The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation (original) (raw)
. Author manuscript; available in PMC: 2016 May 21.
SUMMARY
The circadian timing system synchronizes cellular function by coordinating rhythmic transcription via a transcription-translational feedback loop. How the circadian system regulates gene expression at the translational level remains a mystery. Here, we show that the key circadian transcription factor BMAL1 associates with the translational machinery in the cytosol and promotes protein synthesis. The mTOR-effector kinase, ribosomal S6 protein kinase 1 (S6K1), an important regulator of translation, rhythmically phosphorylates BMAL1 at an evolutionarily conserved site. S6K1-mediated phosphorylation is critical for BMAL1 to both associate with the translational machinery and stimulate protein synthesis. Protein synthesis rates demonstrate circadian oscillations dependent on BMAL1. Thus, in addition to its critical role in circadian transcription, BMAL1 is a translation factor that links circadian timing and the mTOR signaling pathway. More broadly, these results expand the role of the circadian clock to the regulation of protein synthesis.
Graphical Abstract
INTRODUCTION
Circadian timing is a ubiquitous and evolutionarily conserved property of cells and animal behavior (Bass and Takahashi, 2010; Lowrey and Takahashi, 2011). On a molecular level, transcriptional-translational feedback loops are a common organizing principle of circadian clocks across kingdoms (Koike et al., 2012; Ukai and Ueda, 2010). Additionally, epigenetic, translational, and post-translational mechanisms confer both robustness and plasticity to the clock (Eckel-Mahan et al., 2012; Gallego and Virshup, 2007; Lim and Allada, 2013b). In animals, the Per, Arntl, _Sim1_-(PAS-), and basic helix-loop-helix-(bHLH)-domain-containing transcription factors brain muscle aryl nuclear translocase like-1 (BMAL1; ARNTL; CYCLE1; MOP3) and circadian locomotor output cycles kaput (CLOCK) drive transcription of clock-controlled genes (CCGs) (Lowrey and Takahashi, 2011). BMAL1 is unique among core clock proteins because it is essential for circadian transcription and behavior (Bunger et al., 2000). BMAL1 cycles between the nucleus and cytosol, but no cytosolic function for BMAL1 has yet been established (Kwon et al., 2006; Tamaru et al., 2003).
While a majority of molecular circadian research has focused on transcriptional mechanisms, transcription is not required for all circadian phenomena. For example, post-translational rhythms have been demonstrated in enucleated human red blood cells (O'Neill and Reddy, 2011). In addition, several studies have demonstrated that the rhythmicity of an mRNA is not necessarily predictive of the rhythmicity of its cognate protein (Mauvoisin et al., 2014; Reddy et al., 2006). These reports raise fundamental questions about whether and how the circadian clock regulates post-transcriptional gene expression (Lim and Allada, 2013b).
Protein synthesis is a fundamental property of living systems that—like the circadian clock—is both tightly regulated by and responsive to dynamic cellular and environmental changes (Sonenberg and Hinnebusch, 2009). Translation is primarily regulated at the initiation step (Aitken and Lorsch, 2012). Most mRNAs have a 7-methyl guanosine (m7-(GpppG)n) “cap” at their 5′ end. The cap-binding protein, eukaryotic initiation factor 4E (eIF4E), binds this mRNA cap and subsequently recruits multi-protein cap-binding complexes. Cap-dependent translation is promoted by anabolic stimuli transmitted through the mechanistic target of rapamycin (mTOR) signaling pathway. mTOR stimulates translation by phosphorylation of eIF4E-binding proteins (4E-BPs) and S6K1/2 (Ma and Blenis, 2009). Recent studies have reported circadian oscillations in mTOR signaling (Cornu et al., 2014; Jouffe et al., 2013; Khapre et al., 2014); however, little is known about mechanisms underlying the timing of mTOR-mediated protein synthesis in relation to the light-dark cycle.
Supporting a connection between the circadian clock and translational regulation, total mRNA levels for many ribosomal proteins are not rhythmic on circadian timescales yet their polysome-bound fractions are (Jouffe et al., 2013). Moreover, loss of Bmal1 alters the circadian rhythmicity of ribosomal protein expression. Interestingly, BMAL1 is structurally and evolutionarily related to the transcription factor, hypoxia inducible factor-2α (HIF-2α); HIF-2α biochemically interacts with translation factors to regulate hypoxia-dependent translation (McIntosh et al., 2010; Uniacke et al., 2012).
Based on these reports, we hypothesized that BMAL1 regulates post-transcriptional gene expression. We demonstrate that BMAL1 interacts with the translational machinery in the cytosol in response to S6K1-mediated phosphorylation. BMAL1 stimulates translation in cells in a manner independent of its role as a transcription factor. S6K1-mediated phosphorylation places BMAL1 in context of the mTOR pathway, a major cellular regulator of translation. In synchronized cells, protein synthesis rates demonstrate circadian oscillations that are partially BMAL1-dependent. Together, our data demonstrate that BMAL1 is a translation factor that links mTOR-mediated translation to the circadian clock.
RESULTS
BMAL1 Interacts with Translational Regulators in the Cytosol
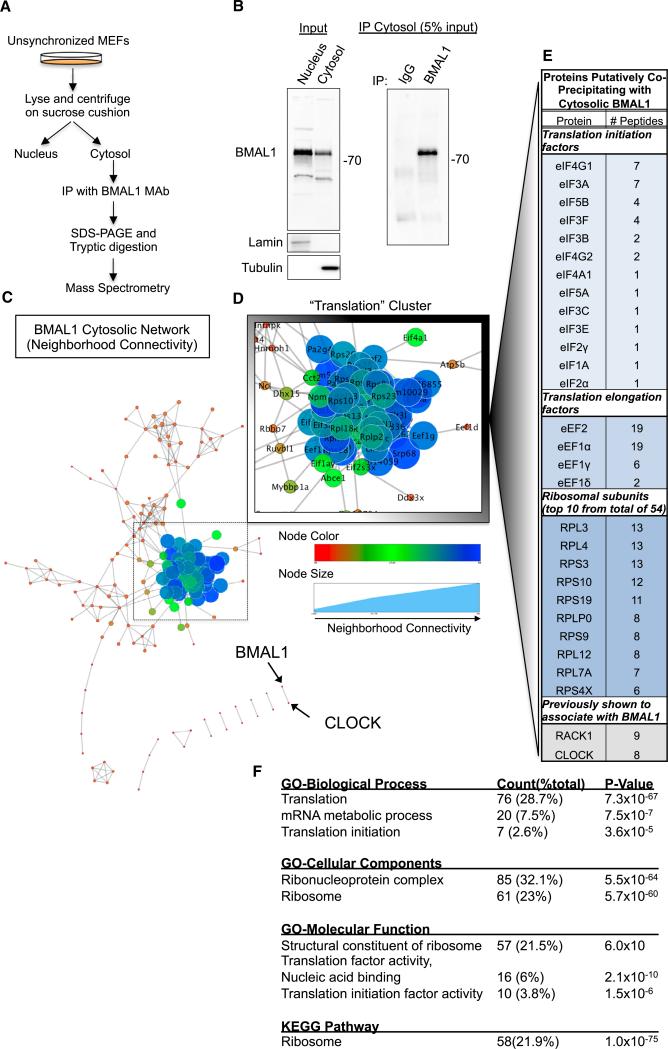
We reasoned that if BMAL1 has a role in post-transcriptional gene expression, characterization of its cytosolic binding partners would yield insights into this potential function. We performed immunoprecipitations of endogenous BMAL1 from cytosolic fractions of immortalized wild-type (WT) mouse embryonic fibroblasts (MEFs) (Figures 1A and 1B). We used unsynchronized cells to eliminate a priori assumptions about when during the circadian cycle BMAL1 might act in the cytosol. We characterized proteins that co-precipitated with BMAL1 by SDS-PAGE followed by mass spectrometry (MS). We retrieved peptides corresponding to 308 annotated mouse proteins (Tables S1A–S1C).
Figure 1. A Screen for BMAL1 Cytosolic Interactions Nominates Translation.
(A) Schematic of experimental plan for cellular fractionation, immunoprecipitation, and tandem mass spectrometry (LC-MS/MS) to isolate proteins that putatively interact with BMAL1 in the cytosol.
(B) Representative immunoblots of WT MEF lysates separated into nuclear and cytosolic fractions probed with indicated antibodies.
(C) BMAL1 Cytosolic Interaction Network demonstrates a single major cluster.
(D) The largest and most connected cluster in the network is the “Translation” Cluster, comprised of 89 proteins including over 50 ribosomal proteins and 15 translation factors. See also Table S1 and Figure S1.
