Mismatch Repair during Homologous and Homeologous Recombination (original) (raw)
Abstract
Homologous recombination (HR) and mismatch repair (MMR) are inextricably linked. HR pairs homologous chromosomes before meiosis I and is ultimately responsible for generating genetic diversity during sexual reproduction. HR is initiated in meiosis by numerous programmed DNA double-strand breaks (DSBs; several hundred in mammals). A characteristic feature of HR is the exchange of DNA strands, which results in the formation of heteroduplex DNA. Mismatched nucleotides arise in heteroduplex DNA because the participating parental chromosomes contain nonidentical sequences. These mismatched nucleotides may be processed by MMR, resulting in nonreciprocal exchange of genetic information (gene conversion). MMR and HR also play prominent roles in mitotic cells during genome duplication; MMR rectifies polymerase misincorporation errors, whereas HR contributes to replication fork maintenance, as well as the repair of spontaneous DSBs and genotoxic lesions that affect both DNA strands. MMR suppresses HR when the heteroduplex DNA contains excessive mismatched nucleotides, termed homeologous recombination. The regulation of homeologous recombination by MMR ensures the accuracy of DSB repair and significantly contributes to species barriers during sexual reproduction. This review discusses the history, genetics, biochemistry, biophysics, and the current state of studies on the role of MMR in homologous and homeologous recombination from bacteria to humans.
The processes of homologous recombination and mismatch repair are linked. They occur in both germline cells (during meiosis) and in somatic cells (during the repair of DNA damage).
Genetic recombination between unrelated parental DNAs during sexual reproduction appears to solve the problem of Muller’s ratchet, a process in which the accumulation of deleterious spontaneous mutations by asexual organisms eventually leads to extinction (Muller 1964; Felsenstein 1974). Recombination of chromosomes during sexual reproduction (meiosis) is considered to generate competitive genetic hybrids from two potentially noncompetitive mutant parents. The cytological observation of chromosome crossovers (COs) in the early days of Drosophila genetics led to their recognition as important intermediates in the formation of genetic hybrids (Muller 1916). These COs were given a molecular framework with the theoretical description of the Holliday junction (Holliday 1964). The fundamental insight of a Holliday junction was that single DNA strands from two parental chromosomes crisscross to form hybrid DNA duplexes. Resolution of the crossed strands of a Holliday junction may then result in CO of genes on either side of the hybrid DNA duplexes. Sequence differences between the parental DNAs were proposed to result in mismatched nucleotides within the hybrid DNA duplex.
The repair of mismatched nucleotides was proposed nearly simultaneously in 1964 by Evelyn Witkin to account for the processing of brominated nucleotides in bacteria and by Robin Holliday to explain gene conversion following recombination (Holliday 1964; Witkin 1964). Mismatch repair (MMR) was proposed to recognize nucleotide mismatches and perform an excision–resynthesis reaction in which one strand is excised within the hybrid DNA duplex and the remaining strand is used as a template for resynthesis. In 1973, a genetic basis for MMR was discovered when the hexA mutation of Pseudomonas was found to be defective in gene conversion (Tiraby and Fox 1973). The HexA gene turned out to be a homolog of the “Siegel mutator” (MutS), originally described by Eli Siegel in 1967 (Siegel and Bryson 1967). The discovery of MutS added to a growing number of genes with historical roots in the 1954 genetic description of the “Treffers mutator” (MutT) (Treffers et al. 1954). Mutation of these Mut genes substantially elevated spontaneous mutation rates in bacteria (termed mutator). Today, most of the Mut genes are known to play a role in the processing of replication misincorporation errors, DNA double-strand breaks (DSBs), and chemical or physical damage to nucleotides (for a review, see Miller 1998; Ciccia and Elledge 2011).
In the early 1980s, a series of clever observations from a number of laboratories showed that a subset of the Escherichia coli mutator genes, MutS, MutL, MutH, and UvrD(MutU), operated in the MMR of polymerase misincorporation errors that occurred frequently during replication (Radman et al. 1980). The recognition of transient undermethylation (hemimethylation) at newly replicated DNA adenine methylation (Dam) GATC sequences was identified as the mechanism for discriminating the error-containing DNA strand (Marinus 1976). Notably, this Dam-directed postreplication MMR mechanism only appears to operate in a subset of γ-proteobacteria, such as E. coli. The mechanism for strand discrimination in Eubacteria, Archea, and Eukaryotes remains speculative. However, as might be expected for important genome maintenance processes, the core MutS homologs (MSHs) and MutL homologs (MLHs)/postmeiotic segregation (PMS) of MMR, have been highly conserved throughout the taxonomic domains (Table 1).
Table 1.
Parallels between the proteins involved in MMR and heteroduplex rejection during recombination
| Escherichia coli | S. cerevisae | Human | Function | Role |
|---|---|---|---|---|
| MutS | ScMsh2-ScMsh6 | HsMSH2-HsMSH6 | Recognition of mismatches and small IDLs; sliding clamp | MMR; ICL repair; gene conversion; heteroduplex rejection |
| ScMsh2-ScMsh3 | HsMSH2-HsMSH3 | Recognition of large IDLs and branched structures; sliding clamp | Postreplicative repair of IDLs; 3′-flap processing; SSA intermediate stabilization | |
| MutL | ScMlh1-ScPms1 | HsMLH1-HsPMS2 | Downstream mediator; endonuclease | MMR; gene conversion |
| ? | ScRad1-ScRad10 | HsXPF-HsERCC1 | Structure-selective nuclease | 3′-flap removal |
| RecJ | ScExo1 | HsEXO1 | 5′ → 3′ exonuclease | MMR; HR |
| ExoVII | Bidirectional exonuclease | MMR | ||
| ExoI, ExoX | 3′ → 5′ exonuclease | |||
| UvrD | ScSgs1 | HsRECQ1 | Structure-selective DNA helicase | MMR; unwinding heteroduplex DNA |
| HsBLM | ||||
| HsWRN |
In 1993, a human MSH (HsMSH2) was found to be associated with the common cancer predisposition syndrome hereditary nonpolyposis colorectal cancer (HNPCC; Fishel et al. 1993). Verification of this association (Leach et al. 1993) and the rapid successive discovery of other MSH and MLH/PMS homolog genes linked with HNPCC and sporadic colorectal cancers have solidified a role for MMR defects in tumor development (Bronner et al. 1994; Nicolaides et al. 1994; Papadopoulos et al. 1994, 1995). These observations also underlined the importance of mutators in driving the enormous numbers of mutations required in the microevolutionary selection processes associated with many forms of tumorigenesis (Loeb 1991, 2001; Fishel 2001).
HOMOLOGOUS RECOMBINATION
In addition to meiosis, genetic recombination plays a significant role in somatic cells during the repair of chemical damage to DNA (particularly chemical cross-links), DSBs introduced by physical damage to the DNA, and in the restoration of damaged replication forks (reviewed in Friedberg et al. 2006). If these lesions persist, chromosomal fragmentation, chromosomal loss (aneuploidy), and genetic rearrangements may occur (Kanaar et al. 1998; Rich et al. 2000). DSBs are repaired via homologous recombination (HR), which includes synthesis-dependent strand annealing (SDSA), single-strand annealing (SSA), and nonhomologous end joining (NHEJ) (see Mehta and Haber 2014). HR and SDSA faithfully repair DSBs without gain or loss of DNA sequences (Jasin and Rothstein 2013). In contrast, SSA is a mutagenic pathway used by cells when the DSB, committed to homology-directed repair, cannot be timely processed by HR or when the DSB occurs between direct repeats (Weinstock et al. 2006; Moynahan and Jasin 2010). NHEJ mends DNA ends, but almost always results in sequence gain or loss surrounding the site of the DSB. Additionally, the NHEJ process has a high risk of inducing chromosomal translocation because it may fuse virtually any broken end (Lieber 2010; Chiruvella et al. 2013). This review focuses entirely on HR and SSA because nucleotide mismatches that arise during these processes account for the outcome of the genetic products, as well as regulate the initiation of genetic exchange.
Studies with fungi have provided a superb model for studying HR. It should be noted that, like MMR, the genes and mechanical processes of HR are highly conserved up to and including humans. The general purpose and outcomes of HR in Saccharomyces cerevisiae are fundamentally different in meiosis and mitosis (reviewed in Andersen and Sekelsky 2010). The main function of HR between paternal and maternal chromosomes in meiosis is to create at least one CO per pair of homologs (reviewed in Page and Hawley 2003; Szekvolgyi and Nicolas 2009). Meiotic COs convert sister chromatid cohesion into homologous chromosome clasps, which ensure accurate segregation in the first meiotic division (meiosis I). The partition of chromosome arms on either side of a CO is the physical basis of Mendelian inheritance and, ultimately, genetic diversity.
The two distinct outcomes of meiotic recombination were first observed following tetrad analysis in S. cerevisiae (Lindegren 1955): (1) conserved crossovers, in which genetic information is exchanged reciprocally between chromosomes without genotype gain or loss; and (2) gene conversion, in which the genetic information of one genotype is replaced with an allelic genotype. The gene conversion process may result in CO or noncrossover (NCO) of markers on either side of the gene conversion event. Asymmetric repair of the mismatched nucleotides contained within the hybrid DNA of the Holliday junction was conceived to account for gene conversion (Holliday 1964). Interestingly, the absence of MMR may lead to PMS of genetic markers contained within the heteroduplex DNA of a Holliday junction, eventually leading to sectored colonies containing both genotypes. This sectored colony phenotype was used in S. cerevisiae to isolate the first eukaryotic MMR gene, the MLH/PMS ScPms1 (Williamson et al. 1985), initiating the beginnings of some muddled nomenclature.
