Inverse Size Scaling of the Nucleolus by a Concentration-Dependent Phase Transition (original) (raw)
. Author manuscript; available in PMC: 2016 Mar 2.
Published in final edited form as: Curr Biol. 2015 Feb 19;25(5):641–646. doi: 10.1016/j.cub.2015.01.012
Summary
Just as organ size typically increases with body size, the size of intracellular structures changes as cells grow and divide. Indeed, many organelles, such as the nucleus [1, 2], mitochondria [3], mitotic spindle [4, 5] and centrosome [6], exhibit size scaling, a phenomenon in which organelle size depends linearly on cell size. However, the mechanisms of organelle size scaling remain unclear. Here, we show that the size of the nucleolus, a membrane-less organelle important for cell size homeostasis [7], is coupled to cell size by an intracellular phase transition. We find that nucleolar size directly scales with cell size in early C. elegans embryos. Surprisingly, however, when embryo size is altered, we observe inverse scaling: nucleolar size increases in small cells and decreases in large cells. We demonstrate that this seemingly contradictory result arises from maternal loading of a fixed number, rather than a fixed concentration of nucleolar components, which condense into nucleoli only above a threshold concentration. Our results suggest that the physics of phase transitions can dictate both whether an organelle assembles, and if so, its size, providing a mechanistic link between organelle assembly and cell size. Since the nucleolus is known to play a key role in cell growth, this biophysical read-out of cell size could provide a novel feedback mechanism for growth control.
Results and Discussion
To characterize nucleolar size as a function of cell size, we utilized the stereotypical changes in cell size resulting from the reductive divisions of developing C. elegans embryos. We acquired 3D time-lapse images of early embryos expressing a GFP fusion of fibrillarin-1 (FIB-1), a well-conserved nucleolar protein [8]. In the earliest stages (1–2 cell), FIB-1::GFP remains diffuse throughout the nucleoplasm and no nucleoli are observed (Figure S1A). Beginning at the 4-cell stage, two discrete foci, corresponding to the two nucleolar organizing regions found in diploid C. elegans, transiently appear in cell EMS and occasionally in P2 (8/30 embryos; Table S1). In subsequent cell cycles, two bright foci assemble and disassemble in every cell except those of the P (germline) lineage (Figure 1A, Movie S1).
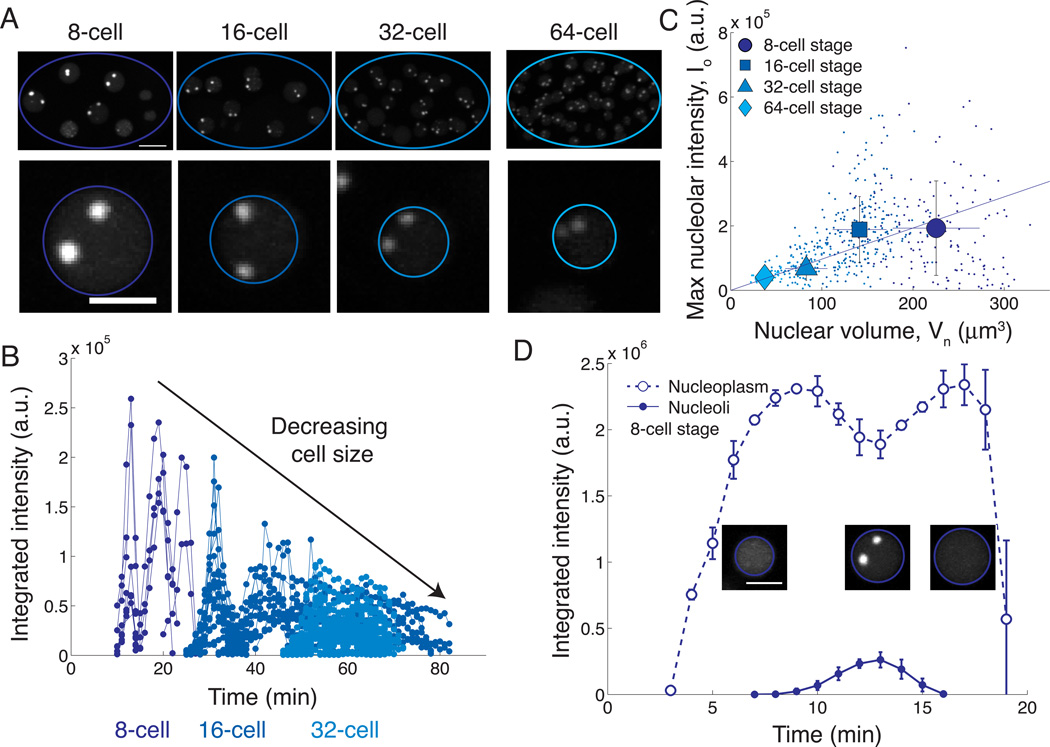
Figure 1.
