Uncleavable Nup98–Nup96 is functional in the fission yeast Schizosaccharomyces pombe (original) (raw)
Graphical abstract
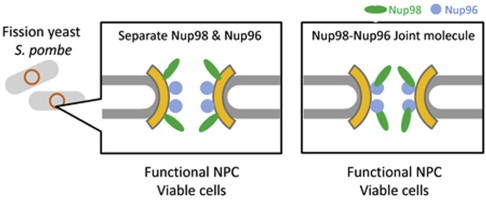
Abbreviations: GLFG repeat, Gly-Leu-Phe-Gly repeat; NE, nuclear envelope; NPC, nuclear pore complex
Keywords: Nucleoporins, Nuclear pore complex, FG nup, Nup107-160 complex, Alternative splicing, Fission yeast
Highlights
- •
Nup98 and Nup96 are produced via alternative splicing and autocleavage in S. pombe. - •
Splicing and autocleavage are both dispensable for S. pombe cell viability. - •
Nup98 and Nup96 expressed from different loci are functional for cell viability. - •
A Nup98–Nup96 joint molecule as a sole protein product of nup189 gene is functional. - •
Autocleavage is essential for Nup98 to remove a part of Nup96 generated by splicing.
Abstract
Essential nucleoporins Nup98 and Nup96 are coded by a single open reading frame, and produced by autopeptidase cleavage. The autocleavage site of Nup98–Nup96 is highly conserved in a wide range of organisms. To understand the importance of autocleavage, we examined a mutant that produces the Nup98–Nup96 joint molecule as a sole protein product of the nup189+ gene in the fission yeast Schizosaccharomyces pombe. Cells expressing only the joint molecule were found to be viable. This result indicates that autocleavage of Nup98–Nup96 is dispensable for cell growth, at least under normal culture conditions in S. pombe.
1. Introduction
In eukaryotic cells, the nucleus is spatially and functionally separated from the cytoplasm by the nuclear envelope (NE). Macromolecule transport across the NE takes place through the nuclear pore embedded in the NE. The nuclear pore is formed by a gigantic protein complex named the nuclear pore complex (NPC), which is estimated to be 125 MDa in vertebrate cells [1] and 50 MDa in yeast cells [2,3]. The NPC is an eightfold symmetrical structure composed of multi-copies of approximately 30 different protein components called nucleoporins.
Among the NPC components, Nup98 and Nup96 are essential nucleoporins that are highly conserved in a wide range of eukaryotic organisms [[4–15]; reviewed in [16]]. Human nucleoporins Nup98 and Nup96 are encoded in a single open reading frame, and Nup98–Nup96 precursor protein is separated by autopeptidase cleavage to produce two proteins [6,17]. A tripeptide sequence is necessary as a target for autocleavage and is evolutionally conserved, suggesting that autocleavage production of Nup98 and Nup96 from a single polypeptide may have a significant role for their functions. In addition, Nup98 is translated from two types of transcripts generated by alternative splicing in humans: one is a large mRNA encoding a large Nup98–Nup96 protein, and another is a short mRNA encoding Nup98 but not Nup96 [6]. These manners of expression of Nup98 and Nup96 are also conserved among many eukaryotes. In human cells, a cleavage-deficient mutant Nup98-Nup96 protein is not localized at the NPC, indicating that autocleavage is necessary for accurate localization of Nup98 and Nup96 [6,18]. In budding yeast Saccharomyces cerevisiae, on the other hand, a cleavage-deficient mutant protein is localized at the NPC, indicating dispensability of autocleavage [4]. Thus the necessity of Nup98–Nup96 autocleavage has been contradictory in human and budding yeast cells.
In S. pombe, Nup98 and Nup96 are coded by a single gene, nup189+. Deletion of the entire nup189+ ORF, which therefore means abolishment of both Nup98 and Nup96, is lethal [7,10], and expression of either Nup98 or Nup96 cannot complement lethality [3]. In this paper, we demonstrate that Nup98 and Nup96, either as separate protein molecules or as a single joint protein molecule, can functionally compensate for the endogenous nup189+ gene product.
2. Materials and methods
2.1. Media and culture condition
Yeast extract with supplements (YES) and Edinburgh minimal medium 2 (EMM2) were used for growth media [19]. Cells were grown at 26 °C or 30 °C.
2.2. Preparation of total RNA
Total RNA was prepared from standard strains (972 and 968) as follows: Cells grown to log phase were harvested, and cell pellet was washed once with ice-cold water and suspended in TES buffer (10 mM Tris–Cl pH7.5, 10 mM EDTA, 0.5% SDS). The cell suspension was mixed with the same volume of acidic phenol–chloroform, and incubated at 65 °C for 1 h. After heating, the sample was placed on ice for 1 min, vortexed, and then centrifuged at 14,000 rpm for 15 min at 4 °C. The aqueous phase was isolated using Phase-Lock Gel Heavy (5 PRIME, Germany) according to the manufacturer’s protocol.
