The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma (original) (raw)
. Author manuscript; available in PMC: 2015 Dec 18.
Published in final edited form as: Nat Cell Biol. 2014 Feb 23;16(3):255–267. doi: 10.1038/ncb2916
Abstract
Pancreatic ductal adenocarcinoma (PDA) develops through distinct precursor lesions, including pancreatic intraepithelial neoplasia (PanIN) and intraductal papillary mucinous neoplasia (IPMN). However, genetic features resulting in IPMN-associated PDA (IPMN–PDA) versus PanIN-associated PDA (PanIN-PDA) are largely unknown. Here we find that loss of Brg1, a core subunit of SWI/SNF chromatin remodelling complexes, cooperates with oncogenic Kras to form cystic neoplastic lesions that resemble human IPMN and progress to PDA. Although Brg1-null IPMN–PDA develops rapidly, it possesses a distinct transcriptional profile compared with PanIN-PDA driven by mutant Kras and hemizygous p53 deletion. IPMN–PDA also is less lethal, mirroring prognostic trends in PDA patients. In addition, Brg1 deletion inhibits Kras-dependent PanIN development from adult acinar cells, but promotes Kras-driven preneoplastic transformation in adult duct cells. Therefore, this study implicates Brg1 as a determinant of context-dependent Kras-driven pancreatic tumorigenesis and suggests that chromatin remodelling may underlie the development of distinct PDA subsets.
PDA is thought to arise from three distinct classes of precursor lesions: PanIN, mucinous cystic neoplasm (MCN) or IPMN. Of the two cystic neoplasms, IPMN has recently received attention due to increasingly frequent diagnosis, probably a consequence of better detection methods1. As a result of a more favourable prognosis compared with PanIN-associated PDA (refs 2–4), IPMN-associated PDA may be biologically distinct5. The properties of transformed cells are lineage-dependent, even in response to the same oncogenic driver6,7, making distinct cells of origin one conceivable possibility for differences between IPMN–PDA and PanIN-PDA. Indeed, whereas considerable functional evidence from mouse models suggests that PanINs may be of acinar origin8–11, the hallmark intraductal location of IPMNs may reflect a ductal origin, although this has not been demonstrated experimentally.
There is growing evidence to support a role for epigenetic regulators in the development and progression of cancer12. Chromatin remodellers are a group of epigenetic regulators that disrupt DNA– protein contacts to regulate gene expression13. The evolutionarily conserved, multi-protein chromatin-remodelling SWI/SNF complexes have been implicated in different human cancers13,14. Mammalian SWI/SNF complexes are composed of two distinct groups, the Brm/Brg1-associated factor (BAF) and Polybromo-associated (PBAF) complexes. The BAF complex is composed of one of two mutually exclusive catalytic ATPase subunits, Brg1 (SMARCA4 in human) or Brm (SMARCA2), whereas the PBAF complex utilizes only Brg1 as the catalytic subunit13. In addition, variant subunits are associated with the complexes13. Inactivating mutations or loss of expression of several SWI/SNF subunits have recently been identified in various human cancers15–19, including pancreatic cancer20. SWI/SNF subunits seem to play a functional role in tumorigenesis as targeted inactivation has been shown to affect cancer formation in different tumour mouse models13,21–23. For PDA, Brg1-inactivating mutations and deletions were found in pancreatic cancer cell lines, and somatic Brg1 mutations were also identified in human PDA (refs 24,25). Moreover, Brg1 expression was noted to be frequently reduced or lost in human IPMN samples26. However, it is unclear whether Brg1 plays a functional role in the specification of PDA precursors and PDA progression. Results from studies presented here establish Brg1 as a bona fide tumour suppressor that constrains oncogenic Kras-induced IPMN–PDA formation and highlight the importance of chromatin remodelling in the development of distinct PDA subsets.
RESULTS
Pancreatic loss of Brg1 in the context of mutant Kras results in formation of cystic neoplasms
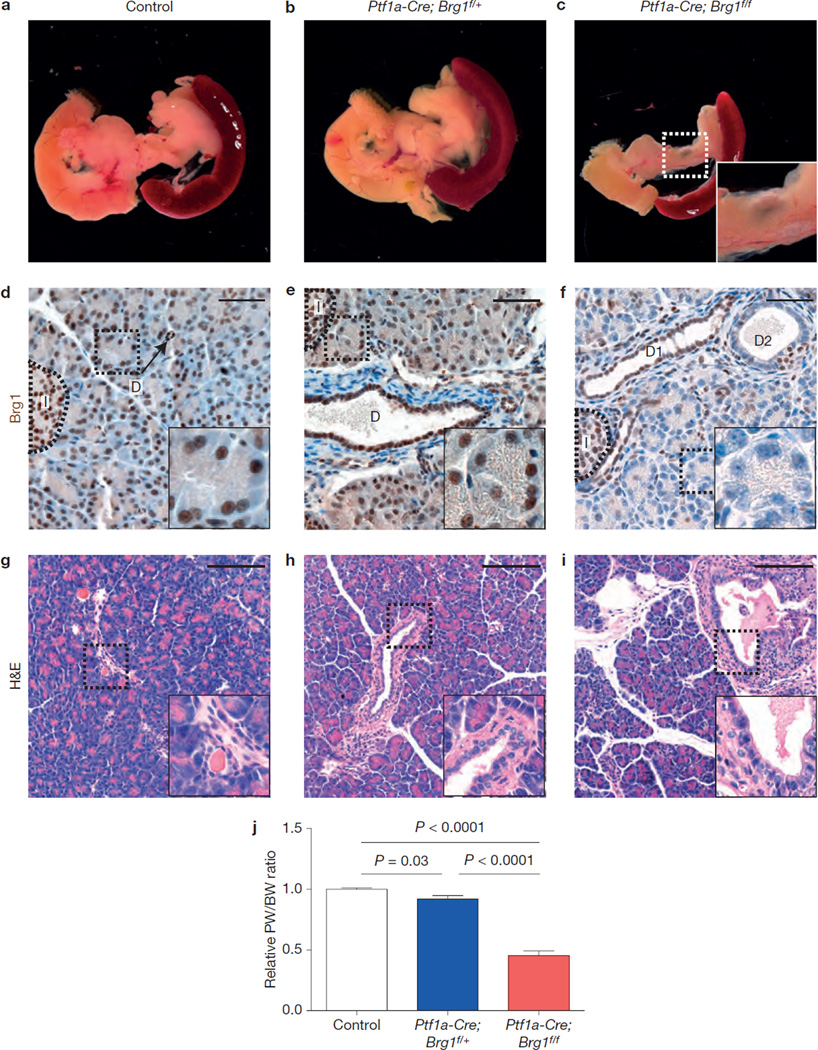
Brg1, a critical ATPase component of chromatin remodelling SWI/SNF complexes, is expressed in all pancreatic cell types in 3 weeks old mice (Fig. 1d). To investigate its functional requirement during pancreas development, we crossed transgenic mice carrying floxed alleles of Brg1 (Brg1f)27 w_ith Pf1a–Cre_ mice28. The Brg1 mutant mice exhibited a hypoplastic pancreas compared to heterozygous or control littermate animals at 3 weeks of age (Fig. 1a–c,j). As expected from the Cre-activity pattern in Ptf1a–Cre mice28,29, almost all acinar cells had lost Brg1 expression, whereas it was maintained in most islet cells (Fig. 1f). In contrast, Brg1 was eliminated only in a subset of duct cells (Fig. 1f). Duct cells retaining Brg1 expression seemed to be normal, whereas ablation of Brg1 resulted in slight dilation of some duct structures at 3 weeks of age (Fig. 1f; ‘D2’). Furthermore, we occasionally detected macroscopic cysts stemming from dilations of the pancreatic ducts in mutant mice (Fig. 1i).
Figure 1.
Loss of Brg1 leads to reduced pancreas size and duct dilations. (a–c) Macroscopic view of a control (a), Ptf1a–Cre; Brg1f/+ (b) and Ptf1a–Cre; Brg1f/f (c) pancreas at the age of 3 weeks. (d–i) Microscopic view of a pancreas of control (d,g), Ptf1a–Cre; Brg1f/+ (e,h) and _Ptf1a_– Cre; Brg1f/f (f,i) mice at the age of 3 weeks. (d–f) Brg1 staining of the indicated genotypes at 3 weeks (D, duct; I, encircled islet; D1 marks a duct with retained Brg1 expression; D2 marks a duct with loss of Brg1 expression). (g–i) Haematoxylin and eosin (H&E) staining at 3 weeks of age (arrow in i marks occasional duct dilatation). (j) Relative pancreatic weight (PW) to body weight (BW) ratio of control (n = 6), Ptf1a_– Cre; Brg1f/+ (n = 7), and Ptf1a–Cre; Brg1_f/f (n = 3) pancreas at the age of 3 weeks. Values are shown as mean ± s.e.m.; unpaired _t_-test was performed to calculate P values. Scale bars: 50µm (d–f) and 250µm (g–i).
