Structure-Function Analysis of STING Activation by c[G(2′,5′)pA(3′,5′)p] and Targeting by Antiviral DMXAA (original) (raw)
. Author manuscript; available in PMC: 2015 Apr 6.
SUMMARY
Binding of dsDNA by cyclic GMP-AMP (cGAMP) synthase (cGAS) triggers formation of the metazoan second messenger c[G(2′,5′)pA(3′,5′)p], which binds the signaling protein STING with subsequent activation of the interferon (IFN) pathway. We show that human hSTINGH232 adopts a ‘‘closed’’ conformation upon binding c[G(2′,5′)pA(3′,5′)p] and its linkage isomer c[G(2′,5′)pA(2′,5′)p], as does mouse mStingR231 on binding c[G(2′,5′)pA(3′,5′)p], c[G(3′,5′)pA(3′,5′)p] and the antiviral agent DMXAA, leading to similar ‘‘closed’’ conformations. Comparing hSTING to mSting, 2′,5′-linkage-containing cGAMP isomers were more specific triggers of the IFN pathway compared to the all-3′,5′-linkage isomer. Guided by structural information, we identified a unique point mutation (S162A) placed within the cyclic-dinucleotide-binding site of hSTING that rendered it sensitive to the otherwise mouse-specific drug DMXAA, a conclusion validated by binding studies. Our structural and functional analysis highlights the unexpected versatility of STING in the recognition of natural and synthetic ligands within a small-molecule pocket created by the dimerization of STING.
INTRODUCTION
The protein TMEM173/STING (stimulator of interferon genes) (Ishikawa and Barber, 2008; Zhong et al., 2008; Sun et al., 2009; Burdette et al., 2011) is a central player in the innate immune response to nucleic acids, particularly cytosolic dsDNA (reviewed in Burdette and Vance, 2013). STING responds to various pathogens, as well as to mitochondrial damage, and its overactivation may contribute or possibly even trigger the onset of autoimmune disorders such as systemic lupus erythematosus (Gall et al., 2012). STING’s role in the immune system is consistent with its higher expression in certain organs such as the thymus, spleen, and placenta. STING is also expressed in THP1 human monocytic cells.
An initial screen designed to discover potential regulators of the type I interferon (IFN) antiviral response identified cyclic GMP-AMP (cGAMP) synthase (MB21D1/cGAS) as a gene with broad antiviral effect (Schoggins et al., 2011). Independently, biochemical fractionation identified cGAS as the metazoan cytosolic DNA sensor and synthase of cGAMP, the endogenous second messenger that activates the type I IFN pathway (Sun et al., 2013; Wu et al., 2013). A structure-function study demonstrated that only one specific isomer of cGAMP, namely c[G(2′,5′) pA(3′,5′)p], was produced by cGAS (Gao et al., 2013). This isomer of the second messenger contained an unanticipated 2′,5′ linkage at the GpA step, a feature subsequently validated by several independent studies (Diner et al., 2013; Ablasser et al., 2013; Zhang et al., 2013). Structures of dsDNA-bound cGAS with ATP and GTP (Gao et al., 2013; Civril et al., 2013), pppGpA dinucleotide intermediate (Gao et al., 2013), and the product c[G(2′,5′) pA(3′,5′)p] (Gao et al., 2013), along with biochemical analysis of reaction intermediates, provided insights into the stepwise conversion of GTP and ATP in the first step to pppGpA (Gao et al., 2013; Ablasser et al., 2013) and subsequent cyclization to c[G(2′,5′)pA(3′,5′)p] (Gao et al., 2013; Ablasser et al., 2013).
The identification of c[G(2′,5′)pA(3′,5′)p] as a novel second messenger generated by dsDNA-bound cGAS in the presence of GTP and ATP (Gao et al., 2013) has prompted studies of the role of c[G(2′,5′)pA(3′,5′)p] in activating the IFN pathway via the downstream receptor STING (Diner et al., 2013; Ablasser et al., 2013; Zhang et al., 2013). Binding of c[G(2′,5′)pA(3′,5′)p] to STING activates a cascade of events whereby STING recruits and activates IκB kinase (IKK) and TANK-binding kinase (TBK1), which, following their phosphorylation, activate nuclear transcription factor κB (NF-κB) and interferon regulatory factor 3 (IRF3), respectively. These activated proteins translocate to the nucleus to induce transcription of the genes encoding type I IFN and cytokines for promoting intercellular host immune defense (reviewed in Keating et al., 2011; Paludan and Bowie, 2013).
Human (h) and mouse (m) STING exhibit 68% amino acid identity and 81% similarity, with distinct sequence alleles reported in humans (Diner et al., 2013). (Note that the numbering systems of human and mouse are offset by one residue, with hSTINGR232 and mStingR231 occupying equivalent positions.) hSTING is composed of a N-terminal transmembrane domain (aa 1–154), a central globular domain (aa 155–341), and a C-terminal tail (aa 342–379). Distinct sequence variants of either mouse or human STING have been studied with different functional assays and outcomes, complicating interpretation. Diner et al. (2013) identified natural variant alleles of STING, namely the R232H variant to the genome reference of human STING (hSTINGH232, reference sequence) and the R231A variant of mouse Sting (mStingA231), which were activated by c[G(2′ ,5′)pA(3′,5′)p] but not by c[di-GMP] or c[G(3′,5′)pA(3′,5′)p]. By contrast, Zhang et al. (2013) proposed based on structural, calorimetric-based binding and cellular assays that c[G(2′,5′)pA(3′,5′)p] is the highest affinity ligand for hSTINGR232, even though their in vitro calorimetric binding measurements did not support their results seen with IFN induction cellular assays. Ablasser et al. (2013) investigated the contribution of 2′ ,5′- and 3′,5′-linkages in cGAMP and concluded that c[G(2′ ,5′)pA(3′,5′)p] was more potent than c[G(3′,5′)pA(3′,5′)] in activating hSTING from human fibroblasts and THP1 cells, and wild-type mStingR231.
Structural studies have been reported on hSTINGH232 in the free and c[di-GMP]-bound states. STING forms a symmetrical dimer in both states with c[di-GMP] bound in a pocket within the dimer interface and anchored by a network of intermolecular hydrogen bonds (reviewed in Burdette and Vance, 2013). For all but one of the c[di-GMP]-hSTINGH232 complexes, the STING dimer adopts the same V-shaped conformation independent of the presence of the ligand and also does not completely surround the bound ligand. In the one exception, c[di-GMP]-bound hSTINGA230/R232 forms an antiparallel b-pleated sheet cap over the binding pocket on complex formation and this conformational change further sequesters the bound ligand (Huang et al., 2012). More recently, the same conformational transition has been reported on formation of the complex between mStingR231 and the antiviral drug CMA (Cavlar et al., 2013) and on formation of the complex between hSTINGR232 and c[G(2′,5′)pA(3′,5′)p] (Zhang et al., 2013).
It is of timely interest to define the structural basis and functional output of ligand binding by hSTING and mSting given STING’s central role in immunoregulation of the antiviral response and in eliciting a macrophage-dependent tumoricidal program (Kim et al., 2013). For example, it remains to be clarified whether hSTINGR232 is as selective for [G(2′,5′)pA(3′,5′)p] as recently claimed (Zhang et al. (2013), or whether its linkage isomers are similarly recognized, and to what extent distinct STING alleles (hSTINGH232 and mStingA231) differentially respond, as proposed by Diner et al. (2013).
DMXAA (5,6-dimethylxanthenone-4-acetic acid, Vadimezan) was initially identified as a small molecule exhibiting immune modulatory activities through induction of cytokines and disrupting tumor vascularization in mouse xenotransplantation models (Baguley and Ching, 2002). DMXAA, in combination with paclitaxel and carboplatin showed promising efficacy and was therefore evaluated in a phase II clinical trial against non-small-cell lung cancer, but subsequently failed in human phase III trials (Lara et al., 2011). Recently, it has been demonstrated that DMXAA-induced IFN production by murine macrophages was impaired by the absence of Sting (Prantner et al., 2012), suggesting that DMXAA targets the STING pathway. There is a high-sequence identity between mSting and hSTING, but DMXAA only activates mSting and has no effect on hSTING (Conlon et al., 2013), and this has presumably hampered further therapeutic development of DMXAA as a human drug.
Here, we have undertaken systematic structure-function studies of mouse and human STING activation by c[G(2′,5′) pA(3′,5′)p], its linkage isomers, c[di-GMP], and antiviral DMXAA ligand. hSTING was more discriminating toward 2′ ,5′-linkage-containing isomers versus the all 3′ ,5′-linkage isomer, and a unique point mutation (S162A) placed at the cyclic-dinucleo-tide-binding site in hSTINGH232 and hSTINGR232 rendered hSTING sensitive to the otherwise mouse-specific drug DMXAA. These findings are critical to guide future mechanistic studies of the cytosolic DNA-sensing pathway and the development of agonists and antagonists of innate immunity as needed for anticancer and vaccine development, as well as therapies for autoimmune disorders.
