Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor (original) (raw)
. Author manuscript; available in PMC: 2015 Mar 12.
Published in final edited form as: Nat Biotechnol. 2014 Apr 20;32(5):490–495. doi: 10.1038/nbt.2886
Abstract
Exosomes show potential for cancer diagnostics because they transport molecular contents of the cells from which they originate. Detection and molecular profiling of exosomes is technically challenging and often requires extensive sample purification and labeling. Here we describe a label-free, high-throughput approach for quantitative analyses of exosomes. Our nano-plasmonic exosome (nPLEX) assay is based on transmission surface plasmon resonance through periodic nanohole arrays. Each array is functionalized with antibodies to enable profiling of exosome surface proteins and proteins present in exosome lysates. We show that this approach offers improved sensitivity over previous methods, enables portable operation when integrated with miniaturized optics and allows retrieval of exosomes for further study. Using nPLEX to analyze ascites samples from ovarian cancer patients, we find that exosomes derived from ovarian cancer cells can be identified by their expression of CD24 and EpCAM, suggesting the potential of exosomes for diagnostics.
Keywords: exosomes, surface plasmon resonance, periodic nanohole arrays, cancer, label-free sensing
Exosomes are membrane-bound phospholipid nanovesicles (50 - 100 nm in diameter) actively secreted by mammalian cells1. Renewed interest in exosomes follows recent reports demonstrating that most types of cancer shed large numbers of exosomes that carry molecular information about the parent tumor2. Capturing this information without biopsying the tumor could be a useful clinical and research tool. Rapid isolation and analysis of exosomes, however, is challenging as ultracentrifugation is time consuming3, and conventional detection standards, such as Western blotting and enzyme-linked immunosorbent assays (ELISA), require large amounts of sample and extensive post-labeling processes for detection2. Given these limitations, current analytical methods for exosomes are often impractical for experiments that require large throughput or in which the exosome concentration is low.
Here we report a surface plasmon resonance (SPR) –based assay for label-free, high-throughput exosome protein analyses. The system is based on extraordinary optical transmission through periodic nanoholes4-7 rather than total internal reflection8,9 as used in commercial SPR systems. We reasoned that plasmonic nanoholes would be an ideal sensing scheme, as their probing depth (< 200 nm) can be readily matched to exosome size for improved detection sensitivity, and the transmission setup allows system miniaturization as well as the construction of highly-packed sensing arrays. We therefore designed a new SPR chip, named nano-plasmonic exosome (nPLEX) sensor, that comprises arrays of periodic nanoholes patterned in a metal film. Each array is functionalized with affinity ligands for different exosomal protein markers. With target-specific exosome binding, the nPLEX sensor displays spectral shifts or intensity changes proportional to target marker protein levels. Compared to conventional methods, the nPLEX technology offers highly sensitive and label-free exosome analyses and enables continuous and real-time monitoring of molecular binding. In order to improve throughput, we further developed a nPLEX imaging system by combining the nanohole chips with a miniaturized imaging setup. This system is readily scalable for massively parallel measurements (105 sensing elements).
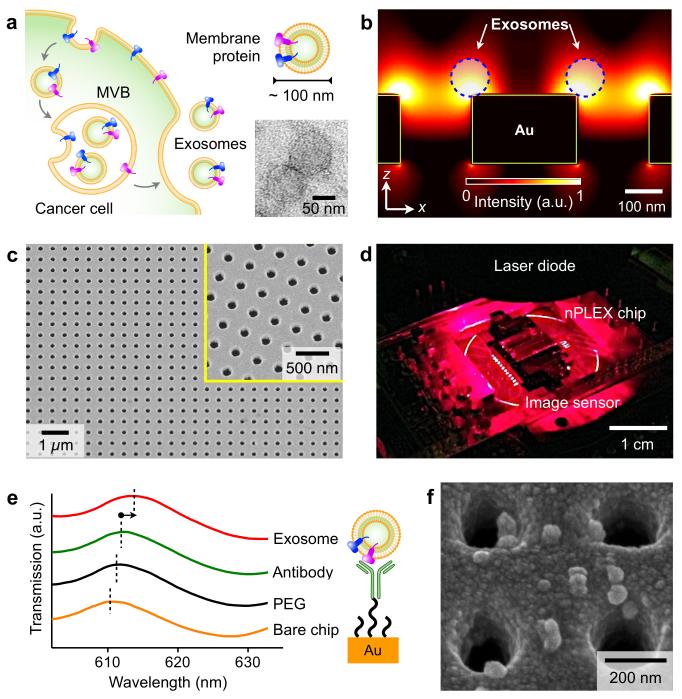
Large quantities of exosomes are actively secreted by cancer cells through fusion of multivesicular endosomes with the plasma membrane, and they circulate in various biofluids (Fig. 1a and Supplementary Figs. 1a, b). Nanoparticle tracking analysis, which determines the nanoparticle size by analyzing its Brownian motion, showed that exosomes have a unimodal size distribution with an average diameter of 100 nm (Supplementary Fig. 1c). We designed the nPLEX sensor to achieve label-free detection of exosomes of this size, where the basic sensing unit consists of a periodic lattice of nanoholes patterned in a gold (Au) film. Simulation studies revealed enhanced electromagnetic fields that were tightly confined within exosome size range (Fig. 1b). We further tuned the field range to overlap with the exosome size by adjusting the nanohole periodicity, thereby maximizing the detection sensitivity (Supplementary Fig. 2). The design had a lattice of 44 × 32 nanoholes per sensing unit; each nanohole had a hole diameter of 200 nm in diameter and a periodicity of 450 nm made in a 200 nm-thick Au film on a glass substrate (Fig. 1c and Supplementary Fig. 2). For high-throughput analyses, we laid out a 12 × 3 array of sensing units with multi-channel microfluidics placed on top (Supplementary Fig. 3). Each channel had a sample volume of 0.3 μL and spanned over three sensing units for triplicate measurements. In a second generation chip, we implemented 1,089 measurement sites (33 × 33 array) for massively parallel detection (Supplementary Fig. 4). Built with the same design parameters, this new chip was fabricated by interference lithography10 at a wafer scale for high-throughput chip production.