2.3. Detection of splicing products by PCR
To detect spliced and unspliced transcripts, cDNA was synthesized from total RNA using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, USA). The cDNA was subjected to PCR using oligonucleotide primers designed upstream and downstream of the intron: 5′-CGACGCGTATGTCTAGATACGGTTTGCTGGATGATG-3′ (underlined region is an artificially added sequence) and 5′-CCTCGTCGACTTTTTTTGCATTATCGTCATAGTCACCTC-3′. The predicted length was 333 bp or 268 bp for DNA amplified from unspliced or spliced product, respectively.
2.4. Construction of mutant genes
To make a cleavage-deficient mutant gene and a splicing-deficient mutant gene, oligonucleotide-directed mutagenesis was applied using Site-Directed Mutagenesis Kit (Stratagene, USA) according to a manufacturer’s protocol. nup189+ ORF was amplified from genomic DNA by PCR and cloned to a conventional plasmid DNA. To make the cleavage-deficient mutant gene, oligonucleotide primers, 5′-GGTTCAGCATTTCGCTAGATACGGTTTGC-3′ and 5′-GCAAACCGTATCTAGCGAAATGCTGAACC-3′, were used. To make the splicing-deficient mutant gene, mutations were introduced at predicted splicing donor and acceptor sites. For the splicing donor mutation, oligonucleotide primers 5′-CAGGTAATTTAAAAAAATACGATCAACCAAACTTG-3′ and 5′-CAAGTTTGGTTGATCGTATTTTTTTAAATTACCTG-3′ were used. For the splicing acceptor mutation, oligonucleotide primers 5′-GTTACTCACCATACACCTGGTGCATTTCCCAAC-3′ and 5′-GTTGGGAAATGCACCAGGTGTATGGTGAGTAAC-3′ were used. For the splicing-deficient mutant, primers were designed to conserve the original amino acid sequence.
2.5. Strains and plasmid constructs
Strains used in this study are listed in Table 2. The construction of Nup98-GFP was described previously [3]. Briefly, we constructed a gene cassette, in which a GFP, a G418 resistance marker gene and the promoter sequence of nup189+ were fused in tandem. This gene cassette was integrated in between the Nup98-coding region and the Nup96-coding region at the genomic nup189+ gene locus by a two-step PCR method. To construct plasmids harboring other nup189+ alleles, mutant genes or control wild type gene were cloned by PCR and inserted to a plasmid harboring GFP or mRFP and either the marker lys1+ or the aureobasidin A-resistant allele of aur1+ (Takara, Japan). All plasmid constructs express proteins with their C-termini fused to GFP. A 312 bp of 5′UTR region of nup189+ was used as promoter. For construction of the mutant strains, endogenous nup189+ was heterozygously deleted with the ura4+ cassette in a diploid strain (namely, harboring one endogenous copy of nup189+), and the diploid cells were transformed with each of plasmids carrying nup189+ for integration at endogenous _lys1_- or aur1+ loci. Transformants were selected by lysine prototrophy or aureobasidin A-resistance, and genomic integration was confirmed by PCR. After the transformation, diploid cells were induced to form spores. The spores were dissected and analyzed for auxotrophy. Clones that were positive for both uracil prototrophy and integration of the marker were used for experiments.
Table 2.
Strains used for this study.
| Names | Genotypes | Sources |
|---|---|---|
| 968 | h90 wild type | laboratory stock |
| 972 | h− wild type | laboratory stock |
| CM1439 | h− ade6-216 ura4-D18 leu1-32 nup189::ura4+ lys1+::Pnup189-nup189+-GFP | This study |
| CM1449 | h− ade6-216 ura4-D18 leu1-32 nup189::ura4+ lys1+::Pnup189-nup189(unspliced)-GFP | This study |
| HA1362-5B | h− ura4-D18 leu1-32 nup189::ura4+ lys1+::Pnup189-nup189+-GFP | This study |
| HA1358-9A | h− ura4-D18 leu1-32 nup189::ura4+ lys1+::Pnup189-nup189(unspliced)-GFP | This study |
| HA1357-2A | h− ura4-D18 leu1-32 nup189::ura4+ lys1+::Pnup189-nup189(uncleavable)-GFP | This study |
| HA1359-10A | h− ura4-D18 leu1-32 nup189::ura4+ lys1+::Pnup189-nup189(unspliced-uncleavable)-GFP | This study |
| HA1623 | h− nup98+-GFP-kanr leu1-32 | [3] |
| HA1361-12B | h− ura4-D18 nup189::ura4+ aur1r::Pnup189-nup98-mRFP lys1+::Pnup189-nup96-GFP | This study |
2.6. Complementation test
To test the nup189 gene function for the splicing-deficient mutation or the cleavage-deficient mutation, or both, the mutant gene was introduced into the ectopic gene locus (lys1) of a diploid strain harboring a heterozygous deletion for nup189 with a ura4+ marker. Tetrad progenies (spores) were dissected from these diploid cells. Viable progenies were tested for uracil and lysine auxotrophy to determine their genotypes.