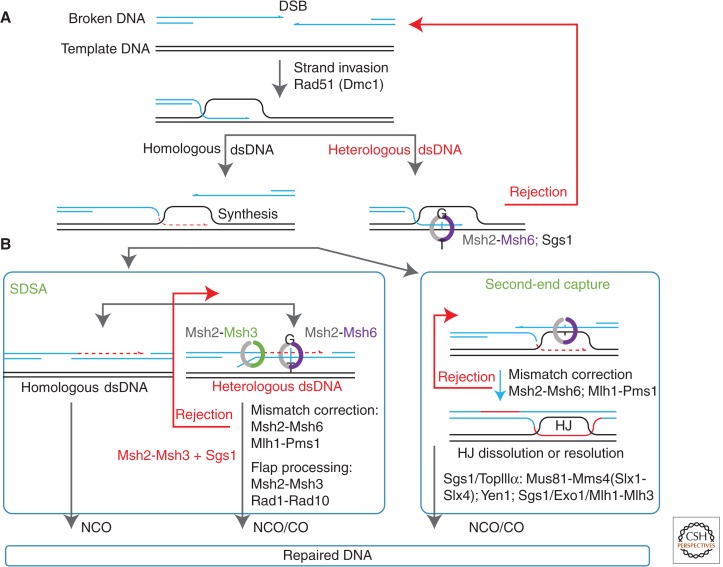
To explain the results of plasmid integration (gap-repair) studies in yeast, the double-strand break repair (DSBR) model was proposed (Szostak et al. 1983). The DSBR model and its variants are now thought to engender the major mechanism(s) associated with HR (Fig. 1) (see Mehta and Haber 2014). DSBR in meiotic recombination is initiated by the introduction of a large number of programmed DSBs catalyzed by the ScSpo11 protein (see Keeney et al. 1997; Lam and Keeney 2015). In mouse and human cells, these programmed DSBs number in the several hundred (Kolas et al. 2005; Lenzi et al. 2005). It is the repair of these numerous DSBs, using sequences donated from a homologous parental chromosome, which ultimately links chromosome homologs before meiosis I. At a molecular level, the DSB ends are first resected to produce 3′-single-stranded DNA (ssDNA) overhangs (reviewed in Symington and Gautier 2011; see Symington 2014). This process is followed by precisely coordinated DNA transactions, which are orchestrated by highly conserved homologs/orthologs of the prototypical RecA/RAD51 DNA homologous pairing and strand exchange proteins (recombinase) (see Li and Heyer 2008; Zelensky et al. 2014; Morrical 2015). In S. cerevisae, and with the help of recombination mediators, the ScRad51 and/or the RecA/RAD51 paralog ScDmc1 form a nucleoprotein filament (NPF) on the 3′-ssDNA overhangs (Sung 1997; New et al. 1998; Shinohara and Ogawa 1998; Gasior et al. 2001; Liu et al. 2010, 2011; Carreira and Kowalczykowski 2011; Kojic et al. 2011; Murayama et al. 2013; Sasanuma et al. 2013; Daley et al. 2014). One of the RecA/RAD51 NPFs assembled on the 3′-ressected ends of the DSB then catalyzes the invasion of a homologous parental double-stranded DNA (dsDNA; donor template) forming a displacement loop (D-loop). DNA synthesis, initiated at the 3′ end of the invading strand, then expands the D-loop until the distal end captures the remaining NPF, ultimately forming a double Holliday junction. Varying lengths of heteroduplex DNA may be produced by branch migration of either or both of the individual Holliday junctions (reviewed in Jasin and Rothstein 2013). These Holliday junctions may then be dissolved or resolved to yield either a CO or NCO (see Bizard and Hickson 2014; Wyatt and West 2014). CO and NCO events are readily distinguished in a plasmid gap-repair assay, in which an NCO outcome yields a repaired autonomously replicating plasmid, whereas a CO event integrates the plasmid into genome (Orr-Weaver and Szostak 1983).
Figure 1.
Multiple roles of the mismatch repair (MMR) machinery in eukaryotic double-strand break (DSB) repair by homologous recombination (HR). The damaged DNA molecule is shown in blue, the template is shown in black, and the strand synthesized during the double-strand break repair (DSBR) process is shown in red. The heteroduplex rejection steps are represented by the red arrows. Major players involved in every step are indicated. (A) Schematic representation of the DSB repair by single-strand annealing (SSA). The direct repeats on either side of the DSB are shown as arrows. (B) Synthesis-dependent strand annealing (SDSA). See text for details. CO, Crossover; dsDNA, double-stranded DNA; HJ, Holliday junction; NCO, noncrossover.
The occurrence of mismatches during meiotic recombination has been documented in genetic experiments via the manifestation of sectored PMS colonies, as well as through physical methods (White et al. 1985; Lichten et al. 1990). Correction of mismatched meiotic recombination intermediates by the MMR machinery results in gene conversion (Alani et al. 1994). Although the precise mechanism(s) associated with the processing of meiotic mismatched (heteroduplex) DNA remain obscure, it is likely that there is significant similarity with postreplication MMR (reviewed in Kolodner and Marsischky 1999; Kolodner et al. 2007).
In contrast to meiosis, mitotic recombination is critical for the repair of spontaneous DSBs and collapsed replication forks. It is significantly biased toward NCOs because the unidirectional transfer of information, which consequently results in the removal of any impediments to chromosome segregation, appears essential for high-fidelity repair. Only a small fraction of mitotic events proceed through the classic DSBR pathway (Bzymek et al. 2010). The majority of DSB repair occurs by an SDSA HR mechanism (Fig. 1) (Andersen and Sekelsky 2010). SDSA begins with DNA strand invasion and D-loop formation similar to classical DSBR. The 3′ end of the invading ScRad51 NPF is then extended by polymerase δ-catalyzed DNA synthesis (Li et al. 2009), which is essential to restore any DNA sequences that might be degraded during the exonucleolytic processing of the DSB. The major difference between SDSA and the DSBR model is that the D-loop is disassembled and the proximal polymerase-extended ssDNA strand is then used to capture the distal ScRad51 NPF (Fig. 1). Any DNA gaps and/or unannealed DNA flaps may then be processed and the remaining strand scissions are ligated to produce an intact duplex DNA. DSB repair by SDSA inevitably results in NCO events.
Three helicases in S. cerevisae have been implicated in regulating HR events. These include the slow growth suppressor of a topoisomerase 3 (Top3) mutation ScSgs1, the suppressor of Rad6 sensitivity ScSrs2, and the mutator phenotype ScMph1 (Ira et al. 2003; Prakash et al. 2009; Heyer et al. 2010). All three helicases have been either directly or genetically shown to dismantle and/or prevent the formation of D-loop structures. The most recent genetic evidence implicates ScMph1 in the disassembly of D-loops, whereas ScSgs1 dissolves intact double Holliday junctions and ScSrs2 dismantles single and double Holliday junctions containing strand scissions (Mitchel et al. 2013). In all cases, dissolution of these recombination intermediates results in NCO events. Additional bias toward NCO events during mitotic recombination, as well as the CO/NCO balance during meiotic recombination, may also be achieved by linking tight regulation of structure-selective nucleases to cell-cycle progression (Matos et al. 2011; Matos and West 2014).
In both meiosis and mitosis, the numerous steps associated with the recombination event must discriminate between identical and nonidentical template sequences and then either repair, process, or reject the ensuing heteroduplex DNA. In contrast to meiosis, which tolerates some degree of sequence heterology, mitotic recombination is extremely sensitive to the presence of mismatched nucleotides. This exquisite sensitivity is generally not caused by the selectivity of the RecA/RAD51 recombinase, but rather depends on the MMR machinery (Datta et al. 1997).
HOMEOLOGOUS RECOMBINATION
In 1989, Radman and colleagues suggested that, in addition to resolving recombination intermediates during gene conversion, MMR introduces a barrier to recombination between species containing excessive sequence heterology (Rayssiguier et al. 1989). The experimental system was relatively simple. E. coli and Salmonella typhimurium are 97% identical at the DNA sequence level. Yet, recombination between well-defined genetic markers contained in their largely identical chromosomes is quite rare; occurring at a frequency of 10−6–10−8 depending on the genetic marker (Rayssiguier et al. 1989; Stambuk and Radman 1998). This is in spite of the fact that F-pillus formation and DNA transfer between these organisms occurs quite readily (Clark and Adelberg 1962; Curtiss 1969). Remarkably, when the recipient bacteria are MMR deficient, the frequency of recombination between several genetic markers increased over 1000-fold (Rayssiguier et al. 1989; Stambuk and Radman 1998). These results were interpreted to suggest that MMR either aborts the formation of recombination intermediates or resolves nascent recombination intermediates containing excess heterology such that genetic recombination rarely occurs. Genetic exchange between partially homologous sequences was termed “homeologous” recombination to distinguish it from HR between fully homologous chromosomal DNA.
THE GENETIC REQUIREMENTS FOR THE SUPPRESSION OF HOMEOLOGOUS RECOMBINATION IN E. Coli
The suppression of homeologous recombination in E. coli required the EcMutS, EcMutL, EcMutH, and EcUvrD genes (Table 1) (Rayssiguier et al. 1989; Stambuk and Radman 1998). However, the relative suppression of homeologous recombination was quite different between these MMR genes. A broad pathway analysis determined that the mutS mutation increased homeologous recombination 735-fold (Stambuk and Radman 1998). Although the requirement for MutL was not examined in this pathway analysis, the initial study suggested that MutS and MutL affected the magnitude of homeologous recombination similarly. In contrast, a mutH mutation only increased homeologous recombination 22-fold, whereas mutation of uvrD increased homeologous recombination fivefold. These observations were qualitatively similar to the initial study (Rayssiguier et al. 1989). Interestingly, the combined mutS uvrD mutation increased homeologous recombination approximately two times over mutS alone, whereas the combined mutH uvrD mutation increased homeologous recombination almost tenfold over mutH alone. Mutation of genes involved in recombination initiation, Holliday junction branch migration, or the up-regulation of damage-inducible genes had little or no effect on homeologous recombination (Rayssiguier et al. 1989). The additive nature of uvrD mutations with MMR mutations suggested that there were likely two redundant pathways capable of aborting homeologous recombination in E. coli: one involving the core MMR genes MutHLS and a second involving the UvrD helicase. The separation of these pathways appeared most convincing in the absence of mutH than mutS. The biochemical properties of these proteins provide a foundation to help explain these distinctions.
