A Molecular Mechanism to Regulate Lysosome Motility for Lysosome Positioning and Tubulation (original) (raw)
. Author manuscript; available in PMC: 2016 Sep 7.
Published in final edited form as: Nat Cell Biol. 2016 Mar 7;18(4):404–417. doi: 10.1038/ncb3324
Abstract
To mediate the degradation of bio-macromolecules, lysosomes must traffic towards cargo-carrying vesicles for subsequent membrane fusion or fission. Mutations of the lysosomal Ca2+ channel TRPML1 cause lysosome storage disease (LSD) characterized by disordered lysosomal membrane trafficking in cells. Here we show that TRPML1 activity is required to promote Ca2+-dependent centripetal movement of lysosomes towards the perinuclear region, where autophagosomes accumulate, upon autophagy induction. ALG-2, an EF-hand-containing protein, serves as a lysosomal Ca2+ sensor that associates physically with the minus-end directed dynactin-dynein motor, while PI(3,5)P2, a lysosome-localized phosphoinositide, acts upstream of TRPML1. Furthermore, the PI(3,5)P2-TRPML1-ALG-2-dynein signaling is necessary for lysosome tubulation and reformation. In contrast, the TRPML1 pathway is not required for the perinuclear accumulation of lysosomes observed in many LSDs, which is instead likely caused by secondary cholesterol accumulation that constitutively activates Rab7-RILP-dependent retrograde transport. Collectively, Ca2+ release from lysosomes provides an on-demand mechanism regulating lysosome motility, positioning, and tubulation.
Keywords: lysosome reformation; TRPML1; phosphoinositide; PI(3,5)P2; cholesterol; membrane trafficking; dynein; ALG-2; nutrient starvation
Introduction
Lysosomes in animal cells accept bio-materials destined for degradation from cargo vesicles, which include endosomes, autophagosomes, and phagosomes1. Their number, size, and positioning are regulated tightly to meet constantly-changing cellular needs. Upon completion of digestion, lysosomes can undergo lysosome exocytosis2,3, lysosome-to-Golgi trafficking, or lysosome reformation, wherein nascent proto-lysosomes bud off from tubular lysosomal structures4. Vesicle trafficking (i.e. fusion and fission between membranous organelles) plays a key role in these cellular processes. Membrane fusion and fission require vesicles to move in a specific direction. Like other organelles, the long-range movement of lysosomes requires microtubule-based motor proteins5,6. Kinesin motors travel centrifugally (i.e. anterogradely, or outwardly) towards the plus-ends of microtubules, which are typically found at the cell periphery5, whereas dynein motors move centripetally (i.e. retrogradely, or inwardly) towards the minus-ends of microtubules, which are typically perinuclear, near the microtubule-organizing center (MTOC)7,8. Kinesin and dynein association determines directions of lysosome transport in response to cellular cues5,6.
Under nutrient deprivation conditions and autophagy induction, autophagosomes form rapidly and accumulate in the perinuclear region of the cell6. Efficient autophagosome-lysosome fusion requires that lysosomes, which are scattered throughout the cytoplasm under resting conditions, also amass rapidly in the perinuclear region6. Starvation may induce cytosolic alkalization, thereby boosting minus-end-directed motility6. Because alkalization of the cytosol as a whole would interfere with the motility of other organelles, it is likely that there are local mechanisms that regulate lysosome mobility selectively.
The motility of intracellular organelles appears to be regulated by Ca2+ 9. For instance, mitochondrial motility is regulated by local increases of cytosolic Ca2+ in an on-demand manner9. The primary Ca2+ channel in the lysosome is believed to be transient receptor potential mucolipin 1 (TRPML1, a.k.a. MCOLN1 or ML1)10,11. TRPML1 participates in late-endocytic membrane trafficking, and human mutations of the gene underlie mucolipidosis type IV (ML-IV)12, a lysosomal storage disease (LSD) characterized by lysosomal storage and autophagic defects13,14. Previous studies have implicated TRPML1 in phagosome-lysosome fusion15, autophagosome-lysosome fusion, and membrane fission from endolysosomes16. However, some of the observed trafficking defects in ML-IV cells may be caused by secondary mechanisms due to chronic lysosome storage16. In the current study, the role of lysosomal Ca2+ and TRPML1 in regulating lysosome motility was examined by manipulating TRPML1 activity acutely through specific synthetic agonists or antagonists.
Results
TRPML1 is necessary for on-demand (acute) retrograde transport of lysosomes towards the MTOC
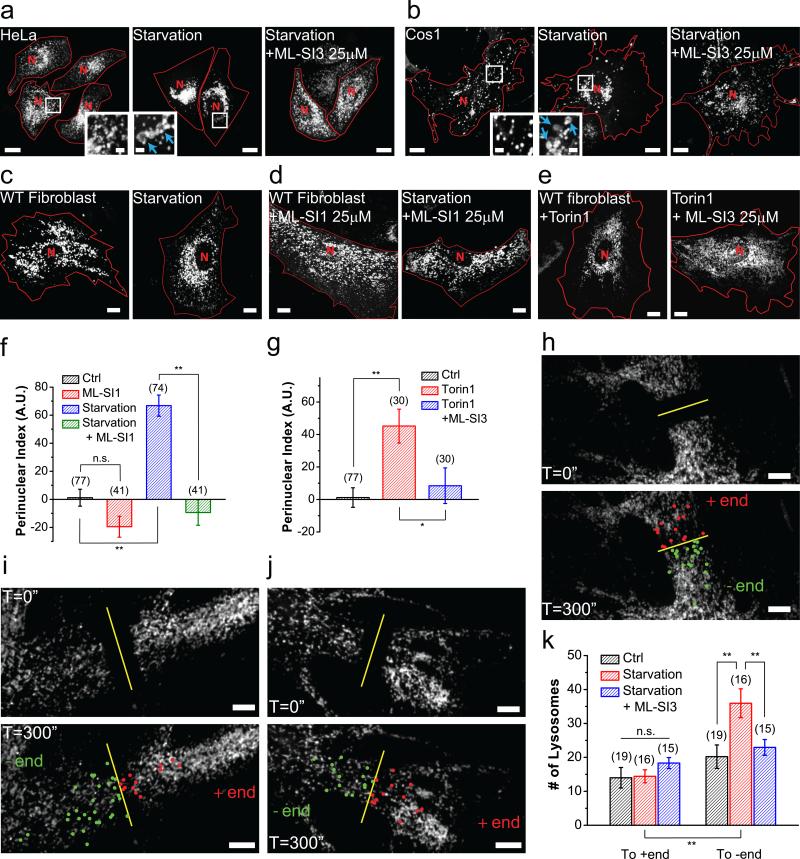
We investigated lysosome positioning in a morphologically diverse series of mammalian cells, including HeLa cells, Cos1 cells, and primary mouse fibroblasts. In cells transfected with Lamp1-mCherry, a marker of late endosomes and lysosomes (LELs, referred to as “lysosomes” for simplicity hereafter), lysosomes were scattered throughout the cytoplasm under normal culture conditions (Fig. 1a-c, Supplementary Figure 1a, 1b). Serum starvation triggered rapid autophagy and generation of autophagosomes, evidenced by the formation of LC3 puncta (Supplementary Figure 1e). Autophagosomes accumulated primarily in the perinuclear region (Supplementary Figure 1e)17,18. Consistent with previous studies6, upon starvation, lysosomes were redistributed rapidly towards the perinuclear region, especially the microtubule-organizing center (MTOC; Fig. 1a-c, 1f; Supplementary Figure 1b-d). It has been hypothesized that this redistribution may promote membrane fusion between lysosomes and autophagosomes17. Indeed, Lamp1-positive compartments in the perinuclear region of starved cells were often larger than those in fed cells (Fig. 1a, 1b, insets), suggesting that they were secondary lysosomes, most likely autolysosomes4. When autophagy was triggered by Torin-1 through inhibition of mTOR19, lysosomes also underwent perinuclear redistribution (Fig. 1e, 1g).
Figure 1. TRPML1 channel activity is required for acute, minus-end-directed retrograde transport of lysosomes.
(a, b) Representative HeLa cells (a) and Cos1 cells (b) transfected with the late-endosome and lysosome marker Lamp1-mCherry under 2 h serum starvation with (right) or without (middle) the TRPML1 inhibitor ML-SI3 (25 μM). Insets illustrate the size of lysosomes with and without starvation (same magnification, blue arrows indicate enlarged lysosomes). (c) Representative WT mouse fibroblasts transfected with Lamp1-mCherry in 2 h starvation condition (right). (d) Lysosome (labelled with Lamp1) distribution in WT fibroblasts in the presence of the TRPML1 inhibitor ML-SI1 (25 μM) with (right) or without (left) starvation. (e) Lysosome distribution in WT fibroblasts treated with mTOR inhibitor Torin 1 (1 μM) or together with ML-SI3 (25 μM) for 2 h. (f) Quantitative analyses of lysosome distribution in the experiments shown in (c) and (d). The intracellular distribution of Lamp1-positive vesicles was quantified as described in Materials & Methods. Fibroblasts were chosen for most quantification analyses for their large cell area, regular shape, and ML1 KO availability. (g) Quantification of groups shown in (e). (h-j) FRAP analysis of lysosome movement in WT fibroblasts without any treatment (h), with 15– 30 min starvation (i), or with starvation in the presence of 25 μM ML-SI3 (j). Snap images immediately after (top panels) or 5 min (bottom panels) after photobleaching are shown. Lysosomes that traveled across the midline of the photobleached region (yellow line) towards the nucleus (retrograde) are labeled green; those moving away from the nucleus (anterograde) are labeled red. (k) Quantification of lysosomes in (h–j) undergoing retrograde or anterograde transport. Red lines in images outline cell boundaries and the red “N” marks the nucleus. Graphed data are presented as means ± SEM, the numbers of cells (n) used for quantification were pooled across at least three independent experiments, and are shown in the parentheses (for each sample group, no sample was excluded). *p < .05, **p < .01 in ANOVA. Scale bars = 10 μm, and 2 μm for insets.