To test the cleavage-deficient Nup98-tail function, the mutant gene was integrated into the aur1+ locus, and the gene corresponding to the Nup96-coding region was integrated into the _lys1_- locus of a diploid strain harboring a heterozygous deletion for nup189 with a ura4+ marker. Tetrad progenies were dissected from these diploid cells. Viable progenies were tested for uracil and lysine auxotrophy and aureobasidin A-resistance to determine their genotypes. The genotypes of inviable progenies were predicted from the segregation of these marker genes in viable progenies. Haploid progenies bearing the mutant Nup98-tail gene and the Nup96-coding region in the nup189Δ background, which are expected to be lys1+, ura4+ and aureobasidin A-resistant, were not obtained among viable progenies.
2.7. Western blot analysis
Whole cell extract was prepared as described previously [3], and 20 μg of whole cell extract was subjected to 7.5% SDS–PAGE. Proteins were transferred to a PVDF membrane in 48 mM Tris, 39 mM glycine, 0.025% SDS and 15% methanol by wet transfer. The membrane was blocked in 5% skim milk and incubated with a rabbit polyclonal anti-GFP antibody (final concentration 0.1 μg/mL; Rockland, USA) or a mouse monoclonal anti-Nup98 antibody, 13C2 (final concentration 0.3 μg/mL) [20]. HRP-conjugated goat anti rabbit IgG or HRP-conjugated goat anti mouse IgG (GE Healthcare, USA) was used as a secondary antibody. For a loading control, endogenous actin was detected on the same membranes after stripping using a mouse monoclonal anti-β-actin antibody, ab8224 (final concentration 0.1 μg/mL; Abcam, USA) and HRP-conjugated goat anti-mouse IgG as described above. Protein bands were detected by chemiluminescence using the ChemiDoc MP imaging system (Bio-Rad, USA).
2.8. Microscopy
Living cells were mounted between coverslips [21]. Images were obtained using a DeltaVision microscope system (GE Healthcare) equipped with a CoolSNAP HQ2 CCD camera (Photometrics, Tucson, USA) through an oil-immersion objective lens (PlanApoN60 × OSC, NA = 1.4, Olympus, Japan). Z-stack images were obtained at 0.4 μm intervals for 10 Z-steps, and subjected to deconvolution to improve images by removing out-of-focus images [22].
3. Results
3.1. Two alternative splice isoforms are coded by nup189+ gene
We determined cDNA sequences of RNA transcribed from S. pombe nup189+, and detected two isoforms of the transcript. Comparison of the two isoforms revealed that in the shorter transcript, a stretch of 65 nucleotides flanked by typical 5′ and 3′ splicing sites was removed from the longer transcript (Fig 1A), suggesting putative splicing. To confirm splicing of the nup189+ transcript, mutations were introduced into the putative intron sequences as indicated in Fig. 1B. The strain harboring the mutations was viable (Table 1). RNA products of wild type and the mutant genes were amplified by RT-PCR using a pair of primers spanning the putative intron sequence. A single band was detected in the mutant strain (Fig. 1C) while three bands were detected in the control, wild-type strain. The size of the single band correlated well with the predicted size of the amplicon from the unspliced transcript (Fig. 1C, arrow labeled “unspliced”). The fastest migrating band in the wild type lane corresponded to the amplicon from the spliced transcript in size (Fig. 1C, arrowhead labeled “spliced”). Amplification of these bands depended on reverse transcription (compare lanes of RT+ with RT- in Fig. 1C), indicating that they were not products from contaminated genomic DNA. The slowest migrating band in the wild type lane (Fig. 1C, indicated by the asterisk) was presumed to be a hybrid duplex of spliced and unspliced products because its PCR amplification reproduced the same three bands in the wild type (data not shown). These results suggest that alternative splicing, which is a rare event in S. pombe, takes place for nup189+. Sequences of the spliced and unspliced isoforms were registered in the DDBJ database (accession numbers: LC043101 and LC037233, respectively).
Fig. 1.