(E) List of a subset of translation factors identified by LC-MS/MS found in the “Translation” Cluster.
(F) The list of annotated putative co-precipitating proteins was loaded into the DAVID algorithm. The only significantly nominated pathway from KEGG analysis was “Ribosome.”
To analyze the putative function of the BMAL1-associated proteins in the cytoplasm, we performed a network clustering analysis using a Markov Clustering Algorithm with the Search Tool for the Retrieval of Interacting Genes/Proteins program (STRING 9.1) (Brohée and van Helden, 2006; Franceschini et al., 2013). Each protein was thereby assigned a “combined neighborhood score” relative to other proteins in the list (Table S1C). A major cluster of 89 proteins was readily apparent within the network whereas other annotated proteins in the network demonstrated relatively weak clustering or no clustering at all (Figures 1C–1E; Table S1D). Proteins within this main cluster included many well-characterized translation factors such as eIF4A, eIF4G, members of the eIF3 ternary complex, eIF5A, eIF5B, eIF2α, polyadenylate binding protein 1 (PABP1), and over 50 ribosomal proteins. We refer to this cluster as the “translation” cluster.
To independently analyze the list of putative BMAL1-interacting proteins, we performed a Functional Annotation Clustering Analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.7) on the original list of annotated peptides (Huang et al., 2009). This analysis demonstrated that protein clusters involved in translation were highly represented in the list of putative BMAL1-interacting proteins. Remarkably, Gene Ontology and KEGG pathway analysis also nominated translation, translational proteins, and the ribosome (Figures 1F and S1A). Together, these data suggest that the translational machinery is preferentially represented in proteins that co-precipitate with cytosolic BMAL1 (see also Table S1D).
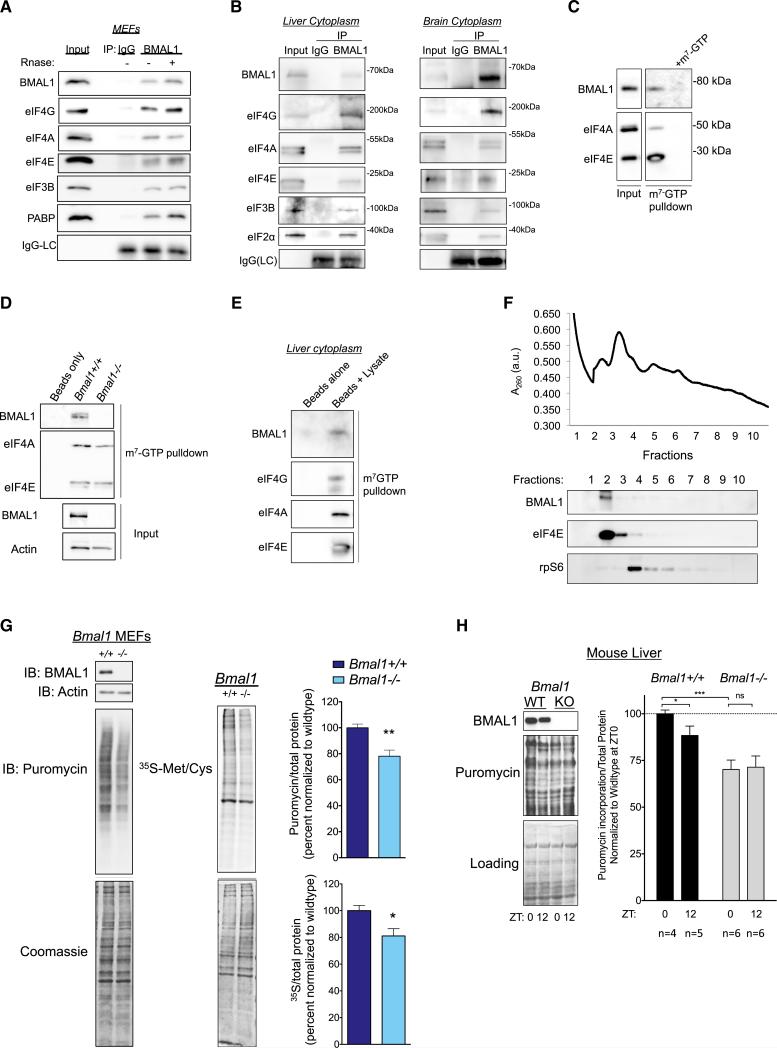
To confirm putative targets identified by MS, we immunoprecipitated endogenous BMAL1 from MEF cytosolic fractions and immunoblotted for translation factors. We found that BMAL1 associated with eIF4E, eIF4A, eIF4G, eIF3B, and PABP (Figure 2A). To test for these interactions in vivo, we prepared cytoplasmic lysates from mouse liver and brain and performed similar immunoprecipitations with BMAL1 or control IgG. BMAL1 co-precipitated with several translation factors in both liver and brain (Figure 2B).
Figure 2. BMAL1 Associates with Translation Initiation Factors and Stimulates Translation in Cells.
(A) Representative western blot of unsynchronized WT MEFs. Cells were fractionated and immunoprecipitations performed with anti-BMAL1 monoclonal antibody or mouse IgG.
(B) BMAL1 associates with translation factors in vivo. Cytoplasmic extract from mouse liver or brain were immunoprecipitated with anti-BMAL1 monoclonal antibody or mouse IgG. Western blots were performed with indicated antibodies.
(C) Cytoplasmic MEF lysates were prepared as in (A) and incubated with m7-GTP-agarose beads. Co-eluting proteins were resolved by SDS-PAGE, and probed as indicated. m7-GTP (1 mM) was used as a cap competitor.
(D) BMAL1 m7-GTP pull-down is specific. Bmal1+/+ or _Bmal1_−/− MEF lysates were prepared as in (A).
(E) BMAL1 associates with CBCs in vivo. Cytoplasmic extracts of mouse liver (as in B) were normalized to total protein and incubated with m7-GTP-agarose beads. Western blots were performed with indicated antibodies.
(F) An extract of Bmal1+/+:GFP MEFs was subjected to sucrose density gradient centrifugation. The absorbance profile at 260 nm is shown. The gradient was fractionated; fractions were analyzed by western blotting. Note that BMAL1 and eIF4E were found in the same ribosomal fractions. Ribosomal protein (rpS6) identifies the small ribosomal subunit.
(G) Primary MEFs (passage 2–4) of indicated genotype were pulsed with 1 μM puromycin for 30 min, and de novo protein synthesis was quantified by detection of mean ratio of puromycin to total protein (Coomassie) ± SEM normalized to WT (n = 7, **p = 0.002, Student's t test). Independent cell lines from at least two mice per genotype were used. The same cell lines were pulsed with 35S-Met/Cys for 30 min followed by lysis and SDS-PAGE. De novo protein synthesis was determined as the mean ratio of 35S incorporation relative to total protein (Coomassie) ± SEM (n = 3, *p = 0.04, Student's t test).
(H) BMAL1 regulates protein synthesis in the liver. Puromycin was injected 30 min before indicated time points into WT or _Bmal1_−/− mice kept on a 12 hr/12 hr light/dark cycle. Livers were harvested and whole cell lysates prepared. Protein synthesis was calculated as the ratio of puromycin/total protein, and the mean ± SEM is shown for the indicated number of mice (p = 0.01 for interaction, ***p < 0.0001 for genotype, *p < 0.02 for time, two-way ANOVA, Tukey post test).
To test whether cap-binding complexes (CBCs) would associate with BMAL1, we performed cap pull-down assays with bead-immobilized m7-GTP, a defined method for isolating CBC components (Sonenberg et al., 1979). BMAL1 was readily detectable in m7-GTP pull-down assays; the addition of competing m7-GTP nucleotide abrogated the signal suggesting that the presence of BMAL1 on the immunoblot was specific to interaction of BMAL1-associated complexes with the cap analog (Figure 2C). To confirm the specificity of the BMAL1 signal, we performed m7-GTP pull-down assays on _Bmal1_−/− MEF lysates and observed no BMAL1 signal (Figure 2D). We confirmed that BMAL1 associates with the CBC in vivo by performing m7-GTP pull-downs from liver cytoplasmic lysates (Figure 2E). Importantly, the addition of RNase to the lysates prior to pull-down assays did not reduce the association of BMAL1 with translation factors in either immunoprecipitations or m7-GTP pull-down assays, implying that BMAL1 associates with the CBC through protein-protein interactions (Figure 2A).
