Global Epistasis Makes Adaptation Predictable Despite Sequence-Level Stochasticity (original) (raw)
. Author manuscript; available in PMC: 2015 Feb 2.
Published in final edited form as: Science. 2014 Jun 27;344(6191):1519–1522. doi: 10.1126/science.1250939
Abstract
Epistatic interactions between mutations can make evolutionary trajectories contingent on the chance occurrence of initial mutations. We used experimental evolution in Saccharomyces cerevisiae to quantify this contingency, finding differences in adaptability between 64 closely related genotypes. Despite these differences, sequencing of 104 evolved clones showed that initial genotype did not constrain future mutational trajectories. Instead, reconstructed combinations of mutations revealed a pattern of diminishing returns epistasis: beneficial mutations have consistently smaller effects in fitter backgrounds. Taken together, these results show that beneficial mutations affecting a variety of biological processes are globally coupled: they interact strongly, but only through their combined effect on fitness. As a consequence, fitness evolution follows a predictable trajectory even though sequence-level adaptation is stochastic.
Epistatic interactions between mutations are pervasive in microbial and viral systems (1-6). In some cases, a single mutation can open up previously unavailable opportunities for a population to colonize a new metabolic niche (2) or survive in a previously intolerable drug concentration (3). Such idiosyncratic epistasis makes evolutionary trajectories dependent on the chance occurrence of initial mutations that constrain or potentiate future adaptation. This historical contingency can render adaptation fundamentally unpredictable (7). However, recent work has also provided evidence for more systematic patterns of epistasis (8-10), which can drive convergent phenotypic evolution (11-13), or lead to parallel adaptation at the sequence level (14). These observations suggest that evolutionary outcomes may be statistically predictable if mutations causing idiosyncratic changes in adaptability are rare, and epistasis instead channels evolution into convergent phenotypic or genotypic pathways.
To test how epistasis and historical contingency affect the predictability of adaptation, we conducted a hierarchical laboratory evolution experiment in S. cerevisiae (Fig. S1). In the first phase of the experiment (“Diversification”), we created 432 independent lines from a single haploid clone (the diversification ancestor, DivAnc) isolated from an earlier long-term evolution experiment (15). We evolved each line independently, half at large and half at small population size, in rich media in 96-well microplates for 240 generations (16). From 64 of these lines we then selected a single clone (“Founders”), chosen to span a range of fitness relative to the DivAnc ((1), Table S1). Founders differ from DivAnc by 4.2 mutations on average (16). In the second phase of the experiment (“Adaptation”), we founded 10 independent replicate populations with each Founder, and allowed each of the resulting 640 lines to adapt at a large population size for 500 generations. This allows us to compare variation among lines descended from the same Founder (which reflects the inherent stochasticity of evolution) to variation between lines descended from different Founders, to assess the extent to which the genetic background influences evolution.
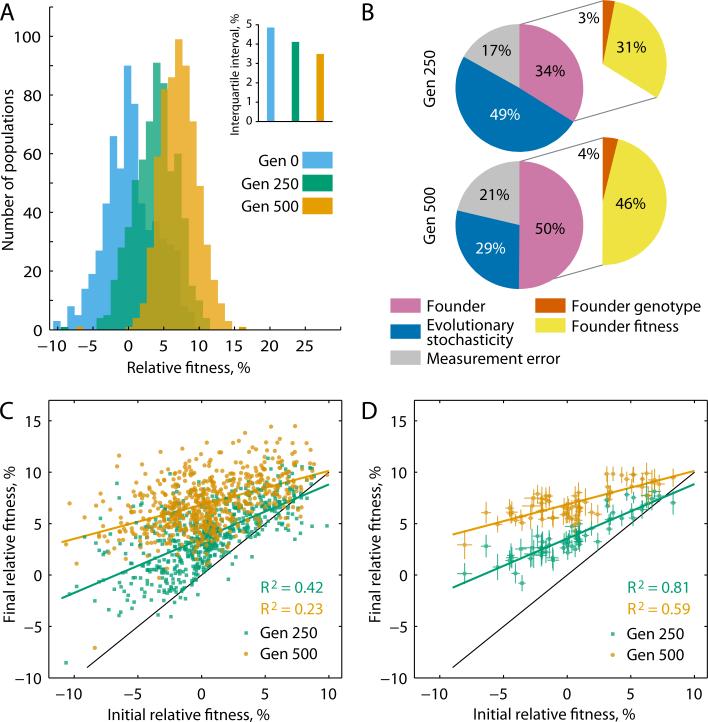
The competitive fitness of each population after 250 and 500 generations of the Adaptation phase increased on average by 3.3% and 6.6%, respectively (Fig. 1A, Table S2). However, not all populations adapted at the same rate. Instead, the initially large variation in fitness between lines declined with time (Fig. 1A). We carried out an analysis of variance (ANOVA) to partition observed variance in fitness increase during the Adaptation phase into contributions from measurement noise, inherent stochasticity of the evolutionary process, and the identity of the Founder (16, 17). After 250 (500) generations of adaptation, inherent stochasticity explains 49% (29%) of the variance in fitness increment, while 17% (21%) is attributed to measurement error and 34% (50%) to the identity of the Founder (Fig. 1B, Table S3). This demonstrates that genetic background is a key determinant of how rapidly a population will adapt.