Figure 1. Label-free detection of exosomes with nPLEX sensor.
(a) Cancer cells secrete a large abundance of exosomes through fusion of multivesicular body (MVB) with cellular plasma membrane. These nanovesicles carry parental proteins in the same topological orientation. High magnification transmission electron micrograph (inset) indicates exosomes from human ovarian cancer cell (CaOV3) culture have a diameter ~100 nm. (b) Finite-difference time-domain (FDTD) simulation shows the enhanced electromagnetic fields tightly confined near a periodic nanohole surface. The field distribution overlaps with the size of exosomes captured onto the sensing surface, maximizing exosome detection sensitivity. (c) A scanning electron micrograph (SEM) of the periodic nanoholes in the nPLEX sensor. The hole diameter is 200 nm with the periodicity of 450 nm. The structure was patterned in a gold film (200 nm thick) deposited on a glass substrate. The inset shows a zoomed-in image. (d) A prototype miniaturized nPLEX imaging system developed for multiplexed and high-throughput analyses of exosomes. The system uses a complementary-metal-oxide-semiconductor (CMOS) imager to record the transmitted light intensity from a nPLEX chip. (e) A representative schematic of changes in transmission spectra showing exosome detection with nPLEX. The gold surface is pre-functionalized by a layer of polyethylene glycol (PEG), and antibody conjugation and specific exosome binding were monitored by transmission spectral shifts as measured by nPLEX (not drawn to scale). (f) SEM indicates specific exosome capture by functionalized nPLEX.
Unlike conventional reflection-based SPR devices, the nPLEX sensor operates in a transmission mode. This scheme made it possible to use a compact collinear optical setup and construct densely packed sensing units (Supplementary Fig. 5a). Specific binding of exosomes to the nPLEX sensor changed its local refractive index, which can be monitored by measuring either wavelength shifts (Δλ) in light spectrum (spectral detection) or intensity changes (Δ_p_) at fixed wavelength11,12 (intensity detection; Supplementary Fig. 5). We employed spectral detection for assay development and optimization and intensity detection with a portable imaging system for point-of-care patient sample analyses (Fig. 1d). Consisting of a laser-diode and a complementary metal–oxide–semiconductor (CMOS) imager, the system offered a large field-of-view (~25 mm2). The entire nPLEX array (36 sensing units) was imaged simultaneously for parallel detection.
To functionalize the SPR surface, we used a multi-step approach. Pre-coating the device surface with a 1:3 mixture of long (MW 1 kDa) and short (MW 0.2 kDa) polyethylene glycol (PEG) polymers minimized non-specific exosome binding (Supplementary Fig. 6) and improved surface hydrophilicity. Following PEG-coating, we grafted monoclonal antibodies onto the long PEG chains for specific exosome capture. All surface modifications were done by flowing reagents through the microfluidic channels while monitoring the spectral shifts from binding (Fig. 1e). The functionalized nPLEX chip shows high specificity for exosome capture, with negligible binding when control antibody was used (Supplementary Fig. 7); these findings were also confirmed by electron microscopy (Fig. 1f) Parallel detection of 12 potential exosomal markers can be accomplished in < 30 min. Furthermore, the sensor can be regenerated for repeated use by eluting attached antibodies and exosomes (Supplementary Fig. 8).
