Differential Toxicity of Nuclear RNA Foci Versus Dipeptide Repeat Proteins in a Drosophila Model of C9ORF72 FTD/ALS (original) (raw)
. Author manuscript; available in PMC: 2016 Sep 23.
SUMMARY
Dipeptide repeat (DPR) proteins are toxic in various models of FTD/ALS with GGGGCC (G4C2) repeat expansion. However, it is unclear whether nuclear G4C2 RNA foci also induce neurotoxicity. Here, we describe a novel Drosophila model expressing 160 G4C2 repeats (160R) flanked by human intronic and exonic sequences. Spliced intronic 160R formed nuclear G4C2 sense RNA foci in glia and neurons about 10 times more abundantly than in human neurons; however, they had little effect on global RNA processing and neuronal survival. In contrast, highly toxic 36R in the context of poly(A)+ mRNA were exported to the cytoplasm where DPR proteins were produced at >100-fold higher level than that in 160R flies. Moreover, the modest toxicity of intronic 160R expressed at higher temperature correlated with increased DPR production but not RNA foci. Thus, nuclear RNA foci are neutral intermediates or possibly neuroprotective through preventing G4C2 RNA export and subsequent DPR production.
Keywords: ALS, C9ORF72, DPR, Drosophila, FTD, repeats, Ran translation, RNA foci
INTRODUCTION
A GGGGCC (G4C2) repeat expansion in the noncoding region of C9ORF72 is the most common mutation in both frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) (DeJesus-Hernandez et al., 2011; Renton et al., 2011). How G4C2 repeat expansion contributes to disease pathogenesis is largely unknown. One potential pathogenic mechanism is mediated by abnormal dipeptide repeat (DPR) proteins arising from repeat-associated non-AUG (RAN) translation (Cleary and Ranum, 2014). DPR proteins, synthesized from both sense and antisense repeat transcripts and adjacent intronic sequences, are found mostly in cytoplasmic inclusions in C9ORF72 patients and a mouse model (Ash et al., 2013; Gendron et al., 2013; Mori et al., 2013, 2013b; Zu et al., 2013; Chew et al., 2015). When overexpressed in cultured cells or Drosophila, different DPR proteins have varying degrees of toxicity (Kwon et al., 2014; May et al., 2014; Mizielinska et al., 2014; Tao et al., 2015; Wen et al., 2014; Yang et al., 2015; Zhang et al., 2014; Zu et al., 2013).
Another potential pathogenic mechanism is RNA-mediated toxicity, in which the formation of RNA foci and sequestration of specific RNA-binding proteins is associated with mis-regulation of RNA processing in repeat-expansion diseases (Mohan et al., 2014). In the case of C9ORF72 FTD/ALS, RNA foci are found mostly in the nucleus of multiple cell types, such as brain cells and fibroblasts from patients and neurons derived from induced pluripotent stem cells (iPSCs) (Almeida et al., 2013; DeJesus-Hernandez et al., 2011; Donnelly et al., 2013; Gendron et al., 2013; Lagier-Tourenne et al., 2013; May et al., 2014; Mizielinska et al., 2014; Sareen et al., 2013; Zu et al., 2013). However, little is known about the pathogenicity of these RNA foci.
To address this question, we generated a novel Drosophila model expressing in a cell-type-specific manner a C9ORF72 minigene containing up to 160 G4C2 repeats (160R) flanked by human intronic and exonic sequences. Overexpressed intronic 160R formed abundant nuclear sense RNA foci in neurons and glial cells but had surprisingly little toxicity in vivo. In contrast, highly toxic 36 G4C2 repeats (36R) expressed in the context of a poly(A)+ mRNA (Mizielinska et al., 2014) were exported to the cytoplasm and translated into DPR proteins at >100-fold higher level than that by intronic 160R. Moreover, 160R expressed at a higher temperature induced a modest toxicity that correlated with increased DPR production but not G4C2 sense RNA foci. Thus, low levels of DPR proteins, but not nuclear RNA foci, are a major source of toxicity in vivo.
