Distinct shifts in microbiota composition during Drosophila aging impair intestinal function and drive mortality (original) (raw)
. Author manuscript; available in PMC: 2015 Sep 10.
Summary
Alterations in the composition of the intestinal microbiota have been correlated with aging and measures of frailty in the elderly. However, the relationships between microbial dynamics, age-related changes in intestinal physiology and organismal health remain poorly understood. Here, we show that dysbiosis of the intestinal microbiota, characterized by an expansion of the Gammaproteobacteria, is tightly linked to age-onset intestinal barrier dysfunction in Drosophila. Indeed, alterations in the microbiota precede and predict the onset of intestinal barrier dysfunction in aged flies. Changes in microbial composition occurring prior to intestinal barrier dysfunction contribute to changes in excretory function and immune gene activation in the aging intestine. In addition, we show that a distinct shift in microbiota composition follows intestinal barrier dysfunction leading to systemic immune activation and organismal death. Our results indicate that alterations in microbiota dynamics could contribute to and also predict varying rates of health decline during aging in mammals.
The composition of the intestinal microbiota co-develops with the host from birth and is subject to a complex interplay involving genetics, diet and lifestyle (Nicholson et al., 2012). Dysbiosis of the microbiota has been implicated in a growing number of human disorders, including inflammatory bowel disease, obesity, cardiovascular disease and neurological disorders (Blumberg and Powrie, 2012; Carding et al., 2015; Khan et al., 2014). Aging is a major risk factor for many of these disorders and recent studies have found that the microbiota of older people is different than that of younger adults (Claesson et al., 2011) and that microbiota composition in the elderly correlates with measures of frailty, comorbidity and inflammation (Claesson et al., 2012). However, fundamental questions remain regarding the relationships between age-related changes in microbiota composition and the pathophysiology of aging.
The fruit fly Drosophila melanogaster is an excellent model to study the interplay between microbial dynamics, intestinal aging and organismal health. In aged flies, excessive proliferation of intestinal stem cells and the accumulation of mis-differentiated cells in the intestinal epithelium result in intestinal dysplasia (Biteau et al., 2008; Choi et al., 2008; Park et al., 2009), which limits organismal lifespan (Biteau et al., 2010; Hur et al., 2013; Rera et al., 2011; Wang et al., 2014). Previous studies have reported increased microbial loads in aged Drosophila populations (Broderick et al., 2014; Buchon et al., 2009; Guo et al., 2014; Ren et al., 2007) and that flies maintained axenically throughout life display reduced levels of dysplasia and other cellular markers of intestinal aging (Broderick et al., 2014; Buchon et al., 2009; Guo et al., 2014). These findings clearly demonstrate that the presence of gut-associated microbes contributes to cellular changes in the aging intestine. However, the nature of age-related alterations in microbiota composition and how microbiota composition relates to changes in intestinal function and fly health, during aging, remains largely unexplored.
A major challenge in determining how age-related changes in microbiota composition relate to the health of the host relates to the sampling of ‘aged’ individuals. In a population of chronologically age-matched animals there exists large variation in physiological health and remaining lifespan (Kirkwood et al., 2005). Hence, it can prove difficult to interpret data describing microbiota composition in a population of aged animals and/or in individual animals without knowledge of health status. Recently, we reported that loss of intestinal barrier function accompanies aging across a range of Drosophila genotypes and, critically, is a harbinger of organismal death (Rera et al., 2012). In the present work, we show that regardless of chronological age, loss of intestinal barrier function is tightly linked to dysbiosis of the microbiota, characterized by early expansion of the Gammaproteobacteria and a concomitant decrease in the proportion of taxa in the phylum Firmicutes. At the organismal level, we demonstrate that age-onset intestinal barrier failure is a prerequisite for the systemic effects of intestinal dysbiosis on immune gene activity and that a dramatic expansion of the commensal population, characterized by an increased proportion of Alphaproteobacteria, follows intestinal barrier failure and drives mortality. Our findings reveal that distinct shifts in microbial dynamics are tightly linked to distinct events in the pathophysiological decline of the aging organism and that preventing age-associated dysbiosis in animals showing intestinal barrier dysfunction can dramatically improve organismal health.
Results
Distinct shifts in microbial dynamics occur before and after age-onset intestinal barrier dysfunction
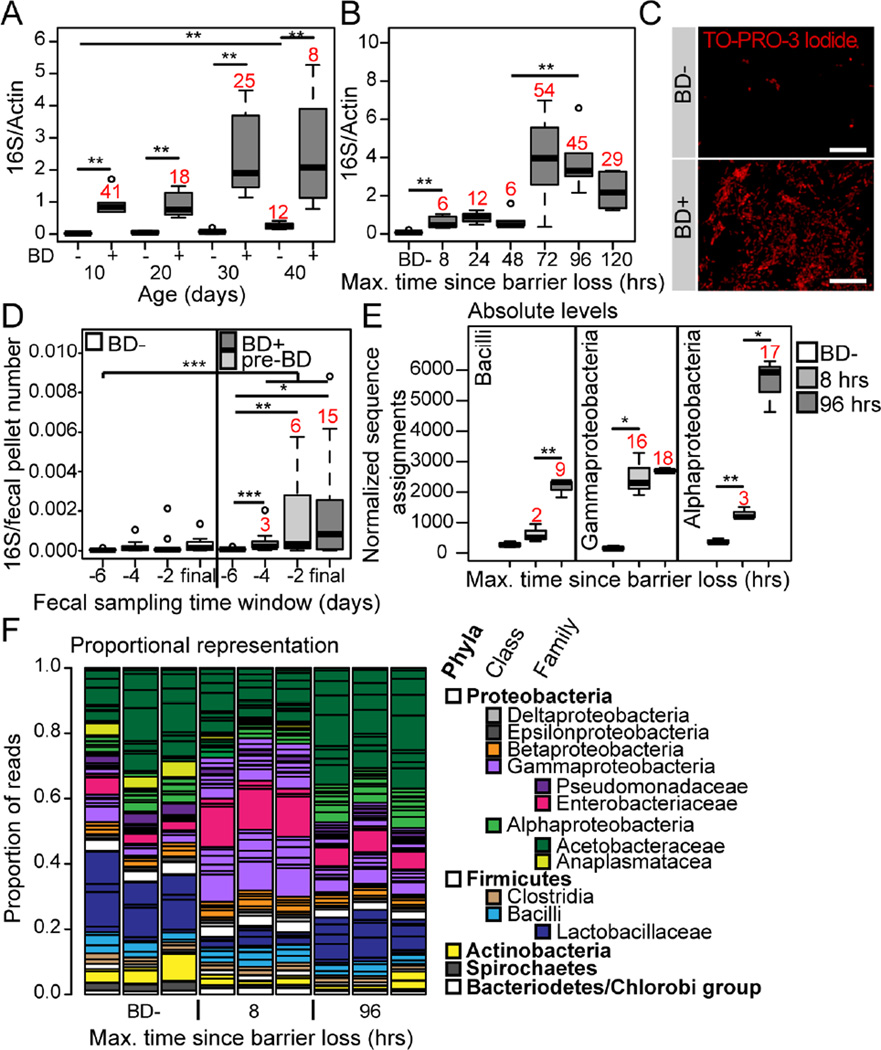
Previously, we reported that flies showing intestinal barrier dysfunction (which we refer to as Smurfs due to the presence of a non-absorbable blue dye outside of the gut post-feeding), in mid-life, had increased internal bacterial loads relative to age-matched control flies (Rera et al., 2012). Here, we utilized universal primers to the bacterial 16S rRNA gene for a qPCR approach to further characterize alterations in microbiota dynamics in relation to aging and age-onset intestinal barrier dysfunction. Across the lifespan of w1118 female flies, dissected intestinal samples from Smurfs consistently showed strikingly higher bacterial loads when compared to age-matched controls (Figure 1A). Therefore, regardless of chronological age, loss of intestinal barrier function is tightly linked to altered microbial dynamics. To assess the kinetics of microbiota changes in relation to intestinal barrier dysfunction, we utilized a large population of 30-day old w1118 female flies to identify groups of individuals that had lost barrier function within a very short 8 or 24 hour time window. An initial small, but significant, increase in bacterial load was detectable in dissected intestinal samples within 8 hours of barrier dysfunction; this was followed by a second, larger increase that occurred three days following barrier loss (Figure 1B). The extent of this later bacterial load increase is highlighted by TO-PRO-3 staining of bacterial cells in the intestinal lumen of 5 day post-Smurf flies, and age-matched non-Smurf controls (Figure 1C).
