CXCL12-producing vascular endothelial niches control acute T cell leukemia maintenance (original) (raw)
. Author manuscript; available in PMC: 2016 Jun 8.
SUMMARY
The role of the microenvironment in T cell acute lymphoblastic leukemia (T-ALL), or any acute leukemia, is poorly understood. Here we demonstrate that T-ALL cells are in direct, stable contact with CXCL12-producing bone marrow stroma. Cxcl12 deletion from vascular endothelial, but not perivascular, cells impeded tumor growth, suggesting a vascular niche for T-ALL. Moreover, genetic targeting of CXCR4 in murine T-ALL after disease onset led to rapid, sustained disease remission, and CXCR4 antagonism suppressed human T-ALL in primary xenografts. Loss of CXCR4 targeted key T-ALL regulators, including the MYC pathway, and decreased leukemia initiating cell activity in vivo. Our data identify a T-ALL niche, and suggest targeting CXCL12/CXCR4 signaling as a powerful therapeutic approach for T-ALL.
INTRODUCTION
Acute lymphoblastic leukemia (ALL) is the most common of childhood cancers, and 15–20% of ALL cases are T lineage (T-ALL) (Pui et al., 2011). A quarter of childhood T-ALL patients relapse within 5 years of treatment and receive a dismal prognosis (Nguyen et al., 2008). Factors predicting poor survival of relapsed childhood ALL patients include T lineage disease and isolated bone marrow involvement, both of which have a less than 25% five year survival rate (Bhojwani and Pui, 2013; Nguyen et al., 2008). Therefore, the search for more effective, less toxic treatments continues. A series of seminal papers has demonstrated that the majority of T3 ALL cases are driven by activating NOTCH1 mutations and activation of downstream pathways, including MYC signaling, which has been shown to be essential for T-ALL cell proliferation and leukemia-initiating cell (LIC) activity (Girard et al., 1996; King et al., 2013; Pear et al., 1996; Roderick et al., 2014; Weng et al., 2004).
Increasing evidence suggests that leukemic stem cells actively engage in crosstalk with the bone marrow microenvironment to regulate their proliferation and survival (Ayala et al., 2009). Similarities between leukemia-initiating cells (LIC) and hematopoietic stem cells (HSC) have raised the hypothesis that LIC require a specialized microenvironment to survive, and that disrupting this niche may be a promising therapeutic strategy (Scadden, 2014). During the last decade, cellular components of the HSC niche have been identified and analyzed (Morrison and Scadden, 2014). Imaging studies showed that HSC tend to localize in the proximity of blood vessels, focusing the field’s attention on the perivascular niche (Sugiyama et al., 2006). In vivo depletion of Nestin+ CXCL12high mesenchymal stem cells (MSC) that surround blood vessels resulted in impaired progenitor cell homing and maintenance (Mendez-Ferrer et al., 2010). Elegant work by Ding et al. and Greenbaum et al. identified endothelial and perivascular populations as distinct and specialized niches supporting HSC homeostasis (Ding and Morrison, 2013; Greenbaum et al., 2013).
Given the functional similarities between HSC and LIC, such as the ability to self-renew and suppress differentiation, we hypothesized that they share dependence on common exogenous signals. In this study, we explore the mechanisms underlying the interaction of leukemia with its microenvironment and investigate the role of CXCL12:CXCR4 signaling in T-ALL pathogenesis.
RESULTS
Visualization of CXCL12-rich T-ALL niches in the bone marrow
We hypothesized that CXCL12 produced by the bone marrow stroma is an important exogenous factor for maintenance of leukemia, analogous to normal HSC and CLP (common lymphocyte progenitors). To model human T-ALL, we generated T-ALL driven by mutated human NOTCH1 (Notch1-ΔE) (Aster et al., 1997). In this model, Lineagenegc-Kit+ bone marrow progenitor cells are transduced with a retrovirus encoding Notch1-ΔE-IRES-GFP and transplanted into lethally irradiated recipient mice. The progenitor cells give rise to GFP+ leukemic blasts with an atypical CD4+CD8+ phenotype in peripheral blood, bone marrow, spleen, thymus, lymph nodes, liver, lung and central nervous system. It was previously suggested that leukemic cells can themselves produce niche factors, augmenting trophic effects (Colmone and Sipkins, 2008). RT-qPCR analysis of mouse T-ALL demonstrated that leukemic cells express undetectable levels of Cxcl12 (Figure S1A). As a second test of whether T-ALL cells can produce CXCL12, we induced T-ALL by transducing bone marrow stem and progenitor cells from _Cxcl12_-DsRed reporter mice or wild-type littermates with Notch1-ΔE-GFP retrovirus, and transplanted them into irradiated syngeneic hosts (Ding and Morrison, 2013). T-ALL cells generated from _Cxcl12_-DsRed donors did not express detectable levels of DsRed, indicating that the tumor cells were not able to produce CXCL12 in an autocrine fashion (Figure S1B–D).
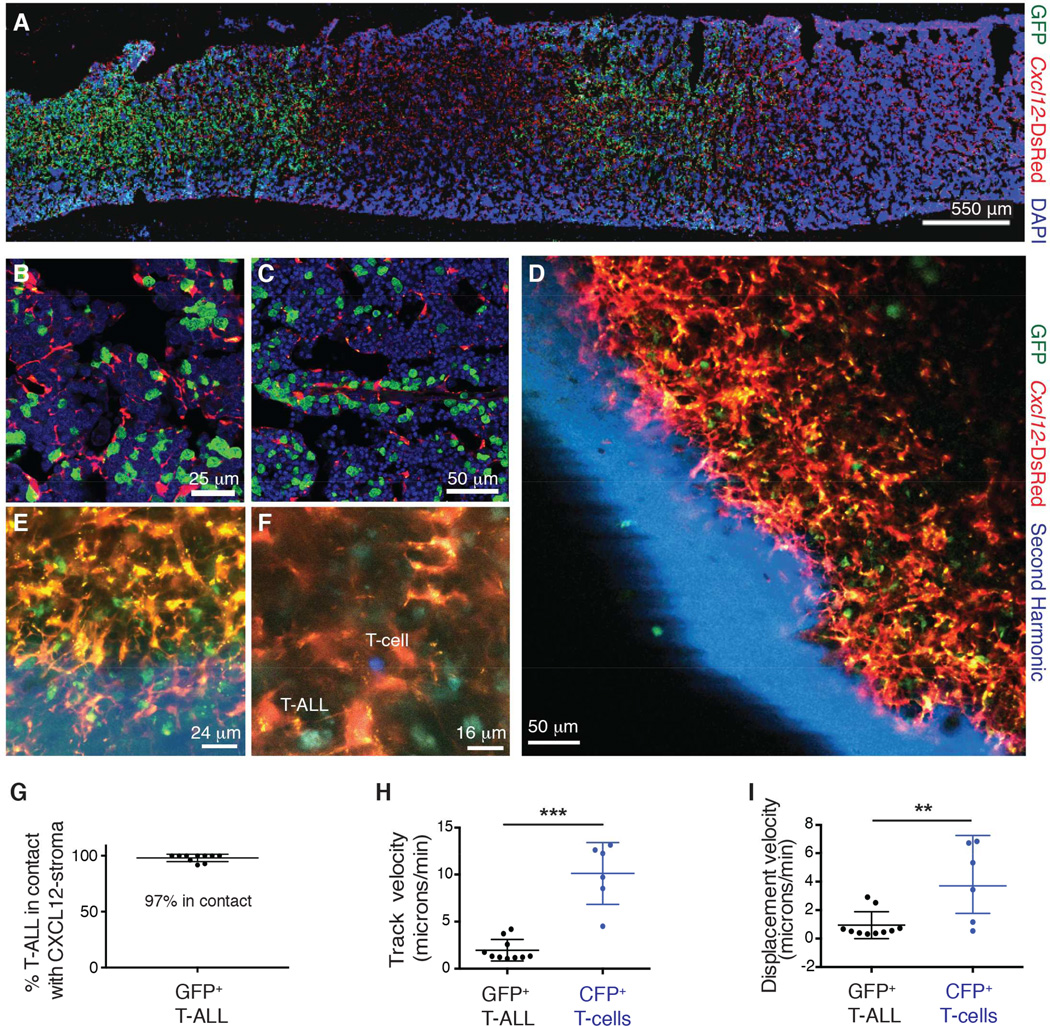
To visualize potential interactions of leukemia cells with the CXCL12-producing microenvironment, GFP+ T-ALL cells isolated from leukemic mice were transplanted into _Cxcl12_-DsRed hosts. Analysis of host bone marrow sections revealed organ-wide dissemination of GFP+ T-ALL cells (Figure 1A), and these cells were observed to be in direct contact with DsRed+ cells (Figure 1B–C). We then performed time-lapse intravital imaging of an intact tibia in vivo. Mice were anesthetized, and the medial or soleus region of the tibial bone was surgically exposed and imaged. Imaging in live animals showed dissemination of leukemic cells throughout the bone marrow (Figure 1D and Movie S1). High-resolution analysis showed leukemic cells to be directly interacting with CXCL12-producing stroma (Figure 1E–G). Time-lapse analysis of _Cxcl12_-DsRed hosts injected concomitantly with GFP+ T-ALL cells and CFP+ normal CD4+ T-cells revealed a dynamic microenvironment, where leukemic cells were highly immotile and stably associated with stromal cells in sharp contrast to motile CD4+ T-cells (Figure 1F and 1H–I and Movie S2). These studies have identified a niche for T-ALL cells and revealed unique adhesion and motility patterns of leukemic cells in the bone marrow during the early onset of disease.