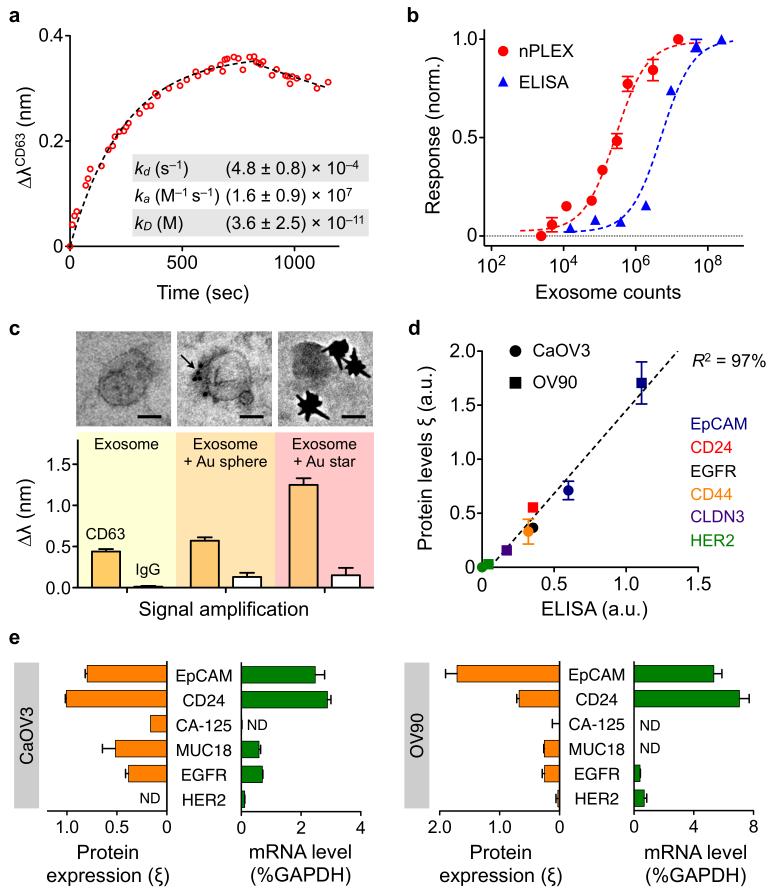
To establish an assay protocol for quantitative exosome analyses, we first used nPLEX to examine exosome binding kinetics. We functionalized the sensor surface with antibodies against CD63, a type III lysosomal membrane protein abundant and characteristic of exosomes13 and introduced exosomes derived from human ovarian cancer cell (CaOV3) culture (Fig. 2a). The observed binding constant was ~36 pM, which was significantly lower than that of individual antigen-antibody binding (~1 nM). Such stable binding of the exosomes could be attributed to the multivalent nature of the nPLEX assay (i.e., multiple binding sites per exosome)14. We next determined the detection sensitivity of the nPLEX assay. We estimated the concentration of exosomes isolated from CaOV3 culture using nanoparticle tracking analysis. Two nPLEX sensors, functionalized with anti-CD63 antibody and control antibody respectively, were used to measure the relative spectral shifts (ΔλCD63) against known exosome counts. The titration experiments established the nPLEX limit of detection (LOD) of ~3000 exosomes (670 aM) (Fig. 2b; see Supplementary Methods for details). The observed sensitivity was 104 fold higher than western blot13 and 102 fold higher than chemiluminescence ELISA (Fig. 2b). The nPLEX platform also facilitated signal amplification through a secondary labeling (Fig. 2c). For instance, when captured exosomes were targeted with spherical Au nanoparticles (diameter, 10 nm), the signal (ΔλCD63) improved by 20%. The signal could be enhanced by 300% by using larger, star-shaped Au nanoparticles (diameter, 50 nm; Fig. 2c and Supplementary Fig. 9)
Figure 2. Exosome quantification and protein profiling with nPLEX.
(a) Real-time kinetic sensorgram of exosome capture. Exosomes isolated from a human ovarian cancer cell line (CaOV3) were introduced onto a nPLEX sensor functionalized with anti-CD63 for exosomal capture (kD ~ 36 pM). (b) Comparison of the detection sensitivity of nPLEX and ELISA. The nPLEX detection limit was determined by titrating a known quantity of exosomes and measuring their associated CD63 signal. The detection threshold for ELISA was independently assessed with chemiluminescence (c) nPLEX signal amplification through secondary labeling. Exosomes captured on the nPLEX sensor were further targeted with anti-CD63 Au nanospheres (arrow) or star-shaped particles to enhance spectral shifts. Scale bar, 50 nm. (d) Correlation between nPLEX and ELISA measurements. Exosomes isolated from human ovarian cancer cell lines (CaOV3 and OV90) were used. The marker protein level (ξ) was determined by normalizing the marker signal with that of anti-CD63, which accounted for variation in exosomal counts across samples. a.u., arbitrary unit. (e) mRNA analysis of exosomes eluted from CaOV3 cells (left) or OV90 cells (right). Following nPLEX protein measurements, captured exosomes were released from the chip and subsequently analyzed for mRNA contents. The mRNA levels were normalized against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels. ND, non-detected. All measurements in b-e were in performed in triplicate and the data is displayed as mean ± s.d. a.u., arbitrary unit.
To quantitatively detect exosome proteins, we functionalized the nPLEX sensors with antibodies against various target and measured the associated spectral shifts (Δλtarget) or intensity changes (Δ_p_target). Next, we defined the protein level (ξtarget) of the target protein by normalizing the target-associated changes to those of CD63 (i.e., ξtarget = Δλtarget/ΔλCD63 = Δ_p_target/Δ_p_CD63). Such normalization accounts for differences in exosome quantities among samples and reports the average level of a target protein per exosome. We applied this method to profile exosomes from two different human ovarian cancer cell lines, CaOV3 and OV90, for various extravesicular markers. Protein levels correlated well (R2 > 98%) between nPLEX and the gold standard ELISA (Fig. 2d), but nPLEX detection was faster, more sensitive and required smaller sample amounts (Fig. 2b). The nPLEX assay can also be used to detect intravesicular markers in exosome lysates (Supplementary Fig. 10) and for downstream genetic analyses by releasing captured exosomes from the device. As an example, we retrieved captured exosomes from the anti-CD63 channel by briefly reducing the pH to elute attached antibodies and exosomes (Supplementary Methods and Supplementary Fig. 8). The collected exosomes were subsequently assayed by fluorescence quantitative real-time polymerase chain reaction (qRT-PCR) to measure mRNA contents (Fig. 2e).