Figure 1. Alterations in microbiota composition are linked to age-onset intestinal barrier dysfunction.
(A and B) Bacterial levels assayed by qPCR of the16S ribosomal RNA gene in dissected intestines from Smurfs (BD+), and non-Smurf controls (BD-), at 10 day intervals to day 40 (A), and during a time course following barrier dysfunction at day 30 (B). n = 6 replicates of 5 dissected intestines. Numbers in red indicate fold change of the median compared to the relevant BD- control. (C) Representative images of bacteria visualized by TO-PRO-3 DNA stain in the posterior midgut lumen of BD- or 5 day post-BD+ (red channel, TO-PRO-3 DNA stain; scale bar represents 10 µm). (D) Time-series data of 16S qPCR in fecal samples of individual 40–50 day old flies, grouped as BD- or pre-BD+ depending on their phenotype during the first 24 hours of the final 48 hour sampling window. n = 12–14 individuals. Numbers in red indicate fold change of the median compared to the −6 pre-BD time point. (E) Normalized number of sequence assignments at the class taxonomic level in 8 and 96 hours post-BD+, and BD−. Only the most abundant groups are shown. n = 3 replicates of 5 dissected intestines at day 30. Numbers in red indicate fold change of the median compared to the relevant BD- control. (F) Proportional sequence assignments for BD-, 8 and 96 hours post-BD+. Each column represents a biological replicate. Horizontal black lines separate the taxonomic units, major groups are color-coded, the legend shows color assignments and taxonomic hierarchy. n = 3 replicates of 5 dissected intestines from day 30. All in w1118 females. BD- = non-Smurf, BD+ = post-Smurf. Boxplots display the first and third quartile, with the horizontal bar at the median. * p<0.05, ** p<0.01, *** p<0.001. Wilcoxon test for panels A, B and D, t-test for panel E.
40 day-old non-Smurfs showed a small, but significant, elevation in bacterial levels when compared to 10-day old non-Smurfs (Figure 1A) indicating that alterations in bacterial load occur prior to barrier failure. To confirm and extend upon this finding, we took a fecal sampling approach to follow bacterial load dynamics over a 10 day time-course in individually housed 40 day old non-Smurf female flies. Fecal samples from flies that lost barrier function toward the end of the 10 day time-course showed significantly increased bacterial loads, apparent as early as 4 days prior to a detectable loss of barrier function, when compared to controls that did not (Figure 1D). Taken together, these data demonstrate that alterations in microbiota dynamics precede and predict age-onset intestinal barrier failure, which is followed by a second larger expansion of the microbiota.
Gammaproteobacteria expansion is associated with intestinal barrier dysfunction
The microbiota of laboratory Drosophila has been well characterized (reviewed in Broderick and Lemaitre, 2012; and Erkosar et al., 2013), and temporal fluctuations in this population occur through the life stages (Wong et al., 2011), and with changes in immune function (Ryu et al., 2008). In order to assess whether changes in microbial composition, in addition to changes in bacterial load, were associated with age-onset barrier dysfunction we took a metagenomics approach. Genomic DNA extractions from dissected intestines of 30 day old w1118 female non-Smurfs, Smurfs that were within 8 hours of barrier dysfunction and Smurfs that were 96 hours following barrier dysfunction were sequenced in a shotgun approach on the Illumina platform. The resulting sequences were assigned to the NCBI taxonomy using MEGAN software (Huson et al., 2007).
Bacterial taxon assignments were largely in agreement with previous studies of the Drosophila microbiota, with members of the Lactobacillales (on average 14%) and Enterobacteriales (8%) orders and the Acetobacteriaceae (23%) family being the most abundant (Figure S1). The Drosophila endosymbiont Wolbachia was also present at low abundance in all of our samples (Figure S1). However, our analysis also identified 21 additional taxa at an abundance equal to, or higher than Wolbachia (Figure S1), each of which was represented by only 0.1–5% of the total bacterial sequences but which together made up, on average, 36% of the total bacterial sequence assignments. A large number of further taxa were represented by a very low number of read assignments, that nevertheless made up, on average, 10% of the total bacterial reads. These taxa were less consistently present across samples, but may reflect additional diversity. Finally, a low abundance of non-bacterial taxa were also identified, including fungi (Figure S1), other microbial eukaryotes, archaea and viral taxa (Figure S2A). To our knowledge, the identification of this level of diversity is unprecedented in a laboratory Drosophila stock. It is unclear whether this reflects differences in genotype or rearing conditions between laboratories, or the use of different methodologies for microbial characterization.
With the exception of the Caudovirales, an order of bacteriophages, the non-bacterial taxa identified did not increase in abundance following intestinal barrier dysfunction (Figure S2A). Given this, in addition to the significantly lower abundance of these taxa when compared to the bacterial component of the microbiota, we chose to focus on the bacterial component in the rest of this study. Most bacterial taxa were present in all samples, and increased in abundance following barrier dysfunction (Figure S1). However, the kinetics of microbial growth varied between taxa, resulting in striking differences in population composition between non-Smurf flies and the early and late time-points post-Smurf. Wolbachia levels were not increased following barrier dysfunction (Figure S1). At the class level, sequence assignments to taxa within the Gammaproteobacteria show a striking increase in animals that have lost barrier function within an 8 hour window (Figure 1E). This is in contrast to taxa within the Bacilli and Alphaproteobacteria, which show the greatest increase at 96 hours post-Smurf (Figure 1E). The proportional representation of each taxon clearly illustrates the Gammaproteobacteria expansion at the 8 hour post-Smurf time-point (Figure 1F). At the 96 hour post-Smurf time-point the proportional representation of the Gammaproteobacteria was similar to that found in the non-Smurf samples; however, the proportion of Alphaproteobacteria was increased and the phylum Firmicutes showed a concomitant decrease (Figure 1F). We also noted shifts in taxon dominance at the species level. For example, L. fructivorans was the most abundant Lactobacillus species in our non-Smurf samples, while L. brevis became dominant at 96 hours post-Smurf (Figure S1).
In order to establish how consistently these changes in the microbial population could be associated with barrier dysfunction, we generated primers to the 16S rRNA gene that would be suitable for quantitative PCR and that were specific to the classes Bacilli, Gammaproteobacteria and Alphaproteobacteria. We confirmed primer specificity by cloning and sequencing a number of 16S amplicons generated by each primer pair (Supplemental Table 1). Quantitative PCR of taxa-specific 16S from samples collected in three independent experiments conducted several months apart consistently showed significantly increased Gammaproteobacteria levels at 8 hours post-Smurf, and increased Bacilli and Alphaproteobacteria at 96 hours post-Smurf (Figure S2B). Taking these data together with the increase in total 16S levels prior to detectable barrier dysfunction, strongly suggests that Gammaproteobacteria expansion begins to occur prior to intestinal barrier failure.
