A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks (original) (raw)
. Author manuscript; available in PMC: 2016 Jul 30.
Abstract
Finding the components of cellular circuits and determining their functions systematically remains a major challenge in mammalian cells. Here, we introduced genome-wide pooled CRISPR-Cas9 libraries into dendritic cells (DCs) to identify genes that control the induction of tumor necrosis factor (Tnf) by bacterial lipopolysaccharide (LPS), a key process in the host response to pathogens, mediated by the Tlr4 pathway. We found many of the known regulators of Tlr4 signaling, as well as dozens of previously unknown candidates that we validated. By measuring protein markers and mRNA profiles in DCs that are deficient in the known or candidate genes, we classified the genes into three functional modules with distinct effects on the canonical responses to LPS, and highlighted functions for the PAF complex and oligosaccharyltransferase (OST) complex. Our findings uncover new facets of innate immune circuits in primary cells, and provide a genetic approach for dissection of mammalian cell circuits.
Introduction
Regulatory circuits that control gene expression in response to extracellular signals perform key information processing roles in mammalian cells, but their systematic unbiased reconstruction remains a fundamental challenge. There are currently two major strategies for associating targets with their putative regulators on a genomic scale (reviewed in (Kim et al., 2009)): (1) observational (correlative) approaches that relate them based on statistical dependencies in their quantities or physical associations; and (2) perturbational (causal) approaches that relate them by the effect that a perturbation in a putative regulator has on its target.
While observational strategies have become a cornerstone of circuit inference from genomic data, perturbational strategies have been more challenging to apply on a genomic scale, especially in primary mammalian cells. RNAi, which until recently was the main tool available in mammals, is limited by off-target effects and lack of sufficient suppression of expression (Echeverri et al., 2006), whereas more effective strategies based on haploid cell lines (Carette et al., 2009) are not applicable to the diversity of primary cell types and their specialized circuitry. As a result, a hybrid approach has emerged (Amit et al., 2011), where genomic profiles (e.g., of mRNAs, protein-DNA binding, protein levels, protein phosphorylation, etc.) are used to build observational models from which a smaller set of dozens of candidate regulators are identified. These candidates are in turn tested by perturbation.
The recent introduction of genome editing in mammalian cells using the clustered, regularly interspaced, short palindromic repeats (CRISPR)-associated nuclease Cas9 system has enabled pooled genome-wide screens of gene function (Gilbert et al., 2014; Konermann et al., 2015; Shalem et al., 2014; Wang et al., 2014). In such screens, pooled libraries are introduced into cell lines and cellular phenotypes are selected based on cell lethality or growth. To expand the biological processes that can be studied, there remains a need to adapt these methods for short-term primary cell cultures and selecting cellular phenotypes based on more versatile molecular markers.
Here, we present a pooled CRISPR strategy to dissect the innate immune response of bone marrow derived dendritic cells (‘BMDCs’ or ‘DCs’) isolated from Cas9-expressing transgenic mice. Building on our recent observation that lentiviruses expressing guide RNAs could be used to knockout genes in these cells (Platt et al., 2014), we infected DCs with a pooled, genome-wide library of lentiviruses, stimulated them with lipopolysaccharide (LPS), and monitored their responses by intra-cellular staining for the inflammatory cytokine Tnf, a major marker of the early response to LPS. We used flow cytometry to isolate cells that failed to fully induce Tnf or that induced it more strongly, and then determined sgRNA abundance by deep sequencing. We recovered many of the key known regulators of TLR signaling, validated dozens of new regulators and identified 3 functional modules of regulators with distinct regulatory effects. Our study identifies new facets in the complex response of immune cells to pathogens, and provides a general strategy for systematically dissecting circuits in other primary mammalian cells.
Results
A system for cell-autonomous, pooled genetic screens in BMDCs derived from Cas9-expressing mice
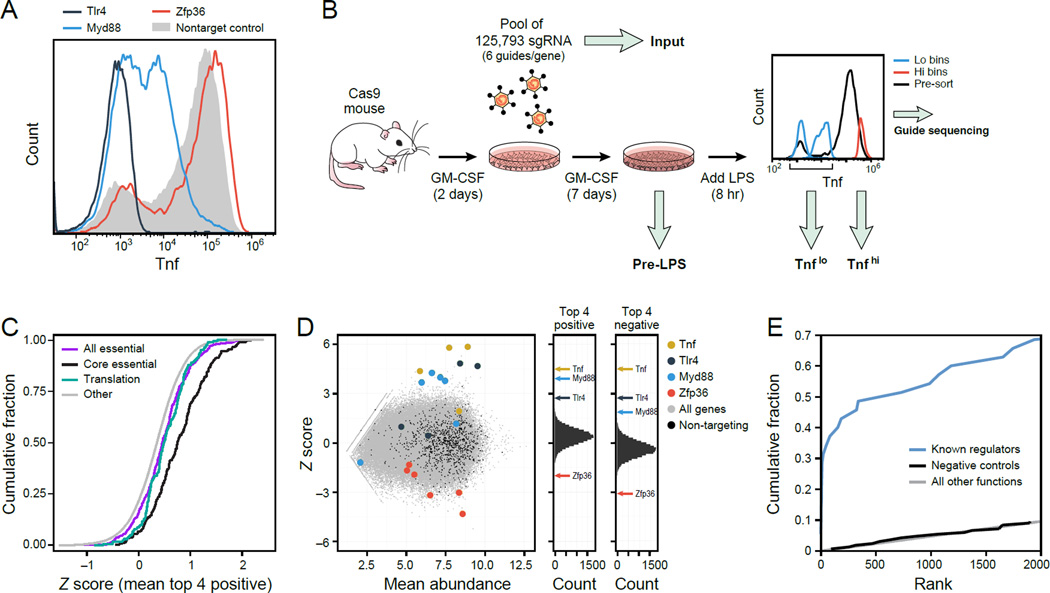
To enable genome-wide pooled genetic screens, we developed a cell autonomous readout of innate immune activation by intracellular staining of a central inflammatory cytokine, Tnf. To test the assay, we individually transduced BMDCs with lentiviruses expressing single guide RNAs (sgRNAs; Experimental Procedures) that target each of three genes: (1) Tlr4, the cell membrane receptor that senses bacterial LPS; (2) Myd88, a key component required for Tlr4 signaling to induce Tnf; and (3) Zfp36 (TTP), an RNA binding protein that destabilizes Tnf mRNA. Following LPS activation, we added Brefeldin A to block Tnf secretion, and at 8 hours post-activation detected Tnf with a fluorescent antibody using flow cytometry. Compared to a non-targeting sgRNA control, sgRNAs targeting Myd88 or Tlr4 strongly reduced Tnf, whereas sgRNAs targeting Zfp36 increased Tnf (Figure 1A). These results provide an experimental system in BMDCs for an autonomous genome-wide pooled screen based on cell sorting.
Figure 1. A genome-wide pooled CRISPR screen in mouse primary DCs.
(A) Flow cytometry of intracellular Tnf levels following 8h of LPS stimulation for single sgRNAs. (B) Design of a genome wide CRISPR screen. (C) Cumulative distribution function (CDF) plots of the gene level Z-score distribution of genes annotated as ‘essential’ (purple) and ‘core essential’ (black) in (Hart et al., 2014), ‘Translation’ (in GO, blue), and all other genes (grey). (D) Left: Binned Z-scores (ZS) of the Tnflo / Tnfhi ratios (y-axis) vs. sgRNA mean abundances in Tnflo and Tnfhi (x-axis). Right: Gene score distribution for positive (ZS) and negative (ZS) regulators (Experimental Procedures). (E) CDFs of gene rank for the 35 genes in the TLR pathway from LPS to Tnf (KEGG, blue), non-targeting controls (black) and all other genes (grey).
