Arf-like GTPase Arl8: Moving from the periphery to the center of lysosomal biology (original) (raw)
Abstract
Lysosomes are dynamic organelles that not only mediate degradation of cellular substrates but also play critical roles in processes such as cholesterol homeostasis, plasma membrane repair, antigen presentation, and cell migration. The small GTPase Arl8, a member of Arf-like (Arl) family of proteins, has recently emerged as a crucial regulator of lysosome positioning and membrane trafficking toward lysosomes. Through interaction with its effector SKIP, the human Arl8 paralog (Arl8b) mediates kinesin-1 dependent motility of lysosomes on microtubule tracks toward the cell periphery. Arl8b-mediated kinesin-driven motility is also implicated in regulating lytic granule polarization in NK cells, lysosome tubulation in macrophages, cell spreading, and migration. Moreover, Arl8b regulates membrane traffic toward lysosomes by recruiting subunits of the HOPS complex, a multi-subunit tethering complex that mediates endo-lysosome fusion. Here we provide a brief review on this recently characterized lysosomal GTPase and summarize the studies focusing on its known functions in regulating lysosomal motility and delivery of endocytic cargo to the lysosomes. We also explore the role of human Arl8b and its orthologs upon infection by intracellular pathogens.
Keywords: autophagy, Arf-like GTPase, Arl8, BORC, HOPS, late endosome, lysosome, membrane trafficking, Motors, SKIP
Abbreviations
Arl
Arf-like
Arl8
Arf-like GTPase 8
BLOC
Biogenesis of lysosome-related organelles
BORC
BLOC-one-related complex
CRISPR
Clustered regularly interspaced short palindromic repeats
EGFR
Epidermal growth factor receptor
GAP
GTPase activating protein
GEF
Guanine nucleotide exchange factor
Gie
GTPases indispensable for equal segregation of chromosomes
HOPS
Homotypic protein sorting complex
LAMP1
Lysosomal-associated membrane protein 1
LRO
Lysosome-related organelle
MTOC
Microtubule-organizing center
mTOR
Mammalian target of rapamycin
NatC
N-acetyl transferase complex C
NK
Natural Killer
RILP
Rab-interacting lysosomal protein
RNAi
RNA interference
SCV
Salmonella containing vacuole
SIF
Salmonella-induced filament
SKIP
Sif-A and kinesin-interacting protein
ToMV
Tomato Mosaic Virus
Vps
Vacuolar protein sorting
Introduction
Lysosomes are generally regarded as the waste disposal and recycling unit of the cell that mediates the turnover of lipids, proteins and other macromolecules. Cargo destined for degradation is trafficked through endocytic, phagocytic or autophagic routes, after which these compartments mature and undergo fusion with lysosomes.1-4 The degradative capacity of lysosomes is crucial for physiological processes such as nutrient provision, termination of signaling pathways, microbial killing and autophagic clearance of protein aggregates. As such, impairment of lysosomal function can manifest in diseased states including cancer, lysosomal storage disorders, and neurodegenerative diseases.5-9 Lysosome function is also required for various other cellular processes including cholesterol homeostasis, plasma membrane repair, antigen presentation, exosome release, and cell migration.10
Members of Rab, Arf and Arf-like (Arl) GTPases of the Ras superfamily are key molecular players that associate with distinct intracellular organelles and regulate membrane trafficking to and from these compartments.11 They act as molecular switches and alternate between GDP-bound (inactive) and GTP-bound (active) conformations–a process tightly regulated by their respective guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs). In the GTP-bound form, these small GTPases mediate membrane recruitment of effector proteins, which in turn facilitates downstream events during membrane trafficking including vesicle tethering, motility, and fusion.
Rab7, a quintessential member of Rab family of GTPases, has a well characterized role in regulating membrane trafficking toward late endosomes and lysosomes.12-16 Rab7 is known to localize primarily on late endosomal membranes, but some reports indicate its localization both to late endosomes and lysosomes. Rab7 interacts with a multitude of effectors to mediate endosomal maturation, late endosome-lysosome fusion and their motility along the cytoskeletal tracks.15-17 We refer the reader to a number of excellent reviews that provide an extensive overview of Rab7 function in membrane trafficking.15,16,18,19
More recently, attention has focused on the role of an Arl GTPase, Arl8–the only known small GTP-binding protein that is present primarily on mature lysosomes.20 Arl8 is emerging as an important regulator of lysosome motility and membrane trafficking in several organisms.20-25 In this review, we provide the first comprehensive overview of the known functions of the Arl8 family, particularly Arl8b, and highlight the interaction of Arl8b with effector proteins and its function in regulating the biology of the lysosome.
Discovery of ADP-ribosylation factor-like protein 8 (Arl8)
Arl8 was identified based on its sequence homology with other Arf family members that share common structural elements involved in an inter-switch toggle mechanism that couples the GDP-GTP transition with the recruitment of the active GTPase onto membranes.26-28 Cloning and characterization of Arl8b was further carried out by Sebald and co-workers who identified Arl8 in a clone-screen from a fetal cartilage cDNA library.29
Arl8 is a primitive GTPase that appeared early during evolution and has been highly conserved from protozoans to metazoans as well as in plants (Fig. 1). The early emergence of Arl8 during eukaryotic evolution is supported by its presence in the protist Giardia, which is thought to be the last common eukaryotic ancestor.26 There is a single gene encoding for Arl8 in several organisms, including fruit fly, worm, protozoa (Dictyostelium, Giardia, Trypanosoma) and some fungal species (Neurospora, Magnoporthe). There is no Arl8 homolog present in yeast, indicating its loss over the course of evolution of these species.26,27,29 In vertebrates, there are 2 closely related paralogs, Arl8a and Arl8b, which share 91% sequence identity to each other.20 Arl8 homologues are also expressed in plants with 4 Arl8-related genes identified in A. thaliana and N. tabacum.27,30
Figure 1.