RESULTS
Crystal Structure of c[G(2′,5′ )pA(3′,5′)p] Bound to hSTINGH232
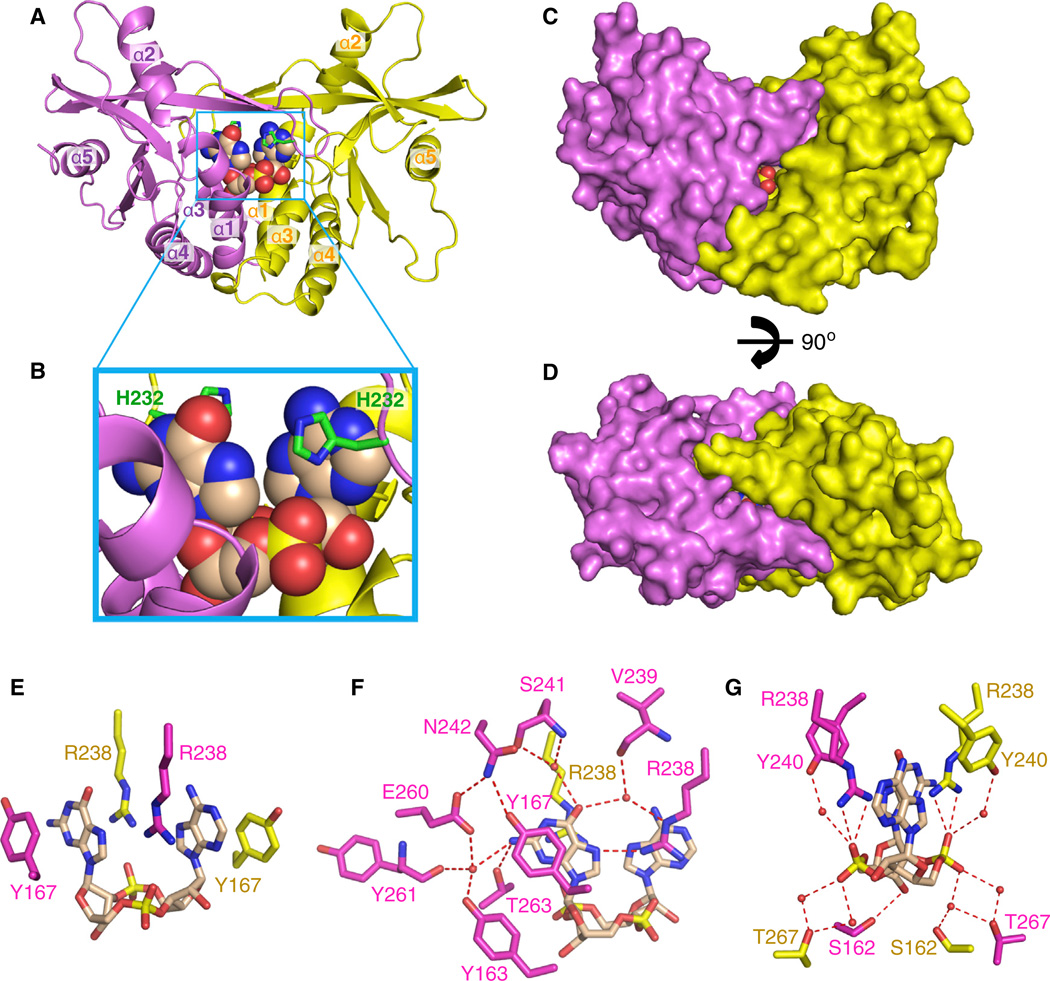
The 2.25 Å crystal structure of c[G(2′,5′ )pA(3′,5′)p] bound to the symmetrical dimer of human STING (G230/H232; aa 155–341; termed hSTINGH232) is shown in Figure 1A (X-ray statistics in Table S1). The individual symmetry-related subunits of STING in a ribbon representation are color-coded in magenta and yellow, whereas the bound c[G(2′,5′)pA(3′,5′)p] is shown in a space-filling representation. The bound ligand is positioned in a deep U-shaped cleft between subunits, with the cyclic sugar-phosphate backbone at the base and the purine rings pointing upward in a parallel alignment (expanded view in Figure 1B). The bound U-shaped ligand is further anchored in place by an overhead cap element formed on complex formation by an antiparallel four-stranded b-pleated sheet (Figure 2A), such that the hSTING dimer completely envelops the bound ligand (Figures 1C and 1D).
Figure 1. Crystal Structure of c[G(2’,5’)pA(3’,5’)p] Bound to hSTINGH232 and Details of Intermolecular Contacts in the Complex.
(A) The 2.25 Å crystal structure of cyclic [G(2’,5’)pA(3’,5’)p] bound to hSTINGH232 (aa 155–341). The symmetrical hSTINGH232 dimer is shown in a ribbon representation, with individual monomers colored in magenta and yellow. α helices are labeled from α1 to α5. The c[G(2’,5’)pA(3’,5’)p] in a space-filling representation is bound in the central cavity at the interface between the two monomers.
(B) An expanded view of the c[G(2’,5’)pA(3’,5’)p]-binding pocket in the complex. The position of H232 (in green) in a stick representation is labeled in this panel.
(C) A surface representation of the structure of the complex shown in (A).
(D) The view in (C) rotated through 90°.
(E-G) Intermolecular contacts in the complex of c[G(2’,5’)pA(3’,5’)p] bound to hSTINGH232. The bound c[G(2’,5’)pA(3’,5’)p] is shown in biscuit color, with individual STING subunits in the symmetrical dimer shown in magenta and yellow. The bracketing of the purine rings of c[G(2’,5’)pA(3’,5’)p] by Y167 is shown in (E), and intermolecular contacts to the base edges and backbone phosphates of the ligand by the subunits of STING are shown in (F) and (G), respectively.
Relates to Figure S1 and Table S1.
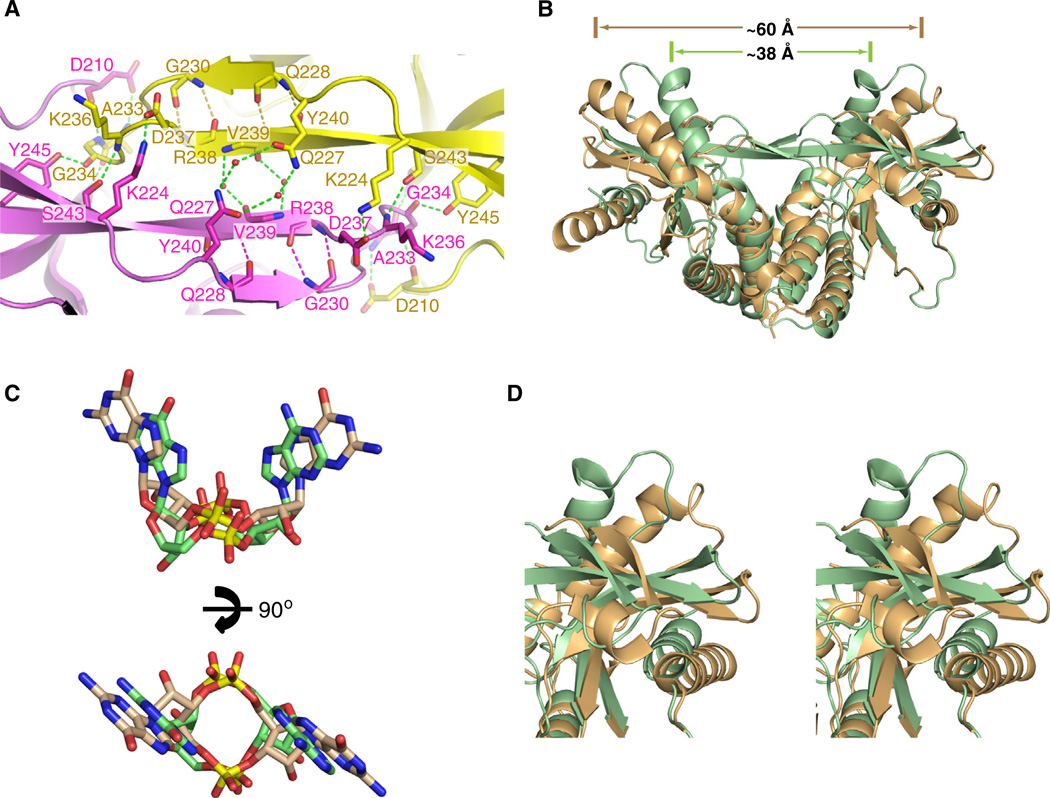
Figure 2. Comparison hSTINGH232 Complexes Bound to c[G(2′,5′)pA(3′ ,5′ )p] and c[di-GMP].
(A) Details related to alignment and hydrogen-bonding patterns within the four-stranded antiparallel β sheet that forms a cap over the binding pocket on formation of the c[G(2′,5′)pA(3′,5′)p]-hSTINGH232 complex.
(B) Superposition of the c[G(2′,5′)pA(3′,5′ )p]-bound structure of hSTINGH232 (aa 155–341) with both subunits in green and c[di-GMP]-bound structure of hSTINGH232 (aa 139–379) with both subunits in beige (PDB: 4EF4).
(C) Superposition of the c[G(2′,5′)pA(3′,5′)p] in green and c[di-GMP] in orange (PDB: 4EF4) in their complexes with hSTINGH232.
(D) An expanded view in stereo of the top right segment of (B) following superposition of the c[G(2′,5′)pA(3′,5′)p]-bound structure of hSTINGH232 (both subunits in green) and c[di-GMP]-bound structure of hSTINGH232 (both subunits in orange) (PDB: 4EF4).
Relates to Figure S1 and Table S1.
The binding pocket is uncharged at its base, whereas both positive- and negative-charged residues line its walls. The bound c[G(2′,5′)pA(3′,5′)p] is anchored by its purine bases being bracketed on either side by Y167 (Figure 1E) and by R238 (whose position is buttressed by Y240), with R238 aligned in the plane and hydrogen bonds to the N7 of one purine, while its guanidinium group stacks over the other purine of the bound cyclic dinucleotide (Figures 1E–1G). The bound ligand is further stabilized through a network of direct and water-mediated hydrogen bonds to the base edges from side chains of hSTINGH232 (Figures 1F and 1G). Amino acids participating in this network and positioned above the bound ligand include N242, S241, and V239, which form water-mediated hydrogen bonds to the O6 of guanosine, whereas Y163, E260, and Y261 form water-mediated hydrogen bonds together with a direct hydrogen bond from T263 to the NH2 group of guanosine (Figure 1F). The edges of the adenosine base are not involved in hydrogen-bond formation (Figure 1F).
The phosphate backbone and ribose hydroxyls of the cyclic dinucleotide ring system are additionally stabilized through hydrogen bonds. Amino acids participating in this network and positioned below the bound ligand include S162 and T267 (Figure 1G), with the 3′-OH group of the guanosine hydrogen-bonded to the side chain hydroxyl of S162, whereas no hydrogen-bonding is observed to the 2′-OH of adenosine of c[G(2′,5′)pA(3′,5′)p] in the complex (Figure 1G). The backbone phosphates of the bound cyclic dinucleotide are recognized by direct contacts from the guanidinium groups of R238 and through water-mediated hydrogen bonds from the hydroxyl groups of T267 and Y240 (Figure 1G).
The four-stranded antiparallel β sheet that forms upon c[G(2′,5′)pA(3′,5′)p]-hSTINGH232 complex formation (Figure 2A) caps the top of the binding pocket and restricts access to it (Figure 1A). In addition to hydrogen-bonding between strands across the four-stranded β-pleated sheet, anchoring at either end of the sheet is achieved by salt bridges (Figure 2A). It should be noted that G230 forms part of the outer β strand of this four-stranded β sheet (Figure 2A).
