A microfluidic device for label-free, physical capture of circulating tumor cell-clusters (original) (raw)
. Author manuscript; available in PMC: 2016 Jan 1.
Published in final edited form as: Nat Methods. 2015 May 18;12(7):685–691. doi: 10.1038/nmeth.3404
Abstract
Cancer cells metastasize through the bloodstream either as single migratory circulating tumor cells (CTCs) or as multicellular groupings (CTC-clusters). Existing technologies for CTC enrichment are designed primarily to isolate single CTCs, and while CTC-clusters are detectable in some cases, their true prevalence and significance remain to be determined. Here, we developed a microchip technology (Cluster-Chip) specifically designed to capture CTC-clusters independent of tumor-specific markers from unprocessed blood. CTC-clusters are isolated through specialized bifurcating traps under low shear-stress conditions that preserve their integrity and even two-cell clusters are captured efficiently. Using the Cluster-Chip, we identify CTC-clusters in 30–40% of patients with metastatic cancers of the breast, prostate and melanoma. RNA sequencing of CTC-clusters confirms their tumor origin and identifies leukocytes within the clusters as tissue-derived macrophages. Together, the development of a device for efficient capture of CTC-clusters will enable detailed characterization of their biological properties and role in cancer metastasis.
Isolation and analysis of rare circulating tumor cells (CTCs) hold great promise in providing insight into the blood-borne metastasis, as well as noninvasive monitoring of cancer response following therapeutic interventions. Considerable progress has been made in the development of devices to capture one tumor cell admixed with a billion normal blood cells1,2. Among these, microfluidic technologies have the important advantage of combining high-throughput processing with low-shear and efficient cell isolation3–6 as well as handling of unfixed cells, which are readily subjected to molecular and functional analyses7.
In addition to single cancer cells acquiring a migratory epithelial-to-mesenchymal transition (EMT) phenotype, cancer metastasis has also been suggested as early as in 1950s to be mediated by groupings of tumor cells that appear to break off from a primary tumor8,9. Such circulating tumor emboli have been reported in both mouse models10 and human blood specimens4,7,11–15, ranging from large thrombi or blood clots carrying tumor cells, to clumps of tumor cells admixed with reactive stromal cells16. Moreover, tumor cell clusters intravenously injected show higher metastasis initiation capability in the mouse compared to otherwise identical single cells8,9. In patients with metastatic cancer, presence of CTC-clusters has recently been associated with a poor prognosis17,18
Existing CTC isolation technologies are designed with a focus on single CTCs and they may lack specificity and overlook sample processing constraints necessary to preserve the integrity of CTC-clusters or to sort them in a reliable manner. Here, we introduce a unique microfluidic chip, the Cluster-Chip, designed to specifically isolate CTC-clusters from unprocessed patient blood samples with high sensitivity. The Cluster-Chip exploits the unique geometries of cellular aggregates to differentiate CTC-clusters from single cells in blood, and hence it does not require antibody coating. This chemistry-free approach enables specific and label-free isolation of CTC-clusters from patients with different cancer types, as well as the release of CTC-clusters following their capture, allowing for downstream molecular and functional assays.
RESULTS
Design of the Cluster-Chip
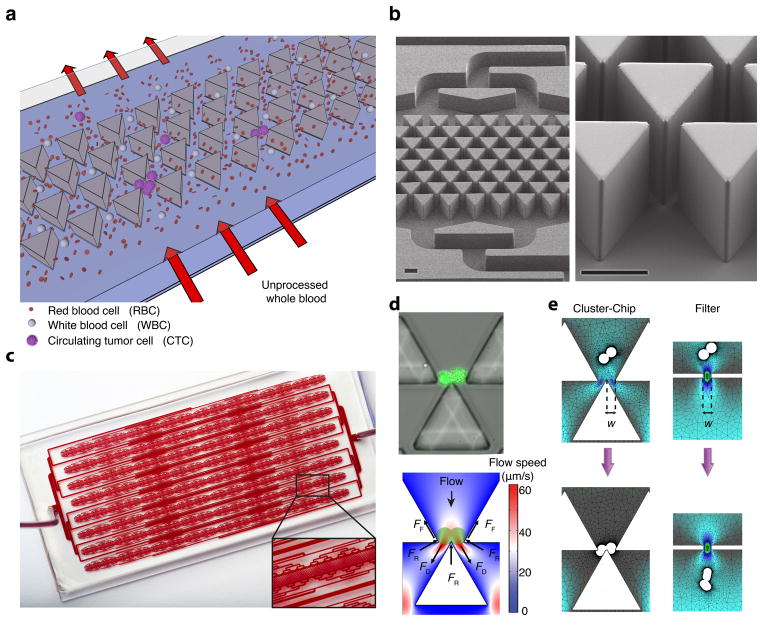
The Cluster-Chip captures CTC-clusters relying on the strength of cell-cell junctions as they flow under physiological flow speed through a set of triangular pillars (Fig. 1a,b). The fundamental building block of the Cluster-Chip is formed by three triangular pillars, two of which form a narrowing channel that funnels the cells into an opening, where the edge of the third pillar is positioned to bifurcate the laminar flow. As blood flows, single blood cells and single CTCs divert to one of the two streamlines at the bifurcation, passing through the 12 μm × 100 μm opening (Fig. 1a). In contrast, CTC-clusters are held by the leading edge of the bifurcating pillar under a dynamic force balance, even if they are deformable enough to squeeze through either one of the openings (Fig. 1a,d). Sharp bifurcating edge retains the captured CTC-cluster in both streamlines simultaneously and under this dynamic balance, cell-cell junctions within a CTC-cluster serve as points of support for a stable equilibrium (not possible for a single cell) while the bifurcating pillar serves as fulcrum (Fig. 1d,e). This building block is repeated in multiple rows for redundancy (Fig. 1b,c).