Also see Figure S1, related to Figure 1.
A genome-wide pooled sgRNA library screen in primary BMDCs
We performed three independent, pooled genome-wide screens using a library of lentiviruses harboring 125,793 sgRNAs targeting 21,786 annotated protein-coding and miRNA mouse genes (Sanjana et al., 2014), as well as 1,000 non-targeting sgRNA as negative controls. In each of the three replicate screens, we infected 60–200 million BMDCs with the library at a multiplicity of infection (MOI) of 1, stimulated cells with LPS and sorted Cd11c+ cells based on high or low Tnf expression levels (~5 million cells/bin, Figure 1B, Experimental Procedures). We then amplified and sequenced sgRNAs from 4 sources (Figure 1B, thick grey arrows): post-LPS cells with (1) high Tnf (“Tnfhi”) or with (2) low Tnf (“Tnflo”), (3) cells from the last day of differentiation prior to LPS stimulation (day 9, “pre-LPS”), and (4) plasmid DNA of the input lentiviral library (“Input”). We reasoned that sgRNAs against positive regulators of Tnf expression would be enriched in Tnflo relative to Tnfhi, that sgRNAs targeting negative regulators will be enriched in Tnfhi relative to Tnflo; and that sgRNAs targeting genes essential for DC viability or differentiation would be depleted in pre-LPS compared to Input. We established two computational methods to address the inherent noise of the screen (Figure S1A): the first using Z scores of the fold change in normalized sgRNA abundance (and then averaging the top 4 sgRNAs per gene), and the second analogous to differential expression (DE) analysis of sequenced RNA (Love et al., 2014), (Experimental Procedures). The top ranked genes substantially overlap between the two approaches (50/100 for positive regulators, 30/100 for negative regulators, P < 10−10, hypergeometric test), and their rankings are well correlated (Figure S1B,C) up to ranks 150 and 50 for positive and negative regulators, respectively (Figure S1D and S1E). While our screen is in principle compatible with discovery of both positive and negative regulators, it was conducted at high (near-saturation) levels of LPS, and is thus likely to be less sensitive for discovery of negative regulators, due to limited dynamic range for observing further Tnf induction.
The screen correctly identifies known regulators of cell viability, differentiation, Tnf expression and Tlr4 signaling
To assess the initial quality of our screen and scoring scheme, we first determined that, as expected, sgRNAs against ‘essential’ genes (Hart et al., 2014) were depleted in pre-LPS samples compared to Input (Figure 1C, Figure S1F and Table S1).
Next, a comparison of sgRNAs between Tnfhi and Tnflo was also consistent with our predictions, with sgRNAs targeting known positive regulators of the response (e.g., Tlr4 and Myd88) were enriched in Tnflo compared to Tnfhi, and those targeting negative regulators (e.g., Zfp36) were depleted in Tnflo (ZS analysis: Figure 1D and Table S1; DE analysis: Figure S1G and Table S1). The top-ranked genes were highly enriched for those annotated as responsive to LPS (the highest scoring category; GOrilla, FDR q-val = 10−12) or assigned to the Tlr4 to Tnf pathway (in KEGG (Kanehisa and Goto, 2000), Figure 1E, Supplemental Experimental Procedures); they were also far more likely to be expressed (Figure S1H, e.g., 78% of the top 169 genes, compared to 44% of all genes; P-value = 10−16, hypergeometric test), at higher levels (p =10−6 Kolmogorov-Smirnov (KS) test), and more likely to be differentially expressed by RNA-Seq following LPS stimulation (Experimental Procedures; Table S1).
The top 10 ranked positive regulator genes were almost exclusively populated by the hallmark members of TLR signaling, with many others among the top 100, showing that an unbiased, genome-wide screen can decipher near-complete pathways (Figure 2B; Table S1). Tnf had the top rank, demonstrating the screen’s quantitative nature. Key regulators of the LPS response with high ranks in our screen included (Figure 2B): Tlr4 (rank 10) and its co-receptors Ly96 (MD2) (rank 2) and Cd14 (rank 3); well-known members of LPS/Tlr4 signaling, including Ticam2 (TRAM, rank 5), Ticam1 (TRIF, rank 8), Myd88 (rank 4), Tirap (rank 9), and Traf6 (rank 13); Rela (rank 11), a component of NFKB, which regulates Tnf transcription; and two regulators of NFKB: Ikbkb and Ikbkg (NEMO) (rank 23 and rank 84, respectively). Other notable known regulators of the immune response and DC function include the DC pioneer transcription factor Cebpb (rank 21), Akirin2 (rank 39), and Rnf31 (rank 42) and Rbck1 (rank 19), two subunits of the linear ubiquitin chain assembly complex (LUBAC) that tags NEMO and enables NFKB activation. Overall, the top 100 ranked genes were highly enriched for central genes in the LPS to Tnf pathway as annotated by KEGG (13/35 annotated genes are in the top 100; P = 10−22, hypergeometric test).
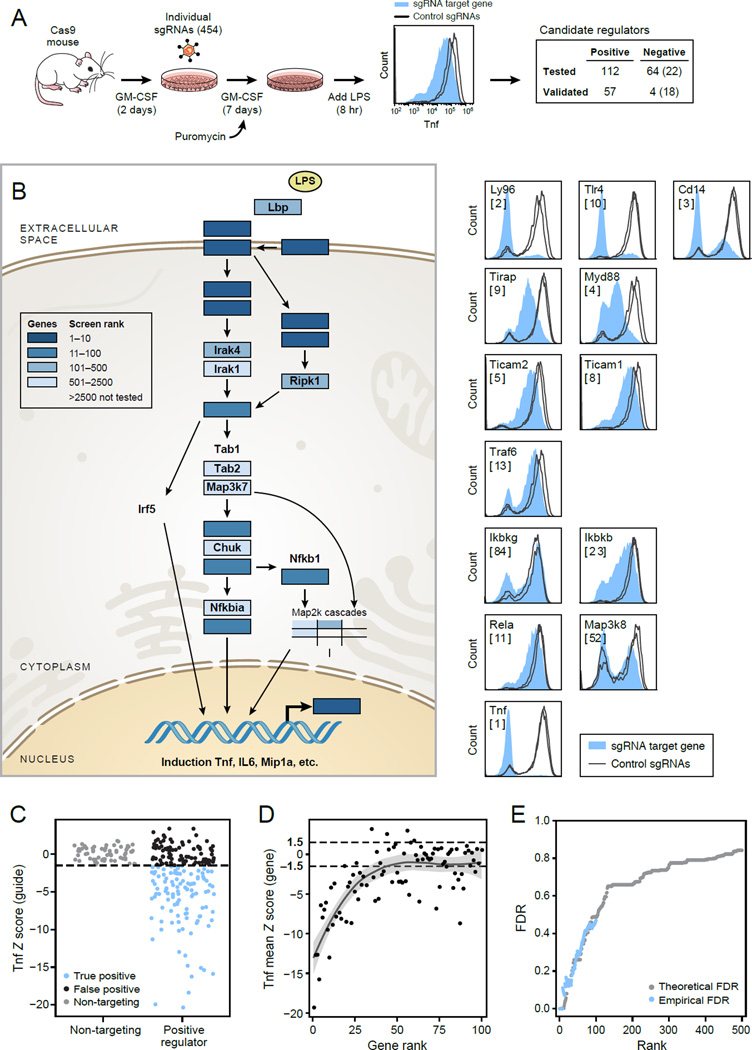
Figure 2. Individual sgRNAs verify dozens of top hits from the pooled screen.