Comparison of Crystal Structures of c[G(2′,5′)pA(3′,5′)p] and c[di-GMP] Bound to hSTINGH232
One of the published crystal structures of c[di-GMP] bound to the symmetrical dimer of hSTINGH232 (aa 139–379) (Protein Database: 4EF4; 2.15 Å resolution) (Ouyang et al., 2012) is shown in Figure S1A (available online) with an expanded view of the ligand-binding pocket shown in Figure S1B. Note that the loops protruding over the binding pockets are disordered over half their lengths in the direction of their tips, whereas the symmetry-related α2-helices form a larger angle in the V-shaped c[di-GMP] complex (Figure S1A), as compared to the U-shaped c[G(2′,5′)pA(3′,5′)p] complex (Figure 1A) with hSTINGH232. Indeed, space filling views of the c[di-GMP] complex indicate that the hSTINGH232 dimer does not encapsulate the bound c[di-GMP] in its complex (Figures S1C and 1D; termed the “open” STING complex), as it does in space filling views of the c[G(2′,5′)pA(3′ ,5′)p] complex (Figures 1C and 1D; termed the “closed” STING complex) with hSTINGH232.
The intermolecular hydrogen bonds stabilizing complex formation in the published structure of the complex of c[di-GMP] with hSTINGH232 (Protein Database [PDB]: 4EF4) are shown in Figures S1E and S1F. Notably, the side chains of R238 are disordered, unlike the key recognition role they play in the complex of c[G(2′,5′)pA(3′,5′)p] with hSTINGH232 (Figure 1F and 1G).
The superposition of c[di-GMP] bound to the “open” hSTING complex (both subunits in beige) with that for c[G(2′,5′)pA(3′,5′)p] bound to the “closed” hSTING complex (both subunits in green) shows large conformational differences in STING between the two complexes (Figure 2B; rmsd = 3.11 Å). Indeed, the separation between the tips of the symmetry-related α2-helices decreases from approx. 60 Å in the “open” complex to approximately 38 Å in the “closed” complex. Furthermore, superposition of the bound c[di-GMP] in beige with that of bound c [G(2′,5′)pA(3′,5′)p] in green establishes that the purine bases are further apart in the former complex compared to the latter complex (Figure 2C), and presumably facilitate switching of the hSTINGH232 from an “open” and more flexible (partly disordered loops positioned over the binding pocket) conformation in the c [di-GMP] complex (Figure S1A) to a “closed” and more compact (well-defined four-stranded b sheet cap over the binding pocket) conformation in the c[G(2′,5′)pA(3′ ,5′ )p] complex (Figure 1A). Furthermore, the bound c[G(2′,5′)pA(3′,5′)p] (in green) is positioned slightly deeper in the binding cleft than its bound c[di-GMP] counterpart (in beige) (Figure 2C, top). Importantly, the conformational changes propagate to the surface of the STING protein as shown in a stereo view (Figure 2D), highlighting the upper right segment of Figure 2B.
Similar “Closed” Conformations Adopted by c[G(2′,5′) pA(3′,5′)p] Bound to mStingR231 and hSTINGH232
We have solved the 1.77 Å crystal structure of c[G(2′,5′)pA(3′,5′)p] bound to the symmetrical dimer of mStingR231 (aa 154–340) (Figure 3A and Table S2). The side chain of R231 of mStingR231 is shown in green in a stick representation (Figure 3A). Given the high resolution of this complex, the network of hydrogen bonds are clearly visible; the 3′-OH of guanosine of bound c[G(2′,5′)pA(3′,5′)p] makes hydrogen bonds with the side chain of S161 and two water molecules (Figure 3B). The guanidinium group of R231 interacts with the backbone phosphates of c[G(2′ ,5′ )pA(3′,5′)p] through a bridging water molecule (Figure 3B). Formation of the four-stranded antiparallel β-pleated sheet acts as a cap over the bound ligand in the complex (Figure 3A).
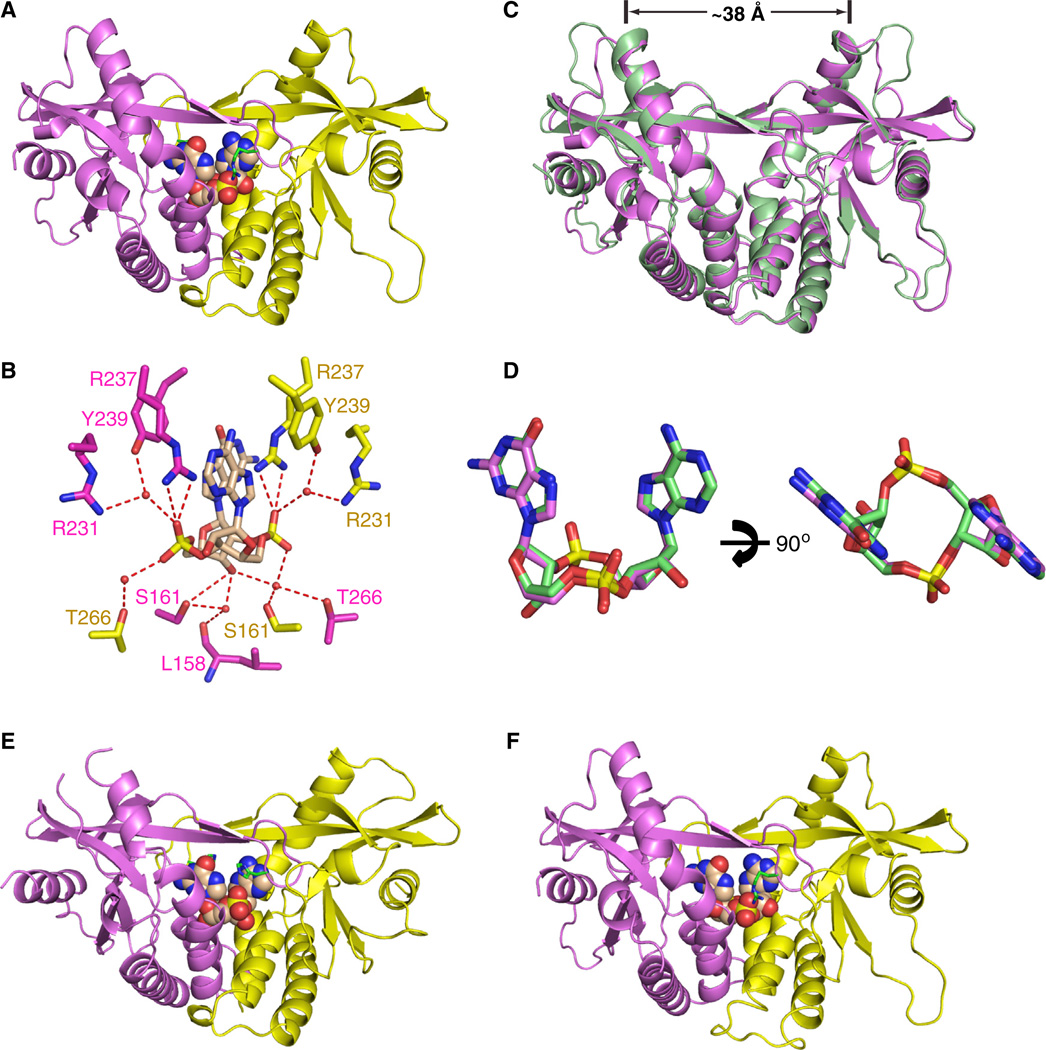
Figure 3. Crystal Structure of c[G(2′,5′)pA(3′,5′)p] Bound to mStingR231 and Comparison of Its Complex with the Same Ligand Bound to hSTINGH232.
(A) The 1.77 Å crystal structure of c[G(2′,5′)pA(3′,5′)p] bound to mStingR231 (aa 154–340).
(B) Intermolecular contacts to the cyclic dinucleotide ring system of the ligand by the subunits of mStingR231.
(C) Superposition of the c[G(2′,5′)pA(3′,5′)p]-bound structures of hSTINGH232 (both subunits in green) and mSTINGR231 (both subunits in magenta).
(D) Superposition of the c[G(2′,5′)pA(3′,5′)p] in its complexes with hSTINGH232 in green and mSTINGR231 in magenta.
(E) The 1.9 Å crystal structure of c[G(2′,5′)pA(2′,5′)p] bound to hSTINGH232 (aa 155–341).
(F) The 2.1 Å crystal structure of c[G(3′,5′)pA(3′,5′)p] bound to mStingR231 (aa 154–340).
Relates to Figure S2 and Tables S1 and S2.
We observe excellent superposition of hSTINGH232 (both subunits in green) and mStingR231 (both subunits in magenta) in its complexes with c[G(2′,5′ )pA(3′ ,5′)p] as shown in Figure 3C (rmsd = 0.84 Å). The separation between the tips of symmetry-related α2-helices are approx. 38 Å for both complexes (Figure 3C). We also observe excellent superposition of the c[G(2′,5′)pA(3′,5′)p]dinucleotide in its complexes with hSTINGH232 and mStingR231 (Figure 3D). Thus, the same c[G(2′,5′)pA(3′,5′)p]-STING complex is observed whether a His or Arg occupies this key position. Indeed, the R238, S162 and T267 in hSTINGH232 (Figure 1H) and their conserved counterparts R237, S161 and T266 in mStingR231 (Figure 3B) are involved in similar hydrogen-bonding interactions with the backbone phosphates and sugar hydroxyl groups in both complexes.
c[G(2′,5′)pA(2′,5′)p]- and c[G(3′,5′)pA(3′,5′)p]-Bound STING Complexes Both Adopt “Closed” Conformations
We also crystallized the two other cGAMP linkage isomers c[G(2′,5′)pA(2′,5′)p] and c[G(3′,5′)pA(3′,5′)p] with hSTINGH232 and mStingR231, respectively. We solved a 1.9 Å crystal structure of c[G(2′,5′)pA(2′ ,5′)p] with hSTINGH232 (aa 155–341) (Figure 3E and Table S1) and a 2.1 Å crystal structure of c[G(3′,5′)pA(3′,5′)p] with mStingR231 (aa 154–340) (Figure 3F and Table S2). The structures of both complexes adopt the “closed” conformation as reflected by the positioning of the ligands in the binding pocket, the separation between the tips of the α2-helices by approximately 38 Å, and the formation of the four-stranded antiparallel β sheet cap over the bound ligands (Figures 3E and 3F).