RESULTS
The Expanded G4C2 Repeat–Containing Intron Is Transcribed and Spliced in iPSC-Derived Human Neurons and Patient Brain Tissues
According to the latest information in the NCBI Database, at least three splice variants of C9ORF72 are expressed in human cells (Figure 1A). In variants 1 and 3 (V1 and V3), G4C2 repeats are located in the first intron. In contrast, transcription of V2, whose expression level is much higher than V1 and V3 (Figure S1), starts downstream of G4C2 repeats (Figure 1A), which has been confirmed experimentally (Sareen et al., 2013). To quantify the effect of expanded G4C2 repeats on the transcription and splicing of the first intron of C9ORF72, we took advantage of a single-nucleotide polymorphism (SNP) in exon 3 (rs10757668) (Figure 1B), which is heterozygous (A/G) in 10–15% of the general population, with the expanded G4C2 repeats associated with the G allele (DeJesus-Hernandez et al., 2011; Sareen et al., 2013). We performed allele-specific expression analysis of the junction between exon 1b and exon 3 in cells and brain tissues carrying the A/G SNP by pyrosequencing, a sequence-based detection technology that allows quantification of a sequence variation such as a SNP (Ronaghi, 2001). We found that the ratio between G- and A-containing V3 alleles was the same in iPSCs derived from three G4C2 repeat expansion carriers and three controls (Figure 1C). A similar result was obtained from postmortem brain tissues (Figure 1D).
Figure 1. Expanded G4C2 Repeats in C9ORF72 Are Fully Transcribed in Patient Cells and Tissues.
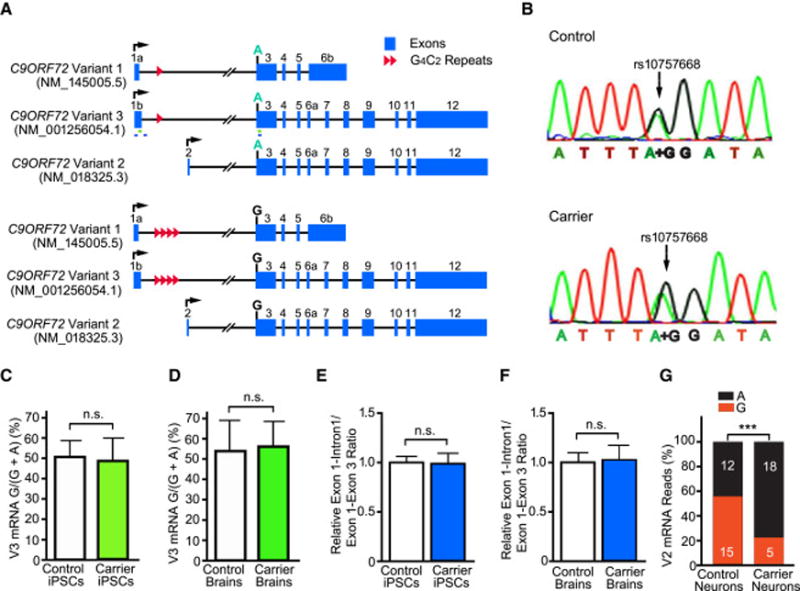
(A) Schematic of the three known transcript variants of wildtype (top) and mutant (bottom) alleles of C9ORF72, named as in PubMed and the UCSC Genome Browser. The locations of primers used in Panels C and D (green) and Panels E and F (blue) are indicated.
(B) The G/A SNP in a control subject and in a carrier of expanded G4C2 repeats.
(C, D) Pyrosequencing analysis of the allele-specific expression of V3 in three C9ORF72 and three control iPSC lines (C) and in brain tissues of three patients and three controls (D).
(E, F) The relative ratio of total V1 and V3 pre-mRNAs (exon 1–intron 1 junction) to mature mRNAs (exon 1–exon 3 junction) in four C9ORF72 and four control iPSC lines (E) and in brain tissues of eight C9ORF72 patients and nine controls (F). n.s.: not statistically significant, by Student’s t test.
(G) RNA-seq analysis reveals a significantly lower V2 level in patient neurons than control neurons. ***: p < 0.001, chi-square test
See also Figure S1.
If expanded G4C2 repeats blocked the full transcription or splicing of the first intron, one would expect that the ratio of unspliced RNA (as measured by the exon 1–intron 1 junction) to mature mRNA (as measured by the exon 1–exon 3 junction) would be higher. The ratio was identical in human neurons differentiated from four C9ORF72 and four control iPSC lines (Figure 1E) and in brain tissues from eight C9ORF72 patients and nine controls (Figure 1F), even though the levels of both intron 1-containing pre-mRNA and exon 1-containng mature mRNA were higher in iPSCs-derived C9ORF72 neurons (Figure S1B) and brain tissues (Figure S1C) than their respective controls. Thus, the expanded G4C2 repeats do not meaningfully affect splicing of the first intron, and most if not all G4C2 sense RNAs in human neurons come from the spliced first intron of C9ORF72, consistent with our earlier finding that intron 1-containing C9ORF72 RNA could not be detected by Northern blot (Almeida et al., 2013). In contrast, RNA-seq analysis revealed that mutant V2 allele (G allele) was expressed at a much lower level in patient iPSC-derived neurons than in controls (Figure 1G), consistent with several reports showing that the total level of V2 expressed from both alleles is lower in C9ORF72 patient samples (Almeida et al., 2013; Belzil et al., 2013; DeJesus-Hernandez et al., 2011; Donnelly et al., 2013; Sareen et al., 2013; Waite et al., 2014).