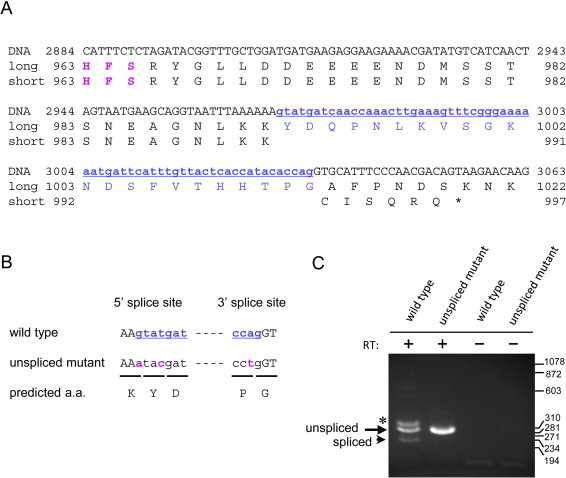
Alternative splicing of the nup189+ transcripts. (A) Nucleotide sequence and the predicted amino acid sequence of S. pombe nup189+. A label of “DNA” represents a partial nucleotide sequence near the intron. The sequences labeled “long” and “short” indicate amino acid sequences predicted from a long cDNA and a short cDNA, respectively. The numbers represent the number of the DNA nucleotide or the amino acid residue. The asterisk indicates the position of the stop codon (TAA). Blue letters indicate intron sequences (small and underlined letters) and the corresponding amino acid sequence (capital letters), identified by comparing long and short cDNA sequences. Magenta “HFS” indicates evolutionary conserved autocleavage site. (B) Construction of splicing-deficient mutant. The letters represent the DNA sequence; black capital and blue small letters represent exon and intron, respectively. Magenta letters indicate mutations introduced at the 5′ and 3′ splice sites. Predicted amino acids from the DNA sequence are shown at the bottom (“predicted a. a.”). (C) Detection of spliced and unspliced transcripts by RT-PCR. Total RNA of a wild type (“wild type”, CM1439) and a splicing-deficient mutant (“unspliced mutant”, CM1449) were treated with reverse transcriptase (labeled as RT+) and amplified by PCR. Positions of unspliced and spliced transcripts are labeled in the left of the panel. The asterisk indicates a hybrid of cDNAs from spliced and unspliced transcripts (see text). RT- indicates samples prepared without reverse transcriptase. Size markers (in bp) are shown in the right of the panel.
Table 1.
Viability of strains expressing nup189+ derivatives.
| Gene constructsa | Viabilities |
|---|---|
| nup189 + -GFP | Viable |
| nup98-nup96 (unspliced)-GFP | Viable |
| nup98-nup96 (uncleavable)-GFP | Viable |
| nup98-nup96 (unspliced and uncleavable)-GFP | Viable |
| nup98-mRFP and nup96-GFP | Viable |
| nup98-tail (uncleavable)-mRFP and nup96-GFP | Lethal |
3.2. Autocleavage of Nup189
Both the spliced and unspliced isoforms are predicted to encode proteins bearing a conserved tripeptide sequence His-Phe-Ser (HFS), a potential autopeptidase cleavage site, between amino acid residues 963–965 (Fig. 2A). To determine if this site is indeed cleaved to generate two separate protein molecules, we constructed a strain bearing the coding region for GFP fused in frame to the 3′-end of the nup189+ coding sequence, and analyzed the protein products by Western blot analysis using antibodies specific to either Nup98 or GFP; this cell strain was expected to produce Nup98 and Nup96–GFP proteins if autocleavage occurred. Using anti-Nup98 antibody, we detected a single band corresponding to the predicted Nup98, and using anti-GFP antibody, we detected a single band corresponding to the predicted Nup96–GFP (Fig. 2B and C, wild type); we failed to detect a band corresponding to the full-length Nup189 fused with GFP (Nup98–Nup96–GFP), indicating that autocleavage occurs with no remains of the joint molecule. The same bands corresponding to Nup98 and Nup96-GFP were also detected in the splicing-defective mutant, as expected (Fig. 2B and C, “unspliced mutant”). Next, we constructed a cleavage-deficient mutant of Nup98–Nup96–GFP by replacing Ser at position 965 with Ala. This mutant strain was viable (Table 1), and used for detection of the mutant protein. The cleavage-deficient mutant produced a protein corresponding to a joint molecule of Nup98–Nup96–GFP when examined by Western blot analysis using either anti-Nup98 or anti-GFP antibodies (Fig. 2B and C, “uncleavable mutant”). The same band, corresponding to the Nup98–Nup96–GFP joint molecule, was detected when analyzing the strain defective in both splicing and autocleavage (Fig. 2B and C, “double mutant”). Anti-Nup98 antibody detected an additional band slightly larger than Nup98, corresponding to Nup98 connected to a tail peptide, an uncleaved product from the spliced nup189+ transcript (Fig. 2B and C, “uncleavable mutant”). These results indicate that autocleavage depends on the HFS sequence in products of S. pombe nup189+, as reported in those of human NUP98–NUP96 [6]. Thus, the spliced isoform is predicted to produce a Nup98 isoform that contains an extended polypeptide tail of 33 amino acids, which undergoes autocleavage to produce Nup98 and the tail. These features of expression, together with the conserved domain structures, are comparable to the human Nup98–Nup96 precursor.
