Microbiota in Allergy and Asthma and the Emerging Relationship with the Gut Microbiome (original) (raw)
. Author manuscript; available in PMC: 2016 May 13.
Published in final edited form as: Cell Host Microbe. 2015 May 13;17(5):592–602. doi: 10.1016/j.chom.2015.04.007
Abstract
Asthma and atopy, classically associated with hyper-activation of the T helper 2 (Th2) arm of adaptive immunity, are amongst the most common chronic illnesses worldwide. Emerging evidence relates atopy and asthma to the composition and function of the human microbiome, the collection of microbes that reside in and on and interact with the human body. The ability to interrogate microbial ecology of the human host is due in large part to recent technological developments that permit identification of microbes and their products using culture-independent molecular detection techniques. In this review we explore the roles of respiratory, gut and environmental microbiomes in asthma and allergic disease development, manifestation and attenuation. Though still a relatively nascent field of research, evidence to date suggests that the airway and/or gut microbiome may represent fertile targets for prevention or management of allergic asthma and other diseases in which adaptive immune dysfunction is a prominent feature.
Keywords: Microbiome, Th2, Allergy, Asthma, T-regulatory cells
The Human Microbiome
The field of microbiology owes its genesis to observations made by Antonie van Leeuwenhoek, a Dutch microscope lens maker, who, in 1676, first described microscopic “animalcules” in dental plaque (Porter, 1976). For much of the ensuing history of the field, research has largely focused on culture-based studies of individual microbial species, with an emphasis on understanding the basis of microbial pathogenesis. However, over the past 30 years, the sub-specialty of microbial ecology has driven development of molecular methods for microbial detection, that obviate the necessity for microbial culture. These efforts have revealed the existence of a vast diversity of microbes that inhabit natural systems and demonstrated that these organisms rarely exist in isolation, but instead occur in multi-species, multi-functional communities.
Pioneered by Dr. Carl Woese, the use of molecular techniques to classify microbes without recourse to laboratory culture-based approaches, led to the subsequent development of the 16S rRNA gene as a widely used bacterial biomarker for profiling bacteria present in a mixed species community. This gene, which is exclusive to and ubiquitous amongst bacteria (Fox et al., 1977; Winker and Woese, 1991; Woese, 1987), is a useful molecular target for bacterial identification due to the presence of regions of sequence that are highly conserved across the majority of known bacteria. These conserved sequences flank hyper-variable regions, reflective of varying evolutionary rates across bacterial species. The conserved 16S rRNA sequence regions permit design of PCR primers and amplification of the gene (or a portion thereof) from the majority of known bacterial species. The amplified hyper-variable region is typically sequenced to permit classification of amplicons into discrete taxonomic groups, generating a fingerprint of bacterial phylotypes within a given community. Application of ecological statistics and theory to these profiles allows for analyses and interpretation of the composition of microbial communities. Distinct biomarkers e.g. the Internal transcribed spacer region 2 (ITS2) of the fungal rRNA gene are used for mycological community profiling, which is performed in a similar manner. More recent advances and declining costs associated with in high-throughput sequencing platforms have permitted more expansive microbial biomarker sequencing efforts, leading to identification of novel microbial species, expansion of the known microbial tree of life and an enhanced appreciation of the diversity of microbes that inhabit natural systems. This increase in sequence-capacity has also led to the development of complementary microbiome analyses tools, including shotgun sequencing approaches to determine the functional genes encoded or expressed by a microbial community (Metagenomics or Metatranscriptomics respectively; Fig. 1). Parallel advances in mass spectroscopy platforms have led to improvements in methods for detecting microbial products, including the dominant proteins (Metaproteomics) or metabolites (Metabolomics) collectively produced by microbial members within a community (Fig. 1). As the field continues to rapidly evolve, efforts are becoming more focused on integration of these approaches to produce a more comprehensive view of microbial community composition and function.
Fig. 1. Molecular tools for interrogating microbiome composition and function.

DNA, RNA, Protein and Metabolite fractions of the samples may be interrogated using next-generation sequencing and mass spectroscopy platforms to assess microbiome composition and function.
Though microbial ecology research has traditionally focused on terrestrial and aquatic ecosystems, more recently, predominantly biomarker-based microbial profiling techniques have been applied to the study of the human microbial ecosystem. The results have led to a more comprehensive view of the healthy human body as a series of ecosystems, each with its own particular microbial composition, when considered at the broadest (phylum) level of classification. It has been estimated that microbial cells outnumber human cells by approximately 10-fold (Savage, 1977), and that healthy humans possess from 500–1000 distinct bacterial phylotypes (Claesson et al., 2009; Frank et al., 2007). Though much research has focused on bacterial communities in the human host, fungi, viruses, and archaea have also been detected in these assemblages. The genetic capacity of such a diverse microbial ecosystem is immense; the pan-genome of the healthy human microbiome is estimated to encode approximately one hundred times the number of genes found in the human genome. Though the genomic capacity of the gut microbiota, where much research has focused to date, is predominantly encoded by bacteria, fungal and archaeal genes are also present in this consortium, though their functional contribution to host health status is as yet, largely undefined (Qin et al., 2010).
Microbial members of the human super-organism contribute critical functions to their host, including enzymic digestion of complex carbohydrates; gastrointestinal microbes digest fermentable fiber to produce short-chain fatty acids (SCFAs; Roediger 1980), an essential energy source for the epithelial cell lining of the gut (Kaneko et al. 1994; Flint et al. 2008). Production of SCFAs also acidifies the local gastrointestinal microenvironment, making it less hospitable for colonization or overgrowth of pathogenic species such as Escherichia coli and Salmonella spp. (Cherrington et al., 1991). Commensal microbes also engage in competitive colonization, which provides further protection against pathogen overgrowth. The gut microbiome also biosynthesizes essential vitamins and hormones (Yatsunenko et al., 2012), as well as a range of anti- inflammatory compounds (Herbst et al., 2011; Mazmanian et al., 2008; Round and Mazmanian, 2010; Sokol et al., 2008). Our reliance on microbial function is evident from the critical role microbes play in mammalian development; germ-free mice, are both immunologically and physiologically aberrant, a phenotype that can only be rescued through introduction of commensal bacteria (Atarashi et al., 2011; Smith et al., 2007). Beyond early-life development, evidence has emerged that the microbiome influences the tone of host immune response, particularly that of the adaptive arm. Specific Clostridium species belonging to clade IV or XIVA as well as members of the Bacteroides, exhibit the capacity to induce T-regulatory (T–reg) cells, which are essential for immune tolerance and abrogation of chronic inflammatory or autoimmune disease (Atarashi et al., 2011; Ochoa-Repáraz et al., 2010). In addition murine studies have demonstrated the capacity of specific species in the gut microbiome to induce discrete arms of the adaptive immune response. Black 6 mice whose ileum is overtly colonized by segmented filamentous bacteria (SFB), exhibit robust proliferation of IL-17 and IL-22 expressing T-helper 17 (Th17) cells (Ivanov et al., 2009). This phenotype could be conferred to Th17 deficient mice by co-housing with SFB colonized mice, or by transfer of fecal material from SFB mono-colonized animals. SFB, an unculturable member of the Clostridiales was identified as the species responsible for induction of Th17 cell proliferation through culture-independent microbiome profiling (Ivanov et al., 2009). However, perhaps more importantly, these studies demonstrate a direct role for microbiome members in the promotion of distinct adaptive immune responses associated with protection against or development of a range of chronic inflammatory and autoimmune diseases. These pertinent observations in mice raise the possibility that diseases associated with overactive arms of adaptive immunity, may owe their genesis to microbiome dysbiosis and related dysfunction. It also raises the possibility that diseases currently associated with “inappropriate immune activation”, may in fact represent perfectly appropriate host immune responses to specific pathogenic microbiome compositions and their related activities that promote immune activation.
