Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin‐binding IFT‐B2 complex (original) (raw)
Abstract
Intraflagellar transport (IFT) relies on the IFT complex and is required for ciliogenesis. The IFT‐B complex consists of 9–10 stably associated core subunits and six “peripheral” subunits that were shown to dissociate from the core structure at moderate salt concentration. We purified the six “peripheral” IFT‐B subunits of Chlamydomonas reinhardtii as recombinant proteins and show that they form a stable complex independently of the IFT‐B core. We suggest a nomenclature of IFT‐B1 (core) and IFT‐B2 (peripheral) for the two IFT‐B subcomplexes. We demonstrate that IFT88, together with the N‐terminal domain of IFT52, is necessary to bridge the interaction between IFT‐B1 and B2. The crystal structure of IFT52N reveals highly conserved residues critical for IFT‐B1/IFT‐B2 complex formation. Furthermore, we show that of the three IFT‐B2 subunits containing a calponin homology (CH) domain (IFT38, 54, and 57), only IFT54 binds αβ‐tubulin as a potential IFT cargo, whereas the CH domains of IFT38 and IFT57 mediate the interaction with IFT80 and IFT172, respectively. Crystal structures of IFT54 CH domains reveal that tubulin binding is mediated by basic surface‐exposed residues.
Keywords: cilia, IFT54, IFT‐B, intraflagellar transport, tubulin transpor
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton
Introduction
Cilia are microtubule (MT)‐based appendages protruding from the surface of many different cell types (Fliegauf et al, 2007). To build and maintain a functional cilium, cells rely on intraflagellar transport (IFT; for general reviews, see Ishikawa & Marshall, 2011; Pedersen & Rosenbaum, 2008; Nachury et al, 2010), a bidirectional motility along the MTs of the ciliary axoneme (Kozminski et al, 1993). IFT in Chlamydomonas reinhardtii is dependent on the heterotrimeric kinesin II motor to transport ciliary cargo from the cell body to the tip of the cilium (anterograde IFT) (Kozminski et al, 1995; Cole et al, 1998; Qin et al, 2004; Hao et al, 2011; Craft et al, 2015), but in C. elegans and most likely also vertebrate cilia the heterotrimeric kinesin II cooperates with a homodimeric kinesin II motor for anterograde transport (Snow et al, 2004; Williams et al, 2014; Prevo et al, 2015). At the ciliary tip, exchange of the cargo and remodeling of the IFT complex take place and dynein 2 transports the IFT complex and recycling products back to the cell body (retrograde IFT) (Pazour et al, 1999; Porter et al, 1999; Signor et al, 1999; Qin et al, 2004). IFT is mediated by the IFT complex containing at least 22 protein subunits (organized into IFT‐A and IFT‐B subcomplexes), which likely mediates the interactions between IFT motors and ciliary cargo (Bhogaraju et al, 2013b). Despite the fact that > 600 different proteins function inside the cilium and in the ciliary membrane (Pazour et al, 2005), only little is known about cargo interaction sites within the IFT complex. However, several ciliary proteins have been shown to move inside cilia with the velocity of IFT and thus likely associate with IFT complexes. These include tubulin, the main axonemal building block, which is a cargo for IFT in both Chlamydomonas reinhardtii (Cr) (Craft et al, 2015) and C. elegans (Hao et al, 2011). In vitro studies suggested that tubulin cargo directly binds to the IFT complex via a dedicated module consisting of a calponin homology (CH) domain of IFT81 and a highly basic domain of IFT74 (Bhogaraju et al, 2013a). However, given that tubulin is the most abundant ciliary protein, it has been suggested that additional tubulin‐binding sites may exist within the IFT complex (Bhogaraju et al, 2014). Other axonemal components known to travel by IFT in Chlamydomonas are the nexin–dynein regulatory complex proteins DRC2 and DRC4 and the central pair protein PF16 (Wren et al, 2013). Outer dynein arms (ODAs) are also likely cargoes for IFT via interaction with IFT46 through the adaptor protein ODA16 (Hou et al, 2007; Ahmed et al, 2008). Several subunits of the BBSome, a cargo adapter for membrane proteins, have also been demonstrated to move by IFT (Blacque et al, 2004; Lechtreck et al, 2009a) as have several integral and associated membrane proteins (Qin et al, 2005; Huang et al, 2007).
The IFT complex organizes into a 6 subunit IFT‐A and a 16 subunit IFT‐B subcomplex that separate in sucrose gradients even at low salt concentrations (Piperno & Mead, 1997; Cole et al, 1998; Lucker et al, 2005; Omori et al, 2008; Follit et al, 2009; Lechtreck et al, 2009b; Fan et al, 2010; Taschner et al, 2011; Ishikawa et al, 2014). Nine of the IFT‐B subunits (IFT88/81/74/70/52/46/27/25/22) were shown to assemble into a stable IFT‐B core (Lucker et al, 2005; Taschner et al, 2014). IFT56 was recently characterized as an IFT‐B complex protein involved in the transport of axonemal motility factors (Ishikawa et al, 2014), and yeast‐2‐hybrid and pull‐down analyses suggest that it may associate with the IFT‐B core complex (Swiderski et al, 2014). The six remaining IFT‐B proteins (IFT172, IFT80, IFT57, IFT54, IFT38, and IFT20) are referred to as “peripheral” subunits as they were shown to dissociate from the core at 300 mM NaCl concentration (Lucker et al, 2005). Despite the label as peripheral, several lines of evidence in the literature suggest that these IFT‐B subunits are as important for IFT and ciliogenesis as the IFT‐B core factors. Mutations in the C. elegans homologs of IFT172 (OSM‐1), IFT80 (CHE‐2), IFT57 (CHE‐13), IFT54 (DYF‐11), and IFT38 (DYF‐3) lead to severe defects in the formation of sensory cilia and associated phenotypes such as deficiencies in dye‐filling, chemosensation, or osmotic avoidance (Perkins et al, 1986; Fujiwara et al, 1999; Haycraft et al, 2003; Murayama et al, 2005; Bacaj et al, 2008; Kunitomo & Iino, 2008; Li et al, 2008). In the ciliate Tetrahymena thermophila, knockout of IFT172 or depletion of IFT80 leads to strong ciliary assembly defects (Beales et al, 2007; Tsao & Gorovsky, 2008), and mutations or depletions of IFT80, IFT172, IFT57, IFT54 (elipsa), and IFT38 (qilin) disrupt ciliogenesis and lead to cystic kidneys and curled body axis in zebrafish (Sun et al, 2004; Beales et al, 2007; Omori et al, 2008; Lunt et al, 2009). Most importantly, knockout mice for IFT172 (wimple), IFT57 (Hippi), IFT54 (Traf3IP/MIP‐T3), IFT38 (Cluap1), IFT20, and IFT80 display embryonic lethality highlighting their essential roles in ciliogenesis in mammals (Huangfu et al, 2003; Houde et al, 2006; Jonassen et al, 2008; Berbari et al, 2011; Rix et al, 2011; Pasek et al, 2012). How the peripheral IFT‐B components associate with the core to form a functional IFT‐B complex is currently unknown.
Here, we purified the six peripheral IFT‐B complex proteins of Chlamydomonas to show that they interact to form a stable subcomplex (IFT‐B2). We map the contacts within IFT‐B2 using direct protein–protein interaction methodology and show that the CH domains of IFT38 and IFT57 are responsible for the association of IFT80 and IFT172, respectively. In contrast, the CH domain of IFT54, of which we present high‐resolution crystal structures, associates with tubulin/MT via conserved basic residues and may constitute a second tubulin‐binding site within the IFT complex. Finally, we demonstrate that IFT57/38 within the IFT‐B2 complex mediates the contacts to the IFT‐B core (IFT‐B1) through association with IFT88 and the N‐terminal domain of IFT52. Crystal structures of the IFT52 N‐terminal domain allow us to pinpoint critical residues required for IFT‐B subcomplex association. The data allow us to present an architectural map of IFT‐B complex assembly.
Results
IFT38/Cluap1 interacts directly with IFT57 to form a heterodimer
IFT38 is an IFT complex protein also known as FAP22 (C. reinhardtii), DYF‐3 (C. elegans), Cluap1 (mammals), Qilin (D. rerio), and PIFTA1 (T. brucei) (Subota et al, 2014). Although IFT38 was not identified in the pioneering characterizations of the IFT‐B complex (Piperno & Mead, 1997; Cole et al, 1998), several lines of evidence suggest that IFT38 is a bona fide IFT‐B complex protein. IFT38 localizes to the cilium and is required for ciliogenesis in a wide range of organisms, and mutations in IFT38 cause cystic kidneys in zebrafish and result in a loss of hedgehog signaling and embryonic lethality in mice (Sun et al, 2004; Stolc et al, 2005; Absalon et al, 2008). Furthermore, IFT38 was shown to undergo IFT in C. elegans (Ou et al, 2005) and co‐precipitated along with other IFT‐B proteins from zebrafish cell lysates using TAP‐IFT54 as a bait (Omori et al, 2008). To investigate how this protein associates with the IFT complex, we co‐expressed _Cr_IFT38 with different IFT‐B components resulting in soluble co‐expression only with the IFT57 subunit (Fig 1A and data not shown). Although IFT38 is insoluble when over‐expressed alone and IFT57 alone partially aggregates, the co‐expressed IFT57/38 complex is highly soluble; co‐purified during affinity, ion‐exchange chromatography, and size‐exclusion chromatography (SEC); and sedimented as a hetero‐dimeric complex in analytical ultracentrifugation (MW of 81 kDa vs. a theoretical MW of 93 kDa, see Appendix Fig S1A). Furthermore, pull‐downs from C. reinhardtii lysates using GST‐tagged IFT20 as bait identified IFT38/FAP22 along with several other IFT‐B proteins, demonstrating that IFT38 is a component of the IFT‐B complex in the green alga (Appendix Fig S1B). We conclude that IFT38 associates with the IFT‐B complex through a direct interaction with IFT57.
Figure 1. IFT172, 80, 57, 54, 38, and 20 form an IFT‐B2 subcomplex.
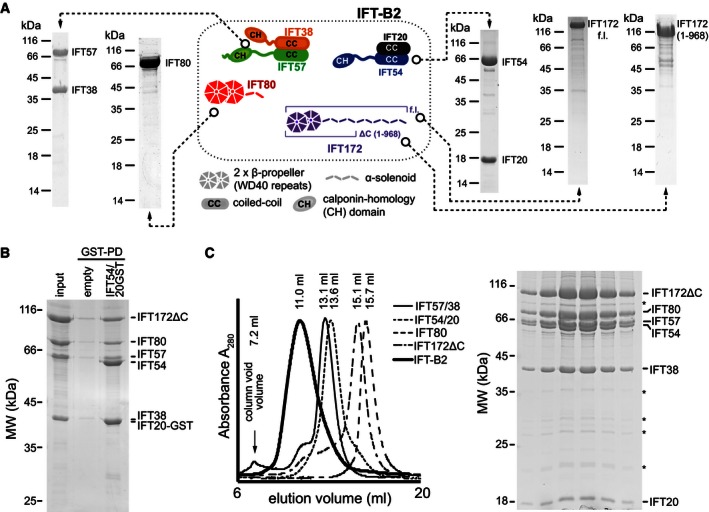
- SDS–PAGE gels stained with Coomassie showing the purity of the Chlamydomonas IFT‐B2 components, over‐expressed in E. coli or insect cells, after the last purification step by SEC. IFT54/20 and IFT57/38 were purified as hetero‐dimeric complexes, whereas IFT80, IFT172, and an N‐terminal IFT172(1‐968) (IFT172ΔC) constructs were purified as single components. A cartoon representation of the predicted domain architecture of IFT‐B2 components using HHPRED (Söding et al, 2005) is shown at the center.
- Pull‐down analysis for the hexameric IFT‐B2 complex. A mixture of IFT172ΔC, IFT80, and the IFT57/38 complex (“input”) shows only weak non‐specific binding to GSH resin (“empty”), but all proteins are pulled down by IFT54/20‐GST complex. Note that the IFT20‐GST and IFT38 proteins co‐migrate on the gel and cannot be properly resolved. However, the presence of the IFT57 protein in the pull‐down sample implies that its binding partner IFT38 is also there.