Fig. 2.
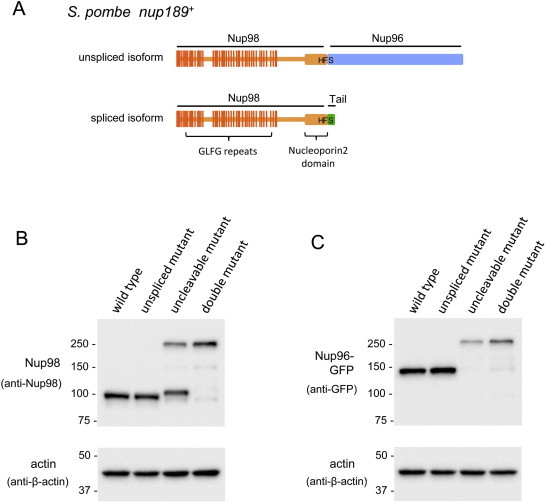
Western blotting of nup189+ gene products. (A) Schematic of predicted precursors of S. pombe nup189+ isoforms. Portions of Nup98 and Nup96 are shown in orange and blue, respectively. The Nup98-tail peptide is shown in green. Vertical lines on Nup98 indicate positions of GLFG repeats. The thick region at the carboxy terminus of Nup98 indicates a Nucleoporin2 domain, a catalytic domain for autocleavage activity. (B and C) Western blotting analysis of Nup98 and Nup96. Whole cell extracts were prepared from the indicated strains. (B) Detection of Nup98 using anti-Nup98 antibody. (C) Detection of Nup96-GFP using anti-GFP antibody. Strains at the top represent wild type (HA1362-5B), cleavage-deficient mutant (“uncleavable mutant”, HA1357-2A), splicing-deficient mutant (“unspliced mutant”, HA1358-9A), and splicing- and cleavage- deficient mutant (“double mutant”, HA1359-10A). Size markers (in kDa) are shown on the left of each panel. Lower panels show actin as loading controls.
3.3. Cleavage-deficient Nup98–Nup96 is functional
We examined a strain simultaneously expressing Nup98 and Nup96 from two separate genomic loci, and found that expression of both Nup98 and Nup96 rescued lethality in nup189Δ cells (Table 1). This result shows that each of the proteins, Nup98 and Nup96, is essential for cell growth. To further understand the significance of autocleavage, we constructed the nup189 deletion strain (nup189Δ) expressing the cleavage- and splicing-defective Nup189 from an ectopic gene (lys1+) locus, which produces the Nup98–Nup96 joint molecule as a sole protein molecule of Nup98 and Nup96, and examined its viability under vegetative culture conditions. The double mutant cell was viable (Table 1). This result suggests that autocleavage to produce Nup98 and Nup96 is not necessary for cell growth when Nup98 and Nup96 are expressed as a joint protein in S. pombe. Fluorescence microscopy indicated that the joint molecule, fused to GFP, (Nup98–Nup96–GFP) was localized at the nuclear periphery as were Nup98–GFP and Nup96–GFP (Fig. 3) while Nup98–GFP showed somewhat clustered localization at the nuclear periphery compared to other expression constructs, as reported previously [3]. Considering that Nup98 can be localized in both the NPC and the nucleoplasm [23], the requirement for Nup98 is due to its function in the NPCs.
Fig. 3.
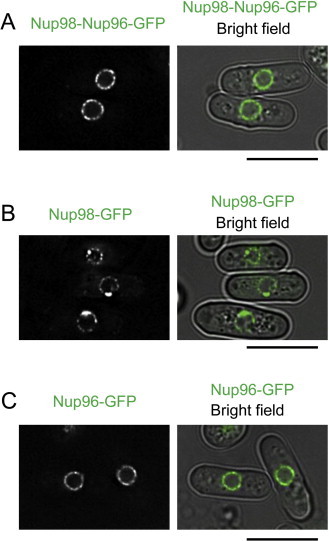
Intracellular localization of cleavage-deficient nup189+ gene product. (A) Fluorescence microscopy of a single joint molecule of Nup98-Nup96-GFP. A strain expressing cleavage- and splicing-deficient mutant Nup189-GFP (HA1359-10A) was observed. Fluorescence image merged with the bright field image is also shown. (B and C) Fluorescence microscopy of Nup98-GFP and Nup96-GFP. A strain expressing Nup98-GFP (HA1623) or Nup96-GFP (HA1362-5B) was observed. Fluorescence images merged with bright field images are also shown. Scale bars, 10 μm.
