microglia populations by FACS. At end-stage, SOD1G93A spinal cords showed greater proportions of CD11b–CD45hi lymphocytes compared to non-Tg controls (2.38% vs. 0.221%). Within CD11b+ myeloid cells, the majority are Ly6C- microglia (99.9% vs. 99.5%), and not Ly6C+ monocytes (0.37% vs. 0.0977%). (F) Quantification of FACS analysis (n=14–16 per group). (G) By qPCR, monocyte markers Ccr2 and Ly6c1 are not upregulated in purified SOD1G93A microglia at end-stage. Relative expression±SEM (**, p<0.01). See also Figure S3.
Resident microglia increase in spinal cords during disease progression while monocytes do not
We carried out FACS analyses to characterize myeloid cell populations in the spinal cords of SOD1G93A mice, and observed an increase in the CD11b+ population with disease progression (Fig. 3A). In addition, we found that the microglia-specific mRNA transcripts Olfml3, Tmem119, and Siglec-H increased over time in whole SOD1G93A spinal cord by RNAseq analysis (Fig. 3B). Immunostaining confirmed that Olfml3 protein was expressed in resting microglia and increased in SOD1G93A spinal cords (Fig. 3C). In the same sections, monocytes known to infiltrate SOD1G93A peripheral ventral roots (Chiu et al., 2009; Graber et al., 2010) were negative for Olfml3, confirming marker specificity for microglia (Fig. 3C). These data indicate that microglia activation may affect the overall spinal cord transcriptional signature. By overlaying the 29 microglia markers onto volcano data plots of spinal cord RNAseq data (p-value vs. fold-change), we detected a significant increase of the microglia signature in SOD1G93A spinal cord with disease progression (Fig. 3D). However, purified SOD1G93A microglia actually showed a decrease in the levels of these microglia transcript markers (Supplemental Fig. S3). These observations suggest that the increase in microglia-specific transcripts observed in whole spinal cords is due to expansion of the number of microglia and not to an increase in the expression of microglia-specific markers on a per cell basis in the spinal cord.
Based on immune cell analysis in SOD1G93A mice, another group recently reported that a significant population of Ly6Chi monocytes are recruited to the spinal cord and become activated microglia during disease progression (Butovsky et al., 2012). However, this conclusion is at odds with the report that microglia in the SOD1G93A mouse spinal cord are resident derived cells, based on analysis of circulation-conjoined GFP parabiosis studies (Ajami et al., 2007). In addition, our FACS analysis of SOD1G93A spinal cord shows that very few (<0.5%) microglia (CD11b+CD45+) are Ly6C+, inconsistent with the findings of Butovsky et al (Butovsky et al., 2012) (Fig. 3E–F). The few Ly6C+ cells we observed also expressed TCRβ, showing that they were T cells and not monocytes (Fig. S3B). We also analyzed monocyte marker expression in transcriptional profiling data and by quantitative PCR (qPCR). Levels for Ly6c1 and Ccr2, two markers of peripheral monocytes (Mildner et al., 2007), did not increase in SOD1G93A spinal cord (Fig S3D) or in purified microglia (Fig 3G). Thus, taken together, our data corroborate experiments by Ajami et al (Ajami et al., 2007), indicating that resident microglia and not peripheral monocytes are major participants in ALS spinal cord innate immune activation.
Microglia co-express potentially neuroprotective and toxic factors in ALS mice
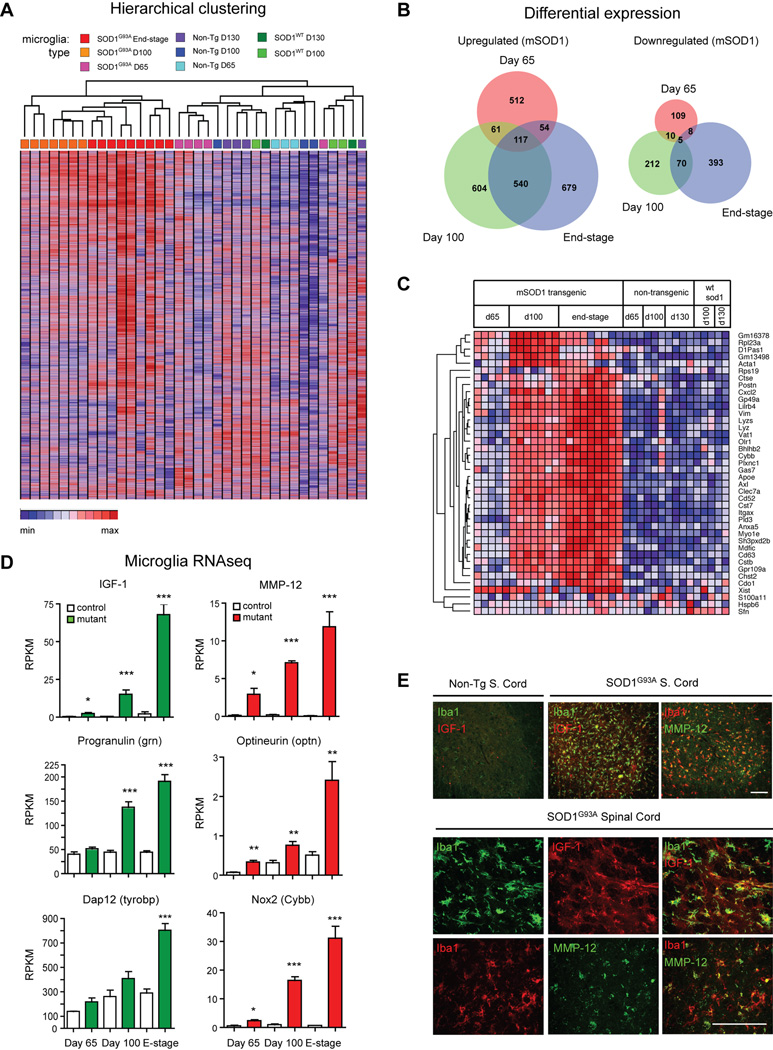
We next analyzed microglia transcriptional changes during neurodegeneration in ALS transgenic mice. Unsupervised hierarchical clustering segregated mutant SOD1G93A microglia groups into distinct clusters that were age-dependent and separate from control microglia (Fig. 4A). SOD1WT microglia and non-Tg microglia showed similar gene expression at day 130 (71 differentially expressed (DE), 2-fold, p<0.01) (Supplemental Fig. S4), indicating that overexpression of the human SOD1 enzyme in mice does not significantly alter microglia gene expression. By contrast, SOD1G93A microglia differed significantly from non-Tg microglia at end-stage/d130 (1041 DE genes, 2-fold, p<0.01) (Fig. S4A). ALS and control microglia differed most significantly at post-symptomatic timepoints (day 65: 956 DE genes; day 100: 1619 DE genes; end-stage: 1866 DE genes; 2-fold, p<0.05) (Fig. S4C). Venn diagrams reveal hundreds of significant transcript changes between SOD1G93A and control microglia at each disease timepoint, with 122 shared transcripts that were up or downregulated at all three timepoints (Fig. 4B, DE gene lists in Table S4), mostly in a progressive, age-dependent manner (Fig. S4D). Principle Components Analysis (PCA) revealed that one principle component (PC1) separated SOD1G93A and control microglia profiles over time (Fig. S4E). These findings are consistent with prior analyses showing that symptom onset marks a transition point for microglia activation (Chiu et al., 2009), and that genetic manipulation of microglia affects the post-symptomatic phase in mutant SOD1 mice (Beers et al., 2006; Boillee et al., 2006).
Fig. 4.