Figure 1. T-ALL cells interact with a CXCL12-producing niche in the bone marrow.
(A–C) Representative immunofluorescence staining of femur sections from _Cxcl12_-DsRed mice transplanted with GFP+ T-ALL cells (n=4).
(D–E) Representative high-resolution two photon images from _Cxcl12_-DsRed mice transplanted with GFP+ T-ALL cells.
(F) Representative high-resolution two-photon image from _Cxcl12_-DsRed mice transplanted with GFP+ T-ALL and CFP+ CD4+ T cells.
(G) Percentage of GFP+ T-ALL cells in contact with _Cxcl12_-DsRed+ cells. Each data point is taken from a different movie or image.
(H) Track velocity and (I) displacement velocity of GFP+ T-ALL cells and CFP+ CD4+ T cells.
Error bars represent +/− SD. Unless otherwise stated each panel reflects data from at least 3 independent experiments. See also Figure S1, Movie S1 and Movie S2.
CXCL12 produced by vascular endothelial cells is necessary for T-ALL progression
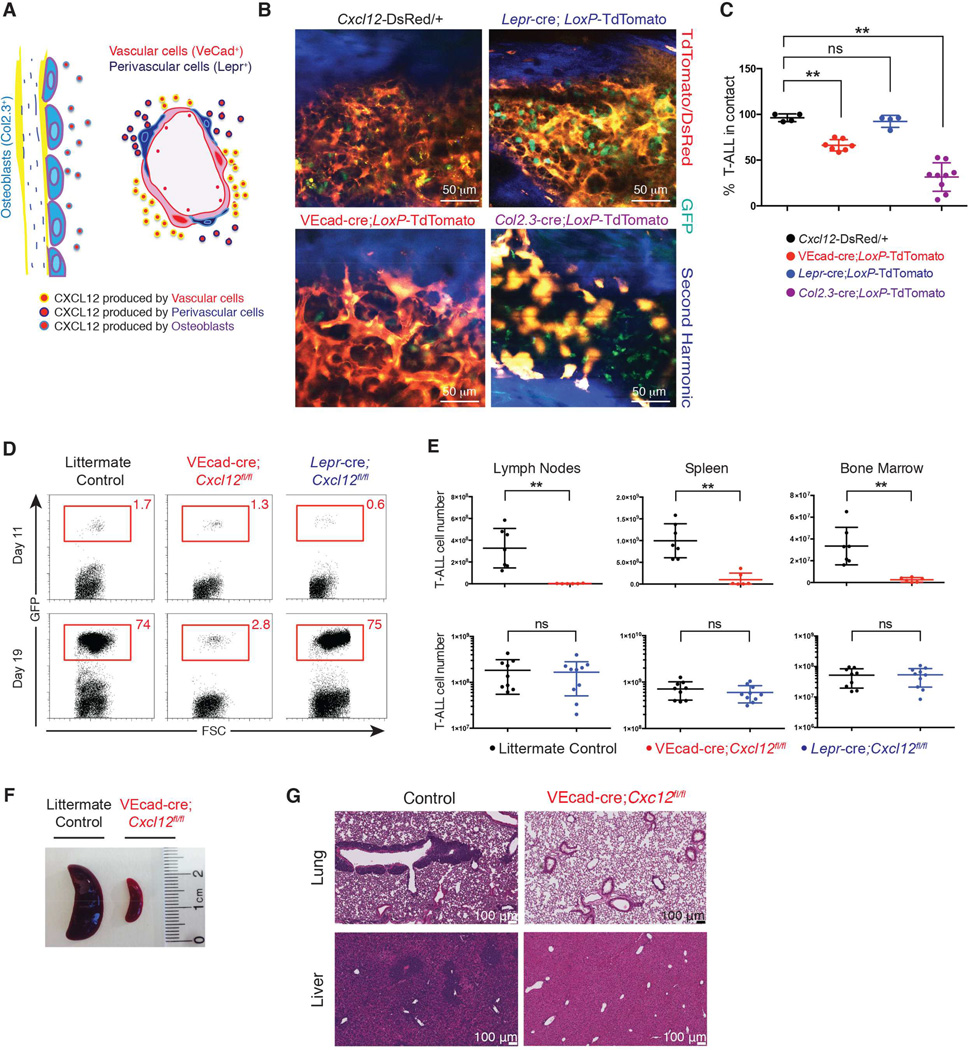
We next tested whether CXCL12 plays an important trophic role in T-ALL. It was previously demonstrated that in the bone marrow, CXCL12 is expressed by numerous lineages including endothelial cells, perivascular cells and osteoblasts (Figure 2A and Figure S2Aa–b) (Ding and Morrison, 2013; Greenbaum et al., 2013; Sugiyama et al., 2006). To visualize these distinct niche populations in vivo, lineage-specific Cre-transgenic mice were crossed to Cre reporter mice, in which tdTomato preceded by a floxed transcriptional stop is knocked into the Rosa26 locus (_LoxP_-tdTomato). _Col2.3_-cre;_LoxP_-tdTomato mice were used to specifically label the endosteal niche, by activating tdTomato in fetal and post-natal osteoblasts. Expression of tdTomato was seen in cells lining the bone in a pattern typical of osteoblasts (Figure S2Ac–d). To label perivascular cells, _Lepr_-cre trangenic mice were crossed to LoxP-tdTomato mice (_Lepr_-cre; _LoxP_-tdTomato) (Figure S2Ae–f), as it was previously demonstrated that Leprin receptor is highly expressed in perivascular stromal cells (Zhou et al., 2014). To visualize the vasculature, VEcad-cre LoxP-tdTomato mice were used. The pattern of tdTomato expression induced by VEcad-cre (Figure S2Ag–h) was identical to the one observed upon intravenous injection of a VE-cadherin specific antibody (data not shown).
Figure 2. CXCL12 production by vascular endothelial cells maintains T-ALL.
(A) Schematic representation of CXCL12 producing populations in bone marrow.
(B) Two photon images of bone marrow from _Cxcl12_-DsRed, VEcad-cre;_LoxP_-tdTomato, _Lepr_-cre; _LoxP_-tdTomato and _Col2.3_-cre;_LoxP_-tdTomato animals 1 week after transfer of GFP+ T-ALL cells.
(C) Frequency of colocalization between GFP+ T-ALL and DsRed/tdTomato niche cells from _Cxcl12_-DsRed, VEcad-cre;_LoxP_-tdTomato, _Lepr_-cre;_LoxP_-tdTomato and _Col2.3_-cre;_LoxP_-tdTomato animals 1 week after transfer of leukemic cells. At least 3 animals were used for each condition. Error bars represent +/− SD.
(D) Representative frequency of T-ALL GFP+ cells in blood 11 days and 19 days post secondary transplantation into VEcad-cre;_Cxcl12fl/fl, Lepr_-cre;Cxcl12fl/fl, or control hosts.
(E) Absolute numbers of T-ALL cells in lymph nodes, spleen, and bone marrow 25 days post secondary transplantation of GFP+ T-ALL cells into VEcad-cre;_Cxcl12fl/fl, Lepr_-cre;Cxcl12fl/fl, or control hosts. Bone marrow numbers represent cells harvested from tibias and femurs. Data is representative of 3 experiments for VEcad-cre;Cxcl12fl/fl (n=6) or littermate sex-matched control animals (n=7) and 2 experiments for _Lepr_-cre;Cxcl12fl/fl (n=9) or control hosts (n=8). Error bars represent +/− SD.
(F) Image of representative spleens from VEcad-cre;Cxcl12fl/fl or control animals.
(G) Histology of lungs and liver from VEcad-cre;Cxcl12fl/fl or control animals.
See also Figure S2.
To investigate whether leukemic cells preferentially localize with osteoblasts or the vasculature (i.e. bone marrow sinusoids) early in disease, VEcad-cre;_LoxP_-tdTomato, _Lepr_-cre;_LoxP_-tdTomato and _Col2.3_-cre;_LoxP_-tdTomato hosts were sublethally irradiated and injected with 1 million GFP+ T-ALL cells. One week later, two photon imaging analysis showed that leukemic cells associated with VE-Cad+ and Lepr+ cells, but not osteoblasts (Figure 2B and 2C). We confirmed these results using two additional types of T-ALL: ETP (_Il7_r 241–242TC mutant) (Treanor et al., 2014) and an ENU-induced mutant carrying G7084 (ins)C7085 in the PEST domain of Notch1 (Figure S2B–C). This was an unexpected finding, as it was previously shown that Col2.3+ osteoblasts constitute a niche for lymphoid progenitor cells in the bone marrow (Ding and Morrison, 2013; Greenbaum et al., 2013).
To assess whether CXCL12 production by vascular endothelial cells or by closely associated perivascular cells regulates T-ALL progression, we deleted Cxcl12 in these populations by crossing Cxcl12fl/fl mice to VEcad-cre (vascular) or Lepr_-cre (perivascular) mice. We then established secondary T-ALL by transplanting GFP+ T-ALL cells (106) into sublethally irradiated VE-Cad-cre;Cxcl12fl/fl, Lepr_-cre;Cxcl12fl/fl, or littermate sex-matched control hosts. Consecutive bleeds to monitor leukemia progression between day 11 and day 19 post-transplantation revealed reduced expansion of T-ALL cells in mice lacking CXCL12 production specifically within the vascular compartment compared to controls (Figure 2D). Moreover, when the mice were sacrificed on day 25 post-transplantation, we observed a significant reduction in tumor burden compared to controls when CXCL12 was absent in vascular cells (Figure 2E). Consistent with this finding, splenomegaly and thymic infiltration were not observed in VEcad-cre;Cxcl12fl/fl mice, in contrast to control animals (Figure 2F and S2D–F). Histo-pathological analysis also showed that T-ALL cells aggressively infiltrated non-hematopoietic tissues such as liver and lungs in control hosts, while these tissues were virtually leukemia-free in VEcad-cre; Cxcl12fl/fl mice (Figure 2G). Meanwhile, leukemia burden in _Lepr_-cre;Cxcl12fl/fl hosts was statistically equivalent to control animals (Figure 2D and 2E). These findings demonstrate that vascular endothelial cells play a key role in leukemia progression through production of CXCL12. These findings contrast with the requirement for both perivascular and endothelial CXCL12 for HSC in normal hematopoiesis.