Figure 1.
Fitness evolution. (A) The distribution of mean population fitness over time, relative to DivAnc. Inset shows inter-population fitness variation over time. (B) Fraction of the variance between lines in fitness increment after 250 and 500 generations of the Adaptation phase that is attributable to each indicated component. All variance components are significant (Table S3). (C) Relationship between Founder fitness and population fitness after 250 and 500 generations of Adaptation. (D) Relationship between Founder fitness and the mean fitness of the 10 independent lines descended from that Founder, after 250 and 500 generations of Adaptation. Error bars show ±1 standard error of the mean (sem).
These differences in adaptability are not random: populations with lower initial fitness systematically adapt more rapidly than populations with higher initial fitness, driving the overall pattern of convergent evolution in fitness (Fig. 1C). We partitioned the variation in fitness increment attributed to Founder identity further and found that after 250 (500) generations of adaptation, 31% (46%) is explained by the fitness of the Founder while only 3% (4%) is determined by its specific genotype (Fig. 1B, Table S3). Thus, the differences in adaptability between Founders are almost entirely predicted by their differences in fitness, and are independent of the specific mutations underlying this fitness. The initial fitness of the Founder therefore predicts the average rate of adaptation in its descendant lines (Fig. 1D). We note, however, that although the effects of specific genotype on adaptability are rare or weak, they are significant (Fig. S2, Tables S3, S4).
A negative correlation between fitness and adaptability has also been observed in prokaryotes (11, 12), and it is consistent with the common observation in evolution experiments that the rate of increase in fitness slows down over time (13, 18). Combined with this earlier work, our results suggest a general “rule of declining adaptability,” which holds for prokaryotes and eukaryotes adapting to rich and minimal media. Further, our observations support a stronger version of this rule: genotypes with lower fitness are more adaptable than those with higher fitness, and distinct genotypes with identical fitness are equally adaptable (up to the rare or weak exceptions noted above). This is consistent with the argument recently presented in (13).
The rule of declining adaptability could arise for two non-exclusive reasons. First, there could only be a few ways to increase in fitness. In this model, high-fitness Founders have lower adaptability because they have already acquired all or most of the possible strong-effect beneficial mutations: they are “running out” of beneficial mutations. In contrast, low-fitness Founders adapt more quickly because they have not yet acquired these mutations. More generally, some groups of mutations may have redundant functional effects (e.g. those that knock out a given pathway). In this case, the number of non-redundant ways to increase fitness would be much smaller than the number of distinct beneficial mutations. We refer to this general form of the running out of mutations hypothesis as the “modular epistasis” model (inspired by (14)): each beneficial mutation improves a single module, mutations within each module are redundant, and high-fitness Founders adapt more slowly because they have fewer remaining modules to improve, especially among those modules that confer the largest fitness gains (16).