A Novel Drosophila Model of Intronic G4C2 Repeat Expansion
To investigate the pathogenicity of intronic G4C2 repeats, we designed and expressed an artificial human C9ORF72 minigene in Drosophila (Figure 2A). This minigene contains exon 1, part of the first intron with synthetic G4C2 repeats of various lengths at the exact same location as in human cells, and exon 3 of human C9ORF72 (Figure 2A). DNA constructs containing uninterrupted 5, 20, 40, 80 and 160 G4C2 repeats (160R) were stable in E. coli (Figure 2B). C9ORF72 minigenes containing 5R, 80R and 160R were then cloned into a transgenic fly vector (Figure 2A). The PhiC31 integrase–mediated site-specific integration system (Bateman et al., 2006) was used to ensure equal transcription of minigenes with different repeat length in flies. Southern blot analysis confirmed the repeat length of the transgenes (Figure S2A). G4C2 repeats with five copies were not detected by southern blot but confirmed by PCR and sequencing analysis. The intronic 160R remained stable in transgenic flies over multiple generations.
Figure 2. A Novel Drosophila Model of FTD/ALS Expressing Expanded G4C2 Repeats.
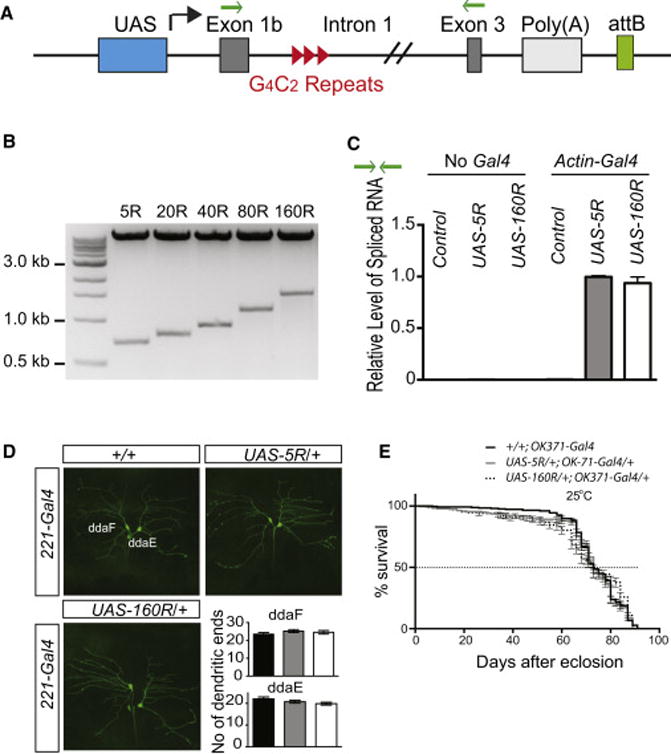
(A) Schematic of human C9ORF72 minigenes containing 5, 80, or 160R, flanked by human intronic and exonic sequences. Green arrows: primer location used in Panel C.
(B) Construction of DNA plasmids containing 5, 20, 40, 80, or 160R. Plasmids were digested with EcoRI.
(C) Quantitative real-time PCR analysis of the relative levels of C9ORF72 minigene spliced RNAs.
(D) Effect of 160R on dendritic branching of dda neurons in third instar larvae. No statistical difference was observed (n >14 flies per genotype).
(E) Kaplan-Meier curves showing the lifespan of transgenic flies expressing 0, 5R or 160R in glutamatergic neurons by OK371-Gal4. No statistical difference between 5R and 160R flies (log-rank test).
See also Figure S2.
To determine whether G4C2 repeats of different length are expressed at the same level, we crossed 5R and 160R transgenic flies carrying the UAS-repeats sequences with Actin5C-Gal4 fly lines and did quantitative real-time PCR (qPCR) analyses. PCR analysis with primers located in exon 1 and exon 3 revealed only a single 120-bp band corresponding to the spliced form (Figure S2B). No difference was observed between 5R and 160R flies when the relative level of the exon 1–exon 3 junction was quantified by with the same set of primers (Figure 2C), suggesting that intronic 160R did not affect transgene expression and splicing, like that in human cells (Figure 1).
