Environment drives selection and function of enhancers controlling tissue-specific macrophage identities (original) (raw)
. Author manuscript; available in PMC: 2015 Dec 4.
SUMMARY
Macrophages reside in essentially all tissues of the body and play key roles in innate and adaptive immune responses. Distinct populations of tissue macrophages also acquire context-specific functions that are important for normal tissue homeostasis. To investigate mechanisms responsible for tissue-specific functions, we analyzed the transcriptomes and enhancer landscapes of brain microglia and resident macrophages of the peritoneal cavity. In addition, we exploited natural genetic variation as a genome-wide ‘mutagenesis’ strategy to identify DNA recognition motifs for transcription factors that promote common or subset-specific binding of the macrophage lineage-determining factor PU.1. We find that distinct tissue environments drive divergent programs of gene expression by differentially activating a common enhancer repertoire and by inducing the expression of divergent secondary transcription factors that collaborate with PU.1 to establish tissue–specific enhancers. These findings provide insights into molecular mechanisms by which tissue environment influences macrophage phenotypes that are likely to be broadly applicable to other cell types.
INTRODUCTION
Macrophages are phagocytic cells of the innate immune system that populate every organ, making key contributions to their development, functions, and protection against infections and injuries (Geissmann et al., 2010; Wynn et al., 2013). Accordingly, each population of tissue macrophages must adapt to its surrounding environment and engage in tissue-specific functions to be effective auxiliary cells. In support of this, recent mRNA profiling studies revealed significant differences between distinct populations of resident tissue macrophages (Gautier et al., 2012; Okabe and Medzhitov, 2014). Thus, in spite of common elements shared across all subtypes of tissue macrophages, including dependency on the transcription factor PU.1 and a strong reliance on signaling downstream of the CSF1 receptor for ontology and survival (Schulz et al., 2012; Wynn et al., 2013), each subset of tissue macrophage possesses its own unique gene expression profile that presumably allows it to function in synergy with the tissue in which it resides.
Accumulating evidence suggests that signaling factors derived from tissue environments play key roles in promoting the ontology and phenotype of the residing macrophage populations. For example, absence of TGF-β1 signaling in the mouse brain impairs the development of the microglia population (Butovsky et al., 2014; Makwana et al., 2007). In the peritoneum, omentum-derived retinoic acid (RA) promotes expression of GATA6 in a subpopulation of local macrophages (Okabe and Medzhitov, 2014). Interestingly, GATA6 expression is exclusive to this particular tissue macrophage population, and decreasing or eliminating its expression interferes with their functions and survival (Gautier et al., 2014; Gautier et al., 2012; Okabe and Medzhitov, 2014; Rosas et al., 2014).
Precisely how these and other signals act on macrophages at the genomic level to promote specialized phenotypes and unique transcriptional signatures remains unknown. However, strong evidence suggests that enhancers, which are fundamental determinants of gene expression, may play a key role in this context (Andersson et al., 2014; Levine, 2010; Shlyueva et al., 2014). Enhancers, in comparison to promoters, exhibit significant enrichment for combinations of DNA recognition motifs that correspond to binding sites for lineage-determining transcription factors (LDTFs), which are required for the development of distinct cell types. Different patterns of LDTF expression drive the selection of cell-specific repertoires of enhancers that are considered to be central to the establishment of cell identity and regulatory potential.
Studies of primary macrophages and B cells indicated that PU.1 acts as an essential LDTF that contributes to the selection of a large fraction of the cell-specific enhancer-like elements in each of these cell types (Barozzi et al., 2014; Ghisletti et al., 2010; Heinz et al., 2010). Macrophage-specific enhancer selection by PU.1 required collaborative interactions with additional macrophage-restricted transcription factors (TFs), including C/EBP and AP-1 factors (Heinz et al., 2013). In contrast, B cell-specific enhancer selection by PU.1 required collaborative interactions with B cell-restricted factors, including EBF and E2A (Heinz et al., 2010).
Pre-existing enhancer landscapes occupied by PU.1 and/or C/EBP factors were shown to be the major sites that bound signal-dependent transcription factors (SDTFs), such as NFκB, nuclear receptors and STAT proteins (Ghisletti et al., 2010; Heinz et al., 2010). The collaborative and hierarchical relationship of LDTFs and SDTFs at pre-existing enhancers was validated at the level of the DNA template by studies of effects of natural genetic variation on enhancer selection and function (Heinz et al., 2013). Mutations in PU.1 motifs causing loss of PU.1 binding resulted in loss of the collaborative binding of C/EBPα. Conversely, mutations in C/EBP motifs causing loss of C/EBPα binding resulted in a loss of collaborative binding of PU.1. Either type of mutation abolished signal dependent binding of NFκB, whereas mutations in NFκB motifs that abolish NFκB binding rarely affected the binding of PU.1 or C/EBPα. However, in contrast to the picture at pre-existing enhancers, NFκB was also shown to be capable of selecting ‘latent’ or ‘de novo’ enhancers by collaborating with PU.1 to bind to genomic locations lacking prior features associated with active enhancers (Kaikkonen et al., 2013; Ostuni et al., 2013). These observations provide an example of an environmentally driven modification of the enhancer repertoire by a broadly expressed SDTF that is nonetheless cell type-specific due to the obligatory participation of PU.1.
Given that each tissue environment is distinguished by a unique combination of signaling factors, it is likely that gene expression in each corresponding macrophage population is under the control of distinct combinations of SDTFs that can modulate the activity of a pre-existing enhancer repertoire to achieve context-dependent gene expression. In addition, it is also possible that environmental signals control the expression and activities of TFs that result in selection of tissue-specific enhancers, analogous to the establishment of ‘latent’ or ‘de novo’ enhancers. Here, we sought to determine the extent to which environment shapes distinct macrophage enhancer repertoires and the underlying mechanisms.
RESULTS
Environment-specific gene expression
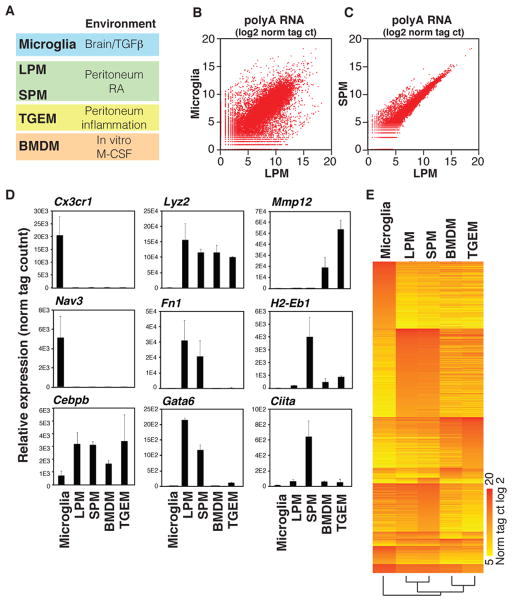
To investigate mechanisms responsible for tissue-specific macrophage phenotypes we isolated microglia (MG; brain macrophages) and two distinct populations of resident peritoneal macrophages (RPMs) that are discriminated by cell surface expression levels of MHCII; Large Peritoneal Macrophages (LPMs – low MHCII) and Small Peritoneal Macrophages (SPMs – high MHCII) by flow cytometry (Figure S1A, B) (Ghosn et al., 2010; Okabe and Medzhitov, 2014) (Figure 1A). These three populations of macrophages allow comparisons of gene expression and epigenetic landscapes in distinct macrophage populations residing in the same environment (i.e., LPMs vs. SPMs) as well as different environments (i.e., LPM vs. MG). In addition, we included Thioglycollate-elicited peritoneal macrophages (TGEMs) and bone marrow-derived macrophages (BMDMs) for comparison, as these macrophages, although maintained in culture conditions, are widely used models of macrophage biology that are derived from different sources (Figure 1A).
Figure 1. Variation in gene expression in different macrophage subsets.