Microbiota changes occurring prior to barrier failure impair excretory function in aging flies
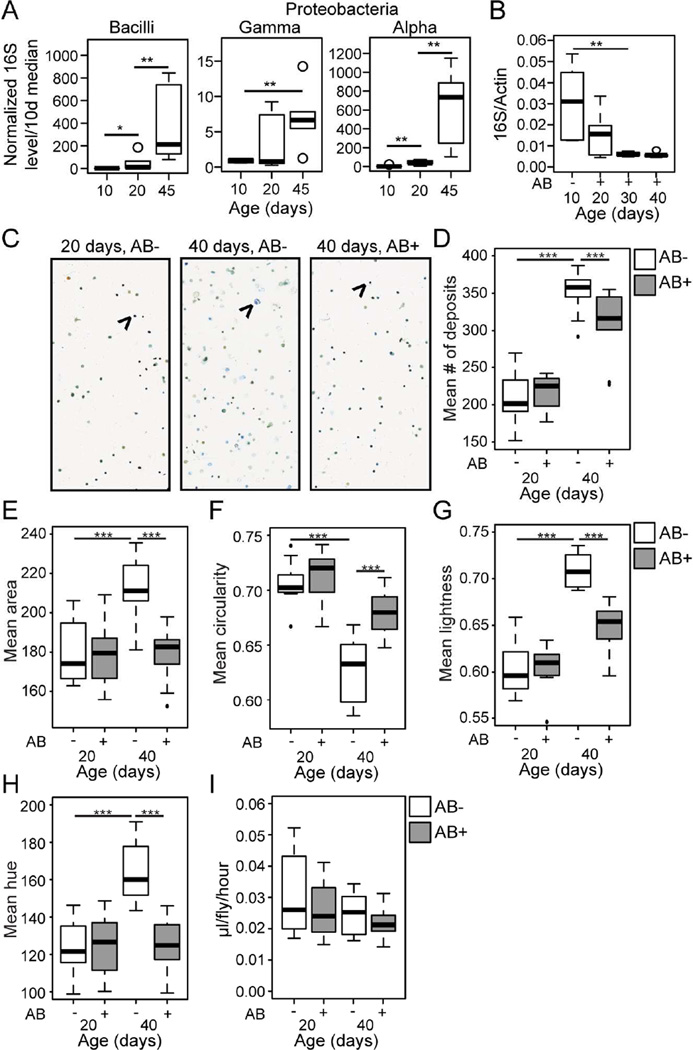
We next utilized our taxa-specific 16S qPCR primers to follow bacterial population dynamics with age in w1118 non-Smurf females. Each bacterial class showed significantly increased levels with age, apparent from midlife (Figure 2A). The Gammaproteobacteria showed a large increase in variability at midlife, suggesting that the proportional representation of these bacterial classes is modified in the microbial population at an early age, in some individuals. Expansion of the Gammaproteobacteria is, therefore, an early event in the development of age-related dysbiosis (Figure 2A). This midlife time-point coincides with when the population begins to show an increased incidence of intestinal barrier dysfunction.
Figure 2. Age-related dysbiosis impacts intestinal physiology.
(A) Bacterial levels assayed by taxon specific qPCR of the 16S ribosomal RNA gene in non-Smurfs at 10, 20 and 45 days of age. n = 6 replicates of 5 dissected intestines. (B) Bacterial levels assayed by qPCR of 16S with universal primers in antibiotic treated and untreated non-Smurfs at 10, 20, 30 and 40 days of age. n = 6 replicates of 5 dissected intestines. (C–H) Representative images (C) and analysis (D–H) of fecal output over a 48 hour period from untreated (AB-) and antibiotic treated (AB+) non-Smurfs, at 20 and 40 days of age. Shown are mean number (D), mean area (E), mean circularity (F), mean lightness (G) and mean hue (H) of deposits. Arrows indicate a representative fecal deposit. n = 10 replicate groups of 10 flies. (I) Food consumption as measured by the CAFE assay, in untreated (AB-) and antibiotic treated (AB+) non-Smurfs, at 20 and 40 days of age. n = 10 replicate groups of 10 flies. All in w1118 females. Antibiotic treatment (AB-/+) was from 10 days of age. Boxplots display the first and third quartile, with the horizontal bar at the median. * p<0.05, ** p<0.01, *** p<0.001, Wilcoxon test.
To address whether dysbiosis that occurs prior to barrier dysfunction is associated with alterations in other aspects of intestinal function, we utilized a non-invasive method to monitor a number of aspects of Drosophila intestinal and excretory physiology through analysis of the graphical features of fecal output in dye-fed flies (Cognigni et al., 2011; Wayland et al., 2014). In order to identify the impact of age-related dysbiosis on the fecal read-outs we utilized a previously published antibiotic cocktail (Brummel et al., 2004) to prevent intestinal bacterial growth from 10 days of adulthood. This antibiotic treatment was effective in preventing bacterial growth, and resulted in intestinal bacterial levels lower than that found in the 10 day old gut (Figure 2B).
Representative images of the fecal output from 20 day old untreated, 40 day untreated, and 40 day antibiotic treated non-Smurfs are shown in Figure 2C. Aged non-Smurfs showed a significant increase in the number of fecal deposits (Figure 2D), significant changes in the size and shape of deposits (Figure 2E–F), and significantly increased lightness (a measure of water content, Figure 2G) and hue (a measure of pH, Figure 2H), when compared to young controls. Antibiotic treatment from day 10 of adulthood significantly reduced, or even prevented these changes (Figure 2D–H), suggesting that age-related dysbiosis plays a significant role in age-related changes in intestinal/excretory function prior to barrier failure. Antibiotic treatment from day 10 had no significant impact on fecal output at 20 days of age (Figure 2D–H), and neither the introduction of antibiotics into the food nor aging had a detectable affect on food intake, as assayed by the CAFE assay (Figure 2I) (Ja et al., 2007).
Intestinal immune activation induces intestinal barrier dysfunction and shortens lifespan
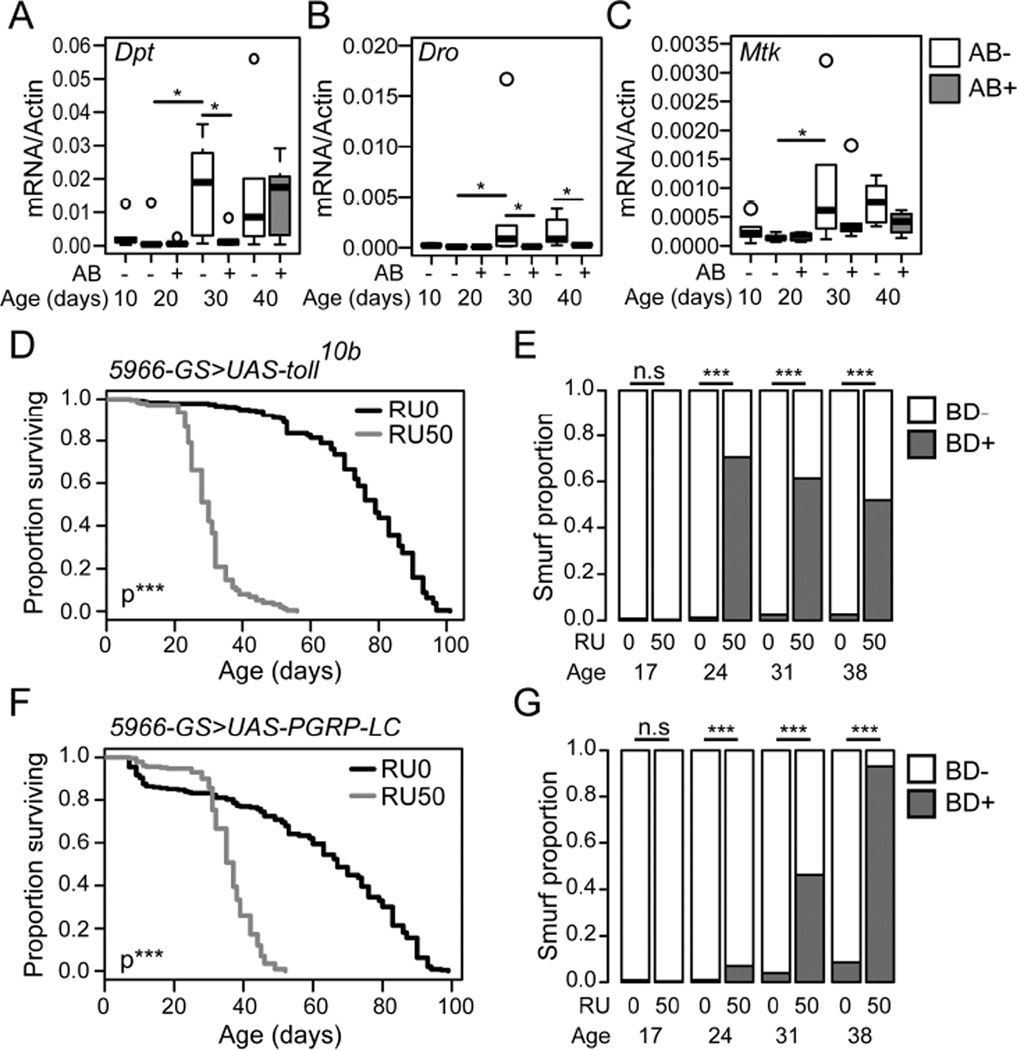
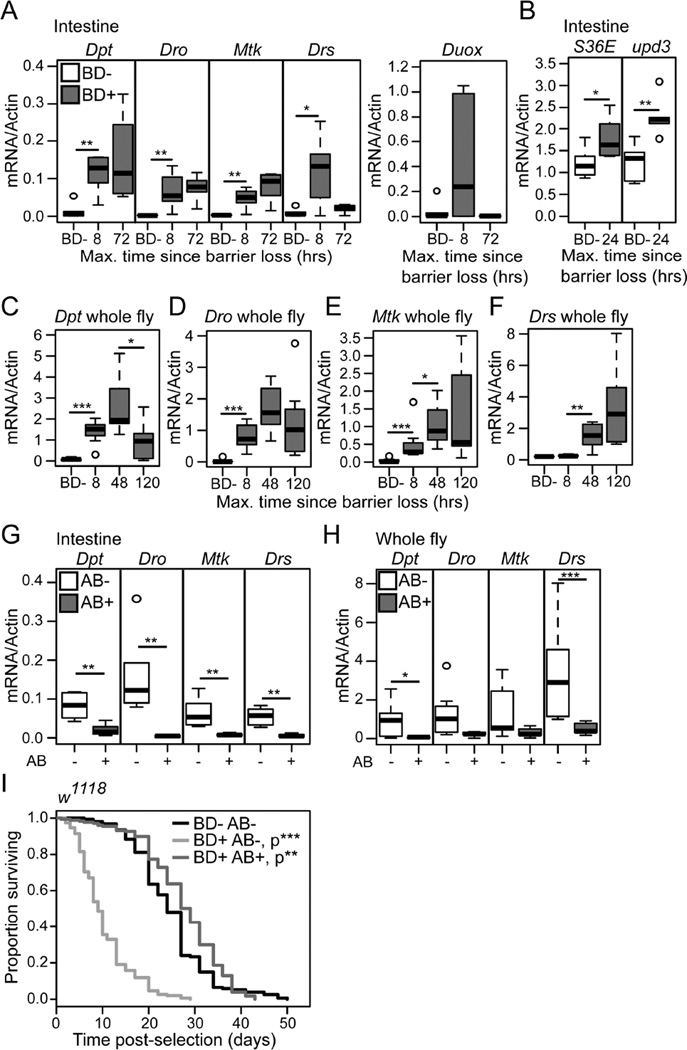