The Asthmatic Airway Microbiome
Asthma is estimated to affect approximately 300 million individuals worldwide, incurs significant health care expenditure (Accordini et al., 2013; Barnett and Nurmagambetov, 2011) and is one of the most common chronic diseases. Given increases in disease prevalence over the last several decades, it is predicted that the number of individuals affected worldwide will increase by 100 million people by 2025 (Masoli et al., 2004). In the United States, the risk of developing asthma is highest for children during the period between birth and four years of age (Jackson et al., 2014); disease prevalence is also higher amongst women, families below the poverty line, and people of multiple races compared to other groups (Akinbami et al., 2009). Though risk alleles have been associated with asthma development (reviewed in Ober and Yao 2011), the rapid increase in prevalence over the recent decades, particularly amongst children points to environmental factors playing a key role in disease development (Cookson and Moffatt, 1997). Atopic sensitization (allergy), characterized by elevated levels of total and allergen-specific IgE in the serum, and typically by positive skin-prick tests to at least one of a panel of common allergens, is considered the strongest risk factor for childhood asthma development in westernized nations (Simpson et al., 2010), and its rise is associated with a parallel increase in asthma prevalence (Masoli et al., 2004). More, recently an unsupervised statistical approach to pediatric subject clustering, based on the range and degree of their atopic sensitization, revealed that subjects that were predominantly multi-sensitized to common food and aero-allergens had the highest risk of subsequently developing asthma in childhood (Havstad et al., 2014; Simpson et al., 2010).
Allergic asthma is a heterogeneous and complex disease, clinically defined as reversible airflow obstruction, and immunologically by hyper-activation of the T-helper 2 (Th2) arm of the adaptive immune response, and the over-expression of the pro-inflammatory cytokines IL-4, IL-5 and IL-13, as well eosinophilia and mast cell infiltration of the airways (Adcock et al., 2008). Asthma is difficult to diagnose because of inherent heterogeneity across asthmatic patient populations and the multiple contributory factors such as risk alleles, environmental exposures and lung function (reviewed in Chung et al. 2014). Patients are commonly stratified using a range clinical parameters including bronchial hyper-responsiveness to methacholine (a drug that promotes bronchoconstriction), as well as medication use and frequency of exacerbation. Recently, unsupervised statistics have been used to improve diagnosing asthma and its severity (Chung et al., 2014).
However, emerging evidence indicates that other distinct immune phenotypes exist amongst discrete asthmatic patient sub-populations; recent studies have demonstrated an association between T-helper 1 (Th1) cells specifically in obesity-related (Rastogi et al., 2012) and smoking induced asthma (Tsoumakidou et al., 2007). Additionally, amongst asthmatic populations, a recent study distinguished two patient subgroups, Th2-high and Th2-low, which evenly divided the cohort under study (n=22, n=20 respectively; Woodruff et al., 2009). Th2-high patients exhibited significantly increased expression of three biomarker genes (POSTN, CLCA1 and SERPINB2), and of the Th2 cytokines IL-5, IL-13, but not IL-4. They exhibited increased mucin stores (as indicated by increased MUC5AC and MUC2 expression) and more severe airway hyper-responsiveness, as measured by the provocative concentration of methacholine necessary to reduce the forced exhibitory volume in one second by 20% from baseline, more commonly known as the PC20 test. Th2-low individuals consist of both asthmatic and non-asthmatic subjects and characteristically exhibit low levels of both Th2 cytokines and the three-gene biomarker expression. While Th2-high and Th2-low asthmatics do not differ in disease severity, they react distinctly to inhaled steroids; only Th2-high asthmatics respond to corticosteroids, a common treatment for asthma. These findings clearly require validation in multiple large cohorts, however they suggest that Th2 heterogeneity exists amongst asthmatics and may represent a critical approach to stratifying patients. Given the existing evidence for microbial activation of T-cell subsets in the gastrointestinal tract, the current hypothesis is that the overall heterogeneity in asthma-associated immune dysfunction is related to the presence of distinct microbial populations in the airways. While several groups have long postulated that asthma is an infectious disease caused by bacterial pathogens (reviewed in Kraft 2000), it has only been the relatively recent application of culture-independent microbiome profiling approaches to asthmatic airway samples that has revealed the diversity of microbes that exist in the asthmatic airway and that relationships do exist between microbial members within these communities and features of the disease.
Multiple studies of asthmatic patient cohorts using distinct microbiome platforms (next-generation sequencing and phylogenetic microarrays) have reported the presence of a diverse microbial community in the airways of these patients. These microbiota are commonly enriched for members of the Proteobacteria (Goleva et al., 2013; Hilty et al., 2010; Huang et al., 2011). When compared to healthy subjects (few of which possessed sufficient bacterial material for 16S rRNA-based microbiome profiling), asthmatics possessed significantly higher bacterial burden and diversity in their lower airways (Huang et al., 2011). The finding of a characteristically low bacterial burden in the lower airways of healthy subjects was supported by a separate study of bronchoscopic lower airway samples, which found that the lower airway microbiome differed from the upper by having lower biomass but being compositionally similar, consisting of members of the Staphylococcaceae, Propinobacteriaceae, Corynebacteriaceae, Streptococcaceae, Veillonellaceae and Prevotellaceae (Charlson et al., 2011). Marri et al. (2013) reported a slightly different community for non-asthmatics based on induced sputum samples that consisted of members of the Lachnospiraceae, Staphylococcaceae, Streptococcaceae, Carnobacteriaceae, Fusobacteriaceae, and Peptostreptococcaceae. This compositional differential between the two studies may be a function of sampling distinct compartments within the lower airways (i.e. induced sputum vs bronchoalveolar lavage (BAL)).
The airway microbiome composition of asthmatic patients is highly correlated with the degree of bronchial hyper-responsiveness (a characteristic feature of allergic airways); thus raising the possibility that specific bacterial members within the asthmatic airway microbiome may potentiate airway allergic responses. Additional evidence of a relationship between airway microbiome composition and disease characteristics comes from a study of corticosteroid-responsive (CS) or -resistant (CR) patients. Goleva and colleagues (2013) demonstrated the presence of distinct lung microbiomes in CS and CR patients. CR patients were enriched in Proteobacteria including Neisseria and Haemophilus which possess lipopolysaccharide (LPS) with short acyl chains known to have high endotoxic activity and an enhanced capacity to trigger Toll-like receptor 4, leading to pro-inflammatory IL-8 production. Conversely CS patient microbiomes were predominantly populated with Bradyrhizobium and Fusobacterium members, possessing longer acyl chain LPS purported to confer relatively lower endotoxicity with reduced capacity to induce innate immune host responses. How these distinct microbiome features relate to steroid responsiveness remains unclear, though it is tempting to hypothesize that discrete functional features of these compositionally distinct airway responsiveness.
Emerging evidence also suggests that the bacterial microbiota that colonizes the respiratory mucosa may also impact host response to viral infection. Severe respiratory viral infection with respiratory syncytial virus (RSV) and rhinovirus (RV) is associated with increased risk of developing asthma and as such, these viruses are considered asthmagenic. Mice nasally exposed to two distinct strains of the commensal bacterial species, Lactobacillus rhamnosus, were protected against subsequent RSV infection. Protection was associated with increased levels of IFN-β, IFN-γ, IL-6, and TNF-α in both BAL and serum samples, which contributed to viral clearance. In parallel animals exhibited elevated levels of the anti-inflammatory cytokine IL-10, which reduced inflammation-associated lung tissue damage (Tomosada et al., 2013). In this study the authors demonstrated that the specific strain of L. rhamonsus mattered; strain Lr05 was found to be more effective in increasing IFN-γ and IL-10 compared with strain Lr06, but that Lr06 was more efficient in increasing IFN-β, TNF-α, and IL-6, thus emphasizing the capacity of distinct strains of the same species to elicit discrete host responses.