Figure 1.
The design and operation of the Cluster-Chip (a) Schematic representation of the Cluster-Chip operation. Cluster-Chip captures CTC clusters from unprocessed whole blood while single cells pass through. (b) SEM micrographs of the Cluster-Chip showing multiple rows of shifted triangular pillars forming consecutive cluster traps (left) and a high magnification image of a cluster trap (right). Scale bars 60 μm. (c) A full image of the Cluster-Chip. The blood from a single inlet is uniformly distributed over 4096 parallel trapping paths and then collected to a single outlet. The inset shows a close up of a CTC cluster-trapping region with part of the microfluidic distribution and collection networks. (d) A spiked 2-cell LNCaP cluster captured on the Cluster-Chip (top) and schematic explaining the dynamic balance that holds it captured (bottom). Forces acting on the cell cluster are drag forces (_F_D) due to fluid flow, reaction forces (_F_R) from the pillars and frictional forces (_F_F) including the effect of cell adhesion. (e) Finite element analysis comparing the cell cluster dynamics in the Cluster-Chip (left) and in a filter with half the opening size (right). The diameters of individual cells are 15 μm and the opening width (w) is 12 μm.
To ensure against dissociation of CTC-clusters, the Cluster-Chip is optimized to handle cellular aggregates with processing flow speeds limited well below the physiological blood flow speed in human capillaries. Therefore, captured CTC clusters are not subjected to shear forces higher than those occur in vivo during circulation. The peak flow speed of ~70 μm/s at the bifurcation (Fig. 1d) is much lower than existing microfluidic and filter-based CTC isolation technologies19,20. Yet the chip can interrogate clinical blood specimens at a rate of 2.5 ml/hr due to its highly parallel architecture (Fig. 1c).
The Cluster-Chip is purposely designed to also capture two-cell clusters because, our analysis shows ~92% of CTC-clusters are of oligoclonal nature, including the majority of two-cell clusters, and characterized by an elevated metastatic potential compared to single CTCs18.
The Cluster-Chip operates fundamentally different from filter-based technologies such as porous membranes or microfluidic traps. The Cluster-Chip captures cell clusters independent of their deformability allowing capture of CTC-clusters that might otherwise squeeze through a smaller pore (Fig. 1e and Supplementary Fig. 1). Moreover, captured clusters are retained under a dynamic force balance unlike filters where cells are primarily retained due to their surface tension and are exposed to damaging stresses20. Finally the Cluster-Chip specifically captures cell clusters and does not trap single cells and this specificity enables clog-free processing of unprocessed whole blood samples.
Chip characterization and optimization using cell lines
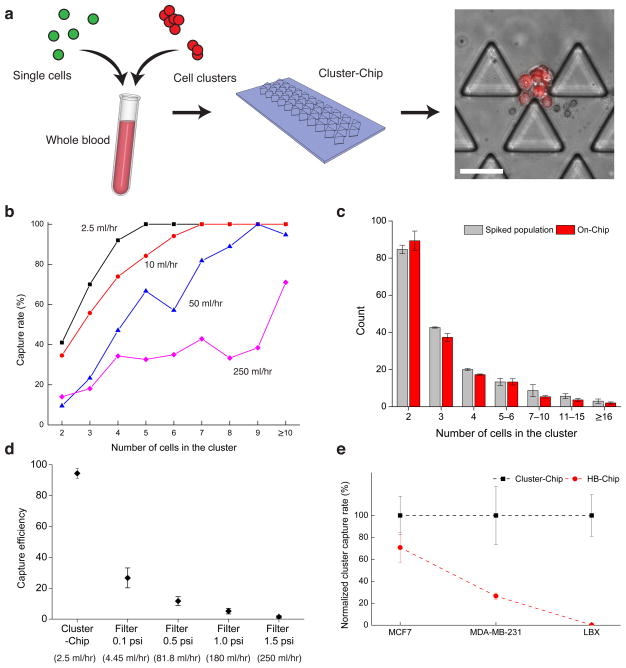
To characterize the device, we first spiked artificially formed clusters of the fluorescently labeled MDA-MB-231 human breast cancer cells into unlabeled blood samples from healthy donors and captured them using the Cluster-Chip. To ensure against any Chip-mediated cell aggregation, we introduced a 1:1 mixture of GFP-tagged single cells and mCherry-tagged clusters (2–30 cells) into whole blood, followed by Cluster-Chip capture (Fig. 2a). All cancer cell clusters captured on the chip were exclusively mCherry positive, and all GFP-tagged single cells flowed unimpeded through the Chip, without adhering to the captured clusters (Fig. 2a). This is consistent with our probabilistic analysis, based on the rarity of CTCs within a blood specimen and the large number of uniformly distributed traps (Supplementary Fig. 2)
Figure 2.