Flies Expressing Intronic 160R Do Not Have Obvious Neurodevelopmental Defects
C9ORF72 patients have age-dependent neurodegeneration, but no early neurodevelopmental defects have been observed. To determine whether flies expressing intronic 160R also develop normally—and are thus a more physiologically faithful model—we traced and quantified larval crawling behavior on an agarose plate (Nichols et al., 2012). In this assay, locomotor activity did not differ between larvae expressing 0, 5R or 160R driven by actin5C-Gal4 (Figure S2C). We then used Drosophila dendritic arborization (dda) neurons as a model system (Li et al., 2004) to study early neuronal defects that might result from RNA toxicity. Again, no significant difference in dendritic branching was observed (Figure 2D). Moreover, cell death was not increased in larval brain cells expressing intronic 160R (Figure S2D). To determine whether expanded G4C2 repeats affect the long-term viability of adult flies, we expressed intronic 5R or 160R in all cells by Actin5C-Gal4 (not shown) or specifically in glutamatergic neurons with the OK371-Gal4 driver. When grown at 25°C, 160R flies did not show a change in their lifespan (Figure 2E).
Overexpressed Intronic 160R Form Numerous Nuclear Sense RNA Foci with Little Effect on rRNA Biogenesis and mRNA Processing
Sense and antisense RNA foci are a key pathological hallmark of C9ORF72 FTD/ALS. In iPSC-derived human neurons, most RNA foci are nuclear although a few are cytoplasmic (Almeida et al., 2013). In 160R flies, no antisense foci were observed; however, on average, 42 sense RNA foci were detected in the nucleus of glutamatergic neurons and 49 in glia expressing intronic 160R; no foci were observed in control flies and flies expressing 5R (Figure 3A, B). The average number of RNA foci per cell was more than 10 times higher than sense RNA foci in iPSC-derived human neurons (Almeida et al., 2013), probably reflecting overexpression of G4C2 repeat by the UAS-Gal4 system.
Figure 3. Overexpressed Intronic 160R Form Abundant Nuclear G4C2 Sense RNA Foci in Glia and Neurons.
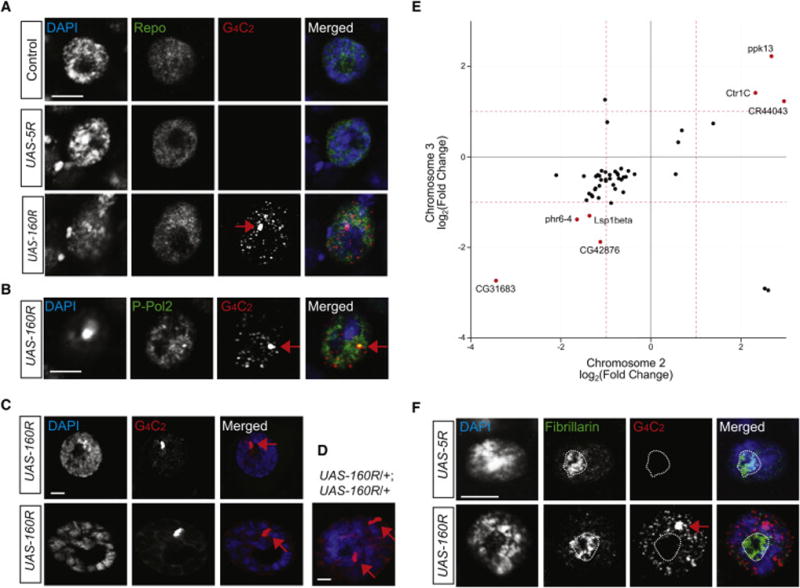
(A, B) The presence of multiple nuclear RNA foci (red) in glial cells (A) and glutamatergic neurons (B). The nucleus was stained with DAPI (blue) and anti-Repo (green) for glia or anti-phospho-RNA polymerase-II for glutamatergic neurons.
(C) RNA FISH in muscle cells (top) and salivary gland cells (bottom) detects one single brightdot (red).
(D) In each salivary gland cell expressing two copies of 160R, two brightdots are present on DNA (blue) at the site of transcription.
(E) Correlation plot showing the 50 DEGs common between flies expressing 160R from the second and the third chromosome with 45 in the same direction. Two-fold change is designated by dashed red lines and genes with over two-fold change in both groups are highlighted in red. The linear regression line has a slope of 0.61, in close agreement with the relative expression levels of the transgenes in these two genetic backgrounds (Table S1).
(F) Multiple RNA foci (red) in glia do not co-localize with the nucleolus-specific marker fibrillarin (green). In all panels, red arrows indicate transgene transcription site and scale bars are 5 μm.