We next analyzed the directional movement of lysosomes using fluorescence recovery after photobleaching (FRAP). Under resting conditions, roughly equal number of lysosomes traveled retrogradely and anterogradely in mouse fibroblasts (Fig. 1h, 1k; Supplementary Video 1). Brief (30 min) starvation resulted in a selective increase in retrograde transport of lysosomes without affecting anterograde transport (Fig. 1i, 1k; Supplementary Video 2). Hence, starvation may upregulate retrograde migration mechanisms, producing a redistribution of lysosomes to the perinuclear area.
Lysosome motility, in both retrograde and anterograde directions, was reduced significantly in mouse fibroblasts that were treated with a membrane-permeable form of the fast Ca2+ chelator BAPTA-AM 16 (Supplementary Video 3), suggesting that Ca2+ plays an essential role in the regulation of lysosome motility. Given the delayed delivery of autophagic substrates to lysosomes and autophagosome accumulation in TRPML1-deficient cells20,21, we investigated the role of TRPML1 in Ca2+-dependent lysosome mobility by manipulating TRPML1 activity acutely with synthetic TRPML1 inhibitors (ML-SIs; see Supplementary Figure 2a)3,22.
The perinuclear accumulation of lysosomes under short-term starvation was blocked by ML-SI1 or ML-SI3, two structurally-unrelated ML-SIs3,23 (Fig. 1a-d, 1f), suggesting that the effects were specific to TRPML1. TRPML1 inhibition under short-term starvation also led to the accumulation of autophagosomes (Supplementary Figure 1f, 1g), suggesting a role of TRPML1 activity in the delivery of lysosomes during autolysosome formation. Consistent with this prediction, FRAP analysis revealed that starvation-induced retrograde migration of lysosomes was reduced by ML-SI3 (Fig. 1j, 1k; Supplementary Video 4). Likewise, when autophagy was triggered by mTOR inhibitors19, the perinuclear redistribution of lysosomes was suppressed upon acute inhibition of TRPML1 (Fig. 1e, 1g).
Starvation has been shown to cause rapid cytosolic alkalization in HeLa cells, which is sufficient to induce perinuclear localization of lysosomes6. Also, whole-endolysosome TRPML1 currents were facilitated by an alkaline cytosolic pH (Supplementary Figure 2b). Interestingly, inhibition of TRPML1 also suppressed retrograde migration of lysosomes induced by acute cytosolic alkalization (Supplementary Figure 2c-f). Hence, TRPML1 is required to drive on-demand retrograde migration of lysosomes in response to autophagy induction and cytosolic alkalization.
TRPML1 activation is sufficient to promote Ca2+-dependent retrograde transport of lysosomes
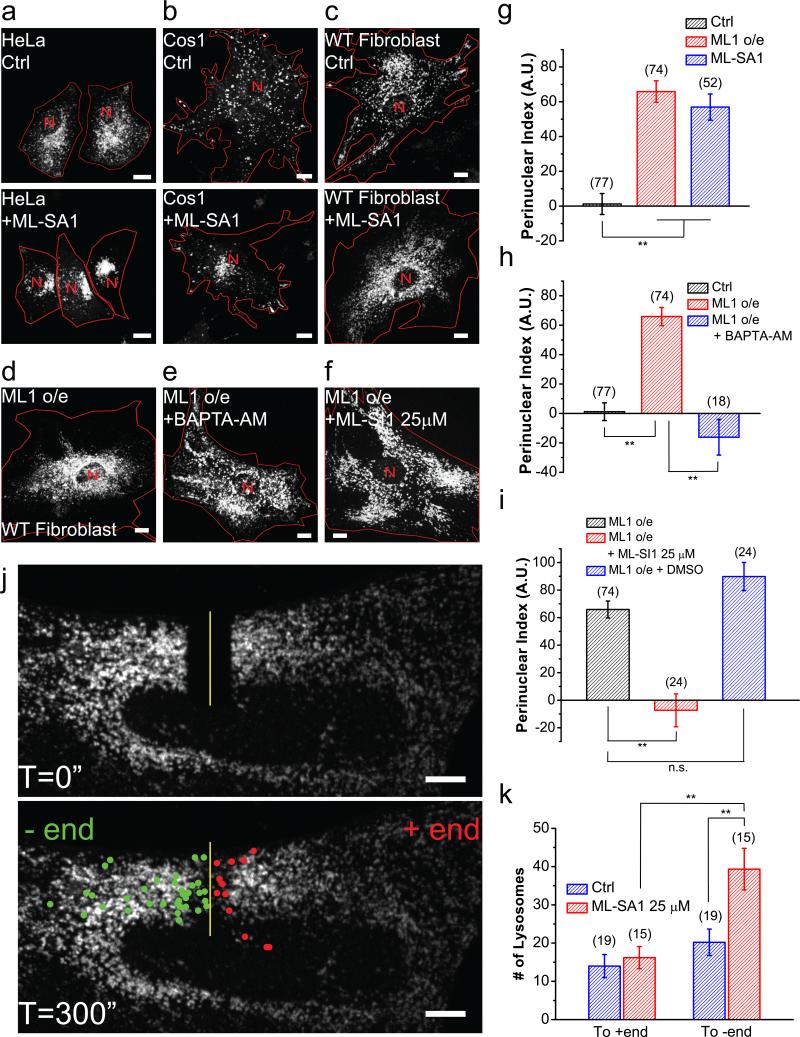
Next, we tested whether artificial activation of TRPML1 is sufficient to induce retrograde migration and perinuclear accumulation of lysosomes. When mouse fibroblasts, Cos1 cells, or HeLa cells were treated with synthetic TRPML1 agonists (ML-SAs) 23,24, perinuclear localization of lysosomes was markedly increased (Fig. 2a-c, 2g; Supplementary Figure 3a). Overexpression of TRPML1, but not the related TRPML2 or TRPML3, also resulted in perinuclear localization (Fig. 2d, 2g; Supplementary Figure 3b-e). The effect of overexpression could be readily reversed by BAPTA-AM (Fig. 2e, 2h) or TRPML1 inhibitors22 (Fig. 2f, 2i; Supplementary Figure 3f, 3g).
Figure 2. Activation of TRPML1 is sufficient to promote Ca2+-dependent retrograde migration of lysosomes.
HeLa cells (a) Cos1 cells (b) and WT fibroblasts (c) with (bottom) and without (top) 25 μM MLSA1 for 2 h. (d-f) Lysosome distribution in TRPML1-overexpessing WT fibroblasts (d) treated with 10 μM BAPTA-AM for 1 h (e) or 25 μM ML-SI1 for 2 h (f). (g) Quantification of lysosome distribution in the experiments shown in (c) and (d). (h) Quantification of experimental group shown in (e). (i) Quantification of experimental group shown in (f). (j) FRAP analysis of lysosome retrograde transport upon ML-SA1 application. (k) Quantification of lysosomes shown in (j) undergoing retrograde and anterograde transport after photobleaching. Red lines delineate cell boundaries and red “N” mark nuclei. Graphed data are presented as means ± SEM, the numbers of cells (n) used for quantification were pooled across at least three independent experiments and are shown in the parentheses (for each sample group, no sample was excluded). *p < .05, **p < .01 in ANOVA. Scale bars = 10 μm.
In FRAP analyses, as well as in time-lapse imaging, acute application of ML-SA1 (≤30 min) increased minus-end directed migration of lysosomes significantly (Fig. 2j, 2k;Supplementary Videos 5-7). In contrast, the distribution of mitochondria was not affected by ML-SA1 application (Supplementary Figure 3h-j). Conversely, time-lapse imaging showed that in TRPML1-overexpressing cells, ML-SI3 application resulted in the dispersal of lysosomes (Supplementary Video 8). Collectively, these results suggest that increasing TRPML1 activity is sufficient to induce perinuclear redistribution and retrograde transport of lysosomes.
Chronic effects of cholesterol accumulation on lysosome distribution
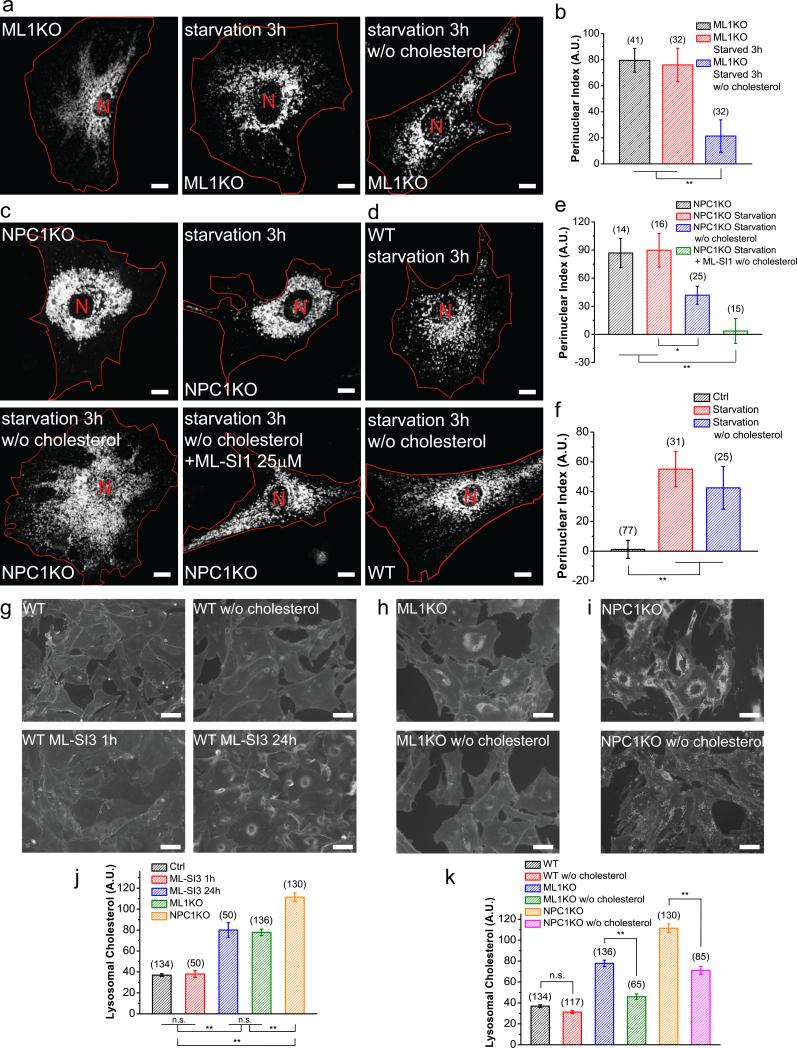
Lysosomes are distributed perinucearly in many LSD cells2,16 , including TRPML1 knockout (ML1 KO) fibroblasts (Fig. 3a-f). This distribution is opposite to that seen with transient TRPML1 inhibition. When we increased the treatment time of the TRPML1 inhibitors to >6 h and up to 48 h, lysosomes became progressively more perinuclear in WT fibroblasts, resembling the distribution in ML1 KO fibroblasts (Supplementary Figure 4a-c). Given the established role of cholesterol in promoting minus-end motility of lysosomes in cooperation with Rab7 and RILP25,26, and given that cholesterol accumulates in lysosomes in a variety of LSDs27 including ML-IV14 (Supplementary Figure 4e), we investigated whether cholesterol accumulation, caused by chronic lysosomal dysfunction, could account for the abnormal lysosome mobility and distribution in ML-IV and other LSDs.