SOD1G93A microglia progressively upregulate potentially neuroprotective and neurotoxic factors. (A) Hierarchical clustering of RNAseq data from SOD1G93A, SOD1WT, and non-Tg microglia at different timepoints shows distinct segregation of mutant from control transcriptomes. (B) Differentially upregulated or downregulated genes (2-fold difference, p<0.05) in mutant microglia compared to control microglia at each timepoint, displayed as Venn diagrams. (C) Heat map displaying the 40 most differentially expressed genes across all datasets (ranked by coefficient of variance), shown by genotype and timepoint. (D) Microglia significantly upregulate potentially neuroprotective (IGF-1, Progranulin, DAP 12) and neurotoxic factors (MMP-12, Optineurin, Nox2) during disease progression (*, p<0.05; **, p<0.01; ***, p<0.001 by t-test). (E) Immunostaining for IGF-1, MMP-12, and microglia (Iba1) in SOD1G93A (end-stage) and non-Tg spinal cords (day 130). Top panels, ventral horn; lower panels, high magnification images of microglia. Scale bars, 100 µm. Error bars, mean±s.e.m. See also Figure S4 and S5.
Analysis of the most DE genes (Fig. 4C) revealed that SOD1G93A microglia show significant induction of potentially neuroprotective and neurotoxic factors concurrently during ALS disease progression. Insulin-like growth factor 1 (Igf1), which activates AKT prosurvival signaling in motor neurons, has been shown to extend lifespan when delivered into ALS mice (Kaspar et al., 2003; Narai et al., 2005). Igf1 was upregulated over time in mutant microglia, showing a 34-fold increase compared to control microglia at end-stage (Fig. 4D–E). Progranulin (Grn) was also strongly upregulated by mutant microglia, (3.9-fold at end-stage, Fig. 4D). Progranulin is a pro-survival factor released by microglia that acts on neurons. Grn mutations are linked to fronto-temporal dementia (Rosen et al., 2011), while genetic ablation experiments have shown that progranulin is neuroprotective in ALS zebrafish models (Laird et al., 2010; Ryan et al., 2009). We found other potentially beneficial factors upregulated by SOD1G93A microglia during disease progression, including Trem2 (Trem2) and its downstream adaptor molecule Dap12 (Tyrobp) (Fig. 4D, Supplemental Fig. S5). Trem2 is a single pass transmembrane protein that triggers myeloid cells that is thought to increase phagocytic activity and suppress cytokine production (Melchior et al.; Thrash et al., 2009). Trem2/Dap12 mutations lead to Nasu-Hakola disease, a cognitive disorder caused by microglia dysfunction that arises from homozygous or compound-heterozygous Trem2 gene deletion, and also includes osteoclast dysfunction (Guerreiro et al., 2012b; Thrash et al., 2009). Trem2 variants were recently shown to increase the risk of Alzheimer’s disease (AD), and its expression is upregulated in an AD mouse model (Guerreiro et al., 2012a).
At the same time, SOD1G93A microglia expressed potentially neurotoxic factors. MMP12 (Mmp12) was among the most significantly induced molecules, an increase of 125-fold compared to control microglia at end-stage (Fig. 4D–E). Inhibiting MMPs has been shown to extend lifespan in ALS mice (Lorenzl et al., 2006), but the particular role of MMP12 has not yet been defined. Optineurin (Optn) is also significantly unregulated in mutant microglia (Fig. 4D). Mutations in Optn are a cause of familial ALS (Maruyama et al., 2010), and expression of mutant optineurin leads to deregulation of interferon signaling, suggesting a deleterious role in disease progression (Sakaguchi et al., 2011). We also analyzed changes in cytokine related gene expression (Fig. S5C). _TNF_-α, _IL_-1β, and _IL_-α levels increased in SOD1G93A microglia, along with receptors for Type 1 interferons (Ifnar1, Ifnar2) and IL-10 (IL-10ra). Several interferon response genes (Ifit1, Ifit3, Ifitm3, Igip30) increased significantly during disease progression in SOD1G93A microglia (Fig. S5C). The pro-inflammatory oxidase Nox2 (Cybb) was also upregulated in SOD1G93A microglia; pharmacological and knockout studies have shown that Nox2 is harmful to ALS mice (Harraz et al., 2008) (Fig. 4D). We also found significant up-regulation of the secreted factor osteopontin (Spp1, Fig, S5A). Osteopontin may be neurotoxic or neuroprotective, as it has been shown to contribute to disease progression in EAE (Chabas et al., 2001) and is upregulated in AD cerebral spinal fluid, but is neuroprotective in animal models of spinal cord injury (Comi et al., 2010; Hashimoto et al., 2007). The robust upregulation of IGF-1, MMP-12, and osteopontin in activated SOD1G93A microglia was confirmed by immunostaining of Iba1+ microglia in spinal cord sections (Fig. 4E, Fig. S5B). Thus, ALS transgenic microglia display a complex transcriptional profile, expressing both potentially neuroprotective and neurotoxic factors concurrently.
SOD1G93A microglia differ significantly from LPS induced microglia and M1 or M2 macrophages
In the periphery, macrophages adopt an M1 phenotype when activated by LPS to combat bacterial pathogens; by contrast, IL-4 activated macrophages adopt an M2 phenotype to combat parasitic infections (Gordon, 2003). In inflammatory conditions, M2 macrophages are involved in wound-healing and regulation of insulin resistance (Odegaard et al., 2007). Despite ontological differences between microglia and peripheral macrophages, microglia have also been categorized as M1 or M2-like (Appel et al., 2011; Beers et al., 2011). During disease progression in SOD1G93A mice, microglia have been reported to adopt an M2 protective phenotype that transitions to an M1 neurotoxic phenotype over time (Beers et al., 2011; Vaknin et al., 2011). However, these analyses involved whole spinal cord expression of a small subset of M1 or M2 macrophage genes (Beers et al., 2011). We addressed the similarities in the ALS microglia profile with M1/M2 activation using two approaches.
First, we compared microglia transcriptomes from SOD1G93A mice with that of microglia activated by LPS injection. We found that these two modes of microglia activation differ significantly (Fig. 5A): SOD1G93A microglia induced expression of Cathepsin D, C1qa, ApoE, and Trem2, while LPS activated microglia showed high expression of Bag5, Stat3, and Adamts1. SOD1G93A microglia transcriptomes (B6 and B6SJL strains) segregated distinctly from LPS activated microglia (Supplemental Fig. S6). Relative to naïve microglia, SOD1G93A activation induced 1158 genes and LPS activation, 499 genes, with 730 shared activation genes (Fig. S6B, 2-fold, p<0.05). Pathway analysis by DAVID (Database for Annotation, Visualization and Integrated Discovery) showed that SOD1G93A microglia upregulated genes in KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways for Mus musculus involving ribosome function (mmu03010), lysosome (mmu04142), and oxidative phosphorylation (mmu00190) (Fig. 5B). Strikingly, ALS microglia were also enriched in the transcripts of genes related to neurodegenerative diseases, including Huntington’s disease (mmu05016), Parkinson’s disease (mmu05012), and Alzheimer’s disease (mmu05010). Conversely, LPS activated microglia were enriched in DNA replication (mmu03030), cell cycle (mmu04110), and innate immune signaling through the RIG-I-like receptor (mmu04622) and Nod-like receptor (mmu04621) (Fig. 5B). We conclude that ALS and LPS microglia display distinct modes of activation.