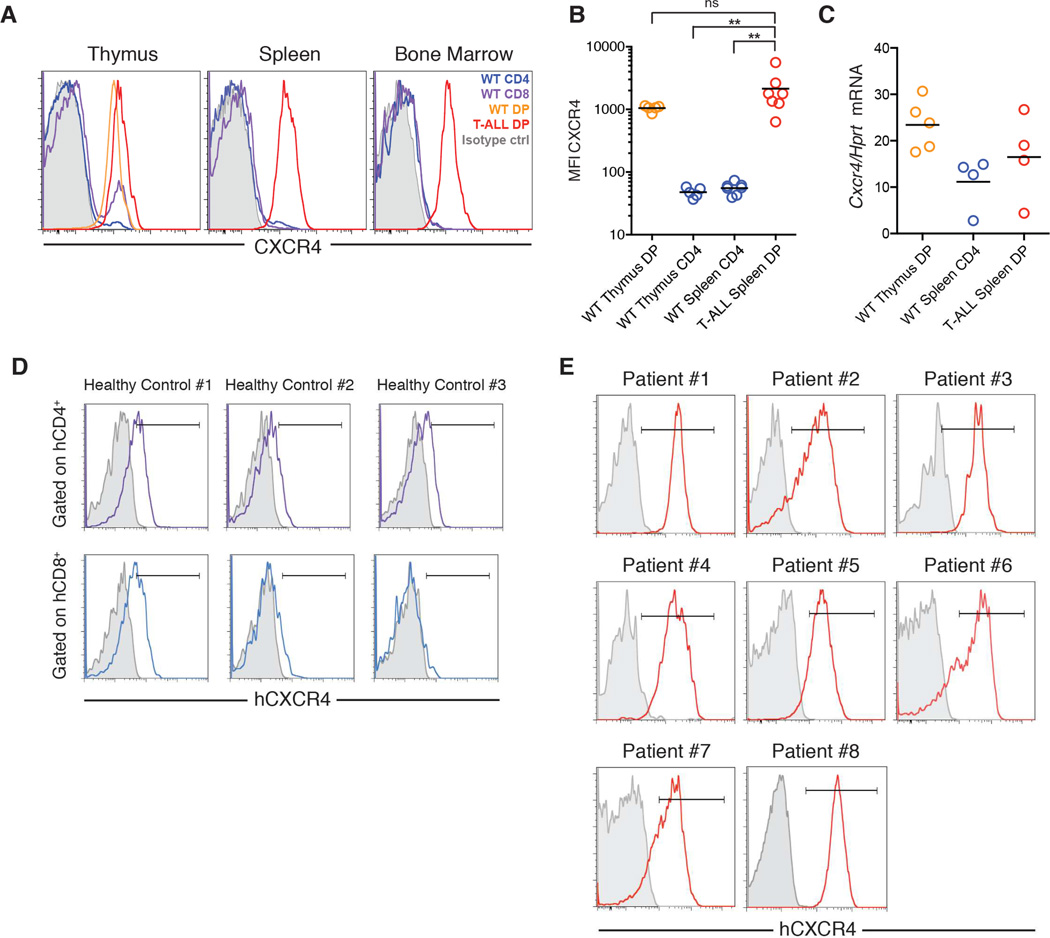
T-ALL cells express high surface levels of CXCR4
Given the importance of CXCL12 for T-ALL progression, we profiled mouse T-ALL cells for surface expression of CXCL12 receptors CXCR4 and CXCR7. We found that primary mouse T-ALL cells express markedly high surface levels of CXCR4, but little surface CXCR7 (Figures 3A and S3A). When we compared CXCR4 staining on the surface of primary mouse T-ALL cells in thymus, spleen and bone marrow to normal T cells from healthy mice in the corresponding tissues, we found that the mean fluorescence intensity (MFI) of CXCR4 on T-ALL cells was 10–50 fold higher than on mature CD4+ T cells and comparable to normal CD4+CD8+ (double-positive; DP) thymocytes (Figure 3B). Notably, this pattern of elevated surface CXCR4 protein expression was not reflected in the level of Cxcr4 mRNA transcripts, as spleen T-ALL cells expressed only marginally higher levels of Cxcr4 mRNA than spleen CD4+ T cells and lower levels than DP thymocytes (Figure 3C).
Figure 3.
CXCR4 is highly expressed on the surface of mouse and human T-ALL cells
(A) Surface CXCR4 expression on T-ALL cells from a representative leukemic mouse and normal T cells from a healthy control. T-ALL cells were identified as CD4+CD8+GFP+.
(B) Surface CXCR4 mean fluorescence intensity (MFI) on T-ALL cells and normal T cells from the indicated organs. Graph pools data from 3–5 pairs of leukemic mice and controls. Bars represent mean MFI.
(C) Abundance of Cxcr4 mRNA expressed relative to Hprt mRNA in purified normal CD4+CD8+ thymocytes, normal spleen CD4+ T cells, and CD4+CD8+ T-ALL cells, measured by RT-qPCR. Bars represent the mean.
(D) Surface CXCR4 expression on human peripheral blood CD4+CD3+ and CD8+CD3+ lymphocytes from healthy controls.
(E) Surface CXCR4 expression on primary bone marrow human biopsies from T-ALL patients expanded in immunodeficient hosts (gated on hCD45) (Patients #1–4, #8; Patient 4 had ETP T-ALL) and primary T-ALL bone marrow biopsies (Patients #5–7). See also Figure S3.
In agreement with these findings, global microarray analysis of T-ALL patient cohorts (Zhang et al., 2012) failed to reveal significant differences in CXCR4 expression when compared to mature human T-cells (Figure S3B). However, when we assessed CXCR4 surface expression on patient T cell leukemia cells and healthy control T cells, we found that T-ALL cells expressed higher levels of CXCR4 than CD4+CD3+ or CD8+CD3+ T cells from the peripheral blood of healthy controls (Figure 3D–E). These studies demonstrate that T-ALL cells express high levels of CXCR4 (when compared to normal peripheral T cells) and suggest that these surface protein levels cannot be explained by mere transcriptional upregulation of CXCR4 mRNA.