We next analyzed the ribosome profile of post-nuclear cell lysates and found that BMAL1 co-fractionated with translation initiation factors (Figure 2F). This is consistent with our finding that BMAL1 interacts with the translation initiation machinery and points to a role for BMAL1 in translation initiation.
BMAL1 Stimulates Translation Independently of Its Transcriptional Activity
The interaction of BMAL1 with the translation machinery raised the question of whether BMAL1 directly affects translation. To test this, we pulse-labeled primary Bmal1+/+ or _Bmal1_−/− MEFs with puromycin followed by immunoblotting with an anti-puromycin antibody (i.e., SuNSET assays) (Schmidt et al., 2009). In parallel, we also pulsed Bmal1+/+ or _Bmal1_−/− cells with 35S-Met/Cys as an independent measurement of de novo protein synthesis. In both experiments, we discovered a reduction in protein synthesis rates in _Bmal1_−/− cells compared to WT controls (Figure 2G). We conclude that BMAL1 stimulates de novo protein synthesis in cells.
We next tested de novo protein synthesis in animals by placing _Bmal1_−/− or WT littermate mice in 12/12 light/dark cycles and injecting them with puromycin intraperitoneally at either ZT0 or ZT12 (Goodman et al., 2011). We found a significant decrease in protein synthesis in _Bmal1_−/− compared to controls at ZT0 (the end of the active period). We also noted a significant difference in protein synthesis between ZT0 and ZT12 (the end of the rest period) in WT mice; this difference was abrogated in _Bmal1_−/− mice (Figure 2H). Together, these data suggest that BMAL1 promotes de novo protein synthesis in liver during the active period.
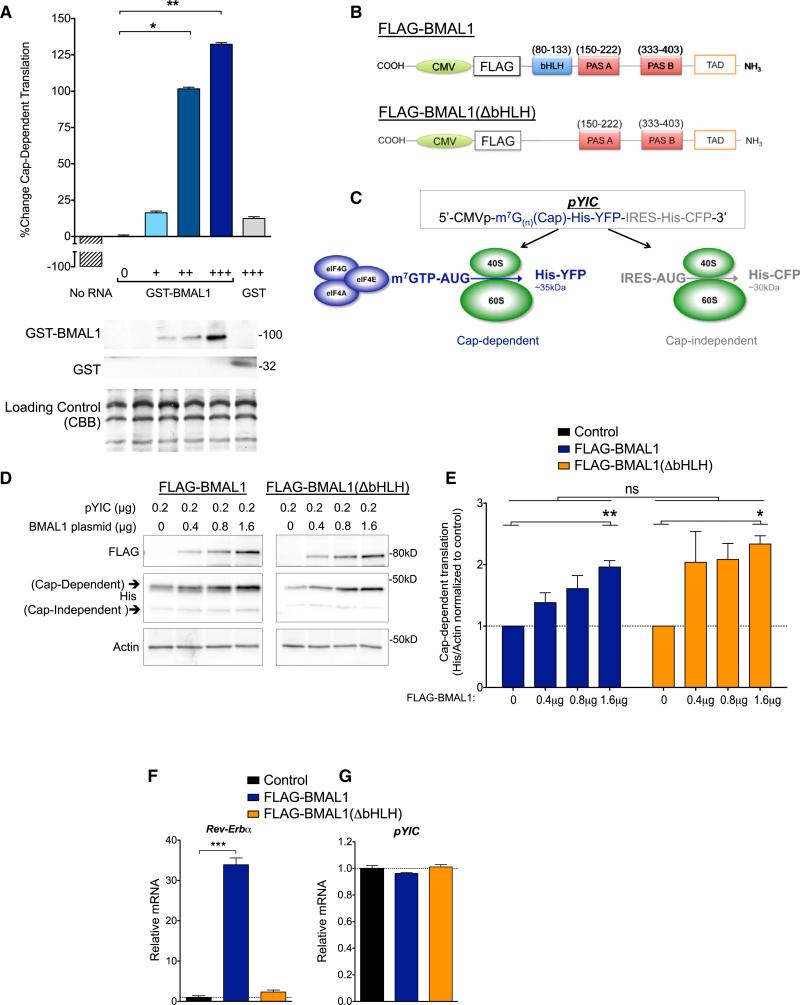
Because BMAL1 is a transcription factor, we tested whether it is capable of stimulating translation independently of transcription. We added a synthetic m7(G)n-capped mRNA to cell-free rabbit reticulocyte lysates in the presence of 35S-Met/Cys and added either recombinant GST-tagged BMAL1 to the reaction, or GST alone as a negative control. We observed that GST-BMAL1 significantly increased translation in a dose-dependent manner whereas GST alone did not (Figure 3A). Thus, BMAL1 can stimulate translation of capped mRNA in a transcription-independent system.
Figure 3. BMAL1 Can Stimulate Translation Independent of Its Role in Transcription.
(A) Recombinant full-length human GST-tagged BMAL1 stimulates translation in vitro in rabbit reticulocyte lysates. Synthetic m7-GTP capped Xef1 mRNA was added to each reaction with increasing amounts of GST-BMAL1 incubated with 35S-Met/Cys. Lysates were resolved by SDS-PAGE, autoradiography performed, and western blots were probed as indicated. Quantification of de novo protein synthesis was performed by calculating the mean volume of 35S compared to control ± SEM (**p = 0.005, *p < 0.05, one-way ANOVA with Tukey post test, n = 3 replicates).
(B) Cartoon of CMV-FLAG-BMAL1 plasmids. CMV-FLAG-BMAL1(ΔbHLH) yields a predicted protein product ~15 kDa smaller than WT.
(C) Cartoon of the pYIC bicistronic translational reporter plasmid (Nie and Htun, 2006). His, 6xHis tag.
(D) Representative western blot of HEK293T cells transfected with pYIC and indicated amounts of FLAG-BMAL1 or FLAG-BMAL1(ΔbHLH) and/or empty vector (pcDNA3.1). Total DNA, 1.8 μg/condition.
(E) Histograms of data from (D). Cap-dependent YFP signal was normalized to actin by densitometry. Mean intensity ± SEM are shown normalized to no BMAL1 plasmid (*p < 0.05; **p < 0.01, amount of BMAL1 plasmid; WT versus ΔbHLH, not significant; two-way ANOVA, n = 4–6 per condition).
(F) BMAL1(ΔbHLH) did not significantly activate the BMAL1 transcriptional target _Rev-Erb_α. HEK293T cells were transfected as in (D) with the addition of the BMAL1 co-activator myc-CLOCK. Data are represented as the mean ± SEM of _Rev-Erb_α RNA normalized to control (***p < 0.0001, one-way ANOVA, Tukey post test; BMAL1(ΔbHLH), not significant).
(G) BMAL1 overexpression has no effect on pYIC transcription in HEK293T cells. Data are represented as the mean of YFP(pYIC) RNA ± SEM (one-way ANOVA not significant, Tukey post test, n = 3 replicates).
To directly test whether BMAL1 stimulates translation in cells independently of its role in transcription, we engineered FLAG-BMAL1(ΔbHLH), which lacks the entire basic-helix-loop helix (bHLH) domain required for both DNA binding and transcription (Figure 3B). We compared the translational function of FLAG-BMAL1 and FLAG-BMAL1(ΔbHLH) in HEK293T cells co-transfected with a bicistronic translational reporter (pYIC) in which a constitutive CMV promoter drives a cap-dependent His-YFP in cis with an IRES-His-CFP (Figure 3C) (Nie and Htun, 2006). Full-length BMAL1 activated cap-dependent translation in a dose-dependent manner (Figures 3D and 3E). Importantly, transcriptionally inept FLAG-BMAL1(ΔbHLH) was commensurate with WT BMAL1 at stimulating cap-dependent translation (Figures 3D–3F). Neither FLAG-BMAL1 nor FLAG-BMAL1(ΔbHLH) had an effect on the transcription of pYIC, confirming that the changes we observed in the reporter are a reflection of changes in translation not transcription (Figure 3G). These data strongly suggest that BMAL1 can activate translation in cells through a mechanism that is independent of transcription.