Characterization of the Cluster-Chip capture using cell lines spiked in whole blood (a) The schematic diagram of the experimental procedure used to ensure against artificial cell aggregation. (b) Cluster-Chip cell cluster capture rate measured using artificial clusters of MDA-MB-231 cell line spiked in whole blood. Capture rate is shown as a function of the number of cells in the cluster at different flow rates. (c) Histograms of the number of cells in the cell clusters within spiked population and cell clusters captured in the Cluster-Chip. Error bars show s.e.m from three independent experiments. (d) Comparison of cluster capture efficiency of Cluster-Chip with membrane filters operated under different pressures using human breast cancer cell line MDA-MB-231. Effective whole blood processing rate for each condition is noted in parentheses on the x-axis. Error bars show s.e.m from three independent experiments. (e) Comparison of cluster capture efficiency of Cluster-Chip with immunoaffinity-based HB-Chip using three human breast cancer cell lines. Surface EpCAM expression is highest in MCF7 and lowest in LBX1. Cluster-Chip has higher cluster capture efficiency for all cases. Error bars show s.e.m from three independent experiments. Scale bars 60 μm.
To quantify the capture efficiency, we imaged and counted clusters captured on the Cluster-Chip and those that flowed through undetected (Supplementary Fig. 3). The capture efficiency increases with the number of cells within the cluster and decreases with increasing flow rates (Fig. 2b). The reduced capture at high flow rates may result from the failure of the dynamic force balancing smaller clusters, as well as the breaking up of captured clusters under higher shear stress. We therefore selected 2.5 ml/hr as the optimal flow rate, providing both high capture efficiency and high throughput (Supplementary Video 1). At 2.5 ml/hr, the Cluster-Chip captured 169/171 (99%) MDA-MB-231 clusters with ≥4 cells (Fig. 2b). For smaller clusters, the Cluster-Chip captured 28/40 (70%) of 3-cell clusters and 48/117 (41%) of 2-cell clusters. Distribution of captured 2-cell and 3-cell clusters to subsequent rows indicates that the capture efficiency for small clusters can further be improved by adding more rows (Supplementary Fig. 4). Also, the effectiveness of the CTC-cluster trap in capturing larger clusters (4-cell or more) at 2.5 ml/hr flow rate was confirmed as captured clusters were predominantly (>80%) found in the first row.
To ensure against possible damage to the captured clusters at 2.5 ml/hr, we compared the cluster size distribution in the spiked population with the captured population on the Cluster-Chip. We characterized the artificial clusters of fluorescently labeled MDA-MB-231 cells using microscopy (Supplementary Fig. 5) followed by gently spiking into whole blood collected from a healthy donor (Online Methods). Comparison of the size distribution before and after processing show that Cluster-Chip does preserve the integrity of captured clusters (Fig. 2c). Moreover, majority (>95%) of captured clusters were found in the first row excluding the possibility of damage due to bifurcating cluster traps.
To compare the Cluster-Chip with filter-based isolation techniques, we processed simulated samples using the Cluster-Chip and membrane filters with 5μm pore-diameter. Simulated samples were prepared by spiking clusters of MDA-MB-231 cells into whole blood collected from healthy donors. We processed whole blood sample using the Cluster-Chip at 2.5 ml/hr at room temperature. The simulated samples processed with filters were diluted 1:10 (v/v) as whole blood samples led to clogging. In direct comparisons, the Cluster-Chip has higher cluster capture efficiency than membrane filters operated at different filtration pressures (0.1–1.5 psi) (Fig. 2d). At 1.5 psi, typically used for filtration, we found that cluster capture is inefficient. At 0.1 psi, which is substantially lower than pressures typically employed for filtration, cluster capture efficiency increased to ~26%. At 0.1 psi, the effective whole blood processing rate of filter is comparable to the Cluster-Chip so further reduction is not practical (Fig. 2d). Compared to single cell capture efficiencies reported for membrane filters19,20, there may be two reasons for the lower cluster capture efficiency in our experiments. First, clusters are more likely to be lost during the wash step compared to single cells, which are partially or fully squeezed in pores. Second, we counted only intact clusters excluding some that were damaged (Supplementary Fig. 6). Nevertheless, the operational conditions for filters were not optimized for clusters in our experiments and we cannot exclude that by changing some of the parameters, capture rate of viable clusters could increase.