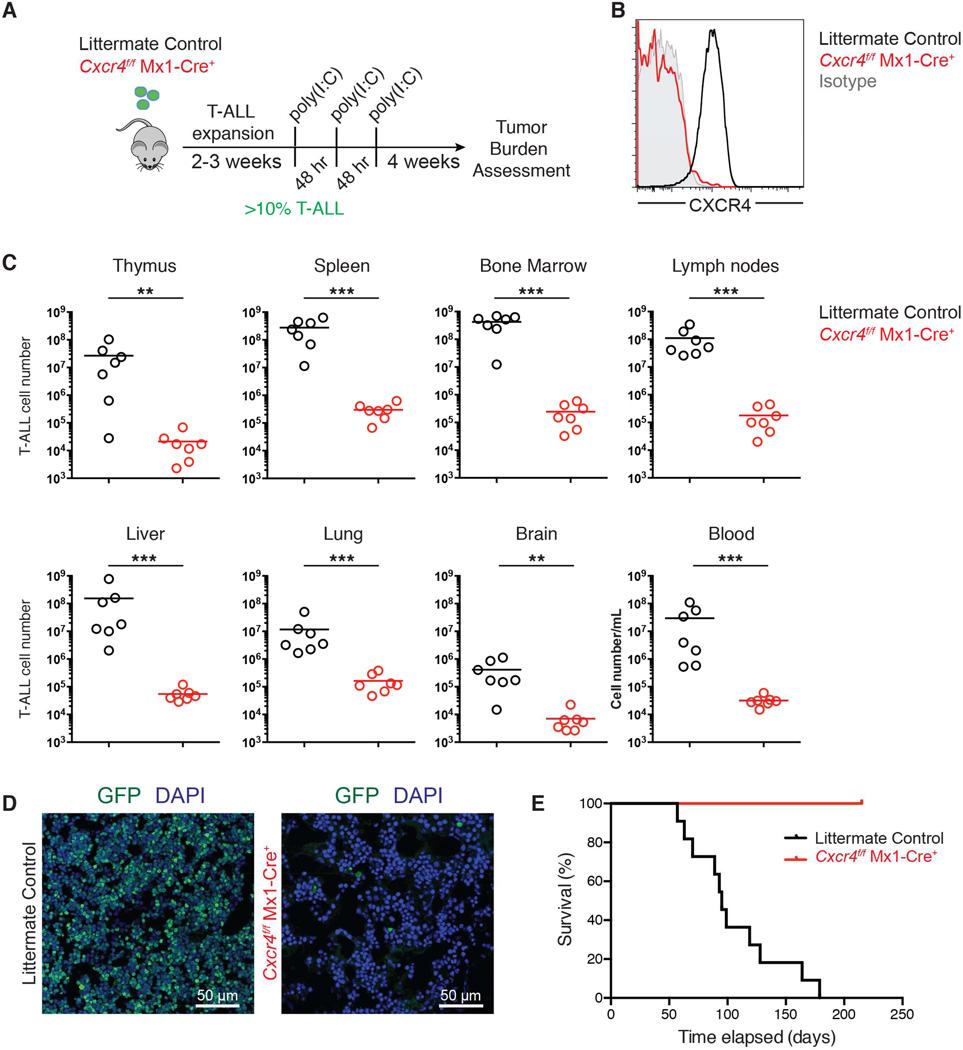
Genetic targeting of Cxcr4 leads to sustained T-ALL remission
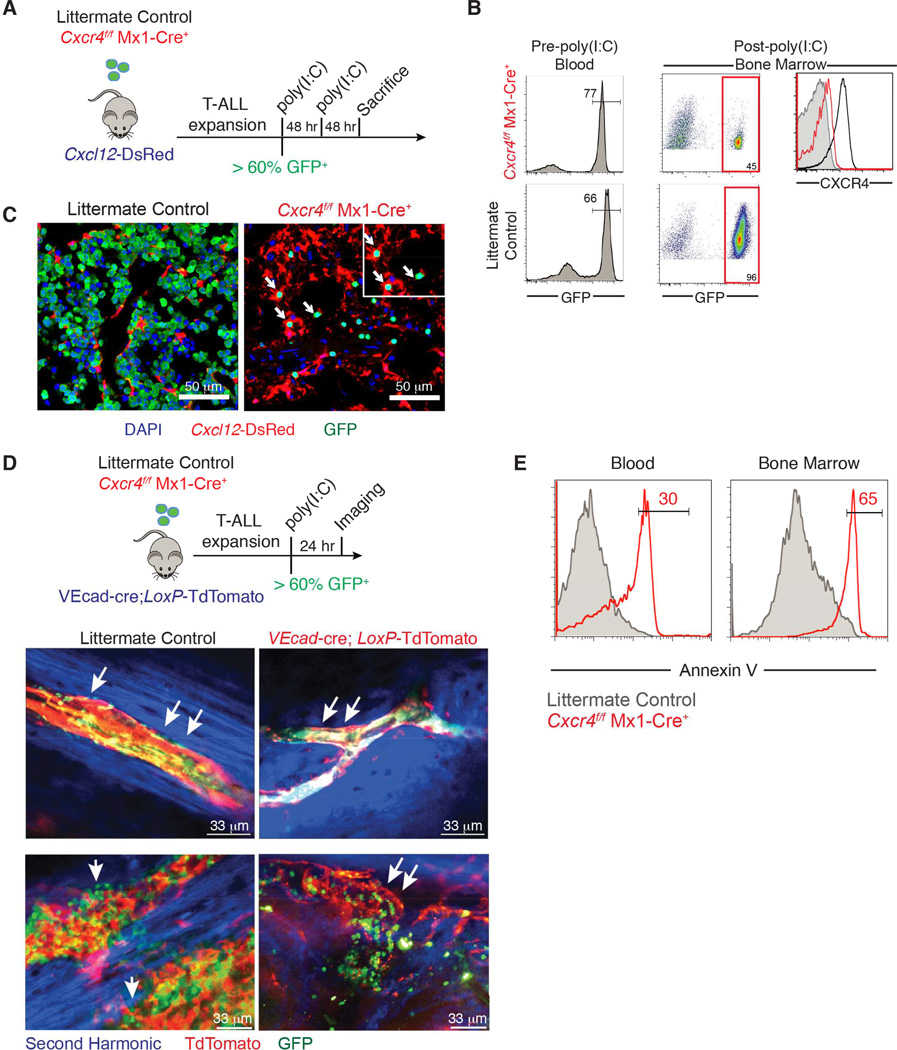
To test the requirement for CXCR4 in T-ALL progression, we examined leukemia burden following genetic ablation of Cxcr4 in vivo. Importantly, we used an inducible Cre recombinase to delete Cxcr4 after disease establishment. Bone marrow progenitor cells derived from _Cxcr4_f/f Mx1-Cre+ mice or littermate control mice (Cxcr4+/+ Mx1-Cre+ or _Cxcr4_f/f) were retrovirally transduced with Notch1-ΔE-IRES-GFP and transplanted into lethally irradiated wild-type recipients. Once GFP+ cells were detected in peripheral blood (>10% of lymphocytes; day 0 (Figure S4A)), both groups were treated with poly(I:C) (days 1, 3 and 5) to induce Cre transcription and Cxcr4 deletion (Kuhn et al., 1995) (Figures 4A and S4B). CXCR4 staining by flow cytometry confirmed that Cxcr4 was deleted in Cxcr4f/f Mx1-Cre+ T-ALL cells (Figure 4B). One month after Cxcr4 deletion, we observed a 3-log reduction in the total number of T-ALL cells, representing a loss in all tissues surveyed, including bone marrow, spleen, blood, lymph nodes, thymus, lung, liver, and brain (Figure 4C–D).
Figure 4. Deletion of Cxcr4 reduces T-ALL burden and significantly prolongs survival.
(A) Schematic representation of experiment design.
(B) Representative surface CXCR4 staining on Cxcr4f/f Mx1-Cre+ or littermate control (Cxcr4+/+ Mx1-Cre+ or Cxcr4f/f) GFP+ T-ALL cells in the spleen one month after they were treated with poly(I:C).
(C) Number of GFP+ Cxcr4f/f Mx1-Cre+ or littermate control (Cxcr4+/+ Mx1-Cre+ or Cxcr4f/f) T-ALL cells in the indicated tissues 1 month after treatment with poly(I:C) (initiated after GFP+ cells represented >10% of peripheral blood lymphocytes). Bars represent the mean. Data are pooled from 2 experiments (n=7–8).
(D) Representative immunofluorescence staining of a femur section from a mouse that received Cxcr4f/f Mx1-Cre+ or littermate control T-ALL, 1 month after poly(I:C) treatment.
(E) Kaplan-Meier survival graph of mice with Cxcr4f/f Mx1-Cre+ (n=10) or littermate control (Cxcr4+/+ Mx1-Cre+, n=11) T-ALL following poly(I:C) treatment (initiated after T-ALL cells reached ~10% of blood lymphocytes; first poly(I:C) injection was defined as day 1). See also Figure S4.
Given the striking reduction in leukemia burden we observed after Cxcr4 deletion, we sought to determine whether survival is prolonged in mice with CXCR4-deficient T-ALL. Indeed, more than 30 weeks after deletion of Cxcr4, 100% (10/10) of mice with Cxcr4f/f Mx1-Cre+ T-ALL cells were alive and appeared healthy, compared to 0% (11/11) of mice with Cxcr4+/+ Mx1-Cre+ T-ALL cells (Figure 4E). Only very low numbers of GFP+ cells could be recovered from spleens and bone marrow of mice with CXCR4-deficient T-ALL when they were sacrificed on day 215 post-treatment with poly(I:C) (Figure S4C). These experiments reveal the ability of Cxcr4 deletion after disease onset to lead to remission in T-ALL, and they further suggest that loss of CXCR4 signaling directly affects T-ALL initiating cell (LIC) function. We will revisit the role of CXCR4 in T-ALL LICs later in the manuscript.
CXCR4 deletion affects leukemic cell localization and survival
To investigate the effects of Cxcr4 deletion on T-ALL cells, we injected secondary _Cxcl12_-DsRed recipients with equal numbers of primary Cxcr4f/f Mx1-Cre+ or Cxcr4f/f GFP+ T-ALL cells. When both cohorts presented 60% ± 10% GFP+ leukemic cells in the bloodstream (Figure 5A–B), animals were injected with two doses of poly(I:C) 48 hours apart. Both cohorts were sacrificed 48 hours after the second poly(I:C) injection and the bone marrow was examined by immunofluorescence analysis. We found that even at this early time point, tumor cells were severely depleted from the bone marrow, and the remaining GFP+ cells (that had some residual surface CXCR4 expression) were localized in proximity to the CXCL12-expressing cells (Figure 5C).
Figure 5. Effects of CXCR4 depletion on leukemic cell localization and survival.
(A) Schematic representation of experiment design.
(B) Left: Frequency of transplanted Cxcr4f/f Mx1-Cre+ or littermate control T-ALL GFP+ cells in the blood prior to poly(I:C) treatment. Right: Frequency of GFP+ leukemic cells and levels of CXCR4 in the bone marrow 48 hr after the second dose of poly(I:C).
(C) Representative immunofluorescence staining of a femur section from _Cxcl12_-DsRed hosts transplanted with GFP+ Cxcr4f/f Mx1-Cre+ or littermate control T-ALL cells 48 hr after poly(I:C) treatment.
(D) Top: Experiment design. Bottom: representative high-resolution two-photon image from VEcad-cre;_LoxP_-tdTomato mice transplanted with Cxcr4f/f Mx1-Cre+ or littermate control T-ALL 24 hr after poly(I:C) treatment. Arrows indicate presence or absence of leukemic cells on top of the vessels.
(E) Annexin V staining on GFP+ Cxcr4f/f Mx1-Cre+ or littermate control T-ALL cells in blood and bone marrow 24 hr after poly(I:C) treatment (n=3). See also Figure S5 and Movies S3-S6.