Nucleolar size scales directly with cell and nuclear volume during development of early C. elegans embryos.
(A) Maximum intensity projections of 3D stacks of a control embryo expressing FIB-1::GFP at various stages. 8, 16 and 32-cell stages were taken from a time-lapse movie of a single embryo; 64-cell stage represents a different embryo. Lower panel shows individual nuclei with assembled nucleoli. Scalebar = 10 µm for whole embryos; 5 µm for individual nuclei.
(B) Integrated intensity in arbitrary units of individual nucleoli as a function of time in a developing control embryo. Colors correspond to cell stage as indicated below. Time was measured relative to nuclear envelope breakdown in cells ABa and ABp.
(C) Direct scaling of maximum nucleolar intensity with nuclear volume for embryos at the 8 to 64-cell stages. Data from time-lapse movies (n = 10) and snapshot images (n = 10–15 per stage) are plotted together. Raw data (points) and mean ± standard deviation for each cell stage (8, circle; 16, square; 32, triangle; 64, diamond) are shown with a linear fit through the origin. r2 = 0.15; p = 9.1 × 10−23 by two-tailed t-test.
(D) The nucleoplasmic pool of FIB-1::GFP is depleted as nucleoli assemble. Mean ± standard deviation of the integrated intensity of the nucleoplasm and nucleoli are plotted as a function of time for the 8-cell stage AB-lineage nuclei in the embryo shown in panel A.
See also Figure S1 and Movie S1.
Nucleoli are typically brightest in the 8-cell stage and their fluorescence intensity subsequently decreases as cells continue dividing (Figure 1A, B). We confirmed that this is not due to photobleaching, as images taken of embryos at different stages give similar intensity values. Since the absolute size of these organelles spans the diffraction limit, we use integrated intensity as a metric for nucleolar size (Figure S1D, F). Nuclear size scales with cell size in C. elegans embryos [11], such that the volume ratio is roughly constant: ξ = Vn/Vcell, where ξ is the karyoplasmic ratio [1, 2] (Figure S1H). Using nuclear size as a proxy for cell size, we find a significant correlation between the maximum nucleolar intensity, Io, summed over all nucleoli in a given nucleus (Figure S1F), and nuclear volume, Vn(Figure S1G) for embryos in the 8- to 64-cell stages (Figure 1C). Thus, for these embryonic stages, nucleoli tend to be larger in larger cells and smaller in smaller cells, consistent with previous reports of direct scaling of nucleolar size with cell size [2, 9, 10].
Previous models of organelle size scaling [4–6, 12, 13] have proposed that finite pools of components can couple organelle size to cell size. This is due to the fact that, for fixed concentrations, small cells have fewer components than large cells, resulting in proportionately smaller organelles. We found that the integrated intensity of FIB-1::GFP in the nucleoplasm decreases as nucleoli begin assembling (Figure 1D). This depletion suggests that the number of FIB-1 molecules in the nucleoplasm may be limiting for nucleolar assembly, consistent with previous models. However, even at the peak of nucleolar assembly, a significant nucleoplasmic pool remains.