The crystal structures of c[G(2′,5′)pA(2′,5′)p] (in red) and c[G(2′,5′)pA(3′,5′)p] (in green) in their bound complexes with hSTINGH232 superimpose with an rmsd of 0.59 Å (Figure S2A), with the 3′-OH groups of bound c[G(2′,5′)pA(2′,5′)p] forming water-mediated hydrogen bonds to the hydroxyls of S162 and T267 (Figure S2B). The crystal structures of c[G(3′,5′)pA(3′,5′)p] (in cyan) and c[G(2′,5′)pA(3′,5′)p] (in magenta) in their bound complexes with mStingR231 superimpose with an rmsd of 0.25 Å (Figure S2C), with the 2′-OH groups of bound c[G(3′,5′) pA(3′,5′)p] forming direct hydrogen bonds to the hydroxyls of T262 (Figure S2D). The bound c[G(2′,5′)pA(2′,5′)p] (in red) is positioned somewhat deeper in the binding pocket than c[G(3′,5′)pA(3′,5′)p] (in cyan) (Figure S2E).
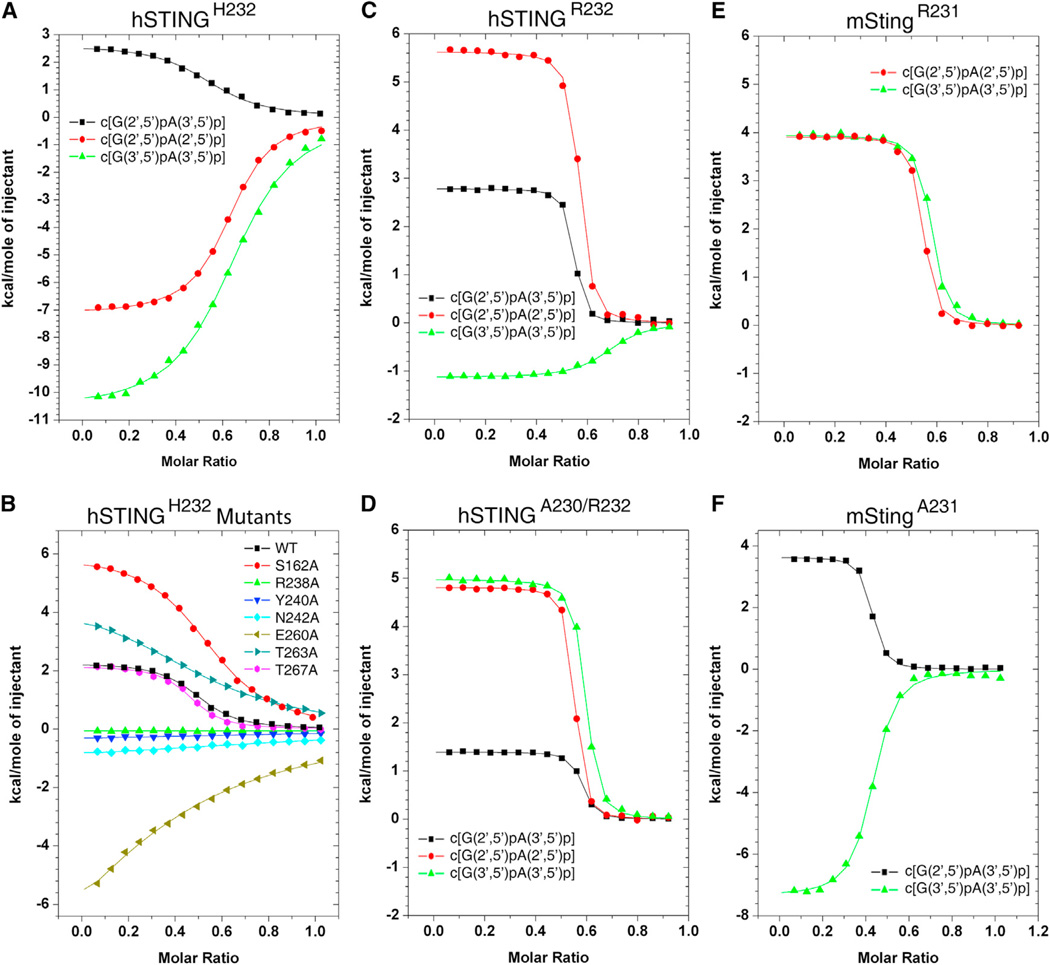
ITC Binding Studies of hSTINGH232 and Its Mutants with Linkage Isomers of cGAMP
We recorded the isothermal titration calorimetric (ITC) binding curves for hSTINGH232 (aa 140–379) with three linkage isomers of cGAMP as shown in Figure 4A. The binding curves are exothermic for complex formation with c[G(2′,5′)pA(2′,5′)p] (red circles) and c[G(3′,5′)pA(3′,5′)p] (green triangles), and endothermic for complex formation with c[G(2′ ,5′)pA(3′,5′)p] (black squares; KD = 5.3 µM). The thermodynamic KD, ΔG, ΔH, and TΔS parameters for these complexes are listed in Table S3.
Figure 4. ITC Data on Binding of cGAMP Linkage Isomers to hSTINGH232 and Its Mutants, as well as to hSTINGR232, hSTINGA230/R232, mStingR231, and mStingA231.
(A and B) ITC binding curves for complex formation between cGAMP linkage isomers bound to hSTINGH232 (aa 140–379) (A) and binding of c[G(2′,5′ )pA(3′,5′)p] to mutants of hSTINGH232 (B).
(C) ITC binding curves for complex formation between cGAMP linkage isomers bound to hSTINGR232 (aa 140–379).
(D) ITC binding curves for complex formation between cGAMP linkage isomers bound to hSTINGA230/R232 (aa 140–379).
(E) ITC binding curves for complex formation between cGAMP linkage isomers bound to mStingR231 (aa 139–378).
(F) ITC binding curves for complex formation between cGAMP linkage isomers bound to mStingA231 (aa 139–378).
Relates to Tables S3 and S4.
We also recorded ITC binding curves for complex formation of c[G(2′,5′)pA(3′,5′)p] with mutants of hSTINGH232 that participate in intermolecular contacts on complex formation. For this linkage isomer, cGAMP binding is completely lost (Figure 4B and Table S4) for the R238A mutant involved in cyclic dinucleotide base N7 and backbone phosphate recognition (Figures 1E, 1F, and 1G), as well as for the Y240A, N242A, and E260A (significantly reduced) mutants involved in water-mediated guanosine base edge recognition (Figure 1F). The impact of mutating the polar Thr and Ser residues involved in intermolecular hydrogen bond formation is more nuanced, with no effect on binding affinity for the T267A mutant, a modest reduction for the S162A mutant and a more pronounced reduction for the T263A mutant (Figure 1B). The thermodynamic parameters are listed in Table S4.
ITC Binding Studies of hSTINGR232 and hSTINGA230/R232 with Linkage Isomers of cGAMP
The corresponding ITC-based thermodynamic parameters were also recorded for complex formation of cGAMP linkage isomers with hSTINGR232 and hSTINGA230/R232 (aa 140–379) (Figures 4C and 4D, respectively). The 2′,5′-containing linkage isomers of cGAMP exhibited increased binding affinity by about an order of magnitude for STING variants with R232 compared to H232, whereas the corresponding binding affinity increase was about 4-fold for the all-3′,5′ linkage isomer of cGAMP (Table S3), with endothermic titration patterns observed for all binding curves, except for an exothermic titration pattern for c[G(3′,5′)pA(3′,5′)p] with hSTINGR232 (Figure 4C). The dissociation constants for c[G(2′,5′)pA(3′,5′)p] bound to hSTINGR232 and hSTINGA230/R232 were KD = 0.11 µM and 0.16 µM, respectively (Table S3).
ITC Binding Studies of mStingR231 and mStingA231 with Linkage Isomers of cGAMP
The corresponding ITC-based thermodynamic parameters for complex formation of cGAMP linkage isomers with mouse StingR231 (I229/R231; aa 139–378) are plotted in Figure 4E, with observed KD values similar to those observed for hSTINGA230/R232 (Table S3). The ITC titrations for mStingA231 with the various cGAMP linkage isomers are plotted in Figure 4F, with the observed KD values listed in Table S3. The dissociation constants for c[G(2′,5′)pA(3′,5′)p] bound to mStingA231 was KD = 0.34 µM (Table S3).
Crystal Structure of DMXAA Bound to mStingR231
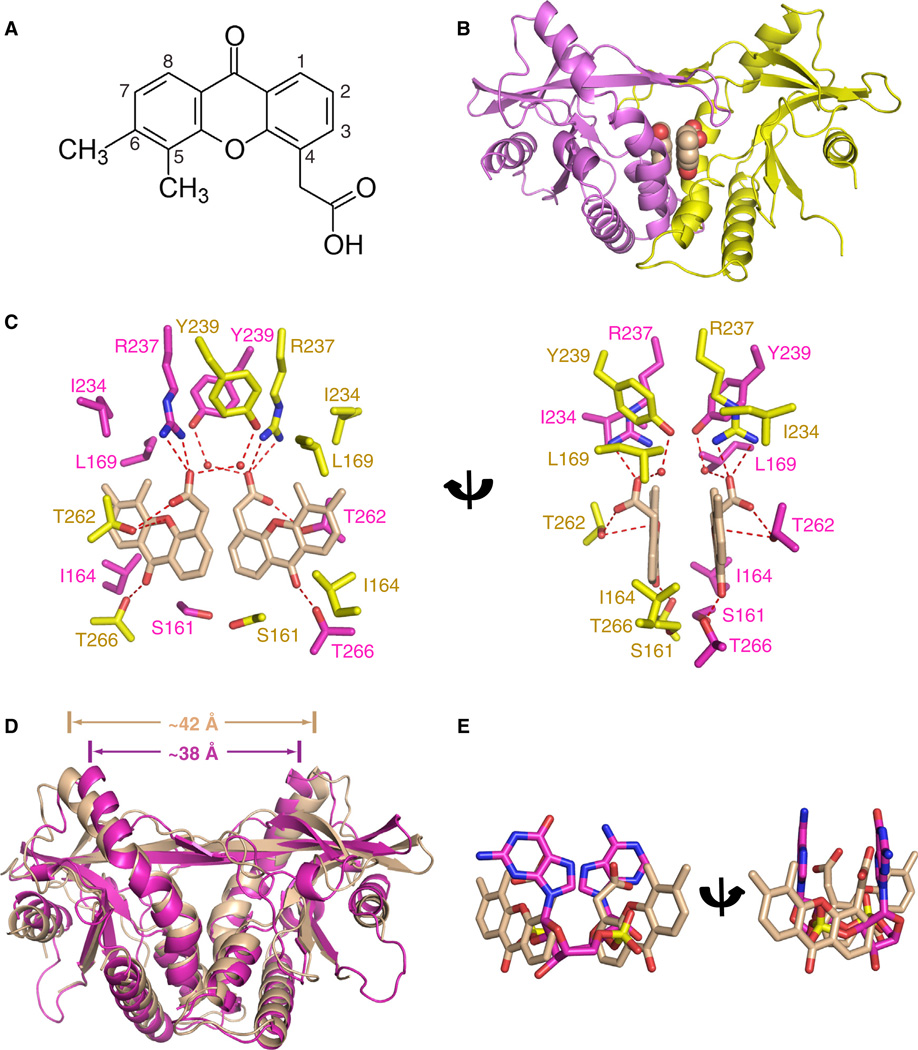
Antiviral small molecules reported to date that target mSting include 5,6-dimethylxanthenone-4-acetic acid (DMXAA) (Figure 5A) (Conlon et al., 2013; Kim et al., 2013) and 10-carboxymethyl-9-acridine (CMA) (Figure S3A) (Cavlar et al., 2013), with DMXAA and CMA showing species specificity for mouse but not human STING. The mode of binding of DMXAA and CMA to mSting is of interest for structure-based design of agonists and antagonists of hSTING with value as anticancer/antiviral vaccine adjuvants and anti-inflammatory compounds, respectively.