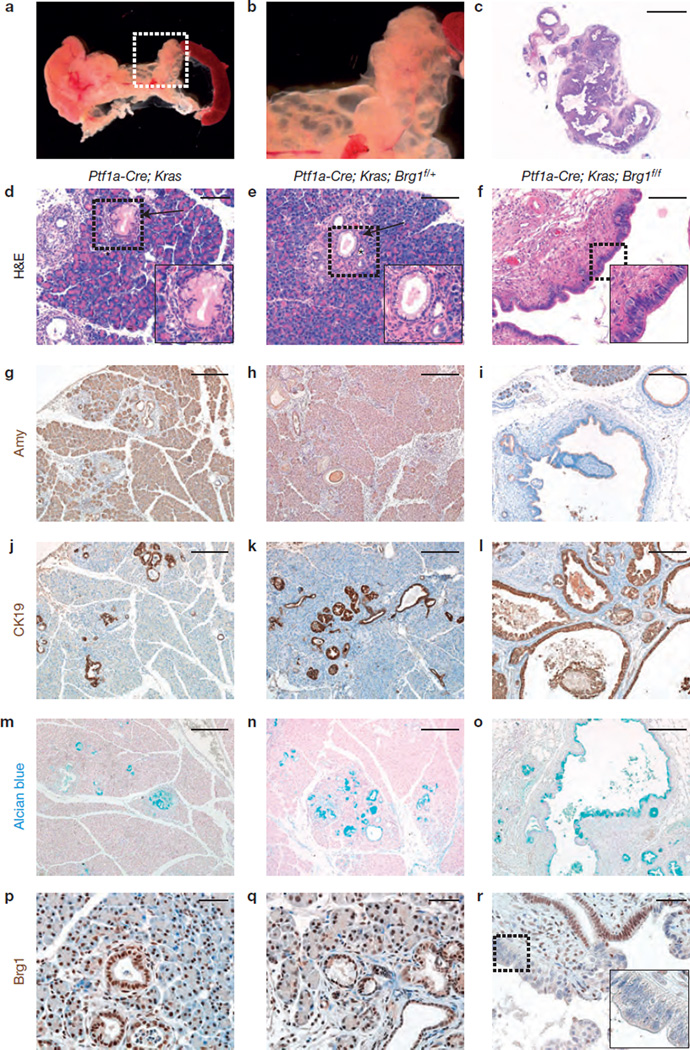
Mutant Kras (KrasG12D) is sufficient to drive the development of PanIN and PDA when targeted to the developing mouse pancreas, and can result in cystic neoplasms in the context of specific tumour suppressor loss30,32,33. To investigate whether Brg1 loss alters Kras-driven pancreatic transformation, we crossed Ptf1a–Cre; Brg1f/f mice with loxP-stop-loxP; KrasG12D (KrasG12D) mice31. Within 9 weeks, Pf1a–Cre; KrasG12D; Brg1f/f mice developed extensive grape-like, multilocular fluid-filled cystic structures throughout the pancreas (Fig. 2a,b). Pf1a–Cre; KrasG12D mice undergo acinar-to-ductal metaplasia and develop low-grade PanINs at 9 weeks of age, lesions that were also observed in Ptf1a–Cre; KrasG12D; Brg1f/+ mice (Fig. 2d,e). In contrast, acinar tissue area was profoundly decreased in Ptf1a–Cre; KrasG12D; Brg1f/f mice (Fig. 2c,i). Instead, the parenchyma of Kras/Brg1 mutant pancreata was mostly replaced by multiple CK19-positive, epithelial-lined cystic lesions of varying size (Fig. 2c,f,l). The cystic epithelium of Pf1a–Cre; KrasG12D; Brg1f/f mice contained columnar, Alcian blue-positive cells indicative of intestinal mucin-production (Fig. 2o). Brg1 was expressed in a mosaic fashion within these cystic ductal structures with dysplastic, Brg1-negative cells often contiguous with Brg1-positive, normal ductal epithelial cells (Fig. 2r). Taken together, the size, cellular morphology and mucinous nature of the predominant cysts in Ptf1a–Cre; KrasG12D; Brg1f/f mice was reminiscent of the two main cystic PDA precursors, MCNs or IPMNs (refs 1,5).
Figure 2.
Loss of Brg1 cooperates with Kras to form neoplastic cystic lesions. (a,b) Macroscopic view of a Ptf1a–Cre; KrasG12D; Brg1f/f pancreas at 9 weeks of age (b shows magnified view of the cysts). The pancreas showed extensive grape-like, multilocular fluid-filled cystic structures throughout, ranging from <1mm up to 5mm in diameter. (c) H&E staining of Ptf1a–Cre; KrasG12D; Brg1f/f pancreas at 9 weeks. (d–r) Histological analysis of a Ptf1a–Cre; KrasG12D; Brg1f/f (f,i,l,o,r) pancreas at 9 weeks of age compared with age-matched Ptf1a–Cre; KrasG12D (d,g,j,m,p) and Ptf1a–Cre; KrasG12D; Brg1f/+ (e,h,k,n,q) pancreata. (d–f) Detailed H&E staining; asterisks mark acinar-to-ductal metaplasia and arrows mark PanINs. Amylase staining (g–i), cytokeratin 19 (CK19) staining (j–l), Alcian blue staining (m–o) and Brg1 staining (p–r) of the indicated genotypes at 9 weeks of age. Scale bars: 2 mm (c); 100 µm (d–f); 250 µm (g–o); 50 µm (p–r).
Cystic neoplasms resemble human pancreatobiliary IPMNs but not MCNs
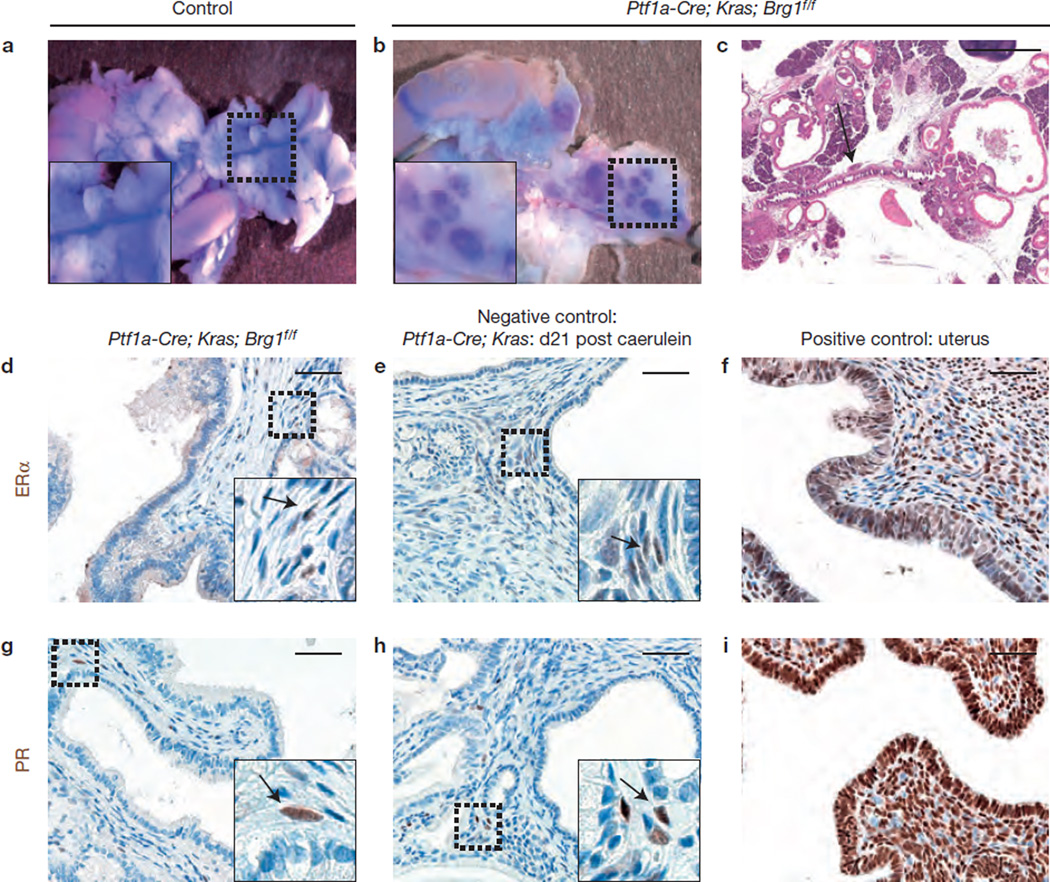
To determine whether the cystic neoplastic lesions observed in Ptf1a–Cre; KrasG12D; Brg1f/f mice mimic human MCNs or IPMNs, we examined several distinguishing criteria, including a connection to the pancreatic ductal system, lack of dense ovarian-like stroma, and absence of oestrogen receptor (ER)- and progesterone receptor (PR)-positive cells in IPMN stroma1,5.
Applying these criteria, we found that the pancreatic duct system had a direct functional connection to the multiple cystic structures (Fig. 3a – c), the cystic lesions lacked ovarian-like stroma characterized by expression of ERα and PR (ref. 5; Fig. 3d – i), and most cystic lesions lacked a MCN-typical thick stroma exhibiting wavy nuclei (Supplementary Fig. 1a). Collectively, the neoplastic cystic lesions observed in Ptf1a–Cre; KrasG12D; Brg1f/f mice were reminiscent of human IPMNs but not MCNs.
Figure 3.
Cystic neoplastic lesions resemble human IPMNs but not MCNs. (a,b) To prove a possible connection of the cystic neoplastic lesions to the pancreatic duct system, blue dye (bromophenolblue) was injected in the common bile duct to allow retrograde filling of the duct system. (c) H&E staining shows a connection of a cystic neoplastic lesions in a Ptf1a–Cre; KrasG12D; Brg1f/f mouse to the pancreatic duct (arrow). (d–i) ERα (oestrogen receptor) and PR (progesterone receptor) staining of cystic lesions of Ptf1a–Cre; KrasG12D; Brg1f/f mice (d,g), Ptf1a–Cre; KrasG12D mice (e,h) that were treated with caerulein to induce pancreatitis (arrows denote positive cells), and of murine uterus that served as a positive control (f,i). The stroma of the cystic lesions showed very little ERα and PR expression comparable to that found in the negative control. Scale bars: 1 mm (c) and 50 µm (d–i).
Further studies, including expression analysis of mucins paired with histological examination demonstrated that the cystic lesions most closely resembled pancreatobiliary IPMNs (Supplementary Fig. 1c). However, papillary projections containing fibrovascular bundles were present but less pronounced compared with human pancreatobiliary IPMN (Supplementary Fig. 1b).