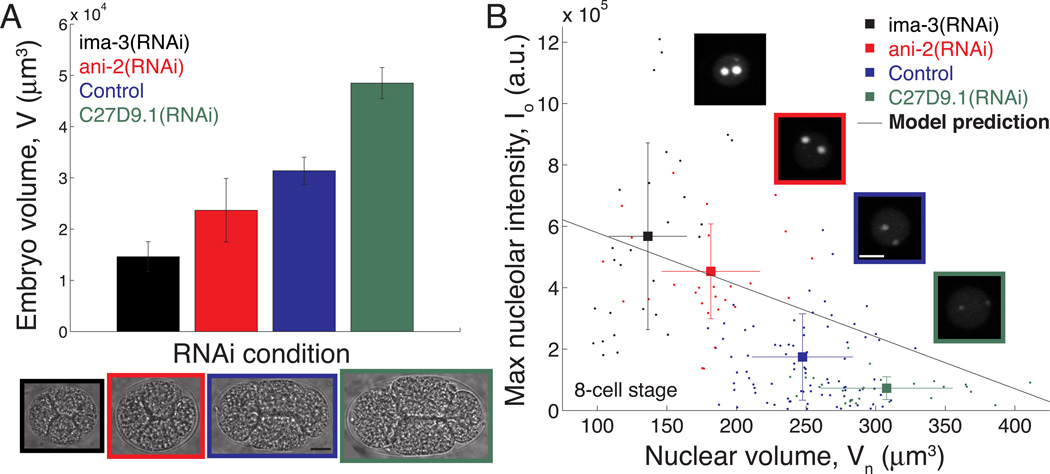
To test whether nucleolar size scaling is indeed a consequence of cell volume changes, we used RNAi to change embryo size (Figure 2A). Following knockdown of the anillin homolog ANI-2 [14], we observed embryos that are ~25% smaller than control embryos. Surprisingly, instead of a corresponding decrease in organelle size, we found a significant increase in maximum nucleolar intensity in small ani-2(RNAi) embryos compared to control embryos at the 8-cell stage (Figure 2B). ANI-2 plays a role in structurally organizing the syncytial gonad [14] and it is possible that this unexpected result arises from this, or some other, function of ANI-2. To rule out this possibility, we tested a different RNAi condition: knockdown of the importin α IMA-3 [15], which produces even smaller embryos (~55% smaller than control). These small embryos also assembled large nucleoli (Figure 2B). We next sought to increase embryo size using RNAi knockdown of the gene C27D9.1 [16], which results in embryos ~55% larger than control. Consistent with the inverse size scaling seen in ani-2(RNAi) and ima-3(RNAi), we found that nucleolar size decreases significantly in large C27D9.1(RNAi) embryos (Figure 2B). We observed similar behavior for DAO-5::GFP, another nucleolar marker [17] (Figure S2A). These RNAi results show that the size of the nucleolus is indeed sensitive to cell volume, but in exactly the opposite manner predicted by a limiting component mechanism of direct size scaling.
Figure 2.
Nucleolar size scales inversely with nuclear volume following RNAi.
(A) RNAi knockdown of select genes produces embryos of different size. n = 25 embryos for control; 10 embryos for each RNAi condition. Images depict 4-cell stage embryos following RNAi. Scalebar = 10 µm.
(B) Inverse scaling of maximum nucleolar intensity with nuclear volume across RNAi conditions at the 8-cell stage. Raw data (points) and mean ± standard deviation for each condition (squares) are shown with the model prediction, Io = α[(N/8)-CsatVn] (solid line). n = 25 embryos for control; 10 embryos for each RNAi condition. The means are all statistically different; p = 1.3 × 10−34 by ANOVA. Representative images of ABal nuclei are shown for each condition. Scalebar = 5 µm.