Figure 5. Crystal Structure of DMXAA Ligand Bound to mStingR231.
(A) Chemical formula of dimethylxanthenone-4-acetic acid (DMXAA).
(B) The 2.4 Å crystal structure of two molecules of DMXAA bound to mStingR231 (aa 154–340).
(C) Intermolecular contacts in the complex of DMXAA bound to mStingR231. The two bound DMXAA molecules are shown in biscuit color, with individual mSting subunits in the symmetrical dimer shown in magenta and yellow. The intermolecular contacts to the polar and nonpolar edges of the DMXAA by the mSting subunits are shown in two alternate views.
(D) Superposition of the 2.4 Å DMXAA-bound structure of mStingR231 (both subunits in biscuit) and of the 1.77 Å c[G(2′,5′)pA(3′,5′)p]-bound structure of mStingR231 (both subunits in magenta).
(E) Superposition of c[G(2′,5′)pA(3′,5′)p] in magenta and DMXAA in biscuit in their complexes with mStingR231.
For dose dependence: Data points were determined in triplicate and are depicted as means ± SEM.
Relates to Figure S3 and Table S2.
We solved the 2.4 Å crystal structure of DMXAA bound to mStingR231 (aa 154–340) (X-ray statistics in Table S2), with the complex containing two molecules of DMXAA per mStingR231 dimer (Figure 5B). The aromatic rings of the two DMXAA moieties are aligned in parallel but are not stacked on each other. The details of the intermolecular contacts in the complex are shown in Figure 5C, with the ketone groups of DMXAA forming direct hydrogen bonds to the side chain of T266, whereas the carboxylate moieties of the ligand are anchored through direct hydrogen bonds to the side chains of R237 and T262. In addition, the adjacent aromatic methyl groups of DMXAA form a hydrophobic patch with side chains of L169 and I234 of mSting, whereas the nonsubstituted aromatic edges (positions 7 and 8) of DMXAA are positioned opposite I164 (Figure 5C). A four-stranded antiparallel β-pleated sheet forms a cap over the binding pocket indicative of formation of a “closed” conformation on complex formation, consistent also with DMXAA exposure leading to type I IFN pathway activation via mSting (Conlon et al., 2013; Kim et al., 2013).
Finally, the mSting moieties bound to DMXAA (both subunits in biscuit) and to CMA (both subunits in yellow) (PDB: 4JC5) superpose well on each other (rmsd = 0.75 Å) representing the “closed” conformation for both complexes (Figure S3B). By contrast, the mSting moieties bound to DMXAA (both subunits in biscuit) and to c[G(2′,5′)pA(3′,5′)p] (both subunits in magenta) show differences upon superposition despite both adopting the “closed” conformation (rmsd = 2.21 Å) with the separation between the tips of the α2-helices increasing from approx. 42 Å in the former complex to approximately 38 Å in the latter complex (Figure 5D). Superposition of the mStingR231-bound DMXAA (in salmon) and c[G(2′,5′)pA(2′,5′)p] (in magenta) ligands emphasize the different orientations adopted by these bound ligands within the same binding pocket (Figure 5E).
cGAMP Isomers Activate Type I IFN Pathway Through the STING/IRF3 Pathway
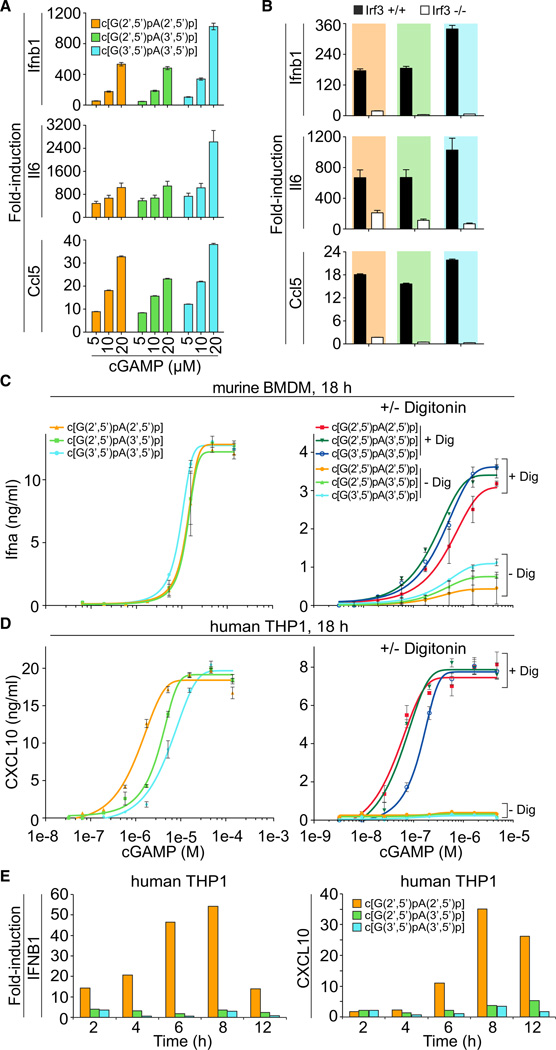
To test whether cGAMP isomers elicit an innate immune response in murine macrophages, we recorded simple dose response of cultured bone-marrow-derived macrophages (BMDMs) by addition of increasing concentrations of c[G(2′,5′) pA(2′,5′ )p], c[G(2′,5′)pA(3′,5′)p] and c[G(3′,5′)pA(3′,5′)p] to the medium, and assaying 4 hr after treatment. Using real-time reverse transcription PCR (qPCR) analysis to monitor induction of Ifnb1, Il6, and Ccl5, we determined that exposure of BMDMs to 20 µM c[G(3′,5′)pA(3′,5′)p] elicited the strongest induction among the three isomers, with Ifnb1, Il6, and Ccl5 mRNA levels increased by 1024-, 2624-, and 38-fold, respectively, relative to mock treatment (Figure 6A). Although c[G(3′,5′)pA(3′,5′)p] was the most potent compound, the differences between it and the other two isomers did not exceed 3-fold. BMDMs isolated from _Irf3_−/− mice had reduced induction of Ifnb1, Il6, and Ccl5 upon exposure to cGAMP isomers, thereby indicating the involvement of Irf3-dependent type I IFN response pathway (Figure 6B).
Figure 6. cGAMP Stimulation of the IFN Pathway in Mouse and Human Cells.
(A) BMDMs (1 × 106) from C57B/6 mice were treated with increasing concentrations (5, 10, and 20 µM) of cGAMP linkage isomers, c[G(2′,5′)pA(2′,5′)p], c[G(2′,5′)pA(3′,5′ )p], and c[G(3′,5′)pA(3′ ,5′)p], and cells were collected at 4 hr after treatment. cGAMP linkage isomers were provided by addition into media. Mock treatment control was included. qPCR analyses of Ifnb1, Il6, and Ccl5 mRNAs were performed.
(B) BMDMs from _Irf3_–/– and age-matched wild-type control mice were generated. Cells (1 × 106) were treated with cGAMP linkage isomers, c[G(2′,5′) pA(2′,5′)p], c[G(2′,5′)pA(3′,5′)p], and c[G(3′,5′) pA(3′,5′)p] at a final concentration of 15 µM. cGAMP linkage isomers were provided by addition into media. qPCR analysis of Ifnb1, Il6, and Ccl5 mRNAs were performed.
(C) Murine BMDMs were incubated in media supplemented with indicated concentrations of cGAMP isomers for 18 hr (left) or for 30 min with and without Digitonin-mediated permeabilization (right). Eighteen hours later, IFN-α concentrations in the supernatant were determined by ELISA.
(D) THP1 cells were incubated in media supplemented with indicated concentrations of cGAMP linkage isomers for 18 hr (left) or for 30 min with and without Digitonin-mediated permeabilization (right). CXCL10 concentrations were determined by ELISA after 18 hr.
(E) Time course of STING-dependent IFN pathway activation by cGAMP linkage isomers. THP1 cells were incubated in media supplemented with cGAMP linkage isomers from 0 to 12 hr without permeabilization. IFNB1 and CXCL10 transcriptional activation was measured by RT-PCR, normalized against TUBA1B and vehicle control.
Data points in (A) and (B) were determined in triplicate and are depicted as the mean ± SEM. A representative of two independently performed experiments is shown. For bar graphs, data points were determined in triplicate and are depicted as means ± SEM. Data points in (C) and (D) were determined in triplicate and are depicted as the mean ± SEM. Related to Figure S4 and Table S5.
Western blot analysis further demonstrated that cGAMP isomers induced phosphorylation of Tbk1 and Irf3 at 2 and 4 hr after treatment of BMDMs (Figure S4). Consistent with our qPCR results, incubation of BMDMs with c[G(3′,5′)pA(3′,5′)p] also showed the highest levels of Tbk1 and Irf3 protein phosphorylation compared to the two other isomers (Figure S4). To test whether mSting is required for cGAMP-induced Tbk1 and Irf3 phosphorylation, we compared type I IFN response in BMDMs derived from wild-type and Goldenticket (Gt) mutant mice (StingGt/Gt), which carry a I199R missense mutation in exon 6 of the mSting gene resulting in no detectable protein by western blot analysis of BMDMs (Rasmussen et al., 2011). We observed that cGAMP-induced Tbk1 and Irf3 phosphorylation was absent from mSting-deficient cells (Figure S4). We conclude that cGAMP treatment of BMDMs triggers type I IFN and proinflammatory cytokine/chemokine via the mSting/Irf3-dependent pathway. The strongest responses for mSting were seen for cGAMP derivatives comprising the nonmetazoan all-3′,5′-linkage isomer produced by bacteria, which was unexpected, considering the recent report suggesting that the natural c[G(2′,5′)pA(3′,5′)p] was the highest-affinity ligand for hSTING (Zhang et al., 2013).