To compare the Cluster-Chip with another microfluidic platform, we processed matched specimens through the herringbone-chip (HB-Chip)4, an antibody-based capturing chamber that we had first employed in detecting such clusters4. We used three cell lines representing varying EpCAM expression levels observable across CTCs: MCF-7, representing high EpCAM-expressing epithelial breast cancer cells; MDA-MB-231, mesenchymal “triple-negative” breast cancer cells with low EpCAM expression, and MCF10A-LBX121, an EMT-induced breast cell line with virtually absent EpCAM expression. In direct comparisons, the Cluster-Chip had 50% and 400% higher efficiency than the anti-EpCAM coated HB-Chip in capturing MCF7 and MDA-MB-231 clusters, respectively (Fig. 2e). MCF10A-LBX1 clusters were not captured by the HB-Chip, but were readily identified in the Cluster-Chip (103 differential capture) (Fig. 2e). Taken together, the reliance on physical properties of clusters rather than on restricted cancer cell surface epitopes to isolate clusters makes the Cluster-Chip uniquely suited to studying cancers across different clinical settings. These include epithelial cancers in which activation of EMT during cancer cell invasion triggers loss of epithelial markers, as well as non-epithelial cancers, such as melanoma.
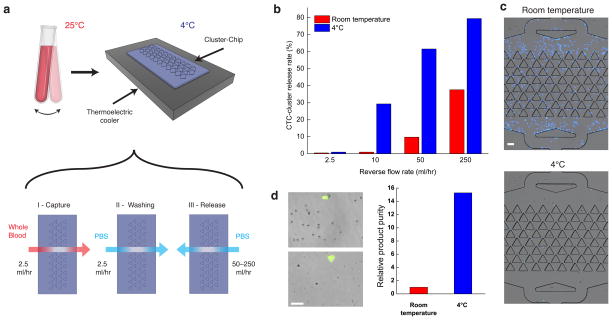
Besides sensitive capture, viable release of captured CTC-clusters is critically important for downstream molecular and functional assays. Despite the fact that clusters are not tethered to the Cluster-Chip through antibody-mediated interactions, we observed that release of captured CTC-clusters is incomplete, especially for large clusters, following reversal of the flow. Using reverse flow rates of 2.5–250 ml/hr only succeeded in releasing 114/308 (37%) of captured clusters. To address this problem, we tested sample processing at lower temperatures, known to reduce non-specific cell adhesion22. We operated the Cluster-Chip on a thermoelectric cooler at 4°C (Fig. 3a), so as to cool the samples transiently, thereby avoiding prolonged cold exposure which is known to activate platelets23. Processing samples at 4°C substantially improved the cluster release efficiency: 188/236 (80%) of captured clusters were released under 250 ml/hr reverse flow. The improvement was particularly evident at low reverse flow speeds, which enabled the use of low shear forces, thereby enhancing release of viable cells (Fig. 3b). Indeed, processing at 4°C and release of clusters under 250 ml/hr reverse flow had no notable effect on cell viability (Supplementary Fig. 7). We note that the Cluster-Chip technology is also compatible with non-adherent coating materials and sacrificial layer approaches24.
Figure 3.
Release of captured clusters from the Cluster-Chip (a) Schematic diagram of the experimental setup (top) and individual steps of the CTC cluster release process (bottom). The bulk of the blood sample is continuously rocked at room temperature and is cooled only when it is being processed by the Cluster-Chip. (b) Release efficiency of MDA-MB-231 clusters from the chip as a function of the reverse flow rate and the processing temperature. (c) Nonspecific binding of leukocytes on the Cluster-Chip when the sample is processed at room temperature (left) and at 4°C (right). The leukocytes were fixed with 4% PFA and stained with DAPI. Fluorescent images were overlaid on bright field images. (d) Images of the product released in solution from the Cluster-Chip operated at room temperature (top-left) and at 4°C (bottom-left). Relative purity of released cell clusters against contaminating blood cells when Cluster-Chip is operated at room temperature and 4°C (right). Scale bars 60 μm.
Processing blood samples at 4°C also proved to substantially reduce non-specific binding by contaminating blood cells. In experiments with blood samples from healthy donors, the temperature-dependent reduction in on-chip leukocyte contamination was as high as 50-fold (Fig. 3c). Moreover, less non-specific binding of leukocytes to the chip translates into a 15-fold higher product purity for cold-processed samples (Fig. 3d), which is particularly important for downstream molecular applications.