Other studies have demonstrated that the presence of specific pathogenic species in the airway potentiates RV infection in children (Kloepfer et al., 2014). Risk of asthma exacerbation significantly increased when RV was found concurrently with airway Moraxella catarrhalis, Streptococcus pneumoniae, or Haemophilus influenzae compared with children with neither bacterial nor viral pathogens. In addition, the odds of experiencing illness symptoms for more than 2 d were significantly higher when Moraxella catarrhalis or Streptococcus pneumoniae was detected concurrently with RV when compared to children who did not possess these bacterial or viral pathogens. Detection of airway pathogens did not predict RV presence, but discovery of RV increased the chances of detecting these bacterial pathogens; RV infection was associated with increased burden of each of three pathogenic bacteria examined, suggesting RV may change the airway environment in a manner that permits outgrowth of these respiratory pathogens which in turn potentiate the effect of this respiratory viral infection. A previous independent study demonstrated that the same three bacterial species and viruses were associated with wheezy episodes in children (Bisgaard et al., 2010). However, this study included eight additional viruses, which may have masked the interactive effect of RV and bacteria.
Development of the nasopharyneal microbiome in the first year of life has been linked to the risk of asthma development (Teo et al., 2015). In a longitudinal study of postnasal aspirates of 234 children in Australia, six distinct bacterial microbiota types were identified – each was characterized by the presence of a particular dominant genus: Moraxella, Streptococcus, Corynebacterium, Staphylococcus, Haemophilus or Alloicoccus. Those genera that have previously been associated with airway disease e.g. Moraxella, Streptococcus, and Haemophilus were, in this study also linked to increased risk of asthma development. Airway communities dominated by these three specific genera during an airway respiratory infection in infants who were later diagnosed with atopy at age 2, was associated with increased risk of lower airway infection and febrile symptoms. Airway microbiota dominated by Streptococcus were significantly associated with both lower respiratory infection at a younger age and chronic wheeze. Also, the age of bacterial colonization appears important, since the risk of developing wheeze at age 5 was highest when a high abundance of Streptococcus was found at age 7 weeks compared to 8 or 9 weeks of age. Moreover, the study demonstrated that the composition of the nasopharyngeal microbiome of these children is related to early life exposures such as day care and antibiotic administration, indicating that airway mucosal colonization, like the gastrointestinal tract (see below), is influenced by local exposures during the critical period of microbiome development in neonatal life and early infancy. This large study of several hundred children demonstrates relationships between patterns of early life bacterial colonization and respiratory infection, recurrent wheeze and asthma, strengthening the hypothesis that microbial mucosal colonization plays a critical role in airway disease susceptibility. A more comprehensive review of the microbiome and respiratory infection is covered by Vissers and colleagues (2014).
While the lower airways do not appear to be overtly colonized in healthy humans, the oropharynx supports a thriving and diverse microbial ecosystem (Huttenhower et al., 2012; Lemon et al., 2010). Early-life perturbations to this community have been linked to childhood asthma development. Neonates whose oropharynx were colonized by a high burden of Streptococcus pneumonia, Haemophilus influenzae, or Moraxella catarrhalis within the first month of life had an increased risk for recurrent wheeze and asthma development, significantly higher counts of eosinophils and elevated levels total serum IgE (Bisgaard et al., 2007). However, infants colonized by these same species at 12 months of age did not incur the same risk for disease development, suggesting that very early-life microbial colonization patterns may strongly influence immune development in a manner that is skewed towards allergic response, and that the activities of these species is contextual, and perhaps dependent on age-dependent changes in the local microbiome in this niche (Fig. 2). Support for this hypothesis comes from early life airway studies in mice. Recently, the lower respiratory microbiome was also shown in a murine model to play a role in the allergic immune response development against house dust mites (HDM; Gollwitzer et al., 2014). Programmed death ligand 1 (PD-L1) is necessary for the development of T regulatory cells which lack expression of the Helio transcription factor (Helio− T-regs). These cells, which reside in the lungs, are inversely related to aeroallergen response to HDM. Within the first two weeks of life, PD-L1 expression peaked in the murine lungs; this occurred in parallel with changes in bacterial burden and composition. However this temporal gene expression pattern is absent in germ-free animals, suggesting that the presence of microbes is necessary for PD-L1 expression and expansion of Helio− T-regs. Collectively these data point to an immune maturation process in the respiratory mucosa that parallels that which occurs in the gut, both of which are intimately dependent on (appropriate) microbial colonization.
Fig. 2. Very early life exposures influence asthma and allergy development.
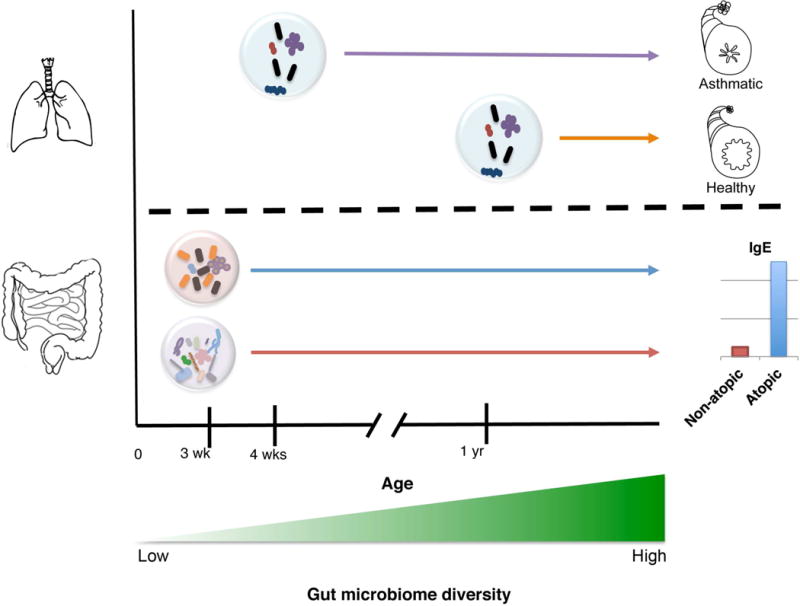
Oropharyngeal colonization of neonates by Streptococcus, Haemophilus or Moraxella species at 4 weeks of age is associated with development of recurrent wheeze. Infants colonized at 12-months with the same species were not at higher risk. Dysbiotic neonatal (3 week old) gut microbiomes enriched with Escherichia coli or Clostridium difficile are at significantly higher risk for childhood atopy development, which is typically characterized by elevated serum IgE levels.
Early Life Gut Microbiome and Childhood Allergic Asthma
While the airway microbiome has naturally formed the focus of asthma studies, a growing body of epidemiological and microbiological literature support the hypothesis that the genesis of allergic disease and by extension asthma development, may lie, at least in part, in the communities of microbes that exist in the gastrointestinal tract. Support for this phenomenon comes from factors that have been identified as related to allergic disease development in childhood. These include early life antimicrobial exposure (Johnson et al., 2005), Caesarian birth (Renz-Polster et al., 2005), formula feeding (Friedman and Zeiger, 2005; Harmsen et al., 2000), lack of maternal exposure to pets or livestock during pregnancy (Ownby et al., 2002) and maternal consumption of antimicrobials during pregnancy (Stensballe et al., 2013). From a microbiological standpoint, these risk factors have the capacity to influence microbiome composition; this has been clearly demonstrated for antimicrobial administration, which acutely depletes gut microbiome diversity (Dethlefsen and Relman, 2011), while formula feeding has been shown to deplete commensal Bifidobacteria populations in the infant gut (Balmer and Wharton, 1989; Bullen et al., 1976; Yoshioka et al., 1983). That the identified risk factors are focused around the pre- and early post-natal period suggests that development of childhood allergic asthma may owe its genesis, at least in part, to microbiome perturbations in early life during the critical period of microbiological and immunological development. In support of this hypothesis, independent studies have demonstrated that three-week old neonates who possessed significantly higher fecal burden of Clostridium difficile and a higher C. difficile to Bifidobacteria ratio, had a significantly higher rate of atopy development (Kalliomäki et al., 2001). In a separate study, infants with a high abundance of fecal Escherichia coli developed IgE-associated eczema (Penders et al., 2006).