Conservation of Arl8 protein across species. Arl8 is a primitive GTPase which is highly conserved from protozoans to metazoans as well in plants. Multiple sequence alignment was constructed using CLC sequence viewer software and NCBI accession numbers of the protein sequences used for sequence comparison were as follows: Homo sapiens (man) Arl8a, NP_620150.1; Homo sapiens (man) Arl8b, NP_060654.1; Pan troglodytes (chimpanzee), XP_001142369.1; Canis lupus familiaris (dog), XP_853013.1; Bos taurus (cattle), NP_001039536.1; Gallus gallus (chicken), XP_414440.3; Mus musculus (mouse), NP_080287.1; Rattus norvegicus (rat), NP_001019503.1; Xenopus tropicalis (frog), NP_001190182.1; Danio rerio (zebrafish), XP_002663861.1; Caenorhabditis elegans (worm), NP_502791.1; Drosophila melanogaster (fruit fly), NP_649769.1; Arabidopsis thaliana (small flowering plant), NP_568553.1; and Oryza sativa (rice plant), NP_001048027.1.
Structure of Arl8
The domain architecture of Arl8 is quite similar to that of the members of the Arf family. Like Arfs, Arl8 also contains a conserved N-terminal amphipathic helix that acts as a membrane anchor and aids in the firm association of the active GTP-bound form of Arl8 to the lipid bilayer.11,20 Analysis of the Arl8 tertiary structure from Homo sapiens (PDB accession number 1ZD9) and Nicotiana tabacum (65% identical in sequence to human Arl8) has accentuated that Arl8, similarly to other Arfs and Arls, is comprised of 2 switch regions (switch 1 and switch 2) that undergo conformational changes upon interacting with the γ-phosphate of GTP. Removal of the N-terminal helix (first 17 residues) from GDP-bound Arl8 leads to a reversible change in conformation to a GTP-like form and the equilibrium depends upon the concentration of Mg2+ which acts to stabilize the GTP-like conformation.31 Similarly to the other Arf-family proteins, structural analysis of Arl8 indicates that toggling of the interswitch region to a GTP-like form must ensue before nucleotide exchange can occur. In the active state (GTP-bound), the N-terminal helix is proposed to interact with the lipid bilayer via its hydrophobic side-chains and the inter-switch region is displaced, bringing the 2 switch regions upward for interaction with GTP.27,31
Mechanism of lysosomal targeting of Arl8b
Early studies reported human Arl8 localization to the mitotic spindle and described its role in regulating chromosome segregation during mitosis.32 Two reports challenged these initial findings and instead identified Arl8 as the first small GTPase that is predominantly localized to lysosomes and regulates its microtubule-dependent transport to the cell periphery.20,24 Arl8 localization as well as its function in regulating lysosome positioning and trafficking has since been found to be conserved across evolution.23,25,33,34
Arl8 is distinct from the other Arf proteins (but similar to the Arls) in that it lacks a conserved glycine residue at the second position crucial for myristoylation–a post-translational lipid modification required for membrane targeting of all Arf proteins.11 The glycine residue is replaced by leucine in case of Arl8b, and by isoleucine in case of Arl8a, rendering the human Arl8 proteins as potential substrates for N-acetyl transferase complex C (NatC). NatC catalyzes acetylation of the N-terminal methionine of its substrate only when the second position is occupied by a large hydrophobic amino acid residue. In cells lacking the catalytic subunit of human NatC complex (hMAK3), Arl8b reportedly showed a reduced lysosomal localization although no direct evidence of Arl8b acetylation by NatC complex was provided.35 Mutations in the hydrophobic face of the N-terminal α-helix of Arl8b also disrupted its membrane association, although the methionine at the N-terminus was still acetylated.20 Together, these findings define Arl8b membrane localization to be dependent upon both the acetylated methionine at the N-terminus and the hydrophobic residues of the amphipathic helix.
Membrane association for the small GTPases is regulated by the action of their respective GEFs and GAPs. Although the identity of the Arl8 GEF and GAP remains unknown, recent studies have identified BORC (BLOC-one-related complex), a multi-subunit protein complex that regulate Arl8b recruitment to lysosomes, and thereby mediate kinesin-dependent lysosome transport toward the cell periphery.36 BORC shares 3 of its 8 subunits with the BLOC-1 complex that is involved in the biogenesis of lysosome-related organelles.37 The clustered regularly interspaced short palindromic repeats (CRISPR)-based knockout of the BORC-specific subunit, myrlysin, was shown to cause Arl8b detachment from lysosomes along with clustering of lysosomes in the perinuclear region (an effect also observed upon Arl8b depletion). These results were further corroborated by siRNA-mediated silencing of other BORC subunits, which yielded similar phenotypes, signifying that BORC-dependent lysosomal recruitment of Arl8b is crucial for the centrifugal movement of lysosomes along microtubules. By contrast, depletion of subunits specific to the BLOC-1 complex did not change Arl8b localization or lysosome positioning, suggesting that BORC and BLOC-1 complex have non-redundant functions. Notably, BORC did not demonstrate GEF activity toward Arl8b. Rather BORC is proposed to act as an adaptor either for the Arl8 GEF, or for a receptor required for targeting Arl8b to lysosomes.