To examine the localization of CXCR4-deficient T-ALL cells in relation to vascular cells, which we found to be the key source of CXCL12, we transplanted VE-Cad-cre LoxP-tdTomato hosts with GFP+ _Cxcr4_f/f Mx1-Cre+ or littermate control _Cxcr4_f/f T-ALL cells. Once both cohorts presented 60% ± 5% GFP+ leukemic cells in the blood, the animals were treated with poly(I:C) and analyzed 24–72 hours later. Time-lapse intravital imaging revealed that while GFP+ _Cxcr4_f/f leukemic cells were present throughout the bone marrow, _Cxcr4_f/f Mx1-Cre+ cells were mainly observed within blood vessels (Figure 5D, Movie S3–6). Moreover, poly(I:C)-treated _Cxcr4_f/f Mx1-Cre+ cells showed significant levels of Annexin V staining in both the bone marrow and peripheral blood (Figure 5E), as did wild-type T-ALL cells isolated from the spleen of VEcad-cre; Cxcl12fl/fl hosts 6 weeks post-transplantation (Figure S5). These results suggested that CXCR4 expression and signaling influences both leukemic cell localization and survival. They also contrast effects between T-ALL and normal stem and progenitor cells, as in the latter populations, loss of CXCL12 leads to cell mobilization and differentiation but has not been reported to induce apoptotic death.
Small molecule CXCR4 inhibitors suppress growth of murine and human T-ALL
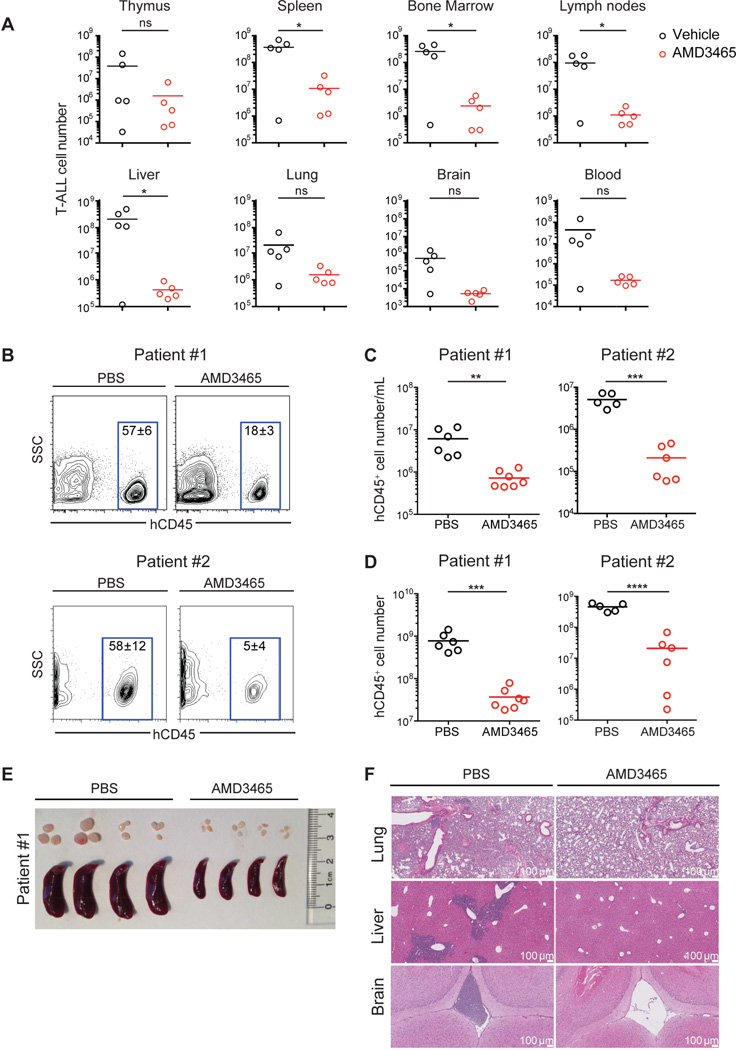
Small molecule CXCR4 antagonists have been developed as a strategy to disrupt the interaction between CXCR4 and CXCL12 and have been used in various settings, including hematopoietic stem cell mobilization (Rettig et al., 2012). Therefore, we examined whether CXCR4 antagonists can recapitulate the effects of Cxcr4 gene ablation and limit T-ALL expansion. To this end, we transplanted two cohorts of lethally irradiated mice with Notch1-ΔE-IRES-GFP+ bone marrow progenitor cells, and after >5% peripheral blood lymphocytes constituted GFP+ leukemic blasts, we administered AMD3465 (20 nmol/hour) or PBS via osmotic pump. After two weeks of treatment, mice treated with AMD3465 showed a substantial reduction in leukemia burden across all tissues surveyed (Figure 6A). These results demonstrate the significant anti-leukemia activity of CXCR4 antagonists, even as single drugs.
Figure 6. Small molecule CXCR4 antagonists efficiently suppress growth of murine and human T-ALL.
(A) T-ALL was induced by transfer of Notch1-ΔE-GFP transduced progenitors. After GFP+ leukemic cells reached 5% of white blood cells, osmotic pumps filled with AMD3465 (20 nmol/hour) or vehicle (PBS) were implanted s.c. Graphs show T-ALL cell number in the indicated tissues 2 weeks after treatment initiation. Bars represent the mean.
(B–F) NOD-SCID mice were injected with leukemia cells from Patient #1 or #2 (represented in Figure 3E). After leukemia reached 5% of white blood cells, osmotic pumps filled with AMD3465 (25 nmol/hr) or vehicle were implanted s.c. Analysis was 2 weeks after treatment initiation.
(B) Representative flow cytometric analysis of human CD45 (hCD45) expression amongst peripheral blood lymphocytes. Values represent the mean frequency of hCD45+ cells +/− SEM.
(C–D) Number of hCD45+ leukemia cells per mL of peripheral blood (C) or in the spleen (D) of animals treated with AMD3465 or vehicle. Bars represent the mean.
(E) Image of representative spleens and lymph nodes (Patient #1 xenograft).
(F) Histology of lungs, liver and brain (Patient #1 xenograft). Data for (B–F) are representative of 6–7 mice per group using Patient #1-derived cells, or 5–6 mice per group using Patient #2-derived cells. See also Figure S6.
To test whether the ability of CXCR4 inhibition to suppress murine leukemia translates to human disease, we assessed the effect of AMD3465 on human xenografts, obtained from primary human bone marrow biopsies (corresponding to Patient #1 and Patient #2 in Figure 3E). We first expanded the primary human leukemia cells in immune-deficient NOD/MrkBomTac-Prkdcscid (NOD-SCID) hosts for 6 weeks. Subsequently, two million human T-ALL cells were transplanted into secondary NOD-SCID hosts and peripheral blood was monitored for the appearance of human CD45+ (hCD45+) leukemic cells. Once leukemia constituted >5% of white blood cells in blood, we began treatment with AMD3465 or vehicle using osmotic pumps. Two weeks later, animals were sacrificed and the leukemia burden was assessed. As a result of AMD3465 treatment, the number of hCD45+ cells in blood and spleen was significantly reduced (Figures 6B–D). Furthermore, splenomegaly was substantially decreased in xenografted mice treated with AMD3465 compared to vehicle (PBS) (Figures 6E and S6). Histo-pathological analysis showed that T-ALL cells in PBS-treated hosts aggressively infiltrated non-hematopoietic tissues, such as brain, liver, and lungs. The same tissues in the AMD3465-treated cohort were virtually leukemia-free, highlighting the ability of AMD3465 to control primary human T-ALL progression in a xenograft model (Figures 6F). These results provide the basis for further testing of this small molecule inhibitor in the clinical setting as a targeted T-ALL therapy.