To understand how these early-life risk factors may be related to disease development, it is first necessary to consider microbiome development in early infancy. Recent studies have described a placental (Aagaard et al., 2014) and cord blood (DiGiulio et al., 2008) associated microbiome, raising the prospect of microbial exposure in utero. However the birthing process appears to represent a strong microbiological influence on the neonate. At birth, vaginally delivered infants are colonized at various sites (skin, mouth, gut) by a microbiome enriched for Snethia and Lactobacillus spp., which is most similar in composition to that found in their mother’s vaginal tract (Dominguez-Bello et al., 2010). In comparison, Caesarian-born infants [who are at significantly higher risk of allergic disease development (Renz-Polster et al., 2005)] typically begin life with a microbiome enriched with Staphylococcus and Streptococcus spp., and a microbiome most similar to that of the maternal skin microbiome (Dominguez-Bello et al., 2010). Ecologically, development of the gut microbiome follows the central tenets of primary succession, a well-established series of events that occur during the initial colonization of a previously pristine environment. Pioneer species, i.e. those that initially colonize and ecosystem, frequently determine ecosystem conditions and dictate the types of organisms that co-colonize the niche. This raises the possibility that early aberrant microbial colonization in the gut, as described by Penders (2006) and Kalliomaki and colleagues (2001), (or indeed, the airway), may lead to dysbiotic microbiomes lacking commensal species necessary for appropriate physiological and immune development and subsequent maintenance of immune homeostasis. Hence, early-life microbiome development may prove a critical factor in dictating both immune maturation and allergic disease development.
Support for this hypothesis comes from studies of mice colonized in early life by a cocktail of 46 spore forming Clostridium clade IV and XIV species (Atarashi et al., 2011) or, in an independent study, by a diversity of bacteria (Cahenzli et al., 2013). In both studies, early life bacterial colonization resulted in significantly decreased circulating IgE concentrations (elevated serum IgE is a hallmark of atopy) in adulthood. However supplementation of adult animals with these same bacterial cocktails did not lead to protection. CD4+ Foxp3 expressing T-reg cells represent an important subset of T-helper cells with respect to allergic asthma, because of their capacity to mitigate pro-inflammatory adaptive immune response via the production of IL-10 (Barnes and Powrie, 2009; Rubtsov et al., 2008). In addition to demonstrating reduced IgE responses, Atarashi, and colleagues (2011), demonstrated, using an IL-10 reporter mouse, that supplementation of mice with Clostridia IV and XIV members promoted T-reg proliferation in the colon and decreased IL-4 concentrations in the airways in response to ovalbumin (OVA) challenge, indicating that appropriate gastrointestinal microbial colonization in early life, leads to induction of immune tolerance (Fig. 3).
Fig. 3. Adaptive immune modulation in the gut and airways through gastrointestinal microbiome manipulation.
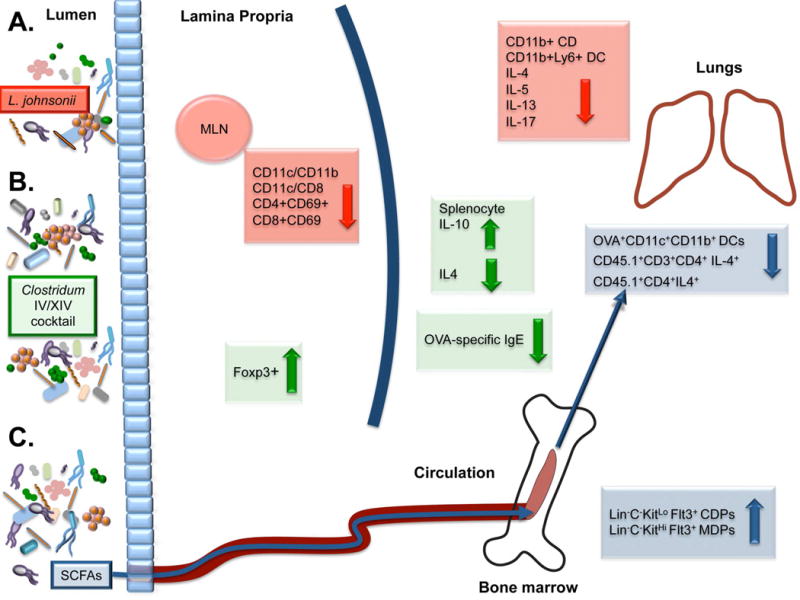
A. Lactobacillus johnsonii supplementation leads to decreased numbers of activated dendritic cells (DC) in the mesenteric lymph nodes (MLN) and a parallel decrease in Th2 cytokine expression IL-4, IL-5, and IL-13, IL-17 and DCs in the lungs. B. Mice supplemented by a cocktail of Clostridium IV and XIV species exhibit an increase in colonic T-regulatory cells and the anti-inflammatory cytokine IL-10, and were protected against ovalbumin-induced allergic diarrhea in part due to decreased levels of serum OVA-specific IgE and IL-4. C. Feeding propionate to mice resulted in circulation of short-chain fatty acids (SCFAs), associated with increased common and macrophage dendritic cell precursor cells (Lin−C−KitLo Flt3+ CDPs and Lin−C−KitHi Flt3+ MDPs respectively), which is associated with reduced DC proliferation and Th2 reactivation in the lungs.
In an independent study, Geuking et al. (2011) also showed that colonization of germ-free mice with Altered Schaedler Flora (ASF), a defined mixture of eight bacterial species (two Clostridium species, ASF356, ASF502; Lactobacillus sp., ASF360; L. murinus, ASF361; Mucispirillum schaedleri, ASF457; Eubacterium plexicaudatum, ASF492; Firmicutes sp., ASF500; and Parabacterioides sp., ASF519; Wannemuehler et al., 2014), induced de novo T-reg proliferation in the colonic lamina propria. T-regs produced in the periphery (not in the thymus) are known as induced T-regs (iTreg) and are principally stimulated in the mesenteric lymph nodes (MLN), Peyer’s patches and lamina propria (LP) of the small and large intestines. Mice deficient in iTregs spontaneously develop Th2 type pathologies characterized by higher percentage of CD4+ T-cells producing IL-4, IL-13, and IL-5 cytokines in the MLN, IL-4 in the LP of the large intestine, and IL-13 and IL-5 in the LP of the small intestine (Josefowicz et al., 2012). iTreg deficient animals possess an altered gut microbiome, enriched with members of the TM7 phylum, a relatively recently described candidate division consisting of uncultivated bacteria, as well as Alistipes species and also exhibit macrophage and neutrophil infiltration in the airways coupled with increased goblet cell numbers, mucin hyper-secretion, smooth muscle hyperplasia, fibrosis and poorer lung function (Josefowicz et al., 2012). That the presence of specific bacterial stimuli have been shown to be critical to the proliferation of this T-helper subsets in mice, suggests that failure to populate the developing gastrointestinal microbiome with such species represents at least one pathway by which allergic disease may develop.
Invariant natural killer T cells (iNKT), represent another important subset of IL-4 and IL-13 producing (Akbari et al., 2003) lymphocytes that accumulate in the colonic LP and lungs of germ free mice. iNKT cells accumulate soon after birth in germ-free animals resulting in persistence of the associated allergic pathology into adulthood. Significantly increased allergic response in response to airway OVA challenge in germ-free mice is only abrogated through maternal colonization (and thus neonatal exposure) when animals are co-housed with specific pathogen free animals (Olszak et al., 2012). Neonatal protection was due, in part, to the capacity of commensal organisms to block methylation of chemokine CXCL16 in the colon and lungs, which is crucial for iNKT recruitment (Olszak et al., 2012). Colonization of adult germ-free mice with commensal bacteria did not abrogate their disease phenotype, indicating that early-life microbial colonization influences epigenetic modification of host DNA in a manner that produces life-long effects on the host immune response. These studies highlight the importance of appropriate early-life microbiome development in Th2 and allergy-associated phenotypes, and suggest that interventions aimed at correcting dysbiotic microbiomes or promoting appropriate colonization in the neonatal phase of life may prove highly effective in preventing childhood allergic disease.