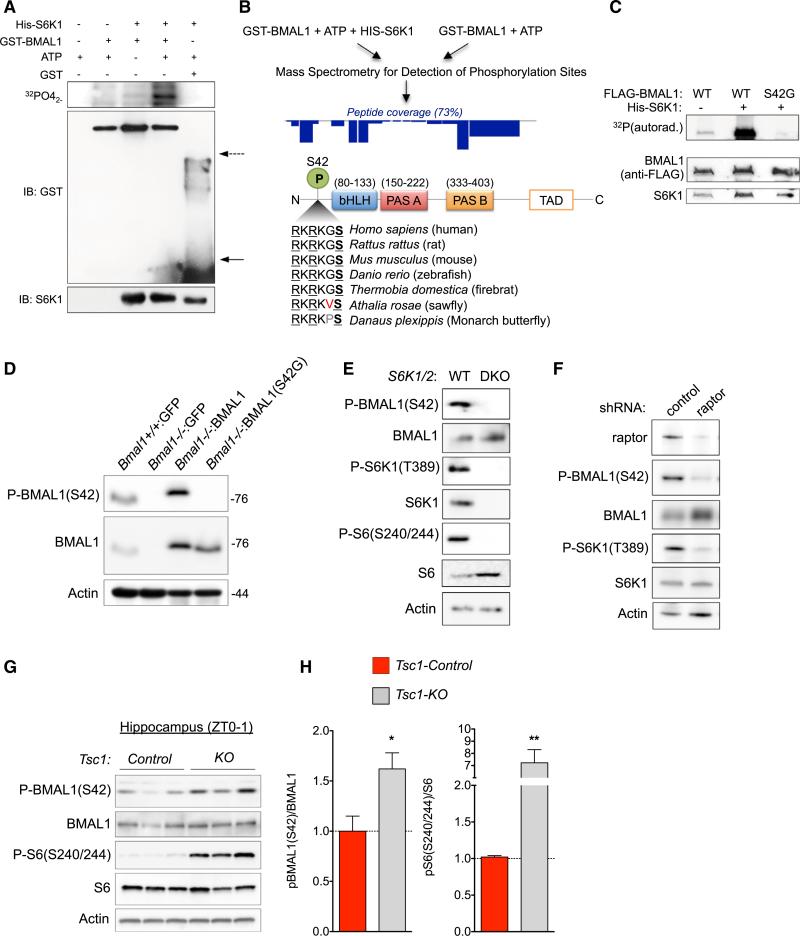
BMAL1 Is a Substrate of S6K1
Translation regulation relies on the modulation of translational factors by integrated signaling responses. The mTOR signaling pathway is a crucial integrator of translational responses with environmental and cellular cues (Ma and Blenis, 2009). In response to anabolic stimuli, mTOR promotes translation by phosphorylation of 4E-BPs and S6K1/2. Several S6K1 substrates such as PDCD4, eIF4B, and eukaryotic elongation factor 2 kinase are, in turn, translational regulators (Dorrello et al., 2006; Raught et al., 2004; Shahbazian et al., 2006; Wang et al., 2001). S6K1 is a member of the AGC kinase family, whose substrates have a well-defined phosphorecognition motif (RxRxxS/T) (Pearce et al., 2010). We observed that BMAL1 has several conserved RxRxxS/T motifs, which led us to hypothesize that BMAL1 is an S6K1 substrate. We performed in vitro kinase assays with activated S6K1 and observed strong transfer of 32P to GST-BMAL1 but not to GST alone (Figure 4A). To identify potential sites of S6K1-mediated phosphorylation, we performed microcapillary-tandem mass spectrometry (LC-MS/MS) comparing gel-purified GST-BMAL1 that had been subjected to kinase assays either with or without the addition of recombinant S6K1 (Bordoli et al., 2014; Breitkopf and Asara, 2012). LC-MS/MS analyses detected phosphorylation at S42, a serine situated within a canonical RxRxxS/T (37-RKRKGS-42) motif and evolutionarily conserved across phyla suggesting that it is functionally important (Figure 4B). To confirm that S42 was an S6K1 site, we repeated kinase assays with WT FLAG-BMAL1 or a phosphorylation-incompetent mutant FLAG-BMAL1(S42G). While 32P was readily incorporated into FLAG-BMAL1, almost none was detected in FLAG-BMAL1(S42G) (Figure 4C). These data demonstrate that S42 is a primary site of S6K1-mediated phosphorylation in vitro.
Figure 4. BMAL1 Is Phosphorylated by S6K1 In Vitro and In Vivo.
(A) Recombinant full-length GST-BMAL1 was incubated with His-S6K1 and 32P-γATP. Reactions were resolved by SDS-PAGE, and autoradiography was performed followed by western blotting. Arrows indicate GST monomers (solid) or GST dimers (dashed).
(B) Outline of workflow for LC-MS/MS experiments to identify sites of S6K1-mediated BMAL1 phosphorylation. Dark blue rectangles approximate the relative representation of BMAL1 peptides in the LC-MS/MS data. Below, alignment of the putative S6K1 phosphorylation site at S42 (in human BMAL1) demonstrates evolutionary conservation across phyla.
(C) S42 is an S6K1 phosphorylation site in vitro. HEK293T cells were transfected with either FLAG-BMAL1 or FLAG-BMAL1(S42G). FLAG immunoprecipitations were performed and washed. Recombinant S6K1 was added in the presence of 32P-γATP. Autoradiography was performed followed by western blotting.
(D) BMAL1 is phosphorylated at S42 in vivo. Western blot of lysates from immortalized Bmal1 cell lines as indicated. Overexpressed BMAL1 is detected at higher level and is larger in size because of FLAG-HA tag.
(E) S6Ks phosphorylate BMAL1 in cells. Whole cell lysates from WT or S6K1/2 double knockout (DKO) cells were probed with indicated antibodies.
(F) BMAL1 is phosphorylated at S42 downstream of mTORC1. WT MEFs were infected with lentivirus expressing a scrambled shRNA (control) or sh-raptor.
(G and H) Hippocampal lysates from SynapsinI-Cre;Tsc1-flox/flox (KO) or SynapsinI-Cre;Tsc1+/+ (control) mice were probed with indicated antibodies. Histogram of quantified western blots from (G). Data are represented as the mean ± SEM relative to WT (n = 3 mice per genotype, t test, *p < 0.05, **p < 0.005).
Using a phospho-specific BMAL1(S42) antibody, we tested whether BMAL1 is phosphorylated at S42 in cells. We confirmed phospho-specificity of the antibody with in vitro kinase assays (Figure S1B). We then produced four immortalized, stably transduced MEF lines: Bmal1+/+:GFP, _Bmal1_−/−:GFP, _Bmal1_−/−:BMAL1 (re-expressing WT BMAL1), and _Bmal1_−/−:BMAL1(S42G) (re-expressing phosphorylation-incompetent BMAL1). Anti-phospho-BMAL1(S42) antibody yielded a signal in both Bmal1+/+:GFP and _Bmal1_−/−:BMAL1 cells but in neither _Bmal1_−/−:GFP nor _Bmal1_−/−:BMAL1(S42G) cells. These data confirm that BMAL1 is phosphorylated at S42 in cells (Figure 4D).
To test whether S42 is a substrate of S6K in vivo, we probed for BMAL1 phosphorylation at S42 in S6K1_−/−;S6K2_−/− (DKO) MEFs and WT MEFs. While WT cells demonstrated a strong phospho-BMAL1(S42) signal, we detected little phosphorylation in DKO cells (Figure 4E). We further confirmed that BMAL1 is a direct substrate of the mTOR pathway, by testing small hairpin RNA (shRNA)-mediated knockdown of the mTORC1-specific component raptor (Laplante and Sabatini, 2012). We observed a marked decrease in phospho-BMAL1(S42) in sh-raptor cells compared to controls (Figure 4F). The TSC1 and TSC2 tumor suppressors inhibit mTORC1 activation; thus, deletion of either protein results in a state of growth factor-independent mTORC1 activation. We observed increased phospho-BMAL1(S42) in hippocampal lysates of mice harboring neuron-specific deletion of Tsc1 compared to those of control littermates (Figures 4G and 4H). Together, these data further support the conclusion that BMAL1 is a S6K substrate downstream of mTORC1.