Arl8b effectors and their functions
In its GTP-bound form, Arl8 interacts with effectors, which then aid in lysosomal motility and function. While Arl8a and Arl8b are both known to localize at the lysosomes and induce their peripheral distribution upon overexpression (Fig. 2), it is not known if the 2 paralogs can compensate each other for their loss-of-function. The hitherto determined roles of the better characterized Arl8 paralog–Arl8b are reviewed here, although an extensive comprehension of the diversity in its function is still underway.
Figure 2.
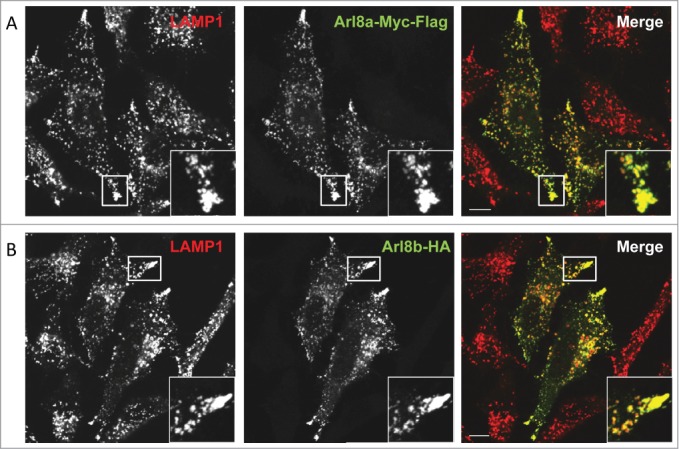
Human Arl8 paralogs (Arl8a and Arl8b) localize to lysosomes. HeLa cells were transfected with either Arl8a-Myc-Flag (A) or Arl8b-HA (B) and analyzed for lysosomal localization by confocal microscopy. The figure indicates the peripheral recruitment of LAMP1-positive endosomes (red) in Arl8a/b transfected cells (green). Colocalized pixels are represented in the inset. Scale bar: 10 μm.
Arl8b regulates lysosomal motility through interaction with SKIP/PLEKHM2
Lysosome positioning is increasingly being recognized as a crucial determinant of physiological processes including plasma membrane repair, cell migration, antigen presentation, and response to nutrient availability.10,38,39 Initial findings on Arl8 described a role for this lysosomal GTPase (inclusive of the 2 paralogs, Arl8a, and Arl8b) in inducing peripheral accumulation of lysosomes in mammalian cells (Figs. 2 and 3). Arl8 overexpression increased the long-range, rapid movement of lysosomes on microtubule tracks, resulting in some lysosomes dissociating from the microtubule at the cell periphery and accumulating in the membrane projections.20,24 In contrast, Arl8b depletion led to lysosome clustering at the microtubule-organizing center (MTOC), suggesting that Arl8b was required for lysosome positioning at the cell periphery (Fig. 3B).23,39
Figure 3.
Human Arl8b governs the distribution of lysosomes in mammalian cells. Control (A), Arl8b-silenced (B), and Arl8b-overexpressing (C) HeLa cells were transfected with GFP-LAMP1 and costained with antibodies to α-tubulin (blue) to label microtubules and pericentrin (red) to label MTOC. Knockdown of Arl8b results in lysosomal clustering at MTOC while its overexpression distributes lysosomes to the cell periphery. Scale bar: 10 μm.
A search for Arl8b effectors using affinity chromatography revealed that Arl8b in its GTP-bound state interacts with the soluble protein SKIP (Sif-A and kinesin-interacting protein, also known as PLEKHM2), which was previously identified as a protein that binds to light chain of kinesin-1. Moreover, depletion of either Arl8b or SKIP resulted in the perinuclear accumulation of lysosomes indicating that both Arl8b and its effector SKIP are crucial for the kinesin-dependent plus-end movement of lysosomes (Fig. 4). A recent report underlined an identical role of Arl8b and SKIP in regulating plus-end lysosomal movement in melanocytes. Surprisingly, while Arl8b dictated peripheral motility of conventional lysosomes in melanocytes, another small GTPase called Rab1a is responsible for anterograde transport of melanosomes (a lysosome-related organelle (LRO) in these cells) with SKIP coordinating movement of both organelles.40 Unlike in melanocytes, Arl8b localized to the LROs/lytic granules in Natural Killer (NK) cells, and along with SKIP and kinesin-1 was required for the polarized secretion of lytic granules toward the immune synapse, leading to NK cell-mediated cytotoxicity.41 Arl8b and SKIP were also found to mediate kinesin-driven lysosome tubulation in lipopolysaccharide-treated macrophages, which facilitates retention of fluid-phase endocytic content as well as phagosomes maturation and acidification in activated macrophages.22,42 A recent study has also elucidated roles for Arl8b and kinesin-1 in cell migration by mediating anterograde transport of the late endosomal protein complex p14-MP1 (MAPK/ERK kinase 1 partner MP1, and its endosomal adaptor protein p14) to focal adhesions, leading to their disassembly and turnover.43 Interestingly in invertebrates that lack a SKIP/PLEKHM2 homolog, Arl8 reportedly displayed a direct interaction with kinesin motor proteins. For instance, in C. elegans, Arl8 regulates the transport and spatial distribution of presynaptic proteins by direct binding to the kinesin motor UNC-04/KIF1A in neuronal cells.21,44 Together, these studies highlight the fact that Arl8b functions in different cell types to regulate the kinesin-dependent movement of lysosomes.