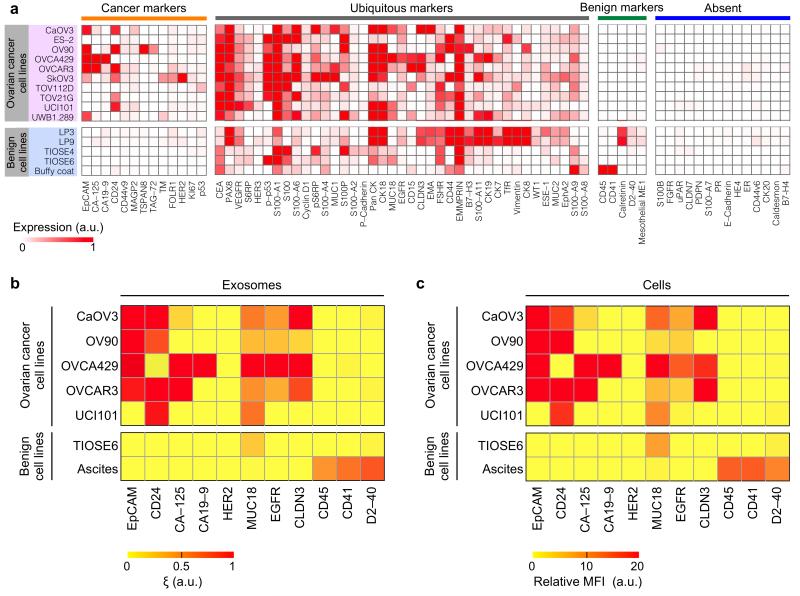
We used the nPLEX assay to molecularly screen exosomes across different ovarian cancer cell lines. We aimed to identify a molecular signature to detect ovarian-cancer exosomes and examine how closely exosomes reflect their cells of origin. We started with antibody profiling of ovarian cancer and other host cell (non-cancer) markers (Fig. 3), chosen based on prior studies and scientific databases15,16. Cluster analysis of the profiling data revealed four protein marker groups that were expressed in ovarian cancer or benign cells. We then chose a subset of markers, favoring extracellular markers present in ovarian cancer cells or benign cells and markers for which consistent antibody targeting methods had been established. (Supplementary Table 1). The following putative cancer markers were thus selected: epithelial cell adhesion molecule (EpCAM)17; CD2417,18; cancer antigen 125 (CA-125)19; cancer antigen 19-9 (CA19-9)20; human epidermal growth factor receptor 2 (HER2)21; mucin 18 (MUC18)22; epidermal growth factor receptor (EGFR)23; claudin 3 (CLDN3)24. For non-cancer cells, these markers were selected: CD45 (leukocyte), CD41 (platelet) and D2-40 (mesothelial cells)25.
Figure 3. Molecular profiling of ovarian cancer protein markers.
(a) Levels of 71 protein markers were determined in ovarian cancer cell lines and benign cells(including mesothelial origin: LP3 and LP9, benign ovarian origin: TIOSE 4 and TIOSE 6 and blood origin: buffy coat). Clustering analyses based on Pearson correlation categorized all markers into four subgroups, from left to right: 1) cancer markers expressed by ovarian cancer cell lines only, 2) ubiquitous markers present in both cancer and benign cells, 3) benign markers expressed by benign cells only and 4) markers absent in both cell types. a.u., arbitrary unit. (b, c) Putative ovarian cancer markers (EpCAM, CD24, CA19-9, CLDN3, CA-125, MUC18, EGFR, HER2), immune host cell markers (CD41, CD45) and a mesothelial marker (D2-40) were profiled on exosomes (b, using nPLEX sensor) and their parental ovarian cell lines (c, using flow cytometry). MFI, mean fluorescence intensity. a.u., arbitrary unit. All measurements were performed in triplicate and the data is displayed as mean values.
We next compared the expression of the aforementioned markers between exosomes (Fig. 3b) and cell lines from which they are derived (Fig. 3c). The exosomal and cellular protein profiles showed excellent correlation (Pearson coefficient > 0.95), which supports the use of exosomes as cellular surrogates for the selected protein markers. In the tested cell lines, levels of EpCAM and CD24 markers helped to distinguish ovarian cancer exosomes from benign cell-derived exosomes.
Based on our findings, we used nPLEX to detect cancer exosomes in patient-derived ascites (i.e., excess fluid accumulation in the peritoneal cavity26). Ascites is common in ovarian cancer patients and is often tapped for symptomatic relief. We hypothesized that the fluid, which is generally discarded, would contain exosomes and thus allow for molecular diagnostics26. We found that native ascites samples indeed contained large quantities of exosomes (> 109 exosomes per mL) sufficient for robust nPLEX detection without further enrichment or signal amplification. As such, we assayed samples directly after collecting exosomes through a membrane filter with a 0.2 μm size cutoff; both size and western blotting analyses confirmed exosome enrichment after filtration (Supplementary Fig. 11).