Figure 3. Cholesterol accumulation causes perinuclear localization of lysosomes in LSDs.
(a) Representative images showing Lamp1-mCherry-transfected ML1 KO fibroblasts in starved cells that were treated with simvastatin and mevalonolactone to deplete cholesterol. (b) Quantification of groups shown in (a). (c) Representative images showing Lamp1-mCherry transfected NPC1 KO fibroblasts (upper left), starved for 3 h (upper right), starved with cholesterol depletion (bottom left), or starved with cholesterol depletion in the presence of 25 μM ML-SI1 (bottom right). (d) Effect of cholesterol depletion on starvation-induced lysosome redistribution in WT fibroblasts. (e) Quantification of groups shown in (c). (f) Quantification of observations shown in (d). (g) Filipin staining of WT fibroblasts (upper left) and WT fibroblasts depleted of cholesterol with simvastatin (upper right), WT fibroblasts treated with 25 μM MLSI3 for 1 h (bottom left) or 24 h (bottom right). (h) ML1 KO fibroblasts with (bottom) or without (upper) cholesterol depletion. (i) NPC1 KO fibroblasts with (bottom) or without (upper) cholesterol depletion. (j) Quantitative comparison of filipin staining in WT fibroblasts treated with different durations of ML-SI3 versus ML1 KO and NPC1 KO fibroblasts. (k) Quantification of filipin stating in WT, ML1 KO, NPC1 KO fibroblasts with or without cholesterol depletion. Red lines outline cell boundaries. Graphed data are presented as means ± SEM, the numbers of cells (n) used for quantification were pooled across at least three independent experiments and are shown in the parentheses. *p < .05, **p < .01 in ANOVA. Scale bars = 10 μm for (a), (c) and (d), and = 50 μm for (g), (h) and (i).
Significant elevation of cholesterol was observed in the lysosomes of ML1 KO fibroblasts, as well as in WT fibroblasts that were treated with ML-SI3 for a prolonged period of time (>6 h), but not in WT cells treated with ML-SI3 for a short (1 h) duration (Fig. 3g, 3h, 3j). Hence cholesterol accumulation in ML1 KO cells might have promoted minus-end motility of lysosomes independent of TRPML126. Indeed, reduction of cholesterol with simvastatin26 (Fig. 3g-i, 3k) resulted in more peripherally-localized lysosomes in ML1 KO fibroblasts (Fig. 3a, 3b), as well as in fibroblasts from NPC1 KO mice (Fig. 3c, 3e), a mouse model of cholesterol storage disease NPC27,28. Taken together, perinuclear lysosome localization observed with long-term loss of TRPML1 activity or in other LSDs may be due to secondary accumulation of cholesterol. Therefore, acute manipulations are needed to investigate the mechanisms of lysosome mobility.
TRPML1 promotes retrograde trafficking independent of the Rab7-RILP pathway
Cholesterol has been previously shown to promote retrograde transport of lysosomes by facilitating the Rab7-RILP pathway through the cholesterol sensor protein ORP1L26,29. In WT fibroblasts, overexpression of the constitutively active form of Rab7 (Rab7-Q67L)30, as well as the Rab7 effector, RILP31, resulted in perinuclear accumulation of lysosomes (Fig. 4a-d). However, ML-SI3 failed to reverse the perinuclear localization under these conditions (Fig. 4ad). Overexpression of dominant negative Rab7 (Rab7-T22N)31,32 did not prevent perinuclear accumulation of lysosomes under acute starvation, or under ML-SA1 application (Fig. 4e, 4g), but readily suppressed the perinuclear accumulation under prolonged inhibition of TRPML1 or in ML1 KO fibroblasts (Fig. 4f, 4h, and Supplementary Figure 4d). Hence, the perinuclear localization of lysosomes observed in ML1 KO cells is likely due to the activation of the Rab7-RILP-ORP1L pathway by cholesterol. Taken together, these results suggest that TRPML1 and cholesterol-Rab7-RILP most likely function in two separate pathways to promote retrograde transport of lysosomes.
Figure 4. TRPML1 promotes retrograde migration of lysosomes independent of the Rab7-RILP pathway.
(a, b) Representative images showing WT fibroblasts overexpressing Lamp1-mCherry and RILP-GFP (a) or Rab7-Q67L-GFP (b) in the presence or absence of 25 μM ML-SI3 for 2 h. (c) Quantification of groups shown in (a). (d) Quantification of groups shown in (b). (e) Representative images showing WT fibroblasts transfected with Lamp1-mCherry and Rab7-T22N-GFP, then left untreated (upper panels), serum starved for 2 h (middle panels), or incubated with 25 μM ML-SA1 for 2 h (bottom panels). (f) Representative images showing ML1 KO fibroblasts overexpressing Lamp1-mCherry and Rab7-T22N-GFP. (g) Quantification of groups shown in (e). (h) Quantification of the group shown in (f). Red lines outline cell boundaries. Graphed data are presented as means ± SEM, the numbers of cells (n) used for quantification were pooled across at least three independent experiments and are shown in the parentheses. *p < .05, **p < .01 in ANOVA. Scale bars = 10 μm.
The role of PI(3,5)P2 in retrograde trafficking of lysosomes
Phosphatidylinositol-3,5-bisphosphate (PI(3,5)P2) is a lysosome-localized phosphoinositide33 that regulates autophagy during nutrient deprivation34 and is the only known endogenous agonist of TRPML12. It binds directly to several positively-charged residues in TRPML1's N-terminus, thereby activating the channel35,36. Prolonged PI(3,5)P2 depletion leads to severe enlargement of lysosomes that occupy the majority of the cytosolic space37,38. Short-term (1 h) treatment with YM 201636 or Apilimod (1 μM; well-established synthetic inhibitors of the PI(3,5)P2 and PI(5)P–synthesizing enzyme PIKfyve37-40) resulted in a small but significant increase in the peripheral distribution of lysosomes in non-starved cells (Supplementary Figure 5a, 5e), and effectively suppressed the retrograde migration of lysosomes in acutely nutrient-deprived cells (Supplementary Figure 5b, 5f) or Torin-1-treated cells (Supplementary Figure 5c, 5g). mTOR inhibition by Torin-1 did not directly activate TRPML1 (see preliminary results in Supplementary Figure 5p, 5q).
Short-term application of ML-SA1 increased perinuclear localization of lysosomes in YM 201636-treated cells (Supplementary Figure 5a, 5e), suggesting that PI(3,5)P2 regulates lysosome distribution through TRPML1. Consistently, overexpression of a PI(3,5)P2-insensitive mutant form, TRPML1-7Q35, did not result in perinuclear lysosome accumulation occurred with WT TRPML1 overexpression (Supplementary Figure 5d, 5h). ML-SA1 treatment, however, increased perinuclear localization of lysosomes reliably in TRPML1-7Q-expressing cells(Supplementary Figure 5d, 5h). Taken together, these results suggest that the PI(3,5)P2 sensitivity of TRPML1 is essential for retrograde lysosome transport under normal physiological conditions, and that synthetic agonists can be used as substitutes for PI(3,5)P2 in this function.