Interestingly, although nucleolar size scales inversely with cell size across RNAi conditions at a particular developmental stage (e.g., 8-cell stage embryos), within each RNAi condition we still find direct scaling of nucleolar size with cell size during development (Figure 3A). However, the slopes of these data are not the same. Small embryos (ima-3 and ani-2 RNAi) have greater slopes than large embryos (C27D9.1 RNAi). This slope is the ratio of maximum nucleolar intensity to nuclear volume, Io/Vn, and thus represents an apparent concentration. When we plot Io/Vn as a function of embryo volume, V, we find a strong inverse relationship (Figure 3B), suggesting that the concentration of nucleolar components may not be fixed, but may instead decrease as embryo size increases.
Figure 3.
Maternal loading of an intact nucleolus results in concentration differences that explain direct and inverse scaling regimes.
(A) Direct scaling of maximum nucleolar intensity with nuclear volume during development in each RNAi condition; inverse scaling across RNAi conditions. Raw data (points) and mean ± standard deviation across 50-µm bins (squares) are shown for embryos at the 8 to 64-cell stages. Raw data for each RNAi condition were fit to a line through the origin to determine the slope, Io/Vn. n = 20–25 embryos per stage for control; 8–15 embryos per stage for each RNAi condition.
(B) Fitted slopes from panel A are plotted as a function of mean embryo volume for each RNAi condition. Error bars are 95% confidence intervals. The master scaling equation, Io/Vn = α[N/(Vξ)-Csat], is plotted with zero free parameters (solid line). Dashed lines represent the range of uncertainty in model parameters.
(C) Nucleoli are loaded into oocytes intact. Integrated intensity (mean ± standard deviation) of nucleoli in the first cellularized oocyte in the hermaphrodite gonad for each RNAi condition (n = 10 oocytes per condition). Wild-type (WT) RNAi conditions are not statistically different; p = 0.73 by ANOVA. ncl-1 is statistically different from all WT RNAi conditions; p = 0.0038 by ANOVA. Image shows WT control gonad expressing fluorescent markers for cell membranes (red) and nucleoli (green). White arrow indicates the intact nucleolus loaded into an oocyte.
(D) Nuclear concentration decreases with increasing embryo volume. Raw data (points) and mean ± standard deviation for each condition (squares) are shown with a fit to the equation Cn = N/(ξV) for WT embryos (filled markers; solid line) and ncl-1 mutant embryos (open markers; dashed line). n = 15 embryos for WT control; 10, WT ima-3(RNAi); 14, WT ani-2(RNAi); 11, WT C27D9.1(RNAi); 10, ncl-1 control; 12, ncl-1 ima-3(RNAi) 16, ncl-1 ani-2(RNAi); 18, ncl-1 C27D9.1(RNAi). (inset) Schematic diagram of nucleoli loaded into oocytes of different size that subsequently disassemble to yield different concentrations in the embryos.
(E) Schematic diagram illustrating the direct and inverse scaling regimes.
See also Figure S2.
Prior to the onset of significant zygotic translation, the concentration of a typical protein in the early embryo is established by the concentration loaded into each oocyte. Oogenesis occurs in the syncytial gonad of C. elegans hermaphrodites by cellularizing gonad cytoplasm [18]. Thus, molecules dissolved in this cytoplasm should give rise to the same concentration in all embryos, regardless of embryo size (Figure S2E). However, using a cross between our FIB-1::GFP line and a line expressing mCherry::PH(PLC1δ1) to visualize cell membranes, we found that the nucleus and a single large nucleolus are loaded into oocytes while they are still fully assembled (Figure 3C). The nucleolus eventually disassembles as the oocyte matures, but it is intact when the oocyte closes off from the syncytium, typically around position 5 in wild-type (WT) control animals (white arrow, Figure 3A). The integrated intensity of the nucleolus loaded into the first cellularized oocyte was the same for all RNAi conditions (ani-2, ima-3 and C27D9.1) (Figure 3C, SF), suggesting that the number of nucleolar components loaded into each oocyte is fixed. This is in contrast to centrosomes and mitotic spindles, which are completely disassembled during oogenesis, leading to a fixed concentration of nucleolar components in each oocyte (Figure S2E). Furthermore, we found that the total fluorescence intensity within an embryo is equal to this maternal load until approximately the 128-cell stage (Figure S2G). This indicates that there is no significant zygotic contribution of FIB-1 protein in these early embryos.