- (left) SEC elution profiles of individual IFT‐B2 components and of the assembled hexameric IFT‐B2 complex. The elution peak of IFT‐B2 is shifted compared to all of the individual components. Note that the peak heights were adjusted for better visualization of the result. (right) Coomassie‐stained SDS–PAGE gel of the six main fractions of the IFT‐B2 SEC elution peak demonstrating the presence of all six IFT‐B2 subunits. Weak contaminating bands are marked with asterisks.
Six of the peripheral IFT‐B subunits form a stable IFT‐B2 complex
In an attempt to map the binding sites for the six “peripheral” IFT‐B components IFT172, 80, 57, 54, 38, and 20 within the IFT‐B complex, we over‐expressed IFT172 and IFT80 as individual subunits and IFT57/38 as a heterodimer in insect cells, and a heterodimer of IFT54/20 in E. coli (Chlamydomonas proteins, Fig 1A). Whereas IFT80, IFT54/20, and IFT57/38 can be purified as full‐length (FL) proteins, FL IFT172 was difficult to purify due to lower expression and a tendency to degrade (Fig 1A). However, an N‐terminal construct of IFT172 lacking 787 residues of the predicted C‐terminal α‐solenoid structure (IFT172ΔC, residues 1–968) expressed well and could be purified (Fig 1A). Surprisingly, incubation of IFT172ΔC, IFT80, and IFT57/38 with GST‐tagged IFT54/20 resulted in the pull‐down of all six components using GSH beads (Fig 1B). Additionally, when IFT172ΔC, IFT80, IFT54/20, and IFT57/38 were mixed, incubated, and subjected to SEC, all six proteins co‐eluted in a peak significantly shifted from the peaks of the individual components, demonstrating the formation of a larger complex (Fig 1C). A similar result was obtained with FL IFT172 although the elution profile was somewhat broader, which is likely a result of partial proteolysis of IFT172 (Appendix Fig S1C). Based on the SEC elution volume, we estimate the molecular weight (MW) of the hexameric complex containing IFT172ΔC to be ~1.4 MDa (calculated MW = 353 kDa). This could suggest a multimer but given the fact that MW determination by SEC is highly shape dependent and the hexameric complex contains four coiled‐coil proteins (IFT57, 54, 38, and 20), and the high estimated MW from SEC could simply be a result of a highly elongated shape of the complex. These data demonstrate that IFT172, 80, 57, 54, 38, and 20 are not subunits peripheral to the IFT‐B core but form a separate stable high‐molecular weight subcomplex. We thus suggest a nomenclature where the IFT‐B core complex is referred to as IFT‐B1 and the complex identified here consisting of previously characterized peripheral subunits as IFT‐B2.
Architecture of the IFT‐B2 complex
To determine the architecture of the IFT‐B2 complex, we carried out pull‐downs and SEC experiments with purified Chlamydomonas proteins. IFT38, IFT54, and IFT57 all share a similar predicted domain organization consisting of an N‐terminal calponin homology (CH) domain followed by C‐terminal coiled‐coil structure (Fig 1A) (Taschner et al, 2012; Schou et al, 2013). IFT172 and IFT80 are both predicted to contain two N‐terminal WD40 β‐propeller domains followed by α‐solenoid structure akin to the domain architecture observed for coatomer subunits (van Dam et al, 2013). Mutations in IFT172 and IFT80 were reported to result in skeletal pattering defects (Beales et al, 2007; Halbritter et al, 2013), and the two components interact genetically (Halbritter et al, 2013) albeit not physically in sucrose gradients (Lucker et al, 2005). We observed no direct interaction between IFT172 and IFT80 in SEC (Appendix Fig S2A and B). However, both IFT172ΔC and IFT80 interacted strongly with IFT57/38 and co‐purified as hetero‐trimeric subcomplexes in SEC experiments (Fig 2A and B). The interactions are not mutually exclusive as the formation of a tetrameric IFT172ΔC/80/57/38 complex was observed in SEC (Fig 2C). To evaluate which domains of the IFT57/38 complex recognize IFT172 and IFT80, a mixture of purified IFT172ΔC and IFT80 was pulled down by GST‐tagged N‐terminal CH domains (IFT38CH or IFT57CH) or C‐terminal coiled‐coil domains (IFT57CC/IFT38CC) (Fig 2D). The results demonstrated that IFT80 interacts with the CH domain of IFT38 and IFT172ΔC with the CH domain of IFT57 (Fig 2D lanes 3–4). No detectable interaction was observed between the coiled‐coil regions of IFT57/38 and IFT80 or IFT172ΔC (Fig 2D, lane 5).
Figure 2. Architecture of IFT‐B2.
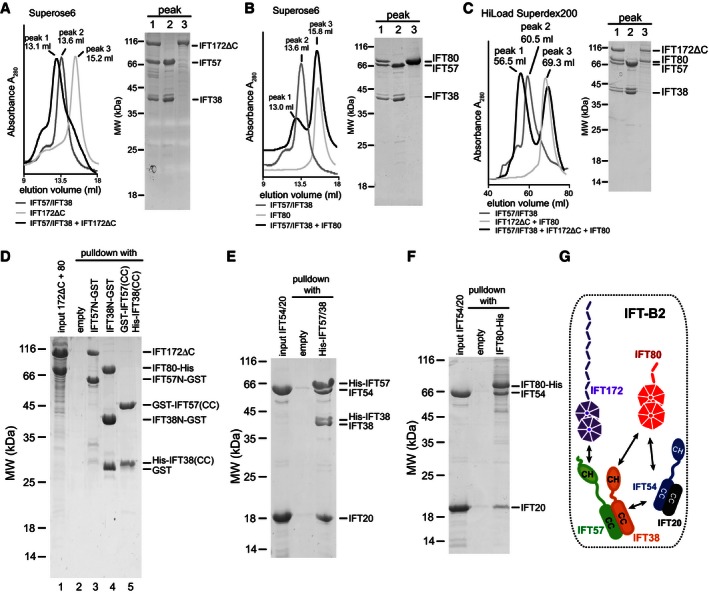
- A–C
SEC elution profiles and corresponding SDS–PAGE gels of IFT172ΔC, IFT57/38, IFT80, IFT54/20 as well as mixtures of IFT172ΔC/57/38, IFT80/57/38, and IFT172ΔC/80/57/38 demonstrating that IFT57/38 associates with both IFT80 and IFT172ΔC. Only the central peak fraction from each SEC experiment is shown on the SDS–PAGE gel. Note that the presence of both IFT172ΔC and IFT80 in a single peak in (C) (peak 3) is due to the fact that these two proteins elute in the same position, and not because they physically interact (see also Appendix Fig S2B). - D
GSH resin pull‐down of a mixture of IFT80 and IFT172ΔC (input, lane 1) with GST‐tagged CH domains of IFT57 or IFT38 (IFT57CH or IFT38CH) or with the GST‐tagged coiled‐coil (CC) region of IFT57 mixed with His‐tagged coiled‐coil region of IFT38. The SDS–PAGE gel shows interaction between IFT57CH‐IFT172ΔC, IFT38CH‐IFT80, and IFT57CC‐IFT38CC. - E
Ni‐NTA resin pull‐down of untagged IFT54/20 with His‐tagged IFT57/38 (both subunits His‐tagged). The two bands for IFT38 on the SDS–PAGE gel correspond to His‐tagged (upper band) and proteolytically cleaved untagged IFT38 (lower band). - F
Ni‐NTA resin pull‐down of untagged IFT54/20 with His‐tagged IFT80. - G
Schematics of the interactions between IFT‐B2 components supported by the data in (A–F) as well as in Appendix Fig S2. Note that it is currently not known whether the coiled‐coil interactions between IFT54, 20, 57, and 38 are parallel or anti‐parallel.
Next, we dissected the interaction within the IFT54/20 and IFT57/38 complexes. Consistent with previously published data (Omori et al, 2008; Follit et al, 2009), we found that purified IFT54 associates with IFT20 via the coiled‐coil domain and not via the CH domain (Appendix Fig S2C and D). Similarly, the coiled‐coil regions of IFT38 and IFT57 were sufficient for IFT57/38 complex formation (Fig 2D, lane 5 and Appendix Fig S2E). Further pull‐down experiments showed that the IFT54/20 complex interacts directly with both the IFT57/38 complex (Fig 2E) and the IFT80 protein (Fig 2F), but individually, these interactions are not strong enough to be detected by SEC (data not shown). We conclude that IFT54/20 is stably incorporated into the IFT‐B2 complex through simultaneous interactions with both IFT57/38 and IFT80. The architecture of the IFT‐B2 complex is summarized in Fig 2G.
IFT‐B2 binds αβ‐tubulin via the CH domain of IFT54
What is the function of IFT‐B2 in cilium formation? We recently showed that the CH domain of IFT81 together with a basic domain of IFT74 (both subunits of the IFT‐B1 complex) forms a tubulin‐binding module that likely functions in the IFT of αβ‐tubulin to the tips of cilia (Bhogaraju et al, 2013a). However, calculations using the kinetics of Chlamydomonas ciliogenesis suggest that each IFT complex likely carries at least two αβ‐tubulin heterodimers to sustain the observed fast initial rate of cilium growth (Bhogaraju et al, 2014). Prime candidates for additional tubulin cargo‐binding sites within the IFT complex are the CH domains of the IFT38, IFT54, and IFT57 subunits of the newly identified IFT‐B2 complex (Taschner et al, 2012; Schou et al, 2013). To test this hypothesis experimentally, we carried out binding experiments where soluble αβ‐tubulin was pulled down with affinity tagged IFT54/20 or IFT57/38 subcomplexes. The results show that IFT54/20 but not IFT57/38 binds tubulin (Fig 3A, lanes 1–2). We determined the affinity of IFT54/20 for soluble αβ‐tubulin to be in the low μM range (K d = 3 ± 1 μM, Appendix Fig S3A), which is similar to the ~1 μM affinity of IFT81/74 for αβ‐tubulin (Bhogaraju et al, 2013a). To assess tubulin binding by IFT54 in context of the IFT‐B2 complex, we reconstituted IFT‐B2 and IFT‐B2Δ54CH lacking the CH domain of IFT54 (Fig 3A, lanes 3–4). Pull‐down experiments showed that IFT‐B2 efficiently co‐precipitated soluble αβ‐tubulin, whereas the IFT‐B2Δ54CH did not (Fig 3A). These data demonstrated that IFT54CH is the only tubulin‐binding domain within the IFT‐B2 complex and suggest that the complete IFT‐B complex likely has two tubulin‐binding sites. The finding that IFT57/38 does not bind tubulin is consistent with a function of the CH domains of IFT38 and IFT57 in mediating the interactions with IFT80 and IFT172, respectively (Fig 2G).
Figure 3. Structure and tubulin binding of IFT54CH .
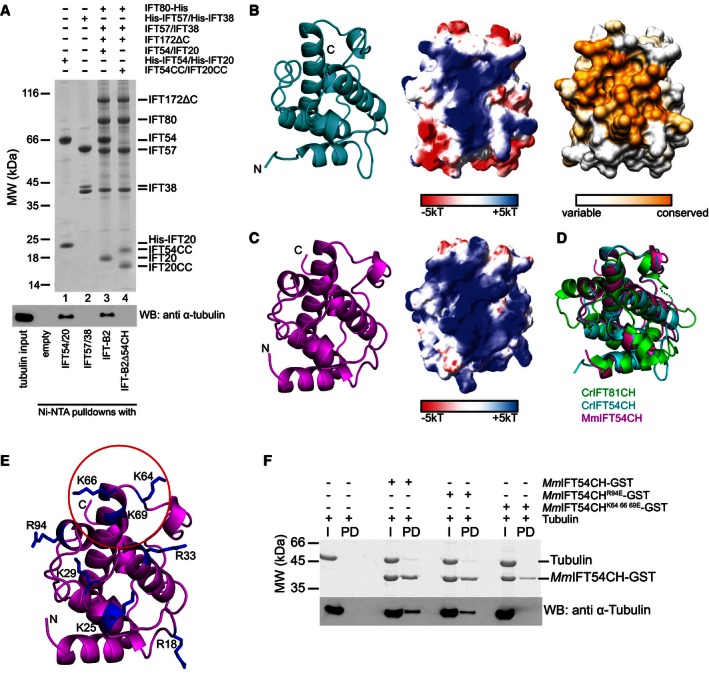
- Ni2+‐NTA affinity pull‐down of unpolymerized bovine αβ‐tubulin with His‐tagged IFT‐B2 (sub)‐complexes. A Coomassie‐stained SDS–PAGE gel shows the IFT‐B2 complexes used for pull‐downs (top), and a Western blot for anti‐α‐tubulin shows co‐precipitated αβ‐tubulin (bottom).