TRPML1-dependent retrograde migration of lysosomes through cytoplasmic dynein motors
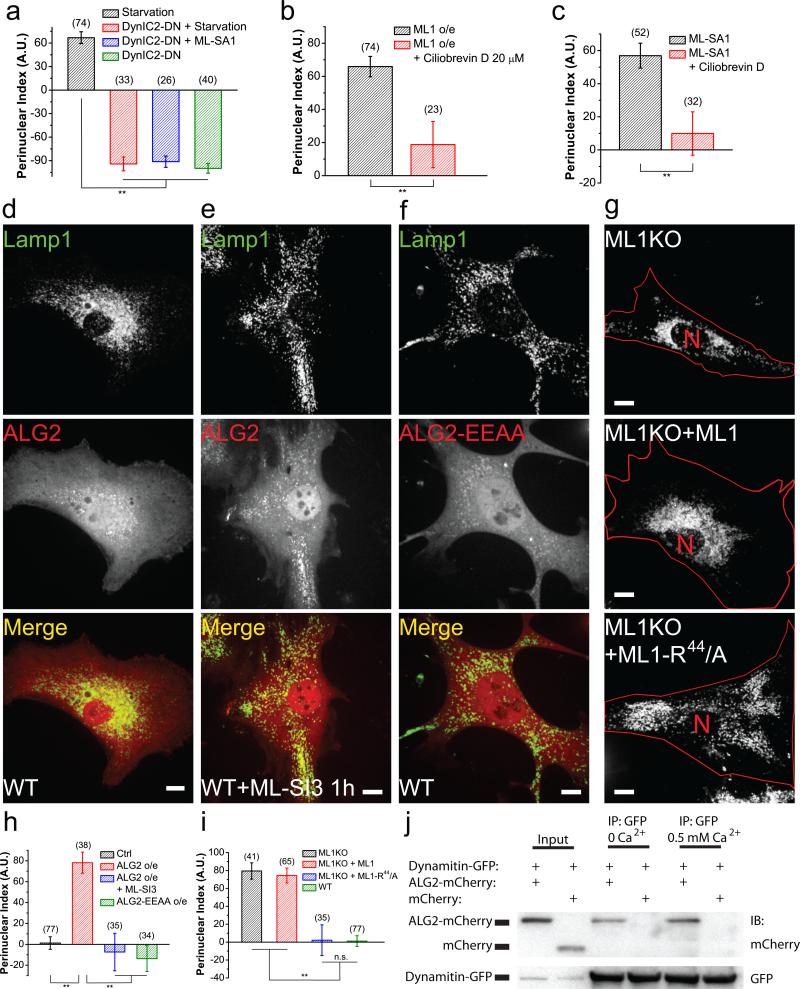
In mammalian cells, long-range transport of organelles is mediated by microtubule-based motor proteins5,29,41, including plus-end-directed kinesins and minus-end-directed dynein complexes. Kif5B and cytoplasmic dynein I (DynC1H1) transport lysosomes in mammalian cells42-45. Overexpression of dominant-negative Kif5B (Kif5B-DN)46 in mouse fibroblasts produced juxtanuclear clustering of lysosomes, whereas overexpression of dominant-negative cytoplasmic dynein intermediate chain 2 (DynIC2-DN)47 resulted in a predominantly peripheral localization of lysosomes (Fig. 5a, Supplementary Figure 5i, 5j, 5o). Expression of DynIC2-DN inhibited starvation-induced or ML-SA1-induced perinuclear lysosomal localization completely (Fig. 5a, Supplementary Figure 5k, 5l). DynIC2-DN overexpression also shifted cholesterol-accumulating puncta structures to the cell periphery in ML1 KO fibroblasts without correcting the cholesterol accumulation phenotype per se (Supplementary Figure 4f). Two-hour exposure to the dynein inhibitor ciliobrevin D48,49 also reversed the perinuclear distribution caused by MLSA1 or TRPML1 overexpression (Fig. 5c, Supplementary Figure 5m, 5n). Collectively, TRPML1-dependent retrograde trafficking of lysosomes may require cytoplasmic dynein activity.
Figure 5. ALG-2 interacts with dynein complexes to mediate TRPML1-dependent minus-end-directed transport of lysosomes.
(a) Quantification of the effect of dominant-negative Dynein Intermediate Chain 2 (DynIC2-DN) on the distribution of lysosomes under different conditions (see Supplementary Figure 5j-l). (b) Quantification of the effect of 20 μM Ciliobrevin D (1 h) on lysosome distribution with or without TRPML1 overexpression (see Supplementary Figure 5m). (c) Quantification of the effect of Ciliobrevin D (1 h) on lysosome distribution in the presence or absence of 25 μM MLSA1 (see Supplementary Figure 5n). (d) Lysosome distribution in WT fibroblasts co-transfected with Lamp1-GFP and mCherry-ALG-2. (e) Lysosome distribution in mCherry-ALG-2-transfected cells treated with ML-SI3 (25 μM) for 1 h. (f) Lysosome distribution in WT fibroblasts co-transfected with Lamp1-GFP and mCherry-ALG-2-EEAA (E47A-E114A). (g) Lysosome distribution in ML1 KO fibroblasts transfected with Lamp1-mCherry alone (top), Lamp1-mCherry + GFP-TRPML1 (middle), or Lamp1-mCherry + GFP-TRPML1-R44-A (bottom). (h) Quantification of lysosome distribution in experiments shown in (d-f). (i) Quantification of groups shown in (g). (j) Co-immunoprecipitation of ALG-2 and dynamitin in Cos-1 cells doubly transfected with mCherry-ALG-2 and GFP-Dynamitin, in the absence or presence of 0.5 mM Ca2+ in the lysis buffer. Cell lysates were directly loaded (input), or immunoprecipitated with either anti-GFP or anti-mCherry antibody, and then blotted against GFP. Red lines in images outline cell boundary and the red “N” marks the nucleus. Graphed data are presented as means ± SEM, the numbers of cells (n) used for quantification were pooled across at least three independent experiments and are shown in the parentheses. *p < .05, **p < .01 in ANOVA. Scale bars = 10 μm. Uncropped western blot images are shown in Supplementary Figure 9.
ALG-2 acts as a downstream effector of TRPML1 in promoting minus-end lysosomal motility
We next investigated whether apoptosis-linked gene 2 (ALG-2), a cytosolic protein with five EF-hand motifs50 that interact directly with TRPML1 in a Ca2+-dependent manner51, might mediate the Ca2+-dependence of lysosome motility. Indeed, ALG-2 overexpression in fibroblasts resulted in a dramatic perinuclear distribution of lysosomes (Fig. 5d, 5h, and Supplementary Figure 6d). In contrast, overexpression of Synaptotagmin-VII (Syt-VII), another lysosomal Ca2+ sensor2, did not affect lysosome distribution (Supplementary Figure 6a-c). Likewise, a Ca2+-binding-defective EF-hand mutant51 (ALG-2-E47A-E114A, or ALG-2-EEAA) also had no effect (Fig. 5f, 5h). Moreover, the ALG-2 effect was abolished by TRPML1 inhibition (Fig. 5e, 5h). On the other hand, activation of TRPML1 with ML-SA1 increased the co-localization of GFP-ALG-2 with Lamp1-mCherry (Supplementary Figure 6d-f). Therefore, ALG-2 may serve as a Ca2+ effector of TRPML1 to promote perinuclear distribution of lysosomes.
ALG-2 interacts with TRPML1 through a stretch of three amino acids (R44, L45 and K46) in the N terminus of TRPML1. Mutations of these residues (e.g., TRPML1-R44-A or TRPML1-R44LK-AAA) attenuate or abolish TRPML1-ALG-2 interaction51. Because TRPML1-R44-A generated whole-endolysosome currents comparable to the WT channel (Supplementary Figure 7a-c), we used TRPML1-R44-A for lysosome mobility studies. As mentioned above, lysosomes in ML1 KO fibroblasts displayed a perinuclear pattern, presumably due to cholesterol-dependent retrograde trafficking (Fig. 3). Overexpression of WT TRPML1 in ML1 KO fibroblasts did not suppress the perinuclear distribution of lysosomes (Fig. 5g, 5i), but rather resulted in a lysosome pattern similar to that produced by overexpression of TRPML1 in WT cells due to the constitutive activity of overexpressed TRPML1 (Supplementary Figure 7d, 7f, 7g). In contrast, overexpression of TRPML1-R44-A readily reversed the perinuclear distribution of lysosomes completely in ML1 KO cells (Fig. 5d, 5f), and rescued cholesterol accumulation (Supplementary Figure 7e). Importantly, ALG-2 pulled down dynamitin, a dynactin complex component29 in coimmunoprecipitation assays in a Ca2+-independent manner (Fig. 5j and Supplementary Figure 7h-j). Hence, ALG-2 may link TRPML1 activation and lysosomal Ca2+ release with dynein motors in retrograde trafficking of lysosomes (see Fig. 8g).
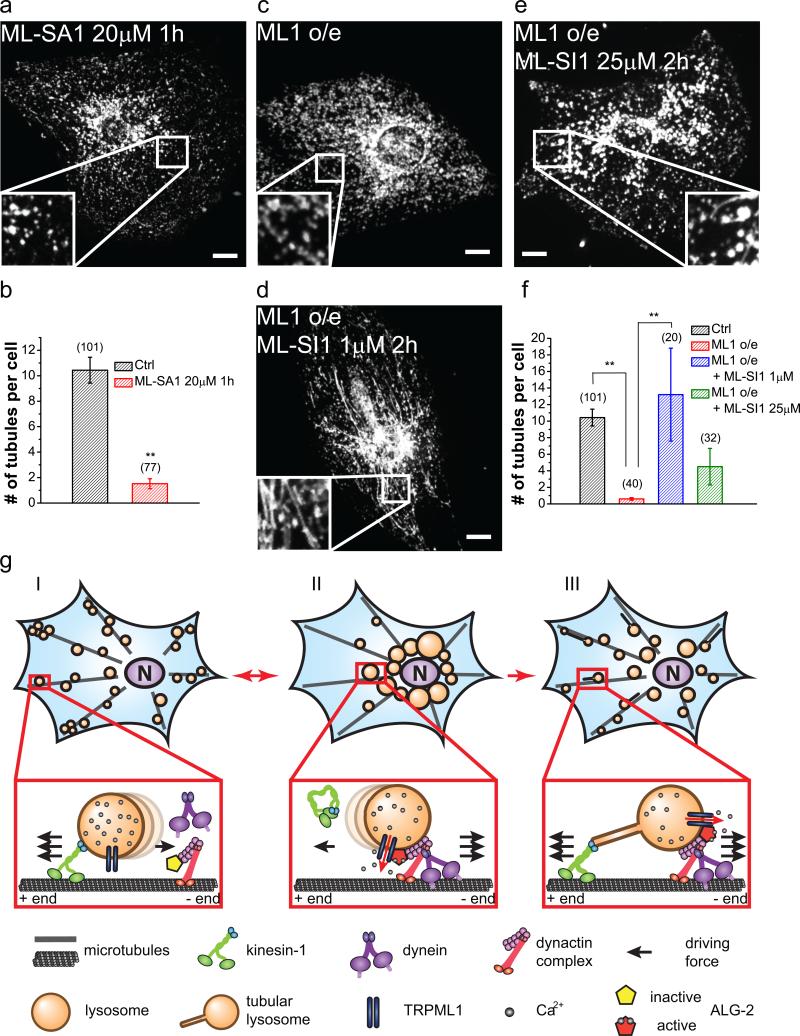
Figure 8. TRPML1 regulates switch between the plus- and minus-end directed lysosome motility.