BIOCHEMICAL ACTIVITIES OF THE MMR PROTEINS
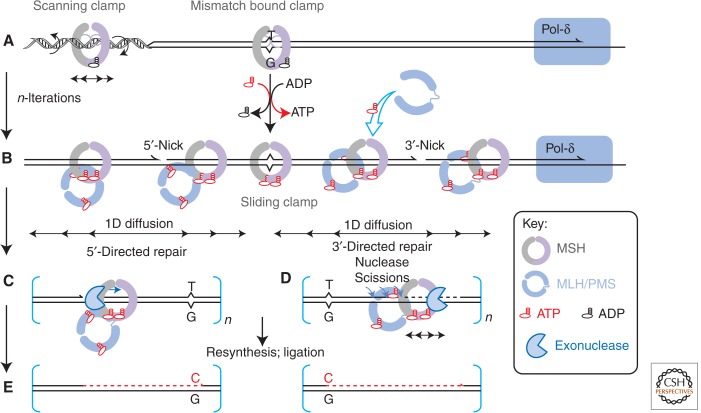
MMR is a mismatch or nucleotide lesion-dependent DNA excision–resynthesis DNA repair process (Modrich 1989, 1997; Kolodner and Marsischky 1999; Fishel et al. 2000). Strand excision may be initiated by several hundred to several thousand base pairs on either the 3′- or 5′-side of the mismatch, and the excision tract extends from this distant site to just past the mismatch (Lahue et al. 1987). A number of models have been proposed to account for the biochemical properties of the individual MMR proteins and the ability to initiate excision bidirectionally at a site significantly distant from the mismatch/lesion (Fig. 2) (Acharya et al. 2003; Kolodner et al. 2007). The mechanism that is most consistent with the data and the Brownian nature of molecular biology appears to be the molecular switch model (Gradia et al. 1997, 1999; Fishel 1998, 2000; Acharya et al. 2003; Jeong et al. 2011; Cho et al. 2012; Gorman et al. 2012; Qiu et al. 2012; Spies 2013). Most, if not all, biochemical discrepancy can be traced to differences in the experimental conditions, an issue that persists today (Hall et al. 2001; Drotschmann et al. 2002; Tessmer et al. 2008; Sass et al. 2010; Tham et al. 2013).
Figure 2.
Molecular switch model for eukaryotic mismatch repair (MMR). (A) An MutS homolog (MSH) scanning clamp binds dsDNA and searches for rare nucleotide misincorporation replication errors by one-dimensional (1D) rotational diffusion while maintaining continuous contact with the DNA duplex. (B) An encounter with a mismatch and subsequent ADP → ATP exchange converts the MSH into a sliding clamp, which dissociates from the mismatch, but still encircles the DNA duplex and moves by 1D diffusion while in discontinuous contact with the DNA backbone. The MutL homolog (MLH)/postmeiotic segregation (PMS) heterodimer associates with the ATP-bound MSH sliding clamp. The interaction between MSH with MLH/PMS, and likely some DNA structure, promotes ATP binding by the MLH/PMS, which activates its intrinsic endonuclease activity. This process is iterative with multiple MSH sliding clamps and MLH/PMS interactions followed by dynamic, likely small, excision events that ultimately release the entire MSH-MLH/PMS complex. Although not entirely known, the strand to be excised and repaired may be distinguished by endonuclease discontinuities and/or a 3′ or 5′ end. The bacterial β-clamp and eukaryotic proliferating cell nuclear antigen (PCNA), along with their respective clamp loading factors γ-complex and replication factor C (RFC), as well as the single-strand binding protein (SSB) and replication protein A (RPA), respectively, play important but not fully defined roles in the processes outlined in A and B. (C,D) The bidirectional nature of MSH-MLH/PMS diffusion ensures that excision may take place bidirectionally on either side of the mismatch in which an appropriate end occurs. (E) The multiple nucleolytic events remove the DNA region from the initial strand scission to just past the mismatch. Once the mismatch is removed, the loading of MSH sliding clamps is halted and the excision reaction ultimately ceases. Mismatch excision requires strand-specific exonucleases (3′ → 5′ ExoI, 3′ → 5′ ExoX, bidirectional ExoVII, or 5′ → 3′ RecJ in Escherichia coli and 5′ → 3′ EXOI in eukaryotes). Resynthesis of the excision gap is thought to be accomplished by the replicative polymerase Pol I in bacteria and POL-δ in eukaryotes.
The complete E. coli MMR reaction in vitro requires EcMutS, EcMutL, EcMutH, EcUvrD, one of four exonucleases, replicative polymerase Pol I complex, and DNA ligase (Lahue et al. 1989; Viswanathan and Lovett 1998). A simplified mismatch-dependent excision reaction only requires EcMutS, EcMutL, EcMutH, EcUvrD, and an exonuclease (Su et al. 1989). The bacterial MutS, MutL, and MutH function as dimeric proteins (Su and Modrich 1986; Welsh et al. 1987; Grilley et al. 1989). In contrast, the eukaryotic MSH and MLH/PMS homologs function as heterodimers (Fishel and Wilson 1997; Kolodner and Marsischky 1999).
MSH proteins are members of the AAA+ ATPase family and contain a highly conserved Walker A/B nucleotide-binding motif (Walker et al. 1982; Iyer et al. 2004). Mismatch nucleotide recognition was first demonstrated in 1986 with the EcMutS (Su and Modrich 1986). Remarkably identical structures of numerous MSH proteins bound to mismatched DNA have appeared in the literature in succeeding years (Lamers et al. 2000; Obmolova et al. 2000; Warren et al. 2007; Gupta et al. 2011). These structures showed that bacterial and human MSHs form a clamp-like configuration around the mismatched DNA with a Phe-residue within a highly conserved Phe-X-Glu motif interrogating the DNA 3′ of the mismatch and inducing a 45°–60° bend. Additional nonspecific electrostatic contacts with the DNA by protein residues surrounding the Phe-X-Glu motif further stabilize the mismatch-bound structure. Only nucleotide-free or ADP-bound structures have been crystallized. Infusion of ATP or ATPγS destroys the crystals (Obmolova et al. 2000), suggesting significant differences in the mismatch-bound and postmismatch conformations.
Multiple functionally distinct forms of the MSH clamp on DNA have been detected using single-molecule biophysical imaging analysis (reviewed in Spies 2013). While searching for a mismatch on duplex DNA, the Thermus aquaticus TaMutS was revealed to form an incipient clamp that underwent facilitated 1D rotational diffusion while in continuous contact with the helical backbone (Fig. 2A) (Cho et al. 2012). A similar mismatch search mechanism has been observed for the ScMsh2-ScMsh6 heterodimer (Gorman et al. 2012) and is likely conserved in all MSH proteins (Heinen et al. 2011). On encountering a mismatch, MutS pauses (presumably forming the clamp structure shown in structural studies), releases bound ADP, and binds ATP (ADP → ATP exchange) (Acharya et al. 2003; Jeong et al. 2011). DNA flexibility surrounding the mismatch has been proposed to distinguish it from the normally smooth DNA backbone triggering the pause in MSH diffusion (Mazurek et al. 2009). Detection of DNA contour alterations, and not the mismatch itself, would explain the wide range of mismatch/lesion recognition properties shown by MSH proteins (Mazurek et al. 2009).
Mismatch, lesion, and/or structure provoked ADP → ATP exchange is a central function of all MSH proteins examined to date (Gradia et al. 1997; Wilson et al. 1999; Acharya et al. 2003; Snowden et al. 2004). ATP binding by the MSH dimer/heterodimer instigates a significant conformational transition (Gradia et al. 1999; Wilson et al. 1999; Acharya et al. 2003; Qiu et al. 2012), which results in an extremely stable (5- to 10-min lifetime) sliding clamp that freely diffuses along the DNA without ATP hydrolysis and in the absence of any stable backbone interactions (Fig. 2A) (Cho et al. 2012). ATP binding and release of the MutS from the mismatch allows the loading of multiple MutS ATP-bound sliding clamps (Fig. 2B) (Gradia et al. 1997; Acharya et al. 2003; Jeong et al. 2011; Cho et al. 2012). 1D diffusion by the ATP-bound MSH sliding clamp(s) has been calculated to easily cover the several thousand nucleotides between the mismatch and the DNA strand scission where excision begins.
Long-lived ATP-bound hydrolysis-independent MSH sliding clamps are the single most critical intermediates in initiating MMR and suppression of homeologous recombination. This conclusion is supported by a number of genetic and biochemical observations. First, ATP-binding or hydrolysis-deficient MSH mutations, located in the consensus Walker ATP-binding motif, retain mismatch-binding activity, but are deficient in both MMR and homeologous recombination (Haber and Walker 1991; Wu and Marinus 1994; Iaccarino et al. 1998; Hess et al. 2002; Junop et al. 2003; Lin et al. 2004; Mendillo et al. 2005; Ollila et al. 2008). Second, the ability to form a sliding clamp strictly correlates with biological function, whereas mismatch/lesion/structure binding is necessary, but not sufficient for biological function (Sia et al. 1997; Genschel et al. 1998; Wilson et al. 1999; Hess et al. 2002; Snowden et al. 2004; Lenzi et al. 2005; Mendillo et al. 2005; Hargreaves et al. 2010). Finally, titration studies suggest that 4–8 MSH molecules are required for a single repair event in vitro (Zhang et al. 2005; Goeliner et al. 2014). Taken together, these observations clearly suggest that for biochemical or biophysical studies to fully reflect the cellular function(s) of MSH proteins, it is important to select conditions in which sliding clamp formation is robust and not dramatically reduced or absent. Besides the obvious prerequisite for ATP or the poorly hydrolysable analog ATPγS (MSH proteins do not readily bind AMP-PNP or AMP-PCP), ionic conditions between 90 mM and 150 mM are essential for the formation of stable mismatch-dependent ATP-bound MSH sliding clamps (Gradia et al. 1999; Wilson et al. 1999; Hess et al. 2002; Acharya et al. 2003; Mendillo et al. 2005; Mazur et al. 2006; Jeong et al. 2011). A requirement for specific ionic conditions is not surprising considering the nonspecific electrostatic contacts observed with MSH-DNA complexes outside the conserved Phe-X-Glu motif.