The loading of a fixed number of components should result in concentration differences between embryos of different size (inset, Figure 3D). Since the total embryonic concentration of a given nucleolar component, C, is equal to the number of molecules, N, divided by the embryo volume, C = N/V, we predict that small embryos will have a high concentration of FIB-1::GFP and large embryos will have a low concentration. To test this prediction, we directly measured FIB-1::GFP intensity in the nucleoplasm of embryonic cells, prior to nucleolar assembly. The average nuclear concentration, Cn, indeed decreases with increasing embryo size across all RNAi conditions (Figure 3D); DAO-5::GFP exhibits a similar, albeit weaker, concentration decrease (Figure S2D). We fit these data to the function Cn = N/(ξV) to determine the number of FIB-1::GFP molecules loaded per embryo, N = 1.66 ± 0.11×105. The factor ξ arises because nucleolar components are concentrated within the nucleus, such that the nuclear concentration is scaled by the karyoplasmic ratio: Cn = C/ξ. These data show that the concentration of nucleolar components varies across different RNAi conditions, violating the underlying assumption of limiting component models of organelle size scaling – namely, that cells of different size have the same component concentration [6, 12, 13].
Nucleoli behave as liquid phase RNA/protein droplets [10, 19] and their assembly could be related to the emerging concept of intracellular phase transitions [20–23]. To test this hypothesis, we developed a simple mathematical model to describe nucleolar assembly, based on the physics of phase transitions [24, 25] (see Supplemental Experimental Procedures). At steady state, we assume that nucleolar components partition between a condensed droplet phase and a soluble pool with concentration Csat. The saturation concentration Csat represents a threshold: if the nuclear concentration is below Csat, nucleoli do not assemble and FIB-1 remains soluble. When the nuclear concentration is above Csat, molecules from the soluble pool condense into nucleolar droplets, depleting the nucleoplasm until its concentration reaches Csat. The final size of the nucleolus is thus determined by the difference between the total concentration in the nucleus and this saturation concentration: Io = α[Cn-Csat]Vn. Here, α is the intensity per molecule.
In a developing embryo, the nucleolus is disassembled during each mitotic cleavage and nucleolar components are allocated proportionately to daughter cells. Therefore, Cn is fixed during early development and the model predicts direct scaling of nucleolar size with nuclear and cell volume: Io ~ Vn ~ Vcell. However, across RNAi conditions, Cn changes (Figure 3D). By expressing the nuclear concentration as Cn = N/(ξV), as above, we obtain a master scaling equation, Io = α[N/(Vξ)-Csat]Vn. Using the relation V ≈ mVcell = mVn/ξ, where the parameter m indicates a particular developmental stage (e.g., 8-cell stage: m=8), we can write Io = α[(N/m)-CsatVn]. Thus, across RNAi conditions, where N is fixed, the model predicts an inverse scaling relationship at a given cell stage, with larger cells/nuclei assembling smaller nucleoli, and vice versa. A schematic diagram illustrating the model’s prediction of direct vs. inverse scaling regimes is shown in Figure 3E.
To quantitatively test this model, we compared both the direct and inverse scaling data with the prediction of our master equation. From independent experiments, we directly measured the value of each parameter: α, N, ξ and Csat (see Supplemental Experimental Procedures). The model predictions show good agreement with experiment for both the direct scaling regime, Io/Vn = α[N/(Vξ)-Csat] (Figure 3B), and the inverse scaling regime, Io = α[(N/m)-CsatVn] where m = 8 (Figure 2B). This agreement is remarkable, given that the prediction involves zero free parameters, rather than a fit to the model.