Figure 4.
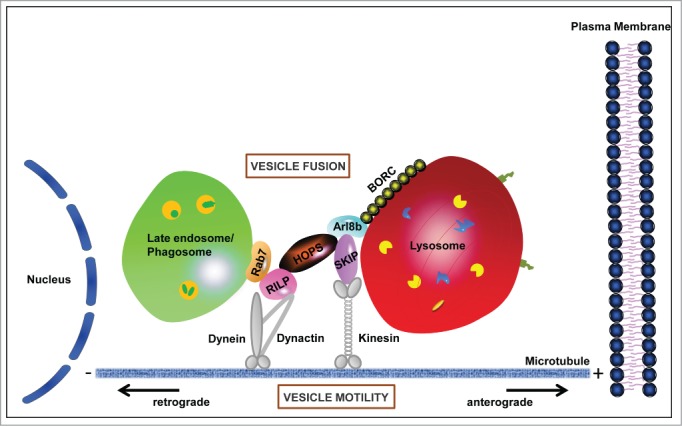
A model depicting the interplay of transport and fusion machinery operating at endosome-lysosome junction. Small GTPase Arl8b is recruited to lysosomal membranes by multi-subunit complex BORC. GTP-bound Arl8b interacts with its effectors: SKIP to mediate kinesin-dependent anterograde lysosomal movement and HOPS to accomplish vesicle (late endosome/phagosome) fusion at lysosomes.
Arl8b-dependent lysosome positioning regulates mammalian target of rapamycin complex 1 (mTORC1) activity and autophagosome-lysosome fusion
As discussed above, acetylation and the protein complex, BORC, play key roles in regulating Arl8b lysosomal localization. Additionally, factors such as nutrient availability to the cell and intracellular pH have also been reported to regulate Arl8b association with lysosomes.39 In the study by Koroluchuk and colleagues, nutrient deprivation and increase in intracellular pH were reported to reduce the levels of Arl8b and motor protein KIF2 in lysosomal fractions. This further led to the perinuclear clustering of lysosomes, resulting in the inactivation of the serine/threonine kinase, mTORC1, and an onset of autophagy. Knockdown of Arl8b also resulted in increased fusion of autophagosomes with lysosomes. Consequently, Arl8b-depleted cells had reduced levels of pathogenic substrates, including huntingtin protein aggregates associated with Huntington's disease and mutant A53T α-synuclein that causes some familial forms of Parkinson's disease.39 These findings suggest that selective inhibition of Arl8 function could act as a basis for design of potential drugs for the treatment of these neurodegenerative disorders.
Arl8b regulates endocytic traffic to lysosomes through interaction with homotypic fusion and protein sorting (HOPS) complex
Arl8b was first described to regulate cargo trafficking to lysosomes in C. elegans. Loss of Arl8 gene in C. elegans increased the number of late endosomal compartments and impaired the degradation of endocytosed macromolecules due to abrogated fusion of late endosomes with lysosomes.34 These findings were further reinforced in mammalian cells, wherein Arl8b depletion impaired the delivery of both fluid-phase (dextran and LDL) and membrane-bound (CD1d and MHC class II) cargo to the lysosomes.23,45 As a result, Arl8b silencing impeded the display of antigen presentation complexes on the surface of dendritic cells and macrophages, and thereby impaired the activation of immune responses against these antigens.23,45 Affinity purification followed by mass spectrometry analysis revealed peptides corresponding to the mammalian orthologs of Saccharomyces cerevisiae HOPS complex as Arl8b interaction partners.23 S. cerevisiae HOPS complex is a tethering factor that mediates fusion at the vacuole (the yeast equivalent of the mammalian lysosomes). However, the protein machinery responsible for recruitment of the yeast HOPS complex to vacuolar membranes differs from that in mammals. In yeast, small GTPase Ypt7 (Rab7 in mammals) directly binds to and recruits Vps41 and Vps39 subunits of HOPS complex to the vacuolar membranes. Recent data has suggested that in metazoan cells, Arl8b directly binds to Vps41 and regulates assembly of the HOPS complex on lysosomal membranes.23,25,46,47 Accordingly, Arl8b-depletion impaired lysosomal localization of Vps41 and other HOPS subunits. Furthermore, Arl8b-binding was required for Vps41 function in epidermal growth factor receptor (EGFR) trafficking to lysosomes and its lysosomal degradation.47 This study also highlighted a cross-talk between the 2 effectors of Arl8, demonstrating that SKIP recruits HOPS complex subunit Vps39 to Arl8b- and kinesin-positive lysosomes.47 Interestingly, 2 recent studies have shown that late endosomal Rab7 effector Rab-interacting lysosomal protein (RILP) that binds to the dynein adaptor, dynactin, also interacts with subunits of the HOPS complex.48,49 These findings suggest a model whereby Rab7-RILP and Arl8b-SKIP both interact with HOPS complex and play opposing roles in regulating positioning of HOPS endosomes by mediating their association with motor proteins (Fig. 4).