See also Figures S2F, Figure S3 and Table S1–4.
These foci were not detected by a DNA probe targeting CUG or CCUG repeats (data not shown) and are made of RNA, as evidenced by their sensitivity to RNase but not DNase treatment (Figure S3A). In these cells, one large focus and numerous smaller foci were invariably present (Figure 3A, B). The small foci were similar to those in human fibroblasts or iPSC-derived neurons (Almeida et al., 2013) but distinct from G4C2 repeats in muscle cells or salivary gland cells (Figure 3C). Salivary gland cells are much larger than neurons and have polytene chromosomes. Thus, expression of intronic 160R was much higher in this cell type; yet, very little was exported to the cytoplasm (Figure S3B). These large “foci” were always present on chromatin and seemed to be located at the site of transgene transcription. Indeed, they were co-stained with an antibody against activated Pol-II (Figure 3B). Moreover, when two copies of intronic 160R were expressed from two different chromosomes, two large dots were invariably observed in each cell (Figure 3D). Thus, signals detected at the site of transcription in transgenic animal models should be distinguished from the actual RNA foci observed in patient cells.
RNA foci formed by intronic 160R did not localize to the nucleolus or grossly alter nucleolar morphology (Figure 3F). Thus, 160R RNAs do not seem to directly impair nucleolar function. Indeed, the level of different rRNA species did not differ between controls and flies overexpressing 5R or 160R (Figure S3C). To determine whether the numerous nuclear RNA foci alter mRNA processing in flies, we sequenced the transcriptomes of male fly heads in which a 5R or 160R transgene was ubiquitously expressed by Actin5C-Gal4. To reduce false-positives caused by the difference in genetic backgrounds and other factors, we used flies bearing these transgenes on either the second or the third chromosome and obtained an average of 32 million nonredundant uniquely mapped reads per library (Table S1).
Gene expression in flies expressing 5R and 160R was highly correlated (Figure S3E, S3F). Because the number of RNA foci was comparable in flies expressing 160R from either the second or the third chromosome (Figure S3D), we analyzed differentially expressed genes (DEGs) from both groups and found 50 common DEGs; 45 of them showed differential expression in the same direction (Figure 3E, Table S2). Gene ontology analysis suggested that proteins with oxidoreductase activities were enriched among these 45 genes (Table S3). Only 7 of these 45 genes showed a change of >2-fold. Moreover, we found only 17 exon-skipping events common to flies expressing 160R from either the second or the third chromosome (Table S4). For example, in the sun gene, the percent-spliced-in (PSI) was 16.5% in 5R flies and 22.3% in 160R flies (Figure S3G). Such a small number of DEGs and exon-skipping changes argues against the hypothesis that those RNA foci are capable of sequestering sufficient RNA binding proteins to exert a global effect on RNA processing in our fly model.
Highly Toxic 36R-poly(A) Are Exported to the Cytoplasm and Intronic 160R Expressed at a Higher Temperature Induces Modest Toxicity and Increased DPR Production
The finding that abundant nuclear G4C2 RNA foci cause little toxicity was surprising, so we examined a fly model in which 36R are highly toxic (Mizielinska et al., 2014). This model and another one expressing 30R (Xu et al., 2013) differ significantly from ours where 160R are transcribed and spliced out as part of human intronic sequence. In those models, G4C2 repeats are transcribed as part of a poly(A)+ mRNA. The steady-state level of 36R-poly(A) was about 2-fold higher than the spliced RNA in 160R flies (Figure S4B), which is likely due to their differential posttranscriptional regulation, since both transgenes were generated using the same site-specific integration system and expressed by the same Gal4 driver. Nonetheless, we examined 160R homozygous flies that expressed the spliced RNA at the same level as the 36R mRNA (Figure S4B) and found no visible eye neurodegenerative phenotype (Figure S4C).
In flies expressing 36R-poly(A) and grown at 25°C, most of the 36R-poly(A) RNA were exported to the cytoplasm of glial cells (Figure 4A) or motor neurons (not shown), as expected for a poly(A)+ mRNA. Yet, in the nucleus of each cell, a bright large dot was still observed which we believe is the site of the transgene transcription as demonstrated earlier (Figure 3B, D). In stark contrast to intronic 160R, ubiquitous expression of 36R-poly(A) by Actin5C-Gal4 resulted in a developmental lethal phenotype. Thus, to quantify DPR levels, we used GMR-Gal4 to express these transgenes in the eye and performed a previously described immunoassay for poly(GP) (Su et al., 2014). A similar quantitative assay is not available for poly(GR), which is highly toxic in flies (Mizielinska et al., 2014; Yang et al., 2014). Therefore, we first performed immunostaining and detected poly(GR) in both neurons and glial cells expressing 36R-poly(A) but not intronic 160R (Figures 4B, S4A). Similarly, the poly(GP) level was >100-fold higher in flies expressing 36R-poly(A) than intronic 160R (Figure 4C). The high level of DPR proteins especially poly(GR) produced from 36R-poly(A) correlates well with the eye degenerative phenotype (Figure S4C) and the increased level of p62 (Figure S4D). Thus, the toxicity of expanded G4C2 repeats is greatly influenced by their subcellular location.