Therefore, we also evaluated hSTING activation and its dependence on cGAMP isomer concentration. We assayed type I IFN and chemokine production in human THP1 cells by ELISA and RT-PCR analysis delivering cGAMP isomers by addition to the medium without or with Digitonin (Dig) permeabilization. In comparison to the murine system, which slightly favored the bacterial c[G(3′,5′)pA(3′,5′)p], the nonnatural c[G(2′,5′) pA(2′,5′)p] stimulated THP1 cells 2.9- or 4.8-fold more effectively than the metazoan second messenger c[G(2′,5′)pA(3′,5′)p] or c[G(3′,5′)pA(3′,5′)p], respectively (Figure 6C and D, left, Table S5). Measuring transcriptional activation of hSTING-dependent IFN response genes by the various cGAMP linkage isomers (Figure 6E) confirmed the ELISA results. As early as 2 hr, we detected by RT-PCR that c[G(2′,5′)pA(2′,5′)p] stimulated expression of IFNB1 and CXCL10 the most. Digitonin-mediated cell permeabilization significantly increased and enhanced sensitivity to all cGAMP isomers in BMDM and THP1 cells (Figures 6C and 6D, right, Table S5). In THP1 cells, there was no difference in the EC50 values obtained for c[G(2′,5′)pA(2′,5′)p] and c[G(2′,5′)pA(3′,5′)p] exposure.
Mutagenesis of hSTING and mSting Identified Amino Acids Critical for Its Ligand-Binding-Induced IFN Pathway Activation
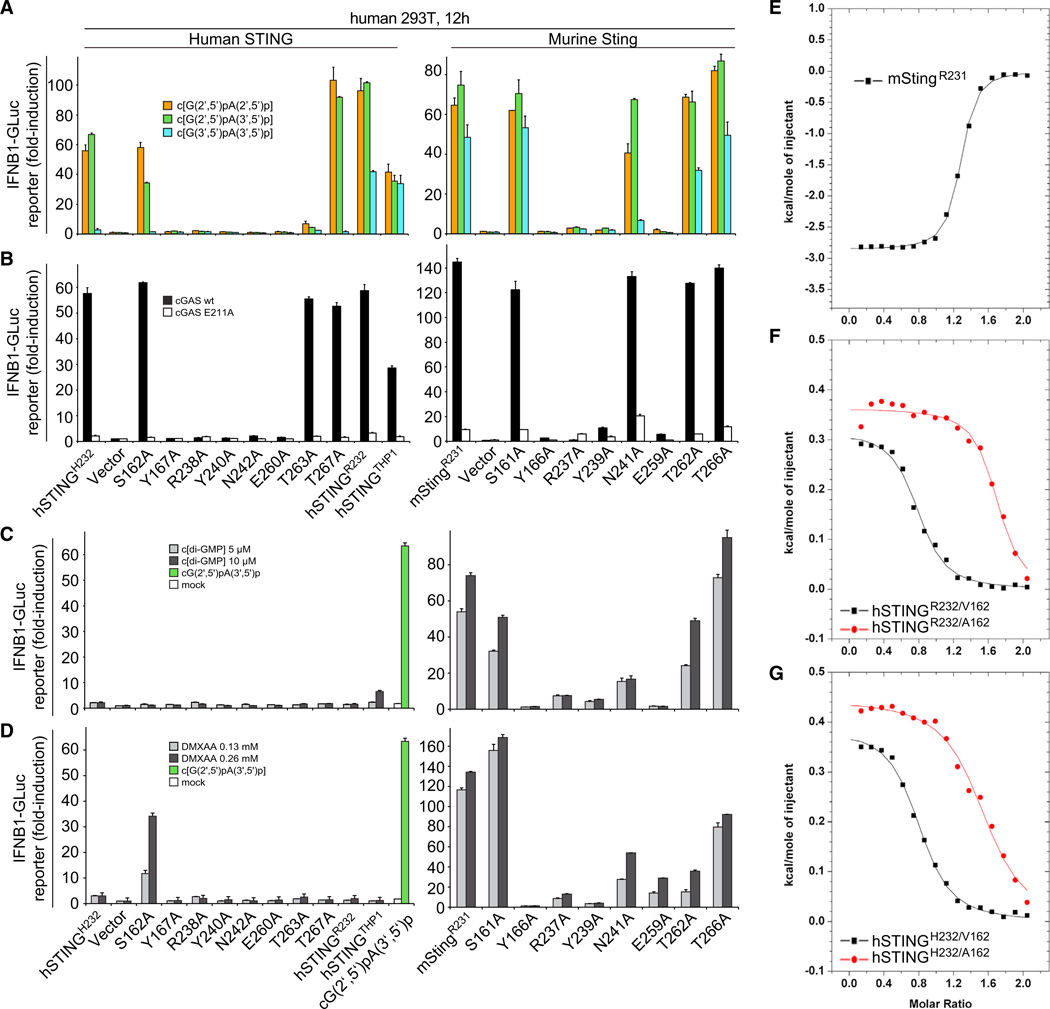
To determine the functional importance of individual amino acids that interacted with c[G(2′,5′)pA(3′,5′)p] in the crystal structure, we generated Ala point mutants of specific residues within hSTINGH232, as well as of the corresponding mutations in mStingR231, and tested their activities in human HEK293T cells using an IFN-sensitive luciferase reporter assay (Burdette et al., 2011; Gao et al., 2013). STING expression plasmids and IFNB1 luciferase reporter constructs were cotransfected followed by Dig-mediated delivery of cGAMP isomers (Figure 7A), or cotransfected with wild-type or catalytic mutant (E211A) cGAS in the absence of exogenous cGAMP addition (Figure 7B) (Gao et al., 2013). Human mutant STING variants with amino acid substitutions of either Y167A, R238A, Y240A, or E260A, as well as their murine counterparts, were no longer able to stimulate the IFN pathway. Notably, hSTINGH232/N242A induced no measurable IFN reporter activity, whereas mStingR231/N241A was only slightly impaired in its response. Mutants S162A and T267A of hSTINGH232 largely retained their normal function, whereas hSTINGH232/T263A showed reduced activation (Figure 7A). Dose-response studies of hSTINGH232 (Figure S5) revealed no significant differences in stimulation between c[G(2′,5′)pA(2′,5′)p] and c[G(2′,5′)pA(3′,5′)p] and a lesser response to c[G(3′ ,5′)pA(3′,5′)p]. However, the hSTINGH232/T263A mutant shows a gain in sensitivity to c[G(3′,5′)pA(3′,5′)p] compared to its response to 2′ ,5′ linkage isomers. The S162A and T267A hSTINGH232 mutants respond similarly to hSTINGH232, also showing a weaker response to c[G(3′,5′)pA(3′ ,5′ )p].
Figure 7. Mouse and Human STING Mutational Analysis.
(A) HEK293T cells were transfected with reporter constructs and human or murine STING expression plasmids as indicated. After 12 hr, cells were digitonin permeabilized to deliver cGAMP linkage isomers (5 µM concentration, 30 min permeabilization) and incubated for an additional 12 hr, followed by luciferase-reporter assay.
(B) To gauge STING mutant stimulation by murine cGAS compared to the inactive cGAS mutant E211A, plasmids containing the indicated human or murine STING variants were cotransfected with either cGAS form and luciferase reporter constructs. Luciferase induction was determined after 30 hr. In this setting, the transfected plasmids provide the dsDNA stimulus for cGAS activation. Activation is expressed as fold induction in relation to control plasmid pMAX-GFP.
(C) HEK293T cells were transfected as in (A) and stimulated with c[di-GMP](5 and 10 µM) following digitonin permeabilization. Luciferase activity was determined 12 hr after stimulation. As negative and positive controls, HEK293T cells transfected with hSTINGH232 were mock-treated (white bar) or stimulated with 5 µM c[G(2′,5′)pA(3′ ,5′)p] following digitonin permeabilization (green bar), respectively.
(D) HEK293T cells were transfected as in (C) and after 12 hr stimulated with medium containing DMXAA (136 and 266 µM). Luciferase activity was measured after additional 12 hr.
(E) ITC binding curves for complex formation between DMXAA and mStingR231.
(F) ITC binding curves for complex formation between DMXAA and hSTINGR232/A162 and hSTINGR232/V162.
(G) ITC binding curves for complex formation between DMXAA and hSTINGH232/A162 and hSTINGH232/V162.
Data points in (A to D) were determined in triplicate and are depicted as the mean ± SEM. We have no explanation at this time for high-stoichiometry N values (∼1.5, instead of the expected 1) for the binding curves for A162 mutants of both hSTINGH232 and hSTINGR232 (F and G).
Related to Figures S6 and S7, and Table S6.
Given the recent report of mSting and hSTING variants with distinct responsiveness (Diner et al., 2013), we sequenced hSTING DNAs derived from eight Caucasians and our THP1 cells. All samples encoded R232 in both alleles; additionally hSTINGTHP1 contained three additional point mutations (H72R, G230A, and R293Q) as reported before. We compared the cGAMP-dependent IFN-response for hSTINGH232, hSTINGR232, hSTINGTHP1, and mStingR231, and found that the prevalent hSTINGR232 was several-fold more responsive to all cGAMP isomers when compared to hSTINGH232, with the order of cGAMP isomer sensitivity remaining the same (Figures 7, S5E, and Figure S5F). hSTINGTHP1 and wild-type mStingR231 displayed slightly reduced overall cGAMP sensitivity compared to hSTINGR232.
To complement our mutagenesis study of the cGAMP isomer response, we tested c[di-GMP]and DMXAA (Figures 7C and 7D). Although both ligands stimulated mSting, nearly all hSTING variants were nonresponsive except for hSTINGR232 and hSTINGTHP-1, which showed a residual response to high concentrations of c[di-GMP] (Figures S6B and 7C, respectively). Moreover, mSting mutants defective for recognition of all cGAMP isomers also failed to recognize c[di-GMP] and DMXAA. Mutants N241A and T262A of mSting were less responsive to c[di-GMP] and DMXAA compared to cGAMPs, whereas mSting (T266A) showed a moderately enhanced recognition of c[di-GMP] over DMXAA.