Phosphorylation at S42 Is Required for BMAL1 to Promote Translation
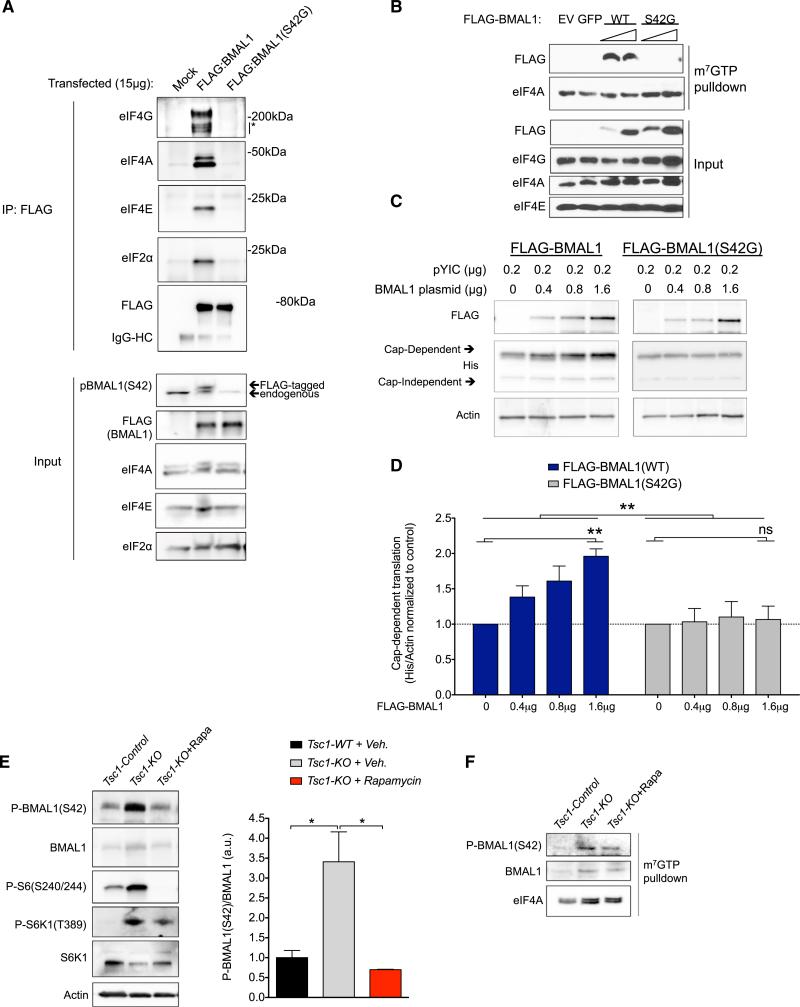
Phosphorylation regulates the activity of several translation factors (Sonenberg and Hinnebusch, 2009). We found that phosphorylated BMAL1 co-fractionated with translation initiation factors at the ribosome (Figure S1C). To directly test whether phosphorylation at S42 is required for BMAL1's association with translation factors, we transfected HEK293T cells with FLAG-BMAL1 or FLAG-BMAL1(S42G), immunoprecipitated with anti-FLAG antibody, and probed immunoprecipitates for CBC proteins. WT BMAL1 strongly associated with eIF4E, eIF4A, eIF4G, and eIF2α. In contrast, BMAL1(S42G) did not (Figure 5A). We performed m7-GTP pull-down assays on similarly transfected cells. Again, WT BMAL1 strongly associated with translation factors in CBC pull-downs whereas BMAL1(S42G) did not (Figure 5B). We conclude that S42 phosphorylation is required for BMAL1 to interact with the CBC.
Figure 5. BMAL1 Phosphorylation at S42 Mediates Its Interaction with Translational Machinery and Promotes Cap-Dependent Translation.
(A) HEK293T cells were transfected with FLAG-BMAL1 or FLAG-BMAL1(S42G). 24 hr later, cells were lysed and immunoprecipitated with FLAG antibody. Western blots were probed with indicated antibodies.
(B) HEK293T cells were transfected with 10 μg or 20 μg of FLAG-BMAL1 or FLAG-BMAL(S42G) for 24 hr. Post-nuclear cell fractions were treated with RNase A followed by m7-GTP pull-down assays. Western blots were probed as indicated.
(C) HEK293T cells were transfected with pYIC and FLAG-BMAL1 or FLAG-BMAL1(S42G) as in Figure 3D. Cell lysates were probed with indicated antibodies. See also Figures S2, S3, and S4.
(D) Quantification of data from (C). His-YFP (cap-dependent) was normalized to actin and the mean ± SEM is represented normalized to control. **p < 0.01, BMAL1 plasmid for WT and non-significant (ns) for S42G. **p < 0.01, WT versus S42G (two-way ANOVA) n = 4– replicates per condition.
(E) Western blots of hippocampal lysates of SynapsinI-Cre;Tsc1+/+ (Tsc1-Control) or SynapsinI-Cre;Tsc1-flox/flox (Tsc1-KO) treated with vehicle or rapamycin. Data are represented as the mean ± SEM relative to WT (n = 2 mice per genotype per condition, one-way ANOVA, Tukey post test, *p < 0.05).
(F) m7-GTP pull-down assays were performed on pooled cytoplasmic lysates from (E) and immunoblots of eluted proteins were performed with indicated antibodies.
We hypothesized that if S42 phosphorylation is required for CBC association, then BMAL1(S42G) would be less efficient at promoting translation when compared to WT BMAL1. We performed bicistronic reporter assays comparing BMAL1 to BMAL1(S42G) (as in Figure 3) and found that, even at highest expression levels, BMAL1(S42G) did not promote cap-dependent translation (Figures 5C and 5D). We confirmed this finding using an independent bicistronic translational reporter (Figure S2).
Phosphorylation at other BMAL1 serine residues has been previously shown to affect BMAL1 stability and nuclear entry (Sahar et al., 2010). We noted that the S42 site (37-RKRKGS-42) is directly adjacent to 38-KR-39 residues previously identified as a nuclear localization signal for BMAL1 (Kwon et al., 2006). Thus, we considered the possibility that BMAL1-S42G might neither interact with the CBC nor activate translation because of changes in its stability or subcellular localization. However, several findings make this unlikely. First, the S42G mutant was more stable than WT BMAL1 (Figures S3A and S3B). Second, we found that more BMAL1-S42G localized to the cytoplasm in MEFs compared to WT BMAL1 (Figure S3C). This effect may be secondary to S6K1-mediated phosphorylation as either WT BMAL1 expressed in _S6K1_−/− cells or BMAL1(S42G) expressed in S6K1+/+ cells demonstrated indistinguishable BMAL1 subcellular localization (Figure S3C). These data make it unlikely that changes in BMAL1(42G) stability or subcellular localization could explain the inability of the phosphorylation-incompetent protein to interact with the translational machinery. Thus, we conclude that BMAL1 phosphorylation at S42 is required for BMAL1 to promote translation by regulating its interaction with the translation machinery.
High levels of S6K1 expression have been recurrently implicated in breast cancer leading us to examine a role for BMAL1-mediated translation in breast cancer cells (Couch et al., 1999; Maruani et al., 2012). We observed that BMAL1(S42) phosphorylation was detectable at high levels in MCF7 cells, in which S6K is active even in low-serum conditions (Figure S4A). Treatment of MCF7 with rapamycin reduced BMAL1 phosphorylation and de novo protein synthesis (Figure S4A). Importantly, overexpression of phosphorylation-competent BMAL1 in MCF7 cells increased the number of metabolically active cells while the phosphorylation-incompetent BMAL1(S42G) had no significant effect (Figure S4B). The effect of BMAL1 on proliferation was dependent on the presence of serum. Together, these data suggest a role for BMAL1 phosphorylation in the promotion of translation and proliferation in breast cancer cells.
To further examine the mTORC1-dependence of BMAL1 phosphorylation in vivo, we tested brain-specific Tsc1 knockouts treated with vehicle or rapamycin. Consistent with our previous data, we noted a marked increase in BMAL1 phosphorylation in Tsc1 mutant hippocampus (Figure 5E). Rapamycin resulted in a decrease in phospho-S6K1 and a decrease in phospho-BMAL1 as well as a decrease in the association of BMAL1 with the m7-GTP cap (Figures 5E and 5F). These data substantiate that S6K1-mediated BMAL1 phosphorylation promotes association of BMAL1 with the translational machinery in vivo.