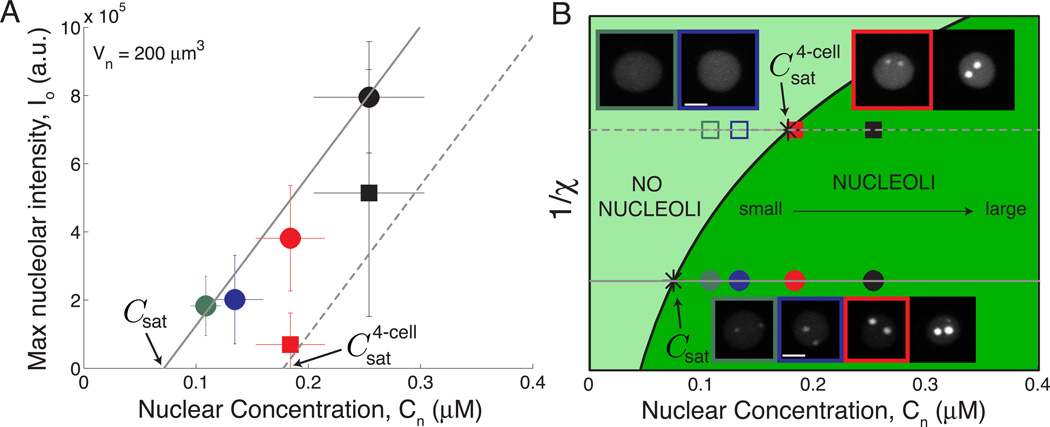
The role of the saturation concentration, Csat, can be highlighted by plotting the maximum nucleolar intensity for a given size cell/nucleus, Io(Vn = 200 µm3), as a function of nuclear concentration for all RNAi conditions. As nuclear concentration decreases and approaches Csat, nucleoli become smaller (circles, Figure 4A).
Figure 4.
A concentration-dependent phase transition controls nucleolar size and assembly.
(A) Maximum nucleolar intensity increases with nuclear concentration above Csat for a given nuclear volume, Vn = 200 µm3. Circles correspond to nucleoli at the 8-cell stage; triangles correspond to nucleoli at the 4-cell stage. Solid line is the model’s prediction, Io = α[Cn-Csat]Vn, for the 8-cell stage. Dashed line is the model’s prediction, Io = α[Cn-Csat4-cell]Vn, for the 4-cell stage.
(B) Phase diagram for nucleolar assembly. Asterisks mark the measured saturation concentration at the 4- and 8-cell stages. Circles correspond to embryos at the 8-cell stage; triangles correspond to embryos at the 4-cell stage. Open symbols indicate no nucleolar assembly. Lines from panel A correspond to horizontal lines on the phase diagram. Representative images of ABal nuclei are shown for 8-cell stage embryos; ABa nuclei are shown for 4-cell stage embryos.
See also Figure S3 and Table S1.
The dependence of nucleolar size and assembly on nuclear concentration can be summarized in a phase diagram (Figure 4B). Here, Csat represents the boundary between nucleoplasm consisting of a single phase of dissolved nucleolar components and nucleoplasm that has phase-separated to form condensed nucleoli that coexist with a dissolved phase of concentration Csat. The nuclear concentration for each RNAi condition falls above Csat, within the phase-separated region, consistent with the fact that nucleoli always assemble in 8- to 64-cell stage embryos. Though we were unable to experimentally reduce nuclear concentration below Csat, our model predicts that nucleoli would not assemble when Cn < Csat and nucleolar components would remain dissolved in the nucleoplasm.
Interestingly, nucleoli indeed do not assemble in very early embryos, suggesting that there is some developmentally regulated parameter, which we call χ (see Supplemental Experimental Procedures), that shifts the phase boundary and thus increases the saturation concentration. Specifically, the anterior cells ABa and ABp do not assemble nucleoli in 4-cell stage control embryos (Figure S1A, Table S1). Remarkably, however, we could induce nucleolar assembly in these early blastomeres by decreasing embryo size and thus increasing the concentration of nucleolar components. Experimental measurements of Csat in the 4-cell stage, Csat4-cell = 0.18 ± 0.04 µM, indicate that the nuclear concentrations in our RNAi conditions span this phase boundary, such that nucleoli do not assemble in embryos where Cn < Csat4-cell (_C27D9.1(RNAi)_ and control), while they do assemble in embryos where Cn > Csat4-cell (ima-3 RNAi) (Figure 4B). The ani-2(RNAi) condition presents an interesting case where Cn ≈ Csat4-cell. Consistent with a close proximity to the phase boundary, nucleoli in ABa cells of ani-2(RNAi) embryos are either very small, or not detected at all (13/28 embryos; Table S1).