Molecular characterization of IPMN-like cystic neoplasms
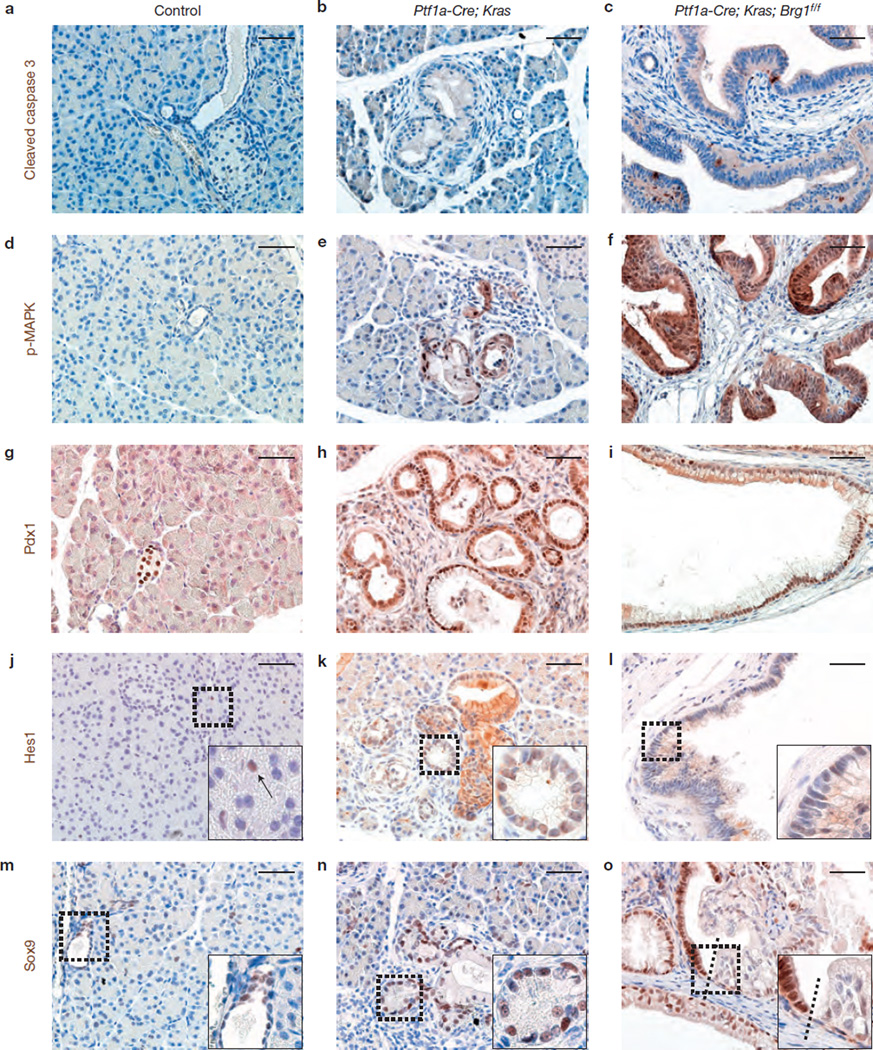
Next, we characterized the molecular features of the IPMN-like cystic neoplasms observed in Ptf1a–Cre; KrasG12D; Brg1f/f mice. Similar to PanINs of Ptf1a–Cre; KrasG12D mice, the IPMN-like cystic neoplasms possessed very few apoptotic cells expressing cleaved caspase 3 (Fig. 4a – c), and expressed phosphorylated MAP kinase (p-MAPK; Fig. 4d – f).
Figure 4.
Molecular characterization of neoplastic cystic lesions. (a–c) The IPMN-like lesions in Ptf1a–Cre; KrasG12D; Brg1f/f mice showed only few apoptotic cells as assessed by cleaved caspase 3 expression similar to PanINs of Ptf1a–Cre; KrasG12D mice. (d–f) Oncogenic Kras is known to promote activation of the MAP-kinase signalling pathway, and we observed phosphorylated (p-MAPK) in PanINs of Ptf1a–Cre; KrasG12D mice and in the cystic lesions in Ptf1a–Cre; KrasG12D; Brg1f/f mice, whereas no p-MAPK was present in control animal tissue. (g–o) Further expression analysis was performed for Pdx1 (g–i), Hes1 (j–l; arrow in j marks centroacinar cell) and Sox9 (m–o) of the indicated genotypes at 9 weeks of age. Dashed lines show the border of Sox9-positive and -negative epithelial cells. Scale bars: 50 µm (a–o).
Initiation of PDA precursors is characterized by reactivation of developmental signalling pathways10, such as up regulation of transcriptional factors including Pdx1, Hes1 and Sox9 in Kras-driven PanINs (Fig. 4h,k,n),34. Similarly, Pdx1 was universally expressed in the cystic lesions of 9-week-old Ptf1a–Cre; KrasG12D; Brg1f/f mice (Fig. 4i). Hes1 expression was absent or substantially reduced in a subset of dysplastic cystic epithelial cells in Ptf1a–Cre; KrasG12D; Brg1f/f mice (Fig. 4l), whereas it was more uniformly found in acinar-to-ductal metaplasia and PanINs of Ptf1a–Cre; KrasG12D mice (Fig. 4k). Moreover, Sox9 expression was mosaic and seemed to be reduced in the dysplastic epithelium of the IPMN-like cystic neoplasms (Fig. 4o). A similar decline in Sox9 expression has also been reported in IPMN progression in humans35.
IPMN-like lesions progress to form PDA
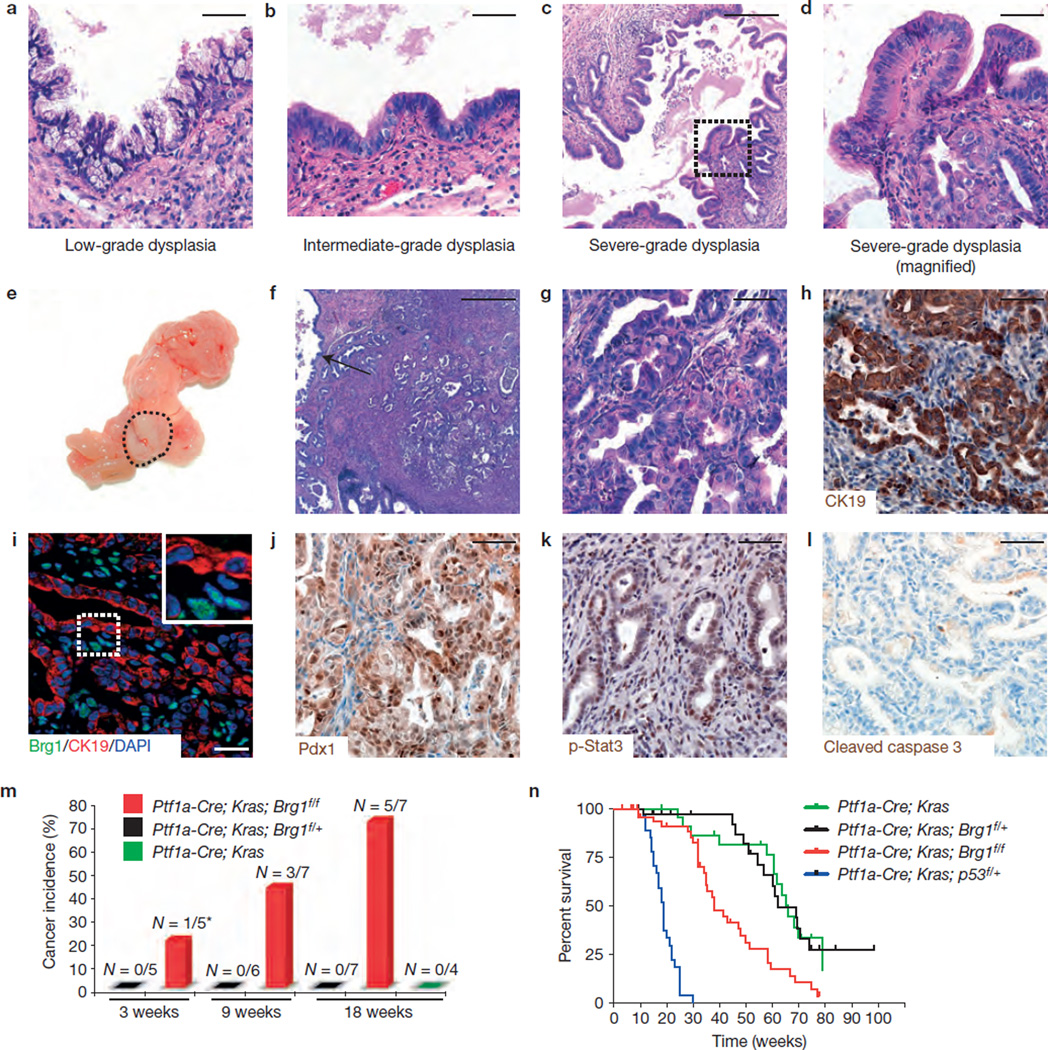
Human IPMNs are precursor lesions for PDA (refs 1,5). We found that the neoplastic cystic lesions observed in Ptf1a–Cre; KrasG12D; Brg1f/f mice at 9 weeks of age contained varying grades of dysplasia (Fig. 5a – d). Brg1 staining confirmed that the advanced dysplastic epithelium had lost Brg1 expression (Supplementary Fig. 2a). In support of the notion that severe dysplasia in the neoplastic cystic epithelium is a precursor to cancer, we observed frank PDA in a subset of Ptf1a–Cre; KrasG12D; Brg1f/f mice (hereafter referred to as IPMN– PDA mice).
Figure 5.
IPMN lesions progress to form PDA with short latency but carry a much better prognosis for survival than PanIN-PDAs. (a–d) H&E staining reveals that cysts in Ptf1a–Cre; KrasG12D; Brg1f/f mice contain varying degrees of dysplasia ranging from low-grade (a) and intermediate-grade (b) to severe dysplasia (c,d shows a higher magnification; carcinoma in situ). (e–g) Staining of PDA in Ptf1a–Cre; KrasG12D; Brg1f/f mice, macroscopic (e) and microscopic (f,g; H&E) view (f: arrow marks adjacent cystic lesion). (h–l) CK19 (h), Brg1/CK19/DAPI co-staining (i; note retained Brg1 expression in non-cancerous stroma cells), Pdx1 (j), phospho-Stat3 (k; p-Stat3) and cleaved caspase 3 staining (l) of Ptf1a–Cre; KrasG12D; Brg1f/f cancer cells. (m) Cancer incidence at indicated time points for each genotype. *At 3 weeks one of the Ptf1a–Cre; KrasG12D; Brg1f/f mice developed an undifferentiated epithelial cancer not PDA (the tumour was CK19-negative but positive for E-cadherin). All other cancers observed were PDA. (n) Overall survival analysis of the indicated mouse genotypes: Ptf1a–Cre; KrasG12D (green, n = 26), Ptf1a–Cre; KrasG12D; Brg1f/+ (black, _n_= 48), Ptf1a–Cre; KrasG12D; Brg1f/f(red, n = 57) and Ptf1a–Cre; KrasG12D; p53f/+ mice (blue, n = 37). Log-rank test revealed a significantly longer survival of Ptf1a–Cre; KrasG12D; Brg1f/f compared with Ptf1a–Cre; KrasG12D; p53f/+ mice (P<0.0001). Overall median survival of PanIN-PDA mice was 19 weeks, compared with 48 weeks in IPMN–PDA mice. Scale bars: 50 µm (a,b,d,g–l); 250 µm (c); 500µm (f).
Histological examination confirmed that PDA was associated with IPMN-like lesions (Fig. 5f). Morphologically, IPMN–PDAs (Fig. 5e – g) were indistinguishable from PanIN-derived PDAs in Ptf1a–Cre; KrasG12D; p53f/+ mice (hereafter referred to as PanIN-PDA mice), an established mouse model of PDA in which one allele of the tumour suppressor p53 is eliminated through Cre recombination in pancreatic epithelial cells in parallel with expression of oncogenic Kras_G12D_30. IPMN–PDA expressed CK19 (Fig. 5h), and immunostaining confirmed that Brg1 expression was lost in these PDA cellsbut was retained inthe surrounding stromal cells (Fig. 5i). To assess whether other signalling pathways characteristic of PDA are still intact, we stained for Pdx1, phospho-Stat3 and p-MAPK (refs 36–38). All of these markers were expressed in the nuclei of IPMN–PDA cells (Fig. 5j,k). Also, we could not detect pronounced apoptosis in CK19-positive PDA cells (Fig. 5l).
