ClinGen — The Clinical Genome Resource (original) (raw)
. Author manuscript; available in PMC: 2015 Dec 4.
Published in final edited form as: N Engl J Med. 2015 May 27;372(23):2235–2242. doi: 10.1056/NEJMsr1406261
Abstract
On autopsy, a patient is found to have hypertrophic cardiomyopathy. The patient’s family pursues genetic testing that shows a “likely pathogenic” variant for the condition on the basis of a study in an original research publication. Given the dominant inheritance of the condition and the risk of sudden cardiac death, other family members are tested for the genetic variant to determine their risk. Several family members test negative and are told that they are not at risk for hypertrophic cardiomyopathy and sudden cardiac death, and those who test positive are told that they need to be regularly monitored for cardiomyopathy on echocardiography. Five years later, during a routine clinic visit of one of the genotype-positive family members, the cardiologist queries a database for current knowledge on the genetic variant and discovers that the variant is now interpreted as “likely benign” by another laboratory that uses more recently derived population-frequency data. A newly available testing panel for additional genes that are implicated in hypertrophic cardiomyopathy is initiated on an affected family member, and a different variant is found that is determined to be pathogenic. Family members are retested, and one member who previously tested negative is now found to be positive for this new variant. An immediate clinical workup detects evidence of cardiomyopathy, and an intracardiac defibrillator is implanted to reduce the risk of sudden cardiac death.
During the past 25 years, major advances in deciphering the genetic bases of human disease have been achieved, and more than 5000 mendelian disorders are now understood at the genetic level.1 Although this is an extraordinarily important achievement in our understanding of the biologic features of human disease, the integration of these findings into clinical care is severely challenged by a lack of publicly available and accurate interpretations of the vast amount of human genetic variation known to exist. More than 80 million genetic variants have been uncovered in the human genome,2 and for the majority, we have no clear understanding of their role in human health and disease. Thus, we are very far from a world in which we can sequence patients’ genomes and easily interpret their risk of disease, even if patients carry a variant in a gene that is associated with a highly penetrant genetic disorder. The rarity of most variants that are identified in mendelian genes (Fig. 1) has made it difficult to decipher the effect of such variants on gene function; most rare variants are labeled a “variant of uncertain significance.” A final factor contributing to our lack of consistent, clear, and clinically relevant annotation of human genetic variation is the so-called silo effect, in which various commercial and academic entities maintain isolated, sometimes proprietary, databases of variant interpretations, thus preventing the sharing of critical knowledge that could benefit patients, families, health care providers, diagnostic laboratories, and payers.
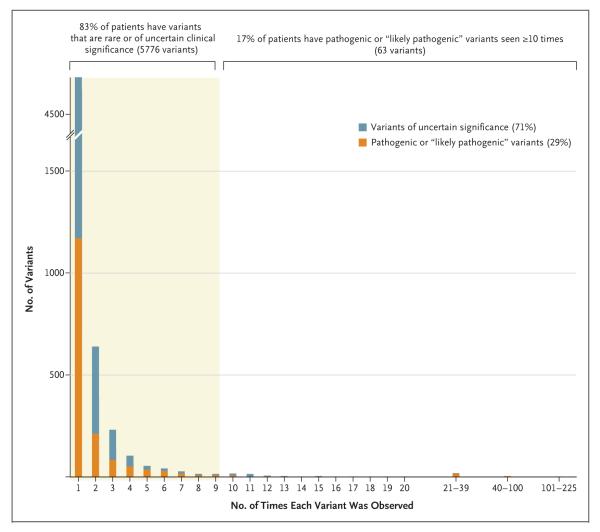
Figure 1. Variant Histogram from Mendelian Disease Testing of 15,000 Probands.