Figure 4. 36R RNAs Expressed in the Context of Poly(A)+ mRNA Are Exported to the Cytoplasm and Produce Much Higher Level of DPRs than Intronic 160R.
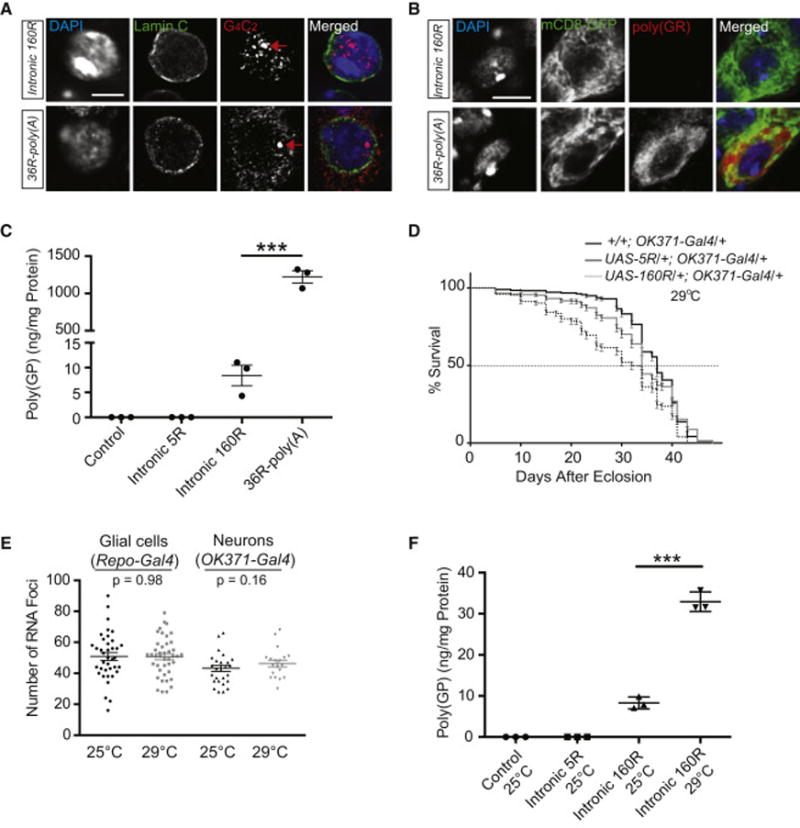
(A) RNA FISH in glial cells detects numerous nuclear RNA foci (red) formed by intronic 160R while 36R-poly(A) RNA is exported to the cytoplasm. Green: nuclear membrane; blue: DAPI staining. Red arrows: transcription sites of transgenes. Scale bar: 5 μm.
(B) Poly(GR) (red) was detected mostly in the cytoplasm of neurons expressing 36R-poly(A) but not in cells expressing intronic 160R. Scale bar: 5 μm.
(C) One of the two experiments to measure poly(GP) in lysates from heads of flies raised at 25°C. Values are mean ± SEM. ***: p < 0.0001 (one-way ANOVA). On average, the poly(GP) level is 122 times higher than that in 36R flies than in 160R flies.
(D) The lifespan of transgenic flies grown at 29°C expressing 0, 5R or 160R in glutamatergic neurons. Statistical difference between 5R and 160R flies was assessed with the log-rank test (p < 0.001).
(E) Quantification of nuclear RNA foci in neurons and glia in flies grown at 25°C and 29°C. No statistical difference was observed (by unpaired nonparametric t test).
(F) Poly(GP) levels in lysates from heads of flies raised at 25°C and 29°C. Values are mean ± SEM. ***: p <0.0001 (one-way ANOVA).
See also Figure S4.
In contrast to flies grown at 25°C (Figure 2E), intronic 160R expression at 29°C induced a modest toxicity as measured by the lifespan assay (Figure 4D). The number of RNA foci in both neurons and glial cells remained the same (Figure 4E). However, DPR production was 4-fold higher in 160R flies grown at 29°C than at 25°C (Figure 4F). These findings suggest that intronic 160R expressed at a higher temperature induces modest toxicity through increased DPR production, further supporting the notion that low levels of DPR proteins, but not nuclear sense RNA foci, are a major source of toxicity in our fly model.