- Cartoon representation of the crystal structure of _Cr_IFT54CH (left), the electrostatic surface potential (middle), and the amino acid conservation mapped onto the surface of the structure (right).
- Cartoon representation of the crystal structure of _Mm_IFT54CH (left) and the electrostatic surface potential (middle).
- Superimposition of the CH domains of _Cr_IFT54CH, _Mm_IFT54CH, and _Cr_IFT81 (pdb: 4VLP).
- Cartoon representation of the crystal structure of _Mm_IFT54CH domain with the residues of the positively charged patch shown as sticks. Three residues whose mutation to glutamate abolished tubulin binding are indicated with a red circle.
- Ni2+‐NTA affinity pull‐down of unpolymerized bovine αβ‐tubulin with WT and mutant _Mm_IFT54CH. (top) Coomassie‐stained SDS–PAGE gel of input (I) and pulled down (PD) proteins and the corresponding Western blot to visualize αβ‐tubulin.
Mammalian IFT54 (known as TRAF3IP1 or MIPT3) and IFT81/74 were previously shown to associate with both soluble αβ‐tubulin and polymerized MTs (Ling & Goeddel, 2000; Bhogaraju et al, 2013a). We find that this is also the case for Chlamydomonas IFT54 as the IFT54/20 subcomplex co‐pelleted with MTs in a sedimentation assay (Appendix Fig S3E). IFT54 was recently implicated in the regulation of the dynamics of cytoplasmic MT stability (Bizet et al, 2015), which is consistent with the MT‐binding activity of IFT54. It remains to be shown whether the in vitro ability of IFT54 to bind αβ‐tubulin (Fig 3) allows it to function in IFT of tubulin to the tips of cilia.
The IFT54 CH domain binds αβ‐tubulin via a conserved basic patch
Tubulin binding by IFT54 was recapitulated using GST‐tagged CH domains of Chlamydomonas (Cr) or mouse (Mm) IFT54 (Appendix Fig S3B). Taken together with the observation that IFT‐B2Δ54CH does not interact with tubulin (Fig 3A), we conclude that the N‐terminal CH domain is responsible for tubulin binding by IFT54. To elucidate the molecular mechanism of tubulin binding by IFT54CH, we determined crystal structures of _Cr_IFT54CH and _Mm_IFT54CH at 1.6 Å resolution (Fig 3B and C and Table 1). The structures of _Cr_‐ and _Mm_IFT54CH are very similar (root‐mean‐square deviation (rmsd) of 0.8 Å) and display a highly basic and well‐conserved surface patch (Fig 3B and C). The overall structure of the IFT54 CH domain is also similar to that of the IFT81 CH domain, although the rmsd of 3.3 Å is somewhat higher (Fig 3D). Significant structural differences between IFT81CH and IFT54CH are found in the two most C‐terminal alpha helices (α4I and α4II) (Figs 3D and 4). Particularly, α4II adopts a different conformation in IFT81CH (where it packs against helix α2) compared to IFT54CH (where it packs against helix α3) (Fig 4B). This difference is significant as helix α4ΙΙ was found to harbor 3 basic residues responsible for tubulin binding in _Hs_IFT81 (Fig 4A) (Bhogaraju et al, 2013a). Although the IFT54 CH domain does not contain the basic residues found to mediate tubulin binding in IFT81, it does display a different Arg/Lys‐rich basic patch, which is highly conserved among IFT54 orthologs (Figs 3B and 4). To experimentally test whether this basic patch is responsible for tubulin binding in IFT54, we mutagenized and purified GST‐tagged _Mm_IFT54CH domains and carried out tubulin pull‐downs (Fig 3E and F, and Appendix Fig S3C). The results show that none of the five tested charge‐reverse single‐point mutations (R18, K25, K29, R33, or R94 to glutamate) was sufficient to disrupt tubulin binding (Fig 3F and Appendix Fig S3C). However, a triple KKK64/66/69EEE‐point mutant located at the edge of the basic surface patch (Fig 3E and F) did abolish tubulin binding suggesting that IFT54CH binds tubulin via these basic residues. Consistent with the observation that IFT57/38 does not associate with tubulin, sequence alignments suggest that the basic residues responsible for tubulin binding in IFT81CH or IFT54CH are not well conserved in IFT57CH or IFT38CH (Fig 4A).
Table 1.
Data collection and refinement statistics
| _Cr_IFT54CH SeMet | _Cr_IFT54CH native | _Mm_IFT54CH native | _Cr_IFT52N SeMet | _Cr_IFT52N native | _Mm_IFT52N native | |
|---|---|---|---|---|---|---|
| PDB code | 5fmt | 5fmu | 5fmr | 5fms | ||
| Data collection and scaling | Anomalous signal from Se to ~2 Å | Anomalous signal from Se to ~3.2 Å | ||||
| Wavelength (Å) | 0.97914 | 1.00000 | 1.00000 | 0.97955 | 1.03320 | 1.00000 |
| Resolution range (Å) | 37–1.60 (1.66–1.60) | 36–1.88 (1.99–1.88) | 37–1.59 (1.69–1.59) | 50–2.60 (2.75–2.60) | 41–2.00 (2.19–2.00) | 48–3.49 (3.60–3.49) |
| Space group | F222 | F222 | P1 | C2 | C2 | P432 |
| Unit cell (Å) | a = 57.83, b = 145.90, c= 148.80,α = β = γ = 90.0 | a = 57.66, b = 145.82, c = 147.87,α = β = γ = 90.0 | a = 38.00, b = 59.80, c = 61.50,α = 108.50, β = 105.40, γ = 90.30 | a = 169.55, b = 81.72, c = 120.16,α = 90, β = 133.73, γ = 90.00 | a = 169.76, b = 81.72, c= 120.48,α = 90, β = 133.97, γ = 90.00 | a = 204.91, b = 204.91, c = 204.91,α = β = γ = 90.0 |
| Total reflections | 556,754 (86,179) | 117,060 (14,884) | 176,087 (26,494) | 298,401 (45,427) | 739,923 (113,495) | 742,488 (116,316) |
| Unique reflections | 80,222 (12,779) | 25,144 (3,712) | 60,996 (9,267) | 71,621 (11,325) | 214,626 (33,965) | 35,358 (5,592) |
| Multiplicity | 6.9 (6.7) | 4.7 (4.0) | 2.9 (2.9) | 4.1 (4.0) | 3.4 (3.3) | 21.0 (20.8) |
| Completeness (%) | 99.7 (98.4) | 98.2 (90.9) | 93.9 (88.4) | 98.8 (96.9) | 98.6 (96.5) | 99.6 (98.0) |
| Mean I/sigma (I) | 15.1 (2.5) | 11.2 (1.7) | 10.5 (1.6) | 11.5 (2.5) | 11.3 (1.9) | 11.3 (1.7) |
| R‐merge | 0.113 (0.563) | 0.173 (0.937) | 0.146 (0.969) | 0.0737 (4.15) | 0.382 (2.48) | |
| CC½ | N/D | 0.997 (0.683) | 0.998 (0.552) | 1.000 (0.49) | 0.997 (0.636) | |
| Refinement | ||||||
| Number of reflections | 41,577 | 25,133 | 60,975 | 217,617 | 35,494 | |
| Protein residues | 265 | 266 | 528 | 732 | 661 | |
| Number of atoms | 4,613 | 2,302 | 4,515 | 6,209 | 4,681 | |
| Protein (non‐solvent) | 2,079 | 2,098 | 4,084 | 5,645 | 4,681 | |
| Water (solvent) | 376 | 204 | 431 | 544 | 0 | |
| R‐work | 0.1844 (0.2334) | 0.1971 (0.2726) | 0.1829 (0.2914) | 0.2009 (0.3242) | 0.2288 (0.3288) | |
| R‐free | 0.2137 (0.2578) | 0.2447 (0.3134) | 0.2144 (0.3126) | 0.2314 (0.3622) | 0.2885 (0.3931) | |
| Ramachandran favored (%) | 99.27 | 98.88 | 98.05 | 91.34 | ||
| Ramachandran outliers (%) | 0.0 | 0.0 | 0.00 | 0.46 | ||
| RMS bonds (Å) | 0.010 | 0.008 | 0.007 | 0.009 | 0.018 | |
| RMS angles (°) | 1.2 | 1.1 | 1.0 | 1.1 | 1.6 | |
| Average B‐Factors (Å2) | 15.3 | 23.5 | 19.5 | 45.4 | 84.1 |
Figure 4. Comparison of IFT54 and IFT81 CH domains.
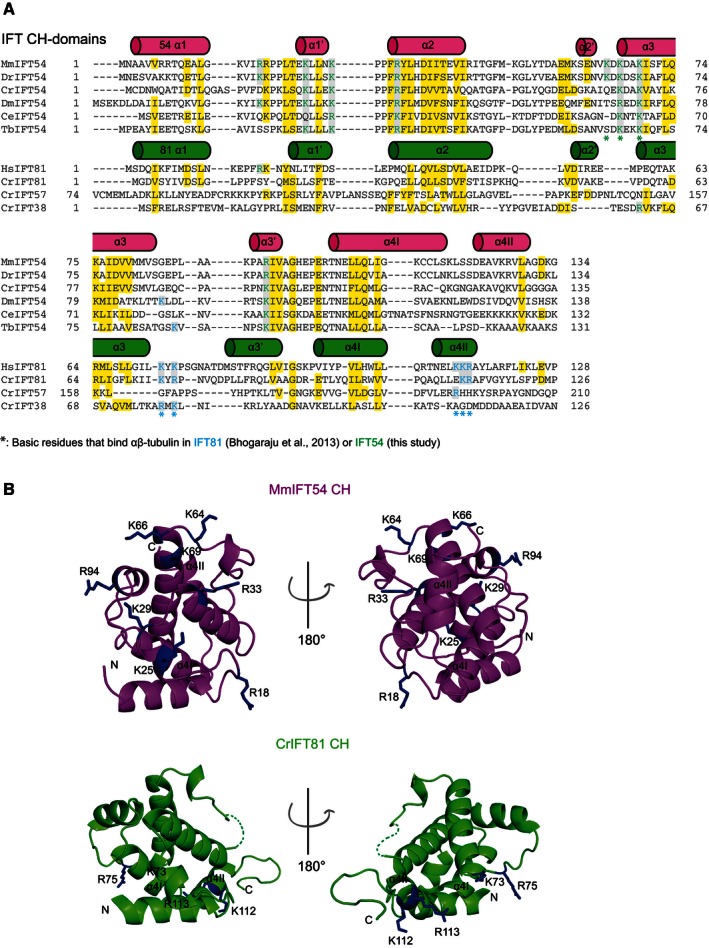
- Multiple sequence alignment of the N‐terminal CH domains of IFT81, IFT57, IFT54, and IFT38 from different species. Conserved residues are highlighted in yellow and residues that bind tubulin are marked blue for IFT81 (Bhogaraju et al, 2013a) and green for IFT54 (this study). The secondary structures for IFT54 (magenta) and IFT81 (green) are displayed above the sequences. Mus musculus (Mm), Danio rerio (Dr), Chlamydomonas reinhardtii (Cr), Homo sapiens (Hs), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), and Trypanosoma brucei (Tb).
- Cartoon representations of the superimposed crystal structures of _Cr_IFT81 and _Mm_IFT54 CH domains in two orientations differing by 180 degrees. The residues mediating tubulin binding are shown in stick representation and are located on different surface areas in IFT81 and IFT54.