Arl8b regulates phagosome-lysosome fusion and microbial pathogenesis
Lysosomes play a crucial role in limiting the replication of intracellular pathogens by fusing with phagosomes that encapsulate the engulfed microbes. Arl8b was identified as an important host factor that regulates phago-lysosome fusion, and thereby clearance of phagocytosed microbes in macrophages.23 A similar function for Arl8 was also discerned in Caenorhabditis elegans, where Arl8 was identified as a novel factor required for delivery of phagocytosed cargo to lysosomes.25 Loss of Arl8 gene in C. elegans resulted in the delayed degradation and subsequent accumulation of apoptotic germ cells due to a defect in the formation of the phago-lysosomal compartment. Consistent with the previous reports in mammalian cells, the Arl8-binding to Vps41 was also found to be conserved in C. elegans.25
Many pathogenic microorganisms that invade eukaryotic cells exploit the host cell machinery for their survival. One such facultative intracellular pathogen, Salmonella enterica serovar typhimurium, establishes a replicative niche in host cells called the _Salmonella_-containing vacuole (SCV), through a process that involves extensive modulation of the host endocytic machinery.50 It has been observed that 4 to 14 hours post infection there is formation of long LAMP1-postive tubules known as _Salmonella_-induced filaments (SIFs) that extend from SCVs toward the cell periphery and are implicated in Salmonella's virulence and pathogenesis. Arl8b has recently been found to associate with SCVs in a screen for GTPases associated with maturing SCVs during Salmonella infection of HeLa cells. Arl8b silencing was observed to remarkably reduce SIF formation. Additionally, Arl8b promoted kinesin-1 recruitment to the SCVs, which in turn enhanced SIF formation. In accordance with its function in regulating anterograde transport of late endocytic compartments along microtubules, Arl8b was required for SCV movement to the cell periphery at later time points (i.e. 20 four hours post infection) which might assist in Salmonella dispersal and spread to the surrounding uninfected cells.51
Arl8's role in mediating host-pathogen response has also been identified in plants, where it was discovered as an important host factor required by the Tomato Mosaic Virus (ToMV) for its replication.30 ToMV replicates its genomic RNA in replication complexes formed on the host's intracellular membranes.52 Arl8 was reported to be present in complex with the ToMV replication proteins and a host 7-pass transmembrane protein, TOM1, in solubilized membranes from ToMV-infected tobacco cells. A lack of either Arl8 or TOM1 hindered the negative-strand RNA synthesis in an in vitro ToMV RNA translation-replication system. Simultaneous mutation in 2 of the 4 Arl8-related genes in Arabidopsis thaliana also abrogated ToMV replication, suggesting that Arl8 was essential for ToMV to establish infection in planta.30 Crystal structure of the helicase domain of ToMV superfamily 1 helicase solved recently has provided key insight into the helicase region required for binding to Arl8 and TOM1.53
Future Perspectives
Arl8 is an ancient Arf-like GTPase that localizes to lysosomes or equivalent compartments in Caenorhabditis elegans, Drosophila melanogaster, Arabidopsis thaliana and mammals and has been found to be highly conserved from protozoans to metazoans.20,30,34 Recent studies in lysosomal trafficking field have revealed key functions of Arl8 ranging from microtubule-dependent motility to cargo degradation to the pathogenesis of intracellular microorganisms. To date, 2 Arl8b effectors (SKIP and HOPS) have been discovered that in conjunction with Arl8b mediate lysosome motility and trafficking (Fig. 4). However, the plausible role of these effectors in regulating other allied functions is yet to be explored. For instance, while Arl8b and SKIP have independently been demonstrated to be regulators of Salmonella pathogenesis, together their role in Salmonella virulence, along with the other Arl8b effector HOPS, has not been elucidated. Given their role in vesicle fusion, these proteins could act as favorable targets for evasion strategies employed by pathogenic microorganisms.
As we have described, a recent study by Pu et al. showed that the multi-subunit complex BORC recruits Arl8b to lysosomes by coupling them to the microtubule plus-end-directed kinesin motor.36 Interestingly, the authors found that cells silenced for BORC subunit-myrlysin were defective in spreading and migration, implicating a conceivable involvement of these proteins during tumor metastasis. Further, the Arl8b-dependent pH control of lysosomal motility could be important for cellular behavior in acidic microenvironments, holding credible relevance to study of Arl8 function in cancer progression.
Autophagy is a fundamental process that regulates cellular survival with implications in a variety of diseases including cancer, neurodegenerative diseases and autoimmune disorders. Given the role of HOPS as a crucial regulator of autophagosome-lysosome fusion in mammals and Drosophila,54-56 the significance of Arl8b-HOPS interaction should be explored regulating the autophagy pathway. Previous reports have also characterized the neuroprotective role of Arl8b effector Vps41 in the clearance of α-synuclein aggregates in C. elegans.57,58 It would be interesting to explore the potential role of Arl8b in HOPS-mediated autophagic removal of protein aggregates, thereby exploring the propensity of Arl8b and its effector proteins to serve as therapeutic targets for various neurodegenerative diseases.