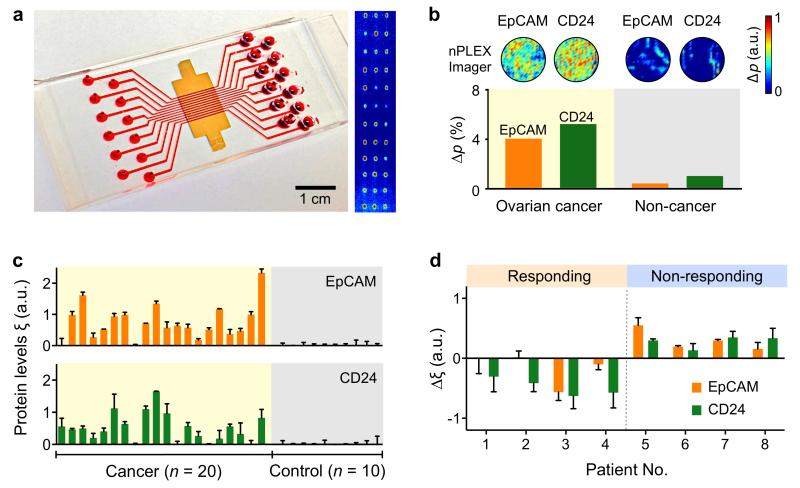
We used the 12-channel nPLEX array, with each channel functionalized with different antibodies for EpCAM, CD24, CD63 and IgG control, and imaged the entire 12 × 3 array using the portable imager system (Fig. 4a and Supplementary Fig. 5). After measuring the diffracted light emitted through the nPLEX sensor, we numerically reconstructed the light intensity at the sensor surface (Supplementary Methods and Fig. 4b) Using cancer-derived ascites, the EpCAM and CD24 arrays displayed significant (P < 0.05; two-tailed t_-test) intensity changes (Δ_p) due to cancer exosome capture; in contrast, changes were negligible in non-cancerous ascites. We corroborated that the protein level (ξ) of exosomal markers measured by the imager were comparable to those by spectral detection (Supplementary Fig. 12).
Figure 4. Profiling of ovarian cancer patient exosomes with nPLEX.
(a) A photograph of nPLEX chip integrated with a multi-channel microfluidic cell for independent and parallel analyses. (Right) Transmission intensities of 12 × 3 nanohole arrays were measured simultaneously using the imaging setup. (b) Ascites-derived exosomes from ovarian cancer and non-cancer patients were evaluated by the nPLEX sensor. Cancer exosomes were captured on EpCAM and CD24-specific sensor sites, which led to intensity changes in the transmitted light. (c) Exosomal protein levels of EpCAM and CD24 in ascites samples from patients were measured by nPLEX. Ovarian cancer patient samples (n = 20) were associated with elevated EpCAM and CD24 levels, while non-cancer patients (n = 10) showed negligible signals. (d) Longitudinal monitoring of treatment responses. Ascites samples were collected from ovarian cancer patients before and after chemotherapy (n = 8) and profiled with nPLEX. The bars represent the changes in CD24 and EpCAM levels per exosome before and after treatment. All measurements in c-d were performed in triplicate and the data is displayed as mean ± s.d. a.u., arbitrary unit.
We then obtained ascites samples from ovarian cancer patients (n = 20), and non-cancerous ascites from cirrhosis patients as controls (n = 10) (Fig. 4c, Supplementary Tables 2 and 3), and profiled them using nPLEX (Fig. 4c). Exosome concentrations estimated by nPLEX using CD63 signal changes were highly heterogeneous among patient and control samples (Supplementary Fig. 13) and could not conclusively differentiate between cancer patients and control subjects (P = 0.11; two-tailed t-test); it is likely that exosome numbers were highly susceptible to sampling variations (e.g., ascitic drainage procedure). The levels of EpCAM and CD24 per exosome, however, were significantly higher in the tested ovarian cancer patient samples (P < 0.001 for both markers; two-tailed _t_-test) than in control groups (Fig. 4c). Analyses with receiver operating characteristic curves (Supplementary Fig. 14a) determine the intrinsic accuracy of 93% for EpCAM and 87% for CD24 (Supplementary Fig. 14b). Pairing expression profiles of EpCAM and CD24 could further increase the diagnostic accuracy to 97%. The early promise of these potential ovarian cancer biomarkers, however, requires validation using much larger cohorts.
Next, we explored exosome profiling to monitor clinical response or progression during treatment. We recruited ovarian cancer patients (n = 8) undergoing standard chemotherapy (Supplementary Tables 2 and 4) and collected their ascites samples before and after treatment. For both time points, we measured exosomal EpCAM and CD24 levels. A board-certified oncologist (C.M.C.), blinded to the nPLEX data, assigned each subject either responder or non-responder status based on accepted clinical, laboratory and/or radiologic metrics. We observed that the levels of exosomal EpCAM, CD24 or both decreased among responding patients, whereas increased levels of these markers were associated with non-responding patients (Fig. 4d). The cohort was too small for these data to obtain statistical significance.
