Robust transcriptome-wide discovery of RNA binding protein binding sites with enhanced CLIP (eCLIP) (original) (raw)
. Author manuscript; available in PMC: 2016 Sep 28.
Published in final edited form as: Nat Methods. 2016 Mar 28;13(6):508–514. doi: 10.1038/nmeth.3810
Abstract
As RNA binding proteins (RBPs) play essential roles in cellular physiology by interacting with target RNAs, binding site identification by UV-crosslinking and immunoprecipitation (CLIP) of ribonucleoprotein complexes is critical to understanding RBP function. However, current CLIP protocols are technically demanding and yield low complexity libraries with high experimental failure rates. We have developed an enhanced CLIP (eCLIP) protocol that decreases requisite amplification by ~1,000-fold, decreasing discarded PCR duplicate reads by ~60% while maintaining single-nucleotide binding resolution. By simplifying the generation of paired IgG and size-matched input controls, eCLIP improves specificity in discovery of authentic binding sites. We generated 102 eCLIP experiments for 73 diverse RBPs in HepG2 and K562 cells (available at https://www.encodeproject.org), demonstrating that eCLIP enables large-scale and robust profiling, with amplification and sample requirements similar to ChIP-seq. eCLIP enables integrative analysis of diverse RBPs to reveal factor-specific profiles, common artifacts for CLIP and RNA-centric perspectives of RBP activity.
Introduction
RNA binding proteins (RBPs) have emerged as critical players in regulating gene expression, controlling when, where, and at what rate RNAs are processed, trafficked and translated within the cell1. These regulatory roles are essential for normal human physiology, as defects in RBP function are associated with diverse genetic and somatic disorders, such as neurodegeneration, auto-immune defects, and cancer2,3. To discover the mechanisms by which RBPs affect RNA processing, technologies such as RNA ImmunoPrecipitation (RIP) and UV Cross-Linking and ImmunoPrecipitation (CLIP) that comprehensively identify the RNA substrates each RBP interacts with are widely used4. Indeed, coupled with high-throughput sequencing (-seq), RNA binding sites can be identified with single-nucleotide level resolution in vivo with improvements in CLIP such as PAR-CLIP5 and iCLIP6. However, current CLIP methods are technically challenging with high experimental failure rates for many users, and sequenced CLIP-seq libraries are often of extremely low complexity: across 279 published CLIP datasets, a median of 83.8% of CLIP-seq reads are discarded as PCR duplicates (Supplementary Fig. 1a, Supplementary Table 1). Further, we observed that the success rate of generating libraries was low for many RBPs, particularly those lacking canonical RNA binding domains, when iCLIP was performed in large scale as part of the ENCODE consortium (Supplementary Fig. 1b). Thus, improved library generation efficiency will save significant sequencing costs, greatly enhance technical and biological reproducibility, and enable RBP binding site identification in limiting samples, for low-abundance RBPs, and those with few RNA targets.
Results
Improved RBP target identification with eCLIP
Here we describe enhanced CLIP (eCLIP), which modifies the iCLIP method to include improvements in library preparation of RNA fragments as described by Shishkin, et al.7. In CLIP, RNA-RBP interactions are covalently linked by UV irradiation, followed by fragmentation of RNA (typically by RNase treatment), immunoprecipitation of a targeted protein along with crosslinked RNA, and conversion of that RNA to dsDNA high-throughput sequencing libraries through adapter ligation and reverse transcription (Fig. 1a, Supplementary Protocol 1). We observed that circular ligation as used in iCLIP is often inefficient, and modified this step to add adapters as two separate steps: an indexed 3′ RNA adapter is ligated to the crosslinked RNA fragment while on the immunoprecipitation beads, and a 3′ ssDNA adapter is ligated after reverse transcription (see Methods). As reverse transcription often terminates at the RBP:RNA crosslink site, the ligation of the ssDNA adapter to the cDNA fragments at their 3′ end maintains the single-nucleotide resolution of iCLIP. Furthermore, the ssDNA adapter (rand3Tr3) contains an inline random-mer (either N5 or N10) to distinguish whether two identical sequenced reads indicate two unique RNA fragments or PCR duplicates of the same RNA fragment. Increased T4 RNA ligase concentration and the addition of high concentrations of polyethylene glycol (PEG8000) and DMSO in these two ligation reactions enables ligation efficiencies of >90% and >70% respectively, decreasing loss of RNA fragments due to failed ligation7. We have also omitted RNA radiolabeling and autoradiographic visualization steps, although these visualizations can identify potential non-antigen contamination for detailed studies of specific factors8. These modifications shorten the hands-on time to as few as 4 days (Fig. 1a), and the unique barcodes incorporated during the RNA ligation on immunoprecipitation beads allow for pooling of samples before SDS-PAGE gel and subsequent library steps (Supplementary Fig. 2a). After 50 nt paired-end sequencing on the Illumina HiSeq 2500 or 4000 platforms, reads are processed using a CLIP-seq pipeline modified to utilize eCLIP-specific adapters (Supplementary Fig. 2b, Supplementary Protocol 2).
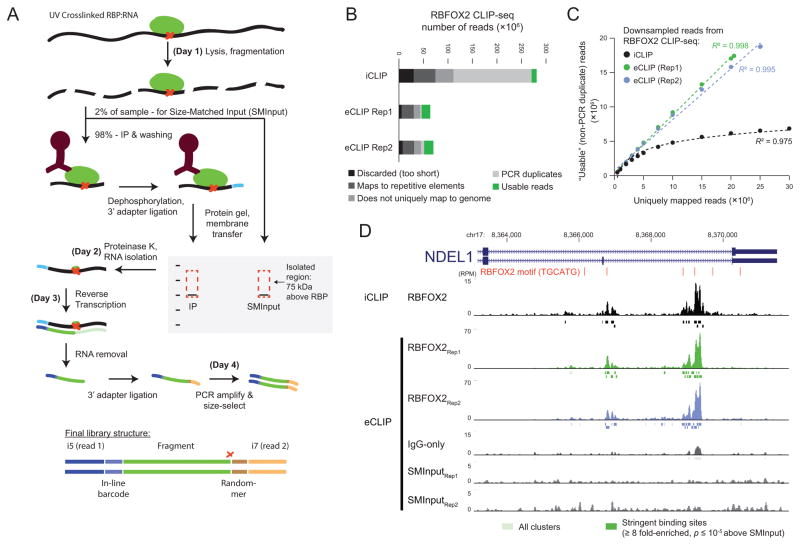
Figure 1. Improved identification of RNA binding protein (RBP) targets by enhanced CrossLinking and ImmunoPrecipitation followed by high-throughput sequencing (eCLIP-seq).