DISCUSSION
In this study, we established a novel Drosophila model in which 160R were flanked by human intronic and exonic sequences, and transcribed and spliced in vivo. We first confirmed that expanded G4C2 repeats, some of them contain more than 1,000 copies (Almeida et al., 2013), are transcribed and spliced as part of the first intron of C9ORF72 in human cells and patient brain tissues. This finding was not unexpected, as the highly evolved human transcription machinery contains multiple factors to perform this complex reaction in vivo (Shilatifard et al., 2003), in contrast to an observed high level of aborted transcription by bacteriophage T7 polymerase alone in an in vitro assay with short G4C2 repeats (Haeusler et al., 2014). Some transcription elongation factors, such as Supt4h, facilitate transcription through long repeat sequences in human cells (Liu et al., 2012).
In general, most spliced introns stay in the nucleus although some are exported to the cytoplasm (Hesselberth, 2013). Indeed, intronic 160R formed abundant nuclear G4C2 RNA foci in both neurons and glia with only a few foci outside of the nucleus. The average number of these nuclear RNA foci was more than 10 times higher than we observed in iPSC-derived neurons (Almeida et al., 2013). However, the transciptome was largely unaffected, rRNA biogenesis was not significantly altered, and nucleolar morphology appeared to be normal. Thus, these foci by themselves do not elicit significant toxicity, at least in this experimental system. Our results comparing flies grown at 25°C vs. 29°C also support the notion that nuclear G4C2 sense RNA foci are not a significant source of toxicity. Whether more abundant antisense foci are neurotoxic remains to be determined.
DPR proteins are mostly found in cytoplasmic inclusions of postmortem brain tissues of C9ORF72 patients and a mouse model (Ash et al., 2013; Gendron et al., 2013; Mori et al., 2013, 2013b; Zu et al., 2013; Chew et al., 2015). We speculate that they are produced from either intronic repeat RNAs and/or unspliced C9ORF72 pre-mRNAs that are transported to the cytoplasm. The poly(A) tail facilitates nuclear export and cytoplasmic translation of mature mRNAs (Weill et al., 2012). Indeed, we found that 36R expressed in the context of poly(A)+ mRNA were exported to the cytoplasm and produced DPRs at a level >100-fold higher than that in 160R flies (Figure 4C).
Many animal models fail to fully recapitulate human neurodegenerative diseases because aging is a major risk factor for these disorders. During aging, a subset of nucleoporins is oxidatively damaged, resulting in increased nuclear permeability (D’Angelo et al., 2009). Thus, aging itself or environmental stress may cause increased leakage of expanded G4C2 repeats in an intron or an unspliced pre-mRNA into the cytoplasm, resulting in increased production of toxic DPRs in some vulnerable human neurons.
DPR proteins of various lengths may have different pathogenic mechanisms. We recently found that in contrast to (GR)20 (Kwon et al., 2014), (GR)80 was located mostly in the cytoplasm and impaired the Notch signaling pathway, whereas (GA)80 recruits (GR)80 into cytoplasmic inclusions thus decreasing (GR)80 toxicity (Yang et al., 2015). Moreover, poly(GR) proteins of different length used in different studies can either directly or indirectly induce nucleolar stress (Kwon et al., 2014; Wen et al., 2014; Yang et al., 2015). Thus, much remains to be learned about the pathogenic mechanisms of DPR proteins of different lengths. Although the life-long presence of sense RNA foci in the nucleus might still be detrimental, our results suggest that they could also be neutral intermediates or even neuroprotective by sequestering G4C2 repeats in the nucleus therefore preventing cytoplasmic DPR production. Thus, inhibition of the toxicity of DPR proteins, especially non-aggregated form, seems to represent a more promising therapeutic approach for C9OFR72 patients.
EXPERIMENTAL PROCEDURES
For details on all methods, please see Supplemental Information.
Supplementary Material
1
2
3
4
5
HIGHLIGHTS.
- Intronic 160R form abundant nuclear sense RNA foci in Drosophila.
- Nuclear RNA foci cause little toxicity and minimal changes in RNA processing.
- Cytoplasmic G4C2 repeats produce >100-fold more poly(GP) than intronic repeats.
- Modest toxicity of 160R at higher temperature correlates with increased DPRs.