Alternatively, mutations arising in higher-fitness backgrounds may be less beneficial than those arising in lower-fitness backgrounds; i.e. diminishing returns epistasis may be pervasive among adaptive mutations, as suggested by (8-10). This epistasis could have two forms. If epistasis is idiosyncratic, mutations may often have widely different effects in different genetic backgrounds (possibly including sign epistasis), but the average effect of a beneficial mutation is smaller in fitter backgrounds. On the other hand, if epistasis is global, each individual beneficial mutation provides a smaller advantage in a fitter genetic background. This latter model implies that the effect of each mutation depends on all other mutations, but only through their combined effect on fitness.
In the modular and idiosyncratic epistasis models, different Founders have different sets of beneficial mutations available to them. Hence in both models we expect lines descended from the same Founder to take more similar mutational trajectories than lines descended from different Founders. In the modular model, we expect each mutation to either confer some fixed advantage (in genotypes lacking mutations in that module) or be neutral (in genotypes that already have a mutation in that module). In the idiosyncratic model, we expect individual mutations to have a variety of different fitness effects in different genetic backgrounds. In contrast to these two models, in the global epistasis model all genotypes acquire beneficial mutations from the same pool, but the advantage conferred by each mutation consistently declines with the fitness of the genetic background. In this model, Founder identity should not affect subsequent mutational trajectories.
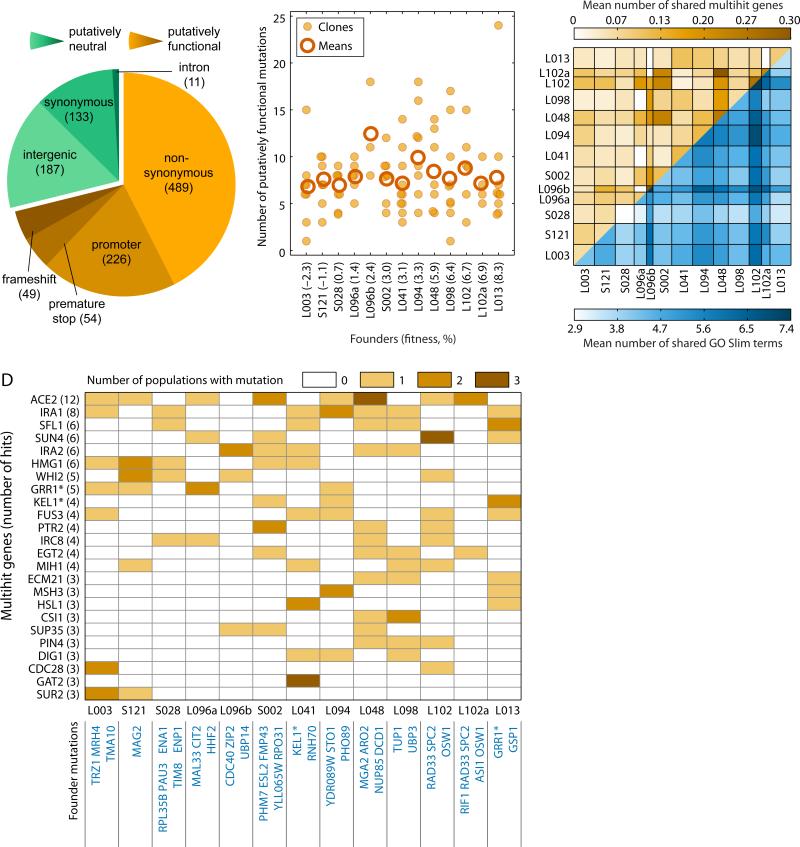
To assess the extent to which these three types of epistasis contribute to the rule of declining adaptability, we sampled one clone from each population descended from 15 Founders at generation 500 of the Adaptation phase and sequenced their complete genomes (16, 19). We found that four sequenced clones acquired a mutator phenotype during the Adaptation phase and two Founders and all their descendants became diploid (Fig. S3). We excluded these from further analysis, leaving a total of 104 sequenced clones descended from 13 Founders (16). We identified a total of 55 mutations that occurred in these Founders during Diversification and 1149 mutations that occurred in their descendants during Adaptation. We annotated each mutation to a gene or intergenic region and classified coding mutations as synonymous or nonsynonymous (Fig. 2A, Table S5). Because most synonymous and intergenic mutations are likely neutral hitchhikers, we restricted analysis to putatively functional nonsense, frameshift, nonsynonymous, and promoter mutations (818 total mutations).