Recent studies have characterized Arl8b function in mediating the subcellular polarization of LROs.41 Acting along with kinesin KIF1A, Arl8b also regulates the transport of synaptic vesicles in C. elegans.21,44 As several diseases arise from defects in the biogenesis and function LROs, it will be important in the future to explore the potential role of Arl8 in transport of LROs and their exocytosis. Additionally, identification of factors that regulate GDP-GTP cycle of Arl8 will reveal mechanistic insights into how this small GTPase is recruited to lysosomal membranes. There is little doubt that future studies of Arl8 functions will contribute to better understanding of the role of lysosome positioning in health and disease.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
M.S. would like to thank Laura Simone for providing helpful editorial suggestions. M.S. would also like to acknowledge her mentors, Drs. Steve Caplan, Naava Naslavsky and Joyce Solheim (University of Nebraska Medical Center) and Dr. Michael B. Brenner (Brigham and Women's Hospital/Harvard Medical School) for their constant support and encouragement.
Funding
D.K. and A.S. acknowledge support from Council of Scientific & Industrial Research (CSIR)-University Grants Commission (UGC) and IISER‐Mohali. M.S. acknowledges financial support from the Wellcome Trust/Department of Biotechnology (DBT) India Alliance [grant number IA/I/12/500523]; and IISER‐Mohali. M.S. is a recipient of the Wellcome Trust/DBT India Alliance Intermediate Fellowship Award.
References
- 1.Luzio JP, Parkinson MD, Gray SR, Bright NA. The delivery of endocytosed cargo to lysosomes. Biochem Soc Trans 2009; 37:1019-21; PMID:19754443; http://dx.doi.org/ 10.1042/BST0371019 [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID:18305538; http://dx.doi.org/ 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol 2012; 22:407-17; PMID:22748206; http://dx.doi.org/ 10.1016/j.tcb.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wartosch L, Bright NA, Luzio JP. Lysosomes. Curr Biol 2015; 25:R315-6; PMID:25898096; http://dx.doi.org/ 10.1016/j.cub.2015.02.027 [DOI] [PubMed] [Google Scholar]
- 5.Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Biol 2004; 5:554-65; PMID:15232573; http://dx.doi.org/ 10.1038/nrm1423 [DOI] [PubMed] [Google Scholar]
- 6.Schultz ML, Tecedor L, Chang M, Davidson BL. Clarifying lysosomal storage diseases. Trends Neurosci 2011; 34:401-10; PMID:21723623; http://dx.doi.org/ 10.1016/j.tins.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 2010; 13:805-11; PMID:20581817; http://dx.doi.org/ 10.1038/nn.2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Usenovic M, Krainc D. Lysosomal dysfunction in neurodegeneration: the role of ATP13A2/PARK9. Autophagy 2012; 8:987-8; PMID:22561922; http://dx.doi.org/ 10.4161/auto.20256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 2013; 14:283-96; PMID:23609508; http://dx.doi.org/ 10.1038/nrm3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 2009; 10:623-35; PMID:19672277; http://dx.doi.org/ 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- 11.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol 2011; 12:362-75; PMID:21587297; http://dx.doi.org/ 10.1038/nrm3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell 2000; 11:467-80; PMID:10679007; http://dx.doi.org/ 10.1091/mbc.11.2.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceresa BP, Bahr SJ. rab7 activity affects epidermal growth factor:epidermal growth factor receptor degradation by regulating endocytic trafficking from the late endosome. J Biol Chem 2006; 281:1099-106; PMID:16282324; http://dx.doi.org/ 10.1074/jbc.M504175200 [DOI] [PubMed] [Google Scholar]
- 14.Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 2004; 117:4837-48; PMID:15340014; http://dx.doi.org/ 10.1242/jcs.01370 [DOI] [PubMed] [Google Scholar]
- 15.Wang T, Ming Z, Xiaochun W, Hong W. Rab7: role of its protein interaction cascades in endo-lysosomal traffic. Cell Signal 2011; 23:516-21; PMID:20851765; http://dx.doi.org/ 10.1016/j.cellsig.2010.09.012 [DOI] [PubMed] [Google Scholar]
- 16.Zhang M, Chen L, Wang S, Wang T. Rab7: roles in membrane trafficking and disease. Biosci Rep 2009; 29:193-209; PMID:19392663; http://dx.doi.org/ 10.1042/BSR20090032 [DOI] [PubMed] [Google Scholar]
- 17.Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol 2001; 11:1680-5; PMID:11696325; http://dx.doi.org/ 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- 18.Hyttinen JM, Niittykoski M, Salminen A, Kaarniranta K. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta 2013; 1833:503-10; PMID:23220125; http://dx.doi.org/ 10.1016/j.bbamcr.2012.11.018 [DOI] [PubMed] [Google Scholar]
- 19.Numrich J, Ungermann C. Endocytic Rabs in membrane trafficking and signaling. Biol Chem 2014; 395:327-33; PMID:24158421; http://dx.doi.org/ 10.1515/hsz-2013-0258 [DOI] [PubMed] [Google Scholar]
- 20.Hofmann I, Munro S. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J Cell Sci 2006; 119:1494-503; PMID:16537643; http://dx.doi.org/ 10.1242/jcs.02958 [DOI] [PubMed] [Google Scholar]