(a) RBP-RNA interactions are stabilized with UV crosslinking, followed by limited RNase I digestion, immunoprecipitation of RBP-RNA complexes with a specific antibody of interest, and stringent washes. After dephosphorylation of RNA fragments, an “inline barcoded” RNA adapter is ligated to the 3′ end. After protein gel electrophoresis and nitrocellulose membrane transfer, a region 75 kDa (~220 nt of RNA) above the protein size is excised and proteinase K treated to isolate RNA. RNA is further prepared into paired-end high-throughput sequencing libraries, where read 1 begins with the inline barcode and read 2 begins with a random-mer sequence (added during the 3′ DNA adapter ligation) followed by sequence corresponding to the 5′ end of the original RNA fragment (which often marks reverse transcriptase termination at the crosslink site (red X)). (b) Bars indicate the number of reads remaining after processing steps. PCR duplicate reads that map to the same genomic position and have the same random-mer as previously considered reads are discarded, leaving only “Usable reads”. (c) Varying numbers of uniquely mapped reads were randomly sampled from RBFOX2 iCLIP and eCLIP experiments and PCR duplicate removal was performed. Points indicate the mean of 100 downsampling experiments (for all, s.e.m. is less than 0.1% of mean value). (d) RBFOX2 read density in reads per million usable (RPM). Shown are iCLIP, two biological replicates for eCLIP with paired size-matched input (SMInput) and IgG-only controls. CLIPper-identified clusters indicated as boxes below, with dark colored boxes indicating binding sites enriched above SMInput.
We evaluated eCLIP improvements using the well-characterized RBP RBFOX2. We performed iCLIP and eCLIP in whole-cell extracts from 20 million HEK293T cells using a validated RBFOX2-specific antibody (A300-864A, Bethyl)9 and size-selected the 50–125 kDa region on the membrane (Supplementary Fig. 3a–b). The iCLIP library required 28 cycles of PCR to obtain 206 femtomoles (fmoles) of library, whereas two biological replicates of eCLIP required only 16 PCR cycles to obtain 66 and 78 fmoles. To simplify comparisons of library yield across experiments, we defined an Extrapolated CT (eCT) value as the number of PCR cycles (assuming 100% PCR efficiency) needed to obtain 100 fmoles (10 nM in 10 uL) of library. This yielded eCT values of 23.6 for iCLIP and 13.0 and 13.3 for eCLIP (Supplementary Fig. 3d), indicating a ~1,000-fold increase in adapter-ligated pre-amplification products.
High-throughput sequencing reads were processed to obtain “usable” reads, defined as reads that uniquely map to the genome and remain after discarding PCR duplicates. We observed that despite similar fractions of uniquely mapped reads, saturation of unique fragments was observed for iCLIP below 10 million uniquely mapped reads (with the remaining increase largely due to sequencing errors in the random-mer, as previously observed10), whereas no observable saturation was seen with eCLIP at 20 million reads (Fig. 1b–c, Supplementary Fig. 3e). At 20 million uniquely mapped reads this translates to a 54.4% increase in usable read fraction (from 30.8% to 85.2% of uniquely mapped reads). We observed a similar decrease in the required amplification and enrichment for unique reads in eCLIP even for the abundantly expressed RBPs IGF2BP1 and IGF2BP2 in K562 cells (Supplementary Fig. 4a–d), confirming that enhanced adapter ligation efficiency significantly improves library complexity for eCLIP experiments.
Examination of individual binding sites revealed comparable read density between iCLIP and eCLIP for RBFOX2 binding sites (Fig. 1d). Using the CLIPper peak-calling algorithm11, we observed that peaks from both iCLIP and eCLIP showed enrichment in proximal and distal introns and were significantly enriched for the RBFOX2 motif (Supplementary Fig. 3f), in agreement with previous RBFOX2 CLIP experiments9,11. We also observed the same stereotypical patterns of read termination (due to reverse transcription termination at the RBP-RNA crosslink site) at two G bases in the canonical UGCAUG as previously described for RBFOX2 (Supplementary Fig. 5a)12, confirming that the dual adapter ligation strategy maintains the single-nucleotide resolution of iCLIP. We confirmed crosslink-dependence of canonical motifs for other factors including TARDBP and PUM2 (Supplementary Fig. 5b–c), and observed enrichment proximal to crosslink sites for others, such as TRA2A (Supplementary Fig. 5d).
To validate that eCLIP identifies functional sites, we designed antisense oligonucleotides with uniform 2′-_O_-methoxyethyl-modified nucleotides and a phosphorothioate backbone against the RBFOX2 motif at three RBFOX2 binding sites flanking RBFOX2-dependent alternatively spliced exons. We observed significantly decreased exon inclusion for at least one oligonucleotide for each region (Supplementary Fig. 6a–f), indicating that RBP-blocking oligonucleotides can validate the functional relevance of eCLIP binding sites.
Radiolabeled detection of RBP-associated RNA is used to validate and optimize fragmentation conditions in traditional CLIP methodology for individual RBPs8. To interrogate the degree to which fragmentation affects eCLIP, we performed eCLIP with various RNase concentrations ranging from 0 U to 2000 U (per mL of lysate) for two RBPs with binding patterns that span the wide spectrum of RNA sizes: RBFOX2, which largely binds to intronic regions within pre-mRNAs which are tens to thousands of kilobases in length, and SLBP, which binds to a canonical hairpin structure in the 3′ end of histone mRNAs which are ~150 nt in length13 (Supplementary Fig. 3c). We found that intronic RBFOX2 binding and motif enrichment were identifiable across a wide range of RNase amounts, and at the 40U concentration which was near-optimal for RBFOX2 (and other intronic binding RBPs), binding of SLBP to short histone RNAs was still significantly enriched (Supplementary Fig. 7a–f). These results along with our experiences with the dozens of RBPs described later in this manuscript indicate that this concentration is appropriate for nearly all RBPs as a robust first pass that identifies region-, peak-, and motif-level signal for both short RNA binding SLBP and intronic pre-mRNA binding RBFOX2. We do note, however, that fragmentation conditions should be initially validated for different cell-lines, tissues, or other sample types, as high endogenous RNase activity (as is the case for pancreatic and liver samples) can lead to over-shearing.