A. Macrophage subsets used for analysis and corresponding environmental factors (See Figure S1A, B for sorting protocols). B, C. Scatterplots illustrating relative gene expression of polyA-selected RNA transcripts in MG compared to LPMs (B), and SPMs compared to LPMs (C) Values are log2 of tag counts normalized to 107 uniquely mapped tags. See Figure S1C for representative replicate. D. Relative gene expression values (mean ±S.D.) for the indicated genes are shown from replicate RNA-Seq experiments. E. Heat map of transcripts exhibiting an expression value of at least 64 normalized tags in at least one subset, and differing in expression by at least 16-fold in at least one other of the indicated subsets. See also Table S1.
Gene expression profiles determined by RNA-Seq from independent biological replicates revealed substantial differences in the patterns of gene expression across the different macrophage populations examined (Figure 1B, C, and Figure S1C), in agreement with previous studies (Gautier et al., 2012; Okabe and Medzhitov, 2014). In particular, ~7000 genes are differently expressed in MG compared to LPMs (p-value < 0.01), with 809 genes being >16-fold more highly expressed in MG and 924 genes being >16-fold more highly expressed in LPMs. On the other hand, LPMs and SPMs share strong similarities (Figure 1C), with SPMs expressing only 108 genes >16-fold higher than LPMs and LPMs expressing only 5 genes >16-fold higher than SPMs. These results corroborate many previous findings, including the highest level of expression of Cx3cr1 in MG and the selective expression of Gata6 in RPMs (Figure 1D) (Cardona et al., 2006; Gautier et al., 2012; Jung et al., 2000; Okabe and Medzhitov, 2014). Interestingly, Ciita, a transcription factor that regulates MHCII expression (Steimle et al., 1993), is preferably expressed in the SPM population (Figure 1D). Finally, gene clustering analyses confirmed that while LPMs and SPMs show highly similar gene expression, MG differ substantially from the other macrophage subsets (Figure 1E). TGEMs and BMDMs are also more similar to one another than either one is to any of the three in vivo subsets, potentially reflecting the similarity of the cell culture environment. Overall, these findings suggest a strong role of environment in determining macrophage gene expression.
Common and distinct macrophage enhancer repertoires
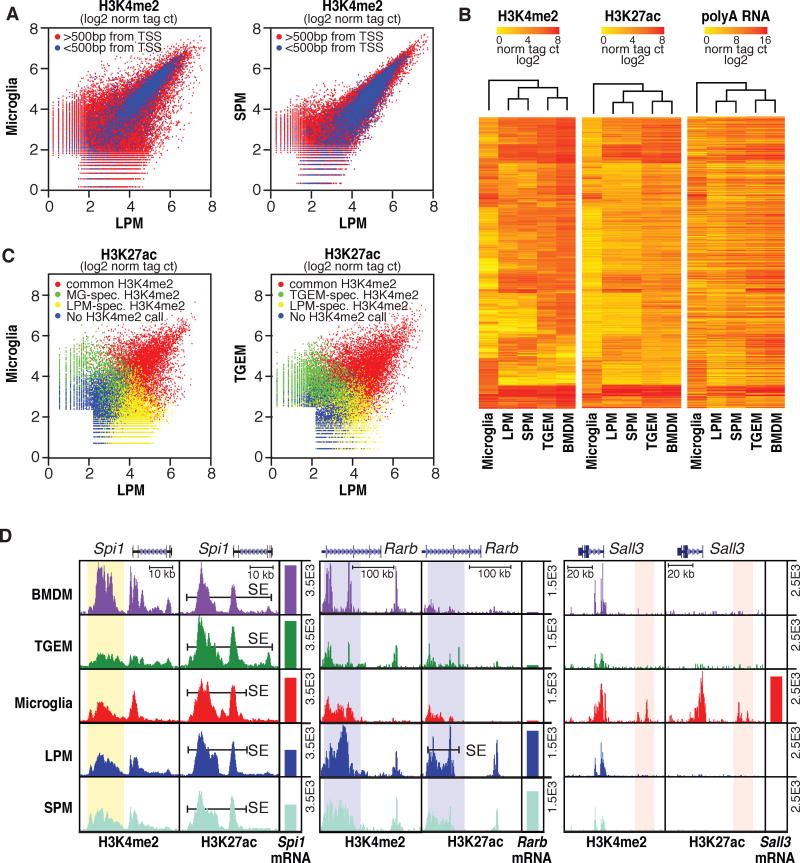
The dissimilarities in gene expression between different macrophage subsets revealed by RNA-Seq analysis imply important differences in how these cells organize and/or use their enhancer repertoires. To examine this, we analyzed di-methylation status of lysine 4 of histone 3 (H3K4me2) and acetylation status of lysine 27 of histone H3 (H3K27ac) by ChIP-Seq in these cells (Tables S2–4). H3K4me2 marks promoters and enhancers (He et al., 2010; Kaikkonen et al., 2013), while H3K27ac correlates positively with transcriptional activity at these elements (Creyghton et al., 2010). Deposition of H3K4me2 results from the binding of LDTFs and other TFs but is not necessarily associated with enhancer activity. We therefore use a heuristic of defining H3K4me2-positive/H3K27ac-negative regions as ‘primed’ and regions positive for both marks as ‘active’. Genomic annotation enabled segregation of these regions into promoters or enhancers by proximity to gene transcriptional start sites (TSS). Notably, the pattern of H3K4me2 deposition in MG substantially differs from that of LPMs (Figure 2A), indicating selection of distinct regulatory landscapes. Of 8,439 promoters marked by H3K4me2 in one or both subsets, 329 exhibit > 4-fold differences (4%), far fewer than the ~1700 mRNAs exhibiting >16-fold differences in expression. Of 50,509 regions >500 bp from TSS marked by H3K4me2 in one or both subsets, 12,449 exhibit >4-fold differences (25%). The vast majority of differential H3K4me2-marked regions are thus distant from promoters and correspond to potential enhancers. In contrast to the comparison of LPMs and MG, both the enhancer and promoter repertoires of the two subsets of RPMs share a much higher degree of similarity (Figure 2A). Furthermore, clustering analyses of the H3K4me2 deposition pattern revealed that MG were more divergent from the other subsets than any two other macrophage subsets are from one another, consistent with gene expression data (Figure 2B).
Figure 2. Variation in enhancer landscapes in different macrophage subsets.
A. Scatterplots of normalized H3K4me2 tag counts at genomic regions marked by significant H3K4me2 tags (>32 tags, 0.1% FDR) in LPMs and/or MG (left) or LPMs and/or SPMs (right). Points colored in blue are within 500 bp of a TSS. See Figure S2 for representative replicates. B. Heatmaps of normalized H3K4me2, H3K27ac and nearest expressed gene RNA-Seq tag counts at genomic locations showing >4-fold pairwise differences in H3K4me2 tag counts between at least two of the five macrophage subtypes. Row order is the same for all three data types. C. Scatterplots of normalized H3K27ac tag counts at genomic regions marked by significant H3K27ac tags (>32 tags, 0.1% FDR) in LPMs and/or MG (left) or LPMs and/or SPMs (right). Points are colored red if genomic locations are also marked by H3K4me2 (>16tags) in both subsets, green if marked by H3K4me2 selectively in MG (left) or SPMs (right), yellow if marked by H3K4me2 selectively in LPMs, or blue if not associated with H3K4me2 in either subset. D. UCSC browser images of selected genomic regions with corresponding RNA-Seq data plotted as bar graphs. Bars labeled SE indicate super enhancers and vertical highlights designate regions of interest for subset-common (Spi1) or subset-specific (Rarb and Sall3) loci. All data are normalized to input and library dimension. See also Tables S2–4.
H3K27ac was present at a large fraction of H3K4me2 marked regions and generally but imperfectly correlated with nearest gene expression (Figure 2B). Overlap of the H3K27ac data with H3K4me2-defined enhancers allowed the identification of common but quantitatively differently activated enhancers as well as activation of enhancers unique to one subset. Figure 2C illustrates such comparisons for LPMs vs MG and LPMs vs TGEMs. Genomic regions marked by H3K4me2 in both subsets are color-coded in red and represent activation of an enhancer landscape that is primed in both subsets. In contrast, regions exclusively marked by H3K4me2 in LPMs, shown in yellow, represent LPM-specific enhancers. Conversely, regions exclusively marked by H3K4me2 in MG or TGEMs, indicated in green, represent MG or TGEM-specific enhancers, respectively. Comparing LPMs vs. MG, 60% of the active enhancers resided at common regions of H3K4me2, 30% at LPM-specific regions and 10% at MG-specific regions. Specific examples are indicated in Figure 2D. As expected, the Spi1 enhancer, controlling expression of PU.1, is marked by H3K4me2 and H3K27ac in all macrophage populations. Interestingly, the RA inducible Rarb gene is also marked by H3K4me2 in all macrophage populations, but high H3K27ac is only observed in LPMs and SPMs, suggesting a role of local RA in enhancer activation. Finally, the Sall3 gene, which is exclusively and highly expressed in MG, is near a genomic region that is exclusively marked by H3K4me2 and H3K27ac in MG. In sum, these analyses provide strong evidence that both differential activation of a common enhancer landscape and the selection of subtype-specific enhancers contribute to the specific transcriptional signature of each subset of macrophages.
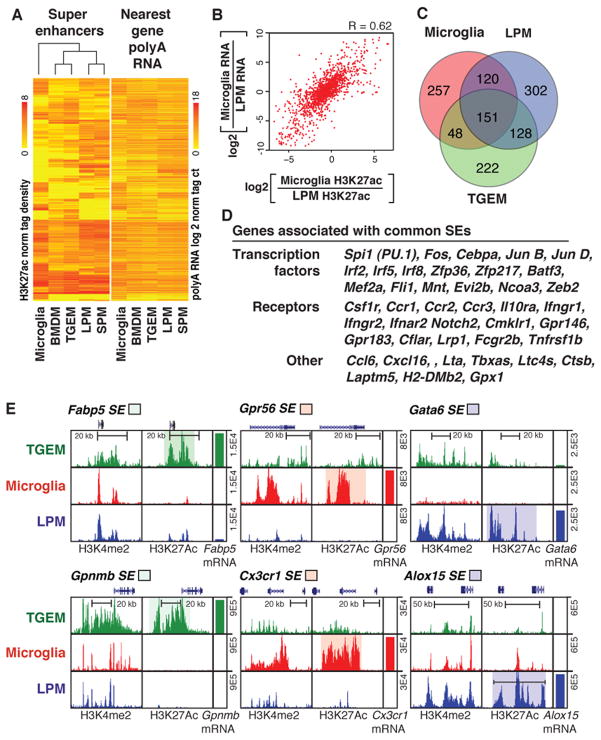
Tissue-specific super enhancers emerge from common enhancer landscapes
Genome-wide analysis of features of active enhancers, including the presence of Mediator and deposition of H3K27ac, indicates marked variation in their local distribution patterns. In all cell types evaluated thus far, ~400–800 regions, representing a small fraction of the genome, exhibit a disproportionately high density of active regulative marks and transcription factor binding (Hnisz et al., 2013; Loven et al., 2013; Whyte et al., 2013). These regions, recently termed super enhancers (SEs), are selected in a cell-specific manner and frequently occur near or encompass genes that play essential roles in defining the identity and function of the corresponding cell type (Hnisz et al., 2013). While LDTFs are enriched in and likely determine cell-specific SE selection, evidence also suggests that the extracellular environment can influence formation of SEs in endothelial cells (Brown et al., 2014). To investigate this relationship in tissue macrophages, we defined SEs in each macrophage subset based on H3K27ac ChIP-Seq. In agreement with previous studies, we observed common and subset-specific SEs, with ~600 to 750 SEs being identified among the five cell types examined. Clustering of these SEs results in the same relationships between subsets as observed using RNA-Seq, H3K4me2 or H3K27ac data (Figure 3A). This analysis also revealed a high concordance between the distribution of SEs genome-wide and the expression level of the nearest genes (Figure 3A). This strong relationship is further illustrated for SEs and nearest gene expression in MG and LPMs, in which the correlation coefficient was 0.62 (Figure 3B), much higher than that observed for the individual enhancer elements not associated with SE regions in these subsets. This may be due to a more likely assignment of SEs to their target genes than conventional enhancers.
Figure 3. Variation in super enhancer landscapes in different macrophage subsets.
A. Heat maps of H3K27ac tag densities at super enhancers and RNA-Seq tag densities at nearest expressed genes. Rows are ordered the same for both plots. B. Scatter plot of the relationship between ratio of MG to LPMs H3K27ac tag density at super enhancers (x-axis) and the ratio of nearest gene expression (y-axis). C. Venn diagram indicating overlap and specificity of super enhancers in MG, LPMs and TGEMs. D. Examples of genes associated with common super enhancers. E. UCSC genome browser images of selected subset-specific super enhancers and associated genes with subsets-specific regions of interest highlighted.
Approximately 40% to 50% of the SEs in a particular macrophage subset are unique to that subset, illustrated by the Venn diagram of LPM, MG and TGEM in Figure 3C. In concert with previous findings (Hnisz et al., 2013; Whyte et al., 2013), common SEs are associated with numerous genes important to macrophage ontology and functions, including Spi1, Cebpa, members of the Irf family, Csf1r, Fcgr2b, Ctsb, etc. (Figure 3D). This pattern is exemplified by the region upstream of Spi1, which is scored as a SE in all five subsets (Figure 2D). In contrast, many SEs are macrophage subset-specific and reside near or surround genes that are highly differentially expressed (Figure 3E). While some SEs exhibit highly specific H3K4me2 and H3K27ac markings, such as the LPM-specific SE upstream of Gata6 (Figure 3E), the majority of SEs are located at regions that are marked by H3K4me2 in multiple macrophage subsets, but only attain SE status in one or a few subsets. For example LPM-specific SEs reside in the vicinity of Rarb (Figure 2D) and Alox15 (Figure 3E) genes, which are selectively expressed in LPMs, but that also exhibit H3K4me2 in other macrophage subsets. Similar relationships are observed for the MG-specific SEs surrounding Gpr56 and Cx3cr1 and the TGEM-specific SEs surrounding Fabp5 and Gpnmb (Figure 3E). These findings suggest that environmental signals play roles in the transition of collections of primed enhancers to genomic regions exhibiting features of SEs.
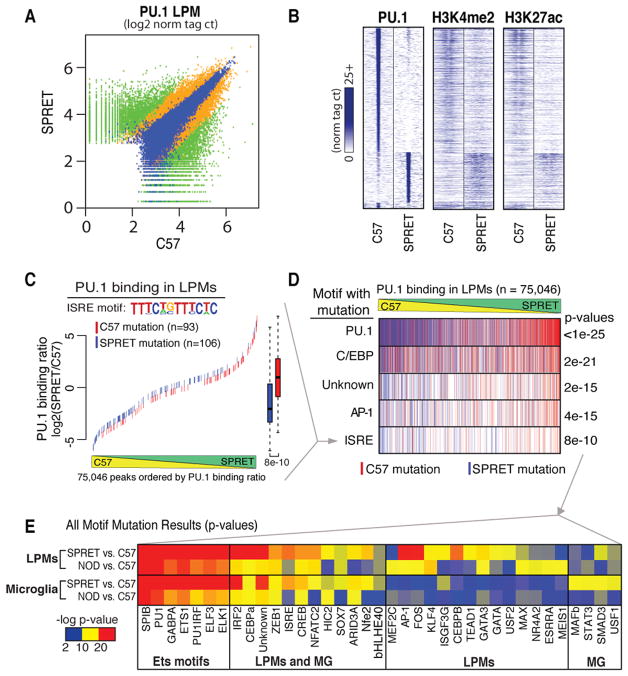
PU.1 co-localizes with distinct TF motifs at subset-specific enhancers
The observation that PU.1 localization to macrophage- or B cell-specific enhancers is dependent on collaborative interactions with alternate LDTFs (Heinz et al., 2010) led us to consider the possibility that an assessment of PU.1 binding in different macrophage subsets might yield insights into the TFs that drive the selection of subset-specific enhancers. We therefore extended existing genome-wide binding profiles for PU.1 to include MG, LPMs and SPMs. These studies indicated that PU.1 bound to both common and subset-specific genomic locations, exemplified for LPMs and MG in Figure 4A (all comparisons in Tables S2, 3). The great majority of subset-specific binding sites were observed at distal regions (>500 bp from an mRNA TSS, Figure 4A), consistent with the patterns of H3K4me2 (Figure 2A). Examples of LPM-specific and MG-specific binding sites for PU.1 in enhancer-like regions vicinal to Msr1 (expressed exclusively in LPMs) and Nav2 (expressed exclusively in MG) genes are illustrated in Figure 4B.
Figure 4. PU.1 binds to subset-specific enhancers.
A. Scatterplot of normalized tag counts for PU.1 peaks in MG versus LPMs. Points colored blue are within 500 bp of the TSS. B. UCSC genome browser images of PU.1 binding in the vicinity of the Msr1 and Nav2 genes in MG and LPMs cells and association with H3K27ac highlighting specific regions. C. Motifs enriched in the vicinity of PU.1 binding sites that are specific for LPMs versus MG using a random GC-corrected genomic background (top) or a background corresponding to MG-specific PU.1 peaks (bottom). D. Motifs enriched in the vicinity of PU.1 binding sites that are specific for MG using a random GC-corrected genomic background. E. Distribution plots of motif frequencies (y-axis) for the indicated motifs within 400bp centered on the PU.1 motif at genomic loci bound specifically by PU.1 in LPMs (blue) or MG (red).
De novo motif enrichment analysis of 200 bp sequences encompassing PU.1 peaks identified the identical PU.1 recognition motif in both LPMs and MG as the most enriched sequence. However, completely different motifs were co-enriched within the two subsets (Figure 4C, D). Using GC content-matched genomic sequence as background, enriched sequences specific to LPMs corresponded to motifs known to bind C/EBP, AP-1, IRF, KLF, and GATA transcription factor family members (Figure 4C). Conversely, MG-specific PU1-binding sequences were co-enriched for a PU.1-IRF composite sequence, and motifs corresponding to CTCFL, HIC2, MEF2, and SMAD TFs (Figure 4D). In addition, by using alternative subset-specific PU.1 binding sites as background, motifs recognized by retinoic acid receptors (e.g., NR2F2), were identified to be co-enriched with PU.1 binding sites in LPMs (Figure 4C).
Previous studies indicated that motifs for collaborative binding partners of PU.1 typically reside within ~100 bp of the PU.1 motif itself (Barozzi et al., 2014; Heinz et al., 2010). We therefore analyzed the genomic distance distribution of enriched motifs (from Figure 4C, D) within a 400 bp window relative to the bound PU.1 motif of LPM- and MG-specific PU.1 peak sets (Figure 4E). This analysis indicated that C/EBP, AP1 and GATA motifs frequently occurred near PU.1 bound motifs in LPMs, but not in MG, indicating that genomic loci containing PU.1 and closely spaced C/EBP, AP-1 or GATA motifs were more likely to become LPM-specific enhancers. The GATA motif was selectively enriched in LPMs relative to MG, suggesting a fundamental difference for the LPM resident population compared to elicited macrophages (Figure 4E). In contrast, the SMAD motif showed MG specificity (Figure 4E), which is consistent with TGFβ signaling in the brain. These findings provide evidence that selection of subset-specific enhancers is in part driven by collaborative interactions between PU.1 and alternative sets of TFs in each subset.
Use of natural genetic variation to validate and discover collaborative TFs
While motif enrichment suggests the identities of TFs that contribute to the function of subset-specific enhancers, this approach does not establish whether or not they are required for collaborative binding. Loss of function strategies are challenging for this purpose, because many of the identified motifs are recognized by multiple members of corresponding TF families. An alternative means to test for collaborative binding is to mutate motifs recognized by the TF family of interest and determine whether this results in loss of binding of a nearby factor. We considered the possibility that this could be accomplished for informative motifs on a genome-wide scale by leveraging the vast degree of natural genetic variation provided by inbred laboratory and wild strains of mice.
To explore the potential of this approach to validate and discover TFs required for collaborative binding and function of PU.1, we determined the genome wide patterns of PU.1, H3K4me2 and H3K27ac in LPMs and MG isolated from NOD/ShiLtJ (NOD), and SPRET/EiJ (SPRET) mice (Table S5). Compared to C57BL/6J (C57) mice, NOD mice have about 5 million SNPs and Indels, while SPRET mice have about 40 million (Keane et al., 2011). This variation is associated with corresponding levels of strain-specific binding of PU.1, illustrated for LPMs derived from C57 and SPRET mice (Figure 5A). Similar observations are made with respect to MG (Table S6). Approximately 8-fold fewer strain-specific PU.1 binding sites were identified in LPMs and MG derived from NOD mice compared to C57, consistent with the lower number of variants between these two strains. Strain-specific binding of PU.1 was associated with corresponding strain-specific H3K4me2 and H3K27ac marks (Figure 5B) suggesting that many strain-specific PU.1 binding sites localize to functional enhancers.
Figure 5. Motif mutations in potential PU.1 collaborating transcription factors confirm cooperative binding for subset-common and subset-specific factor combinations.
A. PU.1 binding between SPRET and C57 is shown at for 200 bp regions where green signifies differential binding (>4 fold, p<1E-4, n = 13,199), blue similar binding (<4 fold, p<1E-4, n = 11,022) and orange in between (n = 12,367). B. Heatmap of 2 kb differentially-bound PU.1 genomic regions (rows) centered on PU.1 binding for ChIP-Seq tags of PU.1, H3K4me2 and H3K27ac between C57 and SPRET (columns). C. An example of motif mutation analysis is shown for the ISRE motif. 200 bp genomic sequence at all PU.1 bound loci (in A) were queried for genetic variants that mutated the ISRE motif matrix in either C57 or SPRET. Mutations were colored according to the genome mutated; red = C57, blue = SPRET. ISRE mutations were plotted according to the PU.1 binding strain ratio (y-axis) as measured in LPMs at that locus and rank-ordered on the x-axis. Boxplots of corresponding color indicate the effect of ISRE motif mutations on PU.1 binding where whiskers extend to data extremes and p-value are from two-sided t-test. D. Results from analyses described in C are vertically compressed and shown in rows for PU.1, C/EBP, Unknown, AP-1, and ISRE motif mutation events. E. Heatmap showing p-values resulting from analysis described in C and D for motif mutations best matching transcription factors indicated on x axis. Each motif was tested for affecting PU.1 binding between C57 and NOD and between C57 and SPRET both in MG and LPMs (y-axis). See also Figure S3 and Tables S5 and S6.
To search for motifs mediating DNA binding by collaborative TFs we analyzed strain-specific binding of PU.1 that was not associated with mutations in PU.1 recognition motifs. This was accomplished by scanning a 200 bp window surrounding PU.1 binding sites lacking PU.1 motif mutations for the presence of the DNA recognition motifs of the 100 most highly expressed TFs in LPMs and MG in C57 or the alternate (NOD or SPRET) genomic sequence. Mutated loci were then queried for a corresponding decrease in PU.1 binding relative to the un-mutated strain. The significant result for ISRE motif mutations affecting PU.1 binding in LPMs is exemplified in Figure 5C. The ISRE was found to be mutated in the vicinity of PU.1 binding sites 93 times in LPMs isolated from C57 mice (indicated by red hash lines in Figure 5C), and 106 times in LPMs isolated from SPRET mice (indicated by blue hash lines in Figure 5C). PU.1 binding strength is rank ordered from most C57-specific at left to most SPRET-specific at right. Many mutations are not associated with strain-specific binding, which is consistent with prior studies indicating that the specific position of the variant (i.e., core vs. periphery of motif), the distance of the motif from the peak center, and presence of additional redundant motifs affect the impact of individual mutations (Heinz et al., 2013). Overall, however, C57 mutations in the ISRE were associated with SPRET-specific binding of PU.1, while SPRET mutations in the ISRE associated with C57-specific binding of PU.1 (p = 2e-21). This strong genetic association implicates factors binding to the ISRE as collaborative partners of PU.1 in LPMs.
This analysis was repeated for each motif of interest in each macrophage subset (LPMs and MG) for the comparisons of C57 vs. NOD and C57 vs. SPRET. Vertical compression of the plot shown in Figure 5C allows stacking of plots for multiple motifs, indicated in Figure 5D. Overall, 37 motifs were found to reach statistical significance in at least one macrophage subset and strain (Figure 5E, Figure S3). Many more motifs were found to be significant in comparisons of macrophages derived from C57 and SPRET mice than C57 and NOD mice, consistent with the much larger number of informative mutations. The most highly significant motifs corresponded to sequences recognized by ETS factors that are similar to motifs recognized by PU.1 itself. Most of these motifs are closely situated to regions of PU.1 binding for which the PU.1 motif itself was considered to be intact. It is therefore unclear at present the extent to which these mutations directly affect PU.1 binding or represent binding sites for collaborative ETS factors.
Twelve non-ETS motifs were identified as being significantly associated with PU.1 binding in both LPMs and MG, including a C/EBPα motif, which is an established collaborative binding partner of PU.1 (Figure 5E). In contrast, fourteen motifs exhibited preferential associations with strain-specific PU.1 binding in LPMs (Figure 5E). This list includes recognition motifs for KLF4, GATA factors and AP-1 factors, independently identifying motifs discovered through de novo motif analysis. Finally, motifs for four factors were preferentially associated with PU.1 binding in MG, including a recognition motif for SMAD3.
Tissue environment regulates collaborative and signal-dependent TF expression
To investigate the importance of tissue environment in maintenance of specific macrophage phenotypes, we placed LPMs and MG into culture under the influence of IL-34 or M-CSF for 7 days. While M-CSF is important to peritoneal macrophages (Witmer-Pack et al., 1993), IL-34 is critical for proper MG ontology and/or survival in vivo (Greter et al., 2012; Wang et al., 2012). This environmental transition resulted in vast changes in gene expression. Comparison of the gene expression program of LPMs freshly purified from the peritoneal cavity with LPMs maintained in M-CSF for 7 days is illustrated in Figure 6A. Data points colored in blue represent genes that are expressed more that 16-fold higher in LPMs than MG, indicating that the LPM-specific program of gene expression is preferentially lost in culture. Comparison of the gene expression program of MG freshly isolated from the brain or maintained in culture in the presence of IL-34 for 7 days is illustrated in Figure 6B. Data points colored in red represent genes that are expressed more than 16-fold higher in MG than LPMs, indicating that the MG-specific program of gene expression is preferentially lost in culture. In both LPMs and MG, many genes exhibiting low levels of expression in vivo are markedly up-regulated in culture.
Figure 6. Environmental influence on gene expression in LPMs and microglia.
A, B. Scatter plots illustrating relative gene expression of RNA transcripts in freshly isolated LPMs compared to LPMs maintained in culture for 7 days (A), and freshly isolated MG compared to MG in culture for 7 days (B). Genes specific to LPMs are colored blue in A and specific to MG are red in B. C. Normalized gene expression values for members of the RAR and TGF-β receptor family members. D. Heatmap showing the fold-change of RNAs for the indicated transcription factors upon removal from the peritoneal cavity and culture with IL-34 or M-CSF. E, F. Effects of chronic stimulation with 2μM ATRA (E) on LPM-Specific or common mRNAs or TGF-β1 (F) on MG-specific or common mRNAs. G. qPCR validation of induced expression by ATRA of key transcription factors in cultured LPMs (error bars = S.D.). See also Figure S4.
In view of recent findings indicating important roles of TGFβ signaling in MG and RA signaling in peritoneal macrophages, we examined the expression of the main receptors for these factors in each macrophage subset. The mRNAs encoding all three RA receptors (Rara, Rarb and Rarg) are highly and selectively expressed in LPMs and SPMs, while mRNAs encoding the TGFβ receptors Tgfbr1 and Tgfbr2 are preferentially expressed in MG (Figure 6C). Interestingly, expression of all three retinoic acid receptors is markedly reduced when LPMs are placed into culture in the presence of M-CSF or IL-34, while the expression of Tgfbr1 is markedly increased under these conditions (Figure 6D). Thus, environment controls the expression of genes responsible for responses to environment-specific signals.
To investigate the extent to which RA and TGFβ influence subset-specific patterns of gene expression, we treated LPMs with RA (ATRA) or TGFβ for 7 days and performed RNA-Seq analysis. RA treatment induced expression of nearly half of the LPM-specific genes by more than 2-fold, while inducing about 8% of genes expressed at similar levels in LPMs and MG (Figure 6E). Conversely, nearly 50% of the genes induced more than 2-fold by TGFβ in LPMs in culture are preferentially expressed by MG in vivo, while only 4% of the genes expressed at similar levels in LPMs and MG were induced by TGFβ in LPMs (Figure 6F). Thus, RA and TGFβ disproportionately regulate genes that specify LPM and MG-specific phenotypes, respectively.
We next evaluated the expression of TFs that recognize motifs identified as putative binding sites for collaborative partners of PU.1 in LPMs through analysis of strain-specific PU.1 binding. Remarkably, expression of the majority of TFs best matched to motifs identified by strains analysis was environment-dependent (Figure 6D). A similar pattern was observed when considering all members of each TF family capable of recognizing these motifs (Figure S4A). RNA-Seq analysis further suggested that several of these factors were inducible by RA. This response was confirmed under M-CSF treatment conditions for Gata6 and Rarb, consistent with previous studies (Okabe and Medzhitov, 2014), as well as for Bhlhe40 and Nfe2 (Figure 6G, S4B). However, expression of Rara, Rarg and most of the other factors illustrated in Figure 6C were not RA-inducible. Thus, the environment modulates in LPMs the expression of collaborative and SDTFs through both RA-dependent and RA-independent mechanisms.
Hierarchical effects of environment on macrophage enhancer landscapes
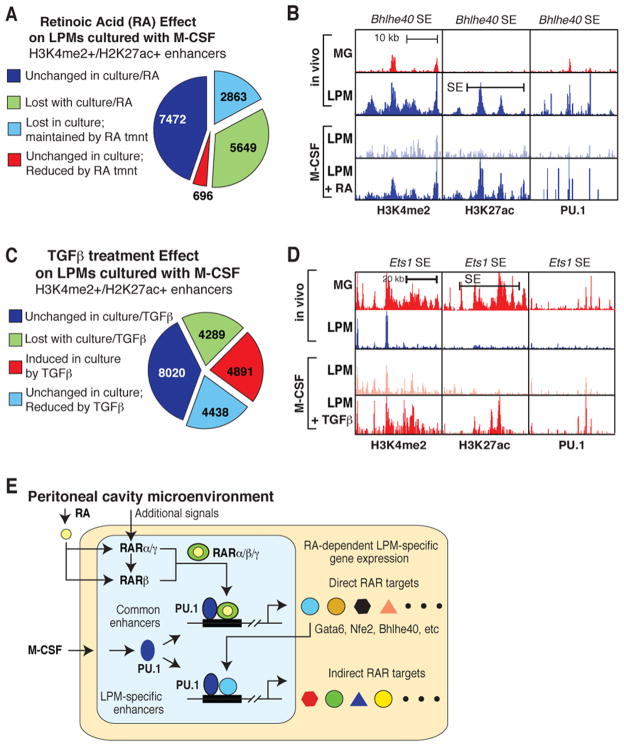
To gain insights into mechanisms underlying effects of environment on macrophage gene expression, we performed ChIP-Seq analysis for H3K4me2, H3K27ac and PU.1 in LPMs maintained in M-CSF and the presence or absence of RA or TGFβ for 7 days. Transition of LPMs from the peritoneal cavity to a tissue culture environment containing M-CSF led to a >2-fold reduction in H3K4me2 and/or H3K27ac at approximately half of the enhancer like regions (Figure 7A). One-third of these lost enhancer elements were maintained by RA treatment (Figure 7A). Of the 302 LPM-specific SEs identified in Figure 3, 223 (74%) no longer met SE criteria, indicating a disproportionate sensitivity to loss of environmental signals. This pattern is exemplified by the SE associated with Bhlhe40, which in addition to substantial reduction in the histone signature of enhancers also exhibits reduced PU.1 binding (Figure 7B). Notably, H3K4me2, H3K27ac and PU.1 binding are largely maintained by RA treatment.
Figure 7. Environmental influence on enhancer landscapes in LPMs and microglia.
A. Effects of culture environment and 2μM ATRA chronic stimulation on the enhancer landscape of LPMs. B. UCSC browser images displaying effects of culture environment and 2μM ATRA chronic stimulation on H3K4me2, H3K27ac, and PU.1 binding at the Bhlhe40 locus in LPMs. C. Effects of culture environment and chronic stimulation with TGF-β1 on the enhancer landscape of LPMs. D. UCSC browser images displaying effects of culture environment and chronic stimulation with TGF-β1 on H3K4me2, H3K27ac, and PU.1 binding at the Ets1 locus in LPMs. E. Hierarchical model for mechanisms by which the peritoneal environment induces the enhancer landscape and gene expression signature of LPMs. See text for details. See also Table S7.
Maintenance of LPMs in M-CSF plus TGFβ resulted in marked changes in the LPM enhancer landscape in comparison to culture in M-CSF alone, consistent with the preferential effects of TGFβ on a MG-specific program of gene expression (Figure 7C). Treatment with TGFβ increased the enhancer signature by more than 2-fold at ~ 25% of pre-existing enhancers. Conversely, TGFβ reduced enhancer signatures at ~25% enhancer-like elements that were stable upon transfer to culture in M-CSF. Induced enhancers are exemplified by a genomic region in the vicinity of the Ets1 gene (Figure 7D) that is preferentially expressed in MG and is highly induced by TGFβ. Culture of LPMs in M-CSF results in appearance of PU.1 binding and H3K4me2 modification, with TGFβ treatment leading to substantial increases in H3K27ac and a marked increase in gene expression.
DISCUSSION
Mechanisms underlying tissue-specific enhancer selection and activation
The present studies provide evidence for a hierarchical model in which the distinct environments of the brain and peritoneal cavity differentially activate a common set of primed enhancers and their target genes that in turn promote the selection and activation of subset-specific enhancer repertoires. The combinatorial activation of both common and subset-specific enhancers enables context-dependent regulation of genes required for specialized functions of MG and RPMs. Aspects of this model as they pertain to the RA-dependent program of gene expression specific to peritoneal macrophages are illustrated in Figure 7E. Common to all macrophage subsets, stimulation of signaling pathways downstream of the M-CSF receptor by M-CSF and/or IL-34, which are present in the environment in a largely tissue non-specific manner, ensures survival and promotes PU.1 expression (Sarrazin et al., 2009). PU.1 is a critical LDTFs required for all macrophage subsets that functions to select common and cell-specific enhancers through collaborative interactions with other TFs. These regions of PU.1 binding in turn serve as subset-specific sites of action for various types of SDTFs.
Within the peritoneal cavity, environment-specific signals control the expression and activities of TFs that act upon primed enhancers that are common to multiple macrophage subsets (Figure 7E). A particularly important signal is omentum-derived RA, which has been shown to be essential for development and function of LPMs through its activation of RARβ and GATA6 (Okabe and Medzhitov, 2014). We find that all three high affinity retinoic acid receptor genes (Rara, Rarb and Rarg) are preferentially expressed in the peritoneal cavity, and that this expression requires continual maintenance by the peritoneal cavity environment. However only Rarb expression is preserved by RA treatment in culture, indicating that expression of Rara and Rarg is under the control of as yet unidentified factors. We speculate that the expression of RARα and RARγ is necessary for full induction of RARβ expression in response to environmental RA, and that this positive feed back loop is important for amplification of the RA signal and activation of direct RA target genes. These findings imply that at least two environmental signals are required for initiating the RA-dependent peritoneal macrophage phenotype, one being RA itself and the second being a signal or signals required for RARα and RARγ expression.
Activated retinoic acid receptors primarily function as SDTFs that act at a common set of primed enhancers established by PU.1 and other LDTFs that are expressed across macrophage subsets. Importantly, direct RA target genes include Gata6, Bhlhe40 and Nfe2, which were identified as putative interacting partners of PU.1 through analysis of effects of natural genetic variation. We propose that RAR-dependent induction of these factors results in collaborative interactions with PU.1 that drive environment-specific selection of LPM-specific enhancers (Figure 7E).
Of note, Gata6, Bhlhe40 and Nfe2, as well as all three retinoic acid receptors, reside in or near peritoneal macrophage-specific SEs that are lost when LPMs are removed from the peritoneal cavity. Our findings suggest that an analogous hierarchy operates in MG, driven in part by TGFβ signaling and SMAD TFs. While the present studies have focused on PU.1, we expect that additional macrophage LDTFs function in an analogous manner to set up macrophage-specific, PU.1-independent enhancers.
Use of natural genetic variation to validate and discover collaborative TFs
Here, we demonstrate the use of the natural genetic variation provided by inbred strains of mice as a powerful means to validate and discover collaborative TFs. By measuring strain-specific binding of PU.1 in macrophages derived from genetically diverse strains of mice, we identified motifs for several different classes of TFs in which strain-specific mutations were highly correlated with the loss of binding of PU.1 to non-mutated PU.1 recognition motifs. Interestingly, the expression of a significant fraction of the TFs recognizing these motifs is dependent on environment.
Many of the motifs identified by analysis of strain-specific binding of PU.1 are recognized by TFs that have well-established roles in macrophage biology. Some, such as C/EBPα and C/EBPβ, are documented to function as factors that enable collaborative binding of PU.1 in macrophages (Heinz et al., 2010), supporting the validity of the approach. Although the biological role of GATA6 in the development and function of LPMs is established (Okabe and Medzhitov, 2014; Rosas et al., 2014), the present studies suggest that a key molecular function of GATA6 is to collaborate with PU.1, and likely other macrophage LDTFs, to drive the selection of LPM-specific enhancers. Bhlhe40 and Nfe2 represent examples of putative collaborative partners of PU.1 that have not as yet been linked to macrophage-specific functions. Bhlhe40, also known as Dec1, Stra13 and Sharp2, has previously been shown to be inducible by RA and to act as both as a repressor and activator (Boudjelal et al., 1997; Ivanova et al., 2004), raising the possibility that it could contribute to selection of LPM-specific enhancers as well as suppress genes that become active when LPMs are removed from the peritoneal cavity. NFE2 is a bZip transcription factor that is broadly expressed in the hematopoietic system and has been established to play important roles in erythropoiesis and megakaryocyte development (Andrews, 1998). The present findings provide a rationale for further investigation of roles of Bhlhe40, NFE2 and other TFs identified as putative collaborative binding partners of PU.1.
The use of natural genetic variation as a strategy for identification of TFs required for enhancer selection can in principle be applied to any cell type in which ChIP-sequencing can be performed for an index LDTF. In addition, although not a focus of the present studies, the variation in enhancer selection and activity observed in macrophages derived from different inbred strains of mice was associated with strain-specific differences in LPM and MG gene expression. Such changes in gene expression are presumably linked to both molecular phenotypes such as eQTLs and to the marked phenotypic differences exhibited by these mice that are influenced by tissue resident macrophage populations, such as relative susceptibility or resistance to metabolic, cardiovascular, infectious and neurodegenerative diseases (Civelek and Lusis, 2014; Threadgill and Churchill, 2012). The principle of collaborative binding, which serves as the basis for the motif discovery method described here, is directly applicable to investigating mechanisms by which non coding variants may exert phenotypic effects in a cell type specific and/or context dependent manner. In concert, these approaches enable insights into gene-by-environment interactions and the genetic architecture of molecular and complex disease traits.
Tuning enhancer landscapes and gene expression to context-specific functions
The present studies reveal that each macrophage subset uniquely posses a distinct sets of active enhancers, including subset-specific SEs, which are associated with strong preferential expression of nearby genes. In LPMs for example, which populate a very potent immunogenic environment, Gbp2b and Alox15 are associated with SEs activity, and we note that the protein products of these genes are critical regulators of immunity, in particular inflammation and tolerance (Pilla et al., 2014; Uderhardt et al., 2012; Yamamoto et al., 2012). In contrast to LPMs, MG reside in the immune-privileged environment of the brain. As with LPMs however, our observations suggest that MG adopt a unique phenotype that is again strongly contributed by distinct enhancers and SEs to accomplish tissue-specific functions required for brain homeostasis. For example, SEs in MG include genomic loci associated with the Cx3cr1 and Gpr56 genes, among others. Interestingly, both genes are highly relevant to brain functions, regulating synaptic pruning and efficient cortical patterning during brain development (Paolicelli et al., 2011; Piao et al., 2004). Together, our studies reveal an intricate relationship between the organization of the genome of tissue macrophage and their surrounding environment.
Divergent macrophage gene expression in a common environment
Distinct macrophage populations can coexist in a similar environment, as illustrated by the co-presence of LPMs and SPMs in the peritoneum. Although these cells are highly concordant with respect to gene expression and organization of their enhancer landscapes, consistent with exposure to common tissue-derived signals, strong points of divergence can nonetheless discriminate the two. These observations raise the possibility that differences in origin and ontology play important roles in determining these later-stage differences (Schulz et al., 2012). Thus, the impact of developmental history on the regulation of enhancer repertoires and gene expression of different tissue macrophages remains a fundamental open question to be addressed in future studies.
EXPERIMENTAL PROCEDURES
Mice
Seven week old C57BL/6J, NOD/ShiLtJ and SPRET/EiJ male mice were purchased from Jackson Labs and used at 8–9 weeks of age. All animal procedures were in accordance with University of California San Diego research guidelines for the care and use of laboratory animals.
Microglia isolation
Mice were anaesthetized with CO2 and quickly perfused intracardially with ice-cold DPBS. Whole brains were removed and gently mechanically homogenized on ice. Cells were fractionated by Percoll gradient centrifugation and microglia-enriched fractions were further purified by cell sorting according to the scheme described in Figure S1A and Extended Experimental Procedures.
Peritoneal macrophage isolation
Following euthanization, peritoneal cells were collected by lavage of the peritoneum with ice-cold staining buffer. LPM and SPM subsets were purified based on relative expression of MHCII and other markers described in Figure S1B and Extended Experimental Procedures.
Thioglycollate-elicited and bone marrow-derived macrophages cultures
TGEMs were harvested by peritoneal lavage with 20 ml ice-cold PBS 4 days after peritoneal injection of 3 ml Thioglycollate broth. Both TGEMs and BMDMs were cultured as described in (Heinz et al., 2010). See also Extended Experimental Procedures.
ChIP-Sequencing
Macrophages were fixed at room temperature with 1% paraformaldehyde/PBS containing 1mM sodium butyrate, for 10 minutes and quenched with glycine. 2.0×105 to 1.0×106 cells were used for ChIPs and samples were processed as previously described (Heinz et al., 2010), with minor modifications noted in Extended Experimental Procedures. Sequencing libraries were prepared as previously described (Heinz et al., 2010).
RNA isolation
For RNA-Seq, TRIzol (Life Technologies) isolated RNA was either PolyA-selected (MicroPoly(A) Purist kit, Ambion) or subjected to RiboZero rRNA removal (Epicentre).
Quantitative PCR, RNA-Seq library preparation and sequencing
Libraries for RNA sequencing were generated as previously described (Heinz et al., 2013). See Extended Experimental Procedures for details and qRT-PCR primer sequences.
Data Analysis
Fastq files from sequencing experiments were mapped to individual genomes for the mouse strain of origin using default parameters for STAR (Dobin et al., 2013) (RNA-Seq) and Bowtie2 (Langmead and Salzberg, 2012) (ChIP-Seq). NOD/ShiLtJ and SPRET/EiJ custom genomes were generated from invariant positions of the mm10 sequence with alleles replaced by those reported in VCF files from the Mouse Genomes Project (Keane et. al. 2011). Mapped data was analyzed with HOMER (Heinz et al., 2010), custom R and Perl scripts. Raw and processed data are provided in GEO Accession GSE62826.
Supplementary Material
1. Table S1: Related to main Figure 1.
PolyA gene expression for all macrophages subsets from male C57BL/6J mice.
10. Table S2: Related to main Figure 2.
Number of intergenic ChIP-Seq peaks identified over input chromatin in the indicated macrophage subset from male C57BL/6J mice.
11. Table S3: Related to main Figure 2.
Summary of macrophage subset- specific (A) and similar (B) peaks in male C57BL/6J mice.
12. Table S4: Related to main Figure 2.
Summary of all primed and active enhancers per macrophage subset (A) and between the different macrophage subsets (B) defined by the existence of peaks in both ChIP-Seq samples (> 16 tags, fold change > 4, p-value < 1E-4) in male C57BL/6J mice.
2. Figure S1: Related to main Figure 1.
A, B. FACS plots showing the gating strategy to isolate (A) MG and (B), LPMs, and SPMs. C. RNA-Seq replicates from two independent MG samples.
3. Figure S2: Related to main Figure 2.
Examples of ChIP-Seq replicates for H3K4me2 for MG and LPMs.
4. Figure S3: Related to main Figure 5.
A. The position weight matrices are shown for the motifs tested in mutational analysis of Figure 5 together with the best guess of corresponding sequence specific TF or TF family. B. PU.1 binding and RNA expression is shown at the tubulin folding cofactor C (Tfcc) locus for MG cells from C57 and NOD. The genetic sequence at the NOD-specific PU.1 binding site is shown for both strains. Here, the NOD sequence satisfies an ISRE motif and the C allele at the overlapping SNO mutates the C57 sequence.
5. Figure S4: Related to main Figure 6. Environmental effects on LPM gene expression.
A. Heatmap showing the fold change of RNAs for (TF) families upon culture with IL-34 or M-CSF. These TFs were identified in genetic variation mutation analysis (Figure 5). B. RT-qPCR validation of maintained expression of genes associated with super enhancers by chronic stimulation with 2 μM ATRA of LPMs in culture. n = 2, error bars = S.D.
6. Table S5: Related to main Figure 5.
Number of ChIP-Seq peaks identified over input chromatin in LPMs and microglia from male C57BL/6J, NOD/ShiLtJ, and SPRET/EiJ mice.
7. Table S6: Related to main Figure 5.
Summary of strain specific (A) and strain similar (B) peaks in LPMs and MG.
8. Table S7: Related to main Figure 7.
RiboZero gene expression for LPM and MG in vivo and in culture.
9
Acknowledgments
The authors would like to thank Lynn Bautista, Andrea Crotti, and Renee F. Glass for valuable assistance with preparation of the manuscript, and Sven Heinz for assistance with the ChIP-Seq library preparation protocol. The authors would also like to thank the UCSD Human Embryonic Stem Cell Core Facility for assistance with cell sorting. These studies were primarily supported by NIH grants DK091183, CA17390, and DK063491 and the San Diego Center for Systems Biology (GM085764). D.G. was supported by a Canadian Institutes of Health Research Fellowship. C.E.R. was supported by the American Heart Association (12POST11760017) and the NIH Pathway to Independence Award (1K99HL12348). F.G. was supported by a Wellcome Trust Senior Investigator award (WT101853MA).