The EcMutH protein uniquely recognizes a hemimethylated Dam (GATC/GAmeTC) site and introduces a single-strand scission between the guanine and unmethylated adenine nucleotides (Welsh et al. 1987). It is this distant strand scission where the MMR excision reaction starts in γ-proteobacteria, such as E. coli. No other biochemical activity for EcMutH has been identified. Moreover, a preexisting strand scission appears to completely eliminate the EcMutH requirement for MMR in vitro (Lahue et al. 1987). EcUvrD is a 3′ → 5′ helicase (Matson 1986; Matson and George 1987), which is activated by EcMutL (Dao and Modrich 1998; Hall et al. 1998). Its likely role in MMR is to unwind the excised DNA strand before and/or in concert with exonuclease digestion (Lee and Yang 2006). One could envisage a number of different mechanistic strategies for the rejection of homeologous recombination intermediates to account for the known genetic requirements. For example, the EcMutH hemimethylated strand scission function might activate MMR strand excision following D-loop-dependent DNA synthesis. Alternatively, EcUvrD helicase might disassemble a homeologous D-loop intermediate. In both of these hypothetical mechanisms, it is likely that EcMutS-mediated mismatch detection would be required to target the homeologous recombination intermediate.
The detailed function of MLH/PMS proteins in MMR remains enigmatic. MLH/PMS proteins contain a GKL ATP-binding domain (Dutta and Inouye 2000), which displays extremely weak ATPase activity (Ban et al. 1999; Spampinato and Modrich 2000; Acharya et al. 2003). Moreover, the yeast MLH/PMS, ScMlh1-ScPms1, appears to undergo conformation contractions between the carboxy-terminal heterodimer interaction domain and the amino-terminal ATP-binding and adenosine nucleotide-dependent dimerization domain (Sacho et al. 2008). The function, if any, of these conformational transitions is unknown. ATP binding by EcMutL significantly enhances the EcMutH endonuclease activity (Acharya et al. 2003). Importantly, EcMutL and MLH/PMS heterodimer proteins form a stable complex with ATP-bound EcMutS and MSH sliding clamps (Fig. 2B) (Acharya et al. 2003; Mendillo et al. 2005). The diffusion characteristics of the MSH-MLH/PMS complex do not appear to be different from the MutS/MSH sliding clamp alone (Gorman et al. 2012). These observations strongly suggest that the ATP-bound MutS/MSH sliding clamp recruits and transports the MutlL/MLH/PMS for downstream MMR functions. Potential functions for EcMutL in bacterial MMR may be to stabilize the EcMutH endonuclease at the hemimethylated GATC site and/or enhance EcUvrD helicase activity during excision. Interestingly, EcMutL and ScMlh1-ScPms1 have been shown to bind ssDNA (Drotschmann et al. 2002; Park et al. 2010). However, EcMutL ssDNA-binding activity is nearly undetectable at physiological ionic strength and unlikely to be significant during MMR (Park et al. 2010).
A major difference between E. coli MutL and virtually all MLH/PMS proteins outside γ-proteobacteria is the presence of an intrinsic endonuclease activity (Kadyrov et al. 2006, 2007; Pillon et al. 2010). It has been suggested that this MLH/PMS intrinsic endonuclease substitutes for the MutH endonuclease activity found only in γ-protobacteria like E. coli (Kadyrov et al. 2006). This parallel seems, at the very least, incomplete because the MLH/PMS endonuclease appears to introduce multiple strand scissions during the MMR reaction (Kadyrov et al. 2006). The MLH/PMS endonuclease appears more efficient in the presence of manganese-divalent cation and can also be stimulated by zinc (Kadyrov et al. 2006, 2007; Pillon et al. 2010). Importantly, the Thermus thermophilus homologs were used to show that the TtMutL ATP-dependent endonuclease is activated only on its association with ATP-bound TtMutS sliding clamps (Shimada et al. 2013).
The molecular switch model, as applied to γ-proteobacteria, suggests that multiple MutS/MutL complexes may diffuse along the DNA to activate MutH incision, UvrD helicase, and exonuclease excision (Fig. 2A,B) (Acharya et al. 2003). The process is proposed to be dynamic such that successive MutS-MutL-UvrD-exonuclease complexes produce redundant small excision tracts, which ultimately discharge the mismatch. For organisms that do not contain a MutH and do not use a helicase during MMR (Eubacteria, Archea, and Eukaryotes), the MLH/ PMS ATP-dependent endonuclease presumably associated with ATP-bound MSH sliding clamps, and an exonuclease could conceivably produce small excision tracts by a similar dynamic and redundant mechanism (Fig. 2C,D) (Fishel et al. 2000). Once the excision tract releases the mismatch, successive loading of MutS sliding clamps ceases and the DNA degradation reaction grinds to a halt. A free 3′ end would allow the polymerase machinery to assemble and complete the resynthesis reaction (Fig. 2E).
THE SUPPRESSION OF HOMEOLOGOUS RECOMBINATION IN YEAST
DSBs and the formation of early recombination intermediates in meiosis are required to establish stable pairing interactions between homologs (Peoples et al. 2002). When formed in unique nonrepetitive sequences, meiotic DSBs initiate recombination between identical allelic positions on homologous chromosomes. DSBs can also form within or near repetitive elements that are found in abundance in all eukaryotes (Lander et al. 2001). Recombination between such repeats carries a risk for deleterious genome rearrangements in the germ line. As with bacteria, in eukaryotes, the MMR machinery is largely responsible for the rejection of heteroallelic homeologous DNA during recombination (Datta et al. 1996). Both the precise mechanism of homeologous recombination rejection and molecular triggers, which signal to either repair or reject these partially homologous sequences, remain unknown.
How many mismatches are required to distinguish homologous from homeologous recombination? A log-linear relationship between sequence divergence and sexual isolation (frequency of recombination) was observed for soil isolates of Bacillus subtilis (Roberts and Cohan 1993). In these studies, as little as 0.3% sequence divergence reduced recombination, thus, increasing sexual isolation. Although a complete answer to this question may vary between organisms, in S. cerevisae, one mismatch in a recombination region of ∼300 bp (0.3% sequence divergence) reduced the recombination rate almost threefold, and that rate decreased logarithmically until ∼1% sequence divergence was reached (Datta et al. 1997). The rate decrease in recombination then becomes linear over three orders of magnitude in sequence divergence (Datta et al. 1997). Remarkably, the impact of MMR in suppressing recombination increased steadily until ∼10% sequence divergence (Datta et al. 1997). The recombination rate between sequences with >10% sequence divergence decreased dramatically and appeared to be largely unaffected by MMR, suggesting that genetic exchange was most likely aborted before any actions by the core MMR machinery. Similar results have been reported for the effect of sequence divergence and MMR on Arabidopsis thalia recombination (Li et al. 2006).
The MMR machinery is required for both recognizing and processing mismatch-containing heteroduplex during homeologous recombination in yeast (Fig. 1). Genetically, msh2, msh3, and msh6 play critical roles in the heteroduplex rejection, suggesting the roles for both ScMsh2-ScMsh3 and ScMsh2-ScMsh6 heterodimers (Fig. 1). The antirecombination activity of ScMsh2 is severalfold greater than that of the ScPms1 or ScMlh1 (Datta et al. 1996). These observations appear qualitatively similar to E. coli and suggest redundancy or bifurcation into subpathways in which both rejection mechanisms depend on ScMsh2, but only one requires both MSH and MLH/PMS. The ScSgs1 and ScSrs2 helicases play additional roles in antagonizing homeologous recombination, but have no known roles in postreplicative MMR (Welz-Voegele and Jinks-Robertson 2008). Notably, deletion of either msh2 or sgs1 increases homeologous recombination to the level of HR (Welz-Voegele and Jinks-Robertson 2008). These observations appear similar to bacteria and suggest the existence of at least two heteroduplex rejection pathways associated with HR (Fig. 1).
HETERODUPLEX REJECTION DURING SSA BETWEEN DIRECT REPEAT SEQUENCES
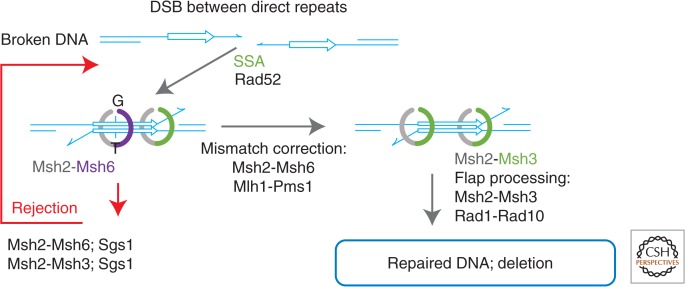
SSA represents the major mechanism for the repair of DSBs that occur between direct repeats (Fig. 3) (Fishman-Lobell et al. 1992). As with the classical DSBR and the SDSA pathways, the ends of the DSB are resected in SSA (Fig. 3, left). If resection covers two complementary regions on either side of the DSB, the exposed ssDNAs are annealed with the help of the ScRad52 protein (Fig. 3, left). Although the SSA pathway is quite robust, it is also utterly mutagenic as it results in the loss of coding sequence between the repeats. SSA has been extensively used to investigate heteroduplex rejection during yeast mitosis because it provides a straightforward process for generating heteroduplex in vivo. As with DSBR and SDSA, SSA is sensitive to the presence of heterology (Sugawara et al. 1997). Heteroduplex rejection in SSA depends on the ScMsh2-ScMsh6 protein, which recognizes mismatches within the heteroduplex, and the Sgs1 helicase, which presumably unwinds heteroduplex DNA strands (Fig. 1B, left) (Sugawara et al. 2004). The SSA heteroduplex rejection process is also significantly reduced in _mlh1_Δ and _pms1_Δ, _mlh2_Δ, _mlh3_Δ triple mutant cells, albeit to a much lesser extent than that observed in the _msh6_Δ strain (Sugawara et al. 2004). These results are consistent with the conclusion that SSA heteroduplex rejection requires ScMsh2-ScMsh6 and one of the three known S. cerevisae MLH/PMS heterodimers (ScMlh1-ScPms1, ScMlh1-ScMlh2, ScMlh1-ScMlh3) (Sugawara et al. 2004).