A number of studies have reported increased immune activation in the intestines of aged flies dependent upon the presence of commensal microbes (Buchon et al., 2009; Guo et al., 2014; Ren et al., 2007). Here, we set out to examine the temporal relationships between age-onset intestinal immune activation and age-onset intestinal barrier failure. Quantitative PCR from dissected intestinal samples of non-Smurf flies identified a number of antimicrobial peptides (AMPs) that showed increased expression from a mid-life time-point (Figure 3A–C). Notably, neither Drosomycin, a reporter of Toll pathway activity, or Dual Oxidase (Duox), an NADPH oxidase responsible for generating microbicidal reactive oxygen species in response to infection, showed increased expression in aged non-Smurf intestines (Figure S3A–B). In order to confirm the impact of age-related dysbiosis on intestinal immune activation, we again utilized antibiotic feeding to prevent intestinal bacterial growth from day 10 of adulthood. Quantitative PCR for AMP expression showed that antibiotic treatment from day 10 of adulthood was effective in reducing, or delaying, immune activation in the intestinal epithelium with age (Figure 3A–C).
Figure 3. Intestinal immune activation shortens life and induces intestinal barrier dysfunction.
(A–C) Antimicrobial gene expression assayed at 10 day intervals by qPCR from dissected intestines of untreated (AB-) or antibiotic treated (AB+) w1118 female non-Smurfs. Antibiotic feeding was from 10 days of age. n = 6 replicates of 5 intestines. Diptericin (Dpt) (A), Drosocin (Dro) (B), Metchnikowin (Mtk) (C). (D–G) Lifespan curves (D and F) and Smurf proportions (E and G) in _UAS-Toll10b/5966_-Geneswitch (D and E) and _UAS-PGRP-LC/5966_-Geneswitch (F and G) female flies drug induced from day 10 of adulthood (RU50), and uninduced controls (RU0). n = >200 flies/condition. BD- = non-Smurf, BD+ = Smurf. Boxplots display the first and third quartile, with the horizontal bar at the median. * p<0.05, ** p<0.01, *** p<0.001. Log Rank test for survival data in panels D and F, Binomial test for Smurf proportions in panels E and G, Wilcoxon test for other data.
Our data supports a model whereby dysbiosis leads to age-onset intestinal immune activation prior to intestinal barrier failure. Therefore, we assessed the impact of immune activation on intestinal barrier function by genetically inducing immune activation in the intestinal epithelium from day 10 of adulthood. Overexpression of either a constitutively active Toll receptor (Toll10b) or the IMD pathway receptor PGRP-LC using a Gene-Switch driver that is expressed in enteroblasts and post mitotic enterocytes (ECs) (5966GS, (Mathur et al., 2010)) resulted in increased Smurf proportions and a greatly shortened lifespan, when compared to uninduced controls (Figure 3D–G, Figure S3C–D).
Both the early-life microbiota and age-related dysbiosis influence lifespan and age-onset barrier failure
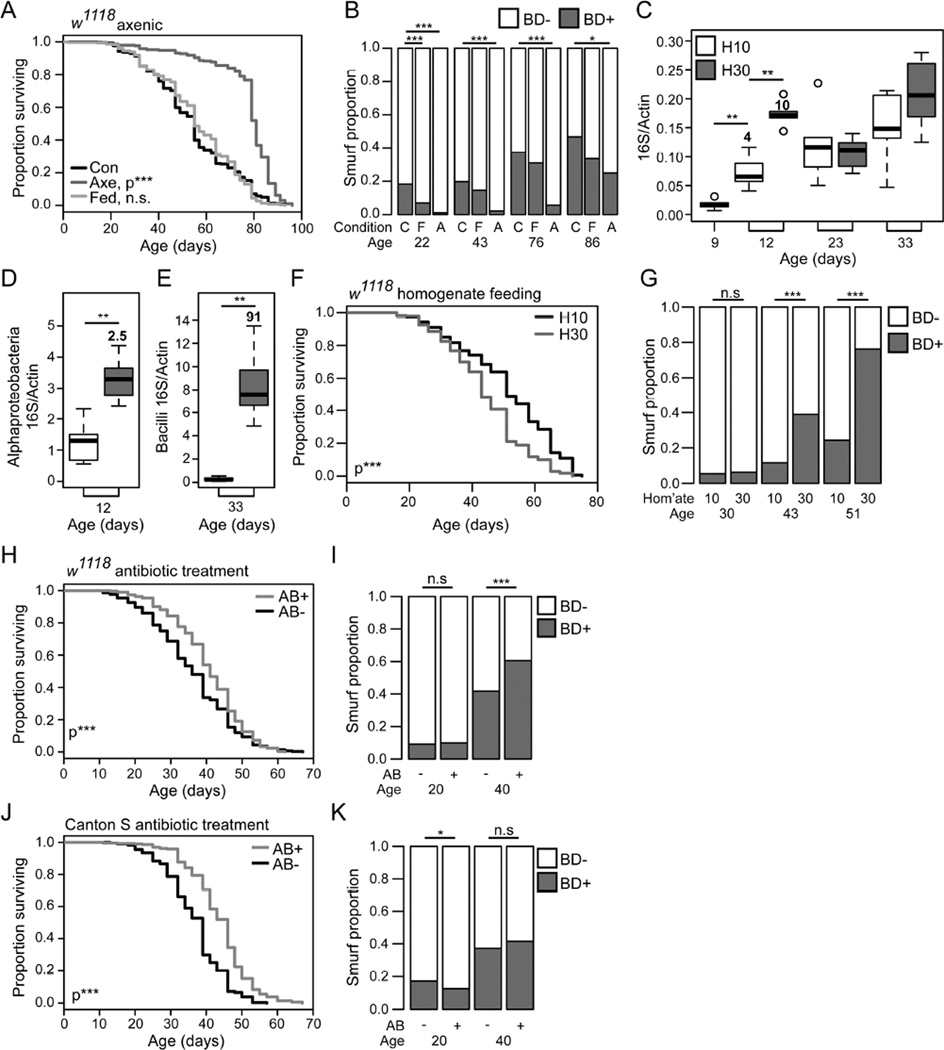
To better understand the relationships between the intestinal microbiota, age-onset intestinal barrier dysfunction and organismal aging, we generated axenic flies. Axenic w1118 females showed an increase in lifespan compared to those that were conventionally reared, and those that were initially axenic but were re-associated with microbes by introducing fly homogenate to embryos (Figure 4A). These animals also showed a very low incidence of intestinal barrier dysfunction that was maintained until the final 10 days of life (Figure 4B). Similar findings were seen in Canton S females (Figure S4A–B). Culturing methods were used to follow the bacterial loads in these animals, and confirmed their axenic status (Figure S4C–D). Consistent with a recent study using antibiotic treatment throughout life (Petkau et al., 2014), these data indicate that the presence of gut microbes influences the onset of intestinal barrier failure during aging.