Input normalization improves CLIP signal-to-noise
The enhanced ligation efficiency allowed the generation of two paired controls that improved specificity to CLIP-seq analysis by removing artifacts, which have often been observed in CLIP experiments14,15. First, we produced an eCLIP library from an IgG isotype-only control which required 16.3 eCT, indicating ~9.5-fold less material than the RBFOX2 eCLIP. Second, we sampled 2% of the pre-immunoprecipitation (post-lysis and fragmentation) sample and prepared libraries identically to the RBFOX2 eCLIP (including the membrane size selection step). This “size-matched input control (SMInput)” serves as a crucial control for non-specific background signal in the identical size range on the membrane as well as any inherent biases in ligations, RT-PCR, gel migration and transfer steps. Incorporation of an input background has long been an essential part of RIP-ChIP and RIP-seq protocols16 and provided significant improvements to ChIP-seq analysis17, suggesting that incorporating SMInput normalization as standard could improve our signal-to-noise in identifying authentic RBP binding sites specific to the factor under study. We found that the IgG control was poorly suited for normalization as it yielded highly PCR duplicated libraries, and thus focused on normalization against SMInput.
To directly assay the effect of SMInput normalization, we first used SLBP as its exclusive binding to histone RNAs13 distinguishes true from false positive signal. Surprisingly, we observed that the RPM (reads per million usable) of most genes was relatively unchanged, with abundant genes such as translation elongation factor EEF2 showing similar read density profiles between SLBP eCLIP and SMInput (Supplementary Fig. 8a–b). However, we observed histone transcripts comprised 43 of 47 significantly enriched 3′UTR and 50 of 54 CDS regions, a >260-fold increase above their transcriptome frequency (Supplementary Fig. 8b). Thus, eCLIP signal at true binding sites is significantly enriched despite the presence of whole-transcriptome background.
To quantify the effect of SMInput normalization at the peak level, we first performed traditional CLIP-seq peak calling with the CLIPper algorithm11. Next, the numbers of reads overlapping each CLIPper-identified binding site in eCLIP and SMInput were used to calculate SMInput-normalized significance and fold-enrichment (Supplementary Fig. 8c–d). Of the 23,034 SLBP clusters identified as significant (p < 0.05) by CLIPper, only 284 (1.2%) were enriched above SMInput (defined as at least 8-fold, p ≤ 10−5 enriched) (Fig. 2a, Supplementary Fig. 8e). 251 of these 284 (88.4%) were located within histone RNAs, demonstrating the specificity of eCLIP. We further observed that ranking peaks by SMInput-normalized _p-_value significantly enriched for peaks overlapping histone RNAs compared to ranking peaks by CLIPper pre-normalized _p-_value, indicating that SMInput normalization accentuates true positive peaks compared to standard CLIP analysis (Fig. 2b).
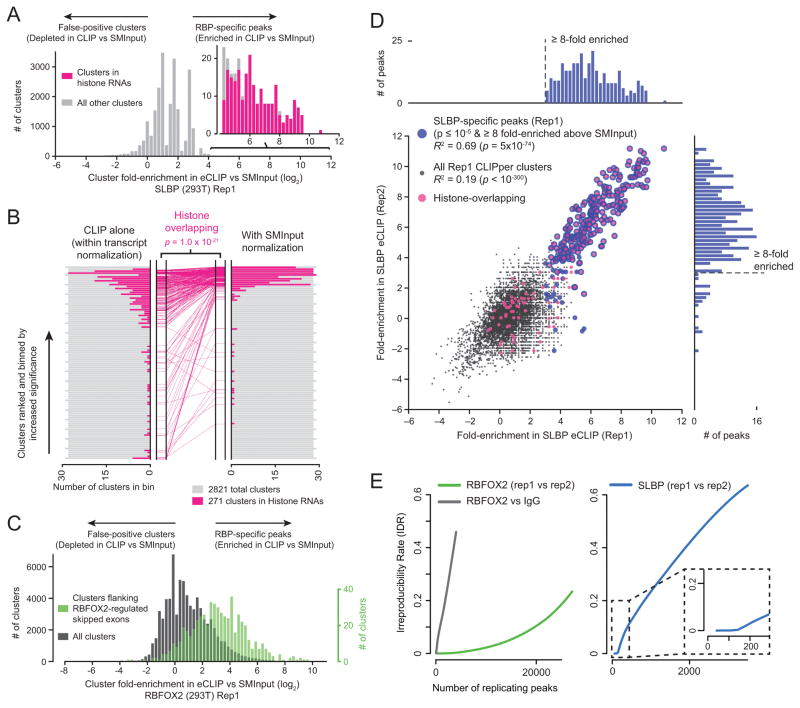
Figure 2. Improved CLIP signal-to-noise and reproducibility by normalization with paired Size-Matched Input (SMInput).
(a) Enrichment for SLBP clusters relative to SMInput was determined for all 23,034 CLIPper-identified clusters. Histograms show the number of clusters with indicated fold-enrichment, with histone-overlapping clusters in pink. (b) The subset of 2,821 SLBP eCLIP clusters with either pre-normalized (CLIPper) or SMInput normalized _p-_value ≤ 10−5 were selected and ranked by (left) pre-normalized CLIPper _p-_value or (right) by SMInput normalization; histograms indicate the number of histone-overlapping binding sites in each bin. (center) For 271 clusters overlapping histone RNAs, pink lines indicate the change in rank, with significance determined by Kolmogorov-Smirnov test. (c) Histogram indicates the number of RBFOX2 eCLIP clusters with indicated fold-enrichment in eCLIP relative to SMInput, with clusters overlapping introns flanking RBFOX2-dependent cassette exons indicated in green. (d) SLBP clusters were identified in Replicate 1, and for each cluster the fold-enrichment was determined for both Replicate 1 and Replicate 2 eCLIP. Histone-overlapping points are indicated in pink, with significantly enriched peaks indicated in blue. Attached histograms show the number of significantly-enriched peaks with specified fold-enrichment in Replicate 1 (top) and Replicate 2 (right). (e) Graphs indicate Irreproducible Discovery Rate (IDR) analysis performed on eCLIP fold-enrichment for (top) RBFOX2 biological replicates and RBFOX2 compared to IgG-only eCLIP, and (bottom) SLBP biological replicates.