Figure 2.
Sequence-level evolution. (A) Mutations from the Adaptation phase arranged by type. (B) Clones descended from different Founders acquired on average about the same number of putatively functional mutations (see also Fig. S5, S6). (C) Convergence and parallelism at the gene (top, orange) and GO Slim (bottom, blue) levels. Cell color represents the average number of mutations shared by two clones descended from the Founders indicated in the row and column headers. Founders are ordered from least-fit (left, bottom) to most-fit (right, top). Row and column width represents the number of clones sequenced. (D) Mutations in multihit genes and the Founder backgrounds in which they were observed (top); putatively functional mutations that determine the Founder background (bottom). Asterisks indicate genes mutated in both Diversification and Adaptation phases.
In contrast to experiments in bacteria and viruses (14, 20), all but 4 mutations are unique at the nucleotide level, consistent with earlier work in S. cerevisiae (21). However, we found significant gene-level convergent evolution. For example, 24 genes had mutations in at least three replicate lines (compared to 2.7 genes expected by chance, multinomial test P < 0.01 (16); Tables S6, S7), indicating that most mutations observed in these “multi-hit” genes are likely beneficial. Moreover, mutations in genes involved in negative regulation of Ras, cell cycle regulation, and filamentous growth were enriched (Table S8), demonstrating convergence at higher levels of biological organization.
We next compared the total number of mutations observed in different evolved lines. Among lines descended from a given Founder, the lines that increased most in fitness acquired more mutations on average in multi-hit genes, as we expect if these mutations are beneficial (Fig. S4). The modular epistasis model predicts that lines descended from high-fitness Founders should acquire fewer beneficial mutations than those descended from low-fitness Founders, because the former have fewer ways to improve. However, this is not the case: the numbers of putatively functional mutations in lines descended from different Founders are not significantly different (Fig. 2B, Table S9). This result is also surprising under the diminishing returns epistasis models, though not strictly inconsistent with them (22). Since neutral hitchhiker mutations could mask differences in numbers of beneficial mutations between lines (23), we repeated this analysis on more restricted sets of “putatively beneficial mutations” (e.g. those in multi-hit genes (24)). We find similar results in all cases (Fig. S5, S6).
In the modular and idiosyncratic epistasis models, many mutations are beneficial only in particular genetic backgrounds. Hence, these models predict that clones descended from the same Founder should on average have more mutations in common (parallelism) than expected by chance given the observed degree of overall convergence. However, this is not the case. Instead, clones descended from the same Founder are not significantly more likely to share mutations than clones descended from different Founders (Figs. 2C, 2D and S7), as expected in the global diminishing returns epistasis model. This pattern holds regardless of the level at which we define parallelism and convergence (genes or GO Slim categories).
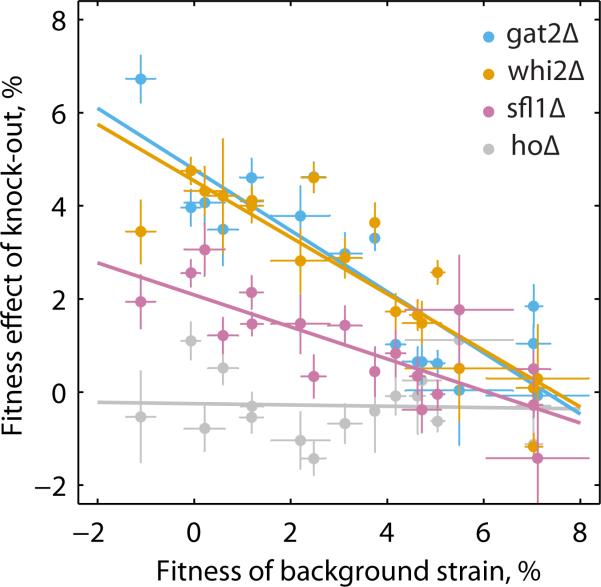
We next selected three genes (SFL1, WHI2, and GAT2) in which we found putative loss-of-function (nonsense or frameshift) mutations in three or more lines, suggesting that knockouts of these genes are beneficial in our system. GAT2 displays the strongest signature of parallel evolution in our data (Fig. 2D), and therefore represents the strongest candidate for idiosyncratic epistasis. We constructed separate targeted knockouts of each of these genes, along with one control gene, HO, in several replicates into all 13 Founders, DivAnc, and four additional clones ((16), Table S1). We measured the fitness effects of each knockout in each background, and found a negative correlation between the fitness effect of the gat2Δ, whi2Δ and sfl1Δ gene deletions and the fitness of the background strain (Fig. 3). Furthermore, there were no idiosyncratic epistatic interactions specific to particular genotypes: up to small deviations, the fitness effect of each knockout depends only on the fitness of the genetic background and not on the specific mutations present in that background.