Capture of CTC-clusters from blood samples of patients
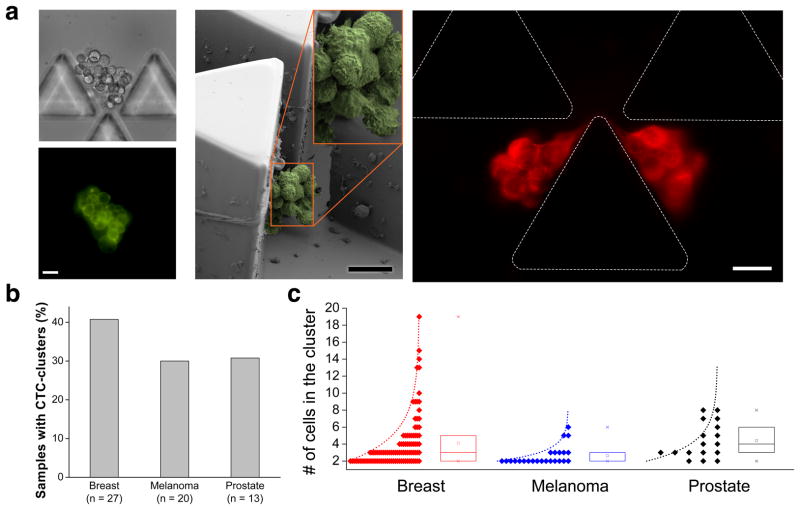
We applied the technology to blood samples collected from patients with metastatic cancer. Patients with breast cancer (n=27), melanoma (n=20) and prostate cancer (n=13) (Supplementary Table 1) provided consent according to an IRB-approved protocol at Massachusetts General Hospital, and 4 ml of blood was directly processed through the Cluster-Chip, followed by immunofluorescence staining (Online Methods). We scored positive those clusters that were a) positive for specific and well-established cancer-associated markers for the disease type, and b) CD45 (leukocyte marker) negative. Previous studies indicate that those specific markers are very accurate for identifying bona fide cancer cells in circulation 4,7,18,25,26. A representative CTC-cluster from a patient with metastatic breast cancer is shown with on-chip capture (Fig. 4a) and subsequent release in solution (Supplementary Fig. 8). In addition to fluorescence microscopic imaging of live cells, we used scanning electron microscopy (SEM) to image fixed CTC-clusters on the chip from the same patient (Fig. 4a). In one case, we observed a CTC-cluster highly strained even under the very low flow speed in the Cluster-Chip. This case shows the extent that a CTC-cluster can deform and also points to the need for elasticity-independent capture mechanism of the Cluster-Chip, which the filter-based technologies fail to achieve. We identified CTC-clusters in 11/27 patients with breast cancer (40.7%, ~0.5 clusters/ml); 6/20 patients with melanoma (30%, ~0.15/ml) and 4/13 patients with prostate cancer (31%, ~0.28/ml) (Fig. 4b). For some patients, multiple (2–3) blood samples were obtained and patients scored positive when a CTC-cluster was observed during at least one time-point. The number of cells within a CTC-cluster ranged from 2 to 19 cells and follows a trend toward exponential distribution (Fig. 4c). In a subset of our patient cohort (n=19), we compared the number of CTC-clusters captured using the Cluster-Chip with the number of single CTCs simultaneously identified using CTC-iChip5 and found no correlation (Supplementary Fig. 9). When processing patient samples, we observed cell debris and fibrins, which did not interfere with chip operation due to large number of traps working in parallel.
Figure 4.
Capture of CTC-clusters from blood samples of patients with metastatic cancer (a) Representative images of CTC-clusters isolated form patients with metastatic breast cancer: (Left) Brightfield and fluorescent images of a live CTC-cluster stained for common breast cancer surface markers; (Middle) SEM micrograph of a fixed CTC-cluster; (Right) Fluorescent image of a highly deformable CTC-cluster stained for cytokeratin. Note that this CTC-cluster is not split but is highly strained even under slow flow in the Cluster-Chip. This particular case clearly justifies the need for the elasticity-independent capture mechanism of the Cluster-Chip. (b) Percentage of patients with CTC-clusters in breast, melanoma and prostate cancer. (c) Size distribution of CTC-clusters isolated from breast, melanoma and prostate cancer patients. The box plots show the 25th, 50th and 75th percentile for each disease type. Scale bars 20 μm.