Similarly, we observed that relative to all 74,902 CLIPper-identified clusters for RBFOX2, the 5,954 significantly enriched peaks were 5.4-fold increased for binding proximal to splicing-array identified alternative splicing events altered upon RBFOX2 depletion (p = 1.6 × 10−109 by Kolmogorov–Smirnov test) (Fig. 2c, Supplementary Fig. 9a and 10a–f), and ranking peaks by SMInput-enrichment improves the ranking of peaks flanking RBFOX2-regulated exons relative to ranking by standard peak significance (Supplementary Fig. 9b). For RBFOX2 as well as three additional RBPs with known binding specificities (PUM2, TARDBP, and TRA2A), we observed enrichment of the known motif in eCLIP-enriched peaks but not in those “depleted” in eCLIP versus SMInput, providing evidence that these depleted clusters are likely false positive clusters (Supplementary Fig. 9c–d, 11a–c). Thus, we conclude that incorporation of SMInput normalization significantly improves signal-to-noise in identifying authentic binding sites.
Reproducibility of eCLIP across replicate experiments
Robust identification of RBP binding sites requires that binding signal is reproducibly detectable above both technical and biological variation across independent samples. First, we observed high correlation for histone RNA enrichment across independent biological replicates for SLBP (R2 = 0.50 for CDS and 0.73 for 3′UTR, both p < 10−300 by standard conversion of r values to t statistics), indicating reproducibility at the level of the entire length of gene regions (Supplementary Fig. 12a). Next, both SLBP and RBFOX2 binding sites identified in one biological replicate show significant correlation in an independent biological replicate (R2 = 0.69 and 0.33 respectively, both p < 10−70) (Fig. 2d, Supplementary Fig. 12b–c). 79% and 93% of significantly enriched peaks for RBFOX2 and SLBP respectively were enriched by at least 8-fold above SMInput in an independent biological replicate, indicating high reproducibility at the level of peaks.
Irreproducible Discovery Rate (IDR) analysis has become routine to assess reproducibility by comparing ranks of ChIP-seq peaks identified across biological samples18. To determine reproducibility of eCLIP, IDR was performed on peaks ranked by eCLIP fold-enrichment over SMInput. For RBFOX2, we observed that 6,751 replicating peaks were identified at an IDR threshold of 0.01, a dramatic increase over the 114 observed when comparing RBFOX2 and IgG eCLIP (Fig. 2e). In contrast to the large number of binding sites observed for RBFOX2, IDR analysis on SLBP replicates identified only 160 reproducible peaks (Fig. 2e), validating the specificity for SLBP in binding only to histone RNAs. These results indicate that eCLIP data is highly reproducible.
eCLIP enables large-scale in vivo RBP target profiling
To demonstrate the reliability and simplicity of the eCLIP protocol we performed 209 eCLIP experiments (each of which includes two biological replicates and one paired SMInput control) to profile the targets of 132 RBPs in K562 chronic myelogenous leukemia and 75 RBPs in HepG2 hepatocellular carcinoma cell lines using antibodies characterized in a large-scale antibody validation effort19, the largest effort to date on profiling RBPs under the same conditions with a standard methodology. Out of these 209 experiments, 199 (95.2%) yielded libraries (Fig. 3a). To obtain a baseline reference point for RBPs with different molecular weights, we performed eCLIP with control IgG for 75 kDa sized regions tiled in 25 kDa increments from 25–100 kDa to 175–250 kDa. We obtained libraries with eCT of 19.3–21.0 for the 25–100 kDa to 150–225 kDa regions, while the largest size region (175–250 kDa) yielded an eCT of 17.7. For 170 RBP eCLIPs (81.3%), both replicates gave libraries with eCT <19.3 (the minimal IgG eCT corresponding to the typical membrane cut size range for most RBPs), indicating that most libraries contain significant RBP-specific signal (Fig. 3b). HNRNPs and other highly abundant RBPs often had eCT < 13, whereas some RBPs with a smaller target repertoire (e.g. SLBP) and many non-canonical RBPs (including those lacking predicted RNA recognition domains) required eCT > 17. 150 replicates were within 2 eCT of each other (71.8%), indicating a high degree of both technical and biological reproducibility regarding the amount of bound RNA recovered and the efficiency of subsequent library preparation (Fig. 3b). We typically observed a ~32-fold decrease in library amount when eCLIP is performed on non-UV crosslinked samples (Supplementary Fig. 13a), indicating that little RNA is recovered if not crosslinked to protein. We observed specific eCLIP profiles for RBPs with known functions throughout RNA transcripts (Supplementary Fig. 13b), confirming that the application of a standardized eCLIP protocol successfully reveals RBP-specific binding profiles.
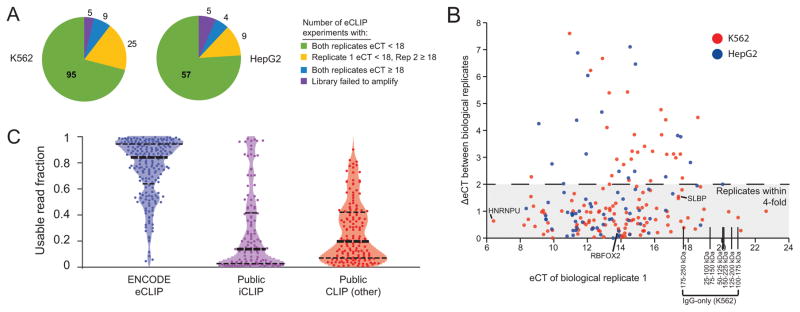
Figure 3. Scalable RBP target identification with eCLIP.
(a) Pie charts indicate experimental success results for 209 eCLIP experiments (each including two biological replicates plus an SMInput control) for which successful immunoprecipitation was performed in K562 (top) or HepG2 (bottom) cell lines, with colors indicating the amount of amplification required to obtain 100 fmoles of library (eCT). 102 experiments pass validation metrics and are deposited at http://www.encodeproject.org (Supplementary Table 2). (b) Each point represents a successful experiment in (a), with the x-axis indicating the eCT of the best replicate (denoted as replicate 1) and the y-axis indicating the increase in eCT between replicate 1 and replicate 2 (indicating decreased efficiency in the second replicate). Seven IgG-only eCLIP experiments are indicated by black lines, covering all 75 kDa intervals from 25 to 250 kDa. (c) The fraction of usable (non-PCR duplicated) reads out of all uniquely mapped reads is shown for eCLIP, public iCLIP experiments (12 performed for the ENCODE consortium as well as 115 published iCLIP datasets) and 152 published CLIP datasets (including PAR-CLIP and HITS-CLIP), shown as points with underlaid kernel density smoothened histogram.