Figure 4. The early life microbiota modulates lifespan and age-onset intestinal barrier function.
(A and B) Lifespan curve (A) and Smurf proportions (B) of w1118 female flies conventionally reared, axenically reared, and axenically treated and exposed to fly homogenate as embryos. n = >250 flies/condition. C - conventional, F - homogenate fed, A - axenic. P-values are compared to conventionally reared. (C–G) w1118 females fed, at the 10 day time-point, homogenate from 10 day old or 30 day old flies (C–E) Bacterial levels assayed by 16S qPCR at multiple time-points with universal primers (C), at 12 days of age with Alphaproteobacteria primers (D), at 33 days of age with Bacilli primers (E). n = 6 replicates of 5 surface sterilized whole flies. Numbers in bold above each box give the fold change in the median relative to the relevant control. (F and G) Lifespan curve (F) and Smurf proportions (G). n = >200 flies/condition. (H–I) Lifespan curve (H) and Smurf proportions (I) of antibiotic treated and untreated w1118 female flies. n = >200 flies/condition. (J and K) Lifespan curve (J) and Smurf proportions (K) of antibiotic treated Canton S female flies, and untreated controls. Antibiotic feeding was from 10 days of age. BD- = non-Smurf, BD+ = Smurf. Boxplots display the first and third quartile, with the horizontal bar at the median. * p<0.05, ** p<0.01, *** p<0.001. Log Rank test for survival data in panels A, F, H and J. Binomial test for Smurf proportions in panels B, G, I and K. Wilcoxon test for other data.
To further assess the impact of the microbiota on intestinal barrier function and lifespan, we fed homogenate from 30-day old non-Smurf w1118 females to 9-day old animals. Three days of homogenate feeding resulted in a transient increase in total 16S levels compared to controls that were fed homogenate from 10-day old animals (Figure 4C). Taxa specific 16S qPCR at this early time-point showed significantly increased Alphaproteobacteria levels (Figure 4D), a small non-significant increase in Bacilli levels, and no increase in Gammaproteobacteria (Figure S4E–F). At later time-points no significant difference was found in Alpha- or Gammaproteobacteria levels; however, flies fed the 30-day homogenate between days 9–12 showed significantly increased Bacilli levels at day 33 (Figure 4E and S4G–H). Flies that were fed aged-fly homogenate also showed significantly decreased lifespan (Figure 4F), and increased incidence in intestinal barrier dysfunction (Figure 4G), compared to flies fed homogenate from young animals.
Finally, we set out to determine whether preventing age-onset dysbiosis in adult flies can affect the onset of intestinal barrier failure. To do so, we again utilized antibiotic feeding to prevent intestinal bacterial growth from day 10 of adulthood. This antibiotic treatment significantly increased the lifespan of w1118 female flies (Figure 4H), regardless of the adult age at which treatment was started (Figure S4I). However, this antibiotic treatment did not reduce the incidence of barrier dysfunction in w1118 females (Figure 4I). Antibiotic treatment of Canton-S females during adulthood also resulted in significant lifespan extension, with the strongest affect when treatment was started from day 10 of adulthood (Figure 4J, and S4J). In Canton-S, this treatment resulted in a small but significant reduction in Smurf proportion at 20 days of age (Figure 4K). Taken together, this data indicates that while preventing intestinal bacterial growth from day 10 of adulthood can prolong lifespan in different laboratory strains, this treatment does not consistently delay the onset of intestinal barrier failure. As axenic flies show a delay in the onset of intestinal barrier dysfunction, one interpretation of our findings is that the presence of gut microbes during development and/or early adult life contributes to age-onset intestinal barrier dysfunction.
Loss of intestinal barrier function is progressive and associated with altered expression of cell junction components
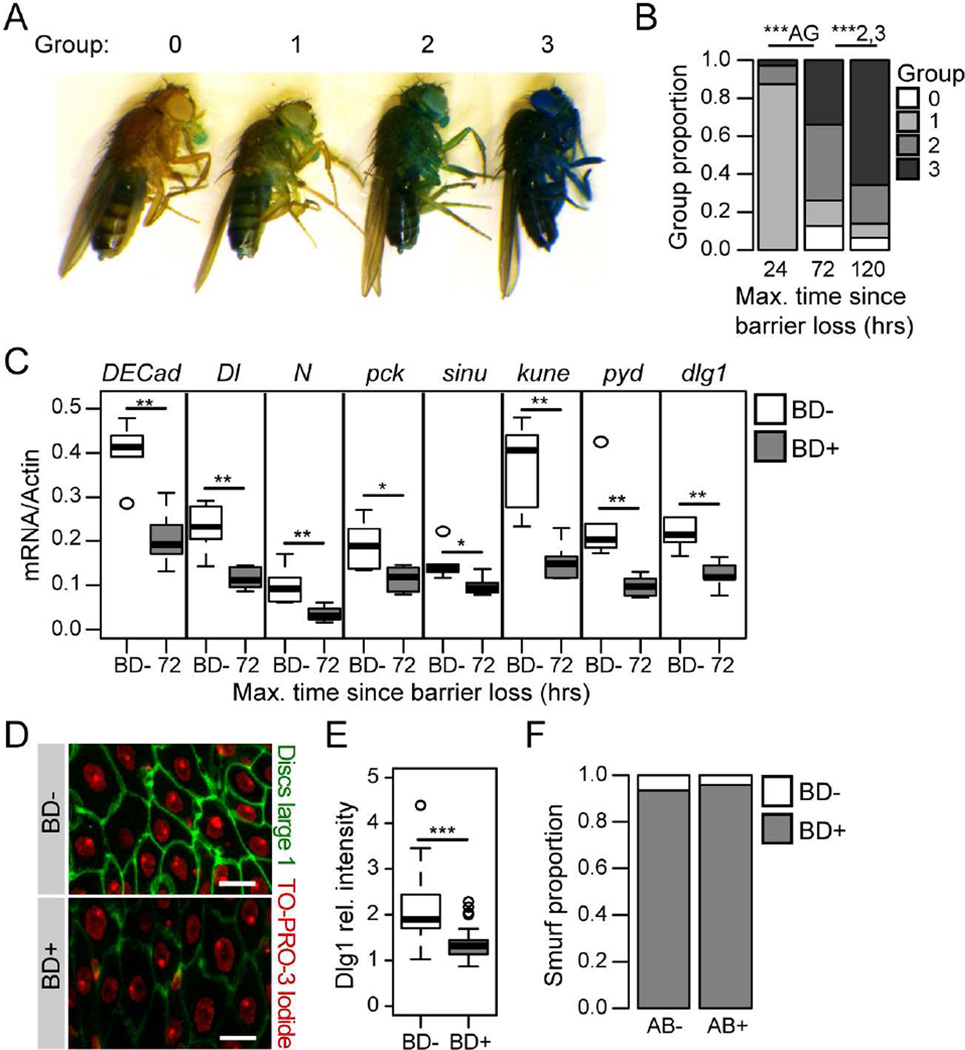
In our previous studies (Rera et al., 2011; Rera et al., 2012), we assessed the Smurf phenotype in a binary fashion; Barrier Dysfunction (BD)- or BD+. Here, we assessed the progression of intestinal barrier dysfunction in BD+ flies. In order to quantify the severity of the phenotype, we identified a number of color classes reflecting the amount of blue dye present in the hemolymph (Figure 5A). The vast majority of flies that were identified as having lost barrier function within a 24-hour time window were within the lightest color class (Figure 5B) and, when removed from the blue food, readily cleared the dye. On re-exposure to the blue dye for a second 24 hours these flies showed a significant increase in the level of blue in the hemolymph, and at 5 days post-Smurf the majority of individuals were assigned to the darkest color class (Figure 5B). This is consistent with an increase in the severity of intestinal permeability and/or a decrease in the efficiency of clearance mechanisms. Animals that were scored in the darkest color class showed high mortality in the subsequent 24 hours, indicating that this level of blue dye in the hemolymph may be toxic (Figure S5A). Similarly, newly Smurf flies that were kept on the blue food showed an accumulation of dye in the hemolymph and a greatly shortened lifespan, compared to newly Smurf flies that were removed from the blue dye (Figure S5A). This demonstrates the importance of identifying animals that have lost barrier function within a short time window to limit hemolymph levels of the dye.