Figure 3.
Diminishing returns epistasis among specific mutations. The fitness effect of knocking out genes gat2, whi2, and sfl1 declines with the fitness of the background strain. The ho knockout is a negative control. Error bars show ±1 sem over biological replicates.
Taken together, these results support the global diminishing returns epistasis model as the dominant explanation for declining adaptability with increasing fitness, and paint a surprisingly simple picture of adaptation in our system. Many mutations scattered across many biological processes appear to be beneficial. Yet despite their lack of apparent functional relationship, these mutations are globally coupled by diminishing returns epistasis – their effects are strongly mediated by background fitness, but are otherwise essentially independent of the specific identity of mutations present in the background. The biological basis of this global coupling remains unknown. Nevertheless, it leads to a striking pattern of convergent evolution, making fitness evolution relatively predictable. Despite this fitness-level convergence, evolution remains highly stochastic at the genotype level, likely because many distinct mutational paths can lead a population to any given fitness.
Supplementary Material
Supplementary Data
Table S1
Table S2
Table S5
Table S8
Acknowledgements
We thank A. Murray, Q. Justman, B. Good, D. van Dyken, M. McDonald for useful discussions; A. Subramaniam, G. Lang, M. Müller, and J. Koschwanez for experimental advice and strains; and P. Rogers and C. Daly for technical support. This work was supported by the Burroughs Wellcome Foundation (S.K.), NSF graduate research fellowships (D.P.R., E.R.J.), and the James S. McDonnell Foundation, the Alfred P. Sloan Foundation, the Harvard Milton Fund, grant PHY 1313638 from the NSF, and grant GM104239 from the NIH (M.M.D.). Sequence data has been deposited to GenBank under BioProject identifier PRJNA242140.
References Cited
- 1.Woods RJ, et al. Second-Order Selection for Evolvability in a Large Escherichia coli Population. Science. 2011;331:1433. doi: 10.1126/science.1198914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blount ZD, Borland CZ, Lenski RE. Historical Contingency and the Evolution of a Key innovation in an Experimental Population of Escherichia coli. PNAS. 2008;105:7899. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom JD, Gong LI, Baltimore D. Permissive Secondary Mutations Enable the Evolution of Influenza Oseltamivir Resistance. Science. 2010;328:1272. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rokyta DR, et al. Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus. PLoS genetics. 2011;7:e1002075. doi: 10.1371/journal.pgen.1002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burch CL, Chao L. Evolvability of an RNA virus is determined by its mutational neighbourhood. Nature. 2000;406:625. doi: 10.1038/35020564. [DOI] [PubMed] [Google Scholar]
- 6.Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian Evolution Can Follow Only Very Few Mutational Paths to Fitter Proteins. Science. 2006;312:111. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 7.Gould SJ. Wonderful Life. Norton; New York: 1989. [Google Scholar]
- 8.Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. Negative Epistasis Between Beneficial Mutations in an Evolving Bacterial Population. Science. 2011;332:1193. doi: 10.1126/science.1203801. [DOI] [PubMed] [Google Scholar]