We observed a high correspondence between eCT and PCR duplicate reads; whereas libraries with eCT < 14 had a median of 91.0% usable (9.0% PCR duplicates), libraries with eCT > 17 cycles had a median of 21.2% usable reads (Supplementary Fig. 13c). Our results with SLBP indicate that RBPs that bind few targets can be profiled accurately from libraries with higher PCR duplication rates, as fewer usable reads are needed to saturate the discovery of true binding events. However, RBPs with more widespread binding may require higher complexity libraries to robustly identify true binding sites. At this time, 102 of these experiments pass additional quality control criteria (specifically, that both samples meet a minimum usable read depth and show reproducible binding signal) and have been deposited for public release at the ENCODE web-portal (https://www.encodeproject.org) (Supplementary Table 2). As a comparison of the eCLIP results against other published CLIP-seq experiments, we collated 127 public iCLIP datasets (including 12 generated in-house as part of the ENCODE efforts) and 152 other public CLIP-seq datasets (including PAR-CLIP and HITS-CLIP) (Supplementary Table 1). We observed dramatic improvement in both the fraction of usable reads and in absolute usable read numbers, with the median usable read percent increasing from 13.9% (iCLIP) and 19.7% (other CLIP) to 84.1% for eCLIP (Fig. 3c, Supplementary Fig. 13d), confirming that eCLIP improves efficiency compared to previously published CLIP data.
CLIP experiments share common artifacts across RBPs
This large set of distinct RBPs offered a unique opportunity to characterize common artifacts in CLIP experiments. We observed that 24,444 of 74,902 (32.6%) RBFOX2 clusters and 1,616 of 21,418 (7%) SLBP clusters identified as significant (p ≤ 0.05) by CLIPper were in fact depleted when compared to SMInput (Fig. 2c), indicating that they are likely to be false-positives in standard CLIP analysis. While 84% of intronic clusters for RBFOX2 were enriched above input, only 36% of clusters within coding exons were similarly enriched above input. Other regions showed even higher rates of these likely false-positive CLIP signals: 86% of clusters mapping to transcripts encoded on the mitochondrial chromosome and 90% of those overlapping snoRNAs were in fact depleted in RBFOX2 eCLIP relative to SMInput (Fig. 4a). Performing similar SMInput-normalization for all 102 experiments, we observed that the identification of CLIP-depleted clusters within mitochondria-encoded RNAs and many classes of ncRNA (including snoRNA, snRNA, and rRNA) was consistent across many datasets (Fig. 4b). These regions often had stereotypical peak shapes and significant read numbers, and may represent either IgG- (or similar) artifacts or simply low-level non-specific signal remaining after IP (Supplementary Fig. 14a-b). Our findings emphasize that CLIP analysis (particularly without proper input or other controls) that focuses on these classes of binding events should be carefully validated due to the high rate of false-positives. However, we observed that RBP-enriched signal could be observed for a small number of RBPs at these loci (including mitochondrial factor AUH20, and known snRNA-binding factors LARP7, SMNDC1, and PRPF8 on snRNAs21,22) (Fig. 4b, Supplementary Fig. 14a–c). Thus, SMInput normalization can identify true RNA binding events even at common false positive regions and help characterize the wide array of RBPs known to directly regulate mitochondrial and small RNA processing and function23,24.
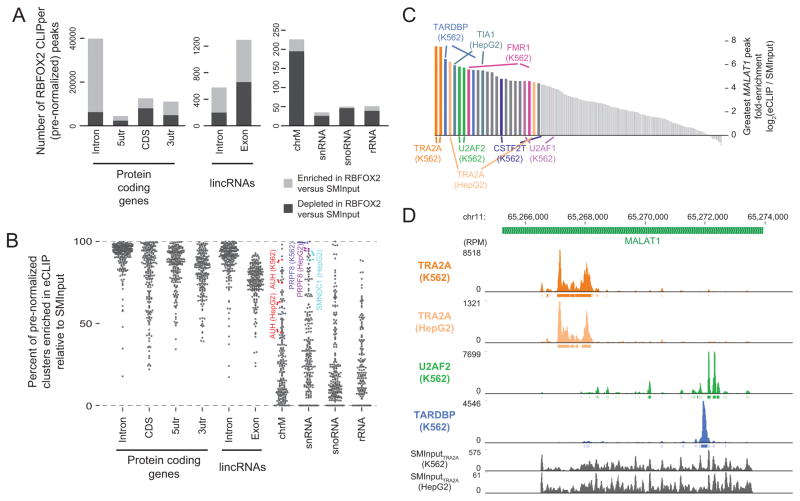
Figure 4. eCLIP enables RNA-centric identification of protein binding to abundant noncoding RNAs.
(a) Bars indicate the distribution of RBFOX2 clusters enriched (eCLIP/SMInput read density > 1) in RBFOX2 eCLIP relative to input (light bar) as compared to those depleted (dark bar). (b) Points indicate the percent of CLIPper-identified clusters identified within given regions that are enriched when compared against the paired SMInput for 102 eCLIP experiments (in biological duplicate) in K562 and HepG2 cells. (c) Bars indicate fold-enrichment of the most enriched peak overlapping lincRNA MALAT1 in each of 204 K562 and HepG2 datasets. Labels indicate biological replicates of RBPs with specific localization patterns. (d) Read density tracks along lincRNA MALAT1 for Replicate 1 of subset of datasets labeled in (c), with others shown in Supplementary Figure 15g.
RNA-centric views of RBP-interactions
Leveraging the scale of eCLIP data generated, we asked whether we can identify RBPs with high specificity and affinity to specific RNAs in an RNA-centric manner for four abundant RNAs: the 7SK snRNA, the histone RNA family, and lncRNAs XIST and MALAT1 that have previously been described as common false-positives in PAR-CLIP data15. Despite 7SK having a median RPM greater than 1000 across all datasets we observed a >21-fold enrichment for LARP7 (a 7SK complex component25) on the 7SK snRNA with no others above 3-fold enriched (Supplementary Fig. 14b, 15a–b). Similarly, SLBP was >71-fold enriched at histone RNAs, with no other RBP greater than 7-fold enriched (Supplementary Fig. 15c). Considering a longer lncRNA, we observed that four RBPs (HNRNPK, PTBP1, HNRNPM, and SRSF1) exhibited a greater than two-fold enrichment on the XIST RNA, with each binding distinct loci along the transcript (Supplementary Fig. 15d–e). These results corroborate recently published RNA affinity purification and mass spectrometry profiling as well as other work on XIST which have identified these four factors26–28, in particular the localized enrichment for SRSF1 at the 5′ A-repeat on XIST28.