Rapid, multiplexed protein analysis of exosomes could improve early disease detection and therapy monitoring. The structure of nPLEX—a periodic array of sub-wavelength apertures in a metal film— generates intense surface plasmons whose extinction depth is comparable to exosome size, making the technology well suited to sensitive, label-free exosome detection. By integrating the system with miniaturized optics, we created a highly portable platform capable of both rapid and large-scale sensing. We established a quantitative assay protocol that reports both exosome concentrations and exosomal protein levels of extra- and intravesicular protein markers, while consuming only small amounts of specimen. The captured exosomes can be readily eluted from the device for downstream analyses, such as genomic profiling. Together, these approaches will facilitate comprehensive exosomal analyses by yielding both proteomic and genetic information.
For research applications, nPLEX could help explore fundamental questions about exosome-mediated intercellular communication and tumor micro-environment27,28. For clinical applications, with further development and validation, nPLEX could be useful for exploring exosomes as a cancer biomarker, for diagnostics and for evaluating tumor response to therapy. While the current study focused on ovarian cancer exosomes in ascites, the nPLEX analysis could readily be extended to exosomes in other bodily fluids (e.g., blood, cerebrospinal fluids and urine).
Several technical modifications could be made to improve nPLEX and accelerate its application for clinical use. First, using light-interference lithography10, we generated a second-generation nPLEX chip that has substantially higher throughput and > 1,000 measurement sites. This chip allows for rapid, wafer-scale nanohole patterning, overcoming the limitations of serial chip processing (i.e., focused-ion beam milling). To implement the next-generation nPLEX chip, we are exploring a molecular printing technique29 (Supplementary Fig. 15) for chip surface modification and developing a new imaging setup for signal readout. The resulting system will be a microarray-type sensor for massively parallel detection. Second, we are currently working to improve signal amplification through secondary labeling with nanoprobes such as gold nanostars of varying size and dimension to further enhance detection sensitivity. This strategy would be particularly useful for on-chip probing of rare exosomal markers (e.g., protein, mRNA, microRNA or DNA) in exosome lysates. Third, large cohorts in prospective trials, such as exosome screening across a spectrum of human illnesses (e.g., other solid tumors, cardiovascular diseases, diabetes, infections), are required to establish the clinical utility of nPLEX. The resulting large datasets could then be critically analyzed as in functional proteomics studies30.
Methods
nPLEX chip fabrication
Standard microscope glass slides were cleaned in a piranha solution (3:1 H2SO4:H2O2) at 80 Δ for 30 min and rinsed with distilled water. The glass slides were then dried under N2 stream and baked on a hotplate at 150 Δ for 15 min. A 200 nm thick Au film with a 2 nm thick Ti adhesion layer was deposited on the glass slides through electron-beam metal evaporation (Denton E-beam evaporator) at deposition rates of 2 Å/sec (Au) and 0.5 Å/sec (Ti). A patterned acrylic sheet was placed on the glass slide as a shadow mask to define a sensing area in the center of the glass slide. Periodic nanohole arrays, wherein each consisted of 44 by 32 apertures with 200 nm diameter and 450 nm periodicity, were fabricated by focused ion-beam milling (Zeiss NVision 40) at 30 keV and 80 pA. The nPLEX chip was integrated with a multi-channel polydimethylsiloxane (PDMS) microfluidic flow-cell fabricated by soft lithography. Detailed information is also available in the Supplementary Information.
Microscope setup
A conventional upright microscope (Nikon Eclipse Ci) was used for spectral measurements. A 100 W halogen lamp illuminated individual nanohole array through a 10× microscope objective, and the transmitted light was collected by an optical fiber placed right underneath the nanohole chip. The transmission spectra were analyzed by a miniature fiber-optic VIS-NIR spectrometer (Ocean optics). The integration time was 2 sec and the spectrum was averaged by 5 times.
Portable imaging setup
An integrated CMOS image sensor (Aptina Imaging) was used for imaging measurements. A laser diode at 638 nm with collimating lens and square pattern diffuser was used for illumination. The beam size was adjusted to cover the entire nanohole arrays. The nPLEX chip was placed above the image sensor with less than a 2 mm distance and fixed by a plastic holder. The intensities of all the arrays were collected simultaneously and analyzed by a custom-built MATLAB program (Detailed information on data analysis is also available in the Supplementary Information). The integration time was approximately 5 msec, and the intensities were averaged by 10 times for each image.
Cell culture
All human ovarian carcinoma cell lines were obtained from American Type Culture Collection (ATCC). CaOV3, OVCAR3, SKOV3 were grown in Dulbecco’s modified essential medium (DMEM, Cellgro). OV90, OVCA429, UCI101 were cultured in RPMI-1640 medium (Cellgro). ES-2 were cultured in McCoy 5a medium, TOV112D and TOV21G in 1:1 mixture of MCDB 105 medium and Medium 199 (Sigma-Aldrich), and UWB1.289 in 1:1 mixture of RPMI-1640 medium and mammary epithelial growth medium (MEGM, Lonza). All media were supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin (Cellgro). Mesothelial cells, LP3 and LP9, were purchased from the Corriell Institute for Medical Research and grown according to protocol. Normal ovarian surface epithelium (NOSE) cell lines were derived from ovarian surface epithelium (OSE) brushings and cultured in 1:1 mixture of MCDB 105 medium and Medium 199 (Sigma-Aldrich) with gentamicin (25 μg/mL) and 15% heat-inactivated serum. TIOSE4 and TIOSE 6 cell lines were then obtained by transfecting NOSE cells with hTERT, and cultured in RPMI-1640 supplemented with 10% FBS and penicillin-streptomycin. All cell lines were tested and free of mycoplasma contamination (MycoAlert Mycoplasma Detection Kit, Lonza, LT07-418).