Shown are data for 5839 variants that have been found in patients with cardiomyopathy, hearing loss, RASopathies (i.e., developmental syndromes caused by germline mutations that alter the Ras subfamily), aortopathies, hereditary cancers, pulmonary disorders, skin disorders, and other genetic syndromes, as tested by the Laboratory for Molecular Medicine at Partners HealthCare. Shown at the top of the chart are the percentages of patients who carry frequently observed pathogenic variants and patients who have variants that are rare or of uncertain significance.
On the basis of an analysis of submissions to the ClinVar variant database of the National Center for Biotechnology Information (NCBI),3 we have discovered that the interpretation of the importance of the same variant by multiple clinical laboratories may differ, so that at least one interpretation must be wrong and could therefore lead to inappropriate medical intervention, as illustrated in the above example. Healthy competition among isolated entities is no longer sufficient to drive our understanding of human variation, and patient care may be compromised when data are not shared. If society is to understand human genomic variation and reap its benefits in clinical care, large collaborative efforts will be the only way to amass sufficient data and distribute responsibility for critical review.
In the past few years, collaborative efforts have shown the effectiveness of submitting data to public databases to advance genetic discovery. For example, the current human reference sequence would not have been possible if public release of data had not been encouraged.4 Similarly, the replication that is critical to validate genomewide association studies5 depended on access to data from larger and larger cohorts to identify rarer and rarer alleles (or common alleles with smaller effect sizes). The field benefited tremendously from a culture of data sharing, and today genetic loci for more than 300 complex traits have been identified and reported in more than 2000 articles, many through highly reproducible genomewide association studies.6-8 The cancer genetics community also organized several large efforts, including the Cancer Genome Atlas9 and the International Cancer Genome Consortium,10 in which the sequencing of genes obtained from both tumors and normal tissue has been implemented and resultant data deposited into databases to identify recurrent variants associated with different types of cancer. Most of these consortia and studies are focused on data obtained exclusively in the research setting with predefined participating entities. To enable medical use of genetic discoveries, it is equally important to improve standards of data collection and sharing from genetic testing and define a systematic method for the clinical annotation and interpretation of genomic and phenotypic variation.
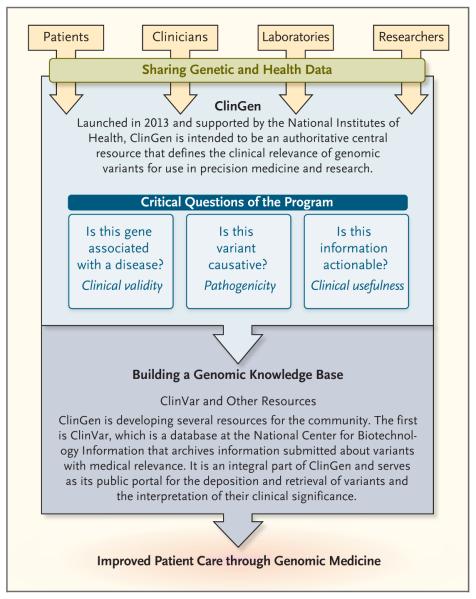
To address these needs, three grants from the National Institutes of Health (NIH) were aligned with the NCBI ClinVar database under the collaborative Clinical Genome Resource (ClinGen) program (Fig. 2). The program was based in part on efforts of the earlier International Standards for Cytogenomic Arrays Consortium, which began collecting data on copy-number variants from chromosomal microarray testing in clinical cytogenetics laboratories in 2007, and was later expanded to include data on sequence variants from clinical molecular laboratories.11 Consistent with its mission, ClinGen is developing interconnected community resources to improve our understanding of genomic variation and improve its use in clinical care. ClinGen represents a strong partnership among public, academic, and private institutions that relies on collaboration between the NIH and academic and commercial laboratories operating in both the research and clinical realms. ClinGen is also engaging numerous entities, including professional societies, to ensure that the resources that are produced meet the expectations of the community. Its goals are outlined in Table 1.
Figure 2. Clinical Genome Resource (ClinGen).
More information on ClinGen is available at www.clinicalgenome.org, and more information on ClinVar is available at www.ncbi.nlm.nih.gov/clinvar.