(A) Experimental design to validate top screen hits by individual sgRNA knockouts. Tnf levels were measured by flow cytometry for each sgRNA (filled) vs. control sgRNAs (lines). Right: the numbers of positive and negative candidate regulators tested and verified using 100ng/mL or, in parentheses, 20ng/mL LPS. (B) Left: All components of the TLR pathway (KEGG) linking LPS and Tnf, and their ranks in the genome wide screen (blue scale). Right: Intracellular Tnf levels for each targeted gene (filled) compared to sgRNA controls (lines). (C) The intracellular Tnf signal (sgRNA Z-score relative to non-targeting sgRNA) of candidate positive regulators (right) and non-targeting controls (left). Blue: validated hits. (D) Mean Tnf Z score for all sgRNAs targeting the same gene at each screen rank. (E) Theoretical (grey) and empirical (blue) FDR by screen rank.
Also see Figure S2, related to Figure 2.
Dozens of positive regulators identified by the screen validated using individually-cloned sgRNAs
To validate the top genes in the ranked list, we next tested 2–3 sgRNAs against each of the top 176 (112 positive and 64 negative) ranked candidate regulators in individual assays, rather than pooled, along with 53 non-targeting controls. We measured intracellular Tnf levels by flow cytometry (Figure 2A), excluding sgRNAs with significant reduction in viability (Table S2, Experimental Procedures).
Overall, we verified 57 positive regulators out of 112 tested: 45 with at least two independent sgRNAs and another 12 genes with one sgRNA (Figure 2C, Table S1), including key known regulators (Figure 2B right). The rate of true positives regulators was in agreement with our predicted FDR (Figure 2E), and the effect size of TNF phenotype was well correlated with the original ranking (Figure 2D), supporting the accuracy of our statistical framework. Notably, 27/57 validated genes are not previously annotated for immune function or Tnf regulation (e.g., the RNA export factor, Ddx39b; Experimental Procedures and Table S1).
We explored the basis for false negatives among the positive regulators by examining 15 known regulators of LPS activation that were not among the top 100 ranked genes in the screen. Using 28 additional sgRNAs, we found that 8 of the 15 known regulators indeed reduced Tnf levels (Figure S2A and S2B; notably these 8 were better ranked (187–4417) than the remaining 7 genes (2,871–18,314) in the original screen), demonstrating that some factors outside our threshold still have functional impact in this complex response.
Optimized characterization of novel negative regulators by analysis at unsaturated levels of Tnf
Only 4 of 64 (Figure S2C, Table S1) putative negative regulators were initially validated by two independent sgRNAs, including: Zfp36 (rank 1 among the ZS negative regulators, Figure 1A), Stat5b (rank 9), Pdcd10 (CCM3, rank 32), and Ppp2r1a (rank 16). Each of these, including Zfp36, our known control, associates with human disease. Stat5b, a transcription factor activated in response to cytokine induction (Darnell, 1997), is important for DC differentiation (Sebastian et al., 2008) (consistent with a low-Cd11c phenotype in its targeted cells; Figure S2E), but was not previously implicated in regulation of Tnf. Pdcd10 (CCM3), was not previously reported to regulate Tnf, and is associated with familial Cerebral Cavernous Malformations (CCM) (Faurobert and Albiges-Rizo, 2010), a vascular pathological condition. Pdcd10 (CCM3) was also found to physically interact with Ppp2r1a, the fourth negative regulator (Goudreault et al., 2009). Interestingly, Calyculin A, a drug that inhibits the protein phosphatase1 and protein phosphatase 2A complexes, of which Ppp2r1a is a member, was previously shown to induce Tnf secretion (Boehringer et al., 1999).
The small proportion of validated negative regulators and their relatively subtle phenotype suggested that our screen, conducted with a high (100 ng/ml) LPS concentration that leads to near-saturated Tnf levels, may be less sensitive for further induction of Tnf when perturbing negative regulators. To increase sensitivity for negative regulators, we reduced LPS by 5-fold, and observed higher Tnf for 24 of 37 (65%) retested sgRNAs targeting 22 genes, including the previously unknown translation initiation factor Eif5 and the Rela-homolog DNA binding protein Dnttip1 (TdIF1) (Yamashita et al., 2001) (Figure S2D). While this test is different from the initial screen and thus cannot assess its FDR, it does provide additional functional regulators.
A deeper secondary pooled screen uncovers additional regulators with greater sensitivity and specificity
To reduce false negatives due to limited cell numbers relative to the size of the sgRNA library or to sgRNA design, we performed a secondary pooled screen targeting the top ranked 2,569 genes from the genome-wide screen with 10 sgRNAs per gene (using the improved design of (Doench et al., 2014)) and 4.9 -fold more cells per sgRNA. The secondary screen showed greater specificity and sensitivity, as reflected in enrichment of the known regulators (Figure S2F and S2G), highly correlated ranking of hits from the Z score and DE analyses (Figure S2H, Table S1) and reduced FDR compared to the genome-wide screen (e.g., FDR=6.7% for top 100 genes, Figure S2I), indicating a reduction in noise and enrichment for true positives. The hits included: Irak4 (ranked 9 in the secondary screen vs. 187 in the primary screen), Irak1 (60 vs. 992), Sharpin (another subunit of the LUBAC complex, ranked 36) and Nedd8 (ranked 52) and its E2 conjugation enzyme, Ube2f (ranked 25). In the secondary screen we found 19 positive regulators with no immune annotation that were not found in the primary screen (Z>1.5 FDR=0.094; Table S1; e.g., Gpatch8). A deeper secondary screen is thus an effective strategy for increasing the rate of true positives when the primary screen is not feasible to expand.