Figure 3.
Roles of the mismatch repair (MMR) proteins in single-strand annealing (SSA). Schematic representation of the double-strand break (DSB) repair by SSA. The direct repeats on either side of the DSB are shown as arrows. See the text for details of the rejection (left) and the mismatch correction (right) mechanisms.
Although mismatch recognition and unwinding of the heteroduplex DNA produced during DSBR and SSA likely share many mechanistic features, the two processes should not be completely equated. Mismatch recognition within a D-loop produced by a ScRad51 NPF may present a unique set of challenges distinct from those associated with the SSA structure annealed by the ScRad52 protein. For example, the three-stranded structure of a D-loop is clearly distinct from the simple heteroduplex produced by ScRad52-dependent annealing of resected stands. Moreover, the heteroduplex region of the D-loop likely remains coated with ScRad51 after the strand exchange step, perhaps until it is removed by ScRad54 (Li and Heyer 2009). Both ScRad52 (Sugiyama et al. 1998) and HsRAD52 after it is activated by phosphorylation (Honda et al. 2011) are exquisitely selective for ssDNA and the homology search mechanism depends on diffusion (Rothenberg et al. 2008). These observations suggest a largely protein-free heteroduplex region following SSA.
THE MMR SYSTEM IN HETEROLOGOUS FLAP PROCESSING
When free homologous ends are available, the RAD51 recombinase NPF forms plectonemic joints in which the 3′ end of the invading strand is paired to the template DNA duplex. This structure then serves as a template for DNA synthesis, as discussed above for SDSA. When fully complementary ends are not available, the exchange of the DNA strands may produce less stable paranemic joints in which the two complementary strands are not topologically interwound. Removal of the protruding heterologous flap by a structure-specific nuclease, such as ScRad1-ScRad10, both converts a paranemic joint into a more stable plectonemic joint and produces a substrate for DNA synthesis (Fig. 3, right). Similarly, nonhomologous flaps must be removed from the ends of the heteroduplex formed during SSA. Because it is likely that the ends of the resected DSB will not be complementary, virtually every SSA event will almost certainly require a flap-processing step. The 3′-protruding end removal in D-loop and SSA intermediates has been extensively studied (Schiestl and Prakash 1990; Fishman-Lobell et al. 1992; Tomkinson et al. 1993). The process appears to depend primarily on the ScMsh2-ScMsh3 heterodimer, which both recognizes and stabilizes flap-containing heteroduplexes (Paques and Haber 1997; Sugawara et al. 1997; Kearney et al. 2001). Stabilization of the flap-containing heteroduplex by ScMsh2-ScMsh3 increases SSA when the DSB occurs in a region flanked by direct repeats with complementary region <1 kb and nonhomologous tails >30 nucleotides (Paques and Haber 1997; Sugawara et al. 1997). Separation-of-function msh2 mutations defective in ScMsh2-ScMsh3-dependent nonhomologous end removal, but functional in ScMsh2-ScMsh6 postreplicative MMR suggest that the 3′ protruding-end removal may involve a distinct DNA-binding mode (Goldfarb and Alani 2005). On ScMsh2-ScMsh3 recognition, flaps appear to be processed by ScRad1-ScRad10 (Fig. 1B, right) (Tomkinson et al. 1993). Moreover, the flap-removal function of ScMsh2-ScMsh3 is independent of ScMsh6, ScMlh1, and ScPms1 (Paques and Haber 1997; Sugawara et al. 1997; Kearney et al. 2001).
PARALLELS BETWEEN YEAST AND MAMMALIAN SYSTEMS
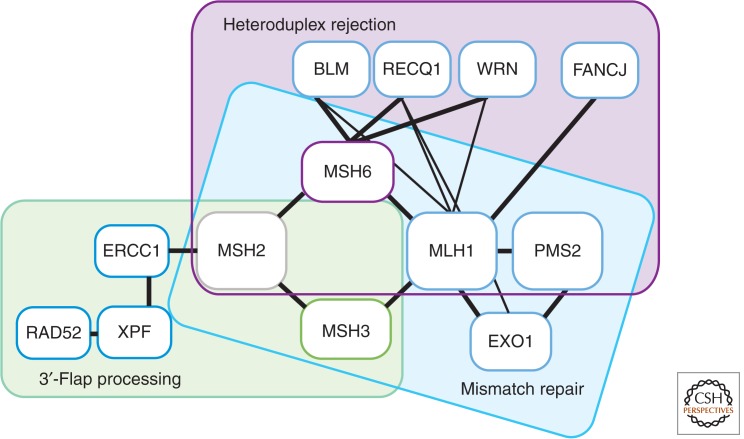
Mammalian MMR proteins participate in processing HR intermediates and suppressing homeologous recombination (Elliott and Jasin 2001). With exception of HsPMS2, which is the ScPms1 homolog, the core MMR machinery that controls recombination retains both the nomenclature and functionalities of their yeast and bacterial counterparts (Fig. 2; Table 1).
Mammalian cells carrying mutations in XPF or ERCC1 are sensitive to DSB-inducing agents (Ahmad et al. 2008). Thus, it is likely that the mammalian homolog of ScRad1-ScRad10, HsXPF-HsERCC1, plays a similar role in processing heterologous flaps during SDSA and SSA in human cells (Fig. 4; Table 1) (Sargent et al. 2000). In addition, HsERCC1 physically interacts with HsMSH2 and HsRAD52 (Lan et al. 2004). Moreover, the interaction with HsRAD52 stimulates HsXPF-HsERCC1 flap-endonuclease activity on 3′ ends (Fig. 4) (Motycka et al. 2004). It is less clear which human helicase homologs play the same role as ScSgs1 (Fig. 4; Table 1). Among the five known human RecQ family helicases, three, HsRECQ1 (Doherty et al. 2005), HsBLM (Pedrazzi et al. 2003), and HsWRN (Saydam et al. 2007), have been shown to interact with the HsMSH2-HsMSH6 heterodimer. HsBLM and HsWRN also interact with the HsMSH2-HsMSH3 heterodimer (Pedrazzi et al. 2003; Saydam et al. 2007). In addition, HsBLM interacts with HsMLH1 and HsExoI and, in conjunction with TopIIIα and both RMI1and RM2, has been shown to dissolve double Holliday junctions (Langland et al. 2001; Pedrazzi et al. 2003; Wu and Hickson 2003; Nimonkar et al. 2011). HsRECQ1 (Doherty et al. 2005) and HsWRN (Saydam et al. 2007) also interact with HsMLH1, and HsRECQ1 as well as with HsExoI (Cui et al. 2004). Furthermore, purified HsRECQ1 forms a subnanomolar affinity complex with HsMSH2-HsMSH6, and HsMSH2-HsMSH6 stimulates HsRECQ1 helicase activity on the 3′-flap structures (Doherty et al. 2005). Finally, both HsMSH2-HsMSH6 and HsMSH2-HsMSH3 stimulate HsWRN helicase activity on forked DNA substrates containing 3′-ssDNA tails (Saydam et al. 2007). Interestingly, the substrate specificity of HsRECQ1, HsBLM, and HsWRN only partially overlaps. Thus, HsRECQ1 readily unwinds immobile Holliday junctions, but does not support the Holliday junction branch migration activity characteristic of HsBLM and HsWRN (Sharma et al. 2005; Popuri et al. 2008).
Figure 4.
Protein–protein interaction network between human mismatch repair (MMR), heteroduplex rejection, and 3′-flap processing components.
BIOCHEMICAL RECONSTITUTIONS OF HETERODUPLEX REJECTION
There have been two approaches toward the biochemical and biophysical analysis of heteroduplex rejection. Both approaches reconstitute the initial steps and/or intermediates in homeologous recombination. One approach has been to determine any inhibitory role for the E. coli MMR machinery on DNA strand exchange and D-loop formation catalyzed by EcRecA (Worth et al. 1994; Tham et al. 2013). Two different recombinase assays have been used to observe homologous pairing and strand exchange in vitro: (1) a three-strand system in which RecA/RAD51 catalyzes the exchange of homologous strands between a double-stranded linear DNA and a large circular ssDNA, and (2) RecA/RAD51-catalyzed formation of a D-loop between a relatively short linear ssDNA and a supercoiled circular DNA (Kowalczykowski and Eggleston 1994). The three-strand reaction generally results in extensive branch migration that progresses through partial strand exchange intermediates into the formation of a terminal circular dsDNA product, whereas the D-loop reaction does not result in extensive branch migration (Kowalczykowski et al. 1994).
Purified EcMutS, EcMutL, EcUvrD, EcSSB, and EcRecA were combined in a recent study to examine the effect of MMR proteins on EcRecA-catalyzed three-strand exchange and D-loop formation with and without homeologous recombination substrates (Tham et al. 2013). The investigators show an apparent mismatch-dependent kinetic effect of MMR proteins in the rejection of heteroduplex recombination intermediates. This was evident from the accumulation of three-stranded intermediates and a reduction in circular dsDNA products. Because EcRecA-catalyzed D-loop formation was not affected by EcMutS or EcMutL, Tham and colleagues concluded that the MMR proteins did not inhibit strand exchange, but rather interfered with the branch migration process. Moreover, the EcUvrD helicase appeared biased toward unwinding heterologous intermediates of the three-strand reaction converting late strand exchange intermediates to early intermediates and, ultimately, into the initial substrates. This activity seemed to depend on EcMutS because, in its absence, EcUvrD appeared unbiased, catalyzing both the forward and reverse reactions equivalently (Tham et al. 2013).