To test whether IFT54CH binds the globular part of tubulin or the glutamate‐rich C‐terminal E‐hooks, we prepared subtilisin‐treated αβ‐tubulin lacking either the β‐tubulin E‐hook or both α‐ and β‐tubulin E‐hooks (Appendix Fig S3D). The results of tubulin pull‐downs demonstrated that _Mm_IFT54CH binds tubulin regardless of the presence of E‐hooks, indicating that IFT54CH, like IFT81CH (Bhogaraju et al, 2013a), recognizes the globular domain of tubulin. It is currently not known whether IFT81CH and IFT54CH recognize the same or different binding sites on αβ‐tubulin.
IFT‐B1/IFT‐B2 association is mediated by IFT88/52 and IFT57/38
How do the IFT‐B1 and B2 subcomplexes interact to form the complete IFT‐B complex? Interaction studies with IFT‐B2 and different IFT‐B1 subcomplexes revealed that IFT88/521–335 is sufficient to pull down IFT‐B2 (Fig 5A, lane 3). No interaction was observed between IFT‐B2 and IFT‐B1 subcomplexes IFT81/74/27/25/22 or IFT70/52/46 (data not shown). Neither the IFT88/52281–335 complex lacking IFT52N (IFT52N equals residues 1–275 of CrIFT52) nor IFT52N alone was sufficient to pull down IFT‐B2 (Fig 5A, lanes 4 and 5). This result suggests that IFT‐B2 recognizes IFT‐B1 via a composite surface area on the IFT88/521–335 subcomplex. To assess the stability of the interaction between IFT88/521–335 and IFT‐B2, GST‐tagged IFT‐B2‐IFT88/521–335 complex was immobilized on GSH beads and washed with increasing NaCl concentrations. Interestingly, only IFT172 was salt‐labile and dissociated from the complex at NaCl concentrations around 150–200 mM, whereas the other subunits remained associated even at 1 M NaCl concentration suggesting significant hydrophobic contacts within the complex (Fig 5B). To validate whether IFT172 is attached to the IFT complex via a salt‐labile link to the N‐terminal part of IFT57 as suggested by the results shown in Figs 2D and 5B, we incubated IFT571–234‐GST immobilized on GSH beads with Chlamydomonas whole‐cell extract and eluted interactors with 250 mM NaCl followed by elution with reduced glutathione (Appendix Fig S4). LC‐MS/MS analysis of eluates demonstrated that IFT172 but not other IFT subunits are significantly enriched vs. the negative control suggesting that IFT571–234‐GST specifically captures IFT172 via a salt‐dependent interaction (Appendix Fig S4). The finding that IFT172 is only loosely attached to the IFT complex is in agreement with previously published results from sucrose gradient centrifugations of natively purified Chlamydomonas IFT‐B complexes (Lucker et al, 2005). On the other hand, our observation that IFT88/52 remains associated with an IFT‐B2 complex consisting of IFT80/57/54/38/20 at even 1 M NaCl is somewhat surprising as the “peripheral” IFT‐B2 subunits were previously reported to dissociate at 300 mM NaCl concentration from the IFT‐B1 complex (Lucker et al, 2005). This difference may be attributed to the use of recombinant rather than endogenous proteins, where post‐translational modifications may alter/regulate complex stability. However, a study using pull‐downs from mouse lysates showed that IFT‐B2 components (IFT57 and IFT20) interacted with endogenous IFT88 (IFT‐B1) even after washing with 1 M NaCl (Baker et al, 2003). We previously reported that the IFT‐B1 complex is stable even at NaCl concentrations of 2 M (Taschner et al, 2014), which when combined with our data on the IFT‐B1/B2 interaction suggests that the IFT‐B complex with the exception of the IFT172 subunit is salt‐stable.
Figure 5. IFT‐B1 and IFT‐B2 form a salt‐stable complex via an (IFT88/52N)‐(IFT57/38) interaction.
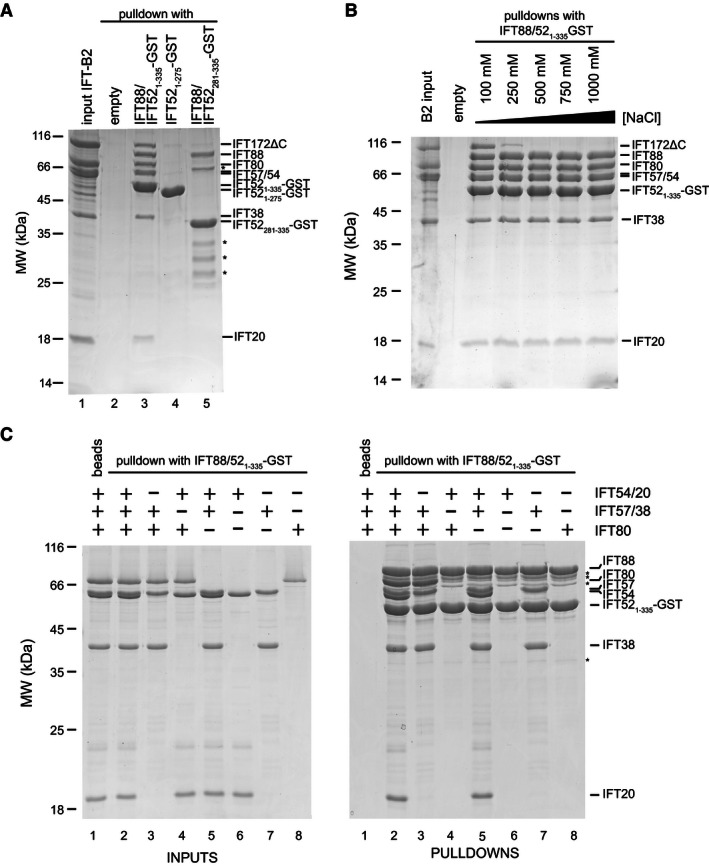
- Coomassie‐stained SDS–PAGE gel showing GST pull‐down experiments between the IFT‐B2 complex and an IFT88/52N subcomplex. The pre‐assembled IFT‐B2 complex (lane 1) shows no non‐specific interaction with empty GSH beads (lane 2), but is efficiently pulled down by IFT88/52N‐GST (with an IFT52(1‐335) construct comprising the N‐terminal domain as well as the IFT88‐binding region) (lane 3). Both IFT88 and IFT52N are required for the interaction with IFT‐B2 because neither the N‐terminal IFT52 domain (residues 1–275; lane 4) alone, nor an IFT88/52 complex lacking the N‐terminal IFT52 domain (with an IFT52(281‐335) construct containing only the IFT88‐binding region) (lane 5) shows efficient pull‐down of IFT‐B2. Contaminants from the purification of the complex shown in lane 5 are marked with asterisks.
- Coomassie‐stained SDS–PAGE gel showing a salt‐stability test of the (IFT88/52N)‐(IFT‐B2) interaction. Pre‐assembled IFT‐B2 complex was bound to IFT88/52N‐GST immobilized on GSH beads. The beads were then divided into five aliquots which were washed with buffers containing increasing concentrations of NaCl. After these washes, the bound material was eluted and analyzed. IFT172ΔC is the only salt‐labile component in this complex and is already washed off at a NaCl concentration of 150–200 mM, whereas all the other IFT‐B2 proteins remain associated even after washing with 1 M NaCl.
- Coomassie‐stained SDS–PAGE gels showing GST pull‐down experiments between IFT88/52N‐GST complex immobilized on GSH beads and various combinations of IFT‐B2 proteins. The left gel shows the IFT‐B2 input mixtures, and the right gel shows the bound material after washing and elution from the beads. A pentameric IFT‐B2 complex (IFT80/57/54/38/20) is efficiently pulled down by IFT88/52N‐GST, but not by empty beads (compare lanes 1 and 2). Omission of IFT54/20 (lane 4) or IFT80 (lane 5) does not influence the binding of any of the other components, but a lack of IFT57/38 in the mixture abolishes detectable pull‐downs of IFT54/20 and IFT80 (lane 4), indicating that IFT57/38 is the direct interaction partner of IFT88/52N. Indeed, IFT57/38 alone (lane 7), but not IFT54/20 (lane 6) or IFT80 (lane 8), is sufficient to be pulled down in this assay. Contaminating bands from the IFT88/52N sample are marked with asterisks.
We next determined which of the IFT‐B2 components is directly contacted by the IFT88/52 module of IFT‐B1, and carried out pull‐downs with immobilized IFT88/521–335‐GST (IFT88/52N‐GST) and various combinations of five IFT‐B2 proteins (IFT57/38, IFT54/20, and IFT80). When all five IFT‐B2 components were mixed, they were efficiently pulled down by immobilized IFT88/52N‐GST but not by empty GSH resin (Fig 5C, compare lanes 1 and 2). Removal of IFT54/20 or IFT80 from the pull‐down reactions did not affect the interaction of IFT88/52N‐GST with the remaining components (Fig 5C, lanes 3 and 5), but when IFT57/38 was omitted, we no longer observed an interaction between IFT88/52N‐GST and IFT80 or IFT54/20 (Fig 5C, lane 4). This result indicates that the IFT88/52N module directly binds to IFT57/38 of the IFT‐B2 complex and that IFT80, IFT54/20, and IFT172 are kept in the complex via their associations with IFT57/38 as shown in Fig 2. Indeed, only IFT57/38 but not IFT80 or IFT54/20 was pulled down by IFT88/52N‐GST (Fig 5C, lanes 6–8). As we can only purify IFT57 and IFT38 in context of the IFT57/38 complex, we are currently unable to dissect whether individual IFT38 or IFT57 subunits are sufficient for the interaction with IFT88/52N. We conclude that IFT‐B complex formation requires a composite interface on IFT88 and IFT52N that mediates the contacts of IFT‐B1 to IFT57/38 of IFT‐B2.
Crystal structures of IFT52N reveal critical residues for IFT‐B1/IFT‐B2 association
To obtain structural information on the interaction between the IFT‐B1 and IFT‐B2 complex, we carried out crystallization trials with the IFT88/521–335 complex, but did not obtain crystals. However, the N‐terminal domains of IFT52 from both Cr (residues 1–275) and Mm (residues 1–268) crystallized and the structures were determined at resolutions of 2 Å (_Cr_IFT52N) and 3.5 Å (_Mm_IFT52N) (see Table 1). IFT52N adopts an aminotransferase fold commonly found in enzymes of purine, pyrimidine, tryptophan, arginine, and folic acid biosynthetic pathways (Tesmer et al, 1996). The structures of _Cr_‐ and _Mm_IFT52 superpose with an rmsd of 1.0 Å and consist of an extended central beta‐sheet flanked on each side by several helices and beta‐hairpins (Fig 6A). As previously predicted based on bioinformatics (Beatson & Ponting, 2004), IFT52N displays structural similarity to β‐galactosidase and superposes onto the B domain of β‐galactosidase with an rmsd of 2.6 Å (Appendix Fig S5A). However, the sugar‐binding domain of β‐galactosidase (Maksimainen et al, 2012) is not present in IFT52 in agreement with the finding that purified IFT52 does not appear to bind sugars (Appendix Fig S5A) (Taschner et al, 2011). Interestingly, the IFT52N structures also superpose well with the glutaminase subunit PDX2 of pyridoxal 5′‐phosphate synthase complex (Strohmeier et al, 2006) (rmsd 2.4 Å), which catalyses the hydrolysis of glutamine. However, IFT52N lacks the catalytic residues of PDX2 and is thus unlikely to be an enzyme (Appendix Fig S5B).
Figure 6. Crystal structure of the N‐terminal domain of IFT52.