Figure 5. Age-onset intestinal barrier dysfunction is progressive and associated with altered levels of cell junction components.
(A) Images of Smurf flies, representing the color classes defined to reflect different amounts of blue dye in the hemolymph. (B) The proportion of Smurf flies in each color class following 24 hours on the blue food, at 24 hours, 72 hours, and 5 days post-Smurf. n = >100 flies/time-point. P-values are given relative to the previous time point. The relevant groups are given for each p-value, AG - all groups. (C) Junction protein gene expression assayed by qPCR from dissected intestines of non-Smurfs (BD-), and 72 hours post-Smurf (BD+). n = 6 replicates of 5 intestines. DECad – Drosophila E-Cadherin, Dl – Delta, N – Notch, pck – pickle, sinu – sinuous, kune – kune-kune, pyd – polychaetoid, dlg1 – discs large 1 (D and E) Representative images (D) and quantification (E) of anti-dlg staining from the posterior midgut of BD- and 5 day post-BD+ (red channel, TO-PRO-3 DNA stain; green channel, anti-dlg; scale bar represents 10 µm). n > 20 confocal stacks from posterior midgut/condition; one fly per replicate stack. (F) Smurf proportions in antibiotic treated and untreated 5 day post-Smurf flies. n = >100 flies/condition. All in 30–35 day old w1118 female flies. antibiotic treatment from 24 hours post-Smurf. BD- = non-Smurf, BD+ = Smurf. Boxplots display the first and third quartile, with the horizontal bar at the median. * p<0.05, ** p<0.01, *** p<0.001, Binomial test for Smurf proportions, Wilcoxon test for other data.
The intestinal barrier is maintained at the level of the cell junctions, consisting of adherens junctions and septate junctions, the invertebrate analog of the tight junction. We next assessed the expression levels of genes encoding protein components of these junctions in the intestinal epithelium in flies showing barrier loss. Flies collected within 8 hours of barrier dysfunction showed a deregulation of junction protein gene expression in the form of high levels of variability in expression levels when compared to controls (Figure S5B). However, by 72 hours following barrier dysfunction cell junction transcripts showed a significant decrease in expression (Figure 5C). We confirmed this result at the protein level through antibody staining to Discs-Large 1 (dlg1) in dissected intestines of 5-day post-Smurf flies and in non-Smurf controls. Antibody staining of the posterior midgut clearly demonstrated a loss of dlg1 protein in flies showing barrier failure (Figure 5D–E). To examine the impact of post-Smurf dysbiosis on intestinal barrier dysfunction, we again utilized antibiotic feeding to prevent bacterial growth. Antibiotic treatment within 24 hours of barrier dysfunction significantly reduced bacterial loads, and prevented further bacterial growth (Figure S5C). However, at 5 days post-Smurf, flies that had received antibiotic treatment for a 4 day period still showed barrier dysfunction (Figure 5F), and antibiotic treatment had no effect on the expression levels of junction protein gene transcripts (Figure S5D). Taken together, these data suggest that a reduction in expression of cell junction proteins may contribute to the progressive worsening of intestinal barrier loss following an as yet unidentified initiating event. This transcriptional reprogramming of the intestinal epithelium may also contribute to the apparently irreversible nature of age-onset barrier dysfunction.
Dysbiosis that follows age-onset intestinal barrier dysfunction induces systemic immune activation and is a primary cause of mortality
Having established the progressive nature of the Smurf phenotype, we next assayed the activity of immune and tissue renewal pathways in post-Smurf intestinal epithelia. Immune activation, assayed by qPCR of AMP expression levels, was apparent in dissected intestines collected from 30 day-old animals within 8 hours of barrier loss. All AMPs assayed showed a significant increase in transcript levels at this time-point (Figure 6A). With the exception of Drosomycin (Drs), an AMP regulated primarily by the Toll pathway, AMP levels were maintained at 72 hours post-Smurf (Figure 6A). Dual Oxidase also showed a highly variable peak in expression at 8 hours post-Smurf (Figure 6A). Increased levels of reactive oxygen species in the gut have been linked to the induction of epithelial renewal in response to infection and the commensal microbiota, via activation of the JAK-STAT pathway (Buchon et al., 2009). We therefore assayed expression levels of the JAK-STAT cytokine unpaired 3 (upd3) and target gene Socs36E, in post-Smurf intestines. Both upd3 and Socs36E showed a transient peak in expression 24 hours following the loss of barrier function (Figure 6B). These data demonstrate a transient induction of epithelial renewal pathways and extended activation of immune signals in the post-Smurf intestinal epithelium.
Figure 6. Preventing dysbiosis post-intestinal barrier dysfunction restores lifespan.
(A and B) Gene expression assayed by qPCR from dissected intestines of non-Smurfs (BD-) and at multiple time-points post-Smurf (BD+). n = 6 replicates of 5 intestines. (C–F) Gene expression assayed by qPCR from whole BD-, 8, 48 and 120 hours post-BD+ flies. n = 6 replicates of 3 whole flies. (G and H) Antimicrobial gene expression assayed by qPCR from dissected intestines (G) or whole fly (H) of antibiotic treated and untreated 5 day post-BD+ flies. n = 6 replicates of 5 intestines, or 3 whole flies. (I) Survival curves of antibiotic treated and untreated 24 hour post-BD+ flies, and age-matched untreated BD- controls. n = >200 flies/condition. P values are compared to BD- controls. Dpt -Diptericin, Dro - Drosocin, Mtk - Metchnikowen, Drs - Drosomycin, Duox – Dual Oxidase. upd3 – unpaired 3, Socs36E – Suppressor of cytokine signaling at 36E. All flies were scored BD- or BD+ between 30–35 days old. All in w1118 female flies. Antibiotic treatment from 24 hours post-BD+. BD- = non-Smurf, BD+ = Smurf. Boxplots display the first and third quartile, with the horizontal bar at the median. * p<0.05, ** p<0.01, *** p<0.001, Log Rank test for survival data in panel I, Wilcoxon test for other data.
Previously, we reported that non-Smurf individuals do not show whole body increases in immunity gene expression during aging (Rera et al., 2012), suggesting that age-related systemic immune activation is dependent upon barrier dysfunction. In order to clarify the relationship between intestinal barrier loss and systemic immune activation we assessed AMP expression levels in whole fly samples during a time-course following barrier loss. Significantly increased systemic levels of AMP expression were apparent within 8 hours of barrier failure in all AMPs assayed except Drs (Figure 6C–F). Drs expression was significantly increased from 48 hours following barrier dysfunction (Figure 6F), a time-point that correlates with the initiation of substantial post-Smurf bacterial growth. In order to assess the impact of post-Smurf immune activation on lifespan, we genetically induced immune activation in the intestine and fatbody of adult female flies. Overexpression of either a constitutively active Toll receptor (Toll10b) or the IMD pathway receptor PGRP-LC with the drug inducible driver S106-Geneswitch, which is expressed in gut/fat body (Poirier et al., 2008), resulted in a dose-dependent decrease in lifespan (Figure S6A–C).