Brg1-null IPMN–PDAs develop with short latency but are less lethal than PanIN-PDAs
To determine the latency of IPMN–PDA in Ptf1a–Cre; KrasG12D; Brg1f/f animals, we analysed cancer incidence at different time points. At 9 weeks of age, we detected invasive PDAs in 43% (3 of 7) of Ptf1a–Cre; KrasG12D; Brg1f/f mice (Fig. 5m). Moreover, at 18 weeks of age the incidence of IPMN–PDA increased to 71% (5 of 7 mice; Fig. 5m). In contrast, Ptf1a–Cre; KrasG12D and Ptf1a–Cre; KrasG12D; Brg1f/+ mice did not develop cancer within the 18 weeks observation period (Fig. 5m). These data suggest that the neoplastic cystic lesions that resemble human IPMNs progress rapidly to form PDA in IPMN–PDA mice.
Next, we performed survival analyses of cohorts of Ptf1a– Cre; KrasG12D, Ptf1a–Cre; KrasG12D; Brg1f/+, Ptf1a–Cre; KrasG12D; Brg1f/f (IPMN–PDA) and Ptf1a–Cre; KrasG12D; p53f/+ (PanIN-PDA) mice. We observed a striking difference in overall survival between IPMN–PDA and the PanIN-PDA mice (Fig. 5n). The most common cause of death in IPMN–PDA mice was pancreatic exocrine insufficiency, in the absence of any indications for death from malignant disease. This was in contrast to the other analysed groups that frequently showed signs of progressive cancer disease (Supplementary Table 1). Thus, similar to the human situation1,3,4, IPMN-derived PDAs carry a much more favourable prognosis for survival compared with PanIN-derived PDAs.
IPMN–PDA cells are intrinsically less proliferative than PanIN-PDA cells and carry a distinct molecular signature
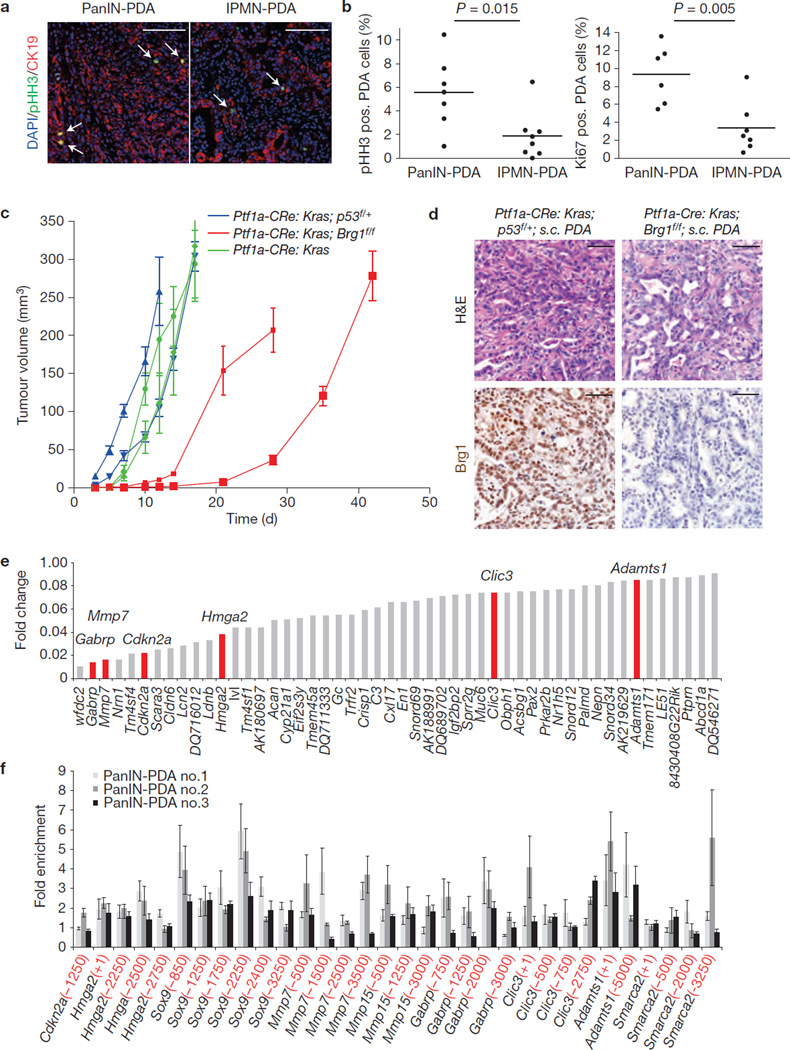
Given increased survival in IPMN–PDA mice compared with PanIN-PDA animals, despite nearly identical morphology and marker expression, we next determined whether tumour cell proliferation was different in PanIN- versus IPMN–PDA. Notably, the percentage of pHH3- or Ki67-positive cells in CK19-positive PDA cells of IPMN– PDA mice was significantly lower than that of PanIN-PDA mice (Fig. 6a,b).
Figure 6.
IPMN–PDA cells are intrinsically less proliferative than PanIN-PDA cells and carry a distinct molecular profile. (a) Co-staining for pHH3 and CK19 with DAPI of PanIN-PDA (Ptf1a–Cre; KrasG12D; p53f/+) and IPMN–PDA (Ptf1a–Cre; KrasG12D; Brg1f/f). Arrows indicate pHH3/CK19 double-positive proliferating cancer cells. (b) Percentage of pHH3- or Ki67-positive PDA cells of PanIN-PDA mice (n = 7 for pHH3 and n = 6 for Ki67) and IPMN–PDA mice (n = 8); unpaired _t_-test was performed to calculate P values. (c) Xenograft model of cancer cell lines derived from Ptf1a–Cre; KrasG12D; Brg1f/f (red lines: 2 independent cancer cell lines) compared with Ptf1a–Cre; KrasG12D; p53f/+ PDAs (blue lines: 2 independent cancer cell lines), and Ptf1a–Cre; KrasG12D PDAs (green lines: 2 independent cancer cell lines) in vivo. Calculated doubling times were 5.1 and 5.6 days for Ptf1a–Cre; KrasG12D; Brg1f/f versus 3.1 and 3.2 days for Ptf1a–Cre; KrasG12D; p53f/+ versus 3.0 and 4.3 days for Ptf1a–Cre; KrasG12D. For each cell line 8 subcutaneous tumours were quantified; values are shown as mean ± s.e.m. (d) H&E and Brg1 staining of subcutaneous tumours of xenograft model of cancer cell lines derived from Ptf1a–Cre; KrasG12D; Brg1f/f and Ptf1a–Cre; KrasG12D; p53f/+ PDAs. (e) Depicted are the 50 most downregulated genes identified by RNA deep sequencing analysis in IPMN–PDAs compared with PanIN-PDAs. (f) Chromatin immunoprecipitation analysis of PanIN-PDA cell lines detailing the association of Brg1 with promoter regions of Cdkn2a,Hmga2,Sox9,MMP7,MMP15,Gabrp,Clic3,Adamts1 and Smarca2. Values are expressed as fold enrichment over IgG control. Numbers in parentheses indicate upstream location of the promoter sequence with regard to the transcriptional start site (1×106 cells per chromatin immunoprecipitation; n = 4 independent experiments), values are shown as mean ± s.e.m. Scale bars: 100 µm (a); 50 µm (d).
To better define the characteristics of IPMN–PDA cells, we next established cancer cell lines derived from PDA that formed in IPMN– PDA, PanIN-PDA and Ptf1a–Cre; KrasG12D mice. PCR and western blot analyses confirmed efficient recombination and loss of Brg1 in cancer cell lines derived from IPMN–PDA tumours (Supplementary Fig. 2b). To examine the intrinsic differences between IPMN- and PanIN-PDA cells, cancer cell lines were subcutaneously transplanted into immunodeficient recipient mice. Although morphologically indistinguishable, tumours derived from Brg1-deficient IPMN–PDA cells grew significantly slower than those derived from PanIN-PDA or Ptf1a–Cre; _KrasG12D_-derived cancer cells (Fig. 6c,d). Interestingly, IPMN–PDA cells were resistant to anoikis, indicating that they were insensitive to anchorage-dependent cues provided by the surrounding extracellular matrix, compared with Ptf1a–Cre; _KrasG12D_-derived cancer cells (Supplementary Fig. 2c).morphologically indistinguishable, tumours derived from Brg1-deficient IPMN–PDA cells grew significantly slower than those derived from PanIN-PDA or Ptf1a–Cre; _KrasG12D_-derived cancer cells (Fig. 6c,d). Interestingly, IPMN–PDA cells were resistant to anoikis, indicating that they were insensitive to anchorage-dependent cues provided by the surrounding extracellular matrix, compared with Ptf1a–Cre; _KrasG12D_-derived cancer cells (Supplementary Fig. 2c).
The rapid development of IPMN–PDA prompted us to examine the expression levels of canonical tumour suppressor genes previously implicated in PDA progression in the context of oncogenic Kras30,39,40. Immunohistochemical analysis of tumour samples revealed that expression levels of p53, p21 and p16 were reduced or almost completely lost in IPMN lesions and IPMN–PDA (Supplementary Fig. 3a – d). These results suggest that altered tumour suppressor gene expression may be a distinguishing hallmark of early Brg1-deficient neoplastic lesions that could contribute to rapid tumour development in Ptf1a–Cre; KrasG12D; Brg1f/f mice.
Surprisingly, the lack of the tumour suppressor gene expression in Brg1-null PDAs does not result in the formation of aggressive cancer. To gain insights into the underlying molecular mechanisms by which PDA malignancy is restricted in IPMN–PDA mice, we performed deep sequencing analysis of RNA isolated from subcutaneous tumours derived from the IPMN- and PanIN-PDA cell lines (Fig. 6e and Supplementary Table 2). Several genes previously shown to support malignancy (including Mmp7, Gabrp, Hmga2, Clic3 and Adamts1) in PDA and other cancers were among the most downregulated genes in IPMN–PDA compared with PanIN-PDA (Fig. 6e and Supplementary Table 2)36,41–46. Analysis of IPMN and PanIN precursor lesions revealed a significant difference in Hmga2 already at these early stages (Supplementary Fig. 3e). Notably, additional pathway analysis revealed that Brg1-null IPMN–PDAs exhibited a decreased gene expression signature for those signalling cascades regulating cellular motility, invasion and metastasis (Supplementary Fig. 4 and Supplementary Table 3). Summarily, the deep sequencing data confirm the physiological differences between IPMN- and PanIN-derived PDAs and provide molecular support for the notion that IPMN–PDA possesses lower malignant potential in vivo. Of note, we also determined expression of GNAS and RNF43, genes recently implicated in human IPMN development47–50, but could not detect any significant changes in expression or the presence of single nucleotide polymorphisms/mutations in GNAS when comparing IPMN- versus PanIN-PDA.