Positive Tnf regulators are organized in functional modules by their impact on RNA and protein expression
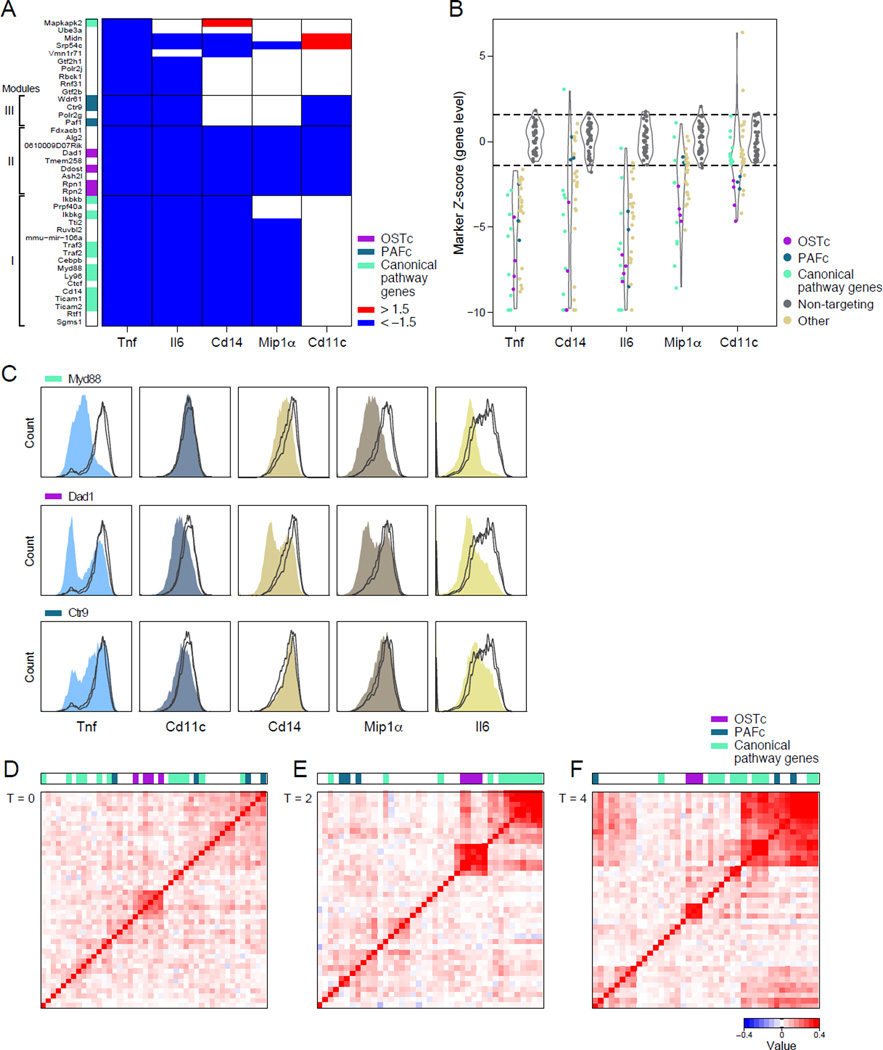
While all the validated regulators affect Tnf levels, the pathways and mechanisms through which they act may be distinct. To help determine those, we first measured the impact of the validated positive regulators on the expression of four additional protein markers (Experimental Procedures), each reflecting distinct facets of DC biology: Cd11c (the defining surface marker of BMDCs), Cd14 (a Tlr4 co-receptor), Mip1a (an induced chemokine), and Il6 (an induced inflammatory cytokine). We statistically tested the effect of each sgRNA on protein expression compared to a set of 6–8 non-targeting controls (Figure S3A, Experimental Procedures), and then grouped genes based on the similarity of their effects (Figure 3A,B, and Table S2). Notably, the Tnf distribution varied from unimodal to bimodal across different targeted genes (Figure 2B); sequencing several target genes showed that in some, but not all, cases this could be explained by the proportion of edited cells (Figure S4A–C).
Figure 3. The validated positive regulators partition into key modules by their effect on protein and RNA expression.
(A) Change in expression (blue, reduced; red, increased) of 5 protein markers (labeled columns) measured by flow staining with antibodies (Experimental Procedures) for cells with sgRNAs targeting the indicated genes (rows). Three modules indicated with brackets, and color bar on left corresponds to legend on right. (B) Violin plots of the distribution of Z scores of true positive regulators of Tnf (left) or of non-targeting control sgRNAs (right) for each marker. Functional groups are colored as in (A). (C) Effects of selected sgRNAs targeting genes in each of three modules on protein markers for true positives (filled) vs. non-targeting controls (lines). (D-F) Correlation of global RNA expression profiles (normalized to non-targeting control values) for verified positive regulators per time point post-LPS, as indicated. Color scale: Pearson correlation coefficient.
Also see Figure S3, related to Figure 3.
The genes are largely partitioned into 3 major modules (Figure 3A): Module I consisted of sgRNAs targeting 17 genes, including 9 canonical regulators validated in the screen, each reducing the levels of Cd14 and Il6, but not Cd11c (Figure 3A–C and Table S2), consistent with the roles of the known regulators in LPS signaling. Additional module members (Figure 3A) included: Ctcf, previously implicated in DC differentiation and activation (Koesters et al., 2007) and Tnf expression (Nikolic et al., 2014); and the miRNA, mmu-mir-106a, a member of the microRNA-17/20a family. Module II included nine regulators whose sgRNAs reduced all four proteins, among them: four subunits of the OST protein glycosylation complex (see below), Alg2, a glycosyltransferase involved in oligosaccharide synthesis (Haeuptle and Hennet, 2009; Huffaker and Robbins, 1983), and genes whose molecular function is currently unknown, such as Tmem258 (Figure 3A). Module III consisted of sgRNAs targeting three subunits of the PAF complex and Pol2rg; each reduced Cd11c and Il6 expression, but had a very minor, albeit consistent, effect on Mip1α (Figure 3B,C) and no effect on CD14. Some genes were not part of the 3 modules, including Midn, which is encoded in a locus associated with ulcerative colitis (based on GWAS studies (Beck et al., 2014)) but has no known molecular function.
Regulators in each of the modules may affect Tnf levels through mechanisms that are shared by members of the same module, but distinct from those of the other modules. To further assess this, we next measured with RNA-Seq the global effects of each regulator on mRNA levels at 0,2,4 and 6 hours post-LPS (without Brefeldin), compared to 12–14 non-targeting sgRNAs per time point (Figure 3D–F and Figure S3B–E, Experimental Procedures). Grouping regulators into modules based on similarity in their profiles, we found that the modules change over time – with the distinctions sharpened earlier in the response, and diminishing at later time points, as they converge through likely indirect effects. Pre-LPS (t=0h, Figure 3D), most regulators show little effect compared to non-targeting controls, except for one group consisting almost entirely of members of the OST complex, as well as Alg2 and Tmem258. At 2 and 4 hours (Figure 3E,F), the regulators partition to several modules – including the known TLR regulators, a PAF complex module, and a module associated with RNA regulators including Akirin2, Polr2g and Pabpc1. Perturbation of the genes in the latter module reduces (p-value <0.001) the expression of genes involved in immune effector processes, and regulation of immune system process (GOrrila qFDR 0.0377 and 0.0246 at t=2h Table S3). By 6 hours (Figure S3E), the transcriptional effect of most regulators is more similar. Notably, addition of Brefeldin in these profiling experiments does not affect Tnf expression, suggesting that the effect of gene perturbation in the screen vs. the profiling experiment is comparable for inflammatory gene expression (Figure S3F).
Taken together, our data suggests three key modules that impact Tnf levels in distinct ways. We next explored these, focusing on the modules of the OST and PAF complexes.