Exosome isolation and quantification
Cells at passages 1-15 were cultured in vesicle-depleted medium (with 5% depleted FBS) for 48 hours. Conditioned medium from ~107 cells was collected, filtered through a 0.2 μm membrane filter (Millipore) and concentrated via differential centrifugation as previously described3,13. For exosome collection from clinical samples, ascites samples were filtered through a 0.2 μm membrane filter (Millipore) to remove cells and debris. The filtrates were used directly for exosomal analysis with the nPLEX sensor. For independent measure of exosome concentrations, we used the nanoparticle tracking analysis (NTA) system (LM10, Nanosight). For the quantification by NTA, exosome concentrations were adjusted to obtain ~ 50 vesicles in the field of view in order to achieve optimal counting. All NTA measurements were performed with identical system settings for consistency.
Sensor surface modification with antibodies
The Au nanohole surface was first coated with a mixture of polyethylene glycol (PEG) containing long active (carboxylated or biotinylated) thiol-PEG and short inactive methylated thio-PEG (Thermo Scientific, Nanocs) (1:3 active: inactive, 10 mM in PBS). After washing, the surface was either briefly activated with EDC/NHS mixture in MES buffer and conjugated to protein A/G (Thermo Scientific, 2 mg/mL) or used directly for binding with neutravidin (Thermo Scientific, 50 μg/mL). The following monoclonal antibodies were used without modification (protein A/G linker) or after biotinylation (neutravidin linker): EpCAM (ABCAM, clone MOC-31); CD24 (eBioscience, clone eBioSN3); CA19-9 (Abcam, clone SPM110); Claudin 3 (R&D Systems, clone 385021); CA-125 (Abcam, clone X75); MUC18 (R&D Systems, clone 128018); EGFR (Abcam, clone EGFR.1); HER2 (Biolegend, clone 24D2); CD41 (Biolegend, clone HI30); CD45 (Biolegend, clone HIP8); D2-40 (Abcam, clone D2-40); HSP90 (Abcam, clone AC88); HSP70 (Biolegend, clone W27); CD63 (BD Biosciences, clone H5C6) and respective IgG isotype controls (Biolegend). Antibodies were diluted in blocking solution (50 ug/mL in 2.5% bovine serum albumin (BSA) solution, Sigma), injected into individual sensor channels and incubated for 1 hour at room temperature. Excess unbound antibodies were removed by rinsing in PBS with 0.5% Tween 20 (PBST). Antibody-conjugated sensors were stored in PBS or dried at 4 °C for subsequent use.
Exosome detection with nPLEX sensor
Before introducing exosomes onto the nPLEX sensor, the fluidic channels were flushed with PBS buffer (3 min), and the baseline spectrum was measured. For in vitro assay with exosomes isolated from cell cultures, exosomes were flown to the device at a flow rate of 0.2 - 2 μL/min. For clinical ascites samples, the filtered ascites were continuously injected at a constant flow rate of 10 μL/min for 15 min. After exosome incubation, the channels were washed with PBST for 5 - 10 min at a flow rate of 10 μL/min followed by another set of measurements. The measured spectra and transmitted intensities were analyzed by a custom-designed program (MATLAB).
Clinical samples
Subjects were recruited according to an Institutional Review Board approved protocol with informed consent. A total of 38 individuals were enrolled. Ascites fluid samples were collected from patients as per routine in Massachusetts General Hospital Abdominal Imaging and Intervention suites. For the profiling study, we obtained clinical ascites samples from ovarian cancer patients (n = 20) as well as non-cancer patients (n = 10) with ascites-generating conditions (i.e. coincidentally, all were from cirrhosis). Cancer diagnoses and subtypes were confirmed by histological examination and clinical imaging. For longitudinal treatment response evaluation, serial ascites samples were collected from each patient (n = 8) during two distinct treatment visits. Responder and non-responder status was independently assigned by a gynecologic oncologist based on commonly used response criteria in ovarian cancer studies: 1) CA-125 based on Gynecologic Cancer Intergroup (GCIG) criteria, 2) scans based on Response Evaluation Criteria In Solid Tumors (RECIST), and /or 3) in cases where such data were not available within a week of collection, the electronic medical record for documented clinical impressions (e.g., palliative care without active therapy due to clinical decline or quality of life changes based on increased/decreased ascites accumulation). All ascites samples were filtered through a 0.2 μm membrane filter (Millipore) to remove cells and debris. Clinical filtrates were used directly for exosomal analyses with the nPLEX sensor.
Supplementary Material
Supplementary
Acknowledgements
The authors thank S. Skates (MGH) for helpful discussion on statistical analyses; K. Joyes for reviewing the manuscript. This work was supported in part by NIH grants R01-HL113156 (H.L.), R01-EB010011 (R.W.), R01-EB00462605A1 (R.W.), T32CA79443 (R.W.), K12CA087723-11A1 (C.M.C) and NHLBI contract HHSN268201000044C (R.W.). The device was fabricated using the facilities at the Center for Nanoscale Systems (CNS) at Harvard University (NSF award ECS-0335765).