(a, b) Effect of ML-SA1 (20 μM, 1 h) on lysosome tubulation in mouse fibroblasts. (c-e) Effects of 1 μM (d) or 25 μM (e) ML-SI1 on lysosome tubulation in TRPML1-overexpressing WT fibroblasts. ML-SIs were applied during the last 2 h of starvation. (f) Quantifications of the groups shown in (c–e). (g) Model illustrating the proposed role of TRPML1 in the regulation of lysosome motibility and tubulation. Under normal growth conditions (I), lysosomes are mostly peripherally distributed. During acute starvation (II), the TRPML1-ALG-2 pathway is activated to increase minus-end-directed motility of lysosomes, resulting in rapid redistribution of lysosomes to the juxtanuclear region, thereby facilitating autophagosome-lysosome fusion. After prolonged starvation (III), reactivation of mTOR turns on the machinery for lysosome tubulation and reformation. While the TRPML1-ALG-2 pathway remains active, the plus-end motility of lysosomes is increased. Subsequently, the “balanced” driving forces on both directions result in the generation of tubular lysosomes. Graphed data are presented as means ± SEM, the numbers of cells (n) used for quantification were pooled across at least three independent experiments and are shown in the. *p < .05, **p < .01 in ANOVA. Scale bars = 10 μm.
We next generated ALG-2 KO HeLa cells using the CRISPR-Cas9 system52 (Fig. 6a, 6b). At resting conditions, ALG-2 KO HeLa cells displayed a dispersed lysosome pattern, similar to WT cells (Fig. 6c, 6h). In a sharp contrast to WT cells, upon ML-SA1 application or acute starvation, the distribution of lysosomes in ALG-2 KO cells remained peripheral (Fig. 6d. 6e, 6h). These data strongly suggest that ALG-2 is required for the TRPM1-dependent retrograde migration of lysosomes. Conversely, perinuclear accumulation of lysosomes caused by RILP overexpression was not blocked by ALG-2 KO (Fig. 6f, 6g). Interestingly, in ALG-2 KO cells, while the basal LC3-II levels were elevated compared to WT cells, starvation-induced degradation of LC3-II was blocked (Fig. 6I, 6j). Collectively, these results suggest that the TRPML1-ALG-2-dependent retrograde transport is required for efficient autophagic clearance.
Figure 6. ALG-2 is required for the TRPML1-promoted acute retrograde migration of lysosomes.
(a) DNA sequencing results of the two ALG-2 CRISPR KO HeLa cell lines, the red “ATG” indicates start codon. Mutant #1 had a 2bp deletion on all chromosomes, while mutant #2 had various lengths of out-of-frame deletions. (b) Western blot confirmation of the ALG-2 KO. (c) Representative images showing Lamp1-mCherry distribution of WT (left) and the two mutant lines in complete medium without any treatment. (d, e) WT (d) and ALG-2 KO (e) cells under 2 h of 25 μM ML-SA1 treatment (left), or under 2 h of serum starvation (right). Mutant #2 was chosen for further studies based on their more extended morphology. (f, g) WT (f) and ALG-2 KO (g) cells overexpressing Lamp-mCherry and RILP-GFP in the presence of 25 μM ML-SI3 for 2h. Red asterisks in RILP panels indicate cells not expressing RILP-GFP. (h) Quantification of the groups shown in (c-e). ( i, j) Western blot analysis of endogenous LC3 in WT or ALG-2 KO HeLa cells upon 2 h starvation. LC3-II over LC3-I ratio normalized to WT control cells from 5 independent experiments were quantified in (j). Red lines in images outline cell boundary and the red “N” marks the nucleus. Graphed data are presented as means ± SEM, the numbers of cells (n) used for quantification were pooled across at least three independent experiments and are shown in the parentheses . *p < .05, **p < .01 in ANOVA. Scale bars = 10 μm. Uncropped western blot images are shown in Supplementary Figure 9.
TRPML1 and lysosomal Ca2+ are required for lysosome tubulation
Lysosome tubules serve as a platform for lysosome reformation, a process through which membrane lipids and proteins are recycled via membrane fission4. Lysosome reformation is especially prominent when lysosomes are undergoing rapid consumption, such as during phagocytosis or prolonged starvation4,53. We found that loss of activity of kinesin or dynein, both of which are implicated in lysosome reformation53, abolished lysosome tubulation during autophagic lysosome reformation (Supplementary Figure 8a).
Lysosome tubulation occurs under various conditions in different cell types, for examples, during prolonged starvation4 in fibroblasts and NRK cells (Fig. 7a, 7b; Supplementary Figure 8b, 8c), during LPS activation in macrophages53 (Fig. 7c, 7d), and constitutively in CV1 cells (Fig. 7e, 7f). Interestingly, ML1 KO or acute inhibition of TRPML1 abolished lysosome tubulation in all conditions (Fig. 7b, 7d, 7f, and Supplementary Figure 8b, 8c). Consistently, BAPTA-AM54 potently inhibited lysosome tubulation in CV cells (Fig. 7e, 7f).
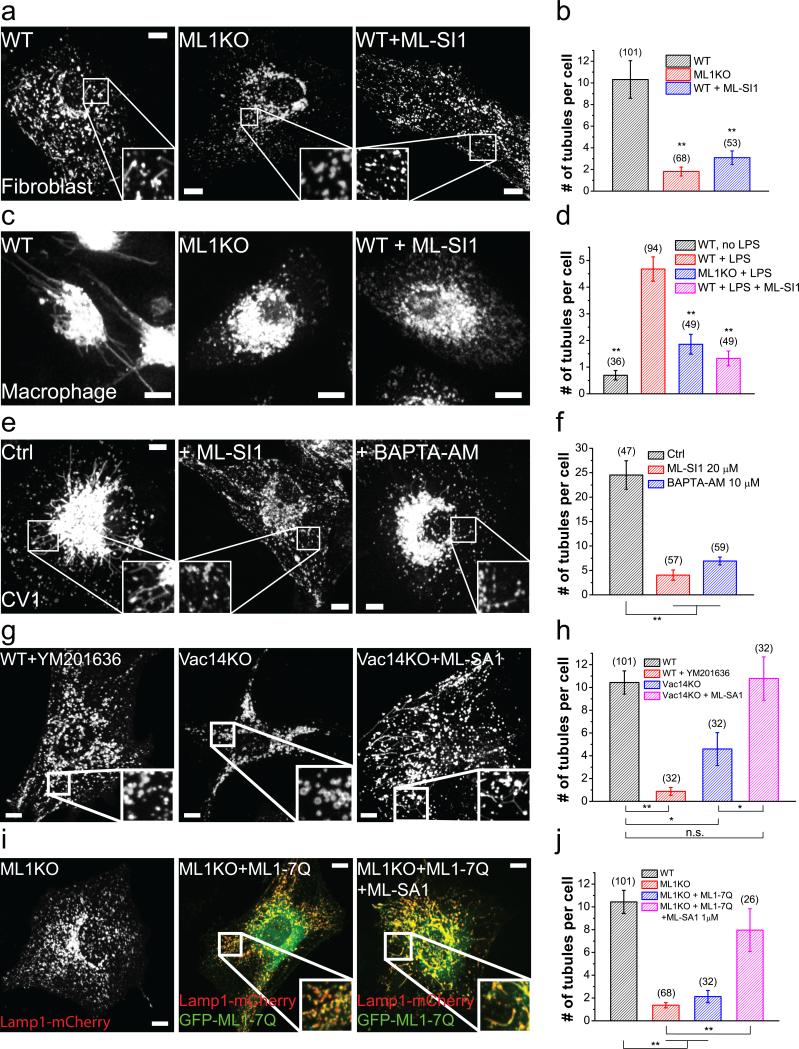
Figure 7. The PI(3,5)P2–TRPML1-Ca2+ pathway is required for lysosome tubulation.
(a) Lamp1-mCherry-transfected fibroblasts were starved for 24 h. A high level of lysosome tubulation was seen in WT fibroblasts, but not in ML1 KO fibroblasts or in WT fibroblasts treated with ML-SI1 (25 μM) during the last hour of starvation. (b) Quantification of lysosome tubules in the groups shown in (a). (c) Lysosome tubulation in macrophages loaded with tetramethylrhodamine-dextran (1 h loading, 2 h chase) and activated with lysopolysaccharides (LPS) for 3 h. Lysosome tubulation was prominent in WT macrophages (left), but not in ML1 KO macrophages (middle) or WT macrophages treated with ML-SI1 (25 μM, 30 min; right). (d) Quantification of the groups shown in (c). (e) Effects of ML-SI1 (25 μM, 1h) or BAPTA-AM (10 μM, 1 h) on spontaneous lysosome tubulation in Lamp1-GFP-expressing CV1 cells. (f) Quantification of groups shown in (e). (g) Representative WT fibroblasts starved for 24 h and treated with 1 μM YM 201636 for 30 min (left), Vac14 KO fibroblasts starved for 24 h (middle), and starved Vac14 KO fibroblasts treated with 10 μM ML-SA1 for 30 min (right). (h) Quantification of the groups shown in (g). (i) Lysosome tubulation after 24 h starvation of ML1 KO fibroblasts transfected with Lamp1-mCherry alone (left), Lamp1-mCherry together with GFP-TRPML1-7Q with (right) or without (middle) a low dose (1 μM) of ML-SA1 for 1 h. (j) Quantification of the groups shown in (i). Graphed data are presented as means ± SEM, the numbers of cells (n) used for quantification were pooled across at least three independent experiments and are shown in the parentheses.*p < .05, **p < .01 in ANOVA. Scale bars = 10 μm.