Finally, we considered binding to lncRNA MALAT1, which has presented a consistent challenge for analysis in many CLIP experiments due to its high abundance (>600 RPM across all 204 experiments (Supplementary Fig. 15f)). By ranking eCLIP datasets according to the peak with the greatest fold-enrichment above SMInput, we observe multiple RBPs with strikingly specific binding to regions of MALAT1 (Fig. 4c–d, Supplementary Fig. 15g). Our results validate previously described binding of TARDBP29, and localized binding of splicing machinery factors U2AF1 and U2AF2 suggests MALAT1 regulation of splicing may extend beyond described roles in modulating serine/arginine-rich splicing regulatory proteins30. These results confirm that properly normalized eCLIP data can robustly distinguish false-positive signals from true binding events even on abundant noncoding RNAs, and show that large-scale eCLIP data permits RNA-centric views of RBP association.
Discussion
In summary, eCLIP provides a robust, standardized framework for large-scale generation of transcriptome-wide binding maps for RBPs. eCLIP maintains the single-nucleotide resolution identification of RBP binding sites from previous methods, dramatically decreases required amplification and greatly enhances the rate of success at generating libraries with high usable read percentages across diverse RBPs. Additionally, the paired size-matched input controls improve the signal-to-noise for discovery of authentic sites. As such, eCLIP empowers large-scale RBPome-wide profiling efforts, while simultaneously allowing binding site identification with decreased sample requirements and high reproducibility for individual studies.
Methods
eCLIP-seq library preparation
(See Supplementary Protocol 1 for detailed Standard Operating Procedures for ENCODE-style eCLIP experiments, including oligonucleotide sequences, catalog numbers for all reagents, and specific details for eCLIP experiments). RNA binding protein (RBP)-RNA interactions were stabilized with UV crosslinking (254 nm, 400 mJ/cm2), followed by lysis in iCLIP lysis buffer, limited digestion with RNase I (Ambion), immunoprecipitation of RBP-RNA complexes with a specific primary antibody of interest using magnetic beads with pre-coupled secondary antibody (typically M-280 Sheep Anti-Rabbit IgG Dynabeads, ThermoFisher Scientific 11204D), and stringent washes. After dephosphorylation with FastAP (ThermoFisher) and T4 PNK (NEB), a barcoded RNA adapter was ligated to the 3′ end (T4 RNA Ligase, NEB) (at this step, multiple replicates of the same RBP, or potentially RBPs of similar size and bound RNA amount, can be uniquely barcoded and pooled after ligation to simplify downstream steps - see Supplementary Fig. 2a). Ligations were performed on-bead (to allow washing away unincorporated adapter) in high concentration of PEG8000, which improves ligation efficiency to > 90%. Samples were then run on standard protein gels and transferred to nitrocellulose membranes, and a region 75 kDa (~150 nt of RNA) above the protein size was isolated and proteinase K (NEB) treated to isolate RNA. RNA was reverse transcribed with AffinityScript (Agilent), and treated with ExoSAP-IT (Affymetrix) to remove excess oligonucleotides. A second DNA adapter (containing a random-mer of 5 (N5) or 10 (N10) random bases at the 5′ end) was then ligated to the cDNA fragment 3′ end (T4 RNA Ligase, NEB), performed with high concentration of PEG8000 (to improve ligation efficiency) and DMSO (to decrease inhibition of ligation due to secondary structure). After cleanup (Dynabeads MyOne Silane, ThermoFisher), an aliquot of each sample was first subjected to qPCR (to identify the proper number of PCR cycles), and then the remainder was PCR amplified (Q5, NEB) and size selected via agarose gel electrophoresis. Samples were sequenced on the Illumina HiSeq 2500 or 4000 platform as two Paired End 50bp (for N5) or 55bp (for N10) reads. All analyses were performed using identical antibody lots for RBFOX2 (A300-864A lot 002, Bethyl), SLBP (RN045P lot 001, MBL International), and IgG Isotype Control (02-6102 lot 32013, Thermo Fisher Scientific). SLBP experiments were performed with 20×106 cells and 10 ug of primary antibody; RBFOX2 experiments were performed with 20×106 cells and 10 ug (eCLIP Rep1 and Rep2) or 10×106 cells and 5 ug (RNase I variation experiments). All experiments in K562 and HepG2 cells were performed with 20×106 cells and 10 ug of indicated primary antibody (Supplementary Table 2). Antibody validation documentation (including Western images of immunoprecipitation and shRNA knockdown19) are available at http://www.encodeproject.org/. Additional experiments performed in K562 and HepG2 cells in which the antibody failed to successfully immunoprecipitate the targeted RBP were excluded from analysis. 293T cells were obtained from Clontech (Lenti-X 293T cell line). K562 and HepG2 cells were purchased from ATCC, and were not independently verified. Cells were routinely tested for mycoplasma using MycoAlert PLUS (Lonza).
eCLIP-seq data processing
(See Supplementary Protocol 2 for detailed description of processing pipeline, including command-line examples, options, and required packages used in basic processing of eCLIP datasets). After standard HiSeq demultiplexing, eCLIP libraries with distinct inline barcodes were demultiplexed using custom scripts, and the random-mer was appended to the read name for later usage. Reads were then adapter trimmed (cutadapt v1.9.dev1) and reads less than 18 bp were discarded (see Supplementary Protocol 2 for adapter sequences used). Mapping was then first performed against human elements in RepBase (v18.05) with STAR (v2.4.0i), repeat-mapping reads were segregated for separate analysis, and all others were then mapped against the full human genome (hg19) including a database of splice junctions with STAR (v 2.4.0i). Uniquely mapping reads were then run through a custom-built PCR duplicate removal script, removing duplicate reads based on sharing identical Read1 start position, Read2 start position, and random-mer sequence to leave “Usable” reads. eCLIP datasets with multiple inline barcodes were merged at the usable read stage, and cluster identification was performed on usable reads using CLIPper11 (available at https://github.com/YeoLab/clipper/releases/tag/1.0) with options –s hg19 –o –bonferroni –superlocal --threshold-method binomial --save-pickle, considering read 2 only (the read that is enriched for termination at the crosslink site). For visualization on the UCSC Genome Browser, all tracks were RPM (reads per million) normalized against the total number of usable reads in that dataset.