We next analysed the contribution of Brg1 in differential gene regulation between PanIN-PDA and IPMN–PDA. To address this question, we determined Brg1 binding to evolutionary conserved regions within promoters of its target genes, starting with those genes previously implicated in PDA and differentially expressed between PanIN-PDA and IPMN–PDA (Fig. 6e and Supplementary Fig. 5). Chromatin immunoprecipitation revealed enrichment for Brg1 on several promoter regions (Fig. 6f). Next, we examined enrichment for H3K4Me3 (active mark) and H3K27Me3 (repressive mark) on these promoters and found enrichment for H3K27Me3 on the promoters of several genes (such as Hmga2 and Mmp7) whose expression was reduced in IPMN–PDA (Supplementary Fig. 6). Thus, the presence of repressive histone marks supports the attenuated expression of these genes in Brg1-depleted IPMN–PDA cells and points to the chromatin modifier as a critical epigenetic regulator of genes important for PanIN and IPMN initiation and progression.
Brg1 blocks IPMN formation from adult pancreatic duct cells but is critical for acinar-derived PanIN formation
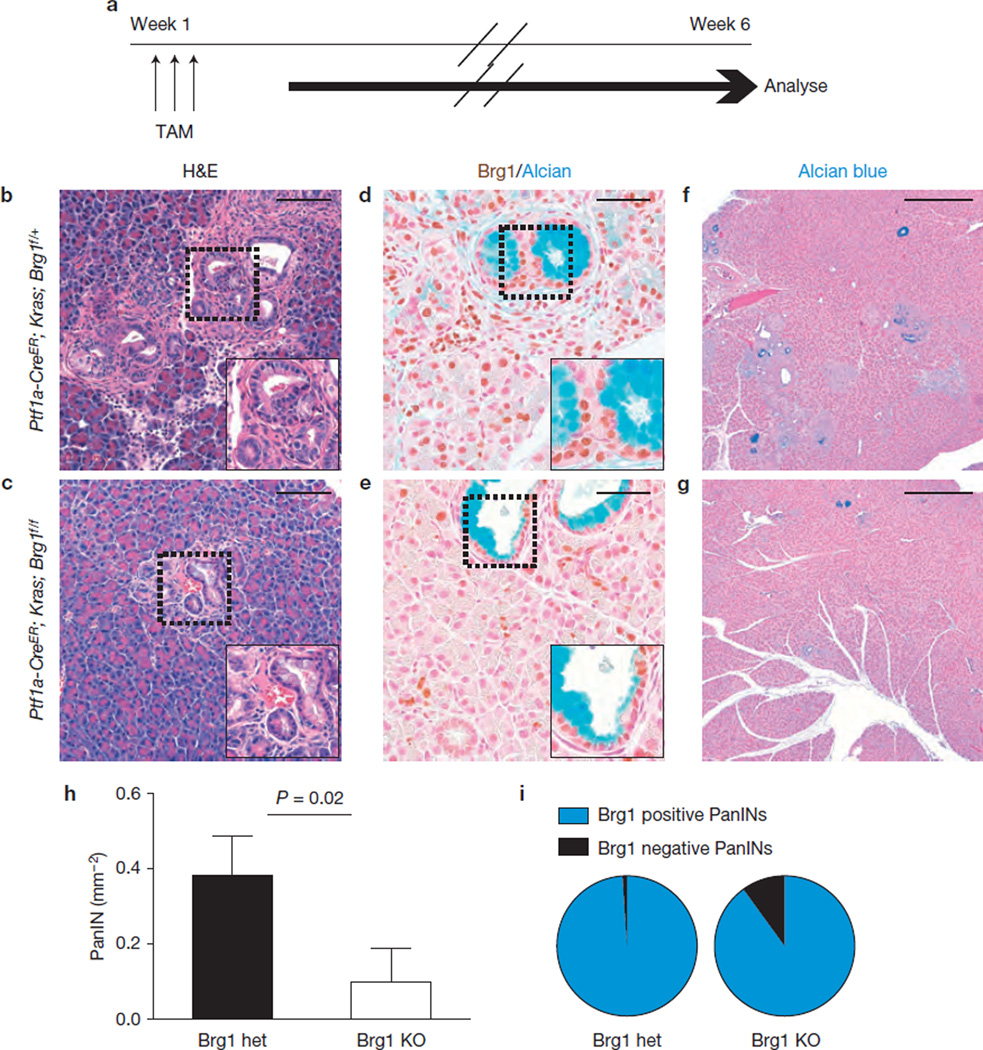
We next examined whether the striking biological differences between PanIN-PDA and IPMN–PDA development and progression might reflect a difference in the cellular origin of the cancer progenitors. We employed Ptf1a–CreER; KrasG12D; Brg1f/f mice permitting expression of mutant Kras and Brg1 deletion specifically in adult acinar cells upon tamoxifen treatment9,51. We predicted that preneoplastic lesions might arise 6 weeks after tamoxifen administration (Fig. 7a). Onmacroscopic and microscopic examination we could not detect any IPMN lesions (Fig. 7b,c). Moreover, the number of PanINs was significantly reduced in Ptf1a–CreER; KrasG12D; Brg1f/f compared with Ptf1a–CreER; KrasG12D; Brg1f/+ mice (Fig. 7b – h), a finding that was confirmed in pancreata isolated 4 months after tamoxifen treatment (Supplementary Fig. 7a). Of note, most of the few PanINs developing in Ptf1a–CreER; KrasG12D; Brg1f/f mice retained Brg1 expression (27 of 30 analysed lesions; Fig. 7i), strongly indicating that Brg1 plays a critical role in acinar-derived PanIN formation.
Figure 7.
Brg1 ablation abrogates PanIN formation from adult acinar cells. (a) Tamoxifen (TAM) administration scheme for experiments using Ptf1a–CreER mice. (b,c) H&E staining. (d,e) Brg1/Alcian blue staining. (f,g) Alcian blue staining. (h) Quantification of PanIN incidence in Ptf1a– CreER; Kras; Brg1f/+ (Brg1 het) and Ptf1a–CreER; Kras; Brg1f/f (Brg1 KO) mice 6 weeks after tamoxifen induction. n = 3 mice per group, 1 entire pancreas section per mouse analysed; values are shown as mean ± s.d., P values were calculated with the unpaired _t_-test. (i) Quantification of Brg1 positive/negative PanINs in Ptf1a–CreER; Kras; Brg1f/+ (Brg1 het; total of 97 PanINs examined) and Ptf1a–CreER; Kras; Brg1f/f (Brg1 KO; total of 30 PanINs examined) mice. Scale bars, 100 µm (b,c), 50 µm (d,e); 500µm (f,g).
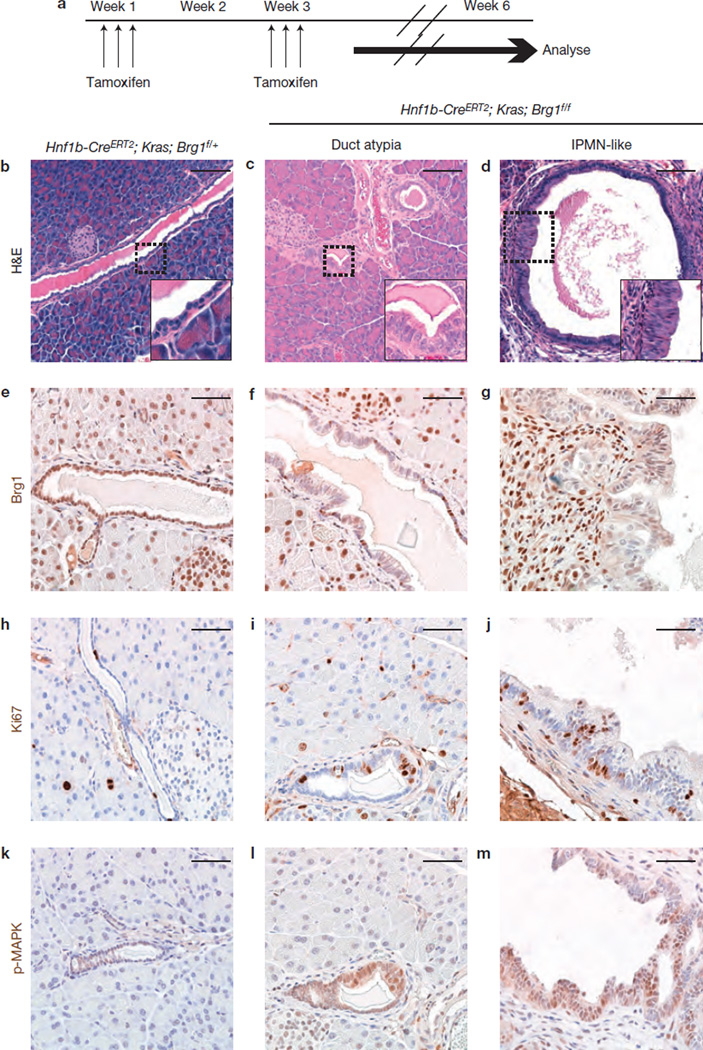
Next we examined, whether IPMN lesions can originate from adult pancreatic duct cells by using the tamoxifen-inducible Hnf1b–CreERT2 line that permits Cre recombination in adult duct cells52. Neither eliminationofBrg1 intheabsenceofoncogenic Kras (Hnf1b–CreERT2; Brg1f/f) nor deletion of only one allele in Hnf1b–CreERT2; KrasG12D; Brg1f/+ mutant mice revealed any signs of duct atypia 6 weeks after tamoxifen induction (Fig. 8a,b and Supplementary Fig. 7b,c). In contrast, atypical duct cells with columnar cell appearance were observed in a subset of pancreatic ducts of all (5/5) induced Hnf1b– CreERT2; KrasG12D; Brg1f/f mutant mice expressing oncogenic Kras (Fig. 8c,f). In addition, 1/5 Hnf1b–CreERT2; KrasG12D; Brg1f/f mice showed the formation of a mucinous dilated duct structure reminiscent of the IPMN lesions (Fig. 8d,g). Supporting the presence of a neoplastic epithelium, we observed increased proliferation and pronounced p-MAPK staining in Hnf1b–CreERT2; KrasG12D; Brg1f/f duct lesions (Fig. 8h – m). Therefore, Brg1 seems to modulate cell-type-specific responses to oncogenic Kras, representing a critical node for acinar to PanIN transformation, but an inhibitor of duct derived neoplasia.