3.4. Cleavage-deficient Nup98-tail protein is non-functional
The significance of autocleavage was also tested for the short, premature form of the protein generated by alternative splicing, which corresponds to Nup98 containing a tail peptide and the autocleavage site. To do so, a cleavage-deficient mutation was first introduced into the cDNA from a spliced isoform. Then, we examined lethality of the cells harboring either this cleavage-deficient Nup98-coding cDNA or the wild type Nup98-coding cDNA in the background of _nup189_Δ. In this experiment, Nup96-coding cDNA was simultaneously expressed in cells to compensate for Nup96 function (see Materials and Methods for details of the complementation test). In combination with Nup96, the wild type cDNA (nup98-mRFP) complemented the lethality of _nup189_Δ, but the cleavage-deficient cDNA (nup98-tail (uncleavable)-mRFP) did not (Table 1; see Section 2 for details of the complementation test). This result suggests that autocleavage, which produces the functional Nup98 by removing the tail polypeptide (corresponding to the N-terminal region of Nup96 when splicing occurs), is necessary for cell growth. Taken together, our results indicate that a cleavage-deficient Nup98-Nup96 is functional if the fusion molecule is expressed as a sole source of Nup98 and Nup96.
4. Discussion
4.1. A Nup98-Nup96 joint molecule is functional in S. pombe
This study demonstrates that the essential nucleoporins Nup98 and Nup96 are functional either as a joint molecule or as separate molecules in S. pombe. According to in vitro experiments, Nup98 forms a complex with Nup96 in the budding yeast S. cerevisiae and in humans [18,24,25]. Analysis of the three-dimension crystal structure of the human Nup98 with the cleaved tail peptide predicted a noncovalent interaction between the C-terminus of Nup98 and the N-terminus of Nup96 [18]. These findings suggest that Nup98 and Nup96 may form a complex even after autocleavage in vivo. Thus, the structure of the cleavage-deficient Nup98–Nup96 may not be critically different from the structure of the complex containing individual molecules of Nup98 and Nup96. As a result, the joint molecule can functionally substitute for Nup98 and Nup96 in the NPCs, at least in S. pombe.
In human cells, the cleavage-deficient protein is localized in the nucleoplasm but not at the nuclear periphery, suggesting that autocleavage is required for targeting Nup98 and Nup96 to the NPC [6,18,24]. These differences in autocleavage requirements for NPC localization between yeasts and humans might reflect the different modes of mitosis among these organisms. In human cells, the NE and the NPCs are disassembled during mitosis, and chromosome segregation proceeds in the absence of the NE (open mitosis), while in yeasts, the mitosis proceeds in the presence of the NE (closed mitosis). In human cells, the NE is reconstructed at telophase, and at the same time, nucleoporins are also reassembled to construct the NPCs in a defined order of protein assembly [26–28] The Nup98–Nup96 joint molecule probably cannot be assembled in the correct order because of an obstruction in structure or functions for NPC assembly with other nucleoporins in human cells. In contrast to human cells, the yeast NPC remains on the NE throughout the cell cycle, and thus, de novo assembly of the NPC proceeds over time, keeping its fundamental structure [29]. This may be the reason why the harmful effects of the Nup98–Nup96 joint molecule are limited in S. pombe.
4.2. Importance of autocleavage in Nup98 and Nup96 functions
Autocleavage may enable cells to regulate Nup98 and Nup96 separately. Indeed, Nup98 and Nup96 have different residence times on the NPCs. Nup98 resides on the NPC of HeLa cells for approximately 3 h [30]. In contrast, Nup96 is predicted to reside for much longer because members of Nup107–Nup160 subcomplex reside on the NPC for approximately 40 h in HeLa cells [30]. In addition, recent studies have indicated that Nup107–Nup160 subcomplex nucleoporins are not turned over in postmitotic cells [31–33]. Thus, differences in the residence times of Nup98 and Nup96 may result from the half-life of each protein, and the turnover rates after cleavage may be differently regulated depending on their specific roles. For organisms with a relatively short generation time, like yeasts, the residence time of Nup98 may be long enough to maintain NPC function, at least while the organisms are in their vegetative growth. This may be one of the reasons why the Nup98-Nup96 joint molecule is functional in S. pombe.