- 21.Klassen MP, Wu YE, Maeder CI, Nakae I, Cueva JG, Lehrman EK, Tada M, Gengyo-Ando K, Wang GJ, Goodman M, et al.. An Arf-like small G protein, ARL-8, promotes the axonal transport of presynaptic cargoes by suppressing vesicle aggregation. Neuron 2010; 66:710-23; PMID:20547129; http://dx.doi.org/ 10.1016/j.neuron.2010.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mrakovic A, Kay JG, Furuya W, Brumell JH, Botelho RJ. Rab7 and Arl8 GTPases are necessary for lysosome tubulation in macrophages. Traffic 2012; 13:1667-79; PMID:22909026; http://dx.doi.org/ 10.1111/tra.12003 [DOI] [PubMed] [Google Scholar]
- 23.Garg S, Sharma M, Ung C, Tuli A, Barral DC, Hava DL, Veerapen N, Besra GS, Hacohen N, Brenner MB. Lysosomal trafficking, antigen presentation, and microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity 2011; 35:182-93; PMID:21802320; http://dx.doi.org/ 10.1016/j.immuni.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagshaw RD, Callahan JW, Mahuran DJ. The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem Biophys Res Commun 2006; 344:1186-91; PMID:16650381; http://dx.doi.org/ 10.1016/j.bbrc.2006.03.221 [DOI] [PubMed] [Google Scholar]
- 25.Sasaki A, Nakae I, Nagasawa M, Hashimoto K, Abe F, Saito K, Fukuyama M, Gengyo-Ando K, Mitani S, Katada T, et al.. Arl8/ARL-8 functions in apoptotic cell removal by mediating phagolysosome formation in Caenorhabditis elegans. Mol Biol Cell 2013; 24:1584-92; PMID:23485564; http://dx.doi.org/ 10.1091/mbc.E12-08-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Kelly WG, Logsdon JM Jr., Schurko AM, Harfe BD, Hill-Harfe KL, Kahn RA. Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J 2004; 18:1834-50; PMID:15576487; http://dx.doi.org/ 10.1096/fj.04-2273com [DOI] [PubMed] [Google Scholar]
- 27.Pasqualato S, Renault L, Cherfils J. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for 'front-back' communication. EMBO Rep 2002; 3:1035-41; PMID:12429613; http://dx.doi.org/ 10.1093/embo-reports/kvf221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahn RA, Cherfils J, Elias M, Lovering RC, Munro S, Schurmann A. Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J Cell Biol 2006; 172:645-50; PMID:16505163; http://dx.doi.org/ 10.1083/jcb.200512057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sebald E, Krueger R, King LM, Cohn DH, Krakow D. Isolation of a new member of the ADP-ribosylation like factor gene family, ARL8, from a cartilage cDNA library. Gene 2003; 311:147-51; PMID:12853149; http://dx.doi.org/ 10.1016/S0378-1119(03)00584-5 [DOI] [PubMed] [Google Scholar]
- 30.Nishikiori M, Mori M, Dohi K, Okamura H, Katoh E, Naito S, Meshi T, Ishikawa M. A host small GTP-binding protein ARL8 plays crucial roles in tobamovirus RNA replication. PLoS Pathog 2011; 7:e1002409; PMID:22174675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okamura H, Nishikiori M, Xiang H, Ishikawa M, Katoh E. Interconversion of two GDP-bound conformations and their selection in an Arf-family small G protein. Structure 2011; 19:988-98; PMID:21742265; http://dx.doi.org/ 10.1016/j.str.2011.04.007 [DOI] [PubMed] [Google Scholar]
- 32.Okai T, Araki Y, Tada M, Tateno T, Kontani K, Katada T. Novel small GTPase subfamily capable of associating with tubulin is required for chromosome segregation. J Cell Sci 2004; 117:4705-15; PMID:15331635; http://dx.doi.org/ 10.1242/jcs.01347 [DOI] [PubMed] [Google Scholar]
- 33.Rosa-Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev Cell 2011; 21:1171-8; PMID:22172677; http://dx.doi.org/ 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakae I, Fujino T, Kobayashi T, Sasaki A, Kikko Y, Fukuyama M, Gengyo-Ando K, Mitani S, Kontani K, Katada T. The arf-like GTPase Arl8 mediates delivery of endocytosed macromolecules to lysosomes in Caenorhabditis elegans. Mol Biol Cell 2010; 21:2434-42; PMID:20484575; http://dx.doi.org/ 10.1091/mbc.E09-12-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starheim KK, Gromyko D, Evjenth R, Ryningen A, Varhaug JE, Lillehaug JR, Arnesen T. Knockdown of human N alpha-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol Cell Biol 2009; 29:3569-81; PMID:19398576; http://dx.doi.org/ 10.1128/MCB.01909-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell 2015; 33:176-88; PMID:25898167; http://dx.doi.org/ 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falcon-Perez JM, Starcevic M, Gautam R, Dell'Angelica EC. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem 2002; 277:28191-9; PMID:12019270; http://dx.doi.org/ 10.1074/jbc.M204011200 [DOI] [PubMed] [Google Scholar]
- 38.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol 2007; 8:622-32; PMID:17637737; http://dx.doi.org/ 10.1038/nrm2217 [DOI] [PubMed] [Google Scholar]