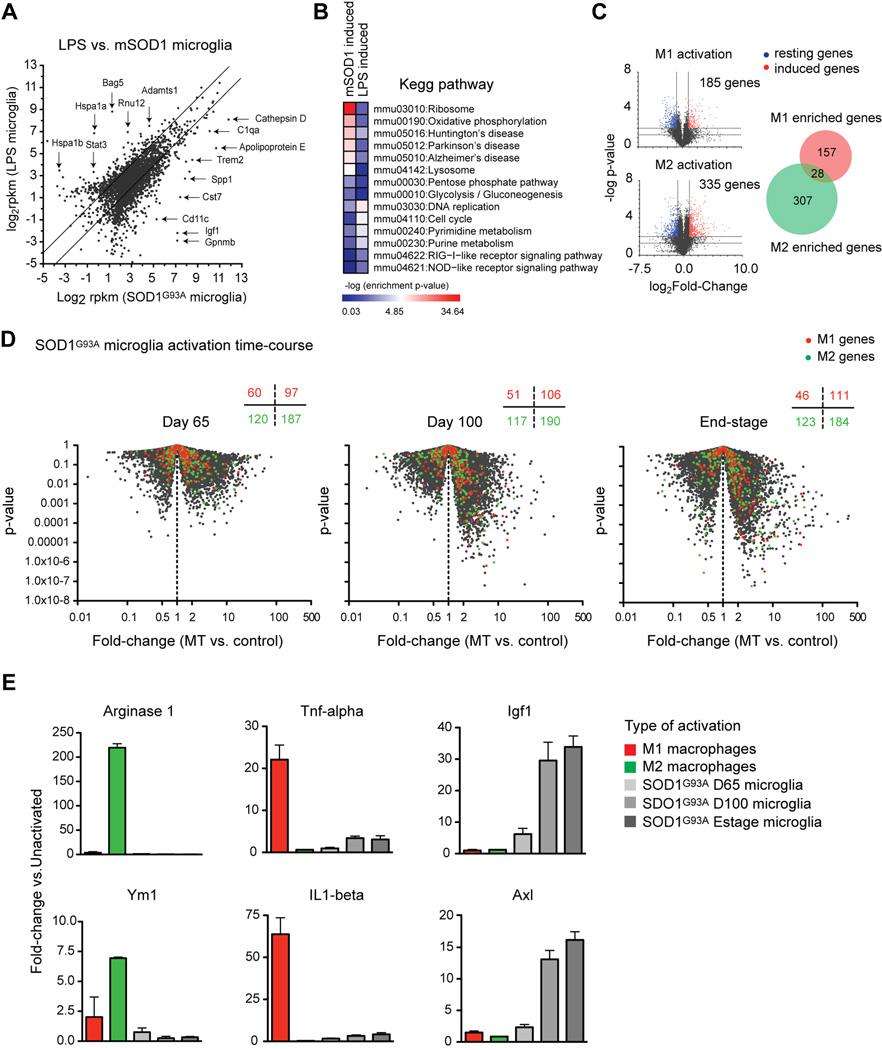
Fig. 5.
ALS spinal cord microglia do not resemble LPS activated microglia or M1 and M2 macrophages. (A) SOD1G93A end-stage microglia profiles were compared to microglia activated by LPS injection. Lines show 2-fold change boundaries, specific enriched genes are highlighted. (B) SOD1G93A or LPS activation induced genes (1.5-fold, p<0.05 compared to naïve mice) show differential KEGG pathway enrichment (heat map, -log p-value). (C) Volcano plots identify M1 activation or M2 activation induced genes (2-fold, p<0.01). Venn diagram shows hallmark genes for each activation modality. (D) M1 or M2 macrophage genes were overlaid onto volcano plots of SOD1G93A vs. control microglia (p-value vs. fold-change). The number of M1 or M2 genes downregulated (left) or upregulated (right) in SOD1G93A microglia at each timepoint is displayed. (E) Fold-change induction for prototypic M2 macrophage genes (Arginase 1, YM1), for M1 genes (TNF-α, IL-1β), and for IGF-1 or Axl, during macrophage or SOD1G93A microglia activation. Error bars, mean±s.e.m. See also Figure S6.
We next mined published microarray datasets of M1 and M2 macrophage activation for genome-wide transcriptional changes that occur for each mode of activation (Shell et al., 2005; Szanto et al., 2010). This analysis yielded 157 M1-enriched and 307 M2-enriched macrophage activation genes (p<0.01, 2-fold induced and non-overlapping, Fig. 5C). These M1 and M2 gene lists were analyzed within purified SOD1G93A vs. control microglia datasets (Fig. 5D). At day 65, the proportion of M1 or M2 genes upregulated in SOD1G93A compared to control microglia were similar (M1 genes: 97/157 = 61.8%; M2 genes: 187/307 = 61.1%). The proportion of M1 activation genes, but not M2 genes, slightly increased in SOD1G93A microglia at day 100 (M1: 106/157 = 67.5%; M2: 190/307 = 62.1%) and at end-stage (M1: 111/157 = 70.7%; M2: 184/307 = 59.80%). This analysis clearly shows that SOD1G93A microglia do not display a significant bias towards either M1 or M2 phenotypes at any time point during disease progression. This conclusion was confirmed by fold-change analysis across macrophage and microglia datasets of known M2 markers (Ym1, Arginase 2) or M1 markers (TNF-α, IL-1p) (Fig. 5E). Moreover, Igf1 and Axl, two highly induced transcripts in SOD1G93A microglia (Fig. 5E), were not enriched in either M1 or M2 activated macrophages. These data unequivocally demonstrate that the microglia phenotype in ALS mice is neurodegeneration-specific and unrelated to M1 or M2 macrophage-like states.
Lysosomal and Alzheimer’s disease pathways are induced in mutant SOD1 microglia
We next analyzed changes in the microglia transcriptome during ALS disease progression using information-theoretic-pathway-level (iPAGE) analysis (Goodarzi et al., 2009). Using a supervised clustering approach, we grouped genes into 15 clusters (C0-C14), where cluster C0 represents a background cluster to which the transcripts without significant differential expression were assigned. The other clusters, while showing distinct expression patterns, are informative of microglia activation. iPAGE identified clusters C3 and C4 as particularly enriched in SOD1G93A microglia (Supplemental Fig. S7). A number of KEGG pathways were significantly enriched in SOD1G93A clusters, in particular, the lysosome (mmu4142), Alzheimer’s disease (mmu5010), and spliceosome (mmu3040) pathways (Fig. 6A). Similar analysis based on GO terms found related biological processes associated with SOD1G93A microglia: lytic vacuole (GO:0000323), RNA splicing (GO:0008380), and organelle inner membrane (GO:0019866) (Fig. S7B). Differential expression gene clusters C2, C8, and C14 correspond to control microglia enriched genes. We found that control microglia were significantly enriched in insulin signaling (KEGG: mmu4910), growth factor binding (GO:0019838), and aromatic compound metabolism (GO:0006725) pathways. As an alternative method to analyze the feature-rich dataset, we also used a timepoint normalization approach for analysis, in which transcripts were clustered based on their relative expression levels across timepoints (Fig. S7C).
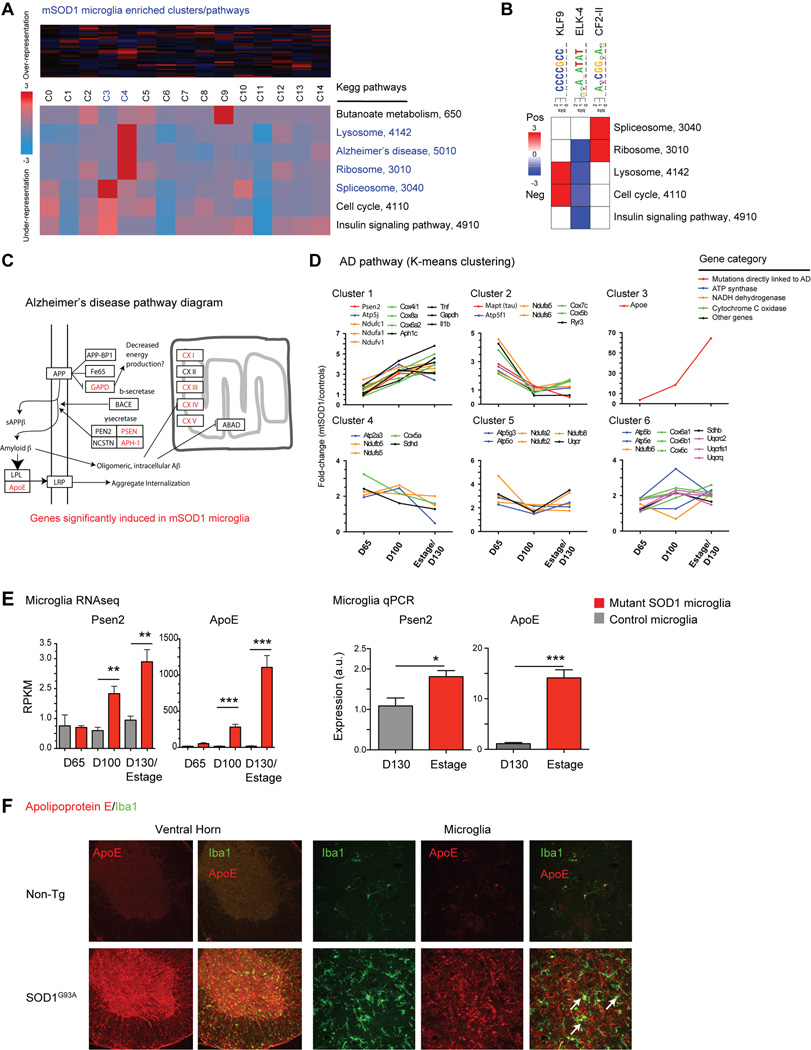
Fig. 6.