Another possible explanation may relate to an evolutional aspect. Despite evolutionary conservation of Nup98–Nup96 homologs among eukaryotes, a Trypanosoma Nup98–Nup96 ortholog, which does not harbor the typical HFS triplet autocleavage site (TbNup158; accession number: EAN79079.1), is uncleaved [12]. Furthermore, Plasmodium, in the Alveolata supergroup, and some organisms in the Kinetoplastids supergroup have predicted Nup98-Nup96 orthologs (accession numbers: CBZ12098.1, CCW71133.1, EAA17057.1), which do not have recognizable HFS triplet cleavage sites (Fig. 4). This suggests that autocleavage is unnecessary for Nup98–Nup96 function in these lower eukaryotes, although it has not yet been experimentally demonstrated that autocleavage does not occur. However, the existence of naturally-uncleavable Nup98–96, as found in Trypanosoma, suggests that the joint Nup98-Nup96 molecule may be an ancient form of Nup98 and Nup96, and that the cleavage-deficient mutant Nup98–Nup96 may perform minimal, conserved functions of Nup98 and Nup96 required for cell viability in S. pombe.
Fig. 4.
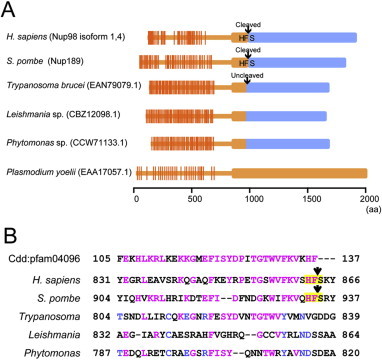
Comparison of Nup98-Nup96 joint sites. (A) Schematic of Nup98-Nup96 molecular structures found in eukaryotes. Orthologs found in H. sapiens, S. pombe, Trypanosoma brucei, Leishmania sp., Phytomonas sp., and Plasmodium yoelii are shown. Portions of Nup98 and Nup96 are shown in orange and blue, respectively. Triplet alphabets shown in the joint of Nup98-Nup96 indicate autocleavage sites or the corresponding sites. The ortholog in P. yoelii have neither an apparent autocleavage site nor a Nup96 sequence. (B) Comparison of amino acid sequences of Nup98 C-termini and Nup98-Nup96 joint sites. A reference sequence shown at the top is pfam04096 in the conserved domain database (CDD). Magenta letters indicate residues identical to the reference; blue letters indicate residues conserved in Kinetoplastids supergroup. Conserved “HFS” found in human and fission yeast sequences are shaded by yellow. Arrows indicate cleavage sites.
4.3. Importance and functions of alternative splicing in Nup98 and Nup96
The results of the present study also indicate that regulation of Nup98–Nup96 gene expression by alternative splicing is conserved in S. pombe as well as in humans. The unspliced transcript generates Nup98 and Nup96, whereas the spliced transcript generates Nup98 and the tail peptide; therefore, the amounts of Nup98 and Nup96 proteins would be different even though they are expressed from the same gene. The excess amount of Nup98 probably compensates for the short residence time of Nup98 at the NPC, as discussed above. In addition, because the functions of the FG-repeat proteins (such as Nup98) are affected by oxidative stress [34], biased expression of Nup98 may be required for maintenance of intact Nup98 at the NPC. According to a comprehensive gene expression analysis, the amount of S. pombe nup189+ mRNA is increased upon oxidative stress [35]. Considering that increased transcription usually promotes splicing in S. pombe [36], splicing of nup189+ mRNA may be required to supply sufficient amounts of Nup98 to adapt to changes in environmental conditions.
Acknowledgments
This study was supported by CREST of JST to T.H., and by JSPS Kakenhi Grant Numbers 26440098 to H.A., 24570227 to M.I., 20114002, 26116511 to Y.H., and 21370094 to T.H.
Contributor Information
Yasushi Hiraoka, Email: hiraoka@fbs.osaka-u.ac.jp.
Tokuko Haraguchi, Email: tokuko@nict.go.jp.