Figure 8.
Brg1 ablation sensitizes adult duct cells to Kras-driven initiation of IPMN-like lesions. (a) Tamoxifen administration scheme for experiments using Hnf1b–CreERT2 mice. The tamoxifen treatment resulted in recombination of the Brg1 allele in most pancreatic duct cells as determined by the number of Brg1-negative duct cells in Hnf1b–CreERT2; KrasG12D; Brg1f/f mice. (b–m) H&E staining (b–d), Brg1 staining (e–g), Ki67 staining (h–j) and p-MAPK staining (k–m) of the indicated genotypes. Scale bars: 100 µm (b–d) and 50 µm (e–m).
Brg1 expression is associated with progression of human PanIN- and IPMN–PDA
Although our data point to Brg1 as an important factor for the transition of acinar cells towards PanIN formation, the gene was reported to be frequently mutated in established human PDA (refs 24,25). To determine whether Brg1 abrogation facilitates PDA progression once PanIN lesions have formed, we tested whether Brg1 expression impacts survival in human PanIN-PDA patients. Interestingly, low Brg1 expression associates with poorer patients’ survival (Supplementary Fig. 8a), suggesting that reduction in Brg1 function promotes PanIN-PDA cancer progression. To determine whether changes in Brg1 expression might also occur during human IPMN–PDA carcinogenesis, we compared IPMN lesions and their associated cancers. Notably, we found that Brg1 expression was higher in carcinoma compared with its precursor IPMN lesions (Supplementary Fig. 8b). In summary, these data indicate that Brg1 might have opposite roles during progression of human PanIN- versus IPMN–PDA.
DISCUSSION
A number of studies have implicated alterations in function and expression of SWI/SNF complexes in human cancer13,15–19,53. In particular, mutations in Brg1 and other members of the SWI/SNF complex are observed in over 30% of human PDA cases25, and decreased Brg1 expression is associated with IPMN precursor lesions26. Here, we show that Brg1 acts as a tumour suppressor in the pancreas in the context of mutant Kras. Brg1 loss seems to change the course of Kras-mediated transformation, promoting the development of mucinous, preneoplastic lesions with some distinguishing molecular attributes compared with Kras-driven PanINs, and the progression of these preneoplastic lesions to PDA. Surprisingly, this rapidly developing Brg1-null PDA is not as lethal as PanIN-derived PDA drivenbymutant Kras and heterozygous p53 loss.
These findings could be explained by a tumour-suppressive role of Brg1 that constrains the biology of Kras-driven IPMN–PDA development, but may be required for establishing the aggressive malignant behaviour characteristic of IPMN-derived PDA. Indeed, several studies have suggested a tumour-suppressive function of Brg1 by its regulation of p21 (refs 54–56) and p16 expression57and the SWI/SNF complex has been suggested to antagonize the Polycomb complex known to suppress p16 expression25. In line with these data, we found p21 and p16 decreased or lost in IPMN lesions and IPMN–PDA of Ptf1a–Cre; KrasG12D; Brg1f/f mice. However, despite the perturbations in p16 and p21, the IPMN–PDA is marked by reduced malignant potential. A possible explanation could be that Brg1 loss impairs the proliferative capacity of cancer cells. This is consistent with previous reports describing that the elimination of Brg1 results in aberrant chromatin organization, mitotic failure and reduced proliferative capacity27,58. Although defects in proliferative capacity can in part explain lower malignant potential, our RNA deep sequencing analysis from Brg1-null cancer cells also indicates impaired expression of targets important for tumour progression. Moreover, the epigenetic analysis revealed that Brg1 loss affects expression of a subset of these pro-tumorigenic factors by modulating their chromatin context.
In addition to the altered expression of cancer-promoting factors, the benign nature of the IPMN–PDA could be related to the fact that it seems to derive from a different progenitor cell than PanIN-PDA. This hypothesis is supported by a recent report that identified PanIN lesions to predominantly arise in the acinar compartment in mice9, whereas the occurrence of IPMN lesions in human patients within main and/or branch duct structures suggests that these lesions originate from the pancreatic ductal compartment. However, the origin of IPMN had not been experimentally proved owing to the lack of appropriate mouse models. Our observations demonstrate that duct cells lacking Brg1 are sensitive to oncogenic Kras-driven neoplastic transformation, and support the hypothesis that duct cells contribute to the formation of IPMN lesions. Notably, Brg1 deletion in adult duct cells did not result in the prominent cystic IPMN lesions observed when the embryonic Ptf1a–Cre line was used. The underlying reasons could be that a longer time period (>6 weeks) might be required for the IPMN lesions to form from adult duct cells or that embryonic duct cells might be more sensitive to neoplastic transformation than adult duct cells. In contrast to duct cells, our studies further suggest that Brg1 deletion inhibits acinar-derived PanIN formation, thus rendering acini less sensitive to Kras-driven neoplasia.
Our mouse model data point to Brg1 as a context-dependent mediator of Kras-driven tumorigenesis in the pancreas. Support for this notion comes from the expression analysis of human cancer tissues that point to opposing roles of Brg1 during IPMN- and PanIN-PDA progression. Future studies are needed to clarify which Brg1-regulated epigenetic and genetic changes contribute to the cell-type-specific effects, and how such differences determine the evolution and character of PDA arising from different preneoplastic lesions with different biological behaviour.
METHODS
Methods and any associated references are available in the online version of the paper.
METHODS
Mouse lines
Experimental animals were generated by crossing Ptf1a–Cre (gift from C. Wright, Vanderbilt University, Nashville, Tennessee, USA (ref. 28)), Ptf1a–CreER (gift from C. Wright, Vanderbilt University, Nashville, Tennessee, USA (ref. 51)) or Hnf1b–Cre _ERT_252 with KrasG12D (gift from D. Tuveson, Cancer Research, UK Cambridge Research Institute, Cambridge, UK)31, Brg1flox (gift from D. Reisman, University of Florida, with permission from P. Chambon)27, and p53flox mice30. Ptf1a–Cre− or Ptf1a–Cre+; Brg1+/+ mice served as a control. Mice were crossed on a mixed background and no selection for a specific gender was done in this study. The age and numbers of the mice are provided in the respective sections. Tamoxifen citrate (TEVA Pharmaceuticals) was administered by oral gavage at a dose of 10 mg per gavage. Time course and amount of gavages are outlined in the respective figure. The UCSF Institutional Care and Use of Animals Committee (IACUC) approved all mouse experiments.
Generation of mouse cancer cell lines
A part of the mouse PDA was cut into small pieces and further digested with collagenaseD (Roche) solution (2.5 mg ml−1). After washing with PBS, tissue suspension was further digested with trypsin-EDTA solution (0.05%). After washing, another digestion step was performed with dispase (Invitrogen) solution (2 U ml−1). Finally, cell suspension was passed through a 40 µm cell strainer and flow through and remnants in the filter were plated on collagen plates (BD BioCoat).
Subcutaneous tumour growth assay
Cells (1 × 106) of the respective cancer cell lines (passage number 10–20) were injected subcutaneously into immune-compromised Nod-SCID gamma mice. Mice were checked for the formation of tumours and tumour volume was determined by measuring tumour length and width (tumour volume = 1/2 × tumour length × (tumour width)2). Mice were euthanized and tumours were taken out when tumours started to show signs of ulceration.
Immunohistochemistry and immunofluorescence
Mouse tissue was fixed overnight in Z-FIX (Anatech), embedded in paraffin, and cut to 5 µm sections. Antigen retrieval was achieved by cooking in antigen retrieval citra solution (biogenex). For primary antibodies, incubation was performed overnight at 4 °C; secondary antibody incubation was for 1 h at room temperature. For immunohisto chemistry, slides were developed using either the ABC and DAB kit (both from Vectastain) or the Envision kit (DAKO) and counterstained with haematoxylin. In some cases, the M.O.M. kit (Vectastain) was used to reduce background for mouse primary antibodies. For immunofluorescence, slides were mounted with DAPI (Vectastain). Primary antibodies used in this study: rabbit anti-Brg1 (Santa Cruz catalogue number sc-10768; dilution 1:500), rabbit anti-amylase (Sigma catalogue number A8273; dilution 1:400), rat anti-CK19 (TROMAIII, Hybridoma Bank Iowa University; dilution 1:200), guinea pig anti-Insulin (Linco catalogue number 4011-01; dilution 1:400), rabbit anti-Sox9 (Millipore catalogue number AB5535; dilution 1:1,000–10,000), mouse anti-p21 (Santa Cruz F-5 catalogue number sc-6246; dilution 1:50), mouse anti-p16 (Santa Cruz F-12 catalogue number sc-1661; dilution 1:400-1,000), rabbit anti-p53 (Vectorlabs catalogue number VP-P956; dilution 1:500), mouse anti-Ki-67 (BD catalogue number 550609; dilution 1:400), Armenian hamster anti-mucin1 (Neomarkers Ab-5 MH-1; catalogue number HM-1630-P1; dilution 1:200), rabbit anti-mucin2 (Santa Cruz catalogue number sc-15334; dilution 1:200), mouse anti-Muc5ac (Neomarkers Ab-1 45M1; catalogue number MS-145-P0; dilution 1:200), rabbit anti-cleaved-caspase 3 (Cell Signaling catalogue number 9661; dilution 1:200), rabbit anti-ER-alpha (Millipore catalogue number 06–935; dilution 1:10,000), rabbit anti-progesteron receptor (Lab vision SP2; catalogue number RM-9102-S0; dilution 1:1,000–10,000), mouse anti-E-cadherin (BD catalogue number 610182; dilution 1:200), rabbit anti-P44/P42-MAPK (Cell Signaling 20G11; catalogue number 4376; dilution 1:200), rabbit anti-Hes1 (gift from T Sudo, Toray, Japan; dilution 1:400), rabbit anti-Pdx1 (Millipore catalogue number 07–696; dilution 1: 400), mouse anti-vimentin (Sigma catalogue number V2258; dilution 1:200) and rabbit anti-phospho-Stat3 (Cell Signaling D3A7; catalogue number 9145; dilution 1:200).
Analysis of PDA proliferation was performed using InCell Analyzer 2000 (GE Healthcare). Co-immunofluorescence images of DAPI, CK19 and either pHH3 or Ki67 were analysed using the Incell Analyzer Developer Toolbox. For quantification of proliferation in PDAs on average 14,328 ± 10,739 cells were analysed per individual PDA.