To further test our model, we sought to change nuclear concentration by manipulating the maternal load of nucleolar components (i.e. the parameter N), in addition to the embryo volume (V). Mutants of the BRAT homologue NCL-1 exhibit enlarged nucleoli throughout the body [26]. Indeed, the size of the nucleolus loaded into ncl-1(e1942) oocytes is nearly 2-fold larger than WT (Figure 3C). The nuclear concentration of FIB-1::GFP in ncl-1 mutant embryos also depends on embryo volume (Fig. 3D), and we fit this data to the equation Cnncl1=Nncl1/(ξV) to determine the number of molecules loaded per embryo: Nncl1 = 4.45 ± 0.45×105, approximately 2.7 times greater than WT embryos. As in WT embryos, nucleolar size peaks at the 8-cell stage, but the maximum integrated intensity in ncl-1 mutants is more than twice that of WT embryos (Figure S1B). Consistent with our finding that decreasing embryo size induces nucleolar assembly in early blastomeres, ABa and ABp assemble nucleoli in ncl-1 embryos of any size. Thus, by increasing nuclear concentration through changing either N or V, we could cross the phase boundary and induce nucleolar assembly (Figure S1A, S3).
Our results demonstrate that nucleoli assemble in a cell size-dependent manner, which has important implications for cell growth and size control. The connection between organelle size and cell size is mediated through the concentration of nucleolar components. Below a threshold concentration, nucleoli do not assemble. Above this threshold, the higher the concentration, the larger the size of the assembled nucleolus. Threshold concentrations are a hallmark of phase transitions [24, 25], strongly suggesting that nucleolar assembly represents an intracellular phase transition. Such concentration-dependent phase transitions may represent a general biophysical framework for understanding organelle assembly and scaling [27].
Experimental Procedures
C. elegans strains were maintained using standard techniques.
Imaging
Embryos were dissected from gravid hermaphrodites and imaged on M9-agarose pads. Images were acquired on a two-photon laser scanning system custom-built around an upright Olympus BX51 microscope. Emitted light was collected with a 40X/NA0.8 water immersion objective and an NA1.3 oil immersion condenser and detected with high quantum efficiency GaAsP photomultiplier tubes (Hammamatsu). 3D volumes were acquired using an objective piezo controlled by ScanImage software [28].
Image analysis
Images were analyzed with custom software in Matlab. Nucleolar intensity was calculated by summing the fluorescence intensity within objects detected with a 3D bandpass filter.
Concentration estimates
Pixel intensities in the nucleoplasm were calibrated using purified His-tagged FIB-1::GFP.
Model Parameters
All model parameters were measured independently to produce a zero-free parameter prediction. For details on the model and parameter estimation, please refer to the Supplemental Experimental Procedures.