References
- 1.Reichelt R., Holzenburg A., Buhle E.L., Jr, Jarnik M., Engel A., Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asakawa H., Yang H.J., Yamamoto T.G., Ohtsuki C., Chikashige Y., Sakata-Sogawa K., Tokunaga M., Iwamoto M., Hiraoka Y., Haraguchi T. Characterization of nuclear pore complex components in fission yeast Schizosaccharomyces pombe. Nucleus. 2014;5:149–162. doi: 10.4161/nucl.28487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teixeira M.T., Siniossoglou S., Podtelejnikov S., Bénichou J.C., Mann M., Dujon B., Hurt E., Fabre E. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 1997;16:5086–5097. doi: 10.1093/emboj/16.16.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powers M.A., Forbes D.J., Dahlberg J.E., Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J. Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fontoura B.M., Blobel G., Matunis M.J. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tange Y., Hirata A., Niwa O. An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J. Cell Sci. 2002;115:4375–4385. doi: 10.1242/jcs.00135. [DOI] [PubMed] [Google Scholar]
- 8.De Souza C.P., Horn K.P., Masker K., Osmani S.A. The SONB(NUP98) nucleoporin interacts with the NIMA kinase in Aspergillus nidulans. Genetics. 2003;165:1071–1081. doi: 10.1093/genetics/165.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baï S.W., Rouquette J., Umeda M., Faigle W., Loew D., Sazer S., Doye V. The fission yeast Nup107-120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol. Cell Biol. 2004;24:6379–6392. doi: 10.1128/MCB.24.14.6379-6392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X.Q., Du X., Liu J., Balasubramanian M.K., Balasundaram D. Identification of genes encoding putative nucleoporins and transport factors in the fission yeast Schizosaccharomyces pombe: a deletion analysis. Yeast. 2004;21:495–509. doi: 10.1002/yea.1115. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Li X. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell. 2005;17:1306–1316. doi: 10.1105/tpc.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeGrasse J.A., DuBois K.N., Devos D., Siegel T.N., Sali A., Field M.C., Rout M.P., Chait B.T. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell Proteomics. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwamoto M., Mori C., Kojidani T., Bunai F., Hori T., Fukagawa T., Hiraoka Y., Haraguchi T. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate Tetrahymena. Curr. Biol. 2009;19:843–847. doi: 10.1016/j.cub.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 14.Neumann N., Lundin D., Poole A.M. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voronina E., Seydoux G. The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development. 2010;137:1441–1450. doi: 10.1242/dev.047654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwamoto M., Asakawa H., Hiraoka Y., Haraguchi T. Nucleoporin Nup98: a gatekeeper in the eukaryotic kingdoms. Genes Cells. 2010;15:661–669. doi: 10.1111/j.1365-2443.2010.01415.x. [DOI] [PubMed] [Google Scholar]
- 17.Rosenblum J.S., Blobel G. Autoproteolysis in nucleoporin biogenesis. Proc. Natl. Acad. Sci. USA. 1999;96:11370–11375. doi: 10.1073/pnas.96.20.11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodel A.E., Hodel M.R., Griffis E.R., Hennig K.A., Ratner G.A., Xu S., Powers M.A. The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol. Cell. 2002;10:347–358. doi: 10.1016/s1097-2765(02)00589-0. [DOI] [PubMed] [Google Scholar]
- 19.Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 20.Iwamoto M., Asakawa H., Ohtsuki C., Osakada H., Koujin T., Hiraoka Y., Haraguchi T. Monoclonal antibodies raised against GLFG repeat of Nucleoporin Nup98 from Tetrahymena, yeast to human. Monoclon. Antib. Immunodiagn. Immunother. 2013;32:81–90. doi: 10.1089/mab.2012.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asakawa H., Hiraoka Y. Live-cell fluorescence imaging of meiotic chromosome dynamics in Schizosaccharomyces pombe. Methods Mol. Biol. 2009;558:53–64. doi: 10.1007/978-1-60761-103-5_4. [DOI] [PubMed] [Google Scholar]
- 22.Agard D.A., Hiraoka Y., Shaw P., Sedat J.W. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- 23.Griffis E.R., Altan N., Lippincott-Schwartz J., Powers M.A. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell. 2002;13:1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffis E.R., Xu S., Powers M.A. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol. Biol. Cell. 2003;14:600–610. doi: 10.1091/mbc.E02-09-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratner G.A., Hodel A.E., Powers M.A. Molecular determinants of binding between Gly-Leu-Phe-Gly nucleoporins and the nuclear pore complex. J. Biol. Chem. 2007;282:33968–33976. doi: 10.1074/jbc.M707911200. [DOI] [PubMed] [Google Scholar]
- 26.Haraguchi T., Koujin T., Hayakawa T., Kaneda T., Tsutsumi C., Imamoto N., Akazawa C., Sukegawa J., Yoneda Y., Hiraoka Y. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 2000;113:779–794. doi: 10.1242/jcs.113.5.779. [DOI] [PubMed] [Google Scholar]
- 27.Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., Sironi L., Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doucet C.M., Talamas J.A., Hetzer M.W. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winey M., Yarar D., Giddings T.H., Jr, Mastronarde D.N. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell. 1997;8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabut G., Doye V., Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 31.D’Angelo M.A., Raices M., Panowski S.H., Hetzer M.W. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savas J.N., Toyama B.H., Xu T., Yates J.R., 3rd, Hetzer M.W. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyama B.H., Savas J.N., Park S.K., Harris M.S., Ingolia N.T., Yates J.R., 3rd, Hetzer M.W. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crampton N., Kodiha M., Shrivastava S., Umar R., Stochaj U. Oxidative stress inhibits nuclear protein export by multiple mechanisms that target FG nucleoporins and Crm1. Mol. Biol. Cell. 2009;20 doi: 10.1091/mbc.E09-05-0397. 5106–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen D., Toone W.M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., Bähler J. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilhelm B., Marguerat S., Watt S., Schubert F., Wood V., Goodhead I., Penkett C.J., Rogers J., Bähler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]