An important caveat in these studies is that EcMutS is incapable of forming stable ATP-bound sliding clamps in the experimental ionic conditions (Acharya et al. 2003). Recall that both genetic and biochemical studies have clearly indicated that ATP-bound MSH sliding clamps are essential for both MMR and heteroduplex rejection (Wu and Marinus 1994; Hess et al. 2002; Acharya et al. 2003; Mendillo et al. 2005; Hargreaves et al. 2010). Moreover, persistent mismatch-bound EcMutS has been shown to abnormally alter MMR functions (Hess et al. 2002). An additional limitation in these studies is that both EcMutS and EcMutL display unusual ssDNA-binding activities at these low experimental ionic strength conditions (Su and Modrich 1986; Gradia et al. 2000; Acharya et al. 2003; Park et al. 2010). It is also well known that ssDNA-binding proteins influence both the DNA interaction(s) and kinetic processes of RecA recombinase activity (Kowalczykowski et al. 1987). Atypical ssDNA-binding activity could also confuse studies that concluded secondary structures in the displaced strand of the three-strand reaction-targeted heteroduplex rejection by EcUvrD (Tham et al. 2013). These issues underscore the complexity of incorporating multiple proteins containing numerous independent and overlapping biochemical activities. Expanding biochemical reconstitutions to include a broader range of experimental conditions compatible with well-known MMR protein functions will likely reveal the nuanced impacts of these DNA repair components in the multifaceted heteroduplex rejection reaction.
In a second approach, the HsMSH2-HsMSH6 heterodimer was shown to bind and form ATP-bound sliding clamps on D-loop intermediates containing a single mismatch in the heteroduplex region (Honda et al. 2014). These studies also showed that HsMSH2-HsMSH6 was capable of recognizing a mismatch within the D-loop heteroduplex when it was bound by the human ssDNA-binding heterotrimer HsRPA and/or the HsRAD51 recombinase (Honda et al. 2014). The results revealed a surprising affinity of MSH proteins for mismatched nucleotides regardless of its surrounding protein and DNA environment. Additional single molecule analysis established that the lifetime of the HsMSH2-HsMSH6 ATP-bound sliding clamp was almost threefold shorter than on a duplex DNA containing a mismatch (Honda et al. 2014). The reduced stability was traced to interactions between HsMSH2-HsMSH6 with the ssDNA–dsDNA junction on either side of the mismatch-containing heteroduplex DNA within the D-loop. This observation appeared similar to studies with the TaMutS in which reduced sliding clamp lifetime was observed when an ssDNA tail was introduced adjacent to the duplex DNA containing a mismatch (Jeong et al. 2011). The instability associated with ssDNA–dsDNA junctions appears contrary to the observations with the E. coli MMR proteins performed under very low ionic strength conditions (Tham et al. 2013). If correct, the shorter lifetime of the MSH sliding clamps on a D-loop would likely reduce the efficiency of MMR and/or heteroduplex rejection. Such a reduced heteroduplex rejection activity during HR would seem important to preserve a small number of gene conversion events as is observed during Mendelian genetics. Clearly, these initial observations will require confirmation along with more complicated biochemical reconstitution containing the remaining MMR machinery, the recombination initiation machinery, relevant recombination substrates, and a wide range of biochemical conditions.
CONCLUDING REMARKS
Despite the importance of cellular processes that balance homologous and homeologous recombination, our understanding of the heteroduplex rejection mechanism, which is at the heart of this balancing act, remains rudimentary. Most of our current knowledge comes from microbial genetic and cellular studies. There is, as yet, only an elementary understanding of the basic biochemical activities of the proteins and enzymes orchestrating different steps in the heteroduplex rejection, as only a few studies have attempted to tackle this important process, producing tantalizing, but still incomplete conclusions (Worth et al. 1994; Tham et al. 2013; Honda et al. 2014). Complete biochemical reconstitution of the heteroduplex rejection reactions occurring during HR and SSA will be necessary to produce a coherent mechanistic description of the intricate molecular choreography of these processes. More thorough and complete biochemical, biophysical, and single-molecule studies performed under a wider range of experimental conditions would seem necessary for a comprehensive understanding of the MMR-dependent heteroduplex rejection process. Finally, all the eukaryotic players of HR, SSA, and MMR appear to be controlled by posttranslational modifications. The effect of these modifications on the heteroduplex rejection and the choice between rejection, processing, or repairing the heteroduplex remains entirely unexplored.
ACKNOWLEDGMENTS
The authors thank Dr. Jong-Bong Lee and laboratory members for many helpful discussions. The work is supported by National Institutes of Health Grants GM108617 (M.S.) and CA67007 (R.F.).
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
REFERENCES
*Reference is also in this collection.
- Acharya S, Foster PL, Brooks P, Fishel R 2003. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell 12: 233–246. [DOI] [PubMed] [Google Scholar]
- Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ 2008. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol 28: 5082–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E, Reenan RA, Kolodner RD 1994. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics 137: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SL, Sekelsky J 2010. Meiotic versus mitotic recombination: Two different routes for double-strand break repair: The different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes. Bioessays 32: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban C, Junop M, Yang W 1999. Transformation of MutL by ATP binding and hydrolysis: A switch in DNA mismatch repair. Cell 97: 85–97. [DOI] [PubMed] [Google Scholar]
- *.Bizard AH, Hickson ID 2014. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol 6: a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, et al. 1994. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 368: 258–261. [DOI] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N 2010. Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira A, Kowalczykowski SC 2011. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci 108: 10448–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiruvella KK, Liang Z, Wilson TE 2013. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol 5: a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WK, Jeong C, Kim D, Chang M, Song KM, Hanne J, Ban C, Fishel R, Lee JB 2012. ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure 20: 1264–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ 2011. The DNA damage response: Making it safe to play with knives. Mol Cell 40: 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Adelberg EA 1962. Bacterial conjugation. Annu Rev Microbiol 16: 289–319. [DOI] [PubMed] [Google Scholar]
- Cui S, Arosio D, Doherty KM, Brosh RM Jr, Falaschi A, Vindigni A 2004. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res 32: 2158–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss R III 1969. Bacterial conjugation. Annu Rev Microbiol 23: 69–136. [DOI] [PubMed] [Google Scholar]
- *.Daley JM, Gaines WA, Kwon YH, Sung P 2014. Regulation of DNA pairing in homologous recombination. Cold Spring Harb Perspect Biol 6: a017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao V, Modrich P 1998. Mismatch-, MutS-, MutL-, and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. J Biol Chem 273: 9202–9207. [DOI] [PubMed] [Google Scholar]
- Datta A, Adjiri A, New L, Crouse GF, Jinks Robertson S 1996. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccaromyces cerevisiae. Mol Cell Biol 16: 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Hendrix M, Lipsitch M, Jinks Robertson S 1997. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci 94: 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty KM, Sharma S, Uzdilla LA, Wilson TM, Cui S, Vindigni A, Brosh RM Jr 2005. RECQ1 helicase interacts with human mismatch repair factors that regulate genetic recombination. J Biol Chem 280: 28085–28094. [DOI] [PubMed] [Google Scholar]
- Drotschmann K, Hall MC, Shcherbakova PV, Wang H, Erie DA, Brownewell FR, Kool ET, Kunkel TA 2002. DNA binding properties of the yeast Msh2-Msh6 and Mlh1-Pms1 heterodimers. Biol Chem 383: 969–975. [DOI] [PubMed] [Google Scholar]
- Dutta R, Inouye M 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci 25: 24–28. [DOI] [PubMed] [Google Scholar]
- Elliott B, Jasin M 2001. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol Cell Biol 21: 2671–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J 1974. The evolutionary advantage of recombination. Genetics 78: 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R 1998. Mismatch repair, molecular switches, and signal transduction. Genes Dev 12: 2096–2101. [DOI] [PubMed] [Google Scholar]
- Fishel R 2001. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: Revising the mutator hypothesis. Cancer Res 61: 7369–7374. [PubMed] [Google Scholar]
- Fishel R, Wilson T 1997. MutS homologs in mammalian cells. Curr Opin Genet Dev 7: 105–113. [DOI] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R 1993. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75: 1027–1038. [DOI] [PubMed] [Google Scholar]
- Fishel R, Acharya S, Berardini M, Bocker T, Charbonneau N, Cranston A, Gradia S, Guerrette S, Heinen CD, Mazurek A, et al. 2000. Signaling mismatch repair: The mechanics of an adenosine-nucleotide molecular switch. Cold Spring Harb Symp Quant Biol 65: 217–224. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J, Rudin N, Haber JE 1992. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol 12: 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T 2006. DNA repair and Mutagenesis. American Society of Microbiology, Washington, DC. [Google Scholar]
- Gasior SL, Olivares H, Ear U, Hari DM, Weichselbaum R, Bishop DK 2001. Assembly of RecA-like recombinases: Distinct roles for mediator proteins in mitosis and meiosis. Proc Natl Acad Sci 98: 8411–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschel J, Littman SJ, Drummond JT, Modrich P 1998. Isolation of MutS-β from human cells and comparison of the mismatch repair specificities of MutS-β and MutS-α. J Biol Chem 273: 19895–19901. [DOI] [PubMed] [Google Scholar]
- Goeliner EM, Smith CE, Campbell CS, Hombauer H, Desai A, Putnam CD, Kolodner RD 2014. PCNA and Msh2-Msh6 activate an Mlh1-Pms1 endonuclease pathway required for Exo1-independent mismatch repair. Mol Cell 55: 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T, Alani E 2005. Distinct roles for the Saccharomyces cerevisiae mismatch repair proteins in heteroduplex rejection, mismatch repair and nonhomologous tail removal. Genetics 169: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, Alani EE, Greene EC 2012. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc Natl Acad Sci 109: E3074–E3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradia S, Acharya S, Fishel R 1997. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell 91: 995–1005. [DOI] [PubMed] [Google Scholar]
- Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R 1999. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell 3: 255–261. [DOI] [PubMed] [Google Scholar]
- Gradia S, Acharya S, Fishel R 2000. The role of mismatched nucleotides in activating the hMSH2-hMSH6 molecular switch. J Biol Chem 275: 3922–3930. [DOI] [PubMed] [Google Scholar]
- Grilley M, Welsh KM, Su SS, Modrich P 1989. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem 264: 1000–1004. [PubMed] [Google Scholar]
- Gupta S, Gellert M, Yang W 2011. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat Struct Mol Biol 19: 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber LT, Walker GC 1991. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J 10: 2707–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Jordan JR, Matson SW 1998. Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. EMBO J 17: 1535–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Wang H, Erie DA, Kunkel TA 2001. High affinity cooperative DNA binding by the yeast Mlh1-Pms1 heterodimer. J Mol Biol 312: 637–647. [DOI] [PubMed] [Google Scholar]
- Hargreaves VV, Shell SS, Mazur DJ, Hess MT, Kolodner RD 2010. Interaction between the Msh2 and Msh6 nucleotide-binding sites in the Saccharomyces cerevisiae Msh2-Msh6 complex. J Biol Chem 285: 9301–9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen CD, Cyr JL, Cook C, Punja N, Sakato M, Forties RA, Lopez JM, Hingorani MM, Fishel R 2011. Human MSH2 (hMSH2) protein controls ATP processing by hMSH2-hMSH6. J Biol Chem 286: 40287–40295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess MT, Gupta RD, Kolodner RD 2002. Dominant Saccharomyces cerevisiae msh6 mutations cause increased mispair binding and decreased dissociation from mispairs by Msh2-Msh6 in the presence of ATP. J Biol Chem 277: 25545–25553. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J 2010. Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44: 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday RA 1964. A mechanism for gene conversion in fungi. Genet Res 5: 282–304. [DOI] [PubMed] [Google Scholar]
- Honda M, Okuno Y, Yoo J, Ha T, Spies M 2011. Tyrosine phosphorylation enhances RAD52-mediated annealing by modulating its DNA binding. EMBO J 30: 3368–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M, Okuno Y, Hengel SR, Martin-Lopez JV, Cook CP, Amunugama R, Soukup RJ, Subramanyam S, Fishel R, Spies M 2014. Mismatch repair protein hMSH2-hMSH6 recognizes mismatches and forms sliding clamps within a D-loop recombination intermediate. Proc Natl Acad Sci 111: E316–E325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaccarino I, Marra G, Palombo F, Jiricny J 1998. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSα. EMBO J 17: 2677–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L 2004. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol 146: 11–31. [DOI] [PubMed] [Google Scholar]
- Jasin M, Rothstein R 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5: a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, Cho WK, Song KM, Cook C, Yoon TY, Ban C, Fishel R, Lee JB 2011. MutS switches between two fundamentally distinct clamps during mismatch repair. Nat Struct Mol Biol 18: 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junop MS, Yang W, Funchain P, Clendenin W, Miller JH 2003. In vitro and in vivo studies of MutS, MutL and MutH mutants: Correlation of mismatch repair and DNA recombination. DNA Repair (Amst) 2: 387–405. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P 2006. Endonucleolytic function of MutLα in human mismatch repair. Cell 126: 297–308. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O’Donnell M, Kunkel TA, Modrich P 2007. Saccharomyces cerevisiae MutLα is a mismatch repair endonuclease. J Biol Chem 282: 37181–37190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JH, van Gent DC 1998. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol 8: 483–489. [DOI] [PubMed] [Google Scholar]
- Kearney HM, Kirkpatrick DT, Gerton JL, Petes TD 2001. Meiotic recombination involving heterozygous large insertions in Saccharomyces cerevisiae: Formation and repair of large, unpaired DNA loops. Genetics 158: 1457–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kojic M, Zhou Q, Fan J, Holloman WK 2011. Mutational analysis of Brh2 reveals requirements for compensating mediator functions. Mol Microbiol 79: 180–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Svetlanov A, Lenzi ML, Macaluso FP, Lipkin SM, Liskay RM, Greally J, Edelmann W, Cohen PE 2005. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J Cell Biol 171: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner RD, Marsischky GT 1999. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev 9: 89–96. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Mendillo ML, Putnam CD 2007. Coupling distant sites in DNA during DNA mismatch repair. Proc Natl Acad Sci 104: 12953–12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Eggleston AK 1994. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem 63: 991–1043. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Clow J, Somani R, Varghese A 1987. Effects of the Escherichia coli SSB protein on the binding of Escherichia coli RecA protein to single-stranded DNA. Demonstration of competitive binding and the lack of a specific protein–protein interaction. J Mol Biol 193: 81–95. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev 58: 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue RS, Su SS, Modrich P 1987. Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proc Natl Acad Sci 84: 1482–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue RS, Au KG, Modrich P 1989. DNA mismatch correction in a defined system. Science 245: 160–164. [DOI] [PubMed] [Google Scholar]
- *.Lam I, Keeney S 2015. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 7: a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK 2000. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature 407: 711–717. [DOI] [PubMed] [Google Scholar]
- Lan L, Hayashi T, Rabeya RM, Nakajima S, Kanno S, Takao M, Matsunaga T, Yoshino M, Ichikawa M, Riele H, et al. 2004. Functional and physical interactions between ERCC1 and MSH2 complexes for resistance to _cis_-diamminedichloroplatinum(II) in mammalian cells. DNA Repair (Amst) 3: 135–143. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Langland G, Kordich J, Creaney J, Goss KH, Lillard-Wetherell K, Bebenek K, Kunkel TA, Groden J 2001. The Bloom’s syndrome protein (BLM) interacts with MLH1 but is not required for DNA mismatch repair. J Biol Chem 276: 30031–30035. [DOI] [PubMed] [Google Scholar]
- Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M, et al. 1993. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 75: 1215–1225. [DOI] [PubMed] [Google Scholar]
- Lee JY, Yang W 2006. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127: 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, Poulos BK, Cohen PE 2005. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis I in human oocytes. Am J Hum Genet 76: 112–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD 2008. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18: 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD 2009. RAD54 controls access to the invading 3′-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res 37: 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Jean M, Belzile F 2006. The impact of sequence divergence and DNA mismatch repair on homeologous recombination in Arabidopsis. Plant J 45: 908–916. [DOI] [PubMed] [Google Scholar]
- Li X, Stith CM, Burgers PM, Heyer WD 2009. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase δ. Mol Cell 36: 704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M, Goyon C, Schultes NP, Treco D, Szostak JW, Haber JE, Nicolas A 1990. Detection of heteroduplex DNA molecules among the products of Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci 87: 7653–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79: 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DP, Wang Y, Scherer SJ, Clark AB, Yang K, Avdievich E, Jin B, Werling U, Parris T, Kurihara N, et al. 2004. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res 64: 517–522. [DOI] [PubMed] [Google Scholar]
- Lindegren CC 1955. Non-Mendelian segregation in a single tetrad of Saccharomyces ascribed to gene conversion. Science 121: 605–607. [DOI] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD 2010. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol 17: 1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD 2011. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479: 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA 1991. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res 51: 3075–3079. [PubMed] [Google Scholar]
- Loeb LA 2001. A mutator phenotype in cancer. Cancer Res 61: 3230–3239. [PubMed] [Google Scholar]
- Marinus MG 1976. Adenine methylation of Okazaki fragments in Escherichia coli. J Bacteriol 128: 853–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, West SC 2014. Holliday junction resolution: Regulation in space and time. DNA Repair (Amst) 19: 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, West SC 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson SW 1986. Escherichia coli helicase II (uvrD gene product) translocates unidirectionally in a 3′ to 5′ direction. J Biol Chem 261: 10169–10175. [PubMed] [Google Scholar]
- Matson SW, George JW 1987. DNA helicase II of Escherichia coli. Characterization of the single-stranded DNA-dependent NTPase and helicase activities. J Biol Chem 262: 2066–2076. [PubMed] [Google Scholar]
- Mazur DJ, Mendillo ML, Kolodner RD 2006. Inhibition of Msh6 ATPase activity by mispaired DNA induces a Msh2(ATP)-Msh6(ATP) state capable of hydrolysis-independent movement along DNA. Mol Cell 22: 39–49. [DOI] [PubMed] [Google Scholar]
- Mazurek A, Johnson CN, Germann MW, Fishel R 2009. Sequence context effect for hMSH2-hMSH6 mismatch-dependent activation. Proc Natl Acad Sci 106: 4177–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Mehta A, Haber JE 2014. Sources of DNA double-strand breaks and models for recombinational DNA repair. Cold Spring Harb Perspect Biol 6: a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo ML, Mazur DJ, Kolodner RD 2005. Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J Biol Chem 280: 22245–22257. [DOI] [PubMed] [Google Scholar]
- Miller JH 1998. Mutators in Escherichia coli. Mutat Res 409: 99–106. [DOI] [PubMed] [Google Scholar]
- Mitchel K, Lehner K, Jinks-Robertson S 2013. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet 9: e1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P 1989. Methyl-directed DNA mismatch correction. J Biol Chem 264: 6597–6600. [PubMed] [Google Scholar]
- Modrich P 1997. Strand-specific mismatch repair in mammalian cells. J Biol Chem 272: 24727–24730. [DOI] [PubMed] [Google Scholar]
- *.Morrical SW 2015. DNA pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motycka TA, Bessho T, Post SM, Sung P, Tomkinson AE 2004. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J Biol Chem 279: 13634–13639. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11: 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ 1916. The mechanism of crossing-over. Am Nat 50: 193–221. [Google Scholar]
- Muller HJ 1964. The relation of recombination to mutational advance. Mutat Res 106: 2–9. [DOI] [PubMed] [Google Scholar]