Immunocytochemical and molecular analysis of patient CTC-clusters
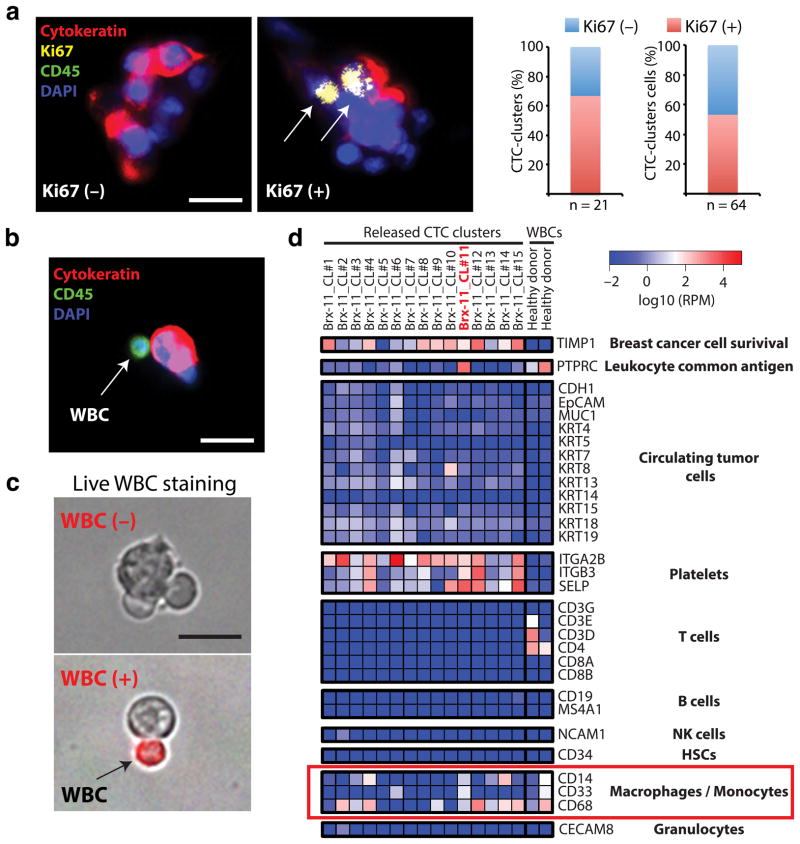
To test the versatility of the Cluster-Chip to address biology of CTC-clusters, we first measured tumor cell proliferation markers to explore intra-tumor cell heterogeneity, and then analyzed non-tumor cells that were adherent to tumor cells within the clusters. Staining for the proliferation marker Ki67 correlates well with invasiveness and poor outcome in comparison with mitotic activity index or phospho-Histone H3 staining27. In a patient with metastatic breast cancer with large numbers of single and clustered CTCs, costaining of CTC-clusters for Ki67, cytokeratin (tumor marker) and CD45 showed no notable difference between the proliferative index of these two cell populations: 34/64 (53%) CTCs within clusters were Ki67-positive, compared with 162/439 (40%) single CTCs (Fig. 5a and Supplementary Fig. 10). Of note, 66% of CTC clusters had at least one Ki67-positive cell (Fig. 5a).
Figure 5.
Immunocytochemical and molecular characterization of patient CTC-clusters (a) Images of a Ki67-negative and a Ki67-positive CTC-clusters stained with CK (red), Ki67 (yellow), CD45 (green) and DAPI (nuclei, blue). The bar graphs show the percentage of Ki67-positive CTC-clusters in this patient (left; n=21) and the percentage of Ki67-positive cells within CTC-clusters (right; n=64). (b) Image of a CTC-cluster associated to a white blood cell (WBC). The cells were stained with CK (red), CD45 (green) and DAPI (nuclei, blue). (c) Images of WBC-negative (top) and WBC-positive (bottom) CTC-clusters released from the Cluster-Chip and live-stained with TexasRed-conjugated antibodies against CD45, CD14 and CD16 (red). (d) Heatmap showing expression levels of transcripts associated to CTCs, macrophages/monocytes, T cells, B cells, natural killer (NK) cells, hematopoietic stem cells (HSCs), granulocytes and platelets in 15 CTC-clusters isolated at a single timepoint from a patient with metastatic breast cancer. RPM: reads per million. Scale bars 20 μm
The low shear stress of the Cluster-Chip also facilitates the identification of heterologous cell types4,16,28 that may be attached to tumor cells. Given recent progress in immunotherapy of cancer, the identification of leukocyte populations adherent to tumor cells in the circulation is of particular interest. Overall, we found non-tumor cells to be rare among clusters captured using the Cluster-Chip (<5% of 60 patients). While these cells consistently expressed of the pan-leukocyte marker CD45 (Fig. 5b), their precise identity is unknown. To address this question, we used the Cluster-Chip to capture CTC-clusters from the blood of a breast cancer patient, released CTC-clusters in solution, stained them against the leukocyte cell surface markers, and then isolated intact CTC-clusters individually using a micromanipulator (Fig. 5c). From a single time-point, we retrieved 15 CTC-clusters, and each of those clusters was individually subjected to RNA-sequencing analysis using a next generation platform (SOLiD 5500). Expression analysis revealed that a) all Brx-11 CTC-clusters expressed low but detectable levels of CTC markers such as keratins, MUC1, EpCAM and/or CDH1, b) 14/15 CTC-clusters expressed high levels of TIMP1, a matrix metalloproteinase widely associated to breast cancer cell survival and absent in WBCs29–32, and c) all Brx-11 CTC-clusters were associated to platelets transcripts, while control WBCs were not, consistent with previous reports18,28 (Fig. 5d). Most CTC-clusters appeared to exist in a hybrid epithelial/mesenchymal state (Supplementary Fig. 11), a phenotype observed in instances of breast CTCs7 and consistent with the possibility of grouped migration. One CTC-cluster associated to a WBC (cluster #11, Fig. 5c) expressed transcriptional signatures of tissue-derived macrophages (e.g. high CD14, CD33 and CD68 expression) as well as CD45, keratins, TIMP1 and platelets transcripts (Fig. 5d). No enrichment for other leukocyte subclasses were observed, including T cells, B cells, natural killer (NK) cells, hematopoietic stem cells (HSCs) and granulocytes (Fig. 5d). Additional transcripts for platelets and epithelial cells were noted, consistent with previous reports28.