Environmental influences on the Developing Microbiome
Due to the rapid increase in prevalence over the past several decades primarily in Westernized nations, allergic asthma is considered a disease that is strongly related to environmental exposures. Emerging evidence suggests that environmental factors play an influential role in shaping human-associated microbial communities and immune responses. A recent study of immune cell population frequencies, cytokine responses, and serum proteins of 210 healthy twins indicated that 77% of these immunological parameters were dominated (>50% of variance) and 58% almost completely determined (>80% of variance) by non-heritable influences (Brodin et al., 2015). The study also demonstrated that immune responses diverged across twin pairs over time (Brodin et al., 2015), supporting the concept of a primarily reactive and adaptive immune response in healthy humans, presumably in response to local environmental exposures.
Although previous studies have reported that family members typically possess gut microbiomes that are more similar to each other than to others (Lee et al., 2011; Turnbaugh et al., 2009; Yatsunenko et al., 2012), their similarity was attributed to environmental factors, since the bacterial composition of monozygotic twins were no more alike than dizygotic twins (Turnbaugh et al., 2009; Yatsunenko et al., 2012). Diet has been shown to play a significant role in shaping gut microbiome composition. A recent murine cross-over study highlighted the strong selective pressure diet exerts on the microbiome. Successively transferring genetically diverse adult mice from high-fat/low fiber, to a low-fat/high fiber diet three times during the course of the study led to similar shifts in microbiome composition despite the genetic diversity of the animal population studied (Carmody et al., 2015). The strong influence of diet on the gut microbiome is also supported by a study of individuals from Malawi, Venezuela and the United States, which demonstrated that the gut microbiome of adult human subjects in the developing nations was significantly distinct from that of residents in the United States. While geography and ethnicity represent confounding factors, diet was considered a primary driver of these compositional and functional differences; developing nation diets were primarily plant polysaccharide based, while those of US participants was enriched in animal products. Functionally, Malawians and Amerindians were enriched for genes encoding glutamate synthase, associated with herbivores, while gut microbiomes of US participants were enriched for the capacity to degrade glutamine a function commonly associated with carnivores. Undoubtedly diet represents a strong selective pressure on the gut microbiome, a factor that likely explains the increased risk of allergy associated with formula feeding. Indeed, studies dating back to 1936 have reported lower incidences of eczema (Grulee and Sanford, 1936) and asthma (Friedman and Zeiger, 2005) in breastfed infants. The lasting effects of breastfeeding has been reported for children 8 years old; infants breastfed for at least 4 months had reduced risk of developing asthma (Kull et al., 2010).
More recently, early-life microbial exposure in the built environment has been linked to allergic asthma development. In western nations where the burden of allergic asthma is greatest, exposures to microbes has decreased substantially, most strikingly 497 in the post-industrial revolution era (Rook, 2010). Proposed in the late 1980’s the hygiene hypothesis encapsulates this altered human exposure to microbes, and posits that a lack of exposure to commensal microbes represents a driving force in the increased prevalence of allergic asthma and other chronic inflammatory disease in Westernized nations (Strachan, 1989). Inhabitants of westernized nations spend almost 90% of their time in the built environment, with approximately 70% of time spent in their personal residences (Klepeis et al., 2001). This represents a significant departure from environment in which the human microbiome evolved, and has led to the hypothesis that residential microbial exposures in early life, influence microbiome development, immune maturation, and consequently, the propensity to develop allergic asthma. In the United States, children raised in inner-city environments are more likely to develop asthma and atopy (Crain et al., 1994; Weiss et al., 1992). This observation was seen to contradict the hygiene hypothesis since the inner city had always been assumed to represent an environment of enhanced microbial exposure (Kay, 2001). However, until recently, no study had comprehensively examined the composition of bacterial communities in inner-city households and whether differences in early-life bacterial exposure related to allergic asthma development in childhood. Using culture-independent profiling, Lynch and colleagues (2014) examined the bacterial composition of house dust samples collected during the first year of life from residences in a study of full-term infants born in low-income, inner-city urban neighborhoods in New York, Baltimore, St. Louis and Boston. Children were followed prospectively and clinical outcomes of atopy and recurrent wheeze were assessed at three years of age. The investigators demonstrated that healthy children in the study (who neither developed atopy nor recurrent wheeze by age 3) were exposed in the first year of life to richer (increased number of bacterial types) and more diverse bacterial communities that were of significantly distinct composition compared to those in households where the children developed either atopy, or both recurrent wheeze and atopy. Consistent with the hygiene hypothesis, healthy inner city children were exposed to a greater diversity of bacterial species in early life, characterized by significant enrichment of approximately eighty bacterial taxa belonging to sixteen distinct families. Amongst those enriched were members of the Bifidobacteriaceae, which are critical commensal colonizers of the human gastrointestinal tract, whose loss in early life is associated with allergy development (Penders et al., 2007).
Further evidence for the role of local household microbial exposures in early life in protecting against allergic disease development comes from an independent study of microbial exposures associated with pet ownership. Exposure to dogs and a lesser extent cats in infancy, protects against allergic disease development in childhood (Ownby et al., 2002). To determine whether a microbial correlate of protection existed in these households, Fujimura et al. (2010) examined both bacterial and fungal communities in house dust collected during the course of a prospective birth cohort study centered around the Detroit metropolitan area. Bacterial communities in household dust from residences with dogs or cats present were significantly richer and more diverse compared to those with no pets present and, as in the inner city studies, a large majority of bacterial taxa enriched in the protective environments have previously been detected in the human gut microbiome (Fujimura et al., 2010). These findings are consistent with previous lower-resolution studies in which environments with no pets present is depleted of bacteria (Gereda et al., 2000; Hesselmar et al., 1999; Ownby et al., 2002). Moreover, the investigators noted that bacteria-impoverished residences that conferred a higher risk of allergic disease development possessed a wider range of fungal species (Fujimura et al., 2010). Since fungal species have long been associated with allergic disease development (Bush and Portnoy, 2001; Gravesen, 1979; Kurup et al., 2000), this study suggests that increased local environmental exposure to fungal species in early life may play a role in shaping the neonatal microbiome and subsequent allergic disease development.
High-resolution microbiome profiling of the residential environment in early life has demonstrated a significant relationship between early-life bacterial and fungal exposure and allergic asthma development in childhood. It has been hypothesized that one mechanism by which these exposures protect against or contribute to disease development is by influencing the composition of the gut microbiome, which in turn influences allergic immune responses. In an attempt to address this hypothesis, Fujimura and colleagues (2014) used a murine model of airway allergen challenge. In the study, groups of mice were supplemented either with house dust from residences with two dogs or no pets present, prior to and during airway challenge with cockroach allergen. Only animals exposed to dust from homes with dogs present exhibited significantly reduced airway expression of Th2 cytokines, IL-4 and IL-13 and the mucin-associated gob-5 gene. Lung histology supported these findings; protected animals exhibited pristine airways with no evidence of inflammatory infiltrate or mucin hyper-secretion. Comparative analysis of the gut microbiome of protected and unprotected animals revealed significantly differences in their taxonomic content. Protected animals possessed a gut microbiome enriched in members of the Firmicutes including taxa belonging to the Lachnospiraceae, Bacillaceae, Peptococcaceae and Lactobacillaceae. Using selective media, the authors isolated and confirmed that the dominant Lactobacillus in protected animals was Lactobacillus johnsonii, a species which, coincidentally, becomes highly enriched in the human vaginal tract immediately prior to birth (Aagaard et al., 2012), and thus transferred to the vaginally born neonate. This suggested that this species may play a crucial role in shaping gut microbiome composition. Using this single species, the authors performed subsequent supplementation experiments using distinct airway allergen challenges (cockroach and OVA) and demonstrated that animals who received L. johnsonii were protected against allergen challenge. Again protection was associated with significant down-regulation of airway Th2 responses (IL-4, IL-5 and IL-13), both at the mRNA and protein level. The study also demonstrated that the protective effect of L. johnsonii supplementation extended to viral infectious agents, a critical finding since children hospitalized due to respiratory infection with respiratory syncytial virus (RSV) have a significantly increased risk of developing allergic asthma (Darville and Yamauchi, 1998; Stein et al., 1999). Using this viral pathogen, the investigators demonstrated that oral supplementation with viable L. johnsonii protected against RSV infection [heat-killed organisms did not confer protection; (Fujimura et al., 2014)]. Protection was again associated with a significant reduction in both bronchial hyperactivity and the expression of Th2 and Th17 cytokines (IL-4, IL-5, IL-13 and IL-17 respectively) in the airways. However these animals did not demonstrate significant differences in airway expression of IFN-γ compared to control animals, suggesting that they maintain the capacity to effectively clear the virus, but significantly down-regulated the adaptive immune responses that are largely responsible for RSV-associated pathology. Protected animals exhibited a significantly distinct gut microbial community (from that of susceptible animals) and significantly reduced activation of dendritic cells in the mesenteric lymph nodes, indicating that compositional (and presumably parallel functional) changes in the gut microbiome influences the capacity of antigen presenting cells to activate local T-cell populations, a mechanism by which allergic responses may be inhibited (Fig. 3).