YM 201636 also potently inhibited lysosome tubulation during prolonged starvation (Fig. 7g, 7h). Vac14 KO fibroblasts, which contain about half of the cellular PI(3,5)P2 found in WT cells55, had moderately reduced lysosome tubulation. Vac14 KO fibroblasts treated with MLSA1 exhibited tubulation approaching that seen in WT cells (Fig. 7g, 7h). Furthermore, although lysosome tubulation defects in ML1 KO fibroblasts could not be rescued with TRPML1-7Q overexpression alone (Fig. 7i, 7j), low-dose of ML-SA1 restored lysosome tubulation to near normal levels in TRPML1-7Q transfected cells (Fig. 7i, 7j). Taken together, these data suggest that PI(3,5)P2-sensitivity of TRPML1 is required for lysosome tubulation.
TRPML1 regulates lysosome tubulation by tuning the balance between minus-end and plus-end motility
Interestingly, lysosome tubulation could be abolished by either loss of TRPML1 activity or hyper-activation of TRPML1 through TRPML1 overexpression or ML-SA1 application (Fig. 8a-c, 8f). We therefore hypothesized that a balance between plus- and minus-end motility may be important for the generation of tubular structures on lysosomes (Fig 8g) and that there may be an inverted-U response curve wherein too much or too little TRPML1 activity would disrupt this balance. Consistent with this hypothesis, we found that, in TRPML1-overexpressing cells, low concentrations of TRPML1 inhibitors restored lysosome tubulation (Fig. 8d, 8f) and high concentrations blocked lysosome tubulation (Fig. 8e, 8f). Likewise, in ALG-2-expressing fibroblasts, a slightly elevated degree of constitutive tubulation was observed under fed conditions, whereas lysosome tubulation was inhibited during prolonged starvation (Supplementary Figure 8d).
Discussion
The association of the microtubule-based motor proteins kinesin and dynein to lysosomes through adaptor proteins dictates the direction of lysosome movement. A series of recent studies established cholesterol as an important regulator of minus-end directed retrograde transport of lysosomes, with the Rab7 effector RILP and the cholesterol sensor ORP1L working together to recruit the dynactin complex to the lysosome26,29. In this study, we report a distinct pathway through which lysosome motility is acutely regulated in an on-demand manner independent of RILP and Rab7. Whereas the cholesterol-regulated Rab7-RILP-ORP1L pathway mediates the housekeeping function of constitutive retrograde transport for endosomal maturation, the PI(3,5)P2-TRPML1-ALG-2 pathway mediates on-demand, acutely regulated transport of lysosomes towards the perinuclear region upon autophagy induction.
What are the potential activation mechanisms of TRPML1 during starvation and autophagy induction? PI(3,5)P2 is the only known endogenous agonist of TRPML1. Although our data showed that PI(3,5)P2 is required for retrograde transport of lysosomes, bulk cellular PI(3,5)P2 levels reportedly drop to about 40% of the control levels under acute starvation33,56. Therefore, PI(3,5)P2 may only play a permissive role in the process. Starvation has been shown to cause mTOR inhibition and induce cytosolic alkalization6, and the latter is shown in the current study to directly trigger retrograde migration of lysosomes. However, TRPML1 channel activity is only slightly increased upon cytosolic alkalization. mTOR was recently reported to suppress TRPML1 function through phosphorylation57, and regulate the subcellular localization of endogenous TRPML1, as suggested by studies on Drosophila TRPML58. Future studies may reveal whether mTOR inhibition plays a role in TRPML1-dependent lysosomal trafficking. However, our electrophysiology recordings do not support the possibility that TRPML1 is activated directly through mTOR inhibition. Finally, unidentified endogenous TRPML1 agonists may be produced during acute starvation and autophagy induction. Therefore, it is likely multiple mechanisms mentioned above may be recruited to locally “activate” TRPML1 in lysosomes upon starvation and autophagy induction.
TRPML1 has been implicated in lysosomal trafficking in previous studies, and both membrane fusion and fission defects have been observed in ML-IV cells16,59. However, the exact role of TRPML1 in those trafficking steps is not clear as most previous studies relied on chronic inhibition of TRPML1 that causes secondary lysosome storage. Our identification of TRPML1 as a regulator in lysosome mobility prompts re-interpretation of the mechanisms underlying some previous observations. In light of the contrasting phenotypes of acute versus chronic TRPML1 inhibition on lysosome positioning, a high-priority experiment would be to verify whether acute inhibition of TRPML1 can affect previously implicated processes. Accordingly, it should be possible to distinguish between phenotypes caused by a primary loss of TRPML1 activity and those caused by chronic storage of lysosomal substances, such as cholesterol, secondary to loss of TRPML1 function.
Previously, we identified TRPML1 as a regulator of lysosome exocytosis through Ca2+ release2. The discrepancy between TRPML1's function in lysosome exocytosis, which happens at the cell periphery, and the retrograde transport of lysosomes towards the MTOC may be due to the involvement of different downstream Ca2+ effectors. Indeed, multiple downstream targets of lysosomal Ca2+ have been identified, including Syt-VII for lysosomal exocytosis2, calcineurin for TFEB activation60, and ALG-2 for lysosome motility in this study. ALG-2 has been shown to be involved in several vesicular trafficking steps, including ER-to-Golgi transport61. By directly interacting with the dynactin complex, ALG-2 may directly promote retrograde transport. The subcellular localization of particular lysosomes and cofactor or substrate availability may determine which Ca2+ effectors are activated for particular Ca2+ release events. For example, Ca2+ release from lysosomes that are docked to the plasma membrane may activate exocytosis, but not retrograde transport.
Lysosome motility is required for many lysosome functions including lysosome tubulation. Lysosome tubulation is a long-known62, but recently-characterized phenomenon that serves as a platform for lysosome reformation4. Under circumstances of heavy lysosome consumption, including prolonged starvation4 and active phagocytosis53, there is an obvious triggering of lysosome tubulation, enabling recycling of lysosomal membranes and membrane proteins and thus, replenishment of functional lysosome pool63. Dysfunctions of various lysosome-related proteins have been shown to inhibit lysosome tubulation64-66, but the mechanisms underlying these effects have not yet been delineated. Our study suggests that the dysfunctions of many of such proteins may affect tubulation through a common mechanism. A shared characteristic of most lysosome-related dysfunctions is the primary and secondary storage of substances in the lysosomes, including cholesterol. The requirement of both kinesin and dynein for lysosome tubulation suggests that tubulation processes require a balance of driving forces towards the two ends. Disruption of the balance will likely disrupt tubulation regardless of the circumstance under which tubulation is triggered. Cholesterol storage under such conditions may disrupt the balance of the motility, leading to the failure of generation or maintenance of tubular structures, as we demonstrated through manipulation of TRPML1 activity. Accordingly, restoration of lysosomal tubulation in disease models may be accomplished simply through fine tuning of lysosomal motility, regardless of the nature of the dysfunction. The present findings demonstrating that TRPML1 functions as a switch in lysosomal motility regulation, together with the availability of specific TRPML1 agonists and antagonists, suggest that TRPML1 may be a valuable target for treating lysosome-related diseases.
Materials and Methods
DNA subcloning
GFP-TRPML1, GFP-TRPML1-7Q, Lamp1-GFP, and Lamp1-mCherry constructs were generated as described previously33. Dominant-negative Kif5B (human Kif5B a.a. residues 592-963) was subcloned to pmCherry-C1 (Clontech) using Kif5B cDNA as the template. Dominant-negative dynein intermediate chain 2 (GFP-DYNIC2-DN) and GFP-dynamitin (both were gifts from Dr. Kristen Verhey at the University of Michigan) have been characterized previously47,67. mCherry-ALG2 and GFP-TRPML1-R44LK-AAA were provided by Dr. Rosa Puertollano (NHLBI, NIH). mCherry-ALG2-E47E114-AA, GFP-TRPML1-R44-A, GFP-TRPML1-L45-A, Rab7-T22N-GFP, Rab7-Q67L-GFP mutants were generated with a site-directed mutagenesis kit. All constructs were verified with sequencing and confirmed with western blotting.
Mouse lines
Characterizations of TRPML114, NPC1, and Vac1455 KO mice were performed as described previously. Animals were used under approved animal protocols and the Institutional Animal Care Guidelines of the University of Michigan.
Generation of ALG-2 KO HeLa cells using CRISPR/Cas9
A SpCas9 plasmid was obtained from Addgene (#48139). Primers for guide RNA generation are as following:
- #1 forward: CACCGAGGGCCGGGGCGGTAAGAGT;
- #1reverse: AAACACTCTTACCGCCCCGGCCCTC;
- #2 forward: CACCGCTCTTACCGCCCCGGCCCTG;
- #2 reverse: AAACCAGGGCCGGGGCGGTAAGAGC.
ALG-2 KO HeLa cells were generated using protocols established previously52. Briefly, primers were annealed and incorporated onto the plasmid using the BpiI site. WT HeLa cells were transfected with the plasmid and followed by a 2-day puromycin selection after transfection for 24 h. The cells were then plated on 96-well plates with an average density of 0.5 cell per well. Single colonies were selected and sequenced. Identified mutant lines were confirmed by western blotting using anti-ALG-2.
Mammalian cell culture and transfection
Mammalian cells were cultured in a 37 °C, 5% CO2 incubator. Mouse fibroblasts35 and macrophages3 were isolated and cultured as previously described. Immortalized cell lines (Cos1, HEK293, HeLa, CV-1, NRK) were originated from ATCC and cultured following standard tissue culture protocols, but were not tested for mycoplasma contaminations. HEK293 cells are on the list of frequently misidentified or cross-contaminated cell lines, but were only used in the Co-IP experiments with endogenous proteins due to antibody specificity. All other cell types used in the study are not listed as misidentified or cross-contaminated cells. Unless otherwise indicated, all cells were cultured in Dulbecco's modified eagle medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Gemini). Macrophages were further supplemented with murine GM-CSF (Peprotech). For macrophages, dextran-red (0.5 mg/ml) was used to visualize lysosomes using a protocol of 1 h loading followed by 2 h of chasing. For fibroblasts, transfection was performed using the Neon electroporation kit (Invitrogen). All other cell types were transfected with Lipofectamine 2000 (Invitrogen). Culture media were refreshed 18–24 h posttransfection, and cells were imaged 48 h post-transfection to allow sufficient recovery from transfection stress. Note that unlike nontransfected cells, lysosomes in transfected cells exhibited perinuclear localization pattern within 24 h of transfection, likely due to cell stress. For starvation, complete medium was replaced with DMEM without supplements through careful and extensive washes.