DISCUSSION
We introduce a novel microfluidic technology that specifically isolates CTC-clusters from unprocessed blood samples of patients with cancer. The dynamic capture of multicellular structures as they are impaled on triangle structures under low flow conditions offers important new capabilities that are not readily achieved with current CTC isolation strategies. Existing technologies primarily target single CTCs and employ non-optimum processing conditions that result in lower sensitivity and specificity. Widely used batch purification techniques13,14,19,20,33 involve multiple processing steps that are likely to disrupt CTC-clusters. High-speed fluorescence imaging of minimally enriched blood samples are efficient for CTC-cluster detection13 but the low purity complicates downstream molecular analysis. Microfluidic devices optimized to isolate single CTCs can also isolate CTC-clusters4,15, although substantial losses of clusters may be associated with the optimization of flow conditions for single CTC capture. Filtering blood samples through membranes with small pores14,19,20 may be effective, but these approaches employ high flow rates resulting in extremely high shear forces. As such, CTC-clusters are likely damaged or even squeeze through relatively smaller pores, as modeled in our computer simulation.
Using the Cluster-Chip, we determined that CTC-clusters are heterogeneous, including both actively proliferating cells as well as apparently quiescent cells. Occasionally, CTC-clusters are also found associated with cells of the immune system, and our RNA sequencing data revealed that these are most likely tumor-associated macrophages. This finding supports increasingly appreciated role played by tumor-associated macrophages in cancer progression34,35. The fact that such tissue-derived macrophages travel with CTC-clusters in the bloodstream has implications for the ability to noninvasively monitor tumor-immune cell interactions, a potentially important benefit, given the increasing use of immune checkpoint blockade in the treatment of multiple different types of cancer36.
The Cluster-Chip enables label-free isolation of unfixed CTC-clusters from unprocessed whole blood specimens from patients with cancer. The reliance on structural properties of CTC-clusters is particularly important, given the variation in tumor epitope expression, as well as the ability of highly flexible clusters to pass through simple pores. The ability to capture CTC-clusters at relatively high frequency in patients with metastatic cancer and to release them for biological studies will enable detailed analyses of the physiological role of these clusters in the progression and metastasis of human cancer.
Supplementary Material
1
2
Acknowledgments
We express our gratitude to all patients and healthy volunteers who participated in this study and contributed blood samples. We thank O. Hurtado, A.J. Aranyosi and L. Libby for coordination of the research laboratories; D.M. Lewis for his help in instrumentation; and L. Nieman, J. Walsh and T. N. Lewis for their help with microscopy. N.A. was supported by the Swiss National Science Foundation and the Human Frontiers Science Program. This work was supported by the US National Institutes of Health (NIH) P41 Resource Center (M.T.); a NIH National Institute of Biomedical Imaging and Bioengineering Quantum Grant (M.T. and D.A.H.); Stand Up to Cancer (D.A.H., M.T. and S.M.); the Howard Hughes Medical Institute (D.A.H.); the Prostate Cancer Foundation and the Charles Evans Foundation (D.A.H. and M.T.); and Johnson and Johnson (M.T. and S.M.).
Footnotes
AUTHOR CONTRIBUTIONS
A.F.S., N.A., S.M., D.A.H and M.T. designed the research, analyzed the data and prepared the manuscript. N.K. performed computer simulations. M.C.D., M.Z. and A.E. processed clinical samples, performed immunofluorescence staining and scanning. B.H. manufactured devices for clinical studies. H.Z. and T.S. performed amplification and RNA sequencing. T.K.S., D.T.M., X.L. and A.B. provided clinical samples. B.S.W. performed statistical analysis on the RNA sequencing data. S.R., D.T.T., S.L.S. and R.K. commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests. A.F.S and M.T. are inventors on a patent MGH filed to protect the Cluster-Chip technology.