Pathway analysis shows dysregulation of Alzheimer’s disease genes in mutant SOD1 microglia. (A) iPAGE analysis finds significant enrichment for ribosomal, lysosome, RNA splicing, and Alzheimer’s disease KEGG pathways in SOD1G93A microglia compared to control microglia (clusters C3, C4). (B) Transcriptional motif enrichment analysis finds 3 positively enriched transcriptional motifs linked to KEGG pathway changes. (C) SOD1G93A microglia show significant dysregulation in Alzheimer’s disease (AD) genes, as shown in this pathway diagram (adapted from KEGG). (D) AD pathway genes were grouped by K-means clustering and plotted over time by fold-difference between SOD1G93A vs. control microglia. Genes whose mutations directly link to AD and components of ATP synthase, NADH dehydrogenase, cytochrome C oxidase, and other AD pathway genes are differentially increased in SOD1G93A microglia (E) Presenilin 2 (Psen2) and Apolipoprotein E (Apoe) levels are upregulated in SOD1G93A microglia by RNAseq and qPCR analysis (*, p<0.05; **, p<0.01; ***, p<0.001 by t-test). (F) Immunostaining of spinal cord sections shows significant upregulation of ApoE throughout the ventral horns of SOD1G93A compared to non-Tg spinal cords. Higher magnification images show microglia (Iba1+) colocalization with ApoE. Scale bars, 50 µm. Error bars, mean±s.e.m.
Using FIRE regulatory element analysis (Elemento et al., 2007), we found significant enrichment for three transcriptional motifs associated with differential regulation of pathways in activated SOD1G93A microglia (Fig. 6B). CCCCGCC, an optimized binding motif for KLF9 (a repressor) and Sp1, which compete for binding sites, was enriched in genes involved in lysosome and cell cycle pathway dysregulation (Fig. 6B). Transcript levels of KLF9 were inversely correlated with motif-enriched clusters (-0.49, p=0.002), which might be KLF9 target genes (Supplemental Fig. S8). AcCGGaAc, a motif for ELK-4, was enriched for spliceosome and ribosome pathway genes, and transcript levels of ELK-4 directly correlated with motif expression in mutant microglia (Fig. 6B and Fig. S8A). These results suggest that KLF9 downregulation and ELK-4 upregulation are potentially linked to induction of lysosome, ribosome, and spliceosome changes in mutant microglia.
The induction of lysosome and Alzheimer’s disease pathways in SOD1G93A microglia may hold functional implications related to protein clearance and neurodegeneration. K-means analysis clustered co-regulated genes in the lysosome pathway (Supplemental Fig. S8B), which include a host of lysosome enzymes (e.g. HexA), membrane markers (e.g. Cd68, Cd63, Lamp1), components of the lysosomal ATPase (e.g. Atp6v0d1), and a large group of cathepsins (Ctsa, Ctsb, Ctsd, Ctsl, Ctss, Ctsz, and Ctse). Cathepsin expression was mostly co-regulated in gene cluster 1 (except Ctse). It remains to be determined if KLF9, as predicted above, plays a role in expressing specific clusters of lysosome genes. Cathepsins are activated by low pH of lysosomes, whose deficiencies have been linked to increased neurodegeneration. Cathepsin B mediates β-amyloid plaque removal in Alzheimer’s disease (Sun et al., 2008), and Cathepsin S degrades antigenic proteins for presentation on MHC class II (Wendt et al., 2008). Cathepsin D deficiency results in CNS lysosomal storage disease with ceroid lipofuscin accumulation (Santambrogio et al., 2001). We confirmed that Cathepsins B and S are expressed in SOD1G93A microglia by spinal cord immunostaining (Fig. S8C).
Alzheimer’s disease (AD) genes were found to be enriched in SOD1G93A microglia, both by targeted pathway analysis (Fig. 5B) and by global transcriptome analysis using iPAGE (Fig. 6A). K-means clustering showed 6 groups of regulated AD genes in mutant vs. control microglia (Fig. 6C), which include components of ATP synthase, Cytochrome C oxidase, and NADH dehydrogenase, enzymes present in the mitochondrial envelope and involved in energy metabolism. Misfolded SOD1 aggregates also localize to the mitochondrial inner membrane (Liu et al., 2004; Pasinelli et al., 2004). Interestingly, SOD1G93A microglia showed significant upregulation of 3 genes whose mutations are directly linked to AD susceptibility – Tau (Mapt), Presenilin 2 (Psen2), and Apoliprotein E (Apoe) (Fig. 6C). Tau was induced at day 65 and downregulated over time, while Psen2 and ApoE were progressively upregulated in SOD1G93A microglia Apoe was one of the most significantly expressed and induced transcripts during disease progression (>1000 RPKM, 64-fold at end-stage) (Fig. 6E). This upregulation was specific to SOD1G93A microglia, as Apoe was not induced by LPS activation or in M1/M2 macrophages (Fig. 5A). Immunostaining revealed robust Apolipoprotein E expression throughout the ventral horn of SOD1G93A and not non-Tg spinal cord, particularly in the cytoplasm of Iba1+ microglia (Fig. 6F). Thus, AD pathway genes are a major component of the microglia activation profile in SOD1G93A mice.
Microglia transcriptome changes relate to surface marker acquisition and T cell infiltration
We performed analysis of spinal cord FACS data in relation to microglia RNAseq from the same mice (Table S8). SOD1G93A microglia were found to up-regulate cell surface expression of CD11c (DC marker), CD86 (co-stimulatory ligand), and CD11b (integrin αM) (Fig. 7A). Surface mean fluorescence intensity (MFI) and transcript levels for these genes increased concurrently (Fig. 7B). However, analysis showed different relationships between surface and transcriptional expression (Fig. 7C). Increased Cd11c (Itgax) mRNA levels correlated with CD11c surface levels by FACS (r2=0.83). On the other hand, CD11b surface levels did not correlate with CD11b (Itgam) transcript levels (r2=0.006). Similarly, CD86 surface levels increased over time to a greater extent than Cd86 mRNA levels (r2=0.16). Thus, microglia CD11c surface expression is directly controlled by transcriptional changes, whereas CD11b and CD86 surface induction occurs through post-transcriptional mechanisms. In dendritic cells, CD86 surface expression is regulated by MARCH1 ubiquitination (Corcoran et al., 2011); it remains to be determined if microglia utilize similar pathways. Thus, microglia functional activation involves transcriptional and post-transcriptional regulation. Whole proteome analyses are required to establish a complete picture of ALS microglia activation.