Table 1.
Goals of the Clinical Genome Resource (ClinGen).
| Goals |
|---|
| Share genomic and phenotypic data provided by clinicians, researchers, and patients through centralized databases for clinical and research use |
| Standardize the clinical annotation and interpretation of genomic variants |
| Implement evidence-based expert consensus for curating genes and variants |
| Improve understanding of variation in diverse populations to realize interpretation of genetic testing on a global scale |
| Develop machine-learning algorithms to improve the throughput of variant interpretation |
| Assess the “medical actionability” of genes and variants |
| Structure and provide access to genomic knowledge for use in electronic health records ecosystems |
| Disseminate the collective knowledge and resources for unrestricted use in the community |
Launched in April 2013, the publicly accessible ClinVar database is a cornerstone of ClinGen. It serves as the primary site for deposition and retrieval of variant data and annotations.3 Variants and supporting evidence can be submitted by researchers, clinical laboratories, expert groups, clinicians, and patients (Fig. 3). Variants can also be reciprocally shared between ClinVar and locus-specific databases that may contain more detailed information specific to certain diseases and that are often maintained by dedicated curators.12 For example, ClinGen-approved expert panels are depositing interpreted variants from databases such as CFTR2 (Clinical and Functional Translation of CFTR, which houses information about specific CFTR mutations),13 InSiGHT (variant database for the International Society for Gastrointestinal Hereditary Tumours),14 and PharmGKB (the Pharmacogenomics Knowledge Base).15 As of May 4, 2015, ClinVar contained 172,055 variant submissions across 22,864 genes (145,311 unique sequence and structural variants) from 314 submitters, including clinical and research laboratories, locus-specific databases, aggregate databases (Online Mendelian Inheritance in Man [OMIM] and GeneReviews), expert consortia, professional organizations, health care providers (e.g., Sharing Clinical Reports Project, at www.sharingclinicalreports.org), and patients (e.g., Free the Data Campaign, at www.free-the-data.org) (Table 2). More than 118,000 of the unique variants in ClinVar have clinical interpretations, although 24,725 of those interpretations (21%) are variants of uncertain significance, which highlights the additional work to be done. Each time a laboratory submits variants for deposition in ClinVar, the submission is analyzed to ensure that all variants are accurately named according to standardized variant nomenclature16 and can be mapped to the human-genome reference sequence and that the terms used for assertions of clinical significance for mendelian disorders conform to those recently approved by the American College of Medical Genetics and Genomics.17 This standardization effort is important for a robust submission and quality-control process. In addition, after deposition, each laboratory receives a report of any differences in interpretation between their submitted variants and those already existing in ClinVar.
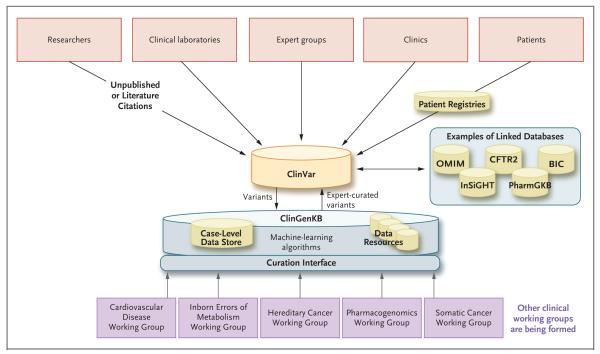
Figure 3. Flow of Data through ClinGen.
Shown is the typical flow of information into ClinVar and ClinGenKB, a new database that is designed to allow for a flexible working environment for curation. Most variants are submitted by external sources and databases directly into ClinVar for immediate access by the community. Variants then flow into ClinGenKB to enable the resolution of differences in interpretation, as well as expert review of variants by the clinical-domain working groups that are shown. Additional sources of data and machine-learning algorithms may be brought into ClinGenKB to aid in the interpretive process. BIC denotes Breast Cancer Information Core, CFTR2 Clinical and Functional Translation of CFTR, InSiGHT the variant database for the International Society for Gastrointestinal Hereditary Tumours, OMIM Online Mendelian Inheritance in Man, and PharmGKB the Pharmacogenomics Knowledge Base.