BMAL1 Rhythmically Associates with the Translational Machinery
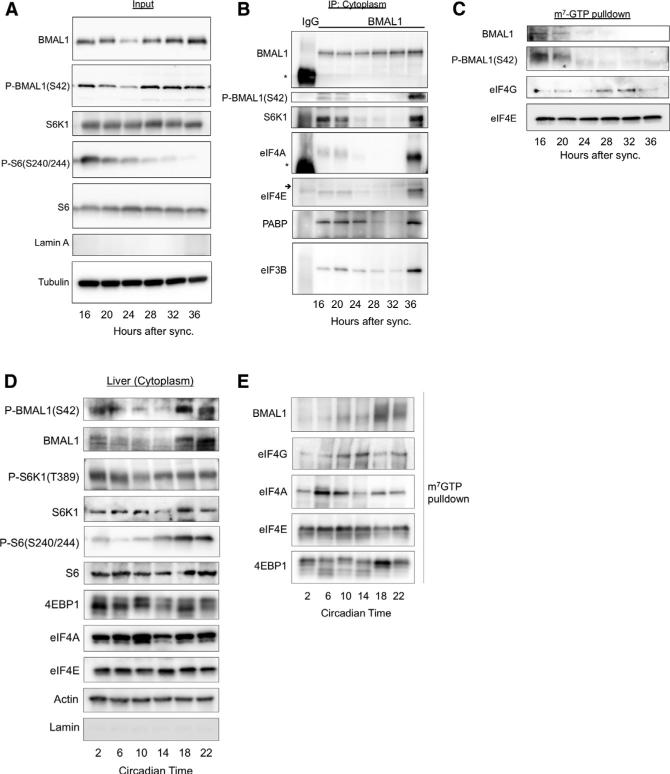
BMAL1 has been previously reported to shuttle between the cytosol and nucleus with a circadian oscillation (Kwon et al., 2006; Tamaru et al., 2003). We asked whether the interaction of BMAL1 with the translational machinery varies during the circadian cycle. We synchronized MEFs with 100 nM dexamethasone and performed subcellular fractionation followed by biochemical assays (Figures 6 and S5A). Levels of cytosolic BMAL1 initially decreased from zeitgeber time (ZT) (i.e., time post-synchronization) 16–24 hr after synchronization and then increased thereafter (Figure 6A). We confirmed this circadian variability by immunocytochemistry, which demonstrated that the cytoplasmic BMAL1 staining was markedly increased at ZT36 compared to ZT24 (Figures S5B and S5C).
Figure 6. BMAL1 Rhythmically Associates with S6K1 and the Translational Machinery.
(A) WT MEFs were synchronized with dexamethasone followed by subcellular fractionation over a sucrose cushion at indicated time points. Representative western blot is shown (n = 4 biological replicates; see Figure S5 for nuclear lysates).
(B and C) BMAL1 immunoprecipitation of normalized cytosolic lysates were treated with RNase and subjected to immunoprecipitation with BMAL1 antibody (B) or pull-down assays with m7-GTP beads (C). Western blots were performed with indicated antibodies. Asterisk corresponds to the IgG-heavy chain and small arrow to IgG-light chain, respectively (see Figure S5).
(D) BMAL1 phosphorylation rises during the active period in liver. Mice were maintained on a 12 hr/12 hr light/dark schedule with ad libitum availability to food prior to release into constant darkness. Tissues were collected at indicated time points. Liver cytoplasmic extracts were collected, normalized to total protein, and immunoblots were performed. Representative western blot from n = 3 mice per time point is shown.
(E) Extracts prepared from (D) were normalized to total protein content, and subjected to m7-GTP pull-down assays followed by immunoblotting as indicated.
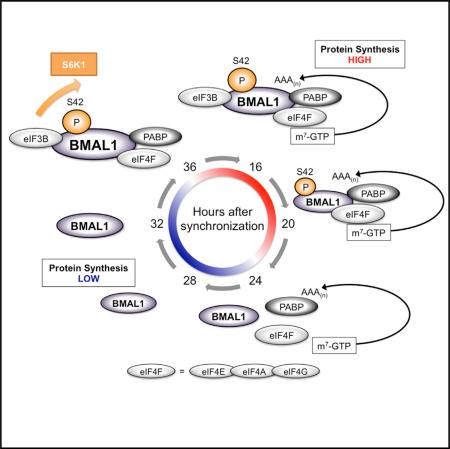
We next immunoprecipitated BMAL1 from the cytosol at different ZTs (Figure 6B). BMAL1 rhythmically associated with S6K1, eIF4E, eIF4A, PABP, and eIF3B in a pattern that followed its phosphorylation at S42. Starting at ZT16 the level of association decreased until ZT28 and then, as both total and phosphorylated levels of BMAL1 increased, association with translation factors also increased.
BMAL1 and phospho-BMAL1(S42) associated with the cap analog m7-GTP in pull-down assays at ZT16-20 and then the association declined precipitously to nearly undetectable levels (Figure 6C). Importantly, we did not observe BMAL1 or phospho-BMAL1(S42) associate with the cap analog at 36 hr after synchronization. One interpretation of these findings is that BMAL1 associates with the translational machinery prior to the assembly of the translational initiation complexes with the mRNA cap.
Previous studies have demonstrated oscillations in the mTOR pathway in mouse liver with peak phospho-S6 levels at the start of the subjective night (the dark period) (Cornu et al., 2014; Jouffe et al., 2013; Khapre et al., 2014). Consistent with these studies, we observed maximal phospho-S6 levels during the active period with a clear variation between circadian night and day. We queried phospho-BMAL1(S42) during the circadian cycle in cytoplasmic extracts from mouse liver harvested in constant darkness. We observed that phosphorylation peaked at the end of the active period (Figure 6D). Similarly, we observed that in m7-GTP pull-down assays from the same lysates that BMAL1 maximally associated with cap binding complexes during the active period (Figure 6E). These data are consistent with our previous finding that de novo protein synthesis is relatively high in the liver at the end of the active period (Figure 2H). Together these data suggest that BMAL1 undergoes circadian phosphorylation by S6K1 during the circadian cycle in phase with its association with the translational machinery in vivo.
Protein Synthesis Rates Demonstrate Circadian Rhythmicity
The rhythmicity of BMAL1 association with the translational machinery and its ability to promote translation led us to investigate whether synchronized cells demonstrate circadian rhythms of overall protein synthesis rates. Rhythms of both protein synthesis in the SCN and polysome quantity in the liver have been reported (Jouffe et al., 2013; Shibata et al., 1992). We tested this question in U2-OS cells stably expressing a Per2:Luciferase transgene (PLuc cells) (Zhang et al., 2009). PLuc cells demonstrated strong circadian rhythms in protein synthesis rate (Figure S6). The observed rhythm is unlikely an artifact of culturing conditions because they persist for 2 days after synchronization, suggesting the involvement of a clock mechanism.
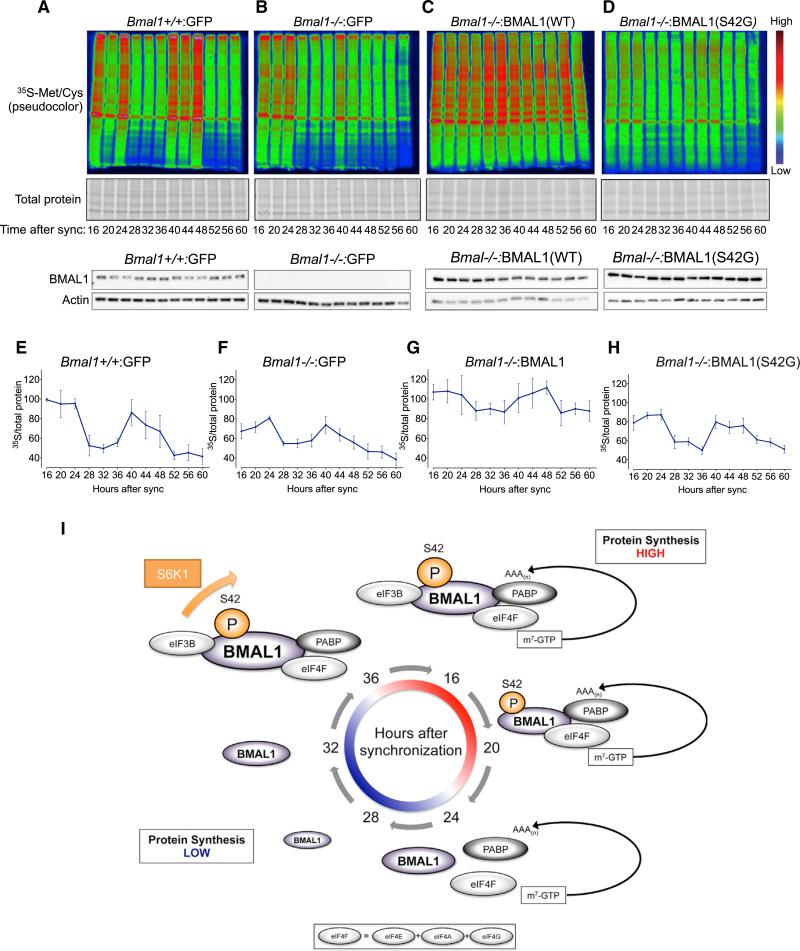
BMAL1 is essential to circadian rhythms in transcription (Bunger et al., 2000). Other groups have demonstrated that the rhythmic expression of ribosomal subunits (with non-rhythmic mRNAs) in the liver is altered in _Bmal1_−/− mice (Jouffe et al., 2013). Thus, we asked whether BMAL1 plays a role in circadian protein synthesis rates using the four immortalized cell lines utilized in Figure 4D. We pulsed cells with 35S-met/cys every 4 hr after synchronization to measure protein synthesis rates (Figures 7A–7H and S7A–S7D). Bmal1+/+:GFP cells demonstrated a clear circadian oscillation in 35S-met/cys incorporation over 2 days after synchronization with high levels from 16–24 and 40–48 hr after synchronization and relatively low levels from 28–36 and 52–60 hr after synchronization (Figures 7A, 7E, and S7E). Importantly, this pattern of de novo protein synthesis correlated well with the association of BMAL1 with the cap complex at 16–20 hr after synchronization (Figure 6C). The observed rhythms persisted for 2 days, suggesting the involvement of an autonomous timing mechanism.