- 39.Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al.. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 2011; 13:453-60; PMID:21394080; http://dx.doi.org/ 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishida M, Ohbayashi N, Fukuda M. Rab1A regulates anterograde melanosome transport by recruiting kinesin-1 to melanosomes through interaction with SKIP. Sci Rep 2015; 5:8238; PMID:25649263; http://dx.doi.org/ 10.1038/srep08238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuli A, Thiery J, James AM, Michelet X, Sharma M, Garg S, Sanborn KB, Orange JS, Lieberman J, Brenner MB. Arf-like GTPase Arl8b regulates lytic granule polarization and natural killer cell-mediated cytotoxicity. Mol Biol Cell 2013; 24:3721-35; PMID:24088571; http://dx.doi.org/ 10.1091/mbc.E13-05-0259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrison RE, Bucci C, Vieira OV, Schroer TA, Grinstein S. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol 2003; 23:6494-506; PMID:12944476; http://dx.doi.org/ 10.1128/MCB.23.18.6494-6506.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schiefermeier N, Scheffler JM, de Araujo ME, Stasyk T, Yordanov T, Ebner HL, Offterdinger M, Munck S, Hess MW, Wickstrom SA, et al.. The late endosomal p14-MP1 (LAMTOR2/3) complex regulates focal adhesion dynamics during cell migration. J Cell Biol 2014; 205:525-40; PMID:24841562; http://dx.doi.org/ 10.1083/jcb.201310043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu YE, Huo L, Maeder CI, Feng W, Shen K. The balance between capture and dissociation of presynaptic proteins controls the spatial distribution of synapses. Neuron 2013; 78:994-1011; PMID:23727120; http://dx.doi.org/ 10.1016/j.neuron.2013.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michelet X, Garg S, Wolf BJ, Tuli A, Ricciardi-Castagnoli P, Brenner MB. MHC class II presentation is controlled by the lysosomal small GTPase, Arl8b. J Immunol 2015; 194:2079-88; PMID:25637027; http://dx.doi.org/ 10.4049/jimmunol.1401072 [DOI] [PubMed] [Google Scholar]
- 46.Gillingham AK, Sinka R, Torres IL, Lilley KS, Munro S. Toward a comprehensive map of the effectors of rab GTPases. Dev Cell 2014; 31:358-73; PMID:25453831; http://dx.doi.org/ 10.1016/j.devcel.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khatter D, Raina VB, Dwivedi D, Sindhwani A, Bahl S, Sharma M. The small GTPase Arl8b regulates assembly of the mammalian HOPS complex on lysosomes. J Cell Sci 2015; 128:1746-61; PMID:25908847; http://dx.doi.org/ 10.1242/jcs.162651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Kant R, Fish A, Janssen L, Janssen H, Krom S, Ho N, Brummelkamp T, Carette J, Rocha N, Neefjes J. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J Cell Sci 2013; 126:3462-74; PMID:23729732; http://dx.doi.org/ 10.1242/jcs.129270 [DOI] [PubMed] [Google Scholar]
- 49.Lin X, Yang T, Wang S, Wang Z, Yun Y, Sun L, Zhou Y, Xu X, Akazawa C, Hong W, et al.. RILP interacts with HOPS complex via VPS41 subunit to regulate endocytic trafficking. Sci Rep 2014; 4:7282; PMID:25445562; http://dx.doi.org/ 10.1038/srep07282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroeder N, Mota LJ, Meresse S. Salmonella-induced tubular networks. Trends Microbiol 2011; 19:268-77; PMID:21353564; http://dx.doi.org/ 10.1016/j.tim.2011.01.006 [DOI] [PubMed] [Google Scholar]
- 51.Kaniuk NA, Canadien V, Bagshaw RD, Bakowski M, Braun V, Landekic M, Mitra S, Huang J, Heo WD, Meyer T, et al.. Salmonella exploits Arl8B-directed kinesin activity to promote endosome tubulation and cell-to-cell transfer. Cell Microbiol 2011; 13:1812-23; PMID:21824248; http://dx.doi.org/ 10.1111/j.1462-5822.2011.01663.x [DOI] [PubMed] [Google Scholar]
- 52.Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell 2002; 9:505-14; PMID:11931759; http://dx.doi.org/ 10.1016/S1097-2765(02)00474-4 [DOI] [PubMed] [Google Scholar]
- 53.Nishikiori M, Sugiyama S, Xiang H, Niiyama M, Ishibashi K, Inoue T, Ishikawa M, Matsumura H, Katoh E. Crystal structure of the superfamily 1 helicase from Tomato mosaic virus. J Virol 2012; 86:7565-76; PMID:22573863; http://dx.doi.org/ 10.1128/JVI.00118-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell 2014; 25:1327-37; PMID:24554770; http://dx.doi.org/ 10.1091/mbc.E13-08-0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takats S, Pircs K, Nagy P, Varga A, Karpati M, Hegedus K, Kramer H, Kovacs AL, Sass M, Juhasz G. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell 2014; 25:1338-54; PMID:24554766; http://dx.doi.org/ 10.1091/mbc.E13-08-0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wartosch L, Gunesdogan U, Graham SC, Luzio JP. Recruitment of VPS33A to HOPS by VPS16 Is Required for Lysosome Fusion with Endosomes and Autophagosomes. Traffic 2015; 16:727-42; PMID:25783203; http://dx.doi.org/ 10.1111/tra.12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamamichi S, Rivas RN, Knight AL, Cao S, Caldwell KA, Caldwell GA. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson's disease model. Proc Natl Acad Sci U S A 2008; 105:728-33; PMID:18182484; http://dx.doi.org/ 10.1073/pnas.0711018105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrington AJ, Yacoubian TA, Slone SR, Caldwell KA, Caldwell GA. Functional analysis of VPS41-mediated neuroprotection in Caenorhabditis elegans and mammalian models of Parkinson's disease. J Neurosci 2012; 32:2142-53; PMID:22323726; http://dx.doi.org/ 10.1523/JNEUROSCI.2606-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]