Table 2.
ClinVar Submitters Who Have Provided More Than 50 Variants with Medical Interpretations.*
| Variable | Variants | Genes |
|---|---|---|
| Submitter | ||
| Expert consortia and professional organizations | ||
| International Society for Gastrointestinal Hereditary Tumours (InSiGHT) | 2,362 | 4 |
| Clinical and Functional Translation of CFTR (CFTR2) | 133 | 1 |
| American College of Medical Genetics and Genomics (ACMG) | 23 | 1 |
| Clinical laboratories | ||
| International Standards for Cytogenomic Arrays Consortium Laboratories | 14,441 | >14,000 |
| Partners HealthCare Laboratory for Molecular Medicine | 12,092 | 302 |
| GeneDx | 11,176 | 613 |
| Ambry Genetics | 9,821 | 50 |
| University of Chicago Genetic Services Laboratory | 7,127 | 622 |
| Emory University Genetics Laboratory | 6,944 | 659 |
| Sharing Clinical Reports Project for BRCA1 and BRCA2 | 2,147 | 2 |
| Invitae | 1,949 | 125 |
| Laboratory Corporation of America (LabCorp) | 1,390 | 160 |
| ARUP Laboratories | 1,374 | 10 |
| Counsyl | 1,136 | 108 |
| Children’s Hospital of Eastern Ontario Molecular Genetics Diagnostic Laboratory | 957 | 21 |
| Blueprint Genetics | 651 | 130 |
| University of Washington CSER Program with Northwest Clinical Genomics Laboratory | 646 | 80 |
| University of Washington Collagen Diagnostic Laboratory | 411 | 2 |
| Children’s National Medical Center GenMed Metabolism Laboratory | 317 | 1 |
| Pathway Genomics | 189 | 18 |
| Baylor College of Medicine Medical Genetics Laboratories | 178 | 12 |
| Greenwood Genetic Center Diagnostic Laboratories | 80 | 19 |
| University of Pennsylvania School of Medicine Genetic Diagnostic Laboratory | 68 | 1 |
| Research programs and locus-specific databases | ||
| Breast Cancer Information Core (BIC) | 3,734 | 2 |
| Royal Brompton Hospital Cardiovascular Biomedical Research Unit | 1,346 | 13 |
| RettBASE | 973 | 5 |
| Muilu Laboratory, Institute for Molecular Medicine Finland | 840 | 43 |
| ClinSeq Project, National Human Genome Research Institute | 425 | 36 |
| Lifton Laboratory, Yale University | 390 | 284 |
| PALB2 Leiden Open Variation Database | 242 | 2 |
| Department of Ophthalmology and Visual Sciences, Kyoto University Hospital | 171 | 59 |
| Developmental Genetics Unit, King Faisal Specialist Hospital and Research Center, Saudi Arabia | 101 | 102 |
| Department of Zoology, M.V. Muthiah Government College, India | 58 | 3 |
| Aggregate databases | ||
| Online Mendelian Inheritance in Man (OMIM) | 25,262 | 3,770 |
| GeneReviews | 4,000 | 488 |
| Totals | ||
| Total variant submissions to ClinVar† | 172,055 | |
| Total unique variants represented† | 145,311 | |
| Total unique variants in ClinVar with clinical assertions† | 118,169 | |
| Total genes in submissions with assertions | ||
| Genes in which variants are confined to the gene† | 7,406 | |
| Genes in which copy-number variants span multiple genes† | 22,864 |
In the past few years, it has become clear that many genetic variants that have been reported in the literature to cause disease have been misinterpreted. Such errors have resulted from insufficient standards for defining the evidence required to link a variant to disease causation and our lack of information on common variation across many populations.18,19 The aggregation of data from many submitters that is enabled by ClinGen permits the identification of some variants that have been misinterpreted, as documented by different interpretations among submitters. Of the 118,169 unique variants with clinical interpretations, 12,895 (11%) have clinical interpretations that have been submitted by more than one laboratory. Of those, 2,229 (17%) are interpreted differently by the submitters, with one- or two-step differences between any of three major levels: “pathogenic or likely pathogenic,” “uncertain significance,” and “likely benign or benign.” For example, one of