Figure 7. Circadian Rhythms of Protein Synthesis Rates Are Partially BMAL1 and Phospho-BMAL1(S42) Dependent.
(A–D) Bmal1+/+:GFP MEFs were synchronized with dexamethasone. At indicated time points, cells were pulsed with 35S-Met/Cys every 4 hr from hours 16–60 after synchronization followed by cell lysis, SDS-PAGE, autoradiography, and western blotting (below). Representative autoradiograms of the same exposure were pseudocolored for visual clarity (see Figures S7A–S7D for originals), n = 3 replicates per cell line. See also Figures S6 and S7.
(E–H) Quantification of (A)–(D). 35S autoradiograms were quantified by densitometry, normalized to Coomassie-stained gels for total protein and normalized to Bmal1+/+:GFP cells at ZT16.
(I) Schematic model of BMAL1 phosphorylation and association with the translational machinery in the cytosol during the circadian cycle.
Although a circadian oscillation in protein synthesis was partially preserved in _Bmal1_−/− :GFP cells, the peak protein synthesis levels during the “positive limb” of the cycle was diminished and overall amplitude reduced (Figures 7B, 7F, S7E, and S7F). Surprisingly, _Bmal1_−/−:BMAL1 cells, which overexpress WT BMAL1, also demonstrated a similar reduction in amplitude of the protein synthesis oscillation over the circadian cycle (Figures 7C, 7G, S7E, and S7F). In contrast, overexpression of BMAL1 in neither PLuc cells nor _Bmal1_−/− MEFs abolished transcriptional rhythms of Per2:Luc; furthermore, Per2:Luc oscillations did not differ in _Bmal1_−/− cells overexpressing either WT BMAL1 or BMAL1(S42G) (Figures S7G and S7H). These findings are consistent with previous reports that have shown that circadian transcriptional rhythms are not lost with BMAL1 overexpression, presumably because of the preservation of negative feedback mechanisms that tolerate high BMAL1 protein levels (Kiyohara et al., 2006). Thus, the diminution of protein synthesis rhythms in _Bmal1_−/−:BMAL1 cells imply first that, in contrast to circadian transcriptional oscillations, the level of BMAL1 protein expression is critical to the expression of circadian rhythms of protein synthesis; second, that rhythms of protein synthesis oscillations and specifically, putative negative feedback mechanisms controlling them, are likely distinct from those involved in negative feedback of circadian transcription. Finally, _Bmal1_−/−:BMAL1(S42G) cells, which overexpress a phospho-incompetent form of BMAL1, demonstrated diminished protein synthesis rates and circadian rhythmicity similar to that of _Bmal1_−/−:GFP cells (Figures 7D, 7H, 7I, and S7D–S7F). These results are consistent with our previous findings that BMAL1 phosphorylation at S42 regulates BMAL1's ability to stimulate translation.
DISCUSSION
In this study, we show that BMAL1 is a translation factor, which in response to phosphorylation by S6K1, regulates rhythms of protein synthesis (Figure 7I). Shuttling of circadian clock proteins between the nucleus and cytoplasm has been known for years; however, biological functions for circadian clock proteins in the cytoplasm have not been widely examined. Our unbiased screen for proteins that interact with BMAL1 in the cytosol identified hundreds of proteins, and those involved in translation and mRNA processing were the most highly represented (Figures 1 and S1A; Tables S1A–S1D). S6K1-mediated BMAL1 phosphorylation at S42 is required for BMAL1 to both interact with the translational machinery and stimulate protein synthesis, placing BMAL1 in context of other S6K1 substrates involved in translation (Dorrello et al., 2006; Raught et al., 2004; Shahbazian et al., 2006). These findings provide a direct molecular link between BMAL1 and the mTOR pathway.
Consistent with others, we have found that the mTOR pathway demonstrates circadian rhythmicity in both synchronized cells and mouse liver. Phospho-S6 levels rise and fall with the circadian cycle, peaking in the active period in mouse liver (Cornu et al., 2014; Jouffe et al., 2013). Similarly, we see levels of phospho-BMAL1 peak in the circadian night and diminish during mid-day. This parallels an increase in BMAL1 association with the mRNA cap during the circadian night and fits well with a relative increase in protein synthesis during the active period (see Figures 2H, 6B, 6C, and 6E). Interestingly, BMAL1 transcriptional activity peaks during the mid-late morning (Koike et al., 2012), suggesting a subcellular segregation of BMAL1 function during the circadian cycle. Thus, phosphorylation of BMAL1 during the circadian cycle by rhythmic activity of S6K1 is a potential mechanism by which temporal variation is conveyed to the translational machinery on circadian time-scales (Figure 7I).
Several studies have argued that transcriptional rhythmicity is not always predictive of rhythmic protein expression (Mauvoisin et al., 2014; Reddy et al., 2006). Thus, circadian transcription is likely not the sole provenance of circadian information in cells (O'Neill and Reddy, 2011). Indeed, inhibiting transcription within four hours of a puromycin pulse did not eliminate rhythms in de novo protein synthesis in synchronized cells (Figure S6C). It is axiomatic that translation is fundamentally dependent on transcription; however, systems-level analysis of protein expression suggests that protein copy number is more dependent on translation rate than mRNA copy number (Schwanhäusser et al., 2011). This points to the pivotal importance of translational control mechanisms at initiation and at the ribosome in the regulation of protein expression (Sonenberg and Hinnebusch, 2009). The question of circadian rhythms in protein synthesis is not new. While contested, circadian rhythms of protein synthesis have been demonstrated in both the SCN and in Drosophila clock neurons (Huang et al., 2013; Scammell et al., 1989; Shibata et al., 1992). Several recent studies have demonstrated a role for translational control of clock proteins (Cao et al., 2013; Lim and Allada, 2013a; Lim et al., 2011). Our study did not assess BMAL1's translational function in the core circadian oscillator. Studies of human erythrocytes and the alga O. tauri have demonstrated a conserved post-translational circadian clock that is transcription-independent (O'Neill and Reddy, 2011; O'Neill et al., 2011). We have found that BMAL1's ability to stimulate translation is independent of its transcriptional function, while S6K1-mediated phosphorylation is essential for BMAL1 to interact with the translational machinery. These molecular details could allow the dissection of the relative transcriptional and translational contributions BMAL1 makes to both the control of circadian timing and pathological states such as cancer, metabolic disease, neurological disease, and aging (Cornu et al., 2013; Lipton and Sahin, 2014; Zoncu et al., 2011).
EXPERIMENTAL PROCEDURES
All animal work was performed according to protocols approved and supervised by the Animal Care Facility of Boston Children's Hospital. Please see Supplemental Experimental Procedures for detailed protocols.
Mass Spectrometry on Cytosolic Fractions
All steps were performed on ice or at 4°C, unless otherwise specified. Cytoplasmic fractions from immortalized WT MEFs were isolated and normalized. Total protein (6 mg) was pre-cleared with 10 μl of pre-washed (2× with PBS and once with lysis buffer [Buffer 1]) DynaBeads Protein G (Invitrogen) and gently agitated for 15 min. Non-immobilized fractions were retrieved using a magnetic separation rack (NEB) and placed in fresh pre-chilled 1.5-ml tube. Immunoprecipitations were performed by adding 3 μg of BMAL1 monoclonal antibody (Santa Cruz #365645)/mg total protein and incubated overnight with gentle end-over-end agitation; “mock” reactions were treated identically, however, no primary antibody was added to the lysate. 16 hr later, 40 μl of fresh, pre-washed Protein G beads were added to each reaction for 1 hr with gentle agitation. Reactions were then placed on magnetic rack separator and beads were washed with Buffer 1 four times. After the final wash, 1 vol of 2× Laemmli sample buffer with β-mercaptoethanol was added to each reaction and boiled at 95°C for 5 min, then placed on ice. The eluates were retrieved on a magnetic rack and placed in a fresh tube. Proteins were separated by SDS-PAGE on 4%–20% gradient gels (Bio-Rad) and stained with Coomassie Blue for 15–20 min followed by de-staining at room temperature for 1 hr. Entire lanes were cut into pieces, washed twice with 50% mass spectrometry grade acetonitrile (Thermo Scientific) followed by two washes with high purity water. Gel slices were submitted for trypsinization and mass spectrometric analysis.