CXCR4 controls a T-ALL-specific gene expression program and modulates leukemia-initiating activity in T-ALL
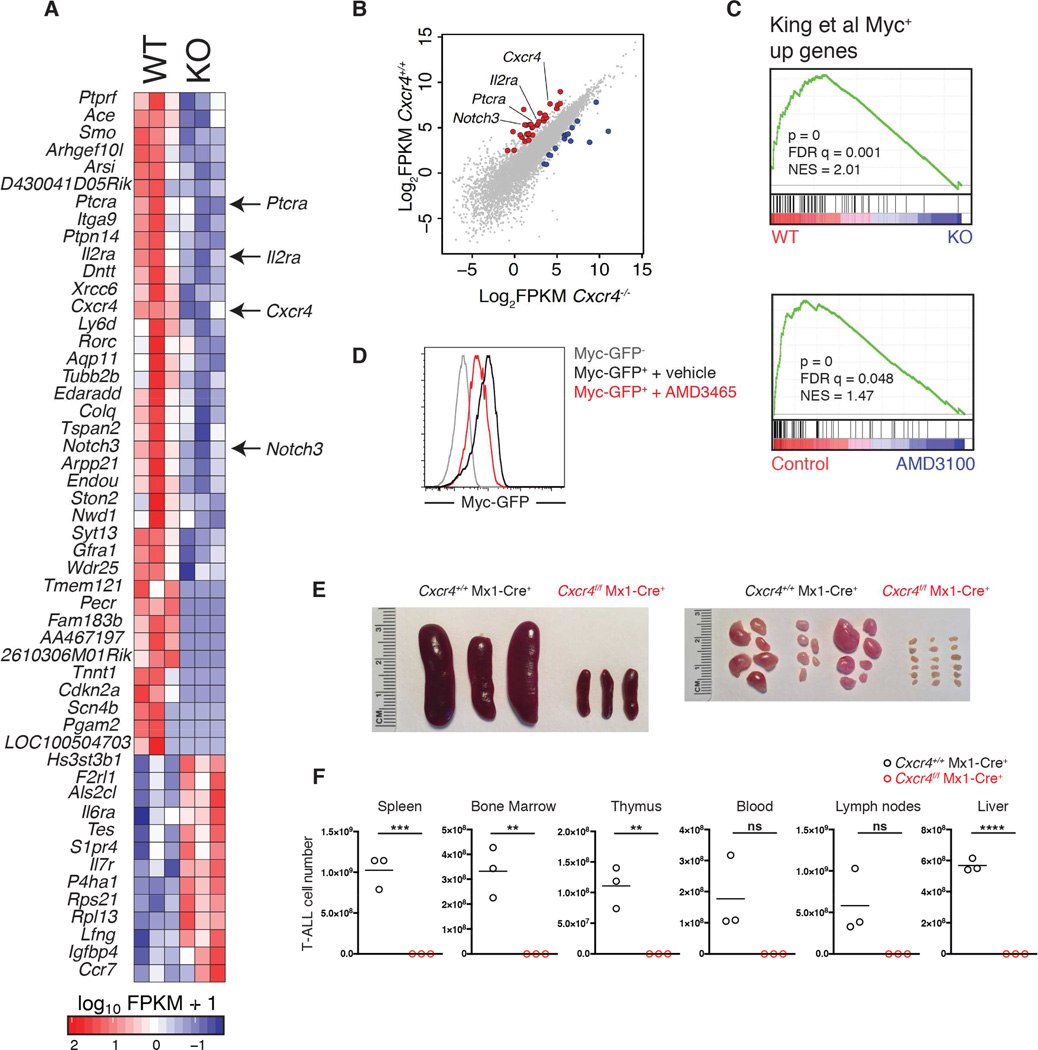
To assess the consequences of CXCR4 signaling loss on T-ALL gene transcription, high-throughput transcriptome sequencing (RNA-Seq) was performed on splenic Cxcr4f/f Mx1-Cre+ and Cxcr4f/f control T-ALL cells 10 days after poly(I:C) treatment to induce Cxcr4 deletion. RNA-Seq was also performed on primary splenic T-ALL cells co-cultured with OP9 cells (which produce CXCL12 (Janas et al., 2010; Trampont et al., 2010)) in the presence of the CXCR4 antagonist AMD3100 or vehicle for 4 days (Figures 7A–C and S7A–B). Initial analysis of the _Cxcr4_-targeted data revealed changes in genes significant for early T cell development and T-ALL induction and progression such as Cdk4, Notch3, Il2ra (CD25), Ptcra, and Cdkn2a. Although altered expression of some of these genes could account for the T-ALL phenotype (i.e. Ptcra expression promotes T-ALL progression), GSEA analysis also demonstrated a significant enrichment of genes that are down-regulated in response to Cxcr4 deletion or AMD3100 treatment in vitro in Myc DNA binding databases (Fig 7C and S7B) (Kim et al., 2008; King et al., 2013; Schuhmacher et al., 2001). Consistent with this, we observed reduced Myc protein levels in T-ALL cells purified after Cxcr4 ablation with poly(I:C) (Figure S7C); after treatment of wild-type T-ALL cells with AMD3465 in vivo (Figure S7D); and in wild-type T-ALL cells isolated from VEcad-cre;Cxcl12fl/fl mice (Figure S7E), which also demonstrated reduced Myc and Myc target gene expression (Figure S7F). Myc over-expression partially restored the proliferation of primary mouse T-ALL cells cultured in the presence of AMD3465, suggesting Myc is an important factor downstream of CXCR4 in T-ALL cells (Figure S7G).
Figure 7. CXCR4 regulates a T cell-specific gene signature and promotes LIC activity in T-ALL.
(A) Heat map displaying differentially expressed genes in Cxcr4f/f and Cxcr4f/f Mx1-Cre T-ALL cells 10 days after administration of poly(I:C).
(B) Scatter plot comparing log2-transformed FPKM expression values for CXCR4-wild-type and -deficient T-ALL cells. Each dot represents an individual gene. Red dots represent genes of interest over-expressed in CXCR4-wild-type (y-axis) compared to CXCR4-deficient T-ALL cells (x-axis).
(C) Myc expression signatures are enriched in T-ALL cells with intact CXCR4 signaling as determined by GSEA. Top: comparison of Cxcr4f/f and Cxcr4f/f Mx1-Cre T-ALL cells. Bottom: comparison of T-ALL cells treated with AMD3100 or vehicle for 4 days in vitro.
(D) MYC-GFP expression by Fbxw7mut Notch1-ΔE LIC expressing a MYC-GFP fusion allele, 4 days after culture on OP9 cells in media with AMD3465 or vehicle, measured by flow cytometry. Graph represents 3 experiments.
(E–F) Cxcr4f/f Mx1-Cre or control Cxcr4+/+ Mx1-Cre Notch1-ΔE+ T-ALL cells treated with poly(I:C) in vivo were isolated from spleen and bone marrow and transferred into sublethally irradiated wild-type mice (1 million cells per mouse; n=3 recipients per genotype). (E) Image shows spleens and lymph nodes harvested from secondary recipients 10 weeks post-transplantation.
(F) Cell number of Cxcr4+/+ Mx1-Cre or Cxcr4f/f Mx1-Cre T-ALL cells in indicated tissues of secondary recipients 10 weeks post-transplantation, assessed by flow cytometry. Bars represent the mean. See also Figure S7.
We have previously shown that T-ALL LIC are characterized by high levels of Myc protein. Furthermore, Myc deletion or silencing in established disease specifically targets LIC and leads to disease remission (King et al., 2013; Roderick et al., 2014). We observed reduced Myc protein expression following AMD3465 treatment of mouse LIC expressing a MYC-GFP fusion allele, cultured in vitro (Figure 7D) (King et al., 2013). To assess the role of CXCR4 signaling in LIC function in vivo, we compared the ability of CXCR4-deficient and wild-type primary T-ALL cells to establish secondary leukemia upon transfer into irradiated recipients. Mice with Cxcr4f/f Mx1-Cre+ or control Cxcr4+/+ Mx1-Cre+ primary T-ALL were treated with 3 doses of poly(I:C), and 3 days after the final dose, T-ALL cells were isolated from spleen and bone marrow and transferred into sublethally irradiated wild-type mice (1 million per mouse, for each genotype). Ten weeks post-transplantation, recipients of poly(I:C)-treated control T-ALL cells displayed severe splenomegaly and lymphadenopathy (Figure 7E) and FACS analysis revealed that the cells had multiplied by more than three orders of magnitude in each of the tissues surveyed (Figure 7F). In contrast, recipients of poly(I:C)-treated Cxcr4f/f Mx1-Cre+ T-ALL cells appeared healthy, with very few or no leukemia cells detected in the same array of tissues. Our data support a critical role for CXCR4 in regulating T-ALL LIC activity, in agreement with our findings showing prolonged survival (Figure 4E) upon deletion of Cxcr4 in established T-ALL and our transcriptome profiling that connected loss of Cxcr4 expression to decreased levels of Myc protein.
DISCUSSION
The importance of CXCR4 in T-ALL pathogenesis would not have been anticipated based on its role in normal T cell maturation. T cell development is profoundly dependent, at different stages, on signaling through Notch1, pre-TCR/TCR, and IL-7R (Ciofani and Zuniga-Pflucker, 2007; Di Santo and Rodewald, 1998; Radtke et al., 1999). All three receptors and components of their signaling pathways have been implicated in T-ALL initiation and progression. By contrast, while CXCR4 signaling increases the efficiency of normal T cell development, CXCR4 is not essential for T cell maturation (Ara et al., 2003; Janas et al., 2010; Trampont et al., 2010). However, our studies demonstrated an essential role for CXCR4 signaling in the progression of T-ALL, suggesting distinct requirements for CXCL12/CXCR4 signaling between physiology and disease.
The efficacy of CXCR4 inhibition would also not have been anticipated based on the precedent of other hematological malignancies, in which CXCR4 inhibition has been promising, but has not had as dramatic effects as a single agent as the ones shown here, including rapid induction of leukemia cell death (Beider et al., 2014; Chen et al., 2013; Kuhne et al., 2013; Tavor et al., 2004; Uy et al., 2012; Welschinger et al., 2013; Zeng et al., 2009). CXCR4 retains normal developing B and myeloid cells in the bone marrow. B and myeloid leukemia cells appear to share this dependence on CXCR4, as CXCR4 antagonism mobilizes these cells out of the bone marrow and into the bloodstream, depriving them of stromal support and exposing them to co-administered chemotherapeutic drugs. Loss of CXCR4 signaling may also inhibit metastasis and predispose these cells to apoptosis (Burger and Peled, 2009). Hence it is surprising that T-ALL appears to be more susceptible to CXCR4 antagonism than B and myeloid cancers; we hypothesize that this may reflect a different dependence of LIC on CXCR4. Finally, although in other tumors (i.e. AML) CXCR4 surface expression is variable, most likely reflecting the heterogeneity of the disease (Mohle et al., 1998), our data suggest that CXCR4 expression on T-ALL is more uniform. Indeed, we have observed high surface CXCR4 on all T-ALL subtypes, including early T precursor (ETP-ALL), a more immature and high-risk disease subtype (Treanor et al., 2014; Zhang et al., 2012). However, we cannot exclude at this point that there are no mechanisms of resistance to CXCR4 signaling inhibition, as T-ALL can be initiated by a large spectrum of mutations, some of them altering signaling pathways (i.e. KRAS, PTEN, JAK3) or epigenetic regulators (i.e. UTX, EZH2, PHF6). It is thus conceivable that inactivation by some of these genes could lead to disease refractory to CXCR4 inhibition.