the initial and ongoing sources of data in ClinVar is the OMIM database (containing nearly 25,000 variants), which catalogues representative pathogenic variants from published studies that define the role of a gene in disease, as well as the spectrum of variant types and phenotypes that are found for a gene.1 Now that ClinVar has already processed many clinically curated submissions, we have identified 220 variants that have been described in research studies and maintained in the public domain in OMIM as pathogenic and that now are being reinterpreted by clinical laboratories as benign, likely benign, or of uncertain significance. Through ClinVar, the curators of OMIM now have a system that can more easily alert them to the need to reevaluate their records of gene–disease relationships. In addition, patients, clinicians, and clinical laboratories now have more robust public access to interpretations of genetic variants, which permits them to better use the information for clinical care decisions.
Of the ClinVar submissions from currently operating clinical laboratories and expert consortia, 415 variants have different assertions of clinical significance of a level that is anticipated to have a differential effect on medical decision making (pathogenic or likely pathogenic vs. uncertain significance, likely benign, or benign). Because a key goal of ClinGen is to resolve these differences, the American College of Medical Genetics and Genomics (a ClinGen grantee) worked with members of the sequence and structural-variant communities to develop new standards for interpreting genetic variants.17,20 ClinGen is now working with laboratories to facilitate adoption of these new standards and openly share the basis of their assertions with respect to pathogenicity. This collaboration has allowed laboratories to resolve differences in interpretation through expert consensus and application of these standardized methods. Furthermore, given the extremely fast pace at which genomic information is now being generated, the use of machine learning (which explores the development of algorithms that can help to make predictions on data) or similar approaches for prioritizing variant curation, along with expert review, are critical for efficient turnaround of results. Thus, a resource that contains variants of uncertain significance and that can be targeted for further research through functional studies will enable improved understanding of genomic variation. This function, combined with the implementation of new standards for the interpretation of variants and the open sharing of assertions with respect to pathogenicity to identify differences, should eventually lead to a stronger reference database and to better health care.
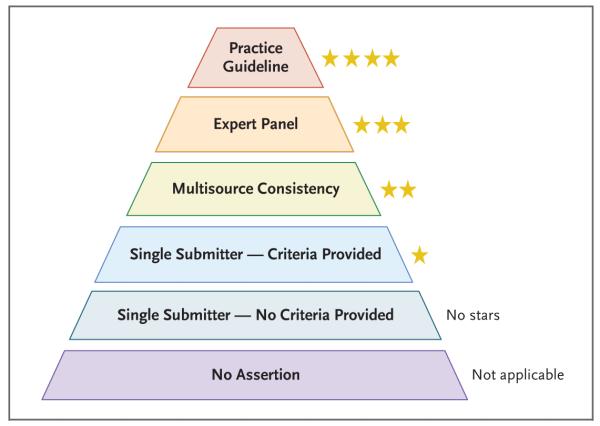
ClinVar requests, but does not require, detailed evidence to support any interpretation of clinical significance. One of the benefits of the ClinGen project, therefore, has been the development of a tiered system to define the type of review by which any variant has been assessed (Fig. 4), as well as rules for aggregating interpretations from multiple sources. In June 2015, the review status, which has always been represented graphically as colored stars, will be modified as follows: no stars if neither an assertion nor a documented method is provided, one star if methods are submitted for an interpretation, two stars if multiple groups with provided methods agree on the interpretation, three stars if the interpretation is provided by a ClinGen-approved expert panel, and four stars if the interpretation is endorsed by published practice guidelines. The review status is a field on which variants can be easily filtered when searching or downloading data from ClinVar, allowing specific subsets of variants to be selected on the basis of the level of review and consensus.