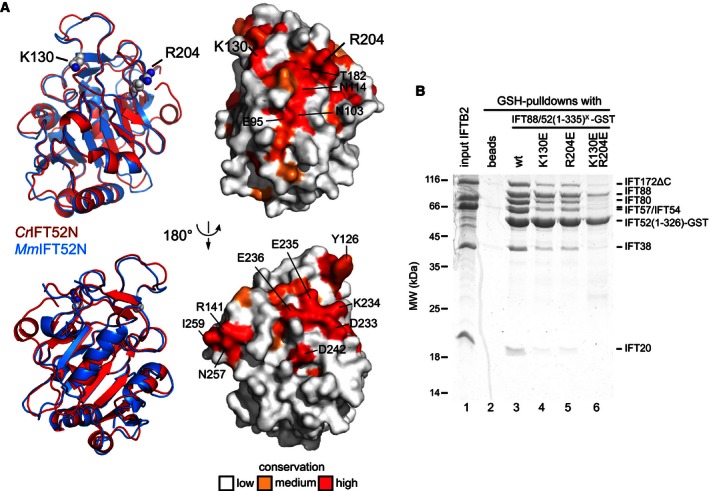
- (top) Cartoon representation and surface plot of a superpositioning of _Cr_IFT52N and _Mm_IFT52N crystal structures. (bottom) Rotated 180 degrees compared to top panel. Conservation is mapped from 5 different species (see alignment in Appendix Fig S5), and conserved residues are labeled (Cr numbering). The two residues (K130 and R204) found to disrupt interaction with IFT‐B2 (B) are labeled in larger font.
- Coomassie‐stained SDS–PAGE gel of a pull‐down of IFT‐B2 using WT or mutant IFT88/521–335‐GST. Whereas single‐point mutations slightly reduced the amounts of pulled down IFT‐B2, the K130E/R204E double mutation reduced it to background levels (compare lanes 2 and 6).
Analysis of conserved surface‐exposed residues between 6 species (Appendix Fig S5C) revealed a number of highly conserved patches on the IFT52N structure (Fig 6A). Given that residues involved in the interaction between the IFT88/52N module and the IFT‐B2 complex (i.e., IFT57/38) are likely conserved across ciliated species, we focussed on two residues (K130 and R204 in the _Cr_IFT52N domain), which are located in the largest conserved and mainly hydrophilic surface patch. K130 and R204 were mutated to glutamic acids either individually or in combination, and the resulting complexes with IFT88 purified for pull‐down analysis with the IFT‐B2 complex. The results showed that whereas the wild‐type IFT88/52N complex readily pulled down IFT‐B2 (Fig 6B, lane 3), the complexes with single‐point mutations were somewhat compromised in their ability to do so (Fig 6B, lanes 4 and 5). This effect was much stronger in the double mutant complex, which pulled down only background levels of IFT‐B2 complex (Fig 6B, lane 6). We conclude that a conserved patch on the surface of IFT52N is required for the interaction between IFT‐B1 and B2.
Reconstitution of a 15‐subunit IFT‐B complex
Finally, we reconstituted the 15‐subunit IFT‐B complex (lacking only IFT56) from previously purified nonameric IFT‐B1 (Taschner et al, 2014) and hexameric IFT‐B2 (this study) subcomplexes. IFT‐B1 and B2 were incubated and subjected to SEC, which resulted in a larger complex as demonstrated by a shifted elution profile compared to IFT‐B1 and B2 subcomplexes (Fig 7A). SDS–PAGE analysis of the peak fraction showed the presence of all 15 components (Fig 7A). Although some of the proteins such as IFT80/81 and IFT70/57/54 run very close together in the gel, these subunits were positively identified by mass‐spec (data not shown). This result showed that the IFT‐B complex can be prepared recombinantly in milligram amounts for structural and in vitro studies. The data presented in this work allow us to propose an architectural model for the IFT‐B complex (Fig 7B) where IFT‐B1 and IFT‐B2 are linked by (IFT88/52N)‐(IFT57/38). In this model of IFT‐B1, the two tubulin/MT‐binding sites of IFT54 and IFT81/74 could operate independently or in cooperation.
Figure 7. Reconstitution of a 15‐subunit IFT‐B complex from recombinant IFT‐B1 and IFT‐B2 subcomplexes.
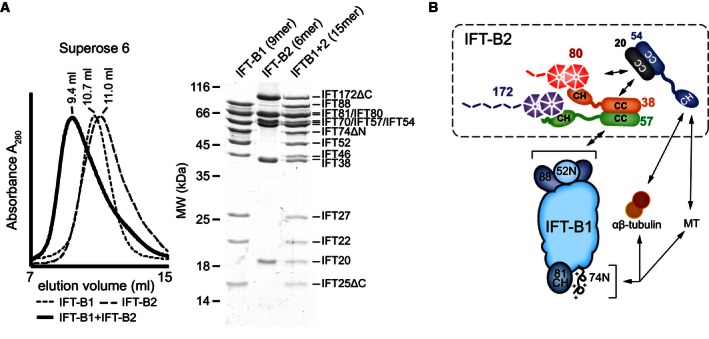
- SEC profiles of the IFT‐B1 and IFT‐B2 complexes as well as the 15‐subunit reconstituted IFT‐B complex. IFT‐B1 and IFT‐B2 elute at similar volumes, but after mixing, the resulting high‐molecular‐weight complex is clearly shifted toward a smaller elution volume. The Coomassie‐stained SDS–PAGE gel on the right shows the peak fractions of the individual runs.
- Schematic representation of the IFT‐B complex summarizing the results presented in this study. The six IFT‐B proteins IFT172, IFT80, IFT57, IFT54, IFT38, and IFT20 form a hexameric complex (IFT‐B2), where an IFT57/38 dimer (mediated by coiled‐coil interactions) binds to IFT172 and IFT80 via its two CH domains, and a dimer of IFT54/20 (again mediated by coiled‐coil interactions) is held in the complex by binding to both IFT80 and IFT57/38. The IFT‐B2 complex binds to the IFT‐B1 complex via an interaction mediated by IFT88/52N (on IFT‐B1) and IFT57/38 (on IFT‐B2). In addition to the previously characterized tubulin/MT‐binding module on IFT‐B1 (IFT81CH/IFT74N), the N‐terminal CH domain of IFT54 is also able to bind tubulin and MTs, giving a total number of two binding sites per IFT‐B complex. Note that several details, such as the exact binding sites for IFT80 and IFT57/38 on the IFT54/20 complex, as well as the directionalities of the coiled‐coils in IFT57/38 and IFT54/20, remain to be determined.
Discussion
Architecture of the IFT‐B complex
Here, we show that the six IFT‐B subunits IFT172, 80, 57, 54, 38, and 20 are not merely components loosely associated with IFT‐B1 but interact with each other to form an IFT‐B2 subcomplex. These two IFT‐B1 (core) and IFT‐B2 (“peripheral”) subcomplexes can be purified independently using recombinant proteins (Fig 1 and Taschner et al, 2014) and when incubated result in the reconstitution of a nearly complete 15‐subunit IFT‐B complex lacking only IFT56 (Fig 7A). We uncovered that the IFT57/38 subcomplex is central to IFT‐B complex assembly. The N‐terminal CH domains of IFT57/38 mediate the contacts to IFT80 and IFT172, whereas the C‐terminal coiled‐coils allow for IFT57/38 hetero‐dimer formation and links to the IFT54/20 hetero‐dimer (Fig 2 and Appendix Fig S2). The interaction between IFT54 and IFT20 is in agreement with previously published data (Omori et al, 2008; Follit et al, 2009), whereas the interaction between IFT57 and IFT38 was not previously reported. Additionally, IFT57/38 mediates the contacts between the two IFT‐B subcomplexes by binding directly to IFT88/52N of IFT‐B1. The importance of IFT57/38 in IFT‐B2 assembly is akin to that of IFT52 in IFT‐B1 assembly. IFT52 mediates contacts to IFT46, IFT70, IFT81/74, and IFT88 within the IFT‐B1 subcomplex as well as contacts to IFT‐B2 thus serving as a central architectural component (Fig 7 and Taschner et al, 2011, 2014).
The only IFT‐B subunit that has so far evaded recombinant purification is IFT56/TTC26/DYF13. Deletion of this subunit in Chlamydomonas does not completely inhibit flagellar assembly but instead leads to slightly shorter flagella with pronounced motility defects, indicating that IFT56 carries out a more specialized role in the transport of motility‐related proteins. This notion was confirmed using proteomic analysis of IFT56 mutant flagella (Ishikawa et al, 2014). A recent study suggested an interaction of mouse TTC26 with IFT46 based on yeast‐2‐hybrid analysis and pull‐downs (Swiderski et al, 2014), making it the tenth member of the IFT‐B1 complex. Although we currently have a good biochemical understanding of how the 15‐subunit IFT‐B complex assembles and have crystal structures of domains and heterodimeric subcomplexes (Figs 3 and 6; Taschner et al, 2014; Bhogaraju et al, 2011), information on how IFT proteins organize the IFT complex in 3‐dimensions is still missing. The reconstitution of the 15‐subunit IFT‐B complex (Fig 7) will pave the way for structural studies of this assembly.
IFT54CH constitutes an additional αβ‐tubulin/MT‐binding domain within the IFT‐B complex
αβ‐tubulin assembles the backbone of the MT‐based axoneme of the cilium and is a known IFT cargo (Craft et al, 2015) that associates directly with IFT81/74 of the IFT‐B1 subcomplex (Bhogaraju et al, 2013a). The CH domain of IFT81 binds the globular part of αβ‐tubulin (K d = 16 μM), and mutations of tubulin‐binding residues significantly impair ciliogenesis in human cells (Bhogaraju et al, 2013a). Although the presence of the basic N‐terminal domain of IFT74 in the form of an IFT81/74 subcomplex increased the affinity for tubulin by ~18‐fold (K d = 0.9 μM) (Bhogaraju et al, 2013a), this domain is not required for tubulin entrance into Chlamydomonas flagella (Brown et al, 2015). However, the deletion of the first 130 residues of IFT74 did result in moderately slower Chlamydomonas flagellum regeneration kinetics and somewhat shorter flagellum length consistent with a reduction in the amounts of tubulin transported to the ciliary tip (Brown et al, 2015). We now show that IFT‐B2 also binds αβ‐tubulin via the CH domain of IFT54 but not via the CH domains of IFT57 or IFT38 (Fig 3). The affinity of IFT54/20 for αβ‐tubulin was measured to be 3 μM (Appendix Fig S3A). The concentration of tubulin in the cell body of Chlamydomonas was estimated to be in the double‐digit μM range (Craft et al, 2015), and K d's of 1–3 μM should thus be sufficient to load tubulin onto IFT complexes in cells. The mechanism by which tubulin cargo loading onto IFT trains is regulated in vivo is currently not known.
The functional relevance of the IFT54 tubulin‐binding activity remains to be determined. Since calculations show that at least two tubulin‐binding sites within the IFT complex are required to sustain the fast initial flagellar assembly rate in Chlamydomonas (Bhogaraju et al, 2014), we favor the possibility that the IFT81/74 module and the IFT54 CH domains both function in IFT of tubulin. Additional data using in vivo systems are required to challenge this hypothesis and to determine whether the tubulin‐binding sites of IFT‐B1 and B2 operate independently to bind two αβ‐tubulin hetero‐dimers, or whether they function together to possibly form one higher‐affinity binding site. Information on the spatial location of IFT54 relative to IFT81/74 within the IFT complex as well as tubulin‐binding experiments with reconstituted IFT‐B complexes should provide further insights into these issues.
Mammalian IFT81/74 and IFT54 were shown to associate with polymerized MT in addition to soluble αβ‐tubulin (Ling & Goeddel, 2000; Bhogaraju et al, 2013a), and we show here that CrIFT54 also associates with MTs (Appendix Fig S3E) raising the possibility of MT‐related functions of IFT54. In support of this notion, mouse IFT54 mutant cell lines showed an accumulation of acetylated MTs in the cytoplasm (Berbari et al, 2011). Additionally, IFT54 mutations in ciliopathy patients facilitated the characterization of an IFT54 function in the regulation of cytoplasmic MT dynamics (Bizet et al, 2015). Interestingly, several IFT proteins cluster at the spindle of dividing cells and are required for proper formation of astral MTs (Delaval et al, 2011). It thus appears likely that IFT54 in addition to its essential function in ciliogenesis has extra‐ciliary functions related to the regulation of the cytoskeleton. Another hypothesis that remains to be explored is that the MT‐binding activities of IFT54 and/or IFT81/74 might function in turnaround from anterograde to retrograde IFT through binding to the MTs at the ciliary tip. A comprehensive functional investigation of the tubulin/MT‐binding activities within the IFT complex still awaits in vivo dissection.