S162A Mutant of hSTING is Activated by DMXAA
Strikingly, we found that the single point mutant S162A enabled recognition of DMXAA by human STING (Figure 7D, left), while not restoring c[di-GMP] responsiveness (Figure 7C, left). In both hSTINGH232 and hSTINGR232 background, the S162A mutation enabled DMXAA recognition with near-identical dose responses (Figures S6C and S6D). In contrast, the differences in recognition of cGAMP isomers and c[di-GMP] were only slightly altered or unchanged compared to the respective S162 variants (Figure S7B). Noticeably, IFN induction was not observed in cellular assays for the S162V and S162I mutants of hSTINGH232 or hSTINGR132, which contain bulkier hydrophobic amino acids compared to Ala at position 162 (Figure S7A).
ITC-based binding studies confirmed that mStingR231-bound DMXAA as reported previously (Figure S7E) (Kim et al., 2013), as did S162A (and S162V) mutants in a hSTINGR232 and hSTINGH232 context-bound DMXAA (Figures 7F and 7G).
DISCUSSION
Recent Structural Studies Established that hSTINGR232 Adopts a “Closed” Conformation in the c[G(2′,5′)pA(3′,5′) p]-Bound State
Recent structural studies established that hSTINGR232 adopts a “closed” conformation upon association with the cGAS-naturally-produced c[G(2′,5′)pA(3′,5′)p] linkage isomer (Zhang et al., 2013). The structural transition from the ‘‘open’’ conformation of the free state of STING (Yin et al., 2012; Ouyang et al., 2012; Shang et al., 2012; Shu et al., 2012) to the “closed” ligand-bound state (Zhang et al., 2013) was manifested in a 22° inward-shift of the pair of symmetrically-related α2-helices of STING and concomitant formation of the four-stranded β sheet cap associated with the “open” to “closed” transition.
STING Complexes with cGAMP Linkage Isomers Adopt a “Closed” Conformation Independent of Allelic Variation at Position 232 of hSTING and Its mSting Equivalent
We observe the same “closed” structures (Figure 3C) for the complexes of c[G(2′,5′)pA(3′,5′)p] bound to hSTINGH232 (Figure 1A) and mStingR231 (Figure 3A), independent of whether a His or Arg occupies this equivalent position. Furthermore, hSTINGH232 adopts the ‘‘closed’’ conformation when in complex with the all-2′,5′-linkage isomer (Figure 3E), as does mStingR231 when in complex with the all-3′,5′-linkage isomer (Figure 3F), indicating that the ligand-dependent conformational transition is not specific to any cGAMP linkage isomer, but distinct from most of the described complexes involving c[di-GMP] and hSTINGH232. Importantly, the ‘‘closed’’ conformation of STING in complex with all cGAMP linkage isomers identically repositions other elements on the outer surface of the STING dimer, such as the α5-helices and b-barrel elements (stereo view in Figure 2D), potentially altering protein-interaction surfaces for potential recognition by regulatory or effector proteins.
Proteins Encoded by Natural hSTING and mSting Alleles Exhibit Micromolar to Submicromolar Binding Affinities for cGAMP Linkage Isomers
Our ITC studies of c[G(2′,5′)pA(3′,5′)p] and its linkage isomers showed no significant differences in their binding affinities with recombinant proteins representing natural alleles of hSTING and mSting (Figures 4A and 4C–4F, and Table S3). The KD values for the cGAMP linkage isomers vary between 2.5 and 5.4 µM for the complexes with hSTINGH232 and between 0.1 and 0.3 µM for all complexes with hSTINGR232 or mStingR231 (Table S3). The difference in the binding affinities of individual STING protein alleles between all cGAMP linkage isomers were largely similar, and even where different, did not exceed 3-fold. The subtle differences between cGAMP isomers in binding and activating STING are recapitulated in our cellular assays of IFN response (Figures 6, 7, and Figure S5), as well as other recently related functional assays (Ablasser et al., 2013; Zhang et al., 2013).
The Contribution of hSTING Positions 230 and 232 to cGAMP Linkage Isomer Recognition and Type I IFN Pathway Activation
Amino acid variation of hSTING at positions 230 and 232 was reported to affect its sensitivity particularly toward c[di-GMP] (Diner et al., 2013). Indeed, c[di-GMP] stimulation of HEK293T cells transfected with hSTINGTHP1, which encodes for A230 and R232, showed reduced IFN-pathway activation (Figure 7C), while retaining response for all cGAMP isomers (Figure 7A). Interestingly, hSTINGR232 or hSTINGH232 preferentially activated with 2′,5′-linkage-containing cGAMP isomers, whereas hSTINGTHP1 responded equally well to all linkage isomers under digitonin-permeabilized conditions (Figure 7A, left). Comparison of the IFN-responses of hSTINGH232 with hSTINGR232, and hSTINGR232 with hSTINGTHP1, suggests that R232 and G230 are critical for optimal response to 2′,5′-linkage-containing natural or unnatural cGAMP isomers. hSTINGH232 was also reported to phenocopy the behavior of mStingR231A regarding its higher sensitivity to 2′,5′-containing cGAMP molecules versus all 3′,5′-cGAMP and c[di-GMP] (Diner et al., 2013). It is interesting to note that G230 is one of the few unconserved residues between human and mouse; the corresponding amino acid in mSting is an isoleucine. This difference might explain why there appears to be no preference for any of the cGAMP linkage isomers or c[di-GMP] with wild-type mStingR231. Position 230 is part of the β sheet cap of the “closed” conformation and side-chains such as Ala or Ile may subtly change its stability or degree of closure. However, molecular modeling of an Ala or Ile substitution for G230 does not indicate any profound structural changes. Thus it remains unclear whether amino acid variation at position 230 affects ligand binding, per se, or STING activation.
Impact of Amino Acid Mutations Surrounding c[G(2′,5′) pA(3′,5′)p] in the “Closed” Conformation of the STING Complex
The ITC binding studies (Figure 4B and Table S4) and IFN functional assays (Figures 7A and 7B) confirmed that Ala substitution of structurally identified amino acids that bracket (Y167), and buttress (Y240) target base edges and backbone phosphates (R238), and recognize guanosine base edges through direct and water-mediated hydrogen bonds (N242 and E260) of the bound c[G(2′,5′)pA(3′,5′)p] abolished ligand binding in vitro and the cellular IFN response. Polar side chains that interact with the sugar-phosphate backbone of the ring scaffold of c[G(2′,5′)pA(3′,5′)p] in its complex with hSTINGH232 exhibit a more nuanced effect in both ITC binding studies (Figure 4B and Table S4) and cellular assays (Figures 7A and 7B). The T267A mutation, with the potential to disrupt water-mediated hydrogen bonding to the backbone phosphates of the ligand (Figure 1G), did not alter in vitro binding affinity (Table S4), while the S162A mutation, with the potential to disrupt the direct hydrogen bond to the 3′-OH of the guanosine of the bound c[G(2′,5′)pA(3′,5′)p] (Figure 1G), resulted in a modest 2-fold reduction of in vitro binding affinity (Table S4). By contrast, the T263A mutation, with the potential to disrupt the direct hydrogen bond to the NH2 group of guanosine of the bound ligand (Figure 1F), resulted in a 10-fold reduction of in vitro binding affinity (Table S4). These data are in agreement with results of cellular assays measuring IFN response (Figures 7A and 7B and S5).
Perturbation of Structurally Identified Amino Acids Coordinating cGAMP Isomers in hSTING Changed Its Specificity and Sensitivity to Cyclic Dinucleotides
The crystal structure of c[G(2′,5′)pA(3′,5′ )p] bound by hSTINGH232 allowed us to identify and functionally confirm the importance of specific amino acids for recognition of the cGAMP isomers. Systematic substitution of Ala at positions Y167, R238, Y240, H242, E260, and T263 of hSTING reduced or abolished cGAMP-isomer-dependent IFN-pathway activation, whereas substitutions at positions S162 and T267 did not substantially affect recognition of c[G(2′,5′)pA(3′ ,5′)p] or the synthetic all-2′,5′-cGAMP isomer (Figure 7A, left, S5B, and S5D). These results were in a hSTINGH232 background that already exhibited a reduced affinity for c[G(3′,5′)pA(3′,5′)p]. However, we also performed parallel experiments using mSTINGR231 that exhibited approximately equivalent response to all cGAMP linkage isomers (Figures 7A, right and S5E). Nearly all amino acid substitutions that attenuated hSTING activity similarly eliminated or reduced induction of the IFN pathway by the corresponding murine mutants in a mStingR231 background. Only mSTINGR231,N241A led to IFN pathway activation but only with cGAMP linkage isomers containing at least one 2′,5′ linkage. Taken together, amino acid variation within the binding pocket of STING leads to functional changes in the recognition of, and response to, exposure to cGAMP linkage isomers. We were able to identify mutations that can reduce or enhance cGAMP-linkage isomer recognition and also distinguish between 2′,5′-linkage-containing cGAMP isomers and the all-3′,5′-cGAMP. However, we could not identify any STING variant, natural or mutant, that could distinguish between c[G(2′,5′)pA(2′,5′)p] or the natural c[G(2′,5′)pA(3′,5′)p] linkage isomer.
Contrasting Positions on Activation of hSTING by 2′,5′-Containing Linkage Isomers of Cgamp
It has been recently proposed by the Z.J. Chen laboratory that c[G(2′,5′)pA(3′,5′)p] is the endogenous high-affinity ligand of STING (Zhang et al., 2013), a conclusion based on three lines of experimental information that are contrasted below in light of data presented in our paper.
First, the Chen laboratory solved the X-ray structure of the c[G(2′,5′)pA(3′,5′)p]-hSTINGR232 (aa 138–379) complex and demonstrated that hSTINGR232 underwent an “open” to “closed” conformational transition that was accompanied by formation of a β sheet lid over the binding pocket (Zhang et al., 2013). They interpreted this transition to be reflective of c[G(2′,5′) pA(3′,5′)p] being a high-affinity ligand of hSTING because the majority of complexes of hSTING with c[di-GMP] retained the “open” conformation. Our position based on structural studies of a series of complexes that include c[G(2′,5′)pA(3′,5′)p]-hSTINGH232 (Figure 1A), c[G(2′,5′)pA(3′,5′)p]-mSTINGR231 (Figure 3A), c[G(2′,5′)pA(2′,5′)p]-hSTINGH232 (Figure 3E), and c[G(3′,5′)pA(3′,5′)p]-mSTINGR231 (Figure 3F) is that all above complexes form “closed” structures and that the observation of an “open” to “closed” transition by itself is not necessarily an indicator for identification of a high-affinity ligand of STING.