Components of the OST complex and the ER folding and translocation pathway are important for Tnf expression in response to LPS
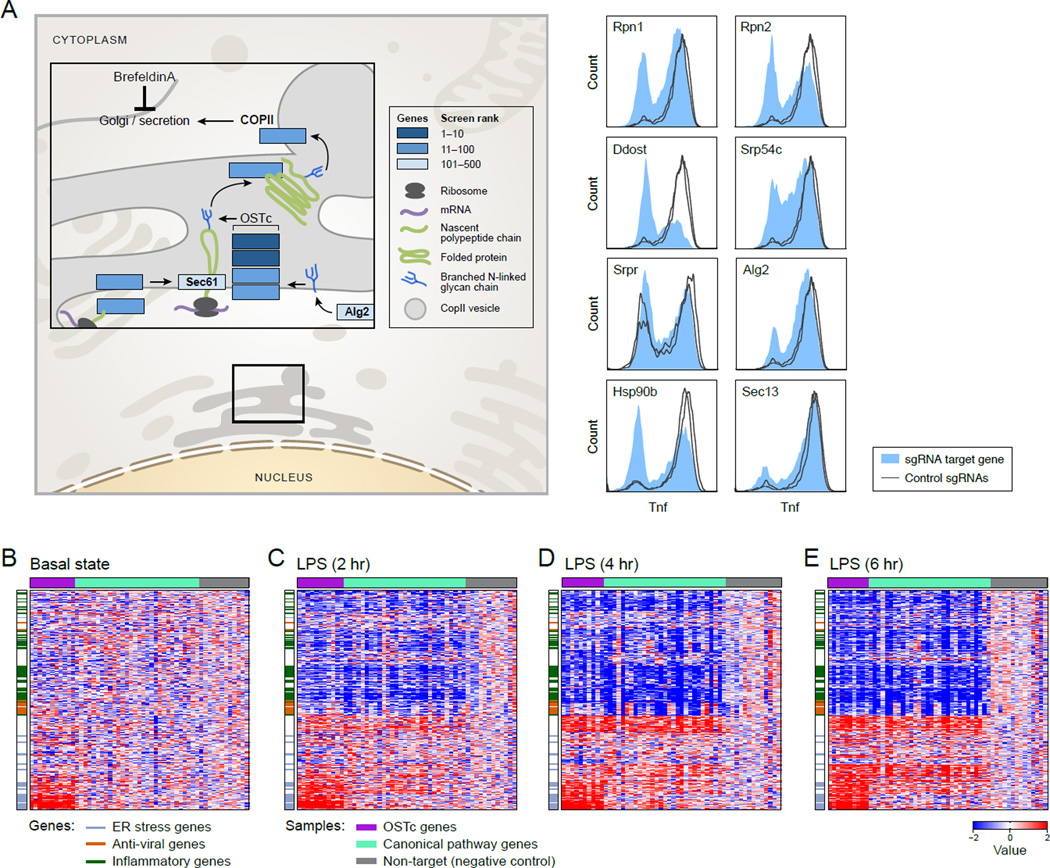
Among the 57 genes that were confirmed individually (Figure 3C and Figure 4) were 4 structural subunits of the 9-protein oligosaccharyltransferase complex (OSTc): Dad1, Ddost/OST48, Rpn1, and Rpn2. Consistent with their physical association, they were all members of the same protein- and RNA-defined modules (Figure 3A; Figure 3D–F). The ER-resident OSTc tags asparagine residues of newly translated proteins with oligosaccharide chains that are critical for protein folding and transport through the ER. At least 6 other genes essential for the ER transport pathway (Alg2, Srpr, Srp54c, Sec61, Hsp90b, Sec13, Figure 4A), upstream or downstream of OSTc, were also among the top-ranking validated positive regulators (although not necessarily in the OSTc module).
Figure 4. The OST complex strongly affects the BMDC inflammatory response.
(A) Left: positive regulators in the context of the secretory pathway; right: intracellular Tnf staining for sgRNAs against each targeted gene (filled) vs. non-targeting controls (lines). (B - E) Impact of OSTc perturbation on gene expression at indicated times post LPS. Heatmaps: row-normalized Z-scores (relative to non-targeting controls) of mRNA levels for each sgRNA-targeted sample (columns). Only mRNAs that are differentially expressed (at least one time point, adjusted p-value <0.001) are shown, in the same order in each panel.
Also see Figure S4, related to Figure 4.
Over 2,300 proteins are known to be N-glycosylated (Zielinska et al., 2010) and knocking out subunits of OSTc may affect Tnf levels directly or indirectly, and – in either case – could reflect a more global effect on N-glycosylated proteins and cell phenotype in LPS-stimulated BMDCs. Since both Ly96 and Tlr4 are N-glycosylated, and Tlr4 transport to the membrane is disrupted in the absence of tagged asparagines (da Silva Correia and Ulevitch, 2002), we hypothesized that OSTc could affect Tnf levels by impacting Tlr4 and/or its signaling. Indeed, targeting any of the four OSTc structural subunits or Alg2 (Figure 3C and Figure S4D) strongly reduced each of the four protein markers (Figure 3A), including CD11c. This general reduction is consistent with either of two hypotheses: (1) the cells are not properly differentiated; or (2) the cells have differentiated properly but their LPS sensing is compromised. In the latter case, OSTc mutants could have either (2a) a global signaling defect (e.g., due to a lack of key membrane receptors) or (2b) a more specific regulatory effect.
To distinguish between these hypotheses, we examined the specific genes whose expression is affected in OSTc-targeted cells, compared to cells targeted by known regulators from the TLR pathway, or in cells with non-targeting sgRNA controls, either before or after LPS stimulation (Figure 4B–E, Experimental Procedures). A global differentiation defect should be apparent in genome-wide expression profiles pre-LPS, a global LPS signaling defect would be apparent post-LPS, while a specific regulatory effect would be manifested as a more specific transcriptional signature.
Pre-LPS (Figure 4B), there were few transcriptional differences between cells in which OSTc is targeted or not (Table S3), except for a group of 60 OST-induced genes that are enriched for the ER stress response (FDR q-value = 5.83X10−16, GOrrila). Furthermore, 42 (p value < 10−10, Hypergeometric test) of these genes are bound by the transcription factor XBP1 at their proximal promoter in bone marrow derived macrophages (Maxim Artomov, Laurie Glimcher, and AR, unpublished results). Thus, OSTc perturbation has a limited and unique pre-LPS effect on ER stress response genes, and the reduction in CD11c is not associated with a differentiation defect. Notably, N-glycosylation and ER stress were previously shown to interact with the TLR pathway (Komura et al., 2013; Martinon et al., 2010), however direct involvement of OSTc was not shown.
The LPS response in DCs has been previously characterized (Shalek et al., 2014) by three distinct co-expression signatures: (1) anti-viral genes (‘Anti viral’), (2) inflammatory genes, including Tnf, whose expression peaks at 2h (‘Peaked Inflammatory’); and (3) inflammatory genes with sustained expression within the 6h time scale (‘Sustained Inflammatory’). While several of the mutants in the known TLR pathway genes were defective in activating all three signatures (Figure 5C–E), targeting OSTc members reduced the inflammatory signatures (sustained: _P_=0.01; peaked: _P_=0.01, T=4h _t_-test), but not the anti-viral signature (_P_=0.24 _t_-test) (Figure 4B–E and Figure 5C–E), suggesting a specific rather than global effect on the Tlr4 response.