The polycystic kidney disease protein IFT88 links IFT‐B1 and IFT‐B2
IFT88 is mutated in the Oak Ridge Polycystic Kidney (ORPK) mouse model for autosomal recessive polycystic kidney disease (Moyer et al, 1994; Lehman et al, 2008) and has since then be used extensively to uncover ciliary involvement in a plethora of processes such as skeletal patterning (Zhang et al, 2003), photoreceptor assembly and maintenance (Pazour et al, 2002), and more (reviewed in Lehman et al, 2008). However, no clear molecular function has so far been assigned to the IFT88 protein. We previously mapped the interaction of IFT88 to a short stretch in the central region of IFT52, but IFT88 was not required for the stability of the IFT‐B1 complex (Richey & Qin, 2012; Taschner et al, 2014). The results presented here do, however, indicate a central role of IFT88 for the stability of the IFT‐B complex by directly linking IFT‐B1 and IFT‐B2. This notion leads to the prediction that IFT88 knockdown would allow for the formation of IFT‐B1(‐IFT88) and IFT‐B2 subcomplexes but not for the formation of a complete IFT‐B complex. Importantly, IFT88 knockout mice die early in embryogenesis due to severe defects in Sonic hedgehog signaling (Huangfu et al, 2003), and the ORPK mouse can thus be considered a hypomorph due to lower IFT88 expression levels caused by an insertion in an intron of the gene. It thus seems likely that the reduced levels of IFT88 in the ORPK mouse are sufficient to maintain a basal level of stable IFT‐B complex and to assemble short cilia that allow the mice to survive until birth, but they later die due to severe kidney cysts. It remains to be shown whether IFT88 has additional functions in IFT such as association with and transport of ciliary cargo.
Materials and Methods
Expression and purification of recombinant proteins from E. coli
Full‐length (FL) Chlamydomonas reinhardtii or Mus musculus IFT cDNAs or smaller fragments were cloned into pEC vectors either only with N‐terminal cleavable tags (6xHis‐TEV or 6xHis‐GST‐TEV) or with an additional C‐terminal non‐cleavable GST‐tag and expressed in E. coli BL21(DE3) as reported previously (Taschner et al, 2011). Cultures of bacteria transformed with the desired plasmid(s) were grown at 37°C in TB medium to an OD600 of 2, the temperature reduced to 18°C, and protein over‐expression induced by addition of 0.5 mM IPTG and continued growth at 18°C overnight. Cells were harvested, resuspended in 2 pellet volumes of lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM β‐mercaptoethanol, 1 mM PMSF, 25 μg/ml DNase I), and lysed by sonication. The extract was clarified by centrifugation (75,000 g, 30 min), loaded on a Ni2+‐NTA column (5 ml, Roche), washed extensively with lysis buffer supplemented with 1 M NaCl, and then eluted with a gradient from 0 to 500 mM imidazole in lysis buffer. The eluted protein was dialyzed overnight against buffer Qa (20 mM Tris–HCl pH 7.5, 50 mM NaCl, 10% glycerol, and 5 mM β‐mercaptoethanol) in the presence of 6xHis‐tagged TEV protease to remove the N‐terminal tag (unless the N‐terminal tag was required for pull‐down experiments), and the resulting solution was again passed over a Ni2+‐NTA column to remove impurities, cleaved tags, and His‐tagged TEV protease. The flowthrough containing the protein of interest was then loaded onto an ion‐exchange column (anion exchanger, 5 ml HiTrapQ‐Sepharose, GE Healthcare) followed by elution with a gradient from buffer Qa to buffer Qb (as buffer Qa, but with 1 M NaCl). In cases where N‐terminal tags were not removed, the Ni2+‐NTA step was omitted and the dialyzed material was directly used for ion‐exchange chromatography. Fractions containing the protein of interest were then concentrated and loaded onto a suitable column for size‐exclusion chromatography (HiLoad Superdex2000 or HiLoad Superdex75) with SEC buffer (10 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT). For expression of seleno‐methionine‐derivatized proteins, cultures were grown in minimal medium containing 50 mg/l seleno‐methionine, shifted to 18°C at an OD600 of 0.6, and then induced overnight with 0.25 mM IPTG. The purification procedure was the same as described for the native proteins.
Expression and purification of recombinant proteins from insect cells
Coding sequences for Chlamydomonas reinhardtii FL cDNAs or smaller fragments were cloned into multiple cloning site(s) of the pFL vector (Fitzgerald et al, 2006; Bieniossek et al, 2008) either with or without tags (for IFT80‐6xHis: cloning into MCS2 via EcoRI/XbaI; for IFT172 fl: cloning into MCS2 via EcoRI/XbaI; for IFT172(1‐968): cloning into MCS1 via SmaI/SphI; for a plasmid containing both IFT57 and IFT38: 6xHis‐TEV‐IFT57 in MCS1 via NcoI/SphI, and 6xHis‐TEV‐IFT38 into MCS2 via EcoRI/XbaI). The resulting plasmids were used to produce recombinant baculoviruses as described previously (Taschner et al, 2014). For protein expression cultures, 3 l of HighFive cells (Invitrogen) was infected with a pre‐determined amount of recombinant virus, harvested after 72 h by centrifugation (1,000 g, 15 min), and resuspended in 5× pellet volume of fractionation buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 250 mM sucrose) freshly supplemented with a protease inhibitor tablet (complete tablet, EDTA‐free; Roche) and 5 mM β‐mercaptoethanol. Cells were lysed with 20 strokes of a dounce homogenizer on ice, followed by incubation on ice for 20 min. Nuclei were pelleted by centrifugation (750 g, 10 min, 4°C) in 50‐ml Falcon tubes, and the supernatant was further clarified by centrifugation (75,000 g, 1 h, 10°C) after addition of NaCl to a final concentration of 150 mM. Ni2+‐NTA chromatography, dialysis and tag removal, ion‐exchange chromatography, and SEC were performed as described for proteins produced in E. coli. For IFT80‐6xHis (uncleavable tag) as well as for IFT172 FL and IFT172(1–968) (which do not have a tag at all and are purified based on their non‐specific interaction with Ni2+‐NTA resin), we routinely skipped the dialysis step and loaded the Ni‐eluate directly on an ion‐exchange column after dilution (3×) in the low‐salt buffer Qa.
Crystallization
All crystals were obtained using the sitting drop vapor diffusion method after mixing protein at a ratio of 1:1 with various precipitants. The CH domain of _Cr_IFT54 (residues 1–134) crystallized at 28 mg/ml after mixing with a precipitant containing 50 mM Tris–HCl pH 8.3, 2% MPD, 80 mM ammonium sulfate, 30% PEG5000, and 5% glycerol followed by incubation at 18°C for 3 days. The seleno‐methionine‐derivatized protein crystallized at 24 mg/ml in a slightly modified precipitant containing 50 mM Tris–HCl pH 8.3, 2% MPD, 80 mM ammonium sulfate, 26% PEG5000, and 7% glycerol followed by incubation at 18°C for 3 days. The CH domain of _Mm_IFT54 (residues 1–133) crystallized at 36 mg/ml after mixing with 50 mM MES pH 5.8, 200 mM ammonium acetate, 4% MPD, and 32% PEG3350 followed by incubation for 2 days at 4°C. _Cr_IFT52N (both native‐ and selenomethionine‐derivatized proteins) were concentrated to 10 mg/ml and crystallized after mixing with a precipitant containing 100 mM Bis‐Tris pH 6.5 and 2 M ammonium sulfate at 18°C. _Mm_IFT52 was also concentrated to 10 mg/ml and crystallized at 18°C after mixing with a precipitant containing 100 mM Bis‐Tris pH 5.5 and 1.8 M ammonium sulfate.
X‐ray diffraction data collection and crystal structure determination
X‐ray diffraction data were acquired at the Swiss Light Source (SLS, Villigen, Switzerland) at PXII and PXIII beam lines, and indexed with the XDS package (Kabsch, 2010) before scaling with Aimless as part of the CCP4 package (Winn et al, 2011). The structures of _Cr_IFT52N and _Cr_IFT54CH were determined from selenomethionine substituted protein crystals. Single anomalous dispersion data were recorded at the Se peak wavelength, and AUTOSOL as part of the PHENIX package (Adams et al, 2010) was used to locate Se sites and calculate experimental phases and electron density. The first round of automatic building was performed with AutoBuild in PHENIX and resulted in 75–90% complete models. These partial models were subsequently used for molecular replacement with the native datasets and the structures finished by iterative cycles of model building in Coot (Emsley et al, 2010) and refinement in PHENIX. The structures of _Mm_IFT52N and _Mm_IFT54CH were determined by molecular replacement using the previously determined structures of _Cr_IFT52N and _Cr_IFT54CH. For data and refinement statistics, see Table 1.
Reconstitution of the hexameric IFT‐B2 complex
IFT57/38 complex (around 5 mg) was diluted into 30 ml of buffer Qa, and excess amounts (roughly 1.5×) of IFT54/20 complex and IFT80 protein were added. After incubation at 4°C for 1 h, the mixture was subjected to ion‐exchange chromatography (Q‐Sepharose column) and the bound material was eluted with a gradient from buffer Qa to buffer Qb (gradient length 20 column volumes). This removed excess amounts of individual components and led to a stoichiometric IFT‐B2 pentamer. After concentration of the pentamer containing fractions, they were diluted 10× with B2‐SEC buffer (10 mM HEPES pH 7.5, 100 mM NaCl, 5% glycerol, 1 mM DTT) and concentrated again after addition of an equimolar amount of IFT172ΔC. The resulting concentrated sample was subjected to SEC in B2‐SEC buffer.
Reconstitution of a 15‐subunit IFT‐B complex
Nonameric IFT‐B1 (IFT‐B core) complex was purified as described previously (Taschner et al, 2014). Equimolar amounts of IFT‐B1 and IFT‐B2 complex were diluted into 2 ml of B2‐SEC buffer, incubated for 1 h at 4°C, and then concentrated to a volume of 400 μl before being subjected to SEC in B2‐SEC buffer.
Interaction experiments using affinity pull‐downs
Tagged protein(s) or protein complex(es) was/were immobilized on affinity beads (Ni2+‐NTA for His‐tagged proteins; GSH resin for GST‐tagged proteins) by incubating 200 μl of 10 μM tagged complex with 15 μl bed volume of affinity resin (volumes correspond to 1 pull‐down reaction). An empty resin control was always included to control for non‐specific interaction with the untagged protein/complex. After incubation for 1 h, the beads were collected and washed once with B2‐SEC buffer. The untagged protein or complex was diluted to 10 μM in B2‐SEC buffer and centrifuged for 5 min (12,000 g, 4°C) to remove any precipitate. An “input” sample was removed, and 200 μl aliquots of the remaining solution was added to each tube with prepared resin (either empty or pre‐loaded with tagged protein). The reactions were incubated for 4–5 h at 4°C before the beads were collected and washed twice with B2‐SEC buffer, and finally, the bound material was eluted with B2‐SEC buffer containing either 500 mM imidazole (for Ni2+‐NTA resin) or 30 mM reduced glutathione (for GSH resin). The input sample and the various elution samples were examined by SDS–PAGE.
Analytical ultracentrifugation (AUC)
Sedimentation velocity experiments were performed on an Optima XL‐I analytical ultracentrifuge (Beckman Inc., Palo Alto, Ca, USA) using a 60 Ti rotor and double‐sector epon centerpieces. The _Cr_IFT57/38 complex (0.56 mg/ml) was in a 10 mM HEPES (pH 7.5) buffer containing 100 mM NaCl, 10% glycerol and 2 mM TCEP. Buffer density and viscosity were measured using a DMA 5000 densitometer and an AMVn viscosimeter, respectively (both Anton Paar, Graz, Austria). Protein concentration distribution was monitored at 280 nm, at 235,000 g and 20°C. Time‐derivative analysis was computed using the SEDFIT software package, version 12.1b (Schuck, 2000), resulting in a c(s) distribution and an estimate for the molecular weight Mf (from the sedimentation coefficient and the diffusion coefficient, as inferred from the broadening of the sedimentation boundary, assuming all observed species share the same frictional coefficient f/f0).