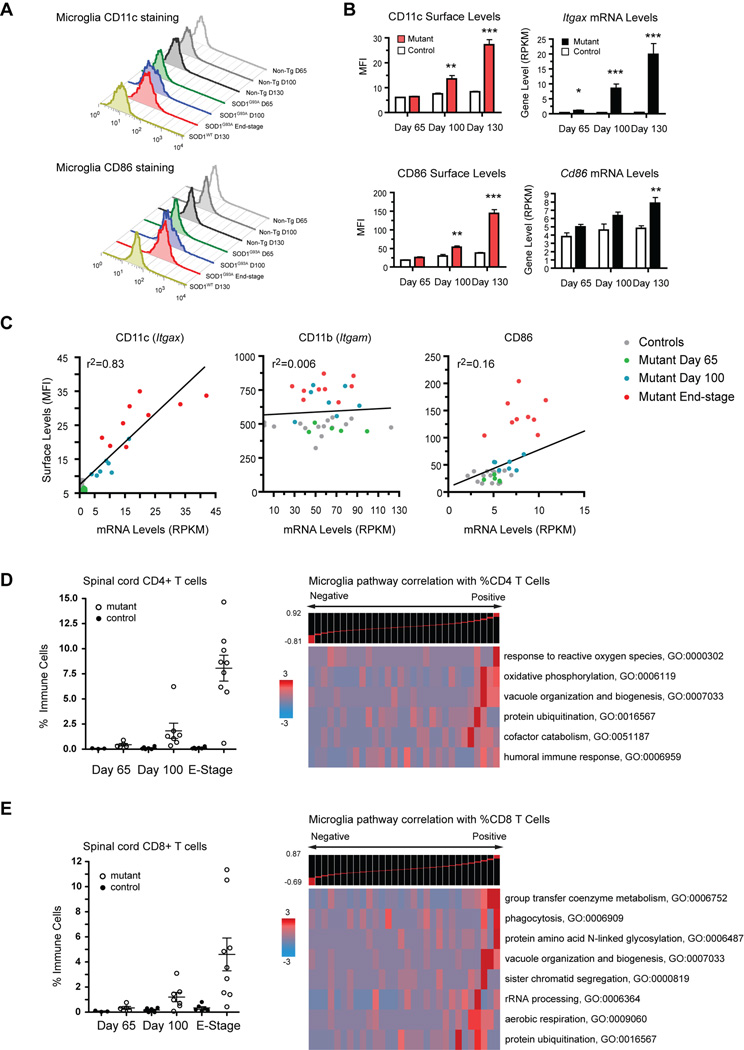
Fig. 7.
Post-transcriptional regulation of microglia surface markers and T cell associated molecular pathways. (A) Representative histograms of CD11c and CD86 surface levels on microglia from SOD1G93A and control spinal cords. (B) Surface MFI and transcript levels (RPKM) for CD11c (Itgax) and CD86. (**, p<0.01; ***, p<0.001 by t-test). (C) FACS-RNAseq correlation shows that surface CD11c is directly regulated by transcription while surface CD11b and CD86 are regulated post-transcriptionally. (D–E) Infiltrating T cells quantified by FACS and cell count of spinal cord samples. Intra-sample comparative analysis with microglia transcriptome data shows specific microglia GO categories correlated to levels of CD4+ or CD8+ T cells (Spearman rank). Error bars, mean±s.e.m.
FACS analyses revealed that the percentage of CD4+ Thelper cells and CD8+ Tcytotoxic cells infiltrating mutant spinal cords increased significantly over time compared to controls (Fig. 7D–E). Spearman rank was used to correlate CD4+ and CD8+ T cell populations to corresponding gene expression levels in microglia at all timepoints (Fig. 7D–E). Both subsets were related to microglia expression of pathways relevant to antigen presentation: vacuole organization (GO:0007033) genes and protein ubiquitination (GO:0016567). Each subset was uniquely linked to different microglia GO categories: CD4+ T cells correlated with microglia expression of reactive oxygen species (GO:0000302), oxidative phosphorylation (GO:0006119), and humoral immune response (GO:0006959) genes. CD8+ T cells were associated with enrichment in microglia phagocytosis (GO:0006909) and coenzyme metabolism (GO:0006752) genes. These data may reflect active T cell-microglia interactions during neurodegeneration.
Discussion
Since their discovery by del Rio Hortega, microglia have been linked to diseases of the CNS (Ransohoff and Perry, 2009). Microglia respond rapidly to injury, transitioning from finely branched, ramified forms to amoeboid morphologies (Frank-Cannon et al., 2009). In this study, we detail transcriptional changes that occur in microglia during activation in ALS transgenic mice and following injection by LPS. We find that SOD1G93A microglia adopt a dual phenotype, expressing factors hypothesized to be both neurotoxic and neuroprotective, the balance of which is likely affected by cell intrinsic factors and the spinal cord micro-environment. Non-specific anti-inflammatory drugs such as minocycline would negate both aspects of microglia, and may explain past failures in clinical trials (Gordon et al., 2007).
Unlike other nervous system cells that derive from the ectoderm, recent work has shown that microglia originate from erythromyeloid progenitors during embryonic hematopoiesis (Alliot et al., 1991; Ginhoux et al., 2010; Kierdorf et al., 2013; Schulz et al., 2012). Due to paucity of specific microglia markers, targeted analysis of neuroinflammation has been difficult. CD11b–Cre and LysM-Cre transgenic mice have been used to study microglia (Boillee et al., 2006; Cho et al., 2008), but these genes are also expressed by peripheral myeloid immune cells. A mixture of monocytes and microglia participate in stroke, multiple sclerosis, and spinal cord injury (Frank-Cannon et al., 2009; Ransohoff and Perry, 2009). Thus, identification of microglia markers would provide useful tools for both genetic and immunohistochemical analysis of these cells. To this end, we used RNAseq and microarray data to find 29 enriched markers expressed by microglia but not by CNS or peripheral myeloid cells. Previous transcriptome analysis comparing related peripheral immune cell-types (B cells, T cells) identified few cell-specific genes (Heng et al., 2008; Painter et al., 2011). By contrast, our analysis suggests that microglia differ substantially from peripheral myeloid cells, which may reflect their early divergence in ontology (Cuadros and Navascués, 1998).
Given transcriptional differences between microglia and peripheral macrophages, it is likely that their transcriptional activation would also differ. Nonetheless, microglia have often been designated as M1 or M2 macrophage-like (Beers et al., 2008; Beers et al., 2011; Hickman et al., 2008; Ransohoff and Perry, 2009). Our analysis using transcriptome data from M1 and M2 macrophage activation revealed that ALS microglia are not similar to either activation modality but rather express a neurodegeneration-specific signature.
A recent report proposed that peripheral monocytes infiltrate the ALS spinal cord and contribute substantially to motor neuron loss (Butovsky et al., 2012). However, this observation is inconsistent with previous parabiosis experiments (Ajami et al., 2007). Our flow cytometry, transcriptional analyses, and qPCR data did not reveal an induction of monocyte markers in SOD1G93A spinal cords or purified CD11b+ microglia at any disease stage. Moreover, bone marrow transplantation, which was used as a second line of evidence (Butovsky et al., 2012), is known to introduce irradiation-induced breakdown of the blood brain barrier and monocyte entry into the CNS (Ajami et al., 2007; Mildner et al., 2007). We are in communication with Butovsky et al to understand these discrepant results. We note that our data do not rule out a role for circulating monocytes in SOD1G93A mice, only that they do not contribute to spinal cord inflammation. We and others have found substantial monocyte infiltration into degenerating peripheral nerves in SOD1G93A mice, which may play a role in affecting motor axon survival (Chiu et al., 2009; Graber et al., 2010; Lincecum et al., 2010).
Our detailed transcriptome analysis reveals that resident ALS microglia play a complex role. We found that microglia induce factors that can suppress neurodegeneration, including IGF-1 (Igf1) and progranulin (Grn), which have been shown to mediate motor neuron protection in ALS animal models (Kaspar et al., 2003; Laird et al., 2010; Ryan et al., 2009). Pathway analyses revealed significant upregulation of lysosomal proteins and enzymes, including cathepsins (Fig. S8). Cathepsins proteolytically degrade and clear phagocytosed debris, including misfolded Aβ aggregates in AD and α-synclein in Parkinson’s disease (Cullen et al., 2009; Sun et al., 2008). We hypothesize that cathepsins may be involved in removal of mutant SOD1 aggregates and neuronal debris in ALS mice, while IGF-1 and progranulin are concurrently released as neuroprotective factors as a wound healing-type response.
ALS microglia also upregulate factors linked to neurotoxicity including Nox2. At the protein level, mutant SOD1 aberrantly associates with Rac1 GTPase, which in turn deregulates NADPH oxidase (Nox2) activity and increases reactive oxygen species (Harraz et al., 2008). It may be that signaling downstream of mutant SOD1 also turns on neurotoxic gene expression. While removal of mutant SOD1 from myeloid cells by conditional deletion ameliorates disease (Boillee et al., 2006), elimination of proliferating microglia (60% of population) in ALS mice does not affect survival (Gowing et al., 2008). These results may reflect the multi-faceted role of microglia in ALS (Henkel et al., 2009).