All these suggest that targeting the CXCL12-expressing microenvironment might have significant implications for the treatment of pediatric and adult T-ALL. Interestingly, recent clinical trial data using AMD3100 (Plerixafor) demonstrated that the drug is absorbed quickly and is well tolerated (McDermott et al., 2014; Uy et al., 2012). It has orphan drug status for the mobilization of HSC and was approved by the US FDA for this indication. While safety needs to be further validated in T-ALL patients and especially children afflicted by the disease, our results in murine and xenograft models provide a strong foundation for the assessment of CXCR4 antagonists in clinical trials, either as a single agent or more likely in combination with established chemotherapy regimens. They also suggest the power of therapeutic targeting of the cancer microenvironment in leukemia.
EXPERIMENTAL PROCEDURES
Animals
Female C57BL/6 mice (6–8 weeks old) were obtained from the National Cancer Institute. Nonobese diabetic/severe combined immunodeficient NOD/MrkBomTac-Prkdcscid (NOD-SCID) mice were from Taconic. Mx1-Cre (Kuhn et al., 1995), VEcad-Cre (B6.FVB-Tg(Cdh5-cre)7Mlia/J) (Alva et al., 2006), _Lepr_-cre (B6.129-Leprtm2(cre)Rck/J) (DeFalco et al., 2001), _Col2.3_-cre (B6.Cg-Tg(Col1a1-cre/ERT2)1Crm/J) (Kim et al., 2004) and _Rosa26_-tdTomato (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J) (Madisen et al., 2010) mice were from Jackson laboratories. Cxcr4f/f (Nie et al., 2004), Cxcl12DsRed knock-in (Ding and Morrison, 2013), _Myc_GFP (Huang et al., 2008) and Fbxw7 knock-in mutant mice (King et al., 2013) have previously been described. To induce deletion by Mx1-Cre, mice received three intraperitoneal injections of poly(I:C) (10 ug/g, GE Heathcare) in PBS, administered every other day. Mice were housed in specific pathogen-free conditions at the Skirball Institute animal facility. All animal experiments were performed in accordance with protocols approved by the New York University Institutional Animal Care and Use Committee.
Bone Marrow Transduction and Transplantation
Hematopoietic stem and progenitor cells were enriched from bone marrow by magnetic selection of cells expressing c-kit (Stemcell Technologies), and cultured in the presence of 50 ng/mL Flt3 ligand, 50 ng/mL SCF, 10 ng/mL IL-7 and 10 ng/mL IL-6 (Peprotech). 24 and 48 hours post-enrichment, c-kit+ cells were infected with concentrated retrovirus. Transduction efficiency was determined by reporter fluorescence at 96 hours. For induction of primary T-ALL, irradiated mice (two rounds of 550 rads) received 50,000 Notch1-ΔE-IRES-GFP+ lineage− cells, together with 500,000 unfractionated bone marrow cells for hemogenic support, by retro-orbital injection. For secondary T-ALL transplantation, recipients were sublethally irradiated (400 rads) and retro-orbitally injected with 1–2 × 106 Notch1-ΔE-GFP+ cells from spleen or bone marrow. For niche experiments all the recipient hosts were sex-matched littermates.
Establishment of Xenografted Human T-ALL
For human xenograft studies, NOD-SCID host mice were used. Primary T-ALL patient samples were collected by Columbia Presbyterian Hospital with informed consent and approved and analyzed under the supervision of the Columbia University Medical Center Institutional Review Board. 1–5 × 106 patient cells were transplanted into immunodeficient mouse strains via retro-orbital injection. Cells from spleen of these primary recipients were used for experiments.
In Vivo Animal Treatments
For inhibition of CXCR4 in primary mouse T-ALL, mice were anaesthetized by isoflurane inhalation and implanted subcutaneously with osmotic pumps (model 2002; Alzet) filled with 40 mM AMD3465 (in PBS; Tocris) or PBS (flow rate 0.5ul/hour). Acute CXCR4 inhibition was achieved with 2 subcutaneous injections of 25 mg/kg AMD3465 (or PBS control) given 4 hours apart. For CXCR4 inhibition in xenografted patient T-ALL, NOD-SCID mice were implanted with osmotic pumps filled with 50 mM AMD3465 or PBS. Mice implanted with osmotic pumps were sacrificed 14 days later for analysis of tumor burden by flow cytometry.
Cell Culture and In Vitro Drug Treatments
Primary mouse T-ALL cells isolated from spleen were maintained on OP9 stromal cells in OptiMEM + GlutaMAX (Invitrogen) supplemented with 10% FBS, 5 ng/ml IL-7, penicillin and streptomycin, and 55 μM β-mercaptoethanol and passaged every 3–4 days onto a fresh feeder layer. OP9 cells were pre-treated with 7.5 ng/mL mitomycin C (Sigma) to prevent feeder cell division. Mouse ETP T-ALL (Treanor et al., 2014) and a T-ALL cell line generated by ENU mutagenesis carrying a Notch1 PEST domain mutation (G7084 (ins)C7085; generated in the lab of A. Ferrando, Columbia University) were maintained on OP9-DLL4 feeders. AMD3100 octahydrochloride hydrate (10 ug/mL in water; Sigma), AMD3465 hexahydrobromide (50 ug/mL in water; Tocris or Cayman Chemical) or vehicle were added to culture media at the start of the culture period.
RNA Extraction and Library Construction for Next Generation Sequencing
Total RNA was extracted from samples using RNeasy Plus Mini Kit (Life Technologies). Samples were then subject to PolyA selection using oligo-dT beads (Life Technologies) according to the manufacturer’s instructions. The resulting RNA samples were then used as input for library construction using the dUTP method as described (Parkhomchuk et al, 2009). RNA libraries were then sequenced on the Illumina HiSeq 2000 or 2500 using 50bp single-end reads. All RNA-Seq data was aligned to mm9 using TopHat v1.4 (Trapnell et al, 2009) with default parameters. We used Cuffdiff v1.3 (Trapnell et al, 2010) for all differential expression (DE) analyses with the RefSeq annotation. In all DE tests, a gene was considered significant if the q-value was less than 0.05 (Cuffdiff default).
Two-Photon Preparation Imaging
Mice were anesthetized using isoflurane and secured on a warming imaging plate in a supine position. The medial or soleus region of the tibial bone was surgically exposed removing soft tissue. The bone was carefully thinned to approximately 200 μm thickness using a microdrill. The leg was imbedded in agarose to stabilize the area and create an immersion well for the microscope objective. Images were collected on an Olympus FV-1000MPE upright laser scanning microscope with 25X 1.05NA water immersion objective using a Spectra-physics DeepSee-MaiTai Ti:sapphire pulsed laser for excitation, with emission filters for the detection of GFP and rhodamine (Tomato or DsRed fluorescent proteins) and qdot 705 (Invitrogen). Images were analyzed on Volocity 6.3 (Improvision). Analysis of cell co-localization is included in Supplemental Experimental Procedures.
Statistical Analysis
Statistical analysis (excluding RNA-Seq experiments) was conducted using the PRISM program (GraphPad). Two groups were compared using an unpaired t test or the Mann-Whitney test. More than two groups were compared using the Kruskal-Wallis test. Significance was defined as *p<0.05, **p<0.01, *** p<0.001, ****p<0.0001.
Supplementary Material
1
2
3
4
5
6
7
SIGNIFICANCE.
While much is known about the cell-intrinsic factors that support leukemia progression, little is understood about the role of the microenvironment. T cell acute lymphoblastic leukemia (TALL) cells have commonly acquired mutations in pathways downstream of surface receptors that regulate differentiation, survival and proliferation in response to environmental cues, and it is unknown whether this frees T-ALL from dependence on a putative leukemic niche. Here we identify CXCL12-producing vascular endothelial cells as a required component of a T-ALL niche. Moreover, we show that targeting CXCL12/CXCR4 signaling after disease onset dramatically reduces T-ALL burden in murine and xenograft models of disease, suggesting a powerful therapeutic approach for this devastating cancer.
ACKNOWLEDGEMENTS
We would like to thank the members of the Schwab and Aifantis laboratories for helpful discussions throughout the duration of the project. We are grateful to Dr. A. Heguy and the NYU Genome Technology Center (supported in part by NIH/NCI P30 CA016087-30 grant) for assistance with sequencing experiments. We are grateful to Dr. C. Loomis and Dr. Z. Dewan from the NYU Histopathology Core (supported by Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center) for assistance with tissue sectioning, and Dr. Cynthia Liu for expert pathological advice. We thank M. Cammer from the NYU Microscopy Core for assistance with image analysis. D.R.F. is supported by a Scholars Award from the Alexandrine and Alexander L. Sinsheimer Fund and is also supported by the Albert Einstein Cancer Center (P30 CA013330). I.A. is supported by the National Institutes of Health (1RO1CA133379, 1RO1CA105129, 1RO1CA149655, 5RO1CA173636, 5RO1CA169784, and 1RO1GM088847). I.A. is also supported by the William Lawrence and Blanche Hughes Foundation, The Leukemia & Lymphoma Society (TRP#6340-11, LLS#6373-13), The Chemotherapy Foundation, The Irma T. Hirschl Trust, The V Foundation for Cancer Research and the St. Baldrick’s Foundation. A.N.T. is a Leukemia and Lymphoma Special Fellow. T.T. is supported by the NIH training grant 5T32CA009161-37. I.A. is a Howard Hughes Medical Institute Early Career Scientist. S.R.S. is supported by the National Institutes of Health (5R01AI085166 and P30CA016087), the Children's Leukemia Research Association, and the Dana Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
L.A.P. and A.N.T. designed, performed and analyzed the majority of the experiments. All microscopy was performed and interpreted by A.N.T. with guidance in the execution and analysis from D.R.F. H.H. provided technical assistance with animal models. T.T. assisted in RNA-Seq library construction and analyzed the data. B.K. provided cell lines and expertise. Y.G. and A.T. analyzed microarray data for human CXCR4 expression. M. S-M. and A.F. provided patient samples, other cell lines and expertise. S.J.M. and D.R.L. provided mice. I.A. and S.R.S. designed and supervised the study. The manuscript was prepared by L.A.P., A.N.T., I.A. and S.R.S.