- 9.Chou H-H, Chiu H-C, Delaney NF, Segrè D, Marx CJ. Diminishing Returns Epistasis Among Beneficial Mutations Decelerates Adaptation. Science. 2011;332:1190. doi: 10.1126/science.1203799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou H-H, Berthet J, Marx CJ. Fast Growth Increases the Selective Advantage of a Mutation Arising Recurrently during Evolution under Metal Limitation. PLoS genetics. 2009;5:e1000652. doi: 10.1371/journal.pgen.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrick JE, Kauth MR, Strelioff CC, Lenski RE. Escherichia coli rpoB Mutants Have Increased Evolvability in Proportion to Their Fitness Defects. Molecular Biology and Evolution. 2010;27:1338. doi: 10.1093/molbev/msq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perfeito L, Sousa A, Bataillon T, Gordo I. Rates of Fitness Decline and Rebound Suggest Pervasive Epistasis. Evolution. 2013;68:150. doi: 10.1111/evo.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiser MJ, Ribeck N, Lenski RE. Long-Term Dynamics of Adaptation in Asexual Populations. Science. 2013;342:1364. doi: 10.1126/science.1243357. [DOI] [PubMed] [Google Scholar]
- 14.Tenaillon O, et al. The Molecular Diversity of Adaptive Convergence. Science. 2012;335:457. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 15.Lang GI, Botstein D, Desai MM. Genetic Variation And the Fate of Individual Beneficial Mutations in Asexual Populations. Genetics. 2011;188:647. doi: 10.1534/genetics.111.128942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Materials and methods are available as supporting material on Science Online.
- 17.Travisano M, Mongold J, Bennett A, Lenski R. Experimental tests of the roles of adaptation, chance, and history in evolution. Science. 1995;267:87. doi: 10.1126/science.7809610. [DOI] [PubMed] [Google Scholar]
- 18.Kryazhimskiy S, Tkačik G, Plotkin JB. The dynamics of adaptation on correlated fitness landscapes. PNAS. 2009;106:18638. doi: 10.1073/pnas.0905497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Note that this includes two Founders inadvertently picked from the same Diversified population (16).
- 20.Bollback JP, Huelsenbeck JP. Clonal Interference Is Alleviated by High Mutation Rates in Large Populations. Mol Biol Evol. 2007;24:1397. doi: 10.1093/molbev/msm056. [DOI] [PubMed] [Google Scholar]
- 21.Lang GI, et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500:571. doi: 10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.This result is also surprising in the diminishing returns models because we expect fewer beneficial mutations to fix in high-fitness backgrounds where they provide a smaller selective advantage. This puzzle is related to the observation by Ref. (25) that fixation rates in long-term evolution of E. coli are constant through time despite a declining rate of fitness increase. However, our result would be consistent with the diminishing returns models if the beneficial mutation rate is also higher in high-fitness backgrounds.
- 23.See (16) for a power analysis.
- 24.These are likely enriched for the most strongly beneficial mutations. Hence if modular epistasis is prevalent it is among these mutations that we expect the strongest trend.
- 25.Barrick JE, et al. Genome Evolution and Adaptation in a Long-Term Experiment with Escherichia Coli. Nature. 2009;461:1243. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 26.Lang GI, Murray AW, Botstein D. The Cost of Gene Expression Underlies a Fitness Trade-Off in Yeast. Proceedings of the National Academy of Sciences. 2009;106:5755. doi: 10.1073/pnas.0901620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman F, Fink G, Lawrence C. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1974. [Google Scholar]
- 28.Smukalla S, et al. FLO1 Is a Variable Green Beard Gene that Drives Biofilm-like Cooperation in Budding Yeast. Cell. 135:726. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breslow DK, et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Meth. 2008;5:711. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meiron H, Nahon E, Raveh D. Identification of the heterothallic mutation in HO-endonuclease of S. cerevisiae using HO/ho chimeric genes. Curr Genet. 1995;28:367. doi: 10.1007/BF00326435. [DOI] [PubMed] [Google Scholar]
- 31.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J. On the evolution of codon volatility. Genetics. 2005;169:495. doi: 10.1534/genetics.104.034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Table S1
Table S2
Table S5
Table S8