Supplementary Material
1
2
3
Acknowledgements
We thank Joel Berry, Chase Broedersz, Mikko Haataja, Tony Hyman, Sravanti Uppaluri and members of the Brangwynne lab for helpful discussions; Stephan Thiberge and Evangelos Gatzogiannis for imaging advice; and Nilesh Vaidya for protein purification. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the NIH Director's New Innovator Award (1DP2GM105437-01), the Searle Scholars Program, an NSF CAREER Award (1253035), and a Damon Runyon Postdoctoral Fellowship (S.C.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Neumann FR, Nurse P. Nuclear size control in fission yeast. J. Cell Biol. 2007;179:593–600. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell. 2007;18:3523–3532. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rafelski SM, Viana MP, Zhang Y, Chan Y-HM, Thorn KS, Yam P, Fung JC, Li H, da F Costa L, Marshall WF. Mitochondrial network size scaling in budding yeast. Science. 2012;338:822–824. doi: 10.1126/science.1225720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science. 2013;342:853–856. doi: 10.1126/science.1243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. Cytoplasmic volume modulates spindle size during embryogenesis. Science. 2013;342:856–860. doi: 10.1126/science.1243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decker M, Jaensch S, Pozniakovsky A, Zinke A, O'Connell KF, Zachariae W, Myers E, Hyman AA. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr. Biol. 2011;21:1259–1267. doi: 10.1016/j.cub.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Jorgensen P, Nishikawa JL, Breitkreutz B-J, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 8.Jansen RP, Hurt EC, Kern H, Lehtonen H, Carmo-Fonseca M, Lapeyre B, Tollervey D. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J. Cell Biol. 1991;113:715–729. doi: 10.1083/jcb.113.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berciano MT, Novell M, Villagra NT, Casafont I, Bengoechea R, Val-Bernal JF, Lafarga M. Cajal body number and nucleolar size correlate with the cell body mass in human sensory ganglia neurons. J. Struct. Biol. 2007;158:410–420. doi: 10.1016/j.jsb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Feric M, Brangwynne CP. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat. Cell Biol. 2013;15:1253–1259. doi: 10.1038/ncb2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hara Y, Kimura A. Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr. Biol. 2009;19:1549–1554. doi: 10.1016/j.cub.2009.07.050. [DOI] [PubMed] [Google Scholar]
- 12.Marshall WF. Centrosome size: scaling without measuring. Curr. Biol. 2011;21:R594–R596. doi: 10.1016/j.cub.2011.06.036. [DOI] [PubMed] [Google Scholar]
- 13.Goehring NW, Hyman AA. Organelle growth control through limiting pools of cytoplasmic components. Curr. Biol. 2012;22:R330–R339. doi: 10.1016/j.cub.2012.03.046. [DOI] [PubMed] [Google Scholar]
- 14.Maddox AS, Habermann B, Desai A, Oegema K. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development. 2005;132:2837–2848. doi: 10.1242/dev.01828. [DOI] [PubMed] [Google Scholar]
- 15.Geles KG, Adam SA. Germline and developmental roles of the nuclear transport factor importin α3 in C. elegans. Development. 2001;128:1817–1830. doi: 10.1242/dev.128.10.1817. [DOI] [PubMed] [Google Scholar]
- 16.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume A-M, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 17.Korčeková D, Gombitová A, Raška I, Cmarko D, Lanctôt C. Nucleologenesis in the Caenorhabditis elegans embryo. PLoS ONE. 2012;7:e40290. doi: 10.1371/journal.pone.0040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolke U, Jezuit EA, Priess JR. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development. 2007;134:2227–2236. doi: 10.1242/dev.004952. [DOI] [PubMed] [Google Scholar]
- 19.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. SciU.SA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 21.Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, ll W, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Hyman AA, Simons K. Beyond oil and water - phase transitions in cells. Science. 2012;337:1047–1049. doi: 10.1126/science.1223728. [DOI] [PubMed] [Google Scholar]
- 24.Lifshitz IM, Slyozov VV. The kinetics of precipitation from supersaturated solid solutions. Journal of Physics and Chemistry of Solids. 1961;19:35–50. [Google Scholar]
- 25.Goldenfeld N. Lectures on phase transitions and the renormalization group (Westview Press) 1992 [Google Scholar]
- 26.Frank DJ, Roth MB. ncl-1 is required for the regulation of cell size and ribosomal RNA synthesis in Caenorhabditis elegans. J. Cell Biol. 1998;140:1321–1329. doi: 10.1083/jcb.140.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 2013;203:875–881. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pologruto TA, Sabatini BL, Svoboda K. ScanImage: Flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3