Accession Numbers
Data generated from RNA-Seq have been deposited in the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE60367.
Supplemental Information
Supplemental Information includes extended Experimental Procedures, Figures S1–S7, Movies S1–S6.
Contributor Information
Iannis Aifantis, Email: Iannis.Aifantis@nyumc.org.
Susan R. Schwab, Email: Susan.Schwab@med.nyu.edu.
REFERENCES
- 1.Alva JA, Zovein AC, Monvoisin A, Murphy T, Salazar A, Harvey NL, Carmeliet P, Iruela-Arispe ML. VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev Dyn. 2006;235:759–767. doi: 10.1002/dvdy.20643. [DOI] [PubMed] [Google Scholar]
- 2.Ara T, Itoi M, Kawabata K, Egawa T, Tokoyoda K, Sugiyama T, Fujii N, Amagai T, Nagasawa T. A role of CXC chemokine ligand 12/stromal cell-derived factor-1/pre-B cell growth stimulating factor and its receptor CXCR4 in fetal and adult T cell development in vivo. J Immunol. 2003;170:4649–4655. doi: 10.4049/jimmunol.170.9.4649. [DOI] [PubMed] [Google Scholar]
- 3.Aster JC, Robertson ES, Hasserjian RP, Turner JR, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 4.Ayala F, Dewar R, Kieran M, Kalluri R. Contribution of bone microenvironment to leukemogenesis and leukemia progression. Leukemia. 2009;23:2233–2241. doi: 10.1038/leu.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beider K, Darash-Yahana M, Blaier O, Koren-Michowitz M, Abraham M, Wald H, Wald O, Galun E, Eizenberg O, Peled A, Nagler A. Combination of imatinib with CXCR4 antagonist BKT140 overcomes the protective effect of stroma and targets CML in vitro and in vivo. Mol Cancer Ther. 2014;13:1155–1169. doi: 10.1158/1535-7163.MCT-13-0410. [DOI] [PubMed] [Google Scholar]
- 6.Bhojwani D, Pui CH. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 2013;14:e205–e217. doi: 10.1016/S1470-2045(12)70580-6. [DOI] [PubMed] [Google Scholar]
- 7.Burger JA, Peled A. CXCR4 antagonists: targeting the microenvironment in leukemia and other cancers. Leukemia. 2009;23:43–52. doi: 10.1038/leu.2008.299. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Jacamo R, Konopleva M, Garzon R, Croce C, Andreeff M. CXCR4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. J Clin Invest. 2013;123:2395–2407. doi: 10.1172/JCI66553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciofani M, Zuniga-Pflucker JC. The thymus as an inductive site for T lymphopoiesis. Annu Rev Cell Dev Biol. 2007;23:463–493. doi: 10.1146/annurev.cellbio.23.090506.123547. [DOI] [PubMed] [Google Scholar]
- 10.Colmone A, Sipkins DA. Beyond angiogenesis: the role of endothelium in the bone marrow vascular niche. Transl Res. 2008;151:1–9. doi: 10.1016/j.trsl.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 11.DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, Enquist L, Friedman JM. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- 12.Di Santo JP, Rodewald HR. In vivo roles of receptor tyrosine kinases and cytokine receptors in early thymocyte development. Curr Opin Immunol. 1998;10:196–207. doi: 10.1016/s0952-7915(98)80249-5. [DOI] [PubMed] [Google Scholar]
- 13.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girard L, Hanna Z, Beaulieu N, Hoemann CD, Simard C, Kozak CA, Jolicoeur P. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 15.Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- 17.Janas ML, Varano G, Gudmundsson K, Noda M, Nagasawa T, Turner M. Thymic development beyond beta-selection requires phosphatidylinositol 3-kinase activation by CXCR4. J Exp Med. 2010;207:247–261. doi: 10.1084/jem.20091430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JE, Nakashima K, de Crombrugghe B. Transgenic mice expressing a ligand-inducible cre recombinase in osteoblasts and odontoblasts: a new tool to examine physiology and disease of postnatal bone and tooth. Am J Pathol. 2004;165:1875–1882. doi: 10.1016/S0002-9440(10)63240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, Aranda-Orgilles B, Perez-Garcia A, Shi J, Vakoc C, et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153:1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 22.Kuhne MR, Mulvey T, Belanger B, Chen S, Pan C, Chong C, Cao F, Niekro W, Kempe T, Henning KA, et al. BMS-936564/MDX-1338: a fully human anti-CXCR4 antibody induces apoptosis in vitro and shows antitumor activity in vivo in hematologic malignancies. Clin Cancer Res. 2013;19:357–366. doi: 10.1158/1078-0432.CCR-12-2333. [DOI] [PubMed] [Google Scholar]
- 23.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDermott DH, Liu Q, Velez D, Lopez L, Anaya-O'Brien S, Ulrick J, Kwatemaa N, Starling J, Fleisher TA, Priel DA, et al. A phase 1 clinical trial of long-term, low-dose treatment of WHIM syndrome with the CXCR4 antagonist plerixafor. Blood. 2014;123:2308–2316. doi: 10.1182/blood-2013-09-527226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma'ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohle R, Bautz F, Rafii S, Moore MA, Brugger W, Kanz L. The chemokine receptor CXCR-4 is expressed on CD34+ hematopoietic progenitors and leukemic cells and mediates transendothelial migration induced by stromal cell-derived factor-1. Blood. 1998;91:4523–4530. [PubMed] [Google Scholar]
- 27.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen K, Devidas M, Cheng SC, La M, Raetz EA, Carroll WL, Winick NJ, Hunger SP, Gaynon PS, Loh ML, Children's Oncology G. Factors influencing survival after relapse from acute lymphoblastic leukemia: a Children's Oncology Group study. Leukemia. 2008;22:2142–2150. doi: 10.1038/leu.2008.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nie Y, Waite J, Brewer F, Sunshine MJ, Littman DR, Zou YR. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J Exp Med. 2004;200:1145–1156. doi: 10.1084/jem.20041185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pui CH, Gajjar AJ, Kane JR, Qaddoumi IA, Pappo AS. Challenging issues in pediatric oncology. Nat Rev Clin Oncol. 2011;8:540–549. doi: 10.1038/nrclinonc.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 33.Rettig MP, Ansstas G, DiPersio JF. Mobilization of hematopoietic stem and progenitor cells using inhibitors of CXCR4 and VLA-4. Leukemia. 2012;26:34–53. doi: 10.1038/leu.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roderick JE, Tesell J, Shultz LD, Brehm MA, Greiner DL, Harris MH, Silverman LB, Sallan SE, Gutierrez A, Look AT, et al. c-Myc inhibition prevents leukemia initiation in mice and impairs the growth of relapsed and induction failure pediatric T-ALL cells. Blood. 2014;123:1040–1050. doi: 10.1182/blood-2013-08-522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scadden DT. Nice neighborhood: emerging concepts of the stem cell niche. Cell. 2014;157:41–50. doi: 10.1016/j.cell.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, Eick D. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 38.Tavor S, Petit I, Porozov S, Avigdor A, Dar A, Leider-Trejo L, Shemtov N, Deutsch V, Naparstek E, Nagler A, Lapidot T. CXCR4 regulates migration and development of human acute myelogenous leukemia stem cells in transplanted NOD/SCID mice. Cancer Res. 2004;64:2817–2824. doi: 10.1158/0008-5472.can-03-3693. [DOI] [PubMed] [Google Scholar]
- 39.Trampont PC, Tosello-Trampont AC, Shen Y, Duley AK, Sutherland AE, Bender TP, Littman DR, Ravichandran KS. CXCR4 acts as a costimulator during thymic beta-selection. Nat Immunol. 2010;11:162–170. doi: 10.1038/ni.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Treanor LM, Zhou S, Janke L, Churchman ML, Ma Z, Lu T, Chen SC, Mullighan CG, Sorrentino BP. Interleukin-7 receptor mutants initiate early T cell precursor leukemia in murine thymocyte progenitors with multipotent potential. J Exp Med. 2014;211:701–713. doi: 10.1084/jem.20122727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uy GL, Rettig MP, Motabi IH, McFarland K, Trinkaus KM, Hladnik LM, Kulkarni S, Abboud CN, Cashen AF, Stockerl-Goldstein KE, et al. A phase 1/2 study of chemosensitization with the CXCR4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood. 2012;119:3917–3924. doi: 10.1182/blood-2011-10-383406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welschinger R, Liedtke F, Basnett J, Dela Pena A, Juarez JG, Bradstock KF, Bendall LJ. Plerixafor (AMD3100) induces prolonged mobilization of acute lymphoblastic leukemia cells and increases the proportion of cycling cells in the blood in mice. Exp Hematol. 2013;41:293–302. e291. doi: 10.1016/j.exphem.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 43.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 44.Zeng Z, Shi YX, Samudio IJ, Wang RY, Ling X, Frolova O, Levis M, Rubin JB, Negrin RR, Estey EH, et al. Targeting the leukemia microenvironment by CXCR4 inhibition overcomes resistance to kinase inhibitors and chemotherapy in AML. Blood. 2009;113:6215–6224. doi: 10.1182/blood-2008-05-158311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5
6
7