An unexpected finding was a robust increase in microglia expression of AD pathway genes during disease progression. Tau, Presenilin 2, and Apoliprotein E, genes whose mutations are directly linked to familial AD, were upregulated. Amyloid precursor protein (APP) processing products sAPP and Aβ accumulate in ALS patient spinal cords and cerebrospinal fluid (Calingasan et al., 2005; Steinacker et al., 2009). Apoε4, the allele linked to early onset AD, is associated with increased risk for the bulbar-onset form of ALS (Praline et al., 2011). Furthermore, Trem2, a microglia receptor upregulated during disease progression, has been recently linked to AD susceptibility (Guerreiro et al., 2012a). What is the potential role of this AD gene induction? Landreth et al showed recently that retinoid X receptor stimulation leads to increased Apoe expression in astrocytes and microglia, which leads to a major beneficial impact on plaque clearance and cognitive performance in APP transgenic mice (Cramer et al., 2012). Furthermore, this mechanism is likely through suppression of pro-inflammatory and induction of antiinflammatory pathways in microglia (Mandrekar-Colucci et al., 2012). It is possible that the microglia transcriptional responses we observe, including Apoe, Igf1, and Grn induction, are programmed responses to neuronal death or injury.
Overall, we conclude that ALS microglia may be a double-edged sword, with SOD1G93A induced negative changes counterbalanced by a neuroprotective response mediated by extrinsic regulatory factors including signals released by dying motor neurons and infiltrating T cells. Cx3cl1 expression on motor neurons was shown to downregulate microglia activity in ALS mice through the fractalkine receptor (Cx3cr1) (Cardona et al., 2006b). T cells also play a key role in regulating microglia function in ALS mice (Beers et al., 2008; Chiu et al., 2009). Using FACS-transcriptome comparisons, we identified potential molecular pathways involved in T-cell microglia cross-talk in ALS mice. Based on these analyses, antigen-processing pathways, which correlate with both CD4+ and CD8+ T cells, may be involved in initiating their activation. CD8+ T cells may then stimulate phagocytosis and CD4+ T cells induce oxidative phosphorylation in microglia (Fig. 7).
This study demonstrates the utility of transcriptome-wide analyses of acutely isolated microglia in neurodegenerative disease models. Our methodology and database captures the complex nature of glial activation in vivo, showing a distinct advantage over culture-based studies of microglia activation, normally involving neonatal brain-derived cells. Elegant transcriptional profiling studies have revealed significant differences between astrocytes acutely isolated from the CNS and cultured astrocytes, which appear to reflect an artifactual activation phenotype (Foo et al., 2011). We have found that activated microglia in ALS mice show a neurodegeneration-specific phenotype that differs from LPS activated microglia and M1 or M2 macrophages. It is likely that the genomic profiles of microglia in each neurological disease context will be unique. Therefore, similar analyses in mouse models of Parkinson’s disease, AD, and Huntington’s disease may reveal differing roles of microglia in each neurological disease context. The data presented here should provide a valuable resource for interpreting these studies.
Experimental Procedures
Mice
C57BL/6, B6.SOD1G93A, B6/SJL.SOD1G93A transgenic, and SOD1WT transgenic mice from Jackson Laboratories (Bar Harbor, ME) were bred and maintained in full-barrier facilities. Onset of motor weakness, including hindlimb tremors and weight loss, and disease end-stage were measured as described (Chiu et al., 2008). For LPS activation, C57BL/6 mice were injected once intra-peritoneally with 5 mg/kg E. coli LPS O55:B55 (Sigma Aldrich), and spinal cord microglia isolated 48 hours later. All studies were conducted according to institutional guidelines for animal use and care at Harvard Medical School, Harvard University, and Columbia University Medical Center.
Microglia purification and spinal cord flow cytometry
Spinal cord immune cells were analyzed by FACS as previously described (Chiu et al., 2008; Chiu et al., 2009). FACS data are given in Table S8. Microglia were purified by sequential percoll gradient centrifugation and magnetic bead isolation (Fig. 1A, Fig. S1B), a procedure modified from previous protocols (Cardona et al., 2006a; Hickman et al., 2008). Detailed FACS and cell purification protocols are in Supplemental Information.
Deep sequencing and expression analysis
RNA was extracted from purified microglia and subjected to paired-end RNA sequencing on the Illumina platform. Read alignment and gene levels were calculated using Bowtie, Cufflinks, and Tophat software. Raw FastQ sequencing data and processed normalized expression data (RPKM) have been uploaded for public access on the NCBI GEO database (Accession No. GSE43366). Expression comparisons and clustering were conducted using GenePattern analysis modules (Broad Institute), pathway analysis through DAVID and iPAGE (Goodarzi et al., 2009), motif analysis through FIRE (Elemento et al., 2007), and T-cell microglia transcriptome analysis through iPAGE. Detailed RNA processing, data analysis, immunostaining and quantitative PCR validation of expression analysis are described in Supplemental Information.
Supplementary Material
01
02
03
04
05
Highlights.
- Identification of specific marker genes for acutely isolated microglia
- Progressive resident microglia transcriptome changes reveal in vivo activation phenotype
- Microglial ALS disease activation signature distinct from M1/M2 macrophages
- Parallel transcriptome and FACS analyses reveal T cell/microglia crosstalk
Acknowledgments
We thank Robert Koffie and Brad Hyman for Apolipoprotein E antibodies, Beth Stevens for critical reading of the manuscript, Michio Painter for advice on bioinformatic analysis, and Ben Barres and Robert H. Brown Jr. for helpful discussions. This work was supported by the ALS Association, ALS Therapy Alliance, and NIH grant 5DP10D003930 to T.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10:1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- Alliot F, Lecain E, Grima B, Pessac B. Microglial progenitors with a high proliferative potential in the embryonic and adult mouse brain. Proc Natl Acad Sci U S A. 1991;88:1541–1545. doi: 10.1073/pnas.88.4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel SH, Zhao W, Beers DR, Henkel JS. The microglial-motoneuron dialogue in ALS. Acta Myol. 2011;30:4–8. [PMC free article] [PubMed] [Google Scholar]
- Beers DR, Henkel JS, Xiao Q, Zhao W, Wang J, Yen AA, Siklos L, McKercher SR, Appel SH. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2006;103:16021–16026. doi: 10.1073/pnas.0607423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers DR, Henkel JS, Zhao W, Wang J, Appel SH. CD4+ T cells support glial neuroprotection, slow disease progression, and modify glial morphology in an animal model of inherited ALS. Proc Natl Acad Sci U S A. 2008;105:15558–15563. doi: 10.1073/pnas.0807419105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers DR, Henkel JS, Zhao W, Wang J, Huang A, Wen S, Liao B, Appel SH. Endogenous regulatory T lymphocytes ameliorate amyotrophic lateral sclerosis in mice correlate with disease progression in patients with amyotrophic lateral sclerosis. Brain. 2011;134:1293–1314. doi: 10.1093/brain/awr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillee S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, Kassiotis G, Kollias G, Cleveland DW. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- Butovsky O, Siddiqui S, Gabriely G, Lanser AJ, Dake B, Murugaiyan G, Doykan CE, Wu PM, Gali RR, Iyer LK, et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J Clin Invest. 2012;122:3063–3087. doi: 10.1172/JCI62636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calingasan NY, Chen J, Kiaei M, Beal MF. Beta-amyloid 42 accumulation in the lumbar spinal cord motor neurons of amyotrophic lateral sclerosis patients. Neurobiol Dis. 2005;19:340–347. doi: 10.1016/j.nbd.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Cardona AE, Huang D, Sasse ME, Ransohoff RM. Isolation of murine microglial cells for RNA analysis or flow cytometry. Nat Protoc. 2006a;1:1947–1951. doi: 10.1038/nprot.2006.327. [DOI] [PubMed] [Google Scholar]