Microscale thermophoresis
The lysine side chains of bovine brain tubulin (Cytoskeleton #TL238) were fluorescently labeled using the Cy3 protein labeling kit from Jena Bioscience according to their protocol. 100 nM of fluorescently labeled tubulin was titrated with purified _Cr_IFT54/20 (200 μM) in a total volume of 20 μl to measure 16 data points. The measurements were recorded on a NanoTemper Monolith NT.115 device (NanoTemper Technologies GmbH) using the following settings: LED power 65% (green), MST power 50%, Laser on 40 s, Laser off 5 s. Raw data were analyzed using the software from NanoTemper to visualize the binding curves and Prism (GraphPad Prism Software) to calculate dissociation constants.
Subtilisin treatment of tubulin
Bovine brain tubulin (Cytoskeleton #TL238) was incubated at 30°C with 1.25 μM subtilisin. Incubation time was either 20 min to remove the C‐terminal tail of β‐tubulin or 130 min to additionally remove the C‐terminal tail of α‐tubulin. 1 mM PMSF was added to stop the reactions. After ultracentrifugation in a Beckmann TLA100 rotor (200,000 g, 30 min, 35°C), the pellet was resuspended in 20 μl of 1× BRB80 buffer. Subtilisin‐treated tubulin was purified using size‐exclusion chromatography (SEC) (Superdex200, GE Healthcare) and subsequently used in pull‐down experiments.
Tubulin pull‐down experiments
For the tubulin pull‐downs shown in Fig 3A and Appendix Fig S3, Ni2+‐NTA beads were blocked overnight at 4°C in tubulin‐binding buffer (10 mM HEPES pH 7.5, 100 mM NaCl, 10% glycerol, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, 20 mM imidazole) containing 1 mg/ml BSA on a rotating wheel. Beads were collected by centrifugation, and 15 μl aliquots was either incubated with 200 μl tubulin‐binding buffer (for the negative control) or with 200 μl tubulin‐binding buffer containing 10 μM of various complexes for 2 h at 4°C. For each complex, two separate tubes were prepared, one was used for SDS–PAGE followed by Coomassie staining to confirm the presence of the desired complex on the beads, and the other was used for tubulin pull‐down followed by Western blot to investigate the tubulin‐binding capabilities of the respective complex. Beads were collected by centrifugation and washed twice with tubulin‐binding buffer. For Coomassie staining, the bound protein was eluted with 30 μl of tubulin‐binding buffer containing 500 mM imidazole, and 5 μl of this elution was analyzed by SDS–PAGE and staining. For tubulin pull‐downs, the collected beads were resuspended in 100 μl of tubulin‐binding buffer containing 4 μM of bovine tubulin (Cytoskeleton #TL238). The reactions were incubated for 1 h at 4°C, and after two washes in tubulin‐binding buffer, the bound material was eluted with 50 μl of tubulin‐binding buffer containing 500 mM imidazole. The various elutions, along with an “input” sample containing only tubulin as a positive control, were separated by SDS–PAGE, blotted on a nitrocellulose membrane and probed with a tubulin antibody (mouse monoclonal anti‐alpha tubulin, Sigma‐Aldrich T9026), followed by a secondary HRP‐linked antibody.
MT co‐sedimentation assay
According to the manufacturers protocol, 1× BRB80 buffer (80 mM PIPES‐KOH, 1 mM MgCl2, 1 mM EGTA, pH 6.8) supplemented with 3 mM GTP was used to solve lyophilized bovine tubulin (Cytoskeleton #TL238). After 45 min of incubation at 37°C, 50 μM Taxol was added to stabilize the polymerization of tubulin, followed by a second incubation for 40 min at 37°C. The MTs were pelleted by ultracentrifugation in a Beckmann TLA100 rotor (200,000 g, 30 min, 37°C) and resuspended in 20 μl of 1× BRB80 supplemented with 20 μM Taxol. 6 μM of _Cr_IFT54/20 complex was mixed with 2 μM of polymerized MTs in a total volume of 50 μl 1× BRB80 buffer containing 5 mM DTT and 20 μM Taxol and incubated for 20 min at room temperature. The reaction mix was carefully pipetted onto 100 μl of cushion buffer (1× BRB80, 50% glycerol, 20 μM Taxol) and ultracentrifuged at 100,000 g for 20 min at 25°C using a Beckmann TLA100 rotor. For SDS–PAGE analyses, a supernatant sample was taken and the pellet was washed 3 × with 1× BRB80 supplemented with 20 μM Taxol and resuspended in 2× SDS loading buffer for SDS–PAGE. Western Blot analysis was carried out as described above. Novagen 70796‐3 anti‐HIS antibody and HRP conjugated antibody (Enzo BML‐SA204‐0100) was used to detect the HIS‐tagged CrIFT54/20 complex.
Proteomics
For each pull‐down reaction, SEC‐purified GST‐IFT20 or IFT571–234‐GST‐tagged protein (baits) or GST protein (negative control) was immobilized on glutathione Sepharose (GSH) beads by incubating 200 μl of 10 μM protein with 20 μl bed volume of beads. After incubation for 1 h, the beads were collected and washed thrice with HMDEK buffer (30 mM HEPES, 5 mM MgSO4, 1 mM DTT, 0.5 mM EGTA, 25 mM KCl). Total cell extract of Chlamydomonas (_Cr_TCL) was prepared by lysing 1 g of freshly harvested Chlamydomonas culture pellet resuspended in 20 ml of cold HMDEK buffer supplemented with protease inhibitor cocktail. C. reinhardtii cells were sonicated by giving 8 s on/off pulses for 2 min with intermittent cooling and subsequently clarified by centrifugation at 20,000 g at 4°C for 15 min. Before pull‐down, 1 ml of lysate (volume corresponding to each reaction) was pre‐cleared by incubating it with 15 μl bed volume of GSH beads for 30 min at 4°C in rotating wheel after which beads were separated from the lysate by centrifugation at 20,000 g for 5 min. For pull‐down assay, 1 ml of pre‐cleared lysate was incubated with GST or GST‐tagged baits (IFT571–234‐GST or GST‐IFT20) at 4°C on a rotating wheel for 3 h. GSH resin along with co‐precipitated protein complexes was separated by low speed centrifugation at 400 g for 2 min. Beads were washed once with cold 20 mM HEPES buffer. Co‐precipitated proteins were eluted from GSH beads with GST elution buffer (10 mM Tris–HCl pH 8, 25 mM reduced glutathione). To observe salt‐dependent interaction between IFT571–234‐GST and its interactors, the co‐precipitated proteins were first eluted with 250 mM NaCl followed by elution with GST elution buffer. Elution fractions corresponding to each of the reaction in triplicates were loaded separately and stacked in a 10% SDS–PAGE gel. Total protein from each lane was digested according to the standard in‐gel digestion protocol (Shevchenko et al, 2006). Digested peptides were purified and concentrated on a C18 StageTip (Rappsilber et al, 2007) and were analyzed using a Q Exactive HF mass spectrometer (ThermoFisher Scientific). The peptides were separated via Easy nLC 1000 system (Thermo Scientific) via 140‐min gradient through a 15‐cm column, 75 μm inner diameter packed with 1.9 μm beads. The column was maintained at a constant temperature of 45°C. Raw data were processed using MaxQuant computational platform (Cox & Mann, 2008), and statistical analysis was done using the Perseus framework.
Accession codes
The coordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 5fmt, 5fmu, 5fmr, and 5fms.
Author contributions
MT and KW carried out the purification and interaction studies of Cr and Mm IFT proteins and complexes. KW determined the crystal structures of IFT54CH and carried out tubulin‐binding experiments with the help of SB. AM carried out the IFT52N purification and crystal structure determination with the help of MV. MA did the proteomics in Appendix Figs S1C and S4. MS helped with cloning and purification experiments. EL supervised the biochemical and structural experiments. MT, KW, and EL wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Review Process File
Acknowledgements
We thank the staff at the Swiss Light Source, Dr. Christian Benda, and Dr. Andreas Bracher for help with X‐ray diffraction data collection, the crystallization facility of the Max Planck Institute of Biochemistry (Munich) for access to crystallization screening, Dr. Stephan Uebel for ultracentrifugation, and Dr. Nagarjuna Nagaraj and Lissy Weyher at the MPI of Biochemistry core facility for mass spectrometric analyses. We are also grateful to Gregory Pazour for providing cDNAs for mouse IFTs. This work was funded by a project grant from the German Research Council (DGF; LO1627/2‐1), the European Research Council (ERC grant 310343), and by the EMBO Young Investigator Program.