Footnotes
Competing financial interest. A provisional patent on the nPLEX sensor technology has been filed and assigned to Massachusetts General Hospital.
References
- 1.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 2.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell. Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3, Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 4.Brolo AG. Plasmonics for future biosensors. Nat. Photonics. 2012;6:709–713. [Google Scholar]
- 5.Gordon R, Sinton D, Kavanagh KL, Brolo AG. A new generation of sensors based on extraordinary optical transmission. Acc. Chem. Res. 2008;41:1049–1057. doi: 10.1021/ar800074d. [DOI] [PubMed] [Google Scholar]
- 6.Im H, Wittenberg NJ, Lesuffleur A, Lindquist NC, Oh S-H. Membrane protein biosensing with plasmonic nanopore arrays and pore-spanning lipid membranes. Chem. Sci. 2010;1:688–696. doi: 10.1039/C0SC00365D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escobedo C. On-chip nanohole array based sensing: a review. Lab Chip. 2013;13:2445–2463. doi: 10.1039/c3lc50107h. [DOI] [PubMed] [Google Scholar]
- 8.Homola J. Surface plasmon resonance sensors for detection of chemical and biological species. Chem. Rev. 2008;108:462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 9.Lee HJ, Nedelkov D, Corn RM. Surface plasmon resonance imaging measurements of antibody arrays for the multiplexed detection of low molecular weight protein biomarkers. Anal. Chem. 2006;78:6504–6510. doi: 10.1021/ac060881d. [DOI] [PubMed] [Google Scholar]
- 10.Campbell MTHRGDM, Sharp DN, Harrison MT, Denning RG, Turberfield AJ. Fabrication of photonic crystals for the visible spectrum by holographic lithography. Nature. 2000;404:53–56. doi: 10.1038/35003523. [DOI] [PubMed] [Google Scholar]
- 11.Im H, Lesuffleur A, Lindquist NC, Oh SH. Plasmonic nanoholes in a multichannel microarray format for parallel kinetic assays and differential sensing. Anal. Chem. 2009;81:2854–2859. doi: 10.1021/ac802276x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanik AA, et al. Seeing protein monolayers with naked eye through plasmonic Fano resonances. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11784–11789. doi: 10.1073/pnas.1101910108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao H, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tassa C, et al. Binding affinity and kinetic analysis of targeted small molecule-modified nanoparticles. Bioconjug. Chem. 2009;21:14–19. doi: 10.1021/bc900438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anglesio MS, et al. Type-specific cell line models for type-specific ovarian cancer research. PLoS One. 2013;8:e72162. doi: 10.1371/journal.pone.0072162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlen M, et al. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 17.Runz S, et al. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol. Oncol. 2007;107:563–571. doi: 10.1016/j.ygyno.2007.08.064. [DOI] [PubMed] [Google Scholar]
- 18.Kristiansen G, et al. CD24 is expressed in ovarian cancer and is a new independent prognostic marker of patient survival. Am. J. Path. 2002;161:1215–1221. doi: 10.1016/S0002-9440(10)64398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bast RC, Jr, et al. CA 125: the past and the future. Int. J. Biol. Markers. 1997;13:179–187. doi: 10.1177/172460089801300402. [DOI] [PubMed] [Google Scholar]
- 20.Canney PA, Wilkinson PM, James RD, Moore M. CA19-9 as a marker for ovarian cancer: alone and in comparison with CA125. Br. J. Cancer. 1985;52:131. doi: 10.1038/bjc.1985.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meden H, Kuhn W. Overexpression of the oncogene c-erbB-2 (HER2/neu) in ovarian cancer: a new prognostic factor. Eur. J. Obstet. Gynecol. Reprod. Biol. 1997;71:173–179. doi: 10.1016/s0301-2115(96)02630-9. [DOI] [PubMed] [Google Scholar]
- 22.Aldovini D, et al. M-CAM expression as marker of poor prognosis in epithelial ovarian cancer. Int. J. Cancer. 2006;119:1920–1926. doi: 10.1002/ijc.22082. [DOI] [PubMed] [Google Scholar]
- 23.Psyrri A, et al. Effect of epidermal growth factor receptor expression level on survival in patients with epithelial ovarian cancer. Clin. Cancer Res. 2005;11:8637–8643. doi: 10.1158/1078-0432.CCR-05-1436. [DOI] [PubMed] [Google Scholar]
- 24.Li J, et al. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer ?2009;9:244. doi: 10.1186/1471-2407-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu AY, Litzky LA, Pasha TL, Acs G, Zhang PJ. Utility of D2-40, a novel mesothelial marker, in the diagnosis of malignant mesothelioma. Mod. Pathol. 2004;18:105–110. doi: 10.1038/modpathol.3800259. [DOI] [PubMed] [Google Scholar]
- 26.Kipps E, Tan DSP, Kaye SB. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat. Rev. Cancer . 2013 doi: 10.1038/nrc3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 28.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 30.Carey MS, et al. Functional proteomic analysis of advanced serous ovarian cancer using reverse phase protein array: TGF-beta pathway signaling indicates response to primary chemotherapy. Clin. Cancer Res. 2010;16:2852–2860. doi: 10.1158/1078-0432.CCR-09-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary