A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish (original) (raw)
. Author manuscript; available in PMC: 2016 Mar 23.
Summary
CRISPR/Cas9 technology of genome editing has greatly facilitated the targeted inactivation of genes in vitro and in vivo in a wide range of organisms. In zebrafish it allows the rapid generation of knock-out lines by simply injecting a guide RNA (gRNA) and Cas9 mRNA into one-cell stage embryos. Here, we report a simple and scalable CRISPR-based vector system for tissue-specific gene inactivation in zebrafish. As proof of principle, we used our vector with the gata1 promoter driving Cas9 expression to silence the urod gene, implicated in heme biosynthesis, specifically in the erythrocytic lineage. Urod targeting yielded red fluorescent erythrocytes in zebrafish embryos, recapitulating the phenotype observed in the yquem mutant. While F0 embryos displayed mosaic gene disruption, the phenotype appeared very penetrant in stable F1 fish. This vector system constitutes a unique tool to spatially control gene knock-out and greatly broadens the scope of loss-of-function studies in zebrafish.
Introduction
The CRISPR/Cas9 technology has recently emerged as a powerful tool for targeted genome editing (Sander and Joung, 2014). Adapted from an immune mechanism in bacteria (Jinek et al., 2012), it takes advantage of the RNA-dependent recognition of specific DNA sequences by Cas9 endonuclease. A chimeric guide RNA (gRNA) was engineered to comprise both a 3’ secondary structure with the ability to interact with Cas9 and a 5’ seed sequence of 20 bases that directs sequence-specific targeting. Cas9 screens the genome and cleaves within sequences complementary to the seed, provided they are immediately followed by the protospacer adjacent motif (PAM) NGG (Sternberg et al., 2014). Double strand breaks are then repaired via homologous recombination or non-homologous end-joining, generally resulting in insertions or deletions (indels) of variable length. Targeting an early exonic sequence frequently leads to gene disruption through frame-shifts or non-sense mutations. Transfection of gRNAs and Cas9 mRNA or DNA into bacteria, human or mouse cells was shown to efficiently inactivate target genes (Cho et al., 2013; Cong et al., 2013; Jiang et al., 2013; Mali et al., 2013). The CRISPR/Cas9 technology was subsequently used in mice to observe in vivo loss-of-function phenotypes and to generate knock-out strains (Wang et al., 2013).
In zebrafish, injection of gRNAs and Cas9 mRNA into one-cell stage embryos similarly yields indels at target sites with relatively high, although variable, frequencies (Gagnon et al., 2014; Hwang et al., 2013). Mutations are inheritable due to mosaic targeting of the germline, allowing rapid establishment of mutant strains. Since gene inactivation by CRISPR/Cas9 is complete and permanent, this technology provides an effective complementary approach to morpholinos for loss-of-function studies in zebrafish, particularly at later stages of development. Mutant lines are invaluable to analyze gene function in both embryos and adults. The global loss of some genes is embryonic lethal, making them challenging to study in adults, and there is a great need in the field to create tissue-specific knockouts. We report here a CRISPR-based vector system that enables stable, tissue-specific gene inactivation in vivo.
Results
Urod gene disruption using CRISPR/Cas9 yields a fluorescent phenotype in zebrafish embryos
To test the efficacy of our tissue-specific CRISPR/Cas9 system, we used an embryonic phenotype resulting from the inactivation of the urod gene in zebrafish. Urod is an enzyme implicated in heme biosynthesis. Mutations in the UROD gene were found in human hepatic cutaneous porphyria, a disorder characterized by defects in iron metabolism in the liver, skin photosensitivity, and reduced erythrocytic heme production (Balwani and Desnick, 2012). A point mutation in urod identified in the yquem mutant zebrafish line was shown to recapitulate these features (Wang et al., 1998). Erythrocytes deficient for urod exhibit strong red fluorescence due to the accumulation of unprocessed porphyrins, which are inherently fluorescent.
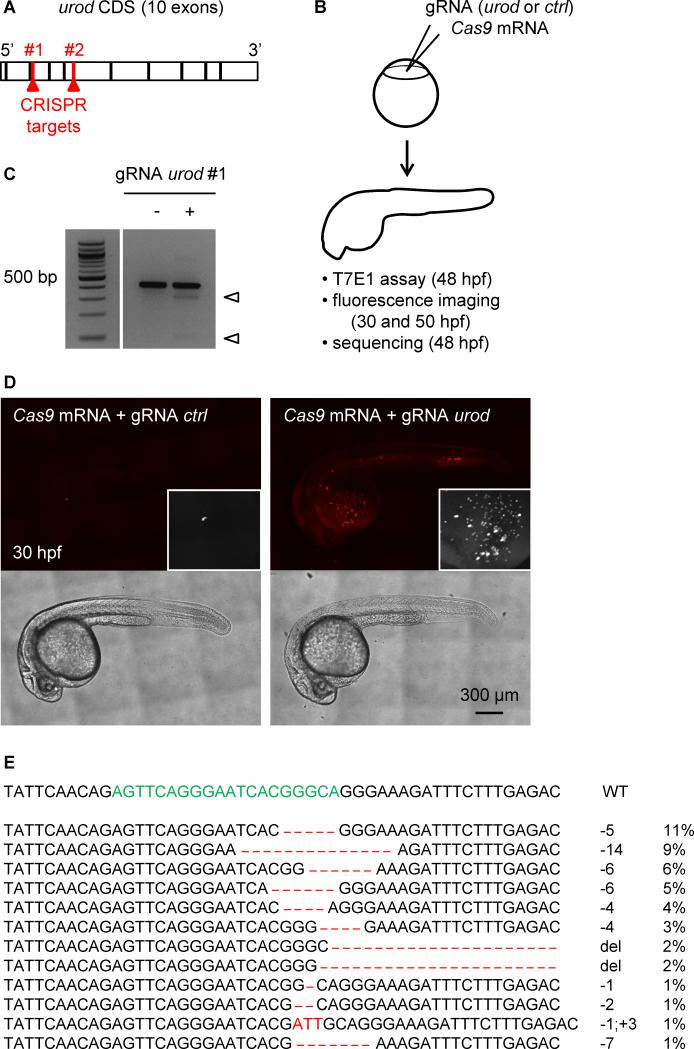
We selected two gRNAs that mutated the third and fifth exons of the urod locus when injected with Cas9 mRNA into single-cell zebrafish embryos, as assessed by the T7E1 mutagenesis assay (Kim et al., 2009) (Figure S1A-B, Fig. 1A-C). As a control, we used a gRNA efficiently targeting an irrelevant gene, p53 (Figure S1A-B). We observed that urod targeting led to the appearance of fluorescent erythrocytes in circulation at 30 hpf (Fig. 1D), mimicking the phenotype seen in yquem mutant fish (Wang et al., 1998). Approximately 70% of injected embryos displayed a strong or intermediate phenotype with 10 or more fluorescent cells (Figure S1C) and the intensity of the phenotype correlated with targeting efficiency (Figure S1D). This phenotype persisted after 2 dpf but was not observed as a consequence of p53 targeting (Figure S1E). As both gRNAs against urod yielded similar results in these early studies, subsequent experiments were performed using gRNA #1 only, unless otherwise noted. Co-injection of urod mRNA partially rescued the fluorescent phenotype, while complementation with an mRNA bearing the yquem mutation did not, demonstrating the specificity of the targeting (Figure S1C). Finally, we sequenced the urod CRISPR locus in injected embryos that displayed a strong fluorescent phenotype. As previously reported (Gagnon et al., 2014), a limited number of different mutations accounted for most mutant alleles, among which deletions were the most frequent (Fig. 1E). More than 80% of detected mutations induced frameshifts and are therefore likely to be disruptive. Some inframe mutations were found that may not affect gene function, but since the CRISPR target sequence is located close to the inactivating point mutation found in the yquem mutant, even missense mutations could affect the activity of the protein. These data demonstrate the accurate targeting of the urod gene by CRISPR technology.
Figure 1. Fluorescent phenotype resulting from CRISPR targeting of urod.
(A) Schematic representation of urod coding sequence (CDS). The position of the CRISPR target sequences at the beginning of exon three and five are indicated.
(B) Outline of the experiment: Cas9 mRNA is injected into one-cell stage embryos along with a gRNA targeting either urod or p53 as a negative control (ctrl). Targeted mutation efficiency is assessed by T7E1 assay and the presence of red fluorescent erythrocytes by confocal microscopy.
(C) T7E1 mutagenesis assay at the CRISPR target site in the urod gene. The assay was performed on genomic DNA from 2 dpf embryos injected at the one-cell stage with Cas9 mRNA and either a gRNA against p53 (negative control) or a gRNA against urod. Cleavage bands (arrowheads) indicate the presence of mutations at the target site. See also Figure S1B.
(D) Confocal images reveal the presence of fluorescent blood cells in urod mosaic knock-out embryos at 30 hpf. The black and white insets show a 2x magnification of the red fluorescent signal in the yolk region. See also Figure S1E.
(E) Most frequent (>1%) mutant urod alleles found by deep-sequencing in whole embryos injected with Cas9 mRNA and urod gRNA. The CRISPR target sequence is in green, mutations in red. The type of mutation and the associated frequency (in % of all mutated alleles) are indicated. del: large deletion.
Construction of an integratable CRISPR vector for gene targeting in zebrafish
The Tol2 technology enables the generation of transgenic lines by transposase-mediated insertion of a plasmid into the genome of zebrafish embryos (Kawakami et al., 2004). In order to create a vector system allowing spatial control of gene inactivation in zebrafish, we engineered a Tol2 integratable vector with three key features (see Figure 2A and Protocol): 1) a zebrafish U6-3 promoter (Halbig et al., 2008) to drive the expression of a gRNA scaffold, into which custom seed sequences can be easily cloned (Figure S2A), 2) a zebrafish codon-optimized Cas9 flanked by two nuclear localization signals (Jao et al., 2013) that can be placed under the control of either the ubiquitous ubiquitin (ubi) promoter (Mosimann et al., 2011), the erythrocyte-specific gata1 promoter (Long et al., 1997) or the muscle-specific mylz2 promoter (Storer et al., 2013) using the Gateway cloning technology (Hartley et al., 2000) and 3) GFP expressed under the control of the heart-specific cmlc2 promoter which serves as a transgenesis marker (Kwan et al., 2007).
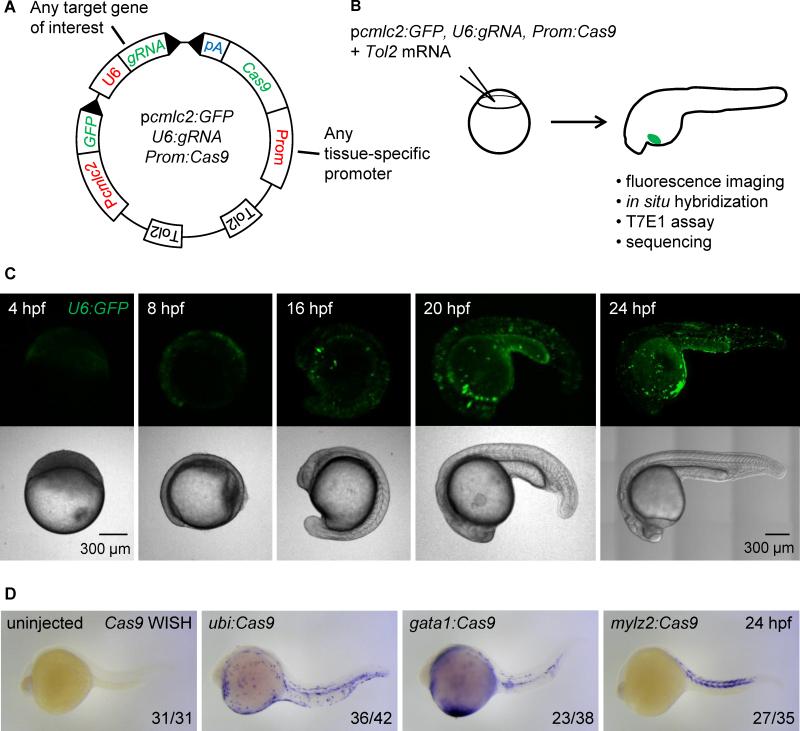
Figure 2. Integratable CRISPR vector for tissue-specific gene targeting.
(A) Schematic representation of the tissue-specific CRISPR vector. Prom denotes any promoter of interest used to drive Cas9 expression in a tissue-restricted manner. GFP expression in the heart of injected embryos is used as a transgenesis marker. Tol2 indicates transposition sites for the Tol2 transposase. pA: SV40 polyA sequence. See Figure S2A for the sequence of U6:gRNA, which comprises two BseRI restriction sites allowing easy cloning of any gene-specific target sequence at the 5’ end of the gRNA.
(B) Outline of the experiment: the CRISPR vector is injected into one-cell stage embryos along with Tol2 mRNA. Embryos with GFP-positive hearts are sorted at 24 hpf and further analyzed. Cas9 expression is assessed by in situ hybridization of Cas9 mRNA, targeted mutation efficiency by T7E1 assay, and the presence of fluorescent red blood cells by confocal microscopy.
(C) Confocal images of embryos injected with Tol2 mRNA and the p_cmlc2:GFP, U6:GFP, gata1:Cas9_ vector at various time points. Note early (4 hpf) and ubiquitous (24 hpf) GFP expression.
(D) Representative images show whole mount in situ hybridization (WISH) using an anti-sense RNA probe against Cas9 mRNA in 24 hpf embryos injected with Tol2 mRNA and p_cmlc2:GFP, U6:gRNA urod_ vectors expressing Cas9 under the control of the indicated promoters. Cas9 expression pattern is governed by the tissue-specificity of the promoters. See also Figure S2B.
We first assessed the spatio-temporal expression of the U6 promoter by replacing the gRNA scaffold in our vector with a sequence encoding GFP. After injection of the resulting p_cmlc2:GFP_, U6:GFP, gata1:Cas9 vector into one-cell stage embryos, we observed green fluorescence as early as 4 hpf (Fig. 2C). Observation at later stages demonstrated the ubiquitous activity of the U6 promoter. Cas9 expression was assessed by in situ hybridization of its mRNA at 24 and 48 hpf. AB embryos were injected with vectors containing the ubi, gata1 or mylz2 promoters, along with Tol2 mRNA, and sorted for GFP-positive hearts at 24 hpf (Fig. 2B). As expected, Cas9 displayed a tissue-restricted expression pattern in a mosaic fashion, as is typical of F0 Tol2 injections (Fig. 2D, Figure S2B). We did not detect any particular toxicity upon expression of our vectors in embryos apart from that usually associated with the microinjection of DNA plasmids at the one-cell stage. These experiments provide functional validation for both tissue-specific promoters and the U6 promoter in our vector setting.
The CRISPR vector allows tissue-specific gene disruption in zebrafish embryos
We first used the T7E1 mutagenesis assay to evaluate gene targeting. In whole embryos significant mutation rates at the urod target locus were only detected with the vector containing a gRNA against urod and the ubi promoter (data not shown). Since erythrocytes only represent a small fraction (less than 2%) of the whole embryo at 2 dpf, the proportion of urod mutations associated with the gata1 promoter was presumably below the detection threshold of the assay. We therefore collected the blood of embryos injected with the vectors containing gRNAs against urod or p53 and driving Cas9 expression under the control of the gata1 promoter, and could detect efficient targeting of both genes (Figure S2C). In order to measure the mutation efficiency of our vectors more quantitatively, we sequenced PCR amplicons of the targeted loci by either deep-sequencing or Sanger sequencing of random clones after TA cloning. We then generated a mutation index calculated as the number of mutated alleles over the total number of sequenced alleles. 25% of alleles were found mutated for both urod and p53 in the blood of embryos injected with the p_cmlc2:GFP_, U6:gRNA, gata1:Cas9 vectors (Table S1). Major mutations detected by deep-sequencing are displayed in Figure S2D. We then injected the p_cmlc2:GFP_, U6:gRNA urod, mylz2:Cas9 construct into Tg(mylz2:mCherry) embryos and similarly observed urod targeting in sorted mCherry-positive cells at 48 hpf (Figure S2E). Seven out of 36 sequenced alleles (19%) displayed mutations at the urod CRISPR locus (data not shown). These results demonstrate that the targeting by our vector system is specific based on the promoter used to drive Cas9 expression.
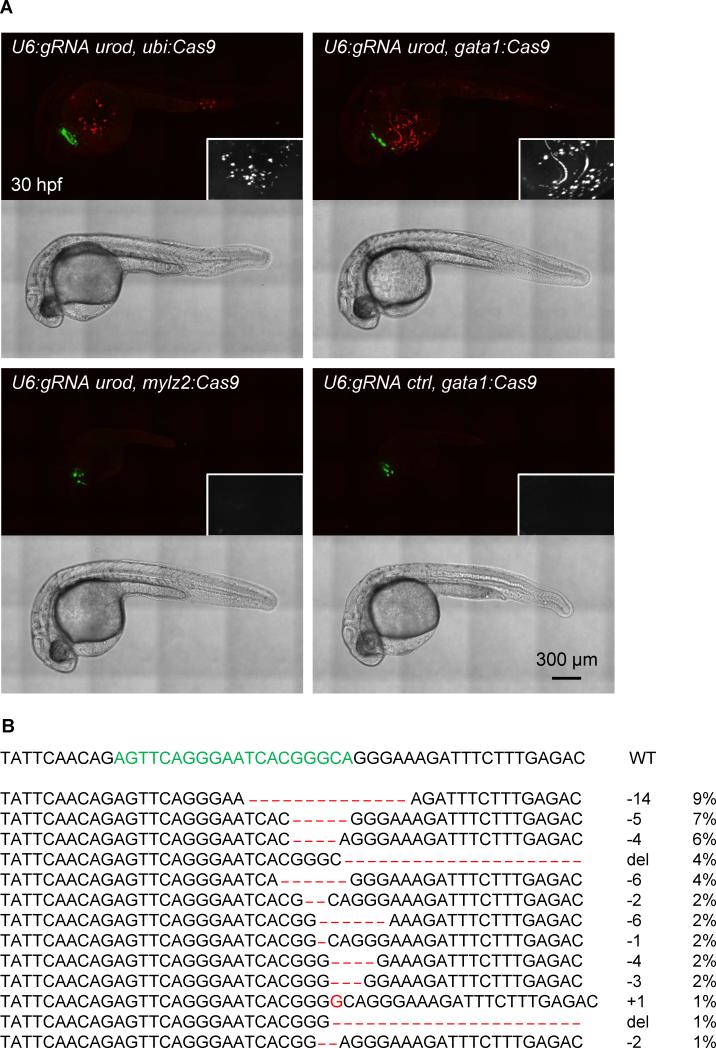
Injection of vectors expressing Cas9 under the control of the ubiquitious ubi or erythrocyte-specific gata1 promoters with the U6 promoter driving the expression of a gRNA against urod recapitulated the fluorescent phenotype associated with urod inactivation (Fig. 3A, Figure S3A, Movie S1). Similar results were obtained with both gRNAs against urod (Fig. 1A, data not shown). In contrast, using a gRNA targeting p53, or a promoter expressed in muscle but not blood, mylz2, to drive Cas9 expression did not result in the appearance of any fluorescent cells. Of note, the vectors containing ubi and gata1 promoters yielded a similar distribution of phenotypes, with approximately a third of injected embryos displaying 50 fluorescent cells or more (Figure S3B). We sorted fluorescent red cells from embryos injected with the p_cmlc2:GFP_, U6:gRNA urod, gata1:Cas9 vector and sequenced the urod CRISPR locus. 43% of alleles were mutant. The spectrum of mutations appeared very similar to that resulting from the co-injection of Cas9 mRNA and urod gRNA (Fig. 3B). The relatively high number of wild-type alleles detected may be due to contamination by non-fluorescent cells, amplified by the fact that most fluorescent cells are dying (see below), which may introduce a bias during the amplification of the low amounts of DNA obtained after sorting. Taken together, these data indicate successful tissue-specific inactivation of the urod gene.
Figure 3. Tissue-specific urod inactivation using the CRISPR vector.
(A) Confocal images of 30 hpf embryos injected with Tol2 mRNA and the indicated vectors. The fluorescent blood phenotype associated with urod knockout is seen when using the ubiquitous ubi promoter or the erythrocyte-specific gata1 promoter with the gRNA targeting urod, but not when using the muscle-specific mylz2 promoter or a gRNA against p53. See also Figure S3A and Movie S1.
(B) Most frequent (>1%) mutant urod alleles found by deep-sequencing in sorted fluorescent red cells from the blood of embryos injected with the p_cmlc2:GFP, U6:gRNA urod, gata1:Cas9_ vector. The CRISPR target sequence is in green, mutations in red. The type of mutation and the associated frequency (in % of all mutated alleles) are indicated.
In an attempt to adapt our system to enable the visualization of mutant cells in F0 mosaics, we engineered our CRISPR vector to express a Cas9-T2A-GFP sequence under the control of any tissue-specific promoter of interest (Figure S3C). We removed the cmlc2:GFP part of the previous vector since GFP marking of the cells that express Cas9 can serve as a transgenesis marker. Injection of the pA2, U6:gRNA urod, gata1:Cas9-T2A-GFP vector yielded GFP-positive blood cells at 30 hpf and, as expected, urod targeting resulted in the appearance of red fluorescent cells. Interestingly, most of the green and red cell populations did not overlap (Figure S3D). Non-red, green cells likely become red with targeting as development proceeds. Some of these green cells may bear non-disruptive urod mutations. Our microscopy showed that most red cells looked fragmented and of various sizes and shapes, frequently smaller than the expected size for erythrocytes. The few red fluorescent cells presenting regular shape and size also appeared of dimer green (Figure S3D). Diffuse red fluorescence was seen in urod mutants only, which presumably resulted from the release of porphyrins after lysis of mutant erythrocytes. This was easily distinguished from the cellular fluorescence. These observations suggest that urod null cells rapidly die from the accumulation of fluorescent heme precursors, consistent with the clinical syndrome of porphyria.
Stable expression of the CRISPR vector induces more penetrant gene inactivation
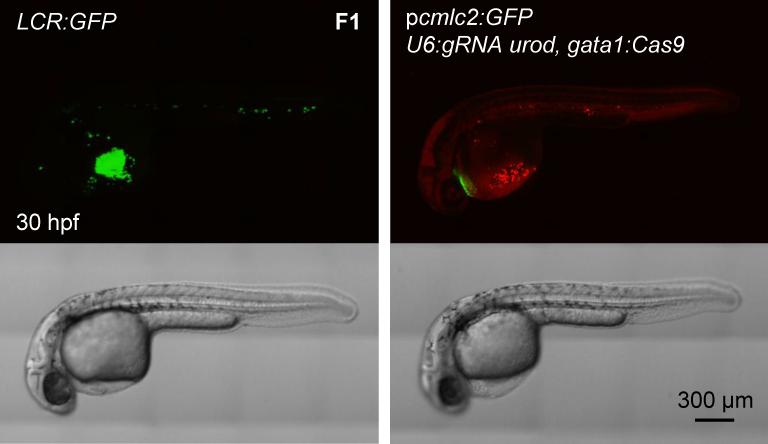
F0 embryos showing mosaic expression of cmlc2:GFP were raised to adulthood. They were then out-crossed to wild-type AB fish. F1 embryos with GFP-positive hearts were sorted and analyzed. The majority of F1 embryos expressing Cas9 under the ubi or gata1 promoters displayed a strong urod null phenotype, i.e. red fluorescent erythrocytes (Fig. 4, Figure S4A). In order to evaluate the proportion of erythrocytes showing red fluorescence, we examined age-matched LCR:GFP transgenic embryos which express GFP in erythrocytes (Ganis et al., 2012). We found that stable p_cmlc2:GFP_, U6:gRNA urod, gata1:Cas9 embryos and LCR:GFP embryos displayed comparable fluorescent phenotypes, although not all erythrocytes appeared fluorescent in urod mutants (Fig. 4). We then collected the blood from F1 embryos stably expressing p_cmlc2:GFP_, U6:gRNA urod, gata1:Cas9 at 36 and 48 hpf and analyzed it by FACS. Approximately 15% of the cells appeared fluorescent. However, about half of them were dying at 36 hpf (Figure S4B) and almost all of them were dead at 48 hpf (data not shown), indicating that the fluorescent cells are extremely fragile and short-lived. This, combined with the fact that not all cells become fluorescent simultaneously, may contribute to strongly underestimate the proportion of urod mutant cells at a given timepoint. Accordingly, the T7E1 assay on F1 blood showed extensive targeting of the locus (Figure S4C). The mutation efficiency observed in F0 was greatly increased in the F1 generation as measured by sequencing of the targeted locus (Figure S4D). Around 70% of alleles were found mutated in the blood of F1 embryos stably expressing the p_cmlc2:GFP_, U6:gRNA urod, gata1:Cas9 vector (Table S1). A fraction of the _cmlc2:GFP-_positive F1 embryos displayed no or few fluorescent erythrocytes (Figure S4E). This lack of urod inactivation was associated with markedly reduced expression of Cas9 as assessed by quantitative PCR in individual ubi:Cas9 embryos (Figure S4F), likely due to silencing of the promoter driving Cas9 expression. These results establish the efficiency of our CRISPR system in stable transgenics.
Figure 4. Generation of transgenic, tissue-specific knockout fish.
Confocal images of 30 hpf embryos of the LCR:GFP transgenic line and F1 generation of fish stably expressing p_cmlc2:GFP, U6:gRNA urod, gata1:Cas9_ vector. See also Figure S4A.
p53 inactivation in erythrocytes rescues hematologic defects in a model of Diamond-Blackfan anemia
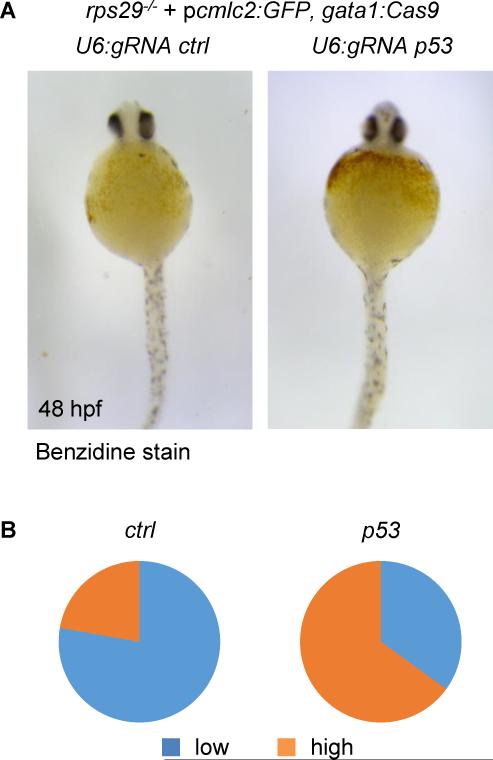
We then wanted to assess the ability of our vector system to correct an existing phenotype in vivo. In Diamond-Blackfan anemia (DBA), erythroid progenitors die of defects in key ribosomal proteins which trigger a robust p53 response. In a zebrafish model of DBA resulting from rps29 deficiency, p53 inactivation was shown to prevent the death of erythroid progenitors and therefore, rescue hemoglobin production (Taylor et al., 2012). We injected embryos from rps29+/− incrosses with the p_cmlc2:GFP_, U6:gRNA p53, gata1:Cas9 vector described above, using the p_cmlc2:GFP_, U6:gRNA urod, gata1:Cas9 vector as a negative control (Figure S2-CD). In rps29-/− embryos, p53 targeting in _gata1_-positive erythroid progenitors restored hemoglobin production as assessed by benzidine staining at 48 hpf (Fig. 5A-B). These results demonstrate the applicability of our CRISPR vector to genetic suppressor screens in vivo.
Figure 5. Rescue of an anemia phenotype by erythrocyte-specific targeting of p53.
(A) Benzidine staining of rps29-/− embryos injected with a p_cmlc2:GFP, U6:gRNA, gata1:Cas9_ vector targeting urod (ctrl) or p53 at 48 hpf.
(B) Quantification of the hemoglobin rescue in rps29-/− embryos. urod (ctrl): n=18; p53: n=20. Pooled results from two independent experiments.
Discussion
In order to greatly expand the potential applications of the powerful CRISPR/Cas technology, we have developed a targeting vector that allows for tissue-specific inactivation of a gene. Using a fluorescent phenotype resulting from the disruption of the urod gene, we show that the CRISPR/Cas knock-out technology can be spatially controlled in the blood lineage in zebrafish. Our vector system allows for the generation of stable zebrafish lines with tissue-specific, inheritable gene knock-out. This approach can be utilized to address cell autonomy in loss-of-function studies, as well as to avoid the embryonic lethality associated with the global knock-out of certain genes, which complicates the analysis of their functions in vivo.
Technically, our system simply requires a promoter to express Cas9. While the ubi promoter (Mosimann et al., 2011) allows efficient ubiquitous gene targeting, any promoter showing tissue-restricted activity can be easily inserted into the vector via the Gateway cloning technology. Further studies will be necessary to determine the efficiency of gene inactivation across different tissue-specific promoters. We have used other promoters successfully with this system such as the mitfa promoter to inactivate genes specifically in melanocytes in our BRAF+/+, p53-/− zebrafish melanoma model (Patton et al., 2005). Yet, the extent to which promoter strength can impact targeting efficiency remains unclear. We did observe promoter silencing in some F1 embryos in our experiments, likely due to positional effects related to the site of integration within the genome and the proximity of the strong cmlc2 promoter. Other transgenic markers than cmlc2:GFP may differentially impact the targeting efficiency of the vector. It is also critical to use a promoter that is truly tissue-specific as low levels of expression in other tissues may lead to inactivation in those tissues.
The U6 promoter that drives expression of the gRNA only requires that the gene-specific target sequence starts with a G so that potential target sites take the form G-(N)20-GG. Thanks to the relatively low sensitivity of Cas9 recognition to mismatches within the 5’ end of the seed sequence of gRNAs (Fu et al., 2013), it is possible to artificially start any seed sequence with a G, as we did for urod, leaving the existence of a PAM as the only requirement for the design of target sequences. The presence of an insertion marker in the vector (here, GFP under the control of a heart-specific promoter) facilitates sorting of transgenic animals, while the usual technique consisting of co-injection of Cas9 mRNA and a gRNA requires genotyping efforts. The overall targeting efficiency of the two methods seems comparable in F0 embryos as evaluated by both T7E1 mutagenesis assays and sequencing of the target locus. The mosaicism of gene targeting associated with Tol2 transposition of the vector may prove higher than when RNA is injected, as per usual procedures, particularly owing to the relatively large size of the vector. However, this disadvantage could be balanced by a greater level of biallelic inactivation by the vector since permanent Cas9 and gRNA expression likely increases the probability of targeting.
The ability to score transient tissue-specific phenotypes using our CRISPR system is a significant advantage. The technique should be ideal for the examination of tumor suppressor genes in cancer models. Other uses for transient expression could be for genetics in which there is a tissue-specific phenotype. For instance, a mutation in PAF1 subunit genes leads to a rescue of the moonshine mutant. It is possible that a tissue-specific CRISPR to PAF1 would rescue the moonshine phenotype. Transient tissue-specific CRISPR could be very helpful for a number of experiments in genetic models. Similar to standard CRISPR targeting, our vector system may have off-target effects. These could be identified by sequencing of predicted off-targets. Validation of an observed phenotype that has not been previously associated with the loss of the gene of interest may require the use of additional, independent gRNAs targeting the same gene.
In each cell of a tissue, Cas9 will potentially hit both alleles of the targeted gene once. Cas9-mediated DNA cleavage does not always result in gene inactivation since indels can yield silent in-frame mutations. Therefore, not all targeted cells completely lose expression of the gene of interest, which likely explains why F1 embryos that stably express the vector targeting urod still display a fraction of non-fluorescent erythrocytes. The resulting phenotype with this technique may not be as strong as a null allele. The efficiency of gene inactivation with our vectors is likely to be lower than in a straight knock-out or a conditional knock-out such as the Cre/Lox system in the mouse. This caveat may be overcome by designing gRNAs directed against essential functional domains of genes in order to increase the disruptive impact of in-frame mutations. Because gene extinction may not be fully penetrant in the targeted cell population, screening for positive phenotypes that arise as a consequence of gene disruption may prove easier than looking for negative phenotypes like the loss of a trait in vivo. Nevertheless, this system allows the observation of a deficiency typically within three months, right after the F0 generation can mate. This speed may have an advantage over the standard CRISPR knockout technique in the zebrafish, as establishing mutant lines requires further characterization to choose the inactivating alleles as well as additional crosses to get homozygous mutants. It is also considerably faster than the time required to generate conditional knock-out mice.
Overall, the flexibility of the tissue-specific CRISPR vector system we describe here makes it a versatile tool that can be adapted to rapidly meet numerous other applications. For example, multiplexing could be achieved through the insertion of several U6:gRNA units into the vector. Cas9 variants (e.g. Cas9 nickase, Cas9 fusions) can also be utilized. Our vector could be further developed to enable temporal control of gene inactivation, and thus serve as a platform to broaden the spectrum of genome editing technologies in zebrafish. High-throughput loss-of-function screens have been performed in vitro using the CRISPR technology (Koike-Yusa et al., 2014; Wang et al., 2014; Zhou et al., 2014). Due to its ease of use, our vector system is amenable to in vivo screening strategies. Finally, since the Tol2 transposon technology has proved effective in cells from a range of vertebrate species including mouse, chicken, Xenopus and medaka fish, our approach may apply to model organisms other than zebrafish.
Experimental Procedures
DNA constructs
We cloned the zebrafish U6-3 promoter (Halbig et al., 2008) from the AB strain, followed by an NheI site, into the p_cmlc2:GFP_ destination vector from the Tol2 Kit (Kwan et al., 2007) using ClaI and KpnI enzymes. The gRNA scaffold used by the Joung lab (Hwang et al., 2013) was modified to replace BsaI enzyme sites by BseRI sites, and inserted at the transcription start site of the U6 promoter using NheI and KpnI enzymes. This resulted in a p_cmlc2:GFP_, U6:gRNA destination vector. The p_cmlc2:GFP_ vector had previously been mutated to eliminate a BseRI site present in the GFP sequence. The two BseRI enzyme sites at the 5’ end of the gRNA scaffold enable easy cloning of any 20-bp target sequence of interest. The same method was employed to generate a p_A2, U6:gRNA_ destination vector.
We then constructed a middle-entry vector containing the Cas9 (codon-optimized for zebrafish) developed by the Chen lab (Jao et al., 2013), in which the BseRI site has also been mutated. The multisite Gateway cloning technology (Hartley et al., 2000) thus allows the introduction of Cas9 with a polyA tail under any promoter of interest into p_cmlc2:GFP_, U6:gRNA in a single cloning step. We similarly constructed a middle entry clone containing Cas9 followed by the sequence of a self-cleaving T2A peptide and GFP in which the BseRI site had been mutated. This Cas9-T2A-GFP sequence can be introduced into the p_A2, U6:gRNA_ vector under any promoter of interest by Gateway cloning.
mRNA and gRNA synthesis
Cas9 mRNA was produced by in vitro transcription from a pCS2 Cas9 vector (Jao et al., 2013) using mMESSAGE mMACHINE SP6 kit (Invitrogen). gRNAs were generated following established methods (Hwang et al., 2013).
urod cDNA was amplified by PCR from a cDNA library established from wild-type AB embryos using the following primers: GAATGGATAAGGACAGTTTCA and GCTTAGCGTTTGAGAAGCT. The cDNA sequence was then cloned into a middle entry clone for Gateway and inserted into the pDest Tol2 pA2 vector between an SP6 promoter and a polyA sequence. The yquem mutation was introduced using Q5 Site-Directed Mutagenesis Kit (NEB) with primers GTGTAGGAGACAGGCTGGA and CACACTGGAATGTGCTCGAT. urodwt and urodyq mRNAs were produced by in vitro transcription using mMESSAGE mMACHINE SP6 kit (Invitrogen).
Microinjections
Zebrafish of AB strain were bred and embryos were collected for microinjection. 20 pg of DNA constructs and 20 pg of Tol2 mRNA were injected into one-cell stage embryos. For typical CRISPR experiments, 600 pg of Cas9 mRNA and 25 pg of gRNA were injected into each embryo. 50 pg of urod mRNA were co-injected for rescue experiments. Post microinjection, embryos were raised in E3 medium at 28.5°C.
Whole-mount in situ hybridization and benzidine staining
We cloned a 713 bp fragment from the 3’ end of Cas9 into pCRII-TOPO vector (Invitrogen) using primers CCCTAAGAAGTATGGAGGCT and GCTGAGACAGGTCAATCCTT, and synthesized both the sense and anti-sense RNA probes by in vitro transcription. In situ hybridization was performed following established protocols (Thisse and Thisse, 2008).
Benzidine staining was performed as previously described (Paffett-Lugassy and Zon, 2005).
Imaging
Embryos were mounted in 0.8% low melting point agarose containing tricaine (160 mg/L) and imaged using Yokogawa CSU-X spinning disk confocal and inverted Nikon Eclipse Ti microscope (Andor Technologies).
T7E1 mutagenesis assay
T7E1 assay was performed as reported (Kim et al., 2009). Briefly, genomic DNA was extracted from 2-days old embryos using the HotSHOT method. A fragment of approximately 400 bp was amplified from genomic DNA using the following primers:
For urod:
1st gRNA: CTTACACAATCACAAGCCTT and CAACATTACATCAACAACCC
2nd gRNA: GTCCCTCAGGTCATTATTGT and CCTTCTTGCAGTGAAGTGAA
For p53: GGCACATATGCAAACAGATT and CAAACACCCAGAAATCTCTA
The PCR amplicons were then purified on a 1% agarose gel. 200 ng of purified DNA was denatured at 95°C for five minutes and slowly reannealed prior to digestion with 10 units of T7E1 enzyme (NEB) for 1 h at 37°C. The digestion product was finally run on a 2.5% agarose gel.
FACS
For fluorescence analysis and/or cell sorting from whole embryos, embryos were dissociated first mechanically using a razor blade, then enzymatically using Liberase at 37°C for 15-20 minutes (Roche).The reaction was stopped by addition of FBS. The cells were pelleted by centrifugation and resuspended in PBS. Finally, the cell suspension was filtered (40 μm) prior to analysis.
FACS analysis was performed using an LSR FORTESSA (BD Biosciences), and FACS sorting was performed using a FACS ARIA II (BD Biosciences). Flow cytometry data was analyzed using FACSDiva and FlowJo softwares. The red fluorescence was best detected in the PE-Cy5 channel. DNA was extracted from sorted cells using QuickExtract solution (Epibio) following manufacturer's instructions.
FACS data are accessible through FlowRepository, ID# FR-FCM-ZZWX.
Deep-sequencing
DNA libraries were prepared from the same PCR amplicons as for the T7E1 assay, using Nextera XT DNA Sample Prep Kit (Illumina). Sequencing was performed on the MiSeq system (Illumina). For the analysis of sequencing data, the raw reads were first trimmed based on FASTQ quality score (>30). Trimmed reads with a minimum length of 100 were aligned with urod sequence -200 to +200 around Cas9 cleavage site using BLAT (Kent, 2002), with tile size 11, no mismatch within tile and minScore of 30. The query sequences that aligned to both the left and right flanking regions of Cas9 cleavage site were finally analyzed to identify SNP, insertion, deletion or other types of mutations.
Quantitative PCR
RNA was extracted from single 50 hpf embryos using RNeasy Mini Kit (Qiagen). cDNA was synthesized using SuperScript III Kit (Invitrogen). qPCR was performed using Sso Fast EvaGreen Supermix and CFX384 Real-Time system (Biorad) with the following primers:
For Cas9: AGGCCCCTGATTGAGACAAA and AGTGGGAGAGTCAAAGCCTC
For β-actin: CGAGCAGGAGATGGGAACC and CAACGGAAACGCTCATTGC
Supplementary Material
1
2
3
Acknowledgements
We thank Charles K. Kaufman and Elliott J. Hagedorn for critical reading of the manuscript. This work was supported by NIH grants R01 CA103846, R01 HL04880, R01 DK53298, PO1 HL32262, P30 DK49216, U01 HL10001 and R24 DK092760. In addition, L. I. Zon is a Howard Hughes Medical Institute Investigator and J. Ablain is supported by an EMBO Long-Term Fellowship (EMBO ALTF 263-2013). L.I.Z. is a founder and stock holder of Fate, Inc. and Scholar Rock.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental information
Supplemental information comprises four supplemental figures, a supplemental table and a supplemental movie.
References
- Balwani M, Desnick RJ. The porphyrias: advances in diagnosis and treatment. Blood. 2012;120:4496–4504. doi: 10.1182/blood-2012-05-423186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Ahkmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganis JJ, Hsia N, Trompouki E, de Jong JL, DiBiase A, Lambert JS, Jia Z, Sabo PJ, Weaver M, Sandstrom R, et al. Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev Biol. 2012;366:185–194. doi: 10.1016/j.ydbio.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbig KM, Lekven AC, Kunkel GR. Zebrafish U6 small nuclear RNA gene promoters contain a SPH element in an unusual location. Gene. 2008;421:89–94. doi: 10.1016/j.gene.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffett-Lugassy NN, Zon LI. Analysis of hematopoietic development in the zebrafish. Methods Mol Med. 2005;105:171–198. doi: 10.1385/1-59259-826-9:171. [DOI] [PubMed] [Google Scholar]
- Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, Berghmans S, Mayhall EA, Traver D, Fletcher CD, et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer NY, White RM, Uong A, Price E, Nielsen GP, Langenau DM, Zon LI. Zebrafish rhabdomyosarcoma reflects the developmental stage of oncogene expression during myogenesis. Development. 2013;140:3040–3050. doi: 10.1242/dev.087858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Humphries JM, White RM, Murphey RD, Burns CE, Zon LI. Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Exp Hematol. 2012;40:228–237. e225. doi: 10.1016/j.exphem.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Wang H, Long Q, Marty SD, Sassa S, Lin S. A zebrafish model for hepatoerythropoietic porphyria. Nat Genet. 1998;20:239–243. doi: 10.1038/3041. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3