Quantification of p16-positive cells was performed on tumour samples stained by immunohistochemistry using the M.O.M kit (Vectastain). Three random high-power magnification fields per tumour sample were analysed (n = 7 individual tumour samples per genotype).
RNA isolation and quantitative real-time PCR
RNA was isolated using the RNA-easy kit (Quiagen). Complementary DNA was synthesized using the Superscript III First-Strand Synthesis Kit (Invitrogen). Quantitative PCR was performed with the Taqman Gene Expression Assays or SYBR-green-based gene expression assays (Applied Biosystems). Expression levels were normalized against mouse cyclophilin A (Genome Analysis Core, UCSF Helen Diller Family Comprehensive Cancer Center).
Primer sequence: Hmga2, forward: 5′-GAGCCCTCTCCTAAGAGACCC-3′; reverse: 5′-TTGGCCGTTTTTCTCCAATGG-3′.
RNA deep sequencing
One microgram of total pooled RNA isolated from 3 individual subcutaneous tumours generated by transplantation of cancer cell lines derived from each 2 Ptf1a–Cre; KrasG12D; p53f/+ and 2 Ptf1a–Cre; KrasG12D; Brg1f/f PDAs was used to generate libraries with the Illumina TruSeq RNA sample preparation kit. Each library was diluted to approximately 10 pM before loading and sequenced using a Hi-seq 2000 instrument generating 50 base pair reads. This produced 199,902,826 reads from Ptf1a–Cre; KrasG12D; p53f/+ tumours and 185,962,412 reads from Ptf1a–Cre; KrasG12D; Brg1f/f tumours. The 50-base-pair (bp) reads were trimmed by five bases at the 5′ end owing to quality scores. The trimmed 45-bp reads were then mapped to the mouse genome (NCBI37/mm9) and exon-exon junctions using TopHat59 version 1.3.1 with default parameters. Genome information was downloaded from the UCSC Genome Browser60. We filtered TopHat alignments and kept reads (and their locations) that could be mapped uniquely. Per gene, we extracted read counts by using samtools61 version 0.1.13. Read counts were normalized to the length of the gene and the total number of mapped reads to obtain a standardized count (RPKM, reads per kilobase of exon model per million mapped reads). Fisher’s exact test was used to search for differentially expressed genes. P values were calculated by comparing the normalized expression level of each gene with the normalized expression level of several housekeeping genes (Actb, Eif2a and Rpl23). The results of the expression comparison are summarized in Supplementary Table 2.
Western blotting
Whole-cell extracts were prepared according to standard protocols using RIPA buffer (Cell Signaling) with PhoSTOP (Roche) and complete mini (Roche). Membranes were incubated overnight with the following primary antibodies: rabbit anti-Brg1 (Santa Cruz sc-10768; dilution 1:1,000) and mouse anti-GAPDH (Santa Cruz sc-32233; dilution 1:2,000). After incubation with the respective HRP-conjugated secondary antibodies for 1 h, membranes were developed using the Supersignal West Pico solution (Thermo).
Genotyping of cancer cell lines
To examine the recombination of the Kras_G12D_ allele a previously published protocol (http://web.mit.edu/jacks-lab/protocols/KrasCond_tablesTWO.html) was used. To determine the recombination of the Brg1 allele the following primers were used as previously reported27: TH185 5′-GTCATACTTATGTCATAGCC-3′, TB82 5′-GATCAGCTCATGCCCTAAGG-3′, and TG57 5′-GCCTTGTCTCAAACTGATAAG-3′.
Anoikis assay
Cells were trypsinized and 1 × 105 cells were seeded onto Poly-Hema (Sigma, catalogue number: P3932)-coated 6-well plates. At 48 h, cells were collected and analysed by Annexin-V/PI staining (Life Technologies, Catalogue number: V13241) followed by FACS.
Chromatin immunoprecipitation
Cells were first crosslinked with 37% formaldehyde (Fisher) to a final concentration of 1% and incubated for 10 min with gentle swirling at room temperature. Crosslinking was stopped by addition of 2.5 M glycine at a final concentration of 0.125 M glycine for 5 min with gentle swirling. Cells were washed twice with ice-cold sterile PBS and then collected by adding 1 ml of ice-cold sterile PBS containing protease inhibitors (Roche). Cells were collected and centrifuged at 2,440_g_ for 5 min. The cell pellet was then re-suspended in sodium dodecyl sulphate (SDS) lysis buffer (1% SDS, 10 mM EDTA and 50 mM Tris, at pH 8.1) and placed on ice for 10 min. Extract was sonicated using a Diagenode Bioruptor, precleared, and immunoprecipitation was carried out with 2 µg of antibody (Brg1, Santa Cruz; H3K4Me3, Millipore; H3K27Me3, Cell Signaling and immunoglobulin G (IgG), Santa Cruz). Crosslinks were reversed on all samples followed by RNase A treatment and proteinase K digestion. DNA was extracted from the digested samples using a PCR purification kit (Qiagen). Extracted DNA was amplified by real-time PCR.
The following primer sequences were used. Cdkn2a: region 1: (−1250 bp); forward: 5′-GGTTCCGAGAGCCTTTATCC-3′; reverse: 5′-CATCCTCGGAAAA GCTGAAC-3′.
Hmga2
region1: (+1 bp); forward: 5′-CATTCCATGTCAACTTGTGC-3′; reverse: 5′-CCGTGGTGTGTGTGTTCAGT-3′; region2: (−2250 bp); forward: 5′-ATCCCAGGCTTCCACTAGGT-3′; reverse: 5′-TGTAAGGGACCACAGAAG AGA-3′; region3: (−2500 bp)-3′; forward: 5′-TCTCAAGAGCCAATGCTCAA-3′; reverse: 5′-GGATGCAAACCAGACCACA-3′; region4: (−2750 bp); forward: 5′-ACAGAGGAATGCCTGCCTTA-3′; reverse: 5′-CTACGGCTCCTGTGTCAGC-3′.
Sox9
region1: (−850 bp); forward: 5′-CAGGAGGCAAGAAGCAGAAC-3′; reverse: 5′-GGGGAATCAATGAAAACCAA-3′; region2: (−1250 bp); forward: 5′-TTGTGAGTTTGTTTGTGGTTCC-3′; reverse: 5′-CACTTAGGTGTCCGTT GCAC-3′; region3: (−1750 bp); forward: 5′-ACGAGTTCCTTTCCCCAAAT-3′; reverse: 5′-TAAAGCGAATCGGCCTGTAT-3′; region4: (−2250 bp); forward: 5′-TCGCAGCACATTATATCTCTCC-3′; reverse: 5′-TGTAATTATCACGTATTCTG GCACA-3′; region5: (−2400 bp); forward: 5′-GACCAGTTTCTGGGCTTTTG-3′; reverse: 5′-TCTTTTCGGGGAGAAACTATT-3′; region6: (−3250 bp); forward: 5′-GCCTGCCCAGGCTTACCT-3′; reverse: 5′-TCCGTGATTTCCATTTTTCG-3′.
Mmp7
region1: (−500 bp); forward: 5′-TGAGAACAATCGCCTGTTGA-3′; reverse: 5′-CAGAGAACCCCGAGTGAGAG-3′; region2: (−1500 bp); forward: 5′-TTTCCCTGGTAAGTGGAACG-3′; reverse: 5′-CCACAGGCTGGGAGAAAATA-3′; region3: (−2500 bp); forward: 5′-CCTGAGTAGCAGCCACTTCC-3′; reverse: 5′-TCCACAGGTGAAGGGATAGG-3′; region3: (−3500 bp); forward: 5′-CGTC ATTGTCACAGGCAAAC-3′; reverse: 5′-TGTGTCTCAGTGTCCTTTCCA-3′.
Mmp15
region1: (−500 bp); forward: 5′-AGCATAGGGTCGAGTCATGG-3′; reverse: 5′-TCTTAAAGGGCCAGTGTGCT-3′; region2: (−1250 bp); forward: 5′-CATCTCCTTTCCTCGAGCTG-3′; reverse: 5′-CTTGAGAACCAGGGCAGAAG-3′; region3: (−3000 bp); forward: 5′-ATGTAAGCCGTGCCTAAGGA-3′; reverse: 5′-GAACAGTTCTTCCCCGAACA-3′.
Gabrp
region1: (−750 bp); forward: 5′-TCCTCAGCTTGTGGACTGC-3′; reverse: 5′-TCCATGGAGGGAGTTGTGTA-3′; region2: (−1250 bp); forward: 5′-CCTTGGAATGGTACAGAAATCA-3′; reverse: 5′-TGCAGAGGTAGGTGGGT CTC-3′; region3: (−2000 bp); forward: 5′-CCTCTGCCTCTTGAGTCTGG-3′; reverse: 5′-GCATCTGGTGCCTTTTCCTA-3′; region4: (−3000 bp); forward: 5′-A AGGACGTTTCCAGATGGTG-3′; reverse: 5′-TTTGTAAGGGACAGGCTGCT-3′.
Clic3
region1: (+1 bp); forward: 5′-GGTAGCACGGACACTCTGGT-3′; reverse: 5′-GCCCAGAACCTTCACAAAGA-3′; region2: (−500 bp); forward: 5′-CTGAGGCATTTCCCACTCAT-3′; reverse: 5′-TGGTTGTGAGGGGAGCTAGT-3′; region3: (−750 bp); forward: 5′-CATCCCCATTTCAGCCACTA-3′; reverse: 5′-GATAAGCCTGGTTTTGGTGTTT-3′; region4: (−2750 bp); forward: 5′-CAC GGTGAAATGCACCTTTA-3′; reverse: 5′-TGGCTCATGTCTCTTTCCTTG-3′.
Adamts1
region1: (+1 bp); forward: 5′-ACTGGTGAGGGAGGCAAATA-3′; reverse: 5′-AAAACAAATGGTGGGCCATA-3′; region2: (−5000 bp); forward: 5′-CCACAAGAGCTGATTCTTTCAG-3′; reverse: 5′-TTTTCCAGTTTTGTCA AGTAGGC-3′.
Smarca2
region1: (+1 bp); forward: 5′-TAAATTCCAGCACGGTCTCC-3′; reverse: 5′-ATGCGTGGATCTGTGTGTGT-3′; region2: (−500 bp); forward: 5′-CAAAAGCAGATCCCATGGTT-3′; reverse: 5′-GCTTCCAAATGCCAGAGAAG-3′; region3: (−2000 bp); forward: 5′-AATTGGGCTTAGCGTTTCTT-3′; reverse: 5′-TGGCTTCCATGTTTTCTGTG-3′; region4: (−3250 bp); forward: 5′-TGCT GCAGCCTTTTATTTCTC-3′; reverse: 5′-ACTTGGCTCCCTTTCACCTC-3′.