Supplementary Material
1
2
3
4
5
6
7
8
Highlights.
- The circadian protein BMAL1 rhythmically interacts with the translational machinery
- BMAL1 is a substrate of the mTOR-effector kinase S6K1
- BMAL1 regulates circadian rhythms of protein synthesis
ACKNOWLEDGMENTS
We are indebted to Drs. T. Scammell, S.L. Pomeroy, E. Henske, D.J. Kwiatkowski, P.T. Tsai, G. Lipton, and members of the M.S. lab for critical reading of the manuscript. We thank W. Lewis for technical assistance and Dr. Susanne Breitkopf and Min Yuan for help with MS experiments. We thank D. Kwiatkowski and W. Harper (Harvard), S. Kozma (University of Cincinnati), J. Hogenesch (UPENN), T. Pappagiannakopoulos (MIT), and R. Rozenfeld (Cell Signaling) for providing critical reagents. This work was supported by AAN Clinical Research Training Program, Tuberous Sclerosis Alliance, William Randolph Hearst Foundation, American Sleep Medicine Foundation Physician Scientist Training Award, the National Institute of Child, Health and Human Development (NICHD) (K08 HD071026 to J.O.L. and P30 HD018655 to M.S.), the National Institute of Neurological Disorders and Stroke (NINDS) (R01NS058956), John Merck Fund, Nancy Lurie Marks Family Foundation, the Children's Hospital Boston Translational Research Program (to M.S.), Heidelberg University Young Investigator Award, the Daimler and Benz Foundation, the Reinhard Frank Foundation (to D.E.-F.), NIH (P01CA120964 and S10RR032861 to J.M.A.), European Molecular Biology Organization (EMBO) (ALTF 1054-2010), and the Human Frontier Science Program (HFSP) (LT001215/2011-L to T.G). J.O.L. dedicates this paper to the memory of Dr. Amiram Lipton.
Footnotes
REFERENCES
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoli MR, Yum J, Breitkopf SB, Thon JN, Italiano JE, Jr., Xiao J, Worby C, Wong S-K, Lin G, Edenius M, et al. A secreted tyrosine kinase acts in the extracellular environment. Cell. 2014;158:1033–1044. doi: 10.1016/j.cell.2014.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkopf SB, Asara JM. Determining in vivo phosphorylation sites using mass spectrometry. Curr. Protoc. Mol. Biol. Chapter. 2012;18:11–27. doi: 10.1002/0471142727.mb1819s98. Unit18.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohée S, van Helden J. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinformatics. 2006;7:488. doi: 10.1186/1471-2105-7-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Robinson B, Xu H, Gkogkas C, Khoutorsky A, Alain T, Yanagiya A, Nevarko T, Liu AC, Amir S, Sonenberg N. Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron. 2013;79:712–724. doi: 10.1016/j.neuron.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 2013;23:53–62. doi: 10.1016/j.gde.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Cornu M, Oppliger W, Albert V, Robitaille AM, Trapani F, Quagliata L, Fuhrer T, Sauer U, Terracciano L, Hall MN. Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21. Proc. Natl. Acad. Sci. USA. 2014;111:11592–11599. doi: 10.1073/pnas.1412047111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FJ, Wang XY, Wu GJ, Qian J, Jenkins RB, James CD. Localization of PS6K to chromosomal region 17q23 and determination of its amplification in breast cancer. Cancer Res. 1999;59:1408–1411. [PubMed] [Google Scholar]
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc. Natl. Acad. Sci. USA. 2012;109:5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, Hornberger TA. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011;25:1028–1039. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang Y, Ainsley JA, Reijmers LG, Jackson FR. Translational profiling of clock cells reveals circadianly synchronized protein synthesis. PLoS Biol. 2013;11:e1001703. doi: 10.1371/journal.pbio.1001703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 2013;11:e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khapre RV, Patel SA, Kondratova AA, Chaudhary A, Velingkaar N, Antoch MP, Kondratov RV. Metabolic clock generates nutrient anticipation rhythms in mTOR signaling. Aging (Albany, N.Y. Online) 2014;6:675–689. doi: 10.18632/aging.100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara YB, Tagao S, Tamanini F, Morita A, Sugisawa Y, Yasuda M, Yamanaka I, Ueda HR, van der Horst GTJ, Kondo T, Yagita K. The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc. Natl. Acad. Sci. USA. 2006;103:10074–10079. doi: 10.1073/pnas.0601416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo S-H, Huang H-C, Kumar V, Lee C, Kim T-K, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Lee J, Chang SH, Jung NC, Lee BJ, Son GH, Kim K, Lee KH. BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol. Cell. Biol. 2006;26:7318–7330. doi: 10.1128/MCB.00337-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Allada R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science. 2013a;340:875–879. doi: 10.1126/science.1234785. [DOI] [PubMed] [Google Scholar]
- Lim C, Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat. Neurosci. 2013b;16:1544–1550. doi: 10.1038/nn.3543. [DOI] [PubMed] [Google Scholar]
- Lim C, Lee J, Choi C, Kilman VL, Kim J, Park SM, Jang SK, Allada R, Choe J. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature. 2011;470:399–403. doi: 10.1038/nature09728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84:275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv. Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Maruani DM, Spiegel TN, Harris EN, Shachter AS, Unger HA, Herrero-González S, Holz MK. Estrogenic regulation of S6K1 expression creates a positive regulatory loop in control of breast cancer cell proliferation. Oncogene. 2012;31:5073–5080. doi: 10.1038/onc.2011.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvoisin D, Wang J, Jouffe C, Martin E, Atger F, Waridel P, Quadroni M, Gachon F, Naef F. Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc. Natl. Acad. Sci. USA. 2014;111:167–172. doi: 10.1073/pnas.1314066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 2010;72:625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- Nie M, Htun H. Different modes and potencies of translational repression by sequence-specific RNA-protein interaction at the 50-UTR. Nucleic Acids Res. 2006;34:5528–5540. doi: 10.1093/nar/gkl584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget F-Y, Reddy AB, Millar AJ. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23:1761–1769. doi: 10.1038/sj.emboj.7600193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O'Neill JS, Wong GKY, Chesham J, Odell M, Lilley KS, et al. Circadian orchestration of the hepatic proteome. Curr. Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Sahar S, Zocchi L, Kinoshita C, Borrelli E, Sassone-Corsi P. Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS ONE. 2010;5:e8561. doi: 10.1371/journal.pone.0008561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scammell TE, Schwartz WJ, Smith CB. No evidence for a circadian rhythm of protein synthesis in the rat suprachiasmatic nuclei. Brain Res. 1989;494:155–158. doi: 10.1016/0006-8993(89)90155-8. [DOI] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- Schwanhaüsser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JW, Blenis J, Pende M, Sonenberg N. The mTOR/ PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25:2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Hamada T, Tominaga K, Watanabe S. An in vitro circadian rhythm of protein synthesis in the rat suprachiasmatic nucleus under tissue culture conditions. Brain Res. 1992;584:251–256. doi: 10.1016/0006-8993(92)90902-l. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Rupprecht KM, Hecht SM, Shatkin AJ. Eukaryotic mRNA cap binding protein: purification by affinity chromatography on sepharose-coupled m7GDP. Proc. Natl. Acad. Sci. USA. 1979;76:4345–4349. doi: 10.1073/pnas.76.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru T, Isojima Y, van der Horst GT, Takei K, Nagai K, Takamatsu K. Nucleocytoplasmic shuttling and phosphorylation of BMAL1 are regulated by circadian clock in cultured fibroblasts. Genes Cells. 2003;8:973–983. doi: 10.1046/j.1365-2443.2003.00686.x. [DOI] [PubMed] [Google Scholar]
- Ukai H, Ueda HR. Systems biology of mammalian circadian clocks. Annu. Rev. Physiol. 2010;72:579–603. doi: 10.1146/annurev-physiol-073109-130051. [DOI] [PubMed] [Google Scholar]
- Uniacke J, Holterman CE, Lachance G, Franovic A, Jacob MD, Fabian MR, Payette J, Holcik M, Pause A, Lee S. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486:126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
7
8