References for main text
- 1.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nature Reviews Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 2.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. The Journal of Cell Biology. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stott SL, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proceedings of the National Academy of Sciences. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozkumur E, et al. Inertial Focusing for Tumor Antigen-Dependent and -Independent Sorting of Rare Circulating Tumor Cells. Science Translational Medicine. 2013;5:179ra47–179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon HJ, et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nature Nanotechnology. 2013;8:735–741. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu M, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe S. The metastasizability of tumor cells. Cancer. 1954;7:215–223. doi: 10.1002/1097-0142(195403)7:2<215::aid-cncr2820070203>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 9.Fidler IJ. The relationship of embolic homogeneity, number, size and viability to the incidence of experimental metastasis. European Journal of Cancer. 1973;9:223–227. doi: 10.1016/s0014-2964(73)80022-2. [DOI] [PubMed] [Google Scholar]
- 10.Yu M, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandt B, et al. Isolation of prostate-derived single cells and cell clusters from human peripheral blood. Cancer Research. 1996;56:4556–4561. [PubMed] [Google Scholar]
- 12.Molnar B, Ladanyi A, Tanko L, Sréter L, Tulassay Z. Circulating tumor cell clusters in the peripheral blood of colorectal cancer patients. Clinical Cancer Research. 2001;7:4080–4085. [PubMed] [Google Scholar]
- 13.Cho EH, et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys Biol. 2012;9:016001. doi: 10.1088/1478-3975/9/1/016001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krebs MG, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and-independent approaches. Journal of Thoracic Oncology. 2012;7:306–315. doi: 10.1097/JTO.0b013e31823c5c16. [DOI] [PubMed] [Google Scholar]
- 15.Hou HW, et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duda DG, et al. Malignant cells facilitate lung metastasis by bringing their own soil. Proceedings of the National Academy of Sciences. 2010;107:21677–21682. doi: 10.1073/pnas.1016234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou JM, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 18.Aceto N, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vona G, et al. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulating tumor cells. The American journal of pathology. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coumans FAW, van Dalum G, Beck M, Terstappen LWMM. Filtration parameters influencing circulating tumor cell enrichment from whole blood. PLoS ONE. 2013;8:e61774. doi: 10.1371/journal.pone.0061774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu M, et al. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes & Development. 2009;23:1737–1742. doi: 10.1101/gad.1809309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rico F, Chu C, Abdulreda MH, Qin Y, Moy VT. Temperature modulation of integrin-mediated cell adhesion. Biophysj. 2010;99:1387–1396. doi: 10.1016/j.bpj.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kattlove HE, Alexander B. The effect of cold on platelets. I. Cold-induced platelet aggregation. Blood. 1971;38:39–48. [PubMed] [Google Scholar]
- 24.Shah AM, et al. Biopolymer system for cell recovery from microfluidic cell capture devices. Anal Chem. 2012;84:3682–3688. doi: 10.1021/ac300190j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo X, et al. Isolation and molecular characterization of circulating melanoma cells. Cell Rep. 2014;7:645–653. doi: 10.1016/j.celrep.2014.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyamoto DT, et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2:995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inwald EC, et al. Ki-67 is a prognostic parameter in breast cancer patients: results of a large population-based cohort of a cancer registry. Breast Cancer Res Treat. 2013;139:539–552. doi: 10.1007/s10549-013-2560-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20:576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuvaja P, et al. Tumor tissue inhibitor of metalloproteinases-1 (TIMP-1) in hormone-independent breast cancer might originate in stromal cells, and improves stratification of prognosis together with nodal status. Exp Cell Res. 2012;318:1094–1103. doi: 10.1016/j.yexcr.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Hekmat O, et al. TIMP-1 increases expression and phosphorylation of proteins associated with drug resistance in breast cancer cells. J Proteome Res. 2013;12:4136–4151. doi: 10.1021/pr400457u. [DOI] [PubMed] [Google Scholar]
- 31.Würtz SO, Schrohl AS, Mouridsen H, Brünner N. TIMP-1 as a tumor marker in breast cancer--an update. Acta Oncol. 2008;47:580–590. doi: 10.1080/02841860802022976. [DOI] [PubMed] [Google Scholar]
- 32.Würtz SØ, et al. Tissue inhibitor of metalloproteinases-1 in breast cancer. Endocr Relat Cancer. 2005;12:215–227. doi: 10.1677/erc.1.00719. [DOI] [PubMed] [Google Scholar]
- 33.Talasaz AH, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proceedings of the National Academy of Sciences. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyckoff JB, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Research. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 36.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nature Reviews Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kojic N, et al. A 3-D model of ligand transport in a deforming extracellular space. Biophysj. 2010;99:3517–3525. doi: 10.1016/j.bpj.2010.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2