- Cardona AE, Pioro EP, Sasse ME, Kostenko V, Cardona SM, Dijkstra IM, Huang D, Kidd G, Dombrowski S, Dutta R, et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006b;9:917–924. doi: 10.1038/nn1715. [DOI] [PubMed] [Google Scholar]
- Chabas D, Baranzini SE, Mitchell D, Bernard CC, Rittling SR, Denhardt DT, Sobel RA, Lock C, Karpuj M, Pedotti R, et al. The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science. 2001;294:1731–1735. doi: 10.1126/science.1062960. [DOI] [PubMed] [Google Scholar]
- Chiu IM, Chen A, Zheng Y, Kosaras B, Tsiftsoglou SA, Vartanian TK, Brown RH, Jr, Carroll MC. T lymphocytes potentiate endogenous neuroprotective inflammation in a mouse model of ALS. Proc Natl Acad Sci U S A. 2008;105:17913–17918. doi: 10.1073/pnas.0804610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu IM, Phatnani H, Kuligowski M, Tapia JC, Carrasco MA, Zhang M, Maniatis T, Carroll MC. Activation of innate and humoral immunity in the peripheral nervous system of ALS transgenic mice. Proc Natl Acad Sci U S A. 2009;106:20960–20965. doi: 10.1073/pnas.0911405106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho IH, Hong J, Suh EC, Kim JH, Lee H, Lee JE, Lee S, Kim CH, Kim DW, Jo EK, et al. Role of microglial IKKbeta in kainic acid-induced hippocampal neuronal cell death. Brain. 2008;131:3019–3033. doi: 10.1093/brain/awn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comi C, Carecchio M, Chiocchetti A, Nicola S, Galimberti D, Fenoglio C, Cappellano G, Monaco F, Scarpini E, Dianzani U. Osteopontin is increased in the cerebrospinal fluid of patients with Alzheimer’s disease and its levels correlate with cognitive decline. J Alzheimers Dis. 2010;19:1143–1148. doi: 10.3233/JAD-2010-1309. [DOI] [PubMed] [Google Scholar]
- Corcoran K, Jabbour M, Bhagwandin C, Deymier MJ, Theisen DL, Lybarger L. Ubiquitin-mediated regulation of CD86 protein expression by the ubiquitin ligase membrane-associated RING-CH-1 (MARCH1) J Biol Chem. 2011;286:37168–37180. doi: 10.1074/jbc.M110.204040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer PE, Cirrito JR, Wesson DW, Lee CY, Karlo JC, Zinn AE, Casali BT, Restivo JL, Goebel WD, James MJ, et al. ApoE-Directed Therapeutics Rapidly Clear beta-Amyloid and Reverse Deficits in AD Mouse Models. Science. 2012 doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros MA, Navascués J. The origin and differentiation of microglial cells during development. Prog Neurobiol. 1998;56:173–189. doi: 10.1016/s0301-0082(98)00035-5. [DOI] [PubMed] [Google Scholar]
- Cullen V, Lindfors M, Ng J, Paetau A, Swinton E, Kolodziej P, Boston H, Saftig P, Woulfe J, Feany MB, et al. Cathepsin D expression level affects alpha-synuclein processing, aggregation, and toxicity in vivo. Mol Brain. 2009;2:5. doi: 10.1186/1756-6606-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim J, Zuo Y, Jung S, Littman D, Dustin M, Gan W. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- Dibaj P, Steffens H, Zschuntzsch J, Nadrigny F, Schomburg ED, Kirchhoff F, Neusch C. In Vivo imaging reveals distinct inflammatory activity of CNS microglia versus PNS macrophages in a mouse model for ALS. PLoS One. 2011;6:e17910. doi: 10.1371/journal.pone.0017910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LC, Allen NJ, Bushong EA, Ventura PB, Chung WS, Zhou L, Cahoy JD, Daneman R, Zong H, Ellisman MH, Barres BA. Development of a method for the purification and culture of rodent astrocytes. Neuron. 2011;71:799–811. doi: 10.1016/j.neuron.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Cannon TC, Alto LT, McAlpine FE, Tansey MG. Does neuroinflammation fan the flame in neurodegenerative diseases? Mol Neurodegener. 2009;4:47. doi: 10.1186/1750-1326-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Elemento O, Tavazoie S. Revealing global regulatory perturbations across human cancers. Mol Cell. 2009;36:900–911. doi: 10.1016/j.molcel.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon PH, Moore DH, Miller RG, Florence JM, Verheijde JL, Doorish C, Hilton JF, Spitalny GM, MacArthur RB, Mitsumoto H, et al. Efficacy of minocycline in patients with amyotrophic lateral sclerosis: a phase III randomised trial. Lancet Neurol. 2007;6:1045–1053. doi: 10.1016/S1474-4422(07)70270-3. [DOI] [PubMed] [Google Scholar]
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- Gowing G, Philips T, Van Wijmeersch B, Audet JN, Dewil M, Van Den Bosch L, Billiau AD, Robberecht W, Julien JP. Ablation of proliferating microglia does not affect motor neuron degeneration in amyotrophic lateral sclerosis caused by mutant superoxide dismutase. J Neurosci. 2008;28:10234–10244. doi: 10.1523/JNEUROSCI.3494-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber DJ, Hickey WF, Harris BT. Progressive changes in microglia and macrophages in spinal cord and peripheral nerve in the transgenic rat model of amyotrophic lateral sclerosis. J Neuroinflammation. 2010;7:8. doi: 10.1186/1742-2094-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, et al. TREM2 Variants in Alzheimer’s Disease. N Engl J Med. 2012a doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro RJ, Lohmann E, Bras JM, Gibbs JR, Rohrer JD, Gurunlian N, Dursun B, Bilgic B, Hanagasi H, Gurvit H, et al. Using Exome Sequencing to Reveal Mutations in TREM2 Presenting as a Frontotemporal Dementia-like Syndrome Without Bone Involvement. Arch Neurol. 2012b:1–7. doi: 10.1001/jamaneurol.2013.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- Hall ED, Oostveen JA, Gurney ME. Relationship of microglial and astrocytic activation to disease onset and progression in a transgenic model of familial ALS. Glia. 1998;23:249–256. doi: 10.1002/(sici)1098-1136(199807)23:3<249::aid-glia7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Harraz MM, Marden JJ, Zhou W, Zhang Y, Williams A, Sharov VS, Nelson K, Luo M, Paulson H, Schöneich C, Engelhardt JF. SOD1 mutations disrupt redox-sensitive Rac regulation of NADPH oxidase in a familial ALS model. J Clin Invest. 2008;118:659–670. doi: 10.1172/JCI34060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Sun D, Rittling SR, Denhardt DT, Young W. Osteopontin-deficient mice exhibit less inflammation, greater tissue damage, and impaired locomotor recovery from spinal cord injury compared with wild-type controls. J Neurosci. 2007;27:3603–3611. doi: 10.1523/JNEUROSCI.4805-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. Immunological GenomeProject C. [DOI] [PubMed] [Google Scholar]
- Henkel JS, Beers DR, Zhao W, Appel SH. Microglia in ALS: the good the bad the resting. J Neuroimmune Pharmacol. 2009;4:389–398. doi: 10.1007/s11481-009-9171-5. [DOI] [PubMed] [Google Scholar]
- Hickman SE, Allison EK, El Khoury J. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer’s disease mice. J Neurosci. 2008;28:8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar BK, Llado J, Sherkat N, Rothstein JD, Gage FH. Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science. 2003;301:839–842. doi: 10.1126/science.1086137. [DOI] [PubMed] [Google Scholar]
- Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Holscher C, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- Laird AS, Van Hoecke A, De Muynck L, Timmers M, Van den Bosch L, Van Damme P, Robberecht W. Progranulin is neurotrophic in vivo and protects against a mutant TDP-43 induced axonopathy. PLoS One. 2010;5:e13368. doi: 10.1371/journal.pone.0013368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincecum JM, Vieira FG, Wang MZ, Thompson K, De Zutter GS, Kidd J, Moreno A, Sanchez R, Carrion IJ, Levine BA, et al. From transcriptome analysis to therapeutic anti-CD40L treatment in the SOD1 model of amyotrophic lateral sclerosis. Nat Genet. 2010;42:392–399. doi: 10.1038/ng.557. [DOI] [PubMed] [Google Scholar]