Second, there are pronounced differences in the ITC-based binding affinities reported by the Chen and our laboratories on complex formation between 2′,5′-linked isomers of cGAMP and hSTING. Thus, the Chen laboratory reported KD values of 3.8 nM for the c[G(2′,5′)pA(3′,5′)p]-hSTINGR232 and 0.28 µM for the c[G(2′ ,5′)pA(2′,5′)p]-hSTINGR232 complexes, a difference approaching two orders of magnitude between these two cGAMP linkage isomers and was interpreted as the strongest evidence for justification of c[G(2′,5′)pA(3′ ,5′)p] being a high-affinity ligand of hSTING (Zhanget al., 2013).By contrast, ITC measurements in our laboratory gave values that differed by less than a factor of two as reflected in KD values of 0.11 µM for the c[G(2′,5′) pA(3′,5′)p]-hSTINGH232 and 0.17 µM for the c[G(2′,5′)pA(2′,5′)p]-hSTINGH232 (Figure 4C and Table S2).
Third, the Chen laboratory found similar EC50 values in their cellular assays of IFN response for binding of hSTINGR232 to the various cGAMP linkage isomers, as we did in our cellular assay measurements, as well as those reported recently from the Hornung laboratory (Ablasser et al., 2013).
Our position is that the available structural data, our ITC-based binding parameters and cellular assays from the Chen, Hornung, and our laboratories are consistent with the 2′,5′ cGAMP linkage isomers c[G(2′,5′ )pA(3′,5′)p] and c[G(2′,5′)pA(2′,5′)p] as equally specific triggers of the hSTING-mediated IFN pathway, and modestly more effective compared to their c[G(3′,5′ )pA(3′,5′)p] linkage isomer counterpart.
Recognition of Antiviral Small Molecules by mStingR231 and the Creation of a hSTING Functional Mutant Sensitive to DMXAA Activation
Our structural study of the DMXAA-mStingR231 complex (Figure 5A) as well as the earlier report on the structure of the CMA-mStingR231 complex (Cavlar et al., 2013) contain two antiviral small molecules bound within the cyclic dinucleotide cleft of mSting. The polar atoms of DMXAA form a network of intermolecular hydrogen bonds involving charged (R237) and polar (S161, T262 and T266) amino acids of mStingR231, while its methyl groups form intermolecular contacts with hydrophobic amino acids (L169 and I234) (Figure 5C), thereby anchoring the antiviral small molecule within the binding cleft. Importantly, a subset of the same conserved amino acid side chains lining the dimeric binding pocket in mStingR231 target a pair of bound DMXAA ligands and bound c[G(2′,5′)pA(3′,5′)p] based on distinctly different intermolecular contacts (Figures 5C and 1E–1G).
The ligand-binding pocket of mStingR231 adopts a “closed” conformation in both the DMXAA (Figure 5B) and c[G(2′,5′) pA(3′,5′)p] (Figure 3A) complexes, though the pocket is more V-shaped in the DMXAA complex (Figure 5D). The more V-shaped “closed” conformation adopted by mStingR231 complex with DMXAA is identical to the one observed for the CMA complex (Figure S3B).
We were surprised to observe that the S162A mutation rendered both hSTINGH232 and hSTINGR232 sensitive to DMXAA binding and activation (Figures 7D and S6). The side chains of S161 in mStingR231 are positioned in the plane and opposite an aromatic edge of DMXAA (Figure 5C) and one would predict that replacement of Ser by Ala in hSTING could favor complex formation due to either enhancement of intermolecular hydrophobic interactions and/or steric constraints. No activation of the IFN response was observed for the S162V and S162I mutations in a hSTINGH232 and hSTINGR232 context (Figure S7A), implying the predominance of the steric constraint contribution. Consistent with this hypothesis, we found that mStingS161A showed an increased response to DMXAA but not to c[G(2′,5′) pA(3′,5′)p] or c[di-GMP] (Figures 7C and 7D, right).
Importantly, the increased IFN response of hSTINGA162 to DMXAA observed in cellular assays (Figure 7D) was confirmed by ITC-based studies, which established binding of DMXAA to S162A (and S162V) mutants in a STINGR232 (Figure 7F) and hSTINGH232 (Figure 7G) context (thermodynamic parameters in Table S6).
CONCLUSIONS
In summary, we provide a comprehensive structural, biophysical, and functional analysis of cGAMP isomer association with hSTING and mSting. Our data are supportive of a preference of hSTING for 2′,5′-linkage-containing cGAMPs that are modestly more effective compared to their c[G(3′,5′) pA(3′,5′)p] linkage isomer counterpart (see also Ablasser et al., 2013).
Our structural and functional results highlight the importance of S162 of hSTING to DMXAA insensitivity, which should potentially enable rational drug design of DMXAA derivatives for the development of human antitumor, antiviral, and vaccine adjuvant applications.
EXPERIMENTAL PROCEDURES
Crystallization and Structure Determination
Crystals were grown using the sitting drop vapor diffusion method and diffraction data collected at synchrotron beam lines. All structures were solved using PHASER, COOT, and REFMAC programs.
Isothermal Titration Calorimetry
The thermodynamic parameters of binding reactions of STING with cGAMP isomers and DMXAA were measured by isothermal titration calorimetry using a MicroCal ITC200 calorimeter at 25°C.
Generation of Bone-Marrow-Derived Macrophages
Female _Irf3_–/–, StingGt/Gt and WT C57B/6 mice were used for the preparation of bone-marrow-derived macrophages. These mice were maintained in the animal facility at the Sloan-Kettering Cancer Institute. Bone marrow cells were cultured in complete medium (CM) in the presence of 5% of supernatant of L929 mouse fibroblasts as conditioned medium. Cells were plated into 6-well plate (1 million cells per well) at day 7, the day before stimulation.
RT-PCR analysis of THP1 Cells
In RT-PCR analyses, 5 × 105 THP1 cells were plated in 12-well dishes and incubated overnight. 12.5 mM of cGAMP isomers were applied to the media and cells were harvested at indicated times. RNA samples were isolated and cDNA libraries were generated. KOD Polymerase was used to PCR amplify regions of IFNB1 and CXCL10 and normalized against TUBA1B.
cGAMP Stimulation of Cells
Bone-marrow-derived murine macrophages and THP-1 cells were stimulated by incubation with cGAMP isomers at indicated concentrations for 18 hr, or by Digitonin permeabilization (30 min). Cytokines in supernatants were determined after 18 hr by ELISA.
Luciferase Assay
HEK293T cells were reverse-transfected with STING expression plasmids and reporter constructs. 12 hr later, DMXAA was added directly, whereas cGAMP isomers and c[di-GMP] were delivered with digitonin permeabilization. Luciferase expression was determined after another 12 hr, or 30 hr when transfected together with a cGAS-expression plasmid. See Extended Experimental Procedures for full details of all above sections.
Supplementary Material
01
ACKNOWLEDGMENTS
We thank the synchrotron beam line staffs at the Brookhaven and Argonne National laboratories for their assistance and Kenneth Breslauer for discussions related to interpretation of ITC experiments. D.J.P is supported by grants from the Abby Rockefeller Mauze Trust and the Maloris and STARR Foundations. T.T. is supported by HHMI. W.B. and G.H. are members of the DFG Excellence Cluster ImmunoSensation. W.B. is supported by DFG grant BA3544/1-1, G.H. by SFB670, and W.B. and G.H. by SFB704. R.J. is supported by NIH grant GM79760, L.D. is supported by NIH grant R56AI095692 and S.S. is supported by NIH grant GM42498. The structural research was performed by P.G. under the supervision of D.J.P., the RT-PCR measurements involving transcriptional activation were performed by M.A. and A.A.S. under the supervision of T.T., the assays describing cGAMP stimulation of BMDM were performed by W.W. and P.D. under the supervision of L.D. and S.S., the cellular assays monitoring STING-dependent interferon induction were performed by T.Z. under the supervision of W.B. and G.H., and chemical synthesis of cGAMP linkage isomers was performed by B.L.G. under the supervision of R.A.J. All participants participated in the writing of the paper and agree with the contents.
Footnotes
ACCESSION NUMBERS
The following coordinates have been deposited in the RCSB Protein Data Bank: c[G(2′,5′)pA(3′,5′)p]-hSTINGH232 complex (4LOH); c[G(2′,5′)pA(2′,5′)p]-hSTINGH232 complex (4LOI); c[G(2′,5′)pA(3′,5′)p]-mStingR231 complex (4LOJ); c[G(3′,5′)pA(3′,5′)p]-mStingR231 complex (4LOK); DMXAA-mStingR231 complex (4LOL).
REFERENCES
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Röhl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguley BC, Ching LM. DMXAA: an antivascular agent with multiple host responses. Int. J. Radiat. Oncol. Biol. Phys. 2002;54:1503–1511. doi: 10.1016/s0360-3016(02)03920-2. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013;32:1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon J, Burdette DL, Sharma S, Bhat N, Thompson M, Jiang Z, Rathinam VA, Monks B, Jin T, Xiao TS, et al. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J. Immunol. 2013;190:5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Jr, Barber GN, Stetson DB. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat. Struct. Mol. Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating SE, Baran M, Bowie AG. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32:574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Kim S, Li L, Maliga Z, Yin Q, Wu H, Mitchison TJ. Anticancer flavonoids are mouse-selective STING agonists. ACS Chem. Biol. 2013 doi: 10.1021/cb400264n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara PN, Jr, Douillard JY, Nakagawa K, von Pawel J, McKeage MJ, Albert I, Losonczy G, Reck M, Heo DS, Fan X, et al. Randomized phase III placebo-controlled trial of carboplatin and paclitaxel with or without the vascular disrupting agent vadimezan (ASA404) in advanced non-small-cell lung cancer. J. Clin. Oncol. 2011;29:2965–2971. doi: 10.1200/JCO.2011.35.0660. [DOI] [PubMed] [Google Scholar]
- Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prantner D, Perkins DJ, Lai W, Williams MS, Sharma S, Fitzgerald KA, Vogel SN. 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) activates stimulator of interferon gene (STING)-dependent innate immune pathways and is regulated by mitochondrial membrane potential. J. Biol. Chem. 2012;287:39776–39788. doi: 10.1074/jbc.M112.382986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SB, Horan KA, Holm CK, Stranks AJ, Mettenleiter TC, Simon AK, Jensen SB, Rixon FJ, He B, Paludan SR. Activation of autophagy by α-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J. Immunol. 2011;187:5268–5276. doi: 10.4049/jimmunol.1100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, Gu L. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat. Struct. Mol. Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High-Affinity Ligand for STING. Mol. Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01