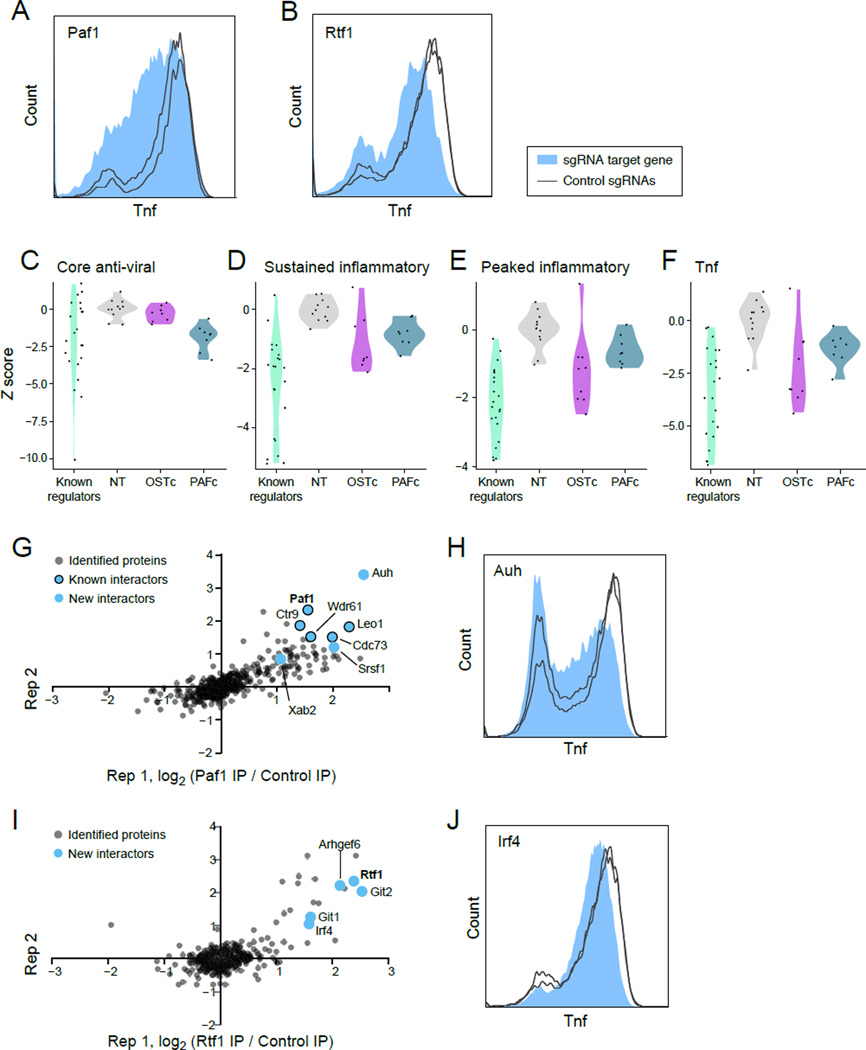
Figure 5. The Paf complex strongly affects the LPS response.
(A,B) Intracellular Tnf staining in cells with sgRNAs targeting Paf1 (A) or Rtf1 (B) (filled), compared to sgRNA controls (lines). (C-F) Violin plots of the distribution of response scores per sgRNA (calculated as an average of all RNA changes relative to non-targeting controls) in cells treated with sgRNAs targeting known regulators, non-targeting controls (NT), OSTc members, and PAFc members for each of 3 response signatures: anti-viral (C, 4h post LPS), sustained inflammatory (D, 4h post LPS), and peaked inflammatory (E, 2h post LPS), as well as Tnf transcript (F, 2h post LPS). Positive and negative values: increased and reduced response, respectively. (G, I) Scatter plots of two independent immunopurifications (IP) of Paf1 (G) or Rtf1 (I) followed by LC-MS/MS. Blue dots: interactors tested by individual sgRNA experiments for an effect on Tnf expression. Bold: IP target . (H, J) Intracellular Tnf staining in cells with sgRNAs targeting Auh (H) or Irf4 (J) (filled), compared to sgRNA controls (lines).
Also see Figure S5, related to Figure 5.
Additional regulators with the same profile as OSTc may regulate Tnf through related pathways. These include Hsp90b and Alg2, known members of the protein folding and secretion pathways (Figure 4A and Figure S4D) and Tmem258, whose human orthologue resides in a locus associated with Crohn’s disease (Franke et al., 2010) and targeted by ANRIL, a long non coding RNA associated with immune and metabolic diseases (Bochenek et al., 2013). Targeting of Tmem258 induced the same ER stress genes pre-LPS (5/14 genes; 7.7X10−5 Q-FDR GOrrila; Table S3), and similar profiles post-LPS.
The PAF complex and its physical interactors form a module that positively regulates Tnf protein expression
Five of six known subunits of the PAF complex (PAFc; Paf1, Ctr9, Wdr61, Rtf1, Leo1), a regulator of transcription elongation and 3’ mRNA processing (Jaehning, 2010), were identified as positive regulators of Tnf expression among the top 100 ranked genes in the primary screen; each was validated individually (Figure 3C, Figure 5A, 5B, and Figure S5A), did not significantly affect cell proliferation (data not shown), had a similar effect on RNA and protein expression, and associated most strongly with a single module (Figure 3, blue). The sixth subunit, Cdc73 (rank 842 in the primary screen) was likely a false negative, since two additionally designed sgRNAs did reduce Tnf expression (Figure S5B). The Ash2l subunit of the MLL complex, previously reported to physically interact with Cdc73 (Rozenblatt-Rosen et al., 2005), was also validated as a positive regulator of Tnf in our screen (rank 41, Figure S5B).
Regulation of transcription elongation was previously shown to be an important key step in the DC transcriptional response (Beaudoin and Jaffrin, 1989; Hargreaves et al., 2009). Prior studies have implicated Paf1 or PAFc in regulation of antiviral gene expression (Marazzi et al., 2012), but PAFc was not previously implicated in Tnf or inflammatory gene expression.
To decipher the specific impact of PAFc, we examined its effect on each of the transcriptional signatures. Targeting PAFc subunits significantly reduces the expression of the anti-viral and sustained inflammatory signatures, and has a weaker, albeit significant, effect on the peaked inflammatory signature, including on Tnf mRNA (Figure 5C–F).
To better understand PAFc’s function we analyzed PAFc interactors by immunopurification of Paf1 from BMDCs followed by mass spectrometry (MS) (Figure 5G and Table S4). We re-identified all known complex components, except Rtf1, and identified interactions with several RNA processing factors (Table S4), including the AU rich RNA binding and leucine metabolism protein AUH (Kurimoto et al., 2009; Nakagawa et al., 1995), an interaction confirmed by Western blot (Figure S5E). Using an individual sgRNA targeting Auh (Figure 5H), we found significant reductions in Tnf levels, whereas Srsf1 did not affect Tnf levels (Figure S5D). Since AUH binds AU-rich motifs in 3’UTRs, and the stability of the Tnf transcript is known to be regulated through an AU rich motif by three other RNA binding proteins (AUF1 (Khabar, 2010), Zfp36 (Carballo et al., 1998) and HuR (Dean et al., 2001; Tiedje et al., 2012)), it would be interesting to test if AUH also interacts with the 3’UTR of Tnf directly to regulate RNA levels.
Although Rtf1 interacts with Paf1 in lower organisms (Mueller and Jaehning, 2002), we did not observe a direct interaction between PAFc and Rtf1, when immunopurifying either Paf1 (Figure 5G) or Rtf1 (Figure 5I and Table S4). Of seven Rtf1 interactors tested (Git1, Arhgef7, Gpx1, Sf3b1, Git2, Arhgef6 and Irf4), only one, Irf4, significantly reduced Tnf expression (Figure 5J) consistent with its ranking in the secondary screen (rank 13). We also found that Irf4 affects Cd11c (Figure S5C), consistent with previous findings (Lehtonen et al., 2005; Tussiwand et al., 2012). The interaction between Irf4 and Rtf1 may suggest that PAFc, Rtf1 and other accessory proteins can perform immune specific transcriptional activation by recruiting sequence-specific transcription factors.
Discussion
We developed a genome-wide genetic screen in primary cells, based on our previous demonstration that the genomes of BMDCs from Cas9-expressing mice could be edited effectively within a relatively short time window ex vivo (Platt et al., 2014). By focusing on a quantitative cellular marker, rather than cell viability, we illustrate the versatility of pooled screens, and provide an effective approach for screening in primary cells derived from the Cas9 transgenic mouse. Our secondary pooled screen illustrates how increases in the number and efficacy of sgRNAs per gene and number of cells infected per sgRNA can substantially improve the specificity and sensitivity of a pooled screen. We thus employed a strategy that uses the results of the primary screen with a relatively permissive FDR threshold, to then guide both a large number of individual sgRNA validation experiments and a secondary screen with a much lower FDR. Using these approaches, we systematically identified previously unrecognized regulators of Tnf in response to LPS, including two conserved protein complexes and many others (e.g., Tti2, Ruvbl2, Tmem258, Midn, Ddx39b, Stat5b and Pdcd10).