Downsampling analysis was performed by 100 iterations of randomly permuting the uniquely mapped reads, selecting the top N reads, and performing PCR duplicate removal to identify usable reads. For iCLIP experiments with multiple libraries generated from different cDNA sizes (low, medium, and high) per sample, only the library with the highest percent usable was used for downsampling analysis. De novo motif finding for RBFOX2 iCLIP and eCLIP were performed with HOMER’s findMotifs program (-p 4 -rna -S 10 -len 5,6,7,8,9), with cluster sequences compared against a set of background ‘clusters’ where three random same-sized regions were selected for each real CLIP cluster corresponding to the same type of genic region (e.g. selected from introns, 3′UTRs, etc.). Cluster location pie charts were determined by counting the total number of bases covered by peaks for each given region type.
For RBFOX2 RNase I shearing analysis, each cluster was annotated according to which genic region it overlapped (using the same priority as above for region-level analysis), and the number of peaks annotated as overlapping each genic region type were identified. For analysis of RBFOX2, PUM2, TARDBP, and TRA2A motif enrichment before and after SMInput normalization, clusters were extended to a minimum of 100 nt centered around the midpoint of the CLIPper-identified clusters, and the sequences in each cluster were randomly shuffled 10 times to generate the sequence background.
Normalization of eCLIP signal against SMInput
To perform peak-level input normalization, SMInput samples were processed identically to eCLIP samples through the usable read stage. The number of eCLIP reads overlapping CLIPper-identified peaks and the number overlapping the identical genomic region in the paired SMInput sample, were counted and used to calculate fold-enrichment (normalized by total usable read counts in each dataset), with enrichment _p-_value calculated by Yates’ Chi-Square test (Perl) (or Fisher Exact Test (calculated in the R statistical computing software) where the observed or expected read number was below 5), which have minimal reportable _p-_values of 10−88 (for Chi-Square) and 2.2 × 10−16 (for Fisher Exact) respectively. Region-level analysis was performed by counting mapped reads along all transcripts in Gencode v19 (“comprehensive”). Reads were then associated with regions with the following priority: coding transcripts (CDS, then 5′ and/or 3′ UTR, then intron), followed by non-coding transcripts (exon, then intron), requiring the majority of the read to overlap that region. A minimum of 10 observed or expected (extrapolated by taking eCLIP RPM and normalizing to SMInput total read depth) reads were required for a gene to be considered in region-based fold-enrichment analyses.
To identify motifs with single-nucleotide resolution, the 5′ end of each usable Read2 was identified, and each k-mer ranging 100 nt on each side of this position was counted for each eCLIP dataset (discarding k-mers with unknown sequence (N’s)), and normalized against the count observed in the paired SMinput dataset (Figures typically show smaller flanking regions to focus on enrichment proximal to crosslink sites). As iCLIP has no paired input, the SMInput from eCLIP (Rep1) was used for normalization in Supplementary Fig. 5a.
IDR Analysis
IDR was performed on both the RBFOX2 and SLBP SMInput normalized peaks by ranking peaks by enrichment _p_-value and performing the 2012 ENCODE IDR Pipeline as documented at https://sites.google.com/site/anshulkundaje/projects/idr.
Public CLIP Database and iCLIP data processing
All data was downloaded directly from the SRA/ERA (listed in Supplementary Table 1), and processed similar to eCLIP-seq, with distinctions described below. Adapter trimming (cutadapt) was only performed once. Datasets with fewer than 100,000 uniquely mapped reads were discarded. PCR duplicate removal was performed according to library preparation: for iCLIP datasets, random-mers (if present) were removed from the reads and used for PCR duplicate removal as described for eCLIP experiments above; for HITS-CLIP and PAR-CLIP datasets, more than one read mapping with the same start position was assumed to be a PCR duplicate and removed. As for eCLIP, only non-duplicate reads were used for peak calling. Usable read density plots and smoothened kernel density histograms were generated in Matlab with the distributionPlot package with default settings.
Blocking RBP binding by antisense oligonucleotide
Antisense oligonucleotide (ASO) treatments were performed by transfecting 293T cells in 24 well format with 1.5 uL of RNAiMax (Thermo Fisher) and 100 uM of antisense oligonucleotide (Isis Pharmaceuticals). Complexes were incubated in 50 uL of OptiMEM (Thermo Fisher) for 30 minutes, added to cells, and incubated for 36 hours, after which RNA was isolated using standard TRIzol extraction (Thermo Fisher). Splicing was assayed by RT-PCR using SuperScript III (Thermo Fisher) and Phusion DNA polymerase (Thermo Fisher), with primers located in flanking constitutive exons. Exon inclusion percentage (PSI) was calculated by ImageJ quantification of agarose gel electrophoresis and imaging of exclusion and inclusion PCR products. Error bars indicate sample standard deviation, with _p-_value calculated by Student’s _t_-test. ASOs targeting different RBFOX2 binding sites were used as controls as follows: ECT2_ASO1/2 for NDEL1 experiments, MPZL1_ASO1/2, EPB41_ASO1/2, and LRRFIP2_ASO1/2 for ECT2 experiments, and ANKRD26_ASO1/2, FAM190Bx_ASO1, and DOCK7_ASO1/2 for EPB41 experiments (Supplementary Table 3).