More mechanistic evidence for how the gut microbiome may prevent allergic response comes from studies demonstrating the ability of the murine gut microbiota to regulate colonic T-cell populations via production of short chain fatty acids (SCFAs) such as butyrate (Furusawa et al., 2013), acetate and propionate (Smith et al., 2013). SCFAs are anti-inflammatory and have been shown to increase significantly with induction of colonic CD103+ Foxp3+ cells and IL-10 production (Furusawa et al., 2013; Smith et al., 2013). SCFAs, which are also an important energy source of gastrointestinal colonocytes, specifically bind the G-protein coupled receptor 43 (GPR43; also known as free fatty acid receptor 2 [FFAR2]), which also mediates their effect on colonic T-regs (Smith et al., 2013). Indeed Gpr−/− mice exhibit lower IL-10 expression compared to wild type controls (Smith et al. 2013) and develop more severe inflammation when their airways are challenged with OVA (Maslowski et al., 2009). Microbial-derived SCFAs are produced through the fermentation of complex carbohydrates. The importance of these microbial fermentation products in allergic disease was recently illustrated by Trompette et al. (2014). Mice fed a low fiber diet followed by nasal exposure to house dust mite (HDM) extract exhibited significantly increased IL-4, IL-5, IL-13 and IL-17A in lung tissue as well as increased airway mucus production and goblet cell hyperplasia. In parallel, these animals possessed higher levels of circulating IgE and significantly higher amounts of total HDM-specific IgG1. In contrast, mice fed a high fiber diet prior to HDM exposure were protected, exhibiting significantly lower cytokine concentrations and a normal mucin phenotype. Fiber intake impacted the gut microbiome of animals; a low fiber diet promoted the abundance of specific members Firmicutes, especially those belonging to the Erysipelotrichaceae family, while animals on a high fiber diet were enriched for Bacteroidaceae and Bifidobacteriaceae. Supplementation of animals with propionate increased Foxp3+CD25+ CD4+ T-reg cell numbers and enhanced hematopoiesis of dendritic cell (DC) precursors. This suggests that SCFAs not only induce iTreg populations, but also modulate bone marrow-derived antigen-presenting cell precursors, indicating a mechanism by gut microbial-derived metabolites may reprogram the immunological tone of the human ecosystem. Collectively these studies provide evidence that gut microbiome composition and activities not only influence local immune activity, but also that at remote mucosal sites (Fig. 3), suggesting that the microbial communities resident in the gut may represent a key target for prevention or management of allergic asthma.
While gastrointestinal microbiome manipulation of mice clearly influences airway immune responses, direct microbial stimulation of the respiratory mucosa has also been shown to be protective against Th2-associated allergic responses. Nembrini and colleagues (2011) demonstrated that inhalation of non-pathogenic Escherichia coli was protective against allergic inflammation (Nembrini et al., 2011). Compared to untreated animals, mice treated nasally with E. coli prior to OVA allergen challenge, exhibited fewer eosinophils in their lower airways, a parallel abrogation of IL-4 and IL-5 production, fewer goblet cells and pristine lungs as evidenced by histology. These animals also exhibited airway induction of γδT cells, which protected mice from airway hyper-responsiveness, but did not appear to impact cell recruitment. In parallel, innate immune responses were also impacted. Protected animals exhibited induction of toll-like receptor 4 (TLR4), which resulted in a suppressive lung environment, altered DC function and reduced Th2 effector function in the airways. Other intranasal instillation studies using a bacterium isolated from cowsheds, Acinetobacter lwoffii, also conferred protection against Th2 allergic airway responses. In this case, protection was associated induction of the Th1 cytokine IL-12 and a concomitant attenuation of Th2 responses following allergen challenge (Debarry et al., 2007). In a separate study, Conrad and colleagues (2009) demonstrated that the capacity of A. lwoffii to protect against allergic response could be passed from mother to offspring. Pregnant mice who received A. lwoffii intranasally during gestation exhibited significantly increased circulating IL-6 in response to this microbial exposure (Conrad et al., 2009). Placental concentrations of this cytokine were reciprocally related to the concentrations found in the maternal circulation and offspring were less susceptible to airway allergen challenge compared to those whose mother was not supplemented with A. lwoffi. In this case, protection appeared to be mediated via innate immune responses, since the offspring of TLR2/3/4/7/9−/− mice supplemented with this species were susceptible to airway OVA challenge. This latter study opens up the possibility that maternal microbial exposures during pregnancy may influence the susceptibility to allergic disease development. An area that clearly merits further intensive study.
In summary, asthma and atopy are complex diseases, which affect and are affected by the communities of microbes that colonize the gut and respiratory tracts, these communities in turn are influenced by an array of environmental influences from diet and antimicrobial administration to early life local microbial exposures. Though many of the studies to date have been performed in mice, they serve as proof of the existence of a gut-airway axis. This opens up the possibility that strategies aimed at manipulating the airway and/or gut microbiome, particularly in the earliest stages of life when the immune response is maturing and the microbiome appears most plastic and receptive to exogenous microbes, may serve as a novel strategy for prevention of allergic asthma, as well as a host of other diseases in which adaptive immune dysfunction is a prominent feature.