Ammonia Ringer's solution
Ammonia Ringer was adapted from that of acidic Ringer's solution33,68 and contained (in mM) 130 HCl, 20 NH4Cl, 5 KCl, 2 CaCl2, 1 MgCl2, 2 NaH2PO4, 10 HEPES, and 10 glucose; after mixing the solution was adjusted to a pH of 7.9 with NaOH.
Immunolabeling
Lamp1-GFP-transfected cells were rinsed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 20 min at room temperature (RT). Fixed cells were washed and blocked with 2% bovine serum albumin in PBS for 2 h, and then incubated overnight with primary antibodies in the blocking solution. Cells were then washed and incubated with secondary antibodies for 1 h before being subjected to fluorescent imaging.
Filipin staining and quantification
Filipin staining was performed as described previously23. Briefly, filipin was dissolved in DMSO at 25 mg/mL as a stock solution. The cells were incubated with filipin working solution (0.05 mg/mL) at RT for 2 h, and washed with PBS four times before imaging. All manipulations involving Filipin were kept strictly in dark until imaging. ImageJ was used to analyze the staining results. For each individual cell, the fluorescence intensities of three randomly selected areas close to the cell border were averaged, subtracted with the image background noise (intensity of void areas), then multiplied by the cell area, and considered intracellular background intensity. Note that because lysosomes are the primary storage sites of unesterified cholesterol under cholesterol storage conditions (see Fig. 3), the puncta filipin signal was presumed to be originated from lysosomal cholesterol69. Lysosomal cholesterol was then normalized as 100 x lysosomal intensity/intracellular background intensity.
Cholesterol depletion
Depletion of cholesterol was performed as described previously26. Briefly, 50 μM of activated simvastatin was applied to DMEM without FBS, and then supplemented with 230 μM mevalonate (to supply essential nonsterol isopreniods) in the absence of cholesterol26. Cells were washed extensively and incubated in the cholesterol-depleting medium for 3 h before imaging.
Fluorescence imaging and image analysis
Live imaging was performed with an Olympus Spindisk Confocal microscope equipped with a heated chamber to maintain the specimen temperature at ~37 °C. Except for FRAP experiments, which were carried out using a 3×1 μm z-stack setting for fast speed, all live imaging were taken using a 0.3 μm step size z-stack setting from top to the bottom of the cells. GFP-tagged proteins were visualized with a 488/515 (excitation/emission in nm) filter set; mCherry-tagged proteins were visualized with a 561/607 filter set. Quantification was performed in ImageJ.
Photobleaching and quantification of lysosome movement
Photobleaching experiments were performed with the FRAP function of Metamorph software in an Olympus spindisk confocal system. Briefly, after cells were incubated at ~37 °C for 10 min, a region (10 μm × 25-30 μm, corresponding to 42 pixels × 106–127 pixels at 60× magnification) was selected for photobleaching with an 800 ms/pulse protocol (561 nm excitation), and then live-imaged for 5 min (1 frame/s) with a z-stack of three 1-μm steps using a 561/607 filter set. The midline of each bleached area was drawn, and the number of lysosomes crossing the midline in each direction during the 5-min imaging time was counted.
Co-immunoprecipitation
Transfected cells from 10 cm dishes were incubated with 1 mL of lysis buffer (1% NP-40, 0.25% Na-deoxycholate, 1 mM Na3VO4, 1 mM NaF, 150 mM NaCl, 0.5 mM CaCl2 in 50 mM Tris-HCl, adjusted to pH 7.4) for 30 min at 4 °C. Lysates were centrifuged at 16,000 ×g for 10 min, and supernatants were incubated with 3 μg of primary antibodies at 4 °C for 1 h. After addition of 30 μl of protein A/G plus-agarose, lysates were incubated at 4 °C overnight with gentle shaking. Agarose beads were then collected through centrifuge at 500 ×g for 5 min. Beads were washed four times in lysis buffer, then heated to 60 °C for 10 min in NuPAGE loading buffer. Proteins were blotted with anti-GFP, anti-mCherry (1:5000), or anti-Dynamitin (1:1000).
Quantification of lysosome distribution
Lysosome distribution was analyzed in fibroblasts with cell areas in the range of 2,500–7,500 μm2, and in HeLa cells ranging 800-2,500 μm2. The nuclear area was excluded during quantification. Average Lamp1 intensities were measured for the whole cell (Itotal), the area within 10 μm of the nucleus (Iperinuclear), and the area >15 μm from the nucleus (Iperipheral). For HeLa cells, the latter two areas were within 5-10 μm of the nucleus, respectively. The perinuclear and peripheral normalized intensities were first calculated and normalized as I<10 = Iperinuclear/Itotal – 100 and I>15 = Iperipheral/Itotal – 100, respectively (I<5 and I>10 for HeLa cells). The perinuclear index was defined as I<10 – I>15 (I<5 – I>10 for HeLa cells). Quantifications were done by researchers blind to the experimental groups presented.
Quantification of lysosome tubulation
Cells were chosen randomly with the criterion that their fluorescence was strong enough to visualize tubular structures. Only tubules longer than 2 μm were included in the analysis to reduce false-positive hits (see Supplementary Figure 8e, 8f). Quantifications were done by researchers blind to the experimental groups presented.
Endolysosomal electrophysiology
Endolysosomal electrophysiology was performed in isolated enlarged endolysosomes with a modified patch-clamp method as described previously24,35,36,70. The pipette (luminal) solution was modified Tyrode's, including (in mM) 145 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES, 10 MES, 10 glucose (pH adjusted with NaOH to pH 4.6). The bath (internal/cytoplasmic) solution contained (in mM) 140 K-gluconate, 4 NaCl, 1 EGTA, 2 MgCl2, 0.39 CaCl2 , 20 HEPES (pH adjusted with KOH to 7.2; free [Ca2+]i ~ 100 nM estimated using MaxChelator software). Data were collected using an Axopatch 2A patch clamp amplifier, Digidata 1440, and pClamp 10.2 software (Axon Instruments). All recordings were analyzed with pClamp 10.2, and Origin 8.0 (OriginLab, Northampton, MA).
Chemicals and reagents
Chemicals were obtained from following vendors: ML-SA1 (Princeton BioMolecular Research), Simvastatin (Sigma), PI(3,5)P2 (Echelon), ML-SI3 (AKOS), BAPTA-AM (Invitrogen), YM 201636 (Symansis), and Ciliobrevin D (EMD Millipore). Antibodies were purchased from Sigma (γ-tubulin, T5326, clone GTU-88, used at 1:500), BD Biosciences (Dynamitin, 611002, clone 25/Dynactin p50, 1:250 for pull-down, and 1:1000 for western blot), Invitrogen (GFP, A11121, clone 11E5, 1:250 for pull-down, and 1:5000 for western blot), and Novus Biologicals (ALG-2, H00010016-M01, clone 2B4, 1:500 for western blot; mCherry, NBP1-96752, clone 1C51, 1:250 for pull-down, and 1:5000 for western blot).
Statistics and reproducibility
Statistical data are presented as means ± standard error of the mean (SEM). Comparisons between the experimental groups were made using analyses of variance (ANOVAs). All data are generated from cells pooled from at least three biologically independent experiments, with the actual sample size (n number) shown on top of each bar in graphs. No sample was excluded.
Supplementary Material
exdata_fig1
video1
video2
video3
video4
video5
video6
video7
vvideo8
exdata_fig2
exdata_fig3
exdata_fig4
exdata_fig5
exdata_fig6
exdata_fig7
exdata_fig8
exdata_fig9
Acknowledgements
This work was supported by NIH grants (NS062792, MH096595, and AR060837 to H.X). The authors are grateful to Dr. Rosa Puertollano (NHLBI, NIH) for providing the mCherry-ALG-2 construct, Dr. Kristen Verhey (University of Michigan) for providing GFP-DYNIC2-DN, GFPDynamitin, and Kif5B cDNA constructs, Dr. Roberto Botelho (Ryerson University, Toronto) for providing the RILP-GFP construct, Dr. Jacques Neefjes (Netherlands Cancer Institute) for providing the ORP1L-GFP construct, and Dr. David Rubinsztein (Cambridge Institute for Medical Research) for providing the LC3 stable cell-line. The authors thank Drs. Richard Hume, Robert Fuller, Kristen Verhey, and Yanzhuang Wang for their suggestions. We appreciate the encouragement and helpful comments from their Xu laboratory colleagues.
Footnotes
Author Contributions
X.L. initiated the project; X.L. and H.X. designed the research; X.L., N.R., A.H., X.Z., J.Y., W.W., Q.G., and X.C. performed the experiments; X.L. generated new reagents; X.L., N.R., A.H., X.Z., J.Y., W.W., Q.G., X.C., and H.X. analyzed and interpreted data; X.L. and H.X. wrote the manuscript with input from all authors.
References
- 1.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nature reviews. Molecular cell biology. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 2.Samie MA, Xu H. Lysosomal exocytosis and lipid storage disorders. Journal of lipid research. 2014;55:995–1009. doi: 10.1194/jlr.R046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samie M, et al. A TRP Channel in the Lysosome Regulates Large Particle Phagocytosis via Focal Exocytosis. Developmental cell. 2013;26:511–524. doi: 10.1016/j.devcel.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akhmanova A, Hammer JA., 3rd Linking molecular motors to membrane cargo. Current opinion in cell biology. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korolchuk VI, et al. Lysosomal positioning coordinates cellular nutrient responses. Nature cell biology. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 8.King SM. The dynein microtubule motor. Biochimica et biophysica acta. 2000;1496:60–75. doi: 10.1016/s0167-4889(00)00009-4. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz TL. Mitochondrial trafficking in neurons. Cold Spring Harbor perspectives in biology. 2013;5 doi: 10.1101/cshperspect.a011304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong XP, et al. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455:992–996. doi: 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng X, Shen D, Samie M, Xu H. Mucolipins: Intracellular TRPML1-3 channels. FEBS letters. 2010;584:2013–2021. doi: 10.1016/j.febslet.2009.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassi MT, et al. Cloning of the gene encoding a novel integral membrane protein, mucolipidin-and identification of the two major founder mutations causing mucolipidosis type IV. American journal of human genetics. 2000;67:1110–1120. doi: 10.1016/s0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CS, Bach G, Pagano RE. Abnormal transport along the lysosomal pathway in mucolipidosis, type IV disease. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6373–6378. doi: 10.1073/pnas.95.11.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venugopal B, et al. Neurologic, gastric, and opthalmologic pathologies in a murine model of mucolipidosis type IV. American journal of human genetics. 2007;81:1070–1083. doi: 10.1086/521954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dayam RM, Saric A, Shilliday RE, Botelho RJ. The Phosphoinositide-Gated Lysosomal Ca(2+) Channel, TRPML1, Is Required for Phagosome Maturation. Traffic. 2015;16:1010–1026. doi: 10.1111/tra.12303. [DOI] [PubMed] [Google Scholar]
- 16.Xu H, Ren D. Lysosomal physiology. Annual review of physiology. 2015;77:57–80. doi: 10.1146/annurev-physiol-021014-071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu M, et al. Regulation of autophagic flux by dynein-mediated autophagosomes trafficking in mouse coronary arterial myocytes. Biochimica et biophysica acta. 2013;1833:3228–3236. doi: 10.1016/j.bbamcr.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell structure and function. 2008;33:109–122. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
- 19.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. The Journal of biological chemistry. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curcio-Morelli C, et al. Macroautophagy is defective in mucolipin-1-deficient mouse neurons. Neurobiology of disease. 2010;40:370–377. doi: 10.1016/j.nbd.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vergarajauregui S, Connelly PS, Daniels MP, Puertollano R. Autophagic dysfunction in mucolipidosis type IV patients. Human molecular genetics. 2008;17:2723–2737. doi: 10.1093/hmg/ddn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng X, et al. The intracellular Ca channel MCOLN1 is required for sarcolemma repair to prevent muscular dystrophy. Nature medicine. 2014 doi: 10.1038/nm.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, et al. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1373–1381. doi: 10.1073/pnas.1419669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen D, et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nature communications. 2012;3:731. doi: 10.1038/ncomms1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Yang J, Low PS, Cheng JX. Cholesterol level regulates endosome motility via Rab proteins. Biophysical journal. 2008;94:1508–1520. doi: 10.1529/biophysj.106.099366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocha N, et al. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. The Journal of cell biology. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwiatkowska K, et al. Visualization of cholesterol deposits in lysosomes of Niemann-Pick type C fibroblasts using recombinant perfringolysin O. Orphanet journal of rare diseases. 2014;9:64. doi: 10.1186/1750-1172-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid PC, Sugii S, Chang TY. Trafficking defects in endogenously synthesized cholesterol in fibroblasts, macrophages, hepatocytes, and glial cells from Niemann-Pick type C1 mice. Journal of lipid research. 2003;44:1010–1019. doi: 10.1194/jlr.M300009-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Schroer TA. Dynactin. Annual review of cell and developmental biology. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 30.Spinosa MR, et al. Functional characterization of Rab7 mutant proteins associated with Charcot-Marie-Tooth type 2B disease. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:1640–1648. doi: 10.1523/JNEUROSCI.3677-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. The EMBO journal. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Molecular biology of the cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, et al. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:21165–21170. doi: 10.1073/pnas.1311864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vicinanza M, et al. PI(5)P regulates autophagosome biogenesis. Molecular cell. 2015;57:219–234. doi: 10.1016/j.molcel.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong XP, et al. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nature communications. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Li X, Xu H. Phosphoinositide isoforms determine compartment-specific ion channel activity. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11384–11389. doi: 10.1073/pnas.1202194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jefferies HB, et al. A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO reports. 2008;9:164–170. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin S, et al. Inhibition of PIKfyve by YM-201636 dysregulates autophagy and leads to apoptosis-independent neuronal cell death. PloS one. 2013;8:e60152. doi: 10.1371/journal.pone.0060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho CY, Choy CH, Wattson CA, Johnson DE, Botelho RJ. The Fab1/PIKfyve Phosphoinositide Phosphate Kinase Is Not Necessary to Maintain the pH of Lysosomes and of the Yeast Vacuole. The Journal of biological chemistry. 2015;290:9919–9928. doi: 10.1074/jbc.M114.613984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai X, et al. PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in Toll-like receptor signaling. Chemistry & biology. 2013;20:912–921. doi: 10.1016/j.chembiol.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verhey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nature reviews. Molecular cell biology. 2009;10:765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 42.Rosa-Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin-1. Developmental cell. 2011;21:1171–1178. doi: 10.1016/j.devcel.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka Y, et al. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- 44.Jordens I, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Current biology : CB. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- 45.Johansson M, et al. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. The Journal of cell biology. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silver KE, Harrison RE. Kinesin 5B is necessary for delivery of membrane and receptors during FcgammaR-mediated phagocytosis. Journal of immunology. 2011;186:816–825. doi: 10.4049/jimmunol.1002161. [DOI] [PubMed] [Google Scholar]
- 47.King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Molecular biology of the cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sainath R, Gallo G. The dynein inhibitor Ciliobrevin D inhibits the bidirectional transport of organelles along sensory axons and impairs NGF-mediated regulation of growth cones and axon branches. Developmental neurobiology. 2014 doi: 10.1002/dneu.22246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Firestone AJ, et al. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 2012;484:125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maki M, Suzuki H, Shibata H. Structure and function of ALG-2, a penta-EF-hand calcium-dependent adaptor protein. Science China. Life sciences. 2011;54:770–779. doi: 10.1007/s11427-011-4204-8. [DOI] [PubMed] [Google Scholar]
- 51.Vergarajauregui S, Martina JA, Puertollano R. Identification of the penta-EF-hand protein ALG-2 as a Ca2+-dependent interactor of mucolipin-1. The Journal of biological chemistry. 2009;284:36357–36366. doi: 10.1074/jbc.M109.047241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cong L, Zhang F. Genome engineering using CRISPR-Cas9 system. Methods in molecular biology. 2015;1239:197–217. doi: 10.1007/978-1-4939-1862-1_10. [DOI] [PubMed] [Google Scholar]
- 53.Mrakovic A, Kay JG, Furuya W, Brumell JH, Botelho RJ. Rab7 and Arl8 GTPases are necessary for lysosome tubulation in macrophages. Traffic. 2012;13:1667–1679. doi: 10.1111/tra.12003. [DOI] [PubMed] [Google Scholar]
- 54.Tsien RY. New calcium indicators and buffers with high selectivity against magnesium and protons: design, synthesis, and properties of prototype structures. Biochemistry. 1980;19:2396–2404. doi: 10.1021/bi00552a018. [DOI] [PubMed] [Google Scholar]
- 55.Jin N, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. The EMBO journal. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zolov SN, et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17472–17477. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Onyenwoke RU, et al. The mucolipidosis IV Ca2+ channel TRPML1 (MCOLN1) is regulated by the TOR kinase. The Biochemical journal. 2015;470:331–342. doi: 10.1042/BJ20150219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong CO, Li R, Montell C, Venkatachalam K. Drosophila TRPML is required for TORC1 activation. Current biology : CB. 2012;22:1616–1621. doi: 10.1016/j.cub.2012.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller A, et al. Mucolipidosis Type IV Protein TRPML1-Dependent Lysosome Formation. Traffic. 2014 doi: 10.1111/tra.12249. [DOI] [PubMed] [Google Scholar]
- 60.Medina DL, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nature cell biology. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Helm JR, et al. Apoptosis-linked gene-2 (ALG-2)/Sec31 interactions regulate endoplasmic reticulum (ER)-to-Golgi transport: a potential effector pathway for luminal calcium. The Journal of biological chemistry. 2014;289:23609–23628. doi: 10.1074/jbc.M114.561829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swanson J, Bushnell A, Silverstein SC. Tubular lysosome morphology and distribution within macrophages depend on the integrity of cytoplasmic microtubules. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:1921–1925. doi: 10.1073/pnas.84.7.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y, Yu L. Autophagic lysosome reformation. Experimental cell research. 2013;319:142–146. doi: 10.1016/j.yexcr.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 64.Rong Y, et al. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nature cell biology. 2012;14:924–934. doi: 10.1038/ncb2557. [DOI] [PubMed] [Google Scholar]
- 65.Rong Y, et al. Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7826–7831. doi: 10.1073/pnas.1013800108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang J, Lee S, Blackstone C. Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. The Journal of clinical investigation. 2014;124:5249–5262. doi: 10.1172/JCI77598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. The Journal of cell biology. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Durchfort N, et al. The enlarged lysosomes in beige j cells result from decreased lysosome fission and not increased lysosome fusion. Traffic. 2012;13:108–119. doi: 10.1111/j.1600-0854.2011.01300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chu BB, et al. Cholesterol transport through lysosome-peroxisome membrane contacts. Cell. 2015;161:291–306. doi: 10.1016/j.cell.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 70.Wang X, et al. TPC proteins are phosphoinositide- activated sodium-selective ion channels in endosomes and lysosomes. Cell. 2012;151:372–383. doi: 10.1016/j.cell.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
exdata_fig1
video1
video2
video3
video4
video5
video6
video7
vvideo8
exdata_fig2
exdata_fig3
exdata_fig4
exdata_fig5
exdata_fig6
exdata_fig7
exdata_fig8
exdata_fig9