Statistical analysis
To determine P values, Student’s _t_-test was performed (unless indicated differently). A P value <0.05 was assumed as statistically significant. For survival curves, Kaplan-Meyer analysis was used. Doubling times for cell lines were calculated by exponential growth function in GraphPad Prism. All statistical analysis were performed with either GraphPad Prism (Version 4.0c) or Microsoft Excel. No statistical method was used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Accession numbers
The gene expression omnibus accession number for the RNA deep sequencing data reported in this paper is GSE52332.
Repeatability of experiments
The following figures show representative images: Fig. 1(a–i), Fig. 2(a–r), Fig. 3(a–i), Fig. 4(a–o), Fig. 5(a–l), Fig. 6(a,d), Fig. 7(b–g), Fig. 8(b–m), Supplementary Fig. 1, Supplementary Fig. 2 (a,b), Supplementary Fig. 3 (a,b), Supplementary Fig. 7. The images are representative of experiments performed on at least 2 mice per group (except for the IPMN-like lesion in Fig. 8 that was present in only 1 (out of 5) mouse).
Supplementary Material
01
ACKNOWLEDGEMENTS
We thank C. Wright for sharing Ptf1a–Cre and Ptf1a–CreER mice, D. Tuveson for KrasG12D mice, and P. Chambon and D. Reisman for Brg1flox mice, respectively. We thank C. Austin and D. Ngow for tissue processing and excellent technical assistance and all M.H. laboratory members for helpful discussion. Work in M.H.’s laboratory was supported by a grant from the NIH (CA112537). G.v.F. was supported by a postdoctoral Research Fellowship from the Deutsche Forschungsgemeinschaft (DFG, FI 1719/1-1) and a Klein Family Foundation Fellowship. A.F. was supported by a post-doctoral Research Fellowship from the Japan Society for the Promotion of Science, a Fellowship from the US National Pancreas Foundation, and a Fellowship from the Kato Memorial Biosciences Foundation. M.E.L. was supported by CIRM training grant TG2 01153. W.F.M., A.B. and K.J.H. were supported by a grant from the NIH (CA149548). Image acquisition was supported by the imaging core of the UCSF Diabetes and Endocrinology Research Center (DERC) NIH grant P30DK63720.
Footnotes
AUTHOR CONTRIBUTIONS
G.V.F., A.F., N.R. and M.E.L. contributed to equal parts. G.V.F., A.F., N.R. and M.E.L. carried out all experiments and were involved together with M.H. in design and analysis of the experiments. G.V.F., A.F., N.R., M.E.L. and M.H. drafted the manuscript. J.PM.IV generated cell lines, contributed to the survival analysis, was involved in experimental analysis, and critically reviewed the manuscript. G.E.K. performed the histopathological analysis including IPMN and tumour identification and tumour grading. H.R. performed quantification of tumour proliferation. J.F. generated the HNF1b–Cre _ERT_2 mice. D.W.D. analysed Brg1 expression on human samples. M.A.F., S.J.M. and J.F. provided intellectual contribution to this study. M.F.W., A.B. and K.J.H. carried out deep sequencing analyses. M.H. conceived the study.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Matthaei H, Schulick RD, Hruban RH, Maitra A. Cystic precursors to invasive pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2011;8:141–150. doi: 10.1038/nrgastro.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthaei H, et al. Clinicopathological characteristics and molecular analyses of multifocal intraductal papillary mucinous neoplasms of the pancreas. Ann. Surg. 2012;255:326–333. doi: 10.1097/SLA.0b013e3182378a18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poultsides GA, et al. Histopathologic basis for the favorable survival after resection of intraductal papillary mucinous neoplasm-associated invasive adenocarcinoma of the pancreas. Ann. Surg. 2010;251:470–476. doi: 10.1097/SLA.0b013e3181cf8a19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mino-Kenudson M, et al. Prognosis of invasive intraductal papillary mucinous neoplasm depends on histological and precursor epithelial subtypes. Gut. 2011;60:1712–1720. doi: 10.1136/gut.2010.232272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi C, Hruban RH. Intraductal papillary mucinous neoplasm. Hum. Pathol. 2012;43:1–16. doi: 10.1016/j.humpath.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Ray KC, et al. Epithelial tissues have varying degrees of susceptibility to Kras(G12D)-initiated tumorigenesis in a mouse model. PLoS One. 2011;6:e16786. doi: 10.1371/journal.pone.0016786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 8.Habbe N, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad. Sci. USA. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopp JL, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris JPt, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat. Rev. Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Figura G, Morris JPt, Wright CV, Hebrok M. Nr5a2 maintains acinar cell differentiation and constrains oncogenic Kras-mediated pancreatic neoplastic initiation. Gut. 2013 doi: 10.1136/gutjnl-2012-304287. http://dx.doi.org/10.1136/gutjnl-2012-304287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna ES, Roberts CW. Epigenetics and cancer without genomic instability. Cell Cycle. 2009;8:23–26. doi: 10.4161/cc.8.1.7290. [DOI] [PubMed] [Google Scholar]
- 13.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 14.Medina PP, Sanchez-Cespedes M. Involvement of the chromatin-remodelling factor BRG1/SMARCA4 in human cancer. Epigenetics. 2008;3:64–68. doi: 10.4161/epi.3.2.6153. [DOI] [PubMed] [Google Scholar]
- 15.Versteege I, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 16.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, et al. Inactivating mutations of the chromatin remodelling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gui Y, et al. Frequent mutations of chromatin remodelling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumour suppressor gene Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- 22.Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumours in mice. Proc Natl Acad. Sci. USA. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glaros S, Cirrincione GM, Palanca A, Metzger D, Reisman D. Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 2008;68:3689–3696. doi: 10.1158/0008-5472.CAN-07-6652. [DOI] [PubMed] [Google Scholar]
- 24.Jones S, et al. Core signalling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.E Shain AH, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumour suppressive complex in pancreatic cancer. Proc Natl Acad. Sci. USA. 2012;109:252–259. doi: 10.1073/pnas.1114817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dal Molin M, et al. Loss of expression of the SWI/SNF chromatin remodelling subunit BRG1/SMARCA4 is frequently observed in intraductal papillary mucinous neoplasms of the pancreas. Hum. Pathol. 2012;43:585–591. doi: 10.1016/j.humpath.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2β-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol. Cell Biol. 1997;17:5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawaguchi Y, et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 29.Heiser PW, Lau J, Taketo MM, Herrera PL, Hebrok M. Stabilization of β-catenin impacts pancreas growth. Development. 2006;133:2023–2032. doi: 10.1242/dev.02366. [DOI] [PubMed] [Google Scholar]
- 30.Bardeesy N, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad. Sci. USA. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 32.Izeradjene K, et al. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11:229–243. doi: 10.1016/j.ccr.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 33.Siveke JT, et al. Concomitant pancreatic activation of Kras(G12D) and Tgfa results in cystic papillary neoplasms reminiscent of human IPMN. Cancer Cell. 2007;12:266–279. doi: 10.1016/j.ccr.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Morris JPt, Cano DA, Sekine S, Wang SC, Hebrok M. β-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J. Clin. Invest. 2010;120:508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka T, et al. Evaluation of SOX9 expression in pancreatic ductal adenocarcinoma and intraductal papillary mucinous neoplasm. Pancreas. 2012;42:488–493. doi: 10.1097/MPA.0b013e318269d281. [DOI] [PubMed] [Google Scholar]
- 36.Fukuda A, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lesina M, et al. Stat3/Socs3 activation by IL-6 transsignalling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Corcoran RB, et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011;71:5020–5029. doi: 10.1158/0008-5472.CAN-11-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aguirre AJ, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morton JP, et al. LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology. 2010;139:586–597. doi: 10.1053/j.gastro.2010.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takehara A, et al. Gamma-aminobutyric acid (GABA) stimulates pancreatic cancer growth through overexpressing GABAA receptor pi subunit. Cancer Res. 2007;67:9704–9712. doi: 10.1158/0008-5472.CAN-07-2099. [DOI] [PubMed] [Google Scholar]
- 42.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 43.Winslow MM, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piscuoglio S, et al. HMGA1 and HMGA2 protein expression correlates with advanced tumour grade and lymph node metastasis in pancreatic adenocarcinoma. Histopathology. 2012;60:397–404. doi: 10.1111/j.1365-2559.2011.04121.x. [DOI] [PubMed] [Google Scholar]
- 45.Dozynkiewicz MA, et al. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev. Cell. 2012;22:131–145. doi: 10.1016/j.devcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masui T, et al. Expression of METH-1 and METH-2 in pancreatic cancer. Clin Cancer Res. 2001;7:3437–3443. [PubMed] [Google Scholar]
- 47.Wu J, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci. Trans. Med. 2011;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J, et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc. Nat. Acad. Sci. USA. 2011;108:21188–21193. doi: 10.1073/pnas.1118046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furukawa T, et al. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci. Rep. 2011;1:161. doi: 10.1038/srep00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanda M, et al. Mutant GNAS detected in duodenal collections of secretin-stimulated pancreatic juice indicates the presence or emergence of pancreatic cysts. Gut. 2013;62:1024–1033. doi: 10.1136/gutjnl-2012-302823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan FC, et al. Spatiotemporal patterns of multipotentiality in Ptf1a–expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development. 2013;140:751–764. doi: 10.1242/dev.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solar M, et al. Pancreatic exocrine duct cells give rise to insulin-producing β cells during embryogenesis but not after birth. Dev. Cell. 2009;17:849–860. doi: 10.1016/j.devcel.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, et al. Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumours. Int. J. Cancer. 2004;112:636–642. doi: 10.1002/ijc.20450. [DOI] [PubMed] [Google Scholar]
- 54.Kang H, Cui K, Zhao K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol. Cell Biol. 2004;24:1188–1199. doi: 10.1128/MCB.24.3.1188-1199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan B, Wang TL, Shih Ie M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodelling, is a tumour suppressor in gynecologic cancers. Cancer Res. 2011;71:6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hendricks KB, Shanahan F, Lees E. Role for BRG1 in cell cycle control and tumour suppression. Mol. Cell Biol. 2004;24:362–376. doi: 10.1128/MCB.24.1.362-376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b–ARF-INK4a locus. Mol. Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bourgo RJ, et al. SWI/SNF deficiency results in aberrant chromatin organization, mitotic failure, and diminished proliferative capacity. Mol. Biol. Cell. 2009;20:3192–3199. doi: 10.1091/mbc.E08-12-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01