RBFOX2-knockdown transcriptome profiling by microarray
To profile RBFOX2-responsive splicing events, 3 independent lentiviral transductions were performed in 293T cells for each of 3 RBFOX2-targeting shRNAs (TRCN0000074544, TRCN0000074546, and TRCN0000074543, Sigma) plus a non-targeting control (SHC016, Sigma). After selection with Puromycin for 10 days, cells were harvested for protein and RNA (isolated by Trizol (Thermo Fisher Scientific) extraction). RBFOX2 protein knockdown was validated by standard western blotting using RBFOX2 (A300-864A, Bethyl) and GAPDH (ab8245, Abcam), imaged using the Odyssey fluorescent imager (LiCor), and quantitated using Image Studio Lite (LiCor) with median local background correction. RNA samples for microarray profiling were prepared using WT Expression Kit (Ambion), and hybridized to Affymetrix HTA2.0 microarrays. After scanning, all probes were RMA-normalized (Affymetrix Expression Console). All probes corresponding to cassette exons profiled on the microarray (comprising exclusion junction, upstream and downstream inclusion junction, and inclusion exonic probes) were identified and normalized against the average signal on a per-gene basis to remove gene expression changes (Supplementary Fig. 10d). Student’s _t-_test was performed on residuals for inclusion probes and exclusion probes separately to identify robust splicing changes; a set of 197 and 217 exons for TRCN0000074544 and TRCN0000074543 respectively met criteria of p ≤ 0.001 for either inclusion or exclusion probes and a combined |SepScore| ≥ 0.5, where SepScore is defined as the normalized change in exclusion minus the normalized change in inclusion. For overlap with eCLIP data, eCLIP peaks were associated with a cassette exon if they were located at any position in the flanking upstream or downstream intron. As all three shRNAs gave similar results (Supplementary Fig. 10e), eCLIP analyses shown use the 197 events identified from TRCN0000074544.
Code availability
Custom code used is available at https://github.com/gpratt/gatk/releases/tag/2.3.2, and described in Supplementary Protocol 2.
Supplementary Material
1
2
Acknowledgments
The authors would like to thank members of the Yeo lab (particularly S. Aigner and S. Markmiller) as well as colleagues J. Van Nostrand, Y. Kobayashi, B.R. Graveley and C.B. Burge for critical reading of the manuscript. This work was supported by grants from the National Institute of Health (HG004659, U54HG007005 and NS075449 to G.W.Y). We would also like to thank Ionis Pharmaceuticals for sharing reagents. E.L.V.N. is a Merck Fellow of the Damon Runyon Cancer Research Foundation (DRG-2172-13). G.W.Y. is an Alfred P. Sloan Research Fellow. G.A.P. is supported by the National Science Foundation Graduate Research Fellowship.
Footnotes
Author Contributions
E.L.V.N., A.A.S., M.G. and G.W.Y. conceived the study. E.L.V.N., A.A.S., and C.S. developed the eCLIP methodology. E.L.V.N., C.G.B. and S.M.B. performed 293T eCLIP and RBFOX2 knockdown experiments. F.R. provided ASOs and M.Y.F. performed ASO experiments. C.G.B., B.S., S.M.B., T.B.N., K.E., and R.S. performed K562 and HepG2 eCLIP experiments. E.L.V.N. and G.A.P. performed computational analyses. E.L.V.N. and G.W.Y. wrote the manuscript.
Competing Financial Interests Statement
F.R. is a paid employee of Ionis Pharmaceuticals, which provided ASO reagents.
Accession codes
All 293T datasets (including SLBP and RBFOX2 eCLIP, RBFOX2 iCLIP, and microarrays profiling RBFOX2 knockdown) have been deposited at the Gene Expression Omnibus (GSE77634). K562 and HepG2 eCLIP datasets have been deposited for public release at the ENCODE Data Coordination Center (https://www.encodeproject.org), with accession identifiers listed in Supplementary Table 2.
References
- 1.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nature reviews. Genetics. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castello A, Fischer B, Hentze MW, Preiss T. RNA-binding proteins in Mendelian disease. Trends in genetics : TIG. 2013;29:318–327. doi: 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Nussbacher JK, Batra R, Lagier-Tourenne C, Yeo GW. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends in neurosciences. 2015;38:226–236. doi: 10.1016/j.tins.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 5.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shishkin AA, et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nature methods. 2015;12:323–325. doi: 10.1038/nmeth.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huppertz I, et al. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nature structural & molecular biology. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovci MT, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nature structural & molecular biology. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weyn-Vanhentenryck SM, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell reports. 2014;6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks L, 3rd, et al. A multiprotein occupancy map of the mRNP on the 3′ end of histone mRNAs. Rna. 2015;21:1943–1965. doi: 10.1261/rna.053389.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyes-Herrera PH, Speck-Hernandez CA, Sierra CA, Herrera S. BackCLIP: a tool to identify common background presence in PAR-CLIP datasets. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv442. [DOI] [PubMed] [Google Scholar]
- 15.Friedersdorf MB, Keene JD. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome biology. 2014;15:R2. doi: 10.1186/gb-2014-15-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nature biotechnology. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li QH, Brown JB, Huang HY, Bickel PJ. Measuring Reproducibility of High-Throughput Experiments. Ann Appl Stat. 2011;5:1752–1779. doi: 10.1214/11-AOAS466. [DOI] [Google Scholar]
- 19.Sundararaman B, Zhan L, Blue S, Stanton R, Elkins K, Olson S, Wei X, Van Nostrand EL, Pratt GA, Huelga SC, et al. Resources for the Comprehensive Discovery of Functional RNA Elements. Molecular cell. 2016 doi: 10.1016/j.molcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richman TR, et al. A bifunctional protein regulates mitochondrial protein synthesis. Nucleic acids research. 2014;42:5483–5494. doi: 10.1093/nar/gku179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. Rna. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rappsilber J, Ajuh P, Lamond AI, Mann M. SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome. The Journal of biological chemistry. 2001;276:31142–31150. doi: 10.1074/jbc.M103620200. [DOI] [PubMed] [Google Scholar]
- 23.Rackham O, Mercer TR, Filipovska A. The human mitochondrial transcriptome and the RNA-binding proteins that regulate its expression. Wiley interdisciplinary reviews. RNA. 2012;3:675–695. doi: 10.1002/wrna.1128. [DOI] [PubMed] [Google Scholar]
- 24.Matera AG, Wang Z. A day in the life of the spliceosome. Nature reviews. Molecular cell biology. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krueger BJ, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic acids research. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu C, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Royce-Tolland ME, et al. The A-repeat links ASF/SF2-dependent Xist RNA processing with random choice during X inactivation. Nature structural & molecular biology. 2010;17:948–954. doi: 10.1038/nsmb.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo F, et al. Regulation of MALAT1 expression by TDP43 controls the migration and invasion of non-small cell lung cancer cells in vitro. Biochemical and biophysical research communications. 2015;465:293–298. doi: 10.1016/j.bbrc.2015.08.027. [DOI] [PubMed] [Google Scholar]
- 30.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2