Following this result, we again utilized antibiotic feeding, in 30 day old animals showing barrier dysfunction, to prevent post-Smurf dysbiosis. Flies treated with antibiotics within 24 hours of barrier loss showed significantly decreased AMP expression levels both in the intestinal epithelium and systemically (Figure 6G–H), demonstrating that immune activation in the gut and in the fatbody is microbe-dependent. To better understand the systemic consequences of post-barrier dysfunction dysbiosis, we examined lifespan in antibiotic treated and untreated animals showing intestinal barrier dysfunction. Importantly, antibiotic treatment within 24 hours of barrier dysfunction resulted in a striking increase in Smurf lifespan, essentially restoring lifespan to that of non-Smurf animals (Figure 6I). Transient antibiotic treatment between 24 and 72 hours post-Smurf was also sufficient to significantly extend Smurf lifespan (Figure S6D). Taken together, these data demonstrate that post-Smurf dysbiosis is a primary factor in the death of animals that have lost barrier function. Our data also demonstrate that intestinal barrier dysfunction alone does not cause death.
Discussion
There is an emerging understanding of the importance of the gut microbiota in host longevity (Heintz and Mair, 2014), yet fundamental questions about microbiota dynamics and the pathophysiology of aging remain unanswered. In the present study, we have used the fruit fly, Drosophila, as a model to study the relationships between microbiota dynamics, the physiological decline of the aging intestine and the ultimate manifestation of the aging process—the death of the animal. To do so, we used a number of non-invasive approaches to assay intestinal function and microbiota dynamics during aging. Studying the physiological decline of individual flies, as opposed to a population of chronologically age-matched animals of variable and unknown health, has allowed us to identify distinct shifts in microbiota composition that are linked to specific pathophysiological events in the aging host organism. Previously, we demonstrated that, regardless of chronological age, intestinal barrier dysfunction is a harbinger of age-onset mortality (Rera et al., 2012). Here, we show that alterations in the microbiota precede and predict the onset of intestinal barrier dysfunction in aged flies and contribute to age-related changes in excretory function. Furthermore, a distinct shift in microbiota composition follows intestinal barrier failure and is a primary cause of organismal death. Taken together, our findings show that microbiota composition is predictive of varying rates of health decline in aging flies and that age-onset dysbiosis promotes the functional decline of the aging intestine and limits lifespan.
Our metagenomics analysis identified distinct shifts in microbial community structure that are in broad agreement with the microbial changes associated with both intestinal inflammatory disorders and aging in the human gastrointestinal tract. Increased proportions of Proteobacteria versus Firmicutes are a signature of both inflammatory bowel disease and human aging (Cheng et al., 2013; Clemente et al., 2012). Increased Proteobacteria levels have also been correlated with inflammation in seniors (Biagi et al., 2010), and increased Enterobacteriaceae levels have been correlated with frailty in the elderly (van Tongeren et al., 2005) and with inflammation in a genetic mouse model of colitis (Carvalho et al., 2012). The family Enterobacteriaceae is the most abundant member of the Gammaproteobacteria represented in our samples. At the same time, intestinal barrier dysfunction is tightly linked to health status and is a common feature of inflammatory and metabolic disorders in mammals (Marchiando et al., 2010). The epithelial barrier is an essential component of the immune system and is essential for immune tolerance (Turner, 2009). Translocation of commensal bacteria, or bacterial products, across the intestinal epithelium has been linked to sepsis following chronic or severe illness (Deitch, 2012; Dillon et al., 2014; Estes et al., 2010), and to the development of systemic metabolic disorders (Amar et al., 2011; Raybould, 2012) These similarities suggest that a set of fundamental principles may underlie intestinal functional decline from Drosophila to humans. In this regard, a pressing challenge will be to determine whether distinct shifts in microbiota composition, linked to age-onset intestinal barrier dysfunction, precede age-related health decline in other organisms. Should this prove to be the case then microbiota sampling would provide an accessible way to monitor health status and predict future health outcomes in aging mammals.
Our work adds to previous Drosophila studies suggesting that increases in microbial load may be an important feature of age-onset dysbiosis (Broderick et al., 2014; Buchon et al., 2009; Guo et al., 2014; Ren et al., 2007). A breakdown of immune regulation in the aged epithelium (Guo et al., 2014), an expansion in the niche available for bacterial growth, or changes in the nutrient environment that supports bacterial growth may all contribute to increased bacterial levels. In any case, we show that the excessive bacterial growth and/or the concomitant immune activation following intestinal barrier failure promote organismal death. Previous reports of the impact of axenic culture on fly lifespan have been inconsistent; indeed, axenic flies have been reported to display shortened lifespan (Brummel et al., 2004), increased lifespan (Petkau et al., 2014) and normal lifespan (Ren et al., 2007). We find that germ-free flies are long-lived, consistent with studies reporting that axenic flies show improved markers of intestinal homeostasis during aging (Broderick et al., 2014; Buchon et al., 2009; Guo et al., 2014) and the growing body of evidence indicating that intestinal homeostasis is a critical determinant of fly lifespan (Rera et al., 2013; Wang et al., 2014). Inconsistent reports of the impact of bacterial exposure on fly lifespan extend to the impact of single bacterial species, with the Drosophila endosymbiont Wolbachia having been reported to have a positive (Aleksandrov et al., 2007; Ikeya et al., 2009), negative (Min and Benzer, 1997), or neutral impact on lifespan (Ikeya et al., 2009), dependent on the strain of Wolbachia and the genotype of the host. While the w111 8 female flies used in this study contain Wolbachia, given the low representation of Wolbachia sequences in our metagenomic data and the absence of an increase in Wolbachia abundance following intestinal barrier dysfunction, it is unlikely that the presence of Wolbachia alone can explain our results. However, these issues highlight the complexity of the host-microbe interaction and its impact on host lifespan. Beneficial contributions of microbes to fly health seem to be highly dependent on nutrient conditions—we recently demonstrated that gut-associated microbes promote amino acid harvest to rescue lifespan during undernutrition (Yamada et al., 2015). Hence, it is possible that the presence of gut-associated microbes can be beneficial or deleterious to host lifespan depending on the nutrient environment. Another intriguing possibility is that different laboratory strains display different alterations in microbiota dynamics during aging and, hence, respond differently to axenic culture.
In this study we have been able to follow the dynamics of the microbiota population throughout the lifespan and with the changing health status of the aging host. The interactions between the microbial species that make up the commensal population, and between the microbial population as a whole and the host organism, result in a highly complex network of interrelated signals. Therefore, establishing the cause and effect relationships between microbial dynamics and a given phenotype is a significant challenge for this research field. While a single study cannot hope to address this complexity, our ability to establish the timing of changes in the microbial population relative to changes in intestinal physiology and health status represents a significant step in that direction. As Drosophila is one of the most important model organisms in aging research, our findings will facilitate a better understanding of the mechanisms by which life-extending manipulations impact pathophysiology and/or microbiota dynamics to improve organismal survival. In addition, our work opens the door to using Drosophila to identify anti-aging interventions that can be differentially applied depending upon the health status of the aged animal.
Experimental procedures
Timed Smurf assays
The Smurf assay was conducted as previously described (Rera et al., 2012) on 30 day old w1118 female flies. Smurf animals were then removed under light nitrogen anesthesia to generate a large population of non-Smurf individuals. New Smurfs were collected within 8 or 24 hours of the prior population clearance and removed from the blue dye. The same number of non-Smurf controls was simultaneously collected from each vial. For later post-Smurf time-points Smurfs collected within the 24 hour window were allowed to age under standard culture conditions prior to sample collection at the specified time-point. The reported affects of husbandry practices on Drosophila bacterial loads (Blum et al., 2013) were controlled for by keeping bacterial sample collection consistent relative to new food transfer and by the collection of non-Smurf controls from the same vials as Smurf individuals.