To determine if the genes that affect Tnf act through different cellular pathways, we quantified how these regulators alter expression of additional protein markers and genome-wide mRNAs, and partitioned the regulators into 3 modules that are dominated by known Tlr4 pathway components, the OST complex or the PAF complex (Figure 3), thus providing clues for the functions of genes within each module. While we do not yet have a molecular model for how OSTc and PAFc impact the TLR pathway, we found that targeting subunits of the OSTc results in baseline ER stress that is likely regulated by XBP1 and may contribute to the reduction in TNF response. Our unbiased approach reveals how conserved cellular processes can have relatively specific effects on a well-defined response, offering a more comprehensive and unified view of how cellular functions are linked within a cell.
Our genome-wide, unbiased approach allowed us to uncover new modules and factors even in a heavily investigated immune pathway, and will be useful across diverse biological systems, especially when coupled with advances in single cell profiling that bridge the gap between genome-wide pooled screens and deep molecular readouts.
Experimental Procedures
Pooled genome-wide CRISPR screens
For the pooled genome-wide CRISPR screen, BMDCs were isolated from six- to eight-week old constitutive Cas9-expressing female mice and used as described previously (Platt et al. 2014). Cells were infected with the pooled lentiviral library at an MOI of 1 at day 2. At day 9, BMDCs were stimulated with 100 ng/ml LPS and after 30min Brefeldin A (GolgiPlug, BD Biosciences) was added. After 8h of LPS stimulation, cells were harvested, fixed and stained for Tnf (Ramirez-Ortiz et al., 2015) and Cd11c, and FACS sorted (Supplemental Experimental Procedures). The genome-wide screens were performed as three independent replicates; in the first screen, 60 million infected cells yielded 350 million cells at day 9, while in the second and third screens, 200 million infected cells yielded 1 billion BMDCs at day 9. The secondary pooled screen (using a reduced library) was done using the same protocol with 200 million infected cells. For individual sgRNA experiments, we used a similar protocol, except BMDCs were infected with high MOI and selected with puromycin (Invitrogen).
Cloning of individual and libraries of sgRNAs and subsequent viral production
For the primary screen, we used the GeCKOv2mouse library in the lentiGuide-Puro vector (Sanjana et al., 2014), For the secondary screen, we designed 10 sgRNAs per gene (Doench et al., 2014) to target the top 2,569 genes in the DE analysis of the primary screen, and added2,500 non targeting sgRNAs (Table S5). For library construction we used a previously published protocol (Shalem et al., 2014). For individual sgRNA cloning, pairs of oligonucleotides (IDT) with BsmBI-compatible overhangs were separately annealed and cloned into the lentiGuide-Puro plasmid (also available at Addgene, plasmid # 52963) using standard protocols. Lentivirus was made using 293T cells transfected with lentiGuide-Puro, psPAX2 (Addgene 12260) and pMD2.G (Addgene 12259) at a 10:10:1 ratio, using Lipofectamine® LTX and plus reagents according to the manufacturer’s instructions.
Amplification and sequencing of sgRNAs from cells
After sorting, DNA was purified using Qiagen DNeasy Blood & Tissue Kit according to the manufacturer’s instruction. PCR was performed as previously described (Shalem et al., 2014) and the PCR products were sequenced on a HiSeq 2500. The reads were aligned to the sgRNAs using Bowtie 1 (Langmead et al., 2009).
Analysis of screen
To score sgRNAs and genes based on their abundance in the different bins we used two strategies: in the first (DE) we normalized the raw reads and averaged on all the sgRNAs per gene and then performed differential expression analysis on three biological repeats using the R package DESeq2 (Love et al., 2014), which fits a negative binomial generalized linear model (GLM). In the second strategy (ZS), we combined all low and high bins from the three experiments, into a single pair of TNFlow and TNFhi bins and fold changes of TNFlow / TNFhi were Z-score normalized. To collapse to gene level, the mean of the top four ranked sgRNAs was taken for positive regulators, and the bottom four ranked sgRNAs for negative regulators. For the secondary screen we used all sgRNAs in both methods. All the ranks in the paper are based on ZS unless otherwise noted.
Analysis of protein and RNA expression
Day 9 differentiated and transduced BMDCs were activated with LPS for 0, 2, 4 and 6 hours for the RNA-seq experiments or for 8 hours before staining with Il-6, MIp1a, CD11c and CD14 antibody. Cells with gene specific sgRNAs were compared to those with non-targeting sgRNAs. For RNA purification we used Qiagen RNAeasy 96 Kit and constructed RNA libraries using the SMART-seq2 protocol (Picelli et al., 2013) in a 96 well plate format followed by Nextera XT DNA Sample Preparation (Illumina) and deep sequencing on a HiSeq 2500.
Protein Immunopurification
For each IP, 20 million unstimulated BMDCs were used. Each Paf1 or Rtf1 IP was always performed in parallel to a control IP and in two independent replicates. In one replicate of the experiment the digested proteins were labeled with iTRAQ and in the second replicate it was labeled with TMT10plex.
For full methods see Supplemental Experimental Procedures.
Supplementary Material
1
9
10
2
3
4
5
6
7
8
11
Highlight.
- Cytokine stain readout for genome-wide CRISPR screen in primary immune cells
- Individual validation of dozens of known and novel regulators of the LPS response
- Regulators cluster into three modules by knockout effect on expression
- General multi-pronged approach to CRISPR screens
A protein marker-based, genome-wide CRISPR screen has been developed in primary immune cells to identify genes that control the induction of tumor necrosis factor. Many of the known regulators, as well as dozens of previously unknown candidates have been identified, individually validated, classified into three functional modules.