Acknowledgments
We thank F. Ladam, P.-H. Wu, E. Ricci, Z. Zhang, S. Ordway, and Gao lab members for comments; the Bloomington Drosophila Stock Center for 36R and other fly lines; and the UMMS Department of Cell and Developmental Biology Confocal Core for use of their microscopes. We also thank L. Ranum for anti-GR antibody, E. Baehrecke for anti-p62 antibody, and W. Seeley (Neurodegenerative Disease Brain Bank at the University of California, San Francisco) for brain tissues of C9ORF72 patients. This work was supported by an ALS Association Milton Safenowitz Postdoctoral Fellowship (H.T.), a grant from ALS Therapy Alliance (F.-B.G), Mayo Clinic Foundation (L.P.), and grants from the Department of Defense (ALSRP AL130125), the ALS Association, Packard Center for ALS Research and Target ALS (F.-B.G. and L.P.), and grants from the National Institutes of Health: R01NS057553, R01NS079725 and R21NS086318 (F.-B.G.); P50AG016574, R21NS084528, R01NS088689, R01NS063964; R01NS077402 and P01NS084974 (LP); R21NS089979 (T.F.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information includes four figures, four tables and Experimental Procedures that can be found with this article online. RNA-seq data are available at the NCBI the Gene Expression Omnibus database (accession number: GSE72771).
AUTHOR CONTRIBUTIONS
H.T. and F.-B.G. conceived the project. H.T. performed all the fly experiments with some help from U.C., Y.L., X.D. and J.A.N. S.A. contributed Figure 1. J.M. and Z.W. performed bioinformatics analysis. T.F.G. and L.P. measured poly(GP) levels. H.T. and F.-B.G. wrote the manuscript with inputs from S.A., J.M., T.F.G. and Z.W.
References
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin WL, DeJesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME, et al. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. 2015;348:1151–1154. doi: 10.1126/science.aaa9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Ranum LPW. Repeat associated non-ATG (RAN) translation: new starts in microsatellite expansion disorders. Curr Opin Genet Dev. 2014;26:6–15. doi: 10.1016/j.gde.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PEA, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR. Lives that introns lead after splicing. Wiley Interdiscip Rev RNA. 2013;4:677–691. doi: 10.1002/wrna.1187. [DOI] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, Jiang J, Watt AT, Chun S, Katz M, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci USA. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang F, Menut L, Gao FB. BTB/POZ-zinc finger protein abrupt suppresses dendritic branching in a neuronal subtype-specific and dosage-dependent manner. Neuron. 2004;43:823–834. doi: 10.1016/j.neuron.2004.08.040. [DOI] [PubMed] [Google Scholar]
- Liu C-R, Chang C-R, Chern Y, Wang T-H, Hsieh W-C, Shen W-C, Chang C-Y, Chu I-C, Deng N, Cohen SN, et al. Spt4 is selectively required for transcription of extended trinucleotide repeats. Cell. 2012;148:690–701. doi: 10.1016/j.cell.2011.12.032. [DOI] [PubMed] [Google Scholar]
- May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, Grässer FA, Mori K, Kremmer E, Banzhaf-Strathmann J, et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014;128:485–503. doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Grönke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IOC, Pietrzyk J, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan A, Goodwin M, Swanson MS. RNA-protein interactions in unstable microsatellite diseases. Brain Res. 2014;1584:3–14. doi: 10.1016/j.brainres.2014.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013a;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Nichols CD, Becnel J, Pandey UB. Methods to assay Drosophila behavior. J Vis Exp. 2012:e3795–e3795. doi: 10.3791/3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- Sareen D, O’Rourke JG, Meera P, Muhammad AKMG, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149–208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Conaway RC, Conaway JW. The RNA polymerase II elongation complex. Annu Rev Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- Tao Z, Wang H, Xia Q, Li K, Li K, Jiang X, Xu G, Wang G, Ying Z. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet. 2015;24:2426–2441. doi: 10.1093/hmg/ddv005. [DOI] [PubMed] [Google Scholar]
- Waite AJ, Bäumer D, East S, Neal J, Morris HR, Ansorge O, Blake DJ. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35:1779.e5–1779.e13. doi: 10.1016/j.neurobiolaging.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill L, Belloc E, Bava FA, Méndez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol. 2012;19:577–585. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83:1043–1050. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci USA. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Abdallah A, Li Z, Lu Y, Almeida S, Gao FB. FTD/ALS associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathol. 2015 Jun;:2. doi: 10.1007/s00401-015-1448-6. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-J, Jansen-West K, Xu Y-F, Gendron TF, Bieniek KF, Lin W-L, Sasaguri H, Caulfield T, Hubbard J, Daughrity L, et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014;128:505–524. doi: 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3
4
5