Figure 4. Review Levels Annotated in ClinVar.
Variants with assertions are rated according to the source and level of review for each submitted variant assertion. Submitters must comply with requirements (www.ncbi.nlm.nih.gov/clinvar/docs/assertion_criteria) for a submission to be assigned one, three, or four stars. Two stars are automatically assigned when multiple one-star submitted assertions are consistent. The distinction between submitters that have provided criteria and those that have not will begin in June 2015.
Overall, ClinGen-related working groups, with membership spanning more than 75 institutions, organizations, and commercial laboratories, have been assembled to tackle many of the key challenges to achieving the goals of ClinGen, including the establishment of standard procedures for evaluating genes, variants, genetic disorders, and phenotypes. For example, the accurate and detailed collection of phenotype information is challenging yet critical to the assessment of human variation. ClinGen is taking a multipronged approach to this problem through support for, and interaction with, researchers, clinical laboratories, clinicians, and patients. The ClinGen Phenotyping Working Group has chosen to use the Human Phenotype Ontology (www.human-phenotype-ontology.org) as its recommended standard for exchanging the phenotypes of patients, though other ontologies are also supported. Tools for the standardized collection of rare-disease phenotypes include PhenoTips21 and PhenoDB22 in addition to a phenotyping survey designed for patients in the ClinGen patient registry (called GenomeConnect), as described below.
The ClinGen Gene Curation Working Group has developed standards for assigning the level of evidence supporting a gene–disease relationship (www.clinicalgenome.org/knowledge-curation/gene-curation), which will be used by expert groups in different disease areas. This framework is particularly relevant as larger gene panels are introduced into genetic testing. Such panels may include genes for which the strength of the data underlying the association between a specific variant or variants in a specific gene and disease is limited. A user interface is being developed to support expert curation of genes and variants within a new database called ClinGenKB, allowing a flexible working environment for curation. Variants that are deposited into ClinVar will be accessible to curators working in ClinGenKB to enable expert review of all variants and resolution of conflicting interpretations. ClinGen has launched a growing number of clinical-domain working groups, with the initial set covering cardiovascular disease, hereditary cancer, somatic cancer, metabolic disease, and pharmacogenomics, with others in the planning stages (Fig. 3). The ClinGen Actionability Working Group is identifying which genes are associated with specific therapeutic or surveillance interventions in persons who do not yet have symptoms of genetic disease. The group has also developed a system for semiquantitative assessment of actionability that includes disease severity and likelihood, as well as the nature and efficacy of interventions. Additional working groups are focusing on new informatics approaches to variant assessment, integration with electronic health records, and outreach to patients through Genome-Connect, which allows patients to upload genetic test results and provide direct phenotypic data to the project. In addition, GenomeConnect enables a system to connect patients with laboratories, research studies, and one another, providing a robust and critical link to the broader community.
It is likely that the hypothetical case that is presented in the introduction to this article has already happened, given that each element of the story has occurred repeatedly. Patients have been receiving clinical genetic test results for hypertrophic cardiomyopathy for more than 10 years, and the American Heart Association has recommended the use of those results for dictating the clinical care of family members.23 The interpretation of many variants in genes associated with hypertrophic cardiomyopathy that have been reported as pathogenic has been challenged,24 and laboratories have had to revise their interpretations and communicate those revisions to patients.25 Fortunately, the ClinVar database is being increasingly used by clinical laboratories, physicians, and even patients, with more than 5000 hits per day. Faced with the challenge of regulating next-generation sequencing tests, the Food and Drug Administration is now looking to ClinGen to provide a possible resource for the clinical interpretation of genetic variation.26 With a system in place to support the open sharing of clinically interpreted genomic data, we are now poised to shepherd in a new era of transparency and advancement in genomic science that has the potential to improve how genomic information will inform the enhanced clinical care of patients.