Acknowledgements
We thank Max Artyomov and Laurie Glimcher for help and data related to Xbp1 targets, and Jonathan Weissman for very helpful discussions on ER stress. We thank the Broad’s Genomic Perturbation Platform for help in design of the secondary library and Broad Technology labs (Tarjei Mikkelssen) for help with synthesis of the secondary library. We thank Dave Gennert, Carl De Boer, Alex Shalek, John Trombetta, Schraga Schwartz, Maxwell Mumbach, Dawn Thompson,Timothy Tickle, Brian Haas, Chloe Villani and all members of the Regev, Hacohen, Carr and Zhang groups for input and discussions. We thank Terry Means for discussions and advice. We thank Leslie Gaffney for help with figure preparation and the Broad Genomics Platform for all sequencing. FACS sorting was performed at the Bauer Core Laboratory, Harvard FAS Center for Systems Biology, Cambridge, MA with great help from Patricia Rogers. This work was supported by NHGRI CEGS P50 HG006193 (A.R, N.H., S.C.) and Broad Institute Funds. A.R. was supported by the Klarman Cell Obervatory and HHMI. N.H. was supported by the MGH Research Scholars Program. M.J. was supported by fellowships of the Swiss National Science Foundation for advanced researchers (SNF) and the Marie Sklodowska-Curie IOF. F.Z. is supported by the National Institutes of Health through (NIMH: 5DP1-MH100706) and (NIDDK: 5R01-DK097768), a Waterman Award from the National Science Foundation, the Keck, New York Stem Cell, Damon Runyon, Searle Scholars, Merkin, and Vallee Foundations, and Bob Metcalfe. F.Z. is a New York Stem Cell Foundation Robertson Investigator. N.E.S. is supported by a Simons Center for the Social Brain Postdoctoral Fellowship and NIH NHGRI award K99-HG008171. O.S. is a fellow of the Klarman Cell Observatory. R.J.P. is supported by a National Science Foundation Graduate Research Fellowship under grant number 1122374‥ I.T. was supported by a Human Frontier Science Program fellowship. A.D supported by the National Space Biomedical Research Institute through NASA NCC 9–58 and the National Defense Science and Engineering Fellowship. R.H.H support from the Herchel Smith Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
O.P., M.J., T.E., N.H., and A.R. conceived and designed the study. O.P., M.J., T.E. conducted the majority of the experiments. In addition OP constructed the sgRNA libraries and performed the RNA-seq experiments, MJ performed the proteomics experiments together with P.M and S.C., and T.E led the protein staining experiments. R.H.H., A.D., and C.J.Y performed most of the large-scale data analyses with help from O.P., R.S., D.P and I.T. OS, NES and FZ provided the library, protocols and guidance in CRISPR screens. R.J.P., F.Z. generated the Cas9 transgenic mouse. R.R. generated and cultured DCs. O.P., M.J., T.E., N.H. and A.R. wrote the manuscript with input from all authors.
References
- Amit I, Regev A, Hacohen N. Strategies to discover regulatory circuits of the mammalian immune system. Nature reviews Immunology. 2011;11:873–880. doi: 10.1038/nri3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin G, Jaffrin MY. Plasma filtration in Couette flow membrane devices. Artificial organs. 1989;13:43–51. doi: 10.1111/j.1525-1594.1989.tb02831.x. [DOI] [PubMed] [Google Scholar]
- Beck T, Hastings RK, Gollapudi S, Free RC, Brookes AJ. GWAS Central: a comprehensive resource for the comparison and interrogation of genome-wide association studies. European journal of human genetics : EJHG. 2014;22:949–952. doi: 10.1038/ejhg.2013.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochenek G, Hasler R, El Mokhtari NE, Konig IR, Loos BG, Jepsen S, Rosenstiel P, Schreiber S, Schaefer AS. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Human molecular genetics. 2013;22:4516–4527. doi: 10.1093/hmg/ddt299. [DOI] [PubMed] [Google Scholar]
- Boehringer N, Hagens G, Songeon F, Isler P, Nicod LP. Differential regulation of tumor necrosing factor-alpha (TNF-alpha) and interleukin-10 (IL-10) secretion by protein kinase and phosphatase inhibitors in human alveolar macrophages. European cytokine network. 1999;10:211–218. [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- da Silva Correia J, Ulevitch RJ. MD-2 and TLR4 N-linked glycosylations are important for a functional lipopolysaccharide receptor. The Journal of biological chemistry. 2002;277:1845–1854. doi: 10.1074/jbc.M109910200. [DOI] [PubMed] [Google Scholar]
- Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Dean JL, Wait R, Mahtani KR, Sully G, Clark AR, Saklatvala J. The 3’ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Molecular and cellular biology. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nature biotechnology. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nature methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- Faurobert E, Albiges-Rizo C. Recent insights into cerebral cavernous malformations: a complex jigsaw puzzle under construction. The FEBS journal. 2010;277:1084–1096. doi: 10.1111/j.1742-4658.2009.07537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nature genetics. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudreault M, D’Ambrosio LM, Kean MJ, Mullin MJ, Larsen BG, Sanchez A, Chaudhry S, Chen GI, Sicheri F, Nesvizhskii AI, et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Molecular & cellular proteomics : MCP. 2009;8:157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeuptle MA, Hennet T. Congenital disorders of glycosylation: an update on defects affecting the biosynthesis of dolichol-linked oligosaccharides. Human mutation. 2009;30:1628–1641. doi: 10.1002/humu.21126. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Molecular systems biology. 2014;10:733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker TC, Robbins PW. Yeast mutants deficient in protein glycosylation. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:7466–7470. doi: 10.1073/pnas.80.24.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochimica et biophysica acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khabar KS. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cellular and molecular life sciences : CMLS. 2010;67:2937–2955. doi: 10.1007/s00018-010-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Shay T, O’Shea EK, Regev A. Transcriptional regulatory circuits: predicting numbers from alphabets. Science. 2009;325:429–432. doi: 10.1126/science.1171347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koesters C, Unger B, Bilic I, Schmidt U, Bluml S, Lichtenberger B, Schreiber M, Stockl J, Ellmeier W. Regulation of dendritic cell differentiation and subset distribution by the zinc finger protein CTCF. Immunology letters. 2007;109:165–174. doi: 10.1016/j.imlet.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Komura T, Sakai Y, Honda M, Takamura T, Wada T, Kaneko S. ER stress induced impaired TLR signaling and macrophage differentiation of human monocytes. Cellular immunology. 2013;282:44–52. doi: 10.1016/j.cellimm.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Kuwasako K, Sandercock AM, Unzai S, Robinson CV, Muto Y, Yokoyama S. AU-rich RNA-binding induces changes in the quaternary structure of AUH. Proteins. 2009;75:360–372. doi: 10.1002/prot.22246. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtonen A, Veckman V, Nikula T, Lahesmaa R, Kinnunen L, Matikainen S, Julkunen I. Differential expression of IFN regulatory factor 4 gene in human monocyte-derived dendritic cells and macrophages. Journal of immunology. 2005;175:6570–6579. doi: 10.4049/jimmunol.175.10.6570. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, et al. Suppression of the antiviral response by an influenza histone mimic. Nature. 2012;483:428–433. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Chen X, Lee AH, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nature immunology. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Molecular and cellular biology. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa J, Waldner H, Meyer-Monard S, Hofsteenge J, Jeno P, Moroni C. AUH, a gene encoding an AU-specific RNA binding protein with intrinsic enoyl-CoA hydratase activity. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:2051–2055. doi: 10.1073/pnas.92.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic T, Movita D, Lambers ME, Ribeiro de Almeida C, Biesta P, Kreefft K, de Bruijn MJ, Bergen I, Galjart N, Boonstra A, et al. The DNA-binding factor Ctcf critically controls gene expression in macrophages. Cellular & molecular immunology. 2014;11:58–70. doi: 10.1038/cmi.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Ortiz ZG, Prasad A, Griffith JW, Pendergraft WF, 3rd, Cowley GS, Root DE, Tai M, Luster AD, El Khoury J, Hacohen N, et al. The receptor TREML4 amplifies TLR7-mediated signaling during antiviral responses and autoimmunity. Nature immunology. 2015;16:495–504. doi: 10.1038/ni.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Molecular and cellular biology. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian C, Serra M, Yeramian A, Serrat N, Lloberas J, Celada A. Deacetylase activity is required for STAT5-dependent GM-CSF functional activity in macrophages and differentiation to dendritic cells. Journal of immunology. 2008;180:5898–5906. doi: 10.4049/jimmunol.180.9.5898. [DOI] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS genetics. 2012;8:e1002977. doi: 10.1371/journal.pgen.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Kc W, Albring JC, Satpathy AT, Rotondo JA, Edelson BT, Kretzer NM, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490:502–507. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
9
10
2
3
4
5
6
7
8
11