Sequencing and Analysis
Indexed paired end libraries were generated from genomic DNA extractions of dissected intestines using Nextera XT DNA sample preparation kit and Nextera XT Index kit (Illumina), with 1 ng of starting material and following the manufacturers’ protocol, but with an extended fragmentation time of 8 minutes. Concentration normalized libraries were then pooled and sequenced on Illumina HiSeq2000 with TruSeq Dual Index Sequencing Primer Kit, Single Read (Illumina). The quality of the sequence dataset was confirmed using fastQC software (Babraham bioinformatics). Over 10,000,000 reads from each sample were then searched against a 2014 version of the NCBI non-redundant database (nr) using RAPSearch version 2.18(Zhao et al., 2012), limiting output to 50 matches per query. RAPSearch results were imported to MEGAN5 (Huson et al., 2011) for taxonomic analysis. Further details for all experimental procedures are available in the supplemental materials.
Supplementary Material
1
2
Acknowledgements
We thank H. Jasper, L. Jones, L. Seroude, M. Dionne and the Drosophila Stock Center (Bloomington) for fly stocks, and the L. Jones, M. Frye and D. Simmons labs for use of their equipment. This work was supported by the UCLA Institute of Genomics and Proteomics (M.P. and S.F-G.) and the National Institutes of Health (R01GM095656, M.M.; R24GM092473, S.F-G.; R00AG030493 and R01AG045036, W.W.J.; R01AG037514, R01AG049157 and R01AG040288, D.W.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
Sequencing data have been deposited in the NCBI Sequence Read Archive and are available under the accession number SRP061446.
References
- Aleksandrov ID, Aleksandrova MV, Goriacheva II, Roshchina NV, Shaǐkevich EV, Zakharov IA. Removing endosymbiotic Wolbachia specifically decreases lifespan of females and competitiveness in a laboratory strain of Drosophila melanogaster. Genetika. 2007;43:1372–1378. [PubMed] [Google Scholar]
- Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermúdez-Humarán LG, Smirnova N, Bergé M, Sulpice T, Lahtinen S, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol. Med. 2011;3:559–572. doi: 10.1002/emmm.201100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkïla J, Monti D, Satokari R, Franceschi C, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 2010;6:e1001159. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum JE, Fischer CN, Miles J, Handelsman J. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio. 2013;4:e00860–e00813. doi: 10.1128/mBio.00860-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci. Transl. Med. 2012;4:137rv137. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in drosophila melanogaster host gene expression and gut morphology. mBio. 2014;5:e01117–e01114. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick NA, Lemaitre B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes. 2012;3:307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummel T, Ching A, Seroude L, Simon AF, Benzer S. Drosophila lifespan enhancement by exogenous bacteria. Proc. Natl. Acad. Sci. USA. 2004;101:12974–12979. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015;26:26191. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, González A, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Palva AM, de Vos WM, Satokari R. Contribution of the intestinal microbiota to human health: from birth to 100 years of age. Curr. Top. Microbiol. Immunol. 2013;358:323–346. doi: 10.1007/82_2011_189. [DOI] [PubMed] [Google Scholar]
- Choi NH, Kim JG, Yang DJ, Kim YS, Yoo MA. Age-related changes in Drosophila midgut are associated with PVF2, a PDGF/VEGF-like growth factor. Aging Cell. 2008;7:318–334. doi: 10.1111/j.1474-9726.2008.00380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA. 2011;1(108 Suppl):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O'Sullivan O, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognigni P, Bailey AP, Miguel-Aliaga I. Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metab. 2011;13:92–104. doi: 10.1016/j.cmet.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitch EA. Gut-origin sepsis: evolution of a concept. Surgeon. 2012;10:350–356. doi: 10.1016/j.surge.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, Gianella S, Siewe B, Smith DM, Landay AL, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkosar B, Storelli G, Defaye A, Leulier F. Host-intestinal microbiota mutualism: "learning on the fly". Cell Host Microbe. 2013;13:8–14. doi: 10.1016/j.chom.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Estes JD, Harris LD, Klatt NR, Tabb B, Pittaluga S, Paiardini M, Barclay GR, Smedley J, Pung R, Oliveira KM, et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 2010;6:e1001052. doi: 10.1371/journal.ppat.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz C, Mair W. You are what you host: microbiome modulation of the aging process. Cell. 2014;156:408–411. doi: 10.1016/j.cell.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JH, Bahadorani S, Graniel J, Koehler CL, Ulgherait M, Rera M, Jones DL, Walker DW. Increased longevity mediated by yeast NDI1 expression in Drosophila intestinal stem and progenitor cells. Aging. 2013;5:662–681. doi: 10.18632/aging.100595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya T, Broughton S, Alic N, Grandison R, Partridge L. The endosymbiont Wolbachia increases insulin/IGF-like signalling in Drosophila. Proc. R. Soc. B. 2009;276:3799–3807. doi: 10.1098/rspb.2009.0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. Prandiology of Drosophila and the CAFE assay. Proc. Natl. Acad. Sci. USA. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MT, Nieuwdorp M, Bäckhed F. Microbial modulation of insulin sensitivity. Cell Metab. 2014;20:753–760. doi: 10.1016/j.cmet.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB, Feder M, Finch CE, Franceschi C, Globerson A, Klingenberg CP, LaMarco K, Omholt S, Westendorp RG. What accounts for the wide variation in life span of genetically identical organisms reared in a constant environment? Mech. Ageing Dev. 2005;126:439–443. doi: 10.1016/j.mad.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Marchiando AM, Graham WV, Turner JR. Epithelial barriers in homeostasis and disease. Annu. Rev. Pathol. 2010;5:119–144. doi: 10.1146/annurev.pathol.4.110807.092135. [DOI] [PubMed] [Google Scholar]
- Mathur D, Bost A, Driver I, Ohlstein B. A transient niche regulates the specification of Drosophila intestinal stem cells. Science. 2010;327:210–213. doi: 10.1126/science.1181958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min KT, Benzer S. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc. Natl. Acad. Sci. USA. 1997;94:10792–10796. doi: 10.1073/pnas.94.20.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Park JS, Kim YS, Yoo MA. The role of p38b MAPK in age-related modulation of intestinal stem cell proliferation and differentiation in Drosophila. Aging. 2009;1:637–651. doi: 10.18632/aging.100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkau K, Parsons BD, Duggal A, Foley E. A deregulated intestinal cell cycle program disrupts tissue homeostasis without affecting longevity in Drosophila. J. Biol. Chem. 2014;289:28719–28729. doi: 10.1074/jbc.M114.578708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier L, Shane A, Zheng J, Seroude L. Characterization of the Drosophila gene-switch system in aging studies: a cautionary tale. Aging Cell. 2008;7:758–770. doi: 10.1111/j.1474-9726.2008.00421.x. [DOI] [PubMed] [Google Scholar]
- Raybould HE. Gut microbiota, epithelial function and derangements in obesity. J. Physiol. 2012;590:441–446. doi: 10.1113/jphysiol.2011.222133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Rera M, Azizi MJ, Walker DW. Organ-specific mediation of lifespan extension: More than a gut feeling? Ageing Res. Rev. 2013;12:436–444. doi: 10.1016/j.arr.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Clark RI, Walker DW. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc. Natl. Acad. Sci. USA. 2012;109:21528–21533. doi: 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- Turner JR. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- van Tongeren SP, Slaets JP, Harmsen HJ, Welling GW. Fecal microbiota composition and frailty. Appl. Environ. Microbiol. 2005;71:6438–6442. doi: 10.1128/AEM.71.10.6438-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Karpac J, Jasper H. Promoting longevity by maintaining metabolic and proliferative homeostasis. J. Exp. Biol. 2014;217:109–118. doi: 10.1242/jeb.089920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayland MT, Defaye A, Rocha J, Jayaram SA, Royet J, Miguel-Aliaga I, Leulier F, Cognigni P. Spotting the differences: Probing host/microbiota interactions with a dedicated software tool for the analysis of faecal outputs in Drosophila. J. Insect Physiol. 2014;69:126–135. doi: 10.1016/j.jinsphys.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CN, Ng P, Douglas AE. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 2011;13:1889–1900. doi: 10.1111/j.1462-2920.2011.02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, Deshpande SA, Bruce KD, Mak EM, Ja WW. Microbes promote amino acid harvest to rescue undernutrition in Drosophila. Cell Rep. 2015;10:865–872. doi: 10.1016/j.celrep.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tang H, Ye Y. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics. 2012;28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2