Acknowledgments
ClinGen is funded by the National Human Genome Research Institute, with additional funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development and the National Cancer Institute (U41 HG006834, U01 HG007436, U01 HG007437, HHSN261200800001E). ClinVar is supported by the Intramural Research Program of the NIH, National Library of Medicine.
We thank all the contributing members of ClinGen at www.clinicalgenome.org/about/people, Scott Goehringer for his assistance in the development of the original Figure 1, and Steven Harrison for ClinVar data analysis.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.McKusick-Nathans Institute of Genetic Medicine. Johns Hopkins Medicine Online Mendelian Inheritance in Man (OMIM) http://omim.org.
- 2.1000 Genomes Project 1000 Genomes: a deep catalog of human genetic variation. http://www.1000genomes.org.
- 3.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Research Council . Sharing publication-related data and materials: responsibilities of authorship in the life sciences. National Academies Press; Washington, DC: 2003. [PubMed] [Google Scholar]
- 5.Chanock SJ, Manolio T, Boehnke M, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–60. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 6.Hindorff LA, MacArthur J, Morales J, et al. A catalog of published genome-wide association studies. http://www.genome.gov/gwastudies.
- 7.Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet. 2013;14:549–58. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
- 8.Paltoo DN, Rodriguez LL, Feolo M, et al. Data use under the NIH GWAS data sharing policy and future directions. Nat Genet. 2014;46:934–8. doi: 10.1038/ng.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Cancer Institute. National Human Genome Research Institute The Cancer Genome Atlas. http://cancergenome.nih.gov.
- 10.International Cancer Genome Consortium home page. https://icgc.org.
- 11.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.den Dunnen JT, Sijmons RH, Andersen PS, et al. Sharing data between LSDBs and central repositories. Hum Mutat. 2009;30:493–5. doi: 10.1002/humu.20977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sosnay PR, Siklosi KR, Van Goor F, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet. 2013;45:1160–7. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson BA, Spurdle AB, Plazzer J-P, et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014;46:107–15. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorn CF, Klein TE, Altman RB. PharmGKB: the Pharmacogenomics Knowledge Base. Methods Mol Biol. 2013;1015:311–20. doi: 10.1007/978-1-62703-435-7_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Human Genome Variation Society Nomenclature for the description of sequence variants. 2013 http://www.hgvs.org/mutnomen.
- 17.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell CJ, Dinwiddie DL, Miller NA, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3:65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg JS, Adams M, Nassar N, et al. An informatics approach to analyzing the incidentalome. Genet Med. 2013;15:36–44. doi: 10.1038/gim.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–5. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- 21.Girdea M, Dumitriu S, Fiume M, et al. PhenoTips: patient phenotyping software for clinical and research use. Hum Mutat. 2013;34:1057–65. doi: 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- 22.Hamosh A, Sobreira N, Hoover-Fong J, et al. PhenoDB: a new Web-based tool for the collection, storage, and analysis of phenotypic features. Hum Mutat. 2013;34:566–71. doi: 10.1002/humu.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gersh BJ, Maron BJ, Bonow RO, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2011;58:2703–38. doi: 10.1016/j.jacc.2011.10.825. [DOI] [PubMed] [Google Scholar]
- 24.Andreasen C, Nielsen JB, Refsgaard L, et al. New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur J Hum Genet. 2013;21:918–28. doi: 10.1038/ejhg.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aronson SJ, Clark EH, Varugheese M, Baxter S, Babb LJ, Rehm HL. Communicating new knowledge on previously reported genetic variants. Genet Med. 2012;14:713–9. doi: 10.1038/gim.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Food and Drug Administration Optimizing FDA’s regulatory oversight of next generation sequencing diagnostic tests — preliminary discussion paper. 2015 Jan 18; http://www.fda.gov/downloads/medicaldevices/newsevents/workshopsconferences/ucm427869.pdf.