The EMBO Journal (2016) 35: 773–790
References
- Absalon S, Blisnick T, Kohl L, Toutirais G, Doré G, Julkowska D, Tavenet A, Bastin P (2008) Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Mol Biol Cell 19: 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L‐W, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR (2008) ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol 183: 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacaj T, Lu Y, Shaham S (2008) The conserved proteins CHE‐12 and DYF‐11 are required for sensory cilium function in Caenorhabditis elegans. Genetics 178: 989–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SA, Freeman K, Luby‐Phelps K, Pazour GJ, Besharse JC (2003) IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J Biol Chem 278: 34211–34218 [DOI] [PubMed] [Google Scholar]
- Beales PL, Bland E, Tobin JL, Bacchelli C, Tuysuz B, Hill J, Rix S, Pearson CG, Kai M, Hartley J, Johnson C, Irving M, Elcioglu N, Winey M, Tada M, Scambler PJ (2007) IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat Genet 39: 727–729 [DOI] [PubMed] [Google Scholar]
- Beatson S, Ponting CP (2004) GIFT domains: linking eukaryotic intraflagellar transport and glycosylation to bacterial gliding. Trends Biochem Sci 29: 396–399 [DOI] [PubMed] [Google Scholar]
- Berbari NF, Kin NW, Sharma N, Michaud EJ, Kesterson RA, Yoder BK (2011) Mutations in Traf3ip1 reveal defects in ciliogenesis, embryonic development, and altered cell size regulation. Dev Biol 360: 66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Taschner M, Morawetz M, Basquin C, Lorentzen E (2011) Crystal structure of the intraflagellar transport complex 25/27. EMBO J 30: 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, Lorentzen E (2013a) Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science 341: 1009–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Engel BD, Lorentzen E (2013b) Intraflagellar transport complex structure and cargo interactions. Cilia 2: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Weber K, Engel BD, Lechtreck K‐F, Lorentzen E (2014) Getting tubulin to the tip of the cilium: one IFT train, many different tubulin cargo‐binding sites? BioEssays 36: 463–467 [DOI] [PubMed] [Google Scholar]
- Bieniossek C, Richmond TJ, Berger I (2008) MultiBac: multigene baculovirus‐based eukaryotic protein complex production. Curr Protoc Protein Sci Chapter 5: Unit 5.20 [DOI] [PubMed] [Google Scholar]
- Bizet AA, Becker‐Heck A, Ryan R, Weber K, Filhol E, Krug P, Halbritter J, Delous M, Lasbennes M‐C, Linghu B, Oakeley EJ, Zarhrate M, Nitschké P, Garfa‐Traore M, Serluca F, Yang F, Bouwmeester T, Pinson L, Cassuto E, Dubot P_et al_ (2015) Mutations in TRAF3IP1/IFT54 reveal a new role for IFT proteins in microtubule stabilization. Nat Commun 6: 8666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque OE, Reardon MJ, Li C, McCarthy J, Mahjoub MR, Ansley SJ, Badano JL, Mah AK, Beales PL, Davidson WS, Johnsen RC, Audeh M, Plasterk RHA, Baillie DL, Katsanis N, Quarmby LM, Wicks SR, Leroux MR (2004) Loss of C. elegans BBS‐7 and BBS‐8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev 18: 1630–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Cochran DA, Craige B, Kubo T, Witman GB (2015) Assembly of IFT trains at the ciliary base depends on IFT74. Curr Biol 25: 1583–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL (1998) Chlamydomonas kinesin‐II‐dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 141: 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Craft JM, Harris JA, Hyman S, Kner P, Lechtreck KF (2015) Tubulin transport by IFT is upregulated during ciliary growth by a cilium‐autonomous mechanism. J Cell Biol 208: 223–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam TJP, Townsend MJ, Turk M, Schlessinger A, Sali A, Field MC, Huynen MA (2013) Evolution of modular intraflagellar transport from a coatomer‐like progenitor. Proc Natl Acad Sci USA 110: 6943–6948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval B, Bright A, Lawson ND, Doxsey S (2011) The cilia protein IFT88 is required for spindle orientation in mitosis. Nat Cell Biol 13: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z‐C, Behal RH, Geimer S, Wang Z, Williamson SM, Zhang H, Cole DG, Qin H (2010) Chlamydomonas IFT70/CrDYF‐1 is a core component of IFT particle complex B and is required for flagellar assembly. Mol Biol Cell 21: 2696–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, Berger I (2006) Protein complex expression by using multigene baculoviral vectors. Nat Methods 3: 1021–1032 [DOI] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H (2007) When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 8: 880–893 [DOI] [PubMed] [Google Scholar]
- Follit JA, Xu F, Keady BT, Pazour GJ (2009) Characterization of mouse IFT complex B. Cell Motil Cytoskeleton 66: 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Ishihara T, Katsura I (1999) A novel WD40 protein, CHE‐2, acts cell‐autonomously in the formation of C. elegans sensory cilia. Development 126: 4839–4848 [DOI] [PubMed] [Google Scholar]
- Halbritter J, Bizet AA, Schmidts M, Porath JD, Braun DA, Gee HY, McInerney‐Leo AM, Krug P, Filhol E, Davis EE, Airik R, Czarnecki PG, Lehman AM, Trnka P, Nitschké P, Bole‐Feysot C, Schueler M, Knebelmann B, Burtey S, Szabó AJ_et al_ (2013) Defects in the IFT‐B component IFT172 Cause Jeune and Mainzer‐Saldino syndromes in humans. Am J Hum Genet 93: 915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Thein M, Brust‐Mascher I, Civelekoglu‐Scholey G, Lu Y, Acar S, Prevo B, Shaham S, Scholey JM (2011) Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat Cell Biol 13: 790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft CJ, Schafer JC, Zhang Q, Taulman PD, Yoder BK (2003) Identification of CHE‐13, a novel intraflagellar transport protein required for cilia formation. Exp Cell Res 284: 251–263 [DOI] [PubMed] [Google Scholar]
- Hou Y, Qin H, Follit JA, Pazour GJ, Rosenbaum JL, Witman GB (2007) Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol 176: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde C, Dickinson RJ, Houtzager VM, Cullum R, Montpetit R, Metzler M, Simpson EM, Roy S, Hayden MR, Hoodless PA, Nicholson DW (2006) Hippi is essential for node cilia assembly and Sonic hedgehog signaling. Dev Biol 300: 523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Diener DR, Mitchell A, Pazour GJ, Witman GB, Rosenbaum JL (2007) Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J Cell Biol 179: 501–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV (2003) Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426: 83–87 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF (2011) Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol 12: 222–234 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ide T, Yagi T, Jiang X, Hirono M, Sasaki H, Yanagisawa H, Wemmer KA, Stainier DY, Qin H, Kamiya R, Marshall WF (2014) TTC26/DYF13 is an intraflagellar transport protein required for transport of motility‐related proteins into flagella. Elife 3: e01566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen JA, San Agustin J, Follit JA, Pazour GJ (2008) Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J Cell Biol 183: 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010) XDS. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL (1993) A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA 90: 5519–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL (1995) The Chlamydomonas kinesin‐like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol 131: 1517–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitomo H, Iino Y (2008) Caenorhabditis elegans DYF‐11, an orthologue of mammalian Traf3ip1/MIP‐T3, is required for sensory cilia formation. Genes Cells 13: 13–25 [DOI] [PubMed] [Google Scholar]
- Lechtreck K‐F, Johnson EC, Sakai T, Cochran D, Ballif BA, Rush J, Pazour GJ, Ikebe M, Witman GB (2009a) The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol 187: 1117–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K‐F, Luro S, Awata J, Witman GB (2009b) HA‐tagging of putative flagellar proteins in Chlamydomonas reinhardtii identifies a novel protein of intraflagellar transport complex B. Cell Motil Cytoskeleton 66: 469–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman JM, Michaud EJ, Schoeb TR, Aydin‐Son Y, Miller M, Yoder BK (2008) The Oak Ridge Polycystic Kidney mouse: modeling ciliopathies of mice and men. Dev Dyn 237: 1960–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Inglis PN, Leitch CC, Efimenko E, Zaghloul NA, Mok CA, Davis EE, Bialas NJ, Healey MP, Héon E, Zhen M, Swoboda P, Katsanis N, Leroux MR (2008) An essential role for DYF‐11/MIP‐T3 in assembling functional intraflagellar transport complexes. PLoS Genet 4: e1000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling L, Goeddel DV (2000) MIP‐T3, a novel protein linking tumor necrosis factor receptor‐associated factor 3 to the microtubule network. J Biol Chem 275: 23852–23860 [DOI] [PubMed] [Google Scholar]
- Lucker BF, Behal RH, Qin H, Siron LC, Taggart WD, Rosenbaum JL, Cole DG (2005) Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J Biol Chem 280: 27688–27696 [DOI] [PubMed] [Google Scholar]
- Lunt SC, Haynes T, Perkins BD (2009) Zebrafish ift57, ift88, and ift172 intraflagellar transport mutants disrupt cilia but do not affect hedgehog signaling. Dev Dyn 238: 1744–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimainen M, Paavilainen S, Hakulinen N, Rouvinen J (2012) Structural analysis, enzymatic characterization, and catalytic mechanisms of β‐galactosidase from Bacillus circulans sp. alkalophilus. FEBS J 279: 1788–1798 [DOI] [PubMed] [Google Scholar]
- Moyer JH, Lee‐Tischler MJ, Kwon HY, Schrick JJ, Avner ED, Sweeney WE, Godfrey VL, Cacheiro NL, Wilkinson JE, Woychik RP (1994) Candidate gene associated with a mutation causing recessive polycystic kidney disease in mice. Science 264: 1329–1333 [DOI] [PubMed] [Google Scholar]
- Murayama T, Toh Y, Ohshima Y, Koga M (2005) The dyf‐3 gene encodes a novel protein required for sensory cilium formation in Caenorhabditis elegans. J Mol Biol 346: 677–687 [DOI] [PubMed] [Google Scholar]
- Nachury MV, Seeley ES, Jin H (2010) Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol 26: 59–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori Y, Zhao C, Saras A, Mukhopadhyay S, Kim W, Furukawa T, Sengupta P, Veraksa A, Malicki J (2008) Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane‐associated small GTPase Rab8. Nat Cell Biol 10: 437–444 [DOI] [PubMed] [Google Scholar]
- Ou G, Qin H, Rosenbaum JL, Scholey JM (2005) The PKD protein qilin undergoes intraflagellar transport. Curr Biol 15: R410–R411 [DOI] [PubMed] [Google Scholar]
- Pasek RC, Berbari NF, Lewis WR, Kesterson RA, Yoder BK (2012) Mammalian Clusterin associated protein 1 is an evolutionarily conserved protein required for ciliogenesis. Cilia 1: 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Witman GB (1999) The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol 144: 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Baker SA, Deane JA, Cole DG, Dickert BL, Rosenbaum JL, Witman GB, Besharse JC (2002) The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J Cell Biol 157: 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB (2005) Proteomic analysis of a eukaryotic cilium. J Cell Biol 170: 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL (2008) Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol 85: 23–61 [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG (1986) Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117: 456–487 [DOI] [PubMed] [Google Scholar]
- Piperno G, Mead K (1997) Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc Natl Acad Sci USA 94: 4457–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Bower R, Knott JA, Byrd P, Dentler W (1999) Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol Biol Cell 10: 693–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevo B, Mangeol P, Oswald F, Scholey JM, Peterman EJG (2015) Functional differentiation of cooperating kinesin‐2 motors orchestrates cargo import and transport in C. elegans cilia. Nat Cell Biol 17: 1536–1545 [DOI] [PubMed] [Google Scholar]
- Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL (2004) Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol 164: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Burnette DT, Bae Y‐K, Forscher P, Barr MM, Rosenbaum JL (2005) Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr Biol 15: 1695–1699 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro‐purification, enrichment, pre‐fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Richey EA, Qin H (2012) Dissecting the sequential assembly and localization of intraflagellar transport particle complex B in chlamydomonas. PLoS One 7: e43118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rix S, Calmont A, Scambler PJ, Beales PL (2011) An Ift80 mouse model of short rib polydactyly syndromes shows defects in hedgehog signalling without loss or malformation of cilia. Hum Mol Genet 20: 1306–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schou KB, Andersen JS, Pedersen LB (2013) A divergent calponin homology (NN‐CH) domain defines a novel family: implications for evolution of ciliary IFT complex B proteins. Bioinformatics 30: 899–902 [DOI] [PubMed] [Google Scholar]
- Schuck P (2000) Size‐distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78: 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In‐gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Signor D, Wedaman KP, Orozco JT, Dwyer ND, Bargmann CI, Rose LS, Scholey JM (1999) Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J Cell Biol 147: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow JJ, Ou G, Gunnarson AL, Walker MRS, Zhou HM, Brust‐Mascher I, Scholey JM (2004) Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol 6: 1109–1113 [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33: W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Samanta MP, Tongprasit W, Marshall WF (2005) Genome‐wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc Natl Acad Sci USA 102: 3703–3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmeier M, Raschle T, Mazurkiewicz J, Rippe K, Sinning I, Fitzpatrick TB, Tews I (2006) Structure of a bacterial pyridoxal 5'‐phosphate synthase complex. Proc Natl Acad Sci USA 103: 19284–19289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subota I, Julkowska D, Vincensini L, Reeg N, Buisson J, Blisnick T, Huet D, Perrot S, Santi‐Rocca J, Duchateau M, Hourdel V, Rousselle J‐C, Cayet N, Namane A, Chamot‐Rooke J, Bastin P (2014) Proteomic analysis of intact flagella of procyclic Trypanosoma brucei cells identifies novel flagellar proteins with unique sub‐localization and dynamics. Mol Cell Proteomics 13: 1769–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N (2004) A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131: 4085–4093 [DOI] [PubMed] [Google Scholar]
- Swiderski RE, Nakano Y, Mullins RF, Seo S, Bánfi B (2014) A mutation in the mouse ttc26 gene leads to impaired hedgehog signaling. PLoS Genet 10: e1004689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Bhogaraju S, Vetter M, Morawetz M, Lorentzen E (2011) Biochemical mapping of interactions within the intraflagellar transport (IFT) B core complex: IFT52 binds directly to four other IFT‐B subunits. J Biol Chem 286: 26344–26352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Bhogaraju S, Lorentzen E (2012) Architecture and function of IFT complex proteins in ciliogenesis. Differentiation 83: S12–S22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Kotsis F, Braeuer P, Kuehn EW, Lorentzen E (2014) Crystal structures of IFT70/52 and IFT52/46 provide insight into intraflagellar transport B core complex assembly. J Cell Biol 207: 269–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer JJ, Klem TJ, Deras ML, Davisson VJ, Smith JL (1996) The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families. Nat Struct Biol 3: 74–86 [DOI] [PubMed] [Google Scholar]
- Tsao C‐C, Gorovsky MA (2008) Different effects of Tetrahymena IFT172 domains on anterograde and retrograde intraflagellar transport. Mol Biol Cell 19: 1450–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CL, McIntyre JC, Norris SR, Jenkins PM, Zhang L, Pei Q, Verhey K, Martens JR (2014) Direct evidence for BBSome‐associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat Commun 5: 5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren KN, Craft JM, Tritschler D, Schauer A, Patel DK, Smith EF, Porter ME, Kner P, Lechtreck KF (2013) A differential cargo‐loading model of ciliary length regulation by IFT. Curr Biol 23: 2463–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Murcia NS, Chittenden LR, Richards WG, Michaud EJ, Woychik RP, Yoder BK (2003) Loss of the Tg737 protein results in skeletal patterning defects. Dev Dyn 227: 78–90 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Review Process File