- Murayama Y, Kurokawa Y, Tsutsui Y, Iwasaki H 2013. Dual regulation of Dmc1-driven DNA strand exchange by Swi5-Sfr1 activation and Rad22 inhibition. Genes Dev 27: 2299–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC 1998. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391: 407–410. [DOI] [PubMed] [Google Scholar]
- Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, et al. 1994. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371: 75–80. [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, Yang W 2000. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407: 703–710. [DOI] [PubMed] [Google Scholar]
- Ollila S, Dermadi Bebek D, Jiricny J, Nystrom M 2008. Mechanisms of pathogenicity in human MSH2 missense mutants. Hum Mutat 29: 1355–1363. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW 1983. Yeast recombination: The association between double-strand gap repair and crossing-over. Proc Natl Acad Sci 80: 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS 2003. Chromosome choreography: The meiotic ballet. Science 301: 785–789. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Nicolaides NC, Wei Y-F, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, et al. 1994. Mutation of a mutL homolog in hereditary colon cancer. Science 263: 1625–1629. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Nicolaides NC, Liu B, Parsons R, Lengauer C, Palombo F, D’Arrigo A, Markowitz S, Willson JKV, Kinzler KW, et al. 1995. Mutations of GTBP in genetically unstable cells. Science 268: 1915–1917. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol 17: 6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Jeon Y, In D, Fishel R, Ban C, Lee JB 2010. Single-molecule analysis reveals the kinetics and physiological relevance of MutL-ssDNA binding. PLoS ONE 5: e15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrazzi G, Bachrati CZ, Selak N, Studer I, Petkovic M, Hickson ID, Jiricny J, Stagljar I 2003. The Bloom’s syndrome helicase interacts directly with the human DNA mismatch repair protein hMSH6. Biol Chem 384: 1155–1164. [DOI] [PubMed] [Google Scholar]
- Peoples TL, Dean E, Gonzalez O, Lambourne L, Burgess SM 2002. Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev 16: 1682–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon MC, Lorenowicz JJ, Uckelmann M, Klocko AD, Mitchell RR, Chung YS, Modrich P, Walker GC, Simmons LA, Friedhoff P, et al. 2010. Structure of the endonuclease domain of MutL: Unlicensed to cut. Mol Cell 39: 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A 2008. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem 283: 17766–17776. [DOI] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. 2009. Yeast Mph1 helicase dissociates Rad51-made D-loops: Implications for crossover control in mitotic recombination. Genes Dev 23: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R, DeRocco VC, Harris C, Sharma A, Hingorani MM, Erie DA, Weninger KR 2012. Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. EMBO J 31: 2528–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M, Wagner RE, Glickman BW, Meselson M 1980. DNA methylation, mismatch correction and genetic stability. In Progress in environmental mutagenesis (ed. Alacevic M), pp. 121–130 Elsevier, Amsterdam. [Google Scholar]
- Rayssiguier C, Thaler DS, Radman M 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342: 396–401. [DOI] [PubMed] [Google Scholar]
- Rich T, Allen RL, Wyllie AH 2000. Defying death after DNA damage. Nature 407: 777–783. [DOI] [PubMed] [Google Scholar]
- Roberts MS, Cohan FM 1993. The effect of DNA sequence divergence on sexual isolation in Bacillus. Genetics 134: 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E, Grimme JM, Spies M, Ha T 2008. Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc Natl Acad Sci 105: 20274–20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacho EJ, Kadyrov FA, Modrich P, Kunkel TA, Erie DA 2008. Direct visualization of asymmetric adenine nucleotide-induced conformational changes in MutLα. Mol Cell 29: 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent RG, Meservy JL, Perkins BD, Kilburn AE, Intody Z, Adair GM, Nairn RS, Wilson JH 2000. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res 28: 3771–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H, Tawaramoto MS, Lao JP, Hosaka H, Sanda E, Suzuki M, Yamashita E, Hunter N, Shinohara M, Nakagawa A, et al. 2013. A new protein complex promoting the assembly of Rad51 filaments. Nat Commun 4: 1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass LE, Lanyi C, Weninger K, Erie DA 2010. Single-molecule FRET TACKLE reveals highly dynamic mismatched DNA-MutS complexes. Biochemistry 49: 3174–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saydam N, Kanagaraj R, Dietschy T, Garcia PL, Pena-Diaz J, Shevelev I, Stagljar I, Janscak P 2007. Physical and functional interactions between Werner syndrome helicase and mismatch-repair initiation factors. Nucleic Acids Res 35: 5706–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Prakash S 1990. RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol Cell Biol 10: 2485–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM Jr 2005. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem 280: 28072–28084. [DOI] [PubMed] [Google Scholar]
- Shimada A, Kawasoe Y, Hata Y, Takahashi TS, Masui R, Kuramitsu S, Fukui K 2013. MutS stimulates the endonuclease activity of MutL in an ATP-hydrolysis-dependent manner. FEBS J 280: 3467–3479. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa T 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391: 404–407. [DOI] [PubMed] [Google Scholar]
- Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD 1997. Microsatellite instability in yeast: Dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol 17: 2851–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel EC, Bryson V 1967. Mutator gene of Escherichia coli B. J Bacteriol 94: 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R 2004. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell 15: 437–451. [DOI] [PubMed] [Google Scholar]
- Spampinato C, Modrich P 2000. The MutL ATPase is required for mismatch repair. J Biol Chem 275: 9863–9869. [DOI] [PubMed] [Google Scholar]
- Spies M 2013. There and back again: New single-molecule insights in the motion of DNA repair proteins. Curr Opin Struct Biol 23: 154–160. [DOI] [PubMed] [Google Scholar]
- Stambuk S, Radman M 1998. Mechanism and control of interspecies recombination in Escherichia coli: I. Mismatch repair, methylation, recombination and replication functions. Genetics 150: 533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S-S, Modrich P 1986. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci 83: 5057–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SS, Grilley M, Thresher R, Griffith J, Modrich P 1989. Gap formation is associated with methyl-directed mismatch correction under conditions of restricted DNA synthesis. Genome 31: 104–111. [DOI] [PubMed] [Google Scholar]
- Sugawara N, Paques F, Colaiacovo M, Haber JE 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci 94: 9214–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc Natl Acad Sci 101: 9315–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, New JH, Kowalczykowski SC 1998. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci 95: 6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev 11: 1111–1121. [DOI] [PubMed] [Google Scholar]
- *.Symington LS 2014. End resection at DNA double-strand breaks: Mechanism and regulation. Cold Spring Harb Perspect Biol 6: a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271. [DOI] [PubMed] [Google Scholar]
- Szekvolgyi L, Nicolas A 2009. From meiosis to postmeiotic events: Homologous recombination is obligatory but flexible. FEBS J 277: 571–589. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW 1983. The double-strand break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Tessmer I, Yang Y, Zhai J, Du C, Hsieh P, Hingorani MM, Erie DA 2008. Mechanism of MutS searching for DNA mismatches and signaling repair. J Biol Chem 283: 36646–36654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham KC, Hermans N, Winterwerp HH, Cox MM, Wyman C, Kanaar R, Lebbink JH 2013. Mismatch repair inhibits homeologous recombination via coordinated directional unwinding of trapped DNA structures. Mol Cell 51: 326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiraby J-G, Fox MS 1973. Marker discrimination in transformation and mutation of pneumococcus. Proc Natl Acad Sci 70: 3541–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson AE, Bardwell AJ, Bardwell L, Tappe NJ, Friedberg EC 1993. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature 362: 860–862. [DOI] [PubMed] [Google Scholar]
- Treffers HP, Spinelli V, Belser NO 1954. A factor (or mutator gene) influencing mutation rates in Escherichia coli. Proc Nat Acad Sci 40: 1064–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M, Lovett ST 1998. Single-strand DNA-specific exonucleases in _Escherichia coli_—Roles in repair and mutation avoidance. Genetics 149: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1: 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS 2007. Structure of the human MutSα DNA lesion recognition complex. Mol Cell 26: 579–592. [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Richardson CA, Elliott B, Jasin M 2006. Modeling oncogenic translocations: Distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst) 5: 1065–1074. [DOI] [PubMed] [Google Scholar]
- Welsh KM, Lu AL, Clark S, Modrich P 1987. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem 262: 15624–15629. [PubMed] [Google Scholar]
- Welz-Voegele C, Jinks-Robertson S 2008. Sequence divergence impedes crossover more than noncrossover events during mitotic gap repair in yeast. Genetics 179: 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JH, Lusnak K, Fogel S 1985. Mismatch-specific post-meiotic segregation frequency in yeast suggests a heteroduplex recombination intermediate. Nature 315: 350–352. [DOI] [PubMed] [Google Scholar]
- Williamson MS, Game JC, Fogel S 1985. Meiotic gene conversion mutants in Saccharomyces cerevisiae: I. Isolation and characterization of pms1–1 and pms1–2. Genetics 110: 609–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T, Guerrette S, Fishel R 1999. Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J Biol Chem 274: 21659–21644. [DOI] [PubMed] [Google Scholar]
- Witkin EM 1964. Pure clones of lactose negative mutants obtained in Escherichia coli after treatment with 5-bromouracil. J Mol Biol 8: 610–613. [DOI] [PubMed] [Google Scholar]
- Worth L Jr, Clark S, Radman M, Modrich P 1994. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci 91: 3238–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874. [DOI] [PubMed] [Google Scholar]
- Wu TH, Marinus MG 1994. Dominant negative mutator mutations in the mutS gene of Escherichia coli. J Bacteriol 176: 5393–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wyatt HDM, West SC 2014. Holliday junction resolvases. Cold Spring Harb Perspect Biol 6: a023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zelensky A, Kanaar R, Wyman C 2014. Mediators of homologous DNA pairing. Cold Spring Harb Perspect Biol 6: a016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM 2005. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122: 693–705. [DOI] [PubMed] [Google Scholar]