Acknowledgments
SVL and KF are supported by the National Institute of Allergy and Infectious Disease grant 1P01AI089473 and the National Institute of Allergy and Infectious Disease program UM1AI114271.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, Raza S, Rosenbaum S, Van den Veyver I, Milosavljevic A, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7:e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accordini S, Corsico AG, Braggion M, Gerbase MW, Gislason D, Gulsvik A, Heinrich J, Janson C, Jarvis D, Jõgi R, et al. The Cost of Persistent Asthma in Europe: An International Population-Based Study in Adults. Int Arch Allergy Immunol. 2013;160:93–101. doi: 10.1159/000338998. [DOI] [PubMed] [Google Scholar]
- Adcock IM, Caramori G, Chung KF. New targets for drug development in asthma. Lancet. 2008;372:1073–1087. doi: 10.1016/S0140-6736(08)61449-X. [DOI] [PubMed] [Google Scholar]
- Akbari O, Stock P, Meyer E, Kronenberg M, Sidobre S, Nakayama T, Taniguchi M, Grusby MJ, DeKruyff RH, Umetsu DT. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med. 2003;9:582–588. doi: 10.1038/nm851. [DOI] [PubMed] [Google Scholar]
- Akinbami LJ, Moorman JE, Garbe PL, Sondik EJ. Status of childhood asthma in the United States, 1980–2007. Pediatrics. 2009;123:S131–S145. doi: 10.1542/peds.2008-2233C. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer S, Wharton B. Diet and faecal flora in the newborn : breast milk and infant formula. Arch Dis Child. 1989;64:1672–1677. doi: 10.1136/adc.64.12.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes MJ, Powrie F. Regulatory T cells reinforce intestinal homeostasis. Immunity. 2009;31:401–411. doi: 10.1016/j.immuni.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Barnett SBL, Nurmagambetov TA. Costs of asthma in the United States: 2002–2007. J Allergy Clin Immunol. 2011;127:145–152. doi: 10.1016/j.jaci.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bonnelykke K, Brasholt M, Heltberg A, Vissing NH, Thorsen SV, et al. of the Airway in Neonates. N Engl J Med. 2007;357:1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- Bisgaard H, Hermansen MN, Bønnelykke K, Stokholm J, Baty F, Skytt NL, Aniscenko J, Kebadze T, Johnston SL. Association of bacteria and viruses with wheezy episodes in young children: prospective birth cohort study. BMJ. 2010;341:c4978. doi: 10.1136/bmj.c4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P, Jojic V, Gao T, Bhattacharya S, Angel CJL, Furman D, Shen-Orr S, Dekker CL, Swan GE, Butte AJ, et al. Variation in the Human Immune System Is Largely Driven by Non-Heritable Influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen CL, Tearle PV, Willis aT. Bifidobacteria in the intestinal tract of infants: an in-vivo study. J Med Microbiol. 1976;9:325–333. doi: 10.1099/00222615-9-3-325. [DOI] [PubMed] [Google Scholar]
- Bush RK, Portnoy JM. The role and abatement of fungal allergens in allergic diseases. J Allergy Clin Immunol. 2001;107:S430–S440. doi: 10.1067/mai.2001.113669. [DOI] [PubMed] [Google Scholar]
- Cahenzli J, Köller Y, Wyss M, Geuking MB, McCoy KD. Intestinal Microbial Diversity during Early-Life Colonization Shapes Long-Term IgE Levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano JM, Jr, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. Diet Dominates Host Genotype in Shaping the Murine Gut Microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical Continuity of Bacterial Populations in the Healthy Human Respiratory Tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington Ca, Hinton M, Pearson GR, Chopra I. Short-chain organic acids at ph 5.0 kill Escherichia coli and Salmonella spp. without causing membrane perturbation. J Appl Bacteriol. 1991;70:161–165. doi: 10.1111/j.1365-2672.1991.tb04442.x. [DOI] [PubMed] [Google Scholar]
- Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, Adcock IM, Bateman ED, Bel EH, Bleecker ER, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43:343–373. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, O’Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O’Toole PW. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One. 2009;4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad ML, Ferstl R, Teich R, Brand S, Blümer N, Yildirim AO, Patrascan CC, Hanuszkiewicz A, Akira S, Wagner H, et al. Maternal TLR signaling is required for prenatal asthma protection by the nonpathogenic microbe Acinetobacter lwoffii F78. J Exp Med. 2009;206:2869–2877. doi: 10.1084/jem.20090845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson WOCM, Moffatt MF. Asthma–An Epidemic in the Absence of Infection? Sci. 1997;275:41–42. doi: 10.1126/science.275.5296.41. [DOI] [PubMed] [Google Scholar]
- Crain EF, Weiss KB, Bijur PE, Hersh M, Westbrook L, Stein RE. An estimate of the prevalence of asthma and wheezing among inner-city children. Pediatrics. 1994;94:356–362. [PubMed] [Google Scholar]
- Darville T, Yamauchi T. Respiratory syncytial virus. Pediatr Rev. 1998;19:55–61. doi: 10.1542/pir.19-2-55. [DOI] [PubMed] [Google Scholar]
- Debarry J, Garn H, Hanuszkiewicz A, Dickgreber N, Blumer N, von Mutius E, Bufe A, Gatermann S, Renz H, Holst O, et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J Allergy Clin Immunol. 2007;119:1514–1521. doi: 10.1016/j.jaci.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Dethlefsen L, Relman Da. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, Kim CJ, Erez O, Edwin S, Relman DA. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. 2008;3:e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Bayer Ea, Rincon MT, Lamed R, White Ba. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Fox GE, Magrum LJ, Balch WE, Wolfe RS, Woese CR. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc Natl Acad Sci U S A. 1977;74:4537–4541. doi: 10.1073/pnas.74.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman NJ, Zeiger RS. The role of breast-feeding in the development of allergies and asthma. J Allergy Clin Immunol. 2005;115:1238–1248. doi: 10.1016/j.jaci.2005.01.069. [DOI] [PubMed] [Google Scholar]
- Fujimura KE, Johnson CC, Ownby DR, Cox MJ, Brodie EL, Havstad SL, Zoratti EM, Woodcroft KJ, Bobbitt KR, Wegienka G, et al. Man’s best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010;126:410–412. doi: 10.1016/j.jaci.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura KE, Demoor T, Rauch M, Faruqi aa, Jang SC, Johnson C, Boushey Ha, Zoratti E, Ownby D, Lukacs NW, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci. 2014;111:805–810. doi: 10.1073/pnas.1310750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo Ta, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gereda JE, Leung DY, Thatayatikom A, Streib JE, Price MR, Klinnert MD, Liu AH. Relation between house-dust endotoxin exposure, type 1 T-cell development, and allergen sensitisation in infants at high risk of asthma. Lancet. 2000;355:1680–1683. doi: 10.1016/s0140-6736(00)02239-x. [DOI] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MaE, Ng DCK, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal Bacterial Colonization Induces Mutualistic Regulatory T Cell Responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Goleva E, Jackson LP, Harris JK, Robertson CE, Sutherland ER, Hall CF, Good JT, Gelfand EW, Martin RJ, Leung DYM. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013;188:1193–1201. doi: 10.1164/rccm.201304-0775OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, Nicod LP, Lloyd CM, Marsland BJ. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. 2014;20:642–647. doi: 10.1038/nm.3568. [DOI] [PubMed] [Google Scholar]
- Gravesen S. Fungi as a cause of allergic disease. Allergy. 1979;34:135–154. doi: 10.1111/j.1398-9995.1979.tb01562.x. [DOI] [PubMed] [Google Scholar]
- Grulee CG, Sanford HN. The influence of breast and artificial feeding oninfantile eczema. J Pediatr. 1936;9:223–225. [Google Scholar]
- Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. Analysis of Intestinal Flora Development in Breast – Fed and Formula – Fed Infants by Using Molecular Identification and Detection Methods. J Pediatr Gastroenterol Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- Havstad S, Johnson CC, Kim H, Levin AM, Zoratti EM, Joseph CLM, Ownby DR, Wegienka G. Atopic phenotypes identified with latent class analyses at age 2 years. J Allergy Clin Immunol. 2014;134:722–727. doi: 10.1016/j.jaci.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst T, Sichelstiel A, Schär C, Yadava K, Bürki K, Cahenzli J, McCoy K, Marsland BJ, Harris NL. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- Hesselmar B, Åberg N, Åberg B, Eriksson B, Björkstén B. Does early exposure to cat or dog protect against later allergy development? Clin Exp Allergy. 1999;29:611–617. doi: 10.1046/j.1365-2222.1999.00534.x. [DOI] [PubMed] [Google Scholar]
- Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, Woyke T, Allgaier M, Bristow J, Wiener-Kronish JP, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–381. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee Ca, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DJ, Hartert TV, Martinez FD, Weiss ST, Fahy JV. Asthma: NHLBI Workshop on the Primary Prevention of Chronic Lung Diseases. Ann Am Thorac Soc. 2014;11:S139–S145. doi: 10.1513/AnnalsATS.201312-448LD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CC, Ownby DR, Alford SH, Havstad SL, Williams LK, Zoratti EM, Peterson EL, Joseph CL. Antibiotic exposure in early infancy and risk for childhood atopy. J Allergy Clin Immunol. 2005;115:1218–1224. doi: 10.1016/j.jaci.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107:129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Mori H, Iwata M, Meguro S. Growth stimulator for bifidobacteria produced by Propionibacterium freudenreichii and several intestinal bacteria. J Dairy Sci. 1994;77:393–404. doi: 10.3168/jds.S0022-0302(94)76965-4. [DOI] [PubMed] [Google Scholar]
- Kay AB. Allergy and allergic diseases. First of two parts. N Engl J Med. 2001;344:30–37. doi: 10.1056/NEJM200101043440106. [DOI] [PubMed] [Google Scholar]
- Klepeis NE, Nelson WC, Ott WR, Robinson JP, Tsang aM, Switzer P, Behar JV, Hern SC, Engelmann WH. The National Human Activity Pattern Survey (NHAPS): a resource for assessing exposure to environmental pollutants. J Expo Anal Environ Epidemiol. 2001;11:231–252. doi: 10.1038/sj.jea.7500165. [DOI] [PubMed] [Google Scholar]
- Kloepfer KM, Lee WM, Pappas TE, Kang TJ, Vrtis RF, Evans MD, Gangnon RE, Bochkov YA, Jackson DJ, Lemanske RF, et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol. 2014;133:1301–1307. 1307.e1–e3. doi: 10.1016/j.jaci.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft M. The role of bacterial infections in asthma. Clin Chest Med. 2000;21:301–313. doi: 10.1016/s0272-5231(05)70268-9. [DOI] [PubMed] [Google Scholar]
- Kull I, Melen E, Alm J, Hallberg J, Svartengren M, van Hage M, Pershagen G, Wickman M, Bergström A. Breast-feeding in relation to asthma, lung function, and sensitization in young schoolchildren. J Allergy Clin Immunol. 2010;125:1013–1019. doi: 10.1016/j.jaci.2010.01.051. [DOI] [PubMed] [Google Scholar]
- Kurup VP, Shen HD, Banerjee B. Respiratory fungal allergy. Microbes Infect. 2000;2:1101–1110. doi: 10.1016/s1286-4579(00)01264-8. [DOI] [PubMed] [Google Scholar]
- Lee S, Sung J, Lee J, Ko G. Comparison of the gut microbiotas of healthy adult twins living in South Korea and the United States. Appl Environ Microbiol. 2011;77:7433–7437. doi: 10.1128/AEM.05490-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. Comparative analyses of the Bacterial microbiota of the human nostril and oropharynx. MBio. 2010;1:4–6. doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SV, Wood RA, Boushey H, Bacharier LB, Bloomberg GR, Kattan M, O’Connor GT, Sandel MT, Calatroni A, Matsui E, et al. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J Allergy Clin Immunol. 2014;134:593–601. doi: 10.1016/j.jaci.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoli M, Fabian D, Holt S, Beasley R. Review article The global burden of asthma : executive summary of the GINA Dissemination Committee Report. Allergy. 2004;59:469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Nembrini C, Sichelstiel A, Kisielow J, Kurrer M, Kopf M, Marsland BJ. Bacterial-induced protection against allergic inflammation through a multicomponent immunoregulatory mechanism. Thorax. 2011;66:755–763. doi: 10.1136/thx.2010.152512. [DOI] [PubMed] [Google Scholar]
- Ober C, Yao TC. The Genetics of Asthma and Allergic Disease: a 21st Century Perpsective. Immunol Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Repáraz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010;185:4101–4108. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ownby DR, Johnson CC, Peterson EL. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. J Am Med Assoc. 2002;288:963–972. doi: 10.1001/jama.288.8.963. [DOI] [PubMed] [Google Scholar]
- Penders J, Stobberingh EE, Thijs C, Adams H, Vink C, Van Ree R, Van Den Brandt Pa. Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin Exp Allergy. 2006;36:1602–1608. doi: 10.1111/j.1365-2222.2006.02599.x. [DOI] [PubMed] [Google Scholar]
- Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. 2007;62:1223–1236. doi: 10.1111/j.1398-9995.2007.01462.x. [DOI] [PubMed] [Google Scholar]
- Porter JR. Antony van Leeuwenhoek: tercentenary of his discovery of bacteria. Bacteriol Rev. 1976;40:260–269. doi: 10.1128/br.40.2.260-269.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi D, Canfield SM, Andrade A, Isasi CR, Hall CB, Rubinstein A, Arens R. Obesity-associated asthma in children: a distinct entity. Chest. 2012;141:895–905. doi: 10.1378/chest.11-0930. [DOI] [PubMed] [Google Scholar]
- Renz-Polster H, David MR, Buist aS, Vollmer WM, O’Connor Ea, Frazier Ea, Wall Ma. Caesarean section delivery and the risk of allergic disorders in childhood. Clin Exp Allergy. 2005;35:1466–1472. doi: 10.1111/j.1365-2222.2005.02356.x. [DOI] [PubMed] [Google Scholar]
- Roediger WEW. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook GaW. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: darwinian medicine and the “hygiene” or “old friends” hypothesis. Clin Exp Immunol. 2010;160:70–79. doi: 10.1111/j.1365-2249.2010.04133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3 + regulatory T-cell development by a commensal bacterium of the intestinal microbiota. PNAS. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Simpson A, Tan VYF, Winn J, Svensén M, Bishop CM, Heckerman DE, Buchan I, Custovic A. Beyond atopy: multiple patterns of sensitization in relation to asthma in a birth cohort study. Am J Respir Crit Care Med. 2010;181:1200–1206. doi: 10.1164/rccm.200907-1101OC. [DOI] [PubMed] [Google Scholar]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RT, Sherrill D, Morgan WJ, Holberg CJ, Halonen M, Taussig LM, Wright AL, Martinez FD. Respiratory syncytial virus in early life and risk of wheeze and allergy by age 13 years. Lancet. 1999;354:541–545. doi: 10.1016/S0140-6736(98)10321-5. [DOI] [PubMed] [Google Scholar]
- Stensballe LG, Simonsen J, Jensen SM, Bønnelykke K, Bisgaard H. Use of antibiotics during pregnancy increases the risk of asthma in early childhood. J Pediatr. 2013;162:832–838. doi: 10.1016/j.jpeds.2012.09.049. [DOI] [PubMed] [Google Scholar]
- Strachan DP. Hay fever, hygiene, and household size. Br Med J. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, et al. The Infant Nasopharyngeal Microbiome Impacts Severity of Lower Respiratory Infection and Risk of Asthma Development. Cell Host Microbe. 2015;17:1–12. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomosada Y, Chiba E, Zelaya H, Takahashi T, Tsukida K, Kitazawa H, Alvarez S, Villena J. Nasally administered Lactobacillus rhamnosus strains differentially modulate respiratory antiviral immune responses and induce protection against respiratory syncytial virus infection. BMC Immunol. 2013;14:40. doi: 10.1186/1471-2172-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- Tsoumakidou M, Elston W, Zhu J, Wang Z, Gamble E, Siafakas NM, Barnes NC, Jeffery PK. Cigarette smoking alters bronchial mucosal immunity in asthma. Am J Respir Crit Care Med. 2007;175:919–925. doi: 10.1164/rccm.200607-908OC. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers M, de Groot R, Ferwerda G. Severe viral respiratory infections: are bugs bugging? Mucosal Immunol. 2014;7:227–238. doi: 10.1038/mi.2013.93. [DOI] [PubMed] [Google Scholar]
- Wannemuehler MJ, Overstreet A, Ward DV, Phillips J. Draft Genome Sequences of the Altered Schaedler Flora, a Defined Bacterial Community from Gnotobiotic Mice. 2014:1–2. doi: 10.1128/genomeA.00287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss KB, Gergen PJ, Crain EF. The Epidemiology of an Emerging US Public Health Concern. Chest. 1992;101:362S–367S. doi: 10.1378/chest.101.6.362s. [DOI] [PubMed] [Google Scholar]
- Winker S, Woese CR. A definition of the domains Archaea, Bacteria and Eucarya in terms of small subunit ribosomal RNA characteristics. Syst Appl Microbiol. 1991;14:305–310. doi: 10.1016/S0723-2020(11)80303-6. [DOI] [PubMed] [Google Scholar]
- Woese CR. Bacterial Evolution Background. Microbiology. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, Arron JR, Koth LL, Fahy JV. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;496:222–228. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H, Iseki K, Fujita K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983;72:317–321. [PubMed] [Google Scholar]