Footnotes
AUTHOR CONTRIBUTIONS
D.G. and C.K.G. designed the study. D.G., G.J.F., D.Z.E., N.J.S., J.D.S., H.B.C. and H.G. performed experiments. D.G., V.L., C.E.R., F.G. and C.K.G. analyzed and interpreted the data. D.G., V.L., C.E.R. and C.K.G. wrote the manuscript.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NC. The NF-E2 transcription factor. The international journal of biochemistry & cell biology. 1998;30:429–432. doi: 10.1016/s1357-2725(97)00135-0. [DOI] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell. 2014;54:844–857. doi: 10.1016/j.molcel.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudjelal M, Taneja R, Matsubara S, Bouillet P, Dolle P, Chambon P. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes & development. 1997;11:2052–2065. doi: 10.1101/gad.11.16.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, Bair S, Newton G, Lichtman AH, Kung AL, et al. NF-kappaB Directs Dynamic Super Enhancer Formation in Inflammation and Atherogenesis. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nature neuroscience. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain DW, O’Koren EG, Kan MJ, Womble M, Sempowski GD, Hopper K, Gunn MD, Kelsoe G. Identification of a tissue-specific, C/EBPbeta-dependent pathway of differentiation for murine peritoneal macrophages. Journal of immunology. 2013;191:4665–4675. doi: 10.4049/jimmunol.1300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona AE, Pioro EP, Sasse ME, Kostenko V, Cardona SM, Dijkstra IM, Huang D, Kidd G, Dombrowski S, Dutta R, et al. Control of microglial neurotoxicity by the fractalkine receptor. Nat Neurosci. 2006;9:917–924. doi: 10.1038/nn1715. [DOI] [PubMed] [Google Scholar]
- Cheng X, Reginato MJ, Andrews NC, Lazar MA. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Molecular and cellular biology. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nature reviews Genetics. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Ivanov S, Williams JW, Huang SC, Marcelin G, Fairfax K, Wang PL, Francis JS, Leone P, Wilson DB, et al. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. J Exp Med. 2014;211:1525–1531. doi: 10.1084/jem.20140570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Ghosn EE, Cassado AA, Govoni GR, Fukuhara T, Yang Y, Monack DM, Bortoluci KR, Almeida SR, Herzenberg LA, Herzenberg LA. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc Natl Acad Sci U S A. 2010;107:2568–2573. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, Kundig TM, Frei K, Ginhoux F, Merad M, et al. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity. 2012;37:1050–1060. doi: 10.1016/j.immuni.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nature genetics. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova AV, Ivanov SV, Zhang X, Ivanov VN, Timofeeva OA, Lerman MI. STRA13 interacts with STAT3 and modulates transcription of STAT3-dependent targets. Journal of molecular biology. 2004;340:641–653. doi: 10.1016/j.jmb.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Molecular cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Current biology: CB. 2010;20:R754–763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makwana M, Jones LL, Cuthill D, Heuer H, Bohatschek M, Hristova M, Friedrichsen S, Ormsby I, Bueringer D, Koppius A, et al. Endogenous transforming growth factor beta 1 suppresses inflammation and promotes survival in adult CNS. J Neurosci. 2007;27:11201–11213. doi: 10.1523/JNEUROSCI.2255-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Piao X, Hill RS, Bodell A, Chang BS, Basel-Vanagaite L, Straussberg R, Dobyns WB, Qasrawi B, Winter RM, Innes AM, et al. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, Ernst RK, Yamamoto M, Miao EA, Coers J. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci U S A. 2014;111:6046–6051. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas M, Davies LC, Giles PJ, Liao CT, Kharfan B, Stone TC, O’Donnell VB, Fraser DJ, Jones SA, Taylor PR. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S, Mossadegh-Keller N, Fukao T, Aziz A, Mourcin F, Vanhille L, Kelly Modis L, Kastner P, Chan S, Duprez E, et al. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell. 2009;138:300–313. doi: 10.1016/j.cell.2009.04.057. [DOI] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nature reviews Genetics. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- Threadgill DW, Churchill GA. Ten years of the collaborative cross. G3. 2012;2:153–156. doi: 10.1534/g3.111.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uderhardt S, Herrmann M, Oskolkova OV, Aschermann S, Bicker W, Ipseiz N, Sarter K, Frey B, Rothe T, Voll R, et al. 12/15-lipoxygenase orchestrates the clearance of apoptotic cells and maintains immunologic tolerance. Immunity. 2012;36:834–846. doi: 10.1016/j.immuni.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, Barrow AD, Diamond MS, Colonna M. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witmer-Pack MD, Hughes DA, Schuler G, Lawson L, McWilliam A, Inaba K, Steinman RM, Gordon S. Identification of macrophages and dendritic cells in the osteopetrotic (op/op) mouse. J Cell Sci. 1993;104(Pt 4):1021–1029. doi: 10.1242/jcs.104.4.1021. [DOI] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, Ohshima J, Sasai M, Kayama H, Okamoto T, et al. A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37:302–313. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Sun L, Chen Z, Whitaker JW, Wang T, Wang W. Predicting enhancer transcription and activity from chromatin modifications. Nucleic acids research. 2013;41:10032–10043. doi: 10.1093/nar/gkt826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. Table S1: Related to main Figure 1.
PolyA gene expression for all macrophages subsets from male C57BL/6J mice.
10. Table S2: Related to main Figure 2.
Number of intergenic ChIP-Seq peaks identified over input chromatin in the indicated macrophage subset from male C57BL/6J mice.
11. Table S3: Related to main Figure 2.
Summary of macrophage subset- specific (A) and similar (B) peaks in male C57BL/6J mice.
12. Table S4: Related to main Figure 2.
Summary of all primed and active enhancers per macrophage subset (A) and between the different macrophage subsets (B) defined by the existence of peaks in both ChIP-Seq samples (> 16 tags, fold change > 4, p-value < 1E-4) in male C57BL/6J mice.
2. Figure S1: Related to main Figure 1.
A, B. FACS plots showing the gating strategy to isolate (A) MG and (B), LPMs, and SPMs. C. RNA-Seq replicates from two independent MG samples.
3. Figure S2: Related to main Figure 2.
Examples of ChIP-Seq replicates for H3K4me2 for MG and LPMs.
4. Figure S3: Related to main Figure 5.
A. The position weight matrices are shown for the motifs tested in mutational analysis of Figure 5 together with the best guess of corresponding sequence specific TF or TF family. B. PU.1 binding and RNA expression is shown at the tubulin folding cofactor C (Tfcc) locus for MG cells from C57 and NOD. The genetic sequence at the NOD-specific PU.1 binding site is shown for both strains. Here, the NOD sequence satisfies an ISRE motif and the C allele at the overlapping SNO mutates the C57 sequence.
5. Figure S4: Related to main Figure 6. Environmental effects on LPM gene expression.
A. Heatmap showing the fold change of RNAs for (TF) families upon culture with IL-34 or M-CSF. These TFs were identified in genetic variation mutation analysis (Figure 5). B. RT-qPCR validation of maintained expression of genes associated with super enhancers by chronic stimulation with 2 μM ATRA of LPMs in culture. n = 2, error bars = S.D.
6. Table S5: Related to main Figure 5.
Number of ChIP-Seq peaks identified over input chromatin in LPMs and microglia from male C57BL/6J, NOD/ShiLtJ, and SPRET/EiJ mice.
7. Table S6: Related to main Figure 5.
Summary of strain specific (A) and strain similar (B) peaks in LPMs and MG.
8. Table S7: Related to main Figure 7.
RiboZero gene expression for LPM and MG in vivo and in culture.
9