- Lorenzl S, Narr S, Angele B, Krell HW, Gregorio J, Kiaei M, Pfister HW, Beal MF. The matrix metalloproteinases inhibitor Ro 28–2653 [correction of Ro 26–2853] extends survival in transgenic ALS mice. Exp Neurol. 2006;200:166–171. doi: 10.1016/j.expneurol.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Loschko J, Heink S, Hackl D, Dudziak D, Reindl W, Korn T, Krug AB. Antigen targeting to plasmacytoid dendritic cells via Siglec-H inhibits Th cell-dependent autoimmunity. J Immunol. 2011;187:6346–6356. doi: 10.4049/jimmunol.1102307. [DOI] [PubMed] [Google Scholar]
- Mandrekar-Colucci S, Karlo JC, Landreth GE. Mechanisms underlying the rapid peroxisome proliferator-activated receptor-gamma-mediated amyloid clearance and reversal of cognitive deficits in a murine model of Alzheimer’s disease. J Neurosci. 2012:10117–10128. doi: 10.1523/JNEUROSCI.5268-11.2012. 32/Users/Isaac/Downloads/pubmed_result-10.txt. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- Mcgeer P, Mcgeer E. Inflammatory processes in amyotrophic lateral sclerosis. Muscle Nerve. 2002;26:459–470. doi: 10.1002/mus.10191. [DOI] [PubMed] [Google Scholar]
- Melchior B, Garcia AE, Hsiung BK, Lo KM, Doose JM, Thrash JC, Stalder AK, Staufenbiel M, Neumann H, Carson MJ. Dual induction of TREM2 and tolerance-related transcript, Tmem176b, in amyloid transgenic mice: implications for vaccine-based therapies for Alzheimer’s disease. ASN Neuro. 2010;2:e00037. doi: 10.1042/AN20100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelucci A, Heurtaux T, Grandbarbe L, Morga E, Heuschling P. Characterization of the microglial phenotype under specific pro-inflammatory and anti-inflammatory conditions: Effects of oligomeric and fibrillar amyloid-beta. J Neuroimmunol. 2009;210:3–12. doi: 10.1016/j.jneuroim.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch UK, Mack M, Heikenwalder M, Brück W, Priller J, Prinz M. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci. 2007;10:1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- Mizutani M, Pino PA, Saederup N, Charo IF, Ransohoff RM, Cardona AE. The fractalkine receptor but not CCR2 is present on microglia from embryonic development throughout adulthood. J Immunol. 2012;188:29–36. doi: 10.4049/jimmunol.1100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narai H, Nagano I, Ilieva H, Shiote M, Nagata T, Hayashi T, Shoji M, Abe K. Prevention of spinal motor neuron death by insulin-like growth factor-1 associating with the signal transduction systems in SODG93A transgenic mice. J Neurosci Res. 2005;82:452–457. doi: 10.1002/jnr.20668. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Odegaard J, Ricardo-Gonzalez R, Goforth M, Morel C, Subramanian V, Mukundan L, Eagle A, Vats D, Brombacher F, Ferrante A, Chawla A. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter MW, Davis S, Hardy RR, Mathis D, Benoist C. Transcriptomes of the B and T lineages compared by multiplatform microarray profiling. J Immunol. 2011;186:3047–3057. doi: 10.4049/jimmunol.1002695. Immunological Genome Project C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praline J, Blasco H, Vourc’h P, Garrigue MA, Gordon PH, Camu W, Corcia P, Andres CR, French ALSSG. APOE epsilon4 allele is associated with an increased risk of bulbar-onset amyotrophic lateral sclerosis in men. Eur J Neurol. 2011;18:1046–1052. doi: 10.1111/j.1468-1331.2010.03330.x. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM, Perry VH. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- Rosen EY, Wexler EM, Versano R, Coppola G, Gao F, Winden KD, Oldham MC, Martens LH, Zhou P, Farese RV, Jr, Geschwind DH. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron. 2011;71:1030–1042. doi: 10.1016/j.neuron.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CL, Baranowski DC, Chitramuthu BP, Malik S, Li Z, Cao M, Minotti S, Durham HD, Kay DG, Shaw CA, et al. Progranulin is expressed within motor neurons and promotes neuronal cell survival. BMC Neurosci. 2009;10:130. doi: 10.1186/1471-2202-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi T, Irie T, Kawabata R, Yoshida A, Maruyama H, Kawakami H. Optineurin with amyotrophic lateral sclerosis-related mutations abrogates inhibition of interferon regulatory factor-3 activation. Neurosci Lett. 2011;505:279–281. doi: 10.1016/j.neulet.2011.10.040. [DOI] [PubMed] [Google Scholar]
- Santambrogio L, Belyanskaya SL, Fischer FR, Cipriani B, Brosnan CF, Ricciardi-Castagnoli P, Stern LJ, Strominger JL, Riese R. Developmental plasticity of CNS microglia. Proc Natl Acad Sci U S A. 2001;98:6295–6300. doi: 10.1073/pnas.111152498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargsyan SA, Blackburn DJ, Barber SC, Grosskreutz J, De Vos KJ, Monk PN, Shaw PJ. A comparison of in vitro properties of resting SOD1 transgenic microglia reveals evidence of reduced neuroprotective function. BMC Neurosci. 2011;12:91. doi: 10.1186/1471-2202-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Sedgwick JD, Schwender S, Imrich H, Dörries R, Butcher GW, ter Meulen V. Isolation and direct characterization of resident microglial cells from the normal and inflamed central nervous system. Proc Natl Acad Sci USA. 1991;88:7438–7442. doi: 10.1073/pnas.88.16.7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell SA, Hesse C, Morris SM, Jr, Milcarek C. Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J Biol Chem. 2005;280:39950–39961. doi: 10.1074/jbc.M508848200. [DOI] [PubMed] [Google Scholar]
- Steinacker P, Hendrich C, Sperfeld AD, Jesse S, Lehnert S, Pabst A, von Arnim CA, Mottaghy FM, Uttner I, Tumani H, et al. Concentrations of beta-amyloid precursor protein processing products in cerebrospinal fluid of patients with amyotrophic lateral sclerosis and frontotemporal lobar degeneration. J Neural Transm. 2009;116:1169–1178. doi: 10.1007/s00702-009-0271-4. [DOI] [PubMed] [Google Scholar]
- Sun B, Zhou Y, Halabisky B, Lo I, Cho SH, Mueller-Steiner S, Devidze N, Wang X, Grubb A, Gan L. Cystatin C-cathepsin B axis regulates amyloid beta levels and associated neuronal deficits in an animal model of Alzheimer’s disease. Neuron. 2008;60:247–257. doi: 10.1016/j.neuron.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, Pap A, Szeles L, Poliska S, Oros M, Evans RM, et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARgamma-regulated gene expression in macrophages and dendritic cells. Immunity. 2010;33:699–712. doi: 10.1016/j.immuni.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash JC, Torbett BE, Carson MJ. Developmental regulation of TREM2 and DAP12 expression in the murine CNS: implications for Nasu-Hakola disease. Neurochem Res. 2009;34:38–45. doi: 10.1007/s11064-008-9657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaknin I, Kunis G, Miller O, Butovsky O, Bukshpan S, Beers DR, Henkel JS, Yoles E, Appel SH, Schwartz M. Excess Circulating Alternatively Activated Myeloid (M2) Cells Accelerate ALS Progression While Inhibiting Experimental Autoimmune Encephalomyelitis. PLoS One. 2011;6:e26921. doi: 10.1371/journal.pone.0026921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt W, Lubbert H, Stichel CC. Upregulation of cathepsin S in the aging pathological nervous system of mice. Brain Res. 2008;1232:7–20. doi: 10.1016/j.brainres.2008.07.067. [DOI] [PubMed] [Google Scholar]
- Xiao Q, Zhao W, Beers DR, Yen AA, Xie W, Henkel JS, Appel SH. Mutant SOD1(G93A) microglia are more neurotoxic relative to wild-type microglia. Journal of neurochemistry. 2007;102:2008–2019. doi: 10